Abstract
Electron bifurcation plays a key role in anaerobic energy metabolism, but it is a relatively new discovery, and only limited mechanistic information is available on the diverse enzymes that employ it. Herein, we focused on the bifurcating electron transfer flavoprotein (ETF) from the hyperthermophilic archaeon Pyrobaculum aerophilum. The EtfABCX enzyme complex couples NADH oxidation to the endergonic reduction of ferredoxin and exergonic reduction of menaquinone. We developed a model for the enzyme structure by using nondenaturing MS, cross-linking, and homology modeling in which EtfA, -B, and -C each contained FAD, whereas EtfX contained two [4Fe-4S] clusters. On the basis of analyses using transient absorption, EPR, and optical titrations with NADH or inorganic reductants with and without NAD+, we propose a catalytic cycle involving formation of an intermediary NAD+-bound complex. A charge transfer signal revealed an intriguing interplay of flavin semiquinones and a protein conformational change that gated electron transfer between the low- and high-potential pathways. We found that despite a common bifurcating flavin site, the proposed EtfABCX catalytic cycle is distinct from that of the genetically unrelated bifurcating NADH-dependent ferredoxin NADP+ oxidoreductase (NfnI). The two enzymes particularly differed in the role of NAD+, the resting and bifurcating-ready states of the enzymes, how electron flow is gated, and the two two-electron cycles constituting the overall four-electron reaction. We conclude that P. aerophilum EtfABCX provides a model catalytic mechanism that builds on and extends previous studies of related bifurcating ETFs and can be applied to the large bifurcating ETF family.
Keywords: bioenergetics, flavin, flavoprotein, radical, electron paramagnetic resonance (EPR), archaea, extreme thermophile, electron transport, bifurcation, charge transfer, semiquinone, EtfABCX, anaerobic physiology
Introduction
Electron-bifurcating enzymes couple exergonic and endergonic reactions, thus maximizing conservation of free energy available from exergonic reactions (1). In this way, electrochemical energy can be captured for cellular metabolism, lowering the demands on transmembrane gradients or substrate-level phosphorylation. Thus, electron bifurcation provides a unifying explanation for many peculiar fermentative pathways found in anaerobic microorganisms, with important implications for understanding anaerobic microbial physiology in general (2–9).
So far, the bifurcating enzymes that have been characterized fall into one of four phylogenetically unrelated groups: electron transfer flavoproteins (EtfAB-containing), [FeFe]-hydrogenase/formate dehydrogenases (HydABC-containing), hetero-disulfide reductases (HdrA-containing), and transhydrogenases (NfnAB-containing) (8, 10). These enzymes catalyze more than a dozen different reactions, most involving the oxidation or reduction of ferredoxin, and are found mainly in anaerobic organisms (reviewed in Refs. 3, 8, 11, and 12). However, some of the EtfAB-containing complexes, such as that described below, can also be found in microaerophiles and aerobes.
Bifurcating ETFs2 are the best-studied bifurcating enzymes, and they form a subset of the large and well-known family of ETFs, which are found in all domains of life (13–15). Whereas the nonbifurcating ETFs contain one FAD and one AMP per AB heterodimer, the bifurcating EtfABs contain two FAD molecules, with the additional FAD replacing the AMP of the canonical ETF (13, 16, 17). The functions of bifurcating ETFs have been established in several strictly anaerobic micoorganisms. For example, the butyryl-CoA dehydrogenase (EtfAB–Bcd) complex from the butyrate-producing Acidaminococcus fermentans accomplishes the endergonic reduction of ferredoxin (Fd) with NADH by driving it with the exergonic reduction of crotonyl-CoA by NADH (16). Likewise, the strict anaerobe Acetobacterium woodii can utilize lactate as a carbon source using the bifurcating LDH–EtfAB complex to drive the endergonic oxidation of lactate to pyruvate with the energy provided by the exergonic electron transfer from reduced Fd to NAD+ (18). A. woodii can also utilize caffeate as an electron acceptor in which EtfAB-containing caffeyl-CoA reductase complex (CarCDE) couples the exergonic reduction of caffeyl-CoA by NADH to the endergonic reduction of Fd by NADH (19). Megasphaera elsdenii similarly contains a lactate dehydrogenase and a Bcd that likely interact with the same EtfAB (20–22).
Bifurcating ETFs are also found in some microorganisms that are capable of fixing atmospheric nitrogen, such as the aerobe Azotobacter vinelandii (13, 23, 24). In A. vinelandii, an enzyme complex containing EtfAB and quinone reductase, termed FixABCX, is associated with nitrogen-fixing activity (13, 24). The FixAB subunits are referred to as bifurcating Etf, whereas the FixCX subunits are referred to as Etf-quinone oxidoreductase (13). FixABCX is thought to provide reduced Fd for nitrogen fixation through the quinone-dependent reduction of Fd with NADH (23, 24). However, genomic analyses reveal that FixABCX-like complexes are not restricted to diazotrophs, indicating a general role extending beyond nitrogen fixation (13). We will refer to such complexes as EtfABCX to recognize their homology with FixABCX while also alluding to broader metabolic roles.
Herein we describe the mechanism for the first EtfABCX to be characterized from a non-nitrogen-fixing respiratory microorganism. Our exemplar was obtained from Pyrobaculum aerophilum, a denitrifying microaerophilic crenarchaeote that grows at 100 °C. P. aerophilum oxidizes organic compounds using oxygen, nitrate, arsenate, selenate, iron, and thiosulfate as terminal electron acceptors (25), but its genome lacks the genes for nitrogen fixation (26). The genes encoding P. aerophilum EtfABCX were expressed in the fermentative hyperthermophile, Pyrococcus furiosus, and both EtfABCX and the EtfAB subcomplex were purified. Like the Fix system (24), P. aerophilum EtfABCX is shown to oxidize NADH and bifurcate electrons to both high-potential (quinone) and low-potential (Fd) acceptors. For the P. aerophilum EtfABCX complex, we refer to each of the flavins in the enzyme in terms of the role that each has been proposed to play in ETF proteins in general (13, 16, 24, 27). Thus, the FAD unique to bifurcating ETFs is denoted the “bifurcating FAD,” or BF-FAD, and this is present in the A-subunit of P. aerophilum EtfABCX (Fig. 1). The electron-transferring FAD or ET-FAD is the FAD that is shared with the structurally homologous nonbifurcating ETFs that appear to specialize in electron transfer between partner proteins (13, 14, 28). By homology, the ET-FAD in P. aerophilum EtfABCX is located in the B subunit, whereas the FAD in the C subunit is referred to as the QR-FAD, as it is likely the site of quinone reduction (Fig. 1).
Figure 1.
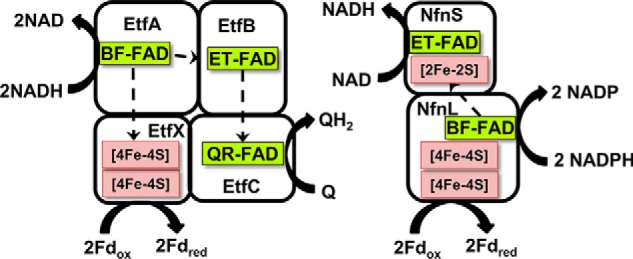
Cofactor content and proposed pathway of electron flow in P. aerophilum EtfABCX and P. furiosus NfnI. Q, quinone; QH2, reduced quinone; Fdox, oxidized ferredoxin; Fdred, reduced ferredoxin.
The primary goal of this work was to elucidate the overall catalytic cycle of P. aerophilum EtfABCX. Structural studies of related ETF complexes have shown that large conformational changes occur during catalysis, and a mechanism involving relatively large domain movement has been proposed (16, 29, 30). However, more information is needed to understand how electron flow to the high- and low-potential acceptors is gated and controlled. It was also important to determine how the catalytic cycle of EtfABCX compared with that recently established for the bifurcating enzyme, NADH-dependent ferredoxin NADP oxidoreductase I (NfnI) from P. furiosus (31). NfnI and EtfABCX have very similar cofactor compositions, even though they are completely unrelated phylogenetically, and both use NAD(P)H as the mid-potential donor to their BF-FADs and use Fd as the low-potential carrier that is reduced or oxidized by a pair of [4Fe-4S] clusters (Fig. 1). However, herein we have identified key catalytic intermediates of P. aerophilum EtfABCX, including a novel NAD+-dependent charge transfer complex, and describe a catalytic cycle that is very distinct from that established for NfnI. Moreover, the P. aerophilum EtfABCX catalytic cycle is consistent with previous structural observations and mechanisms of related EtfAB-containing complexes (8, 9, 13, 24, 27). We propose that the unifying principles proposed here can be applied to all bifurcating members of the large ETF family.
Results
Structure and catalytic activity of EtfAB and EtfABCX
The His-tagged form of P. aerophilum EtfABCX was produced in P. furiosus. Size-exclusion chromatography of the purified enzyme resolved two peaks that corresponded (by MS analysis) to EtfABCX, eluting near 450 kDa, and EtfAB, eluting near 50 kDa. The former is consistent with a tetrameric form of the EtfABCX complex (with a calculated mass of 125 kDa for the heterotetramer), whereas the latter peak corresponds to the EtfAB heterodimer (with a calculated mass of 66 kDa). Although EtfABCX is thought to interact with the cell membrane in vivo and to reduce quinone, the enzyme was purified from the cytoplasmic fraction of P. furiosus, is stable in solution, and, in contrast to the FixABCX system (24), does not require detergent to maintain solubility. Analyses of P. aerophilum EtfAB by native MS gave the expected mass with two FAD bound (measured: 69,562.45 Da; calculated: 69,566.17 Da; Fig. S1). The A and B subunits were not separated by the collision energy during MS analysis, by reverse phase LC under acidic conditions, or by the presence of an organic solvent, implying great stability for the heterodimer. The EtfC subunit was also identified by MS with a mass consistent with a bound FAD cofactor (Fig. S2). Based on MS analysis conducted under native and denaturing conditions, we conclude that as purified, both EtfAB and the EtfABCX holoenzyme have full flavin occupancy.
To predict the most probable three-dimensional structure of individual subunits, the protein fold recognition tool Phyre2 was used (32). To determine the tertiary structure of the protein of interest, this relies on sequence alignment, secondary structure prediction, and finally the match of predicted folds to homologous proteins with known protein structure. In the next step, the energy-minimized homology models of P. aerophilum Etf were overlaid on the structure of the recently published model of FixABCX from Azotobacter vinelandii (24). Once the quaternary structure was in place, cofactor-binding sites were modeled using individually refined models of P. aerophilum EtfAB, -C, or -X, generated based on templates 1O97.pdb (EtfAB domain II), 4L2I.pdb (EtfAB, domains I and III), 2GMH.pdb (EtfC), and 2PVZ.pdb (EtfX) in Swiss Modeler, followed by manual adjustment of a few side chains and unstructured strands and then energy minimization.
To validate the structural model of the complete P. aerophilum EtfABCX complex, three complementary experimental approaches were employed: 1) noncovalent MS to establish the mass of the entire complex in its native form, confirm the presence of cofactors, and determine cofactor and protein component stoichiometry; 2) chemical cross-linking and peptide-based MS to ascertain protein–protein interaction networks and distance constraints; and 3) surface labeling coupled to MS to map the solvent-exposed surfaces. Iterative modeling was then used, in which data from approaches 1–3 were used to test a model, identify inconsistencies, adapt the model, and repeat (24, 33–39). To preserve noncovalent interactions at protein interfaces, the homobifunctional cross-linking reagent, bis(sulfosuccinimidyl)suberate (BS3) was used. BS3 can probe a wide distribution of distances and bridge spatially close primary amine– and hydroxyl–containing amino acid residues (40). To probe the solvent-accessible surface of the intact complex and to obtain the most complementary results, two surface-labeling reagents, glycine ethyl ester (GEE) and dansyl chloride (DnsCl), were used. GEE and DnsCl target carboxyl, primary amine, and hydroxyl groups, which collectively are the most prevalent groups on protein surfaces, allowing for a detailed mapping of solvent-exposed residues. The model shown in Fig. 2 was in agreement with 37% of identified connections, including inter- and intrasubunit cross-links. Taking into account the distance a cross-linker can cover and the conformations a residue can adopt in solution, it is considered a good result if 30% of all generated cross-links within the complex are satisfied (33). Our best model was also in agreement with about 87% of the surface labels. The complete information on the generated cross-links and surface labels satisfying the P. aerophilum EtfABCX complex model can be found in Table S1 and Fig. S3, respectively.
Figure 2.
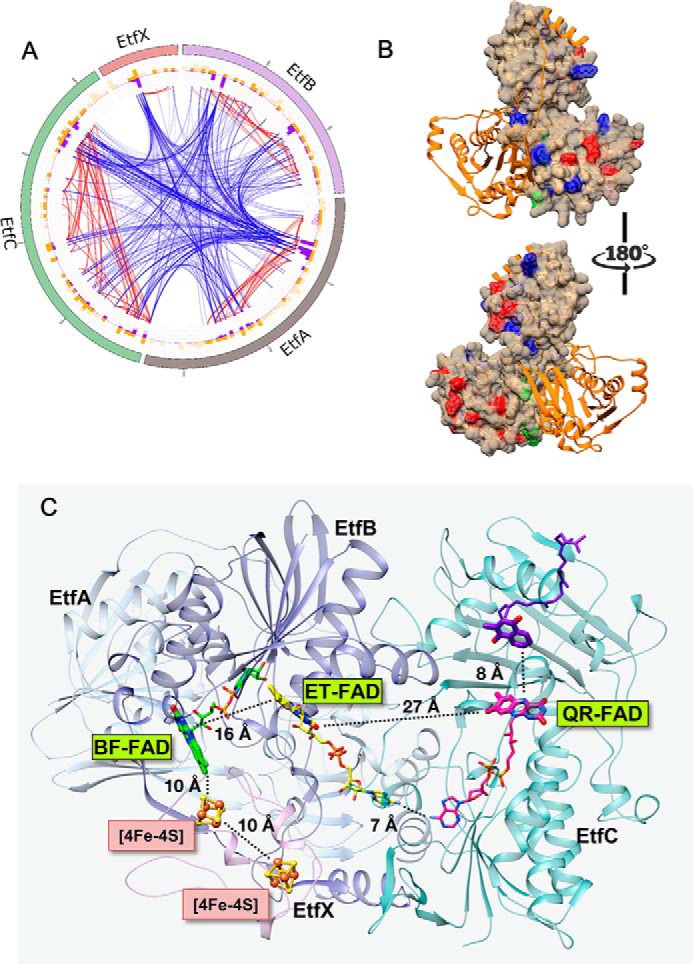
A, the multilayer circular plot presents interactions within the P. aerophilum EtfABCX complex captured by cross-linking (BS3). The most outer layer represents the protein sequence: EtfA is shown in gray, EtfB is purple, EtfC is green, and EtfX is pink. Yellow and purple histograms show the location of BS3 target residues (Lys and Ser) and the density of cross-links, respectively. Blue lines represent connections between subunits (intersubunit cross-links), and red lines highlight interactions within a single unit (intrasubunit cross-links). Cross-links maintaining less than 20 ppm error and a score higher than 3 (a score >2 is considered significant) are displayed. B, protein surface map of the complete P. aerophilum EtfABCX complex (for simplicity, only the AB dimer is presented) shows GEE (modification sites shown in red) and DnsCl (modification sites shown in blue) labels incorporated in to the native complex during short exposure to the two labeling reagents. Residues at the dimer interface highlighted in green correspond to labels integrated into the structure exclusively in the latest time points. Individual proteins are color-coded as follows: EtfB in orange and EtfA in tan. C, ribbon diagram of the MS-validated EtfABCX model. EtfA and EtfB are shown in light and dark blue, respectively, EtfC is teal, EtfX is magenta, the BF-FAD is green, the ET-FAD is yellow, and the QR-FAD is pink. Organic cofactors are shown using thick sticks to emphasize the redox-active portions and thin sticks to complete the structures. Iron–sulfur clusters are shown in orange (Fe) and yellow (S), and the menaquinone (purple) was modeled using the 2GMH (porcine electron transfer flavoprotein-ubiquinone oxidoreductase) structure with bound ubiquinone.
The EtfABCX model shows that one of the [4Fe-4S] clusters in EtfX is within 10 Å of the BF-FAD, which should support rapid direct electron transfer. Although the BF- and ET-FAD are 16 Å apart and the distance between the isoalloxazine rings of the ET- and the QR-flavins is almost 30 Å, similar distances have been reported for the analogous flavins in the bifurcating EtfAB–Bcd and CarCDE complexes (29, 30). Both of these complexes undergo a large conformational change as part of the catalytic cycle, and such changes are also incorporated as part of the mechanism of P. aerophilum EtfABCX proposed herein.
P. aerophilum EtfABCX is thought to couple the reversible oxidation of NADH to the reduction of Fd and menaquinone (MQ), by analogy with FixABCX, according to Reaction 1.
The primary Fd in P. aerophilum contains a single [4Fe-4S] cluster (41) and transfers a single electron (Reaction 1), whereas MQ is thought to be the natural quinone component of the P. aerophilum respiratory system (42). The purified forms of both EtfAB and EtfABCX catalyzed the oxidation of NADH and reduced the artificial electron carrier iodonitrotetrazolium with specific activities of 41.8 ± 6.5 and 15.8 ± 2.7 μmol of NADH oxidized/min/mg (at 80 °C), respectively. Although EtfC is proposed to reduce MQ in vivo, both EtfAB and EtfABCX also oxidized NADH and reduced the water-soluble MQ analog, menadione. The specific activities were 8.8 ± 0.8 and 5.1 ± 1.2 μmol of NADH oxidized/min/mg (at 80 °C), respectively. The lack of specificity with respect to the quinone acceptor makes it difficult to directly measure the bifurcation activity of EtfABCX, which is further complicated by the fact that menadione and NADH have similar absorption spectra (24). Bifurcation activity of EtfABCX was therefore assessed by coupling the oxidation of reduced menadione and reduced Fd to the reduction of NAD+, which was followed by the fluorescence of the NADH that was generated. The Fd used in this reaction was that of P. furiosus, which also contains a single [4Fe-4S] cluster (43). Importantly, reduction of NAD+ by EtfABCX depended upon the presence of both reduced Fd and reduced menadione, and no bifurcation activity was detected using EtfAB (Fig. S4). With the FixABCX from N2-fixing A. vinelandii, a 25% increase in activity was observed when all substrates (NADH, quinone, and Fd) were present relative to when only NADH and quinone were present (24), whereas in the case of P. aerophilum EtfABCX, an ∼2-fold increase was observed (Fig. S4).
The reduction potentials of the flavins in EtfAB
The optical signatures of the two flavins of EtfAB were monitored over the course of stepwise reduction with sodium dithionite or titanium citrate in the absence of mediators (Figs. S5A and S6). The initial increase in absorbance at 374 nm, where the anionic semiquinone (ASQ) of flavin absorbs maximally, was accompanied by a decrease at 454 nm (absorption maximum of oxidized flavin), indicating that an FAD undergoes a 1 e− reduction to ASQ (Fig. S5, phase 1). We assigned this to the ET-FAD, as it is expected to be more readily reduced than the BF-FAD (17, 27). Further addition of reductant resulted in only a slight intensity loss at 374 nm, suggesting little change in the amount of ASQ (Fig. S5, phase 2, staggered or nested mechanisms). As discussed further below, to account for these observations, we propose that a radical is present on either the ET-FAD or the BF-FAD, leading to the observed optical signal. This ASQ signal also persisted upon the addition of excess titanium citrate (this was also true with excess dithionite, although less clear; see Figs. S5A and S6).
Titrations in the presence of redox mediators showed that one flavin undergoes reduction from OX to ASQ to HQ before the other is reduced from OX to HQ (Fig. S5C, sequential). Thus, in Fig. S7A, the initial negative difference absorbance near 374 nm clearly indicates that ASQ is formed, whereas the positive intensity at 454 nm represents the OX signature lost in the course of this reaction (OX at the beginning exceeds OX at the end; Fig. S5A). Conversion of the ET-FAD from OX to ASQ occurred with an Em of 33 ± 6 mV (Fig. 3). The slope of the line near 0.5 confirms that the reaction in question is a 1 e− reaction and therefore rules out reduction to HQ. Difference spectra associated with phase 2 of the reductive titration (Fig. S5B) indicate that it corresponds to net reduction of an ASQ to HQ (Fig. S7B). These data clearly show net loss of the ASQ features. The slope of the Nernst log/log plot in Fig. 3 confirms this event as a 1 e− reaction. The intercept of the Nernst log/log plot yields an Em value for ET-FAD (ASQ/HQ) of −94 ± 7 mV. Titration of the lower-potential FAD (Fig. S7C, phase 3) reveals no significant contribution from ASQ, and the slope of 1 of the Nernst log/log plot confirms that this phase corresponds to a 2 e− reduction (Fig. 3A). An Em value for BF-FAD (OX/HQ) of −285 ± 5 mV was determined, which is close to the value of −279 mV reported for the bifurcating EtfAB from M. elsdenii (21). Thus, we conclude that the higher-potential FAD, ET-FAD, is reduced via two sequential 1 e− events to HQ but that the lower potential FAD, BF-FAD, undergoes reduction directly to the HQ state in a 2 e− event under equilibrium conditions (Fig. 3B). The values measured for P. aerophilum EtfAB of Em(OX/ASQ) = +33 mV and Em(ASQ/HQ) = −94 mV fall well within values reported for other EtfABs (21, 27), and the high Em(OX/ASQ) of +33 mV for P. aerophilum EtfAB indicates that the resting state of the EtfAB dimer possesses ET-FAD in its ASQ state.
Figure 3.
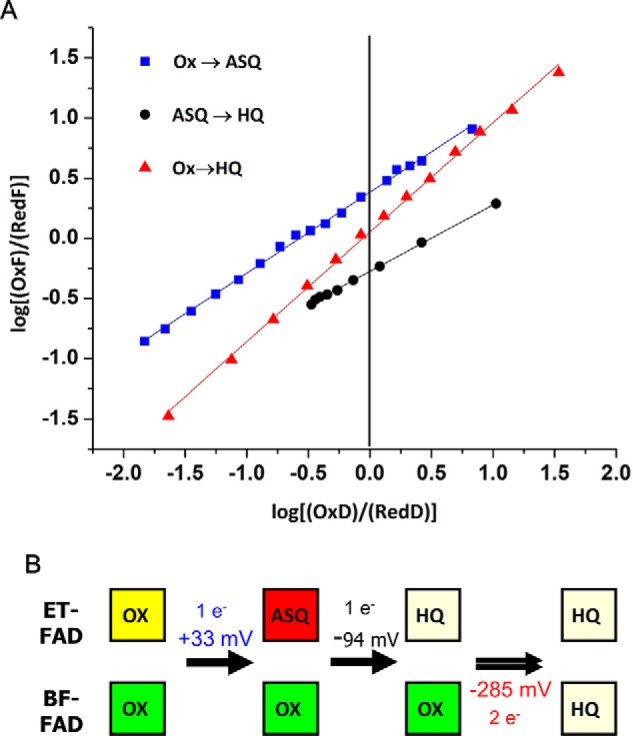
A, slopes of Nernst plots confirm the 1 e− nature of events in phases 1 and 2 and the 2 e− nature of phase 3. Best linear fits are as follows: phase 1 (blue), 0.66x + 0.55 (R2 = 0.997); phase 2 (black), 0.55x − 0.27 (R2 = 0.999), and phase 3 (red), 0.91x + 0.05 (R2 = 0.998). For the data in blue the “oxidized” flavin is OX, “reduced” flavin is the ASQ, and the dye is thionine. For the data in black the oxidized flavin is ASQ, reduced flavin is the HQ, and the dye is Nile blue. For the data in red, oxidized flavin is OX, reduced flavin is HQ, and the dye is safranin-O. B, proposed reduction events for each FAD cofactor in EtfAB and the measured potentials.
Transient absorption spectroscopy of EtfAB and EtfABCX
The SQ state of the BF-FAD was not observed in steady-state titrations, so transient absorption spectroscopy (TAS) was used to probe for short-lived flavin species. Formation of an ASQ with loss of absorption from the oxidized flavin was observed for both EtfAB and EtfABCX (Fig. S8). In each, the ASQ kinetic trace was best fit with two components corresponding to lifetimes of 60 and 1080 ps, suggesting that they arise from distinct flavins that are present in both EtfAB and EtfABCX. We therefore assigned the 60- and 1080-ps SQ components to the BF- and ET-FAD, respectively, based on the titration results that show the BF-FAD generates a more thermodynamically unstable ASQ than ET-FAD. In addition to the ASQ signal, we also observed the presence of a signal at ∼570 nm, suggesting that there is emission from FAD OX, the origin of which is unknown (Fig. S8A). The formation of flavin SQ states was further studied by visible and EPR spectroscopy, as discussed below.
Reductive titrations of EtfAB
The spectral features around 374 nm of as-purified EtfAB were assigned to an ASQ, and this was converted to the fully oxidized form by exposure to air (Fig. 4A and Fig. S6). Stepwise reduction of air-oxidized EtfAB with NADH regenerated the ASQ seen in the as-purified complex (prior to air oxidation), as indicated by spectral features at 374 nm with a shoulder at 398 nm. These reached a maximum with the addition of about half an equivalent of NADH (Fig. 4A and Fig. S9). To explain how a two-electron hydride transfer from NADH generates a singly reduced ASQ state on the ET-FAD, we propose intermolecular electron transfer between two EtfAB molecules, as was suggested for the NADH-induced ASQ state of the related EtfAB–Bcd complex of A. fermentans (16, 29). These data with P. aerophilum EtfAB therefore confirm the conclusion from the equilibrium titrations that the ET-FAD rests in the ASQ state when the complex is “ready” for bifurcation. EtfAB appeared fully reduced upon further addition of NADH to more than 2 eq, as shown by the lack of absorption at 454 nm. In addition, a strong charge transfer (CT) band feature is seen from 550 to 1000 nm (Fig. 4A and Fig. S9). The reduction of A. fermentans EtfAB with NADH showed a similar absorption spectrum, although the CT band was not as obvious (16). The CT band has to arise from the interaction of NAD+ with the BF-FAD because the ET-FAD does not interact with NAD(H) (13). Considering that NADH adds two electrons to the EtfAB core, there must be a flavin radical present (with three electrons distributed between two FAD cofactors); however, this is not obvious from the UV-visible spectrum.
Figure 4.
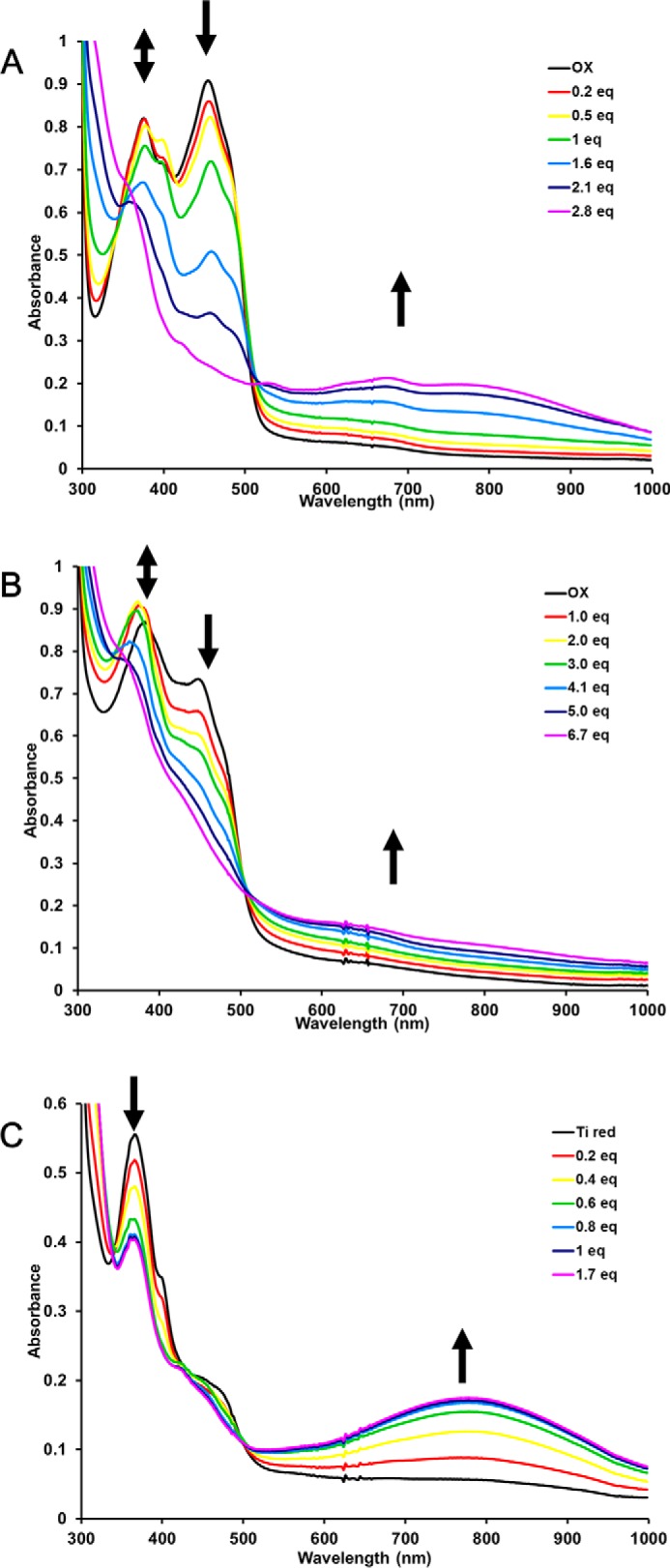
Titration of P. aerophilum EtfAB (74 μm) (A) and EtfABCX (15 μm) (B) with NADH. Arrows (left to right) indicate formation and disappearance of the ASQ, disappearance of OX flavin absorbance, and the formation of a broad CT complex. C, anaerobic titration with NAD+ of P. aerophilum EtfAB (74 μm) reduced with excess titanium citrate. Arrows (left to right) indicate the disappearance of ASQ and the formation of a broad CT complex.
EPR spectroscopy was used to directly probe for the presence and nature of radical species (44, 45). P. aerophilum EtfAB, as purified anaerobically in the absence of reductants, displayed a weak radical centered at g = 2.006 (0.1 spin mol−1), and this increased slightly (to 0.2 spin mol−1) upon the addition of NADH (Fig. 5). In contrast, reduction of EtfAB with excess sodium dithionite produced an intense anisotropic radical signal centered at g = 2.007 representing ∼1.0 spin mol−1. The narrow line width (14 G) is indicative of an ASQ radical (46), in agreement with the absorption data (see below) indicating that an ASQ radical is the source of this signal; however, it is not clear whether the radical resides on the BF- or ET-FAD. The addition of NAD+ to the dithionite-reduced sample decreased the EPR absorption by 50% and broadened the line width of the radical signal (from 14 to 20 G). Although change in line width is suggestive of a neutral semiquinone (NSQ) flavin radical (45), this broadening of the radical signal more likely reflects interactions of the BF-FAD with NAD+ affecting the optical and paramagnetic signal of the ASQ radical.
Figure 5.
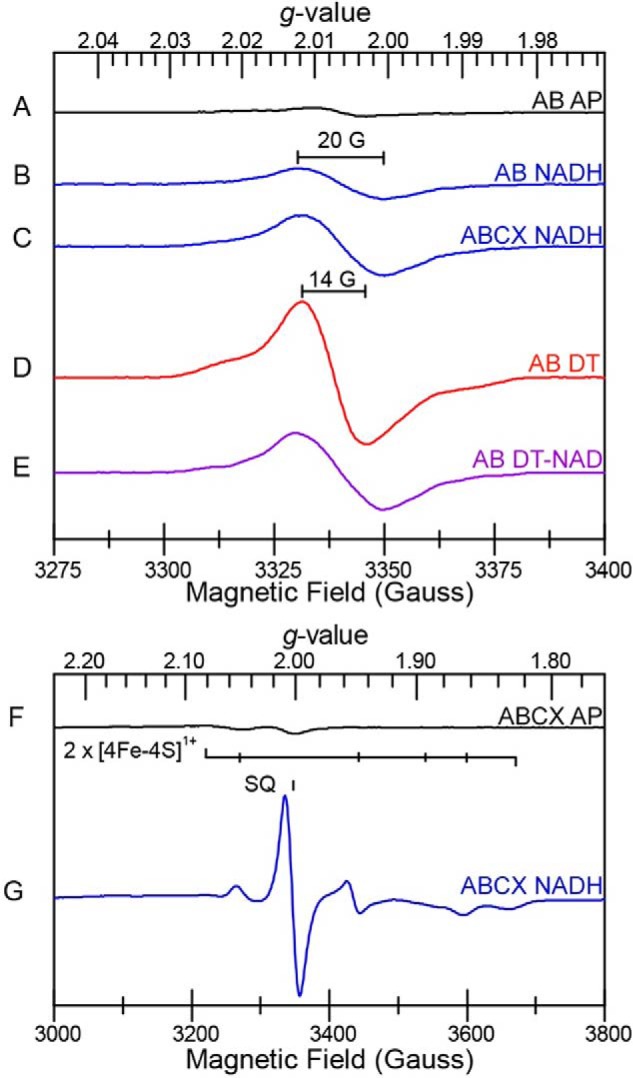
EPR spectra (9.38 GHz CW) of P. aerophilum EtfAB and EtfABCX. The samples are as follows. AB AP, as-purified EtfAB; AB NADH, EtfAB treated with NADH; ABCX NADH, EtfABCX treated with NADH; AB DT, EtfAB treated with sodium dithionite; AB DT-NAD, EtfAB sequentially treated with sodium dithionite and NAD+; ABCX AP, EtfABCX as purified; ABCX NADH, EtfABCX treated with NADH. Top spectra (A–E) were recorded at 77 K, and bottom spectra (F and G) were recorded at 10 K.
Similarly, treatment of P. aerophilum EtfAB with the chemical reductant titanium citrate (Fig. S6) generated an ASQ state that persisted even in the presence of excess reductant (Fig. 4C). We propose that this corresponds to the 3 e− reduced state of EtfAB. However, upon the addition of NAD+ to titanium-reduced EtfAB, the optical ASQ signature is lost, and a CT complex is generated (Fig. 4C and Fig. S10). It should be noted that this CT signature is slightly different (less absorbance in the 680-nm region) from that observed after treatment of EtfAB with excess NADH, where NAD+ is generated in situ (compare Figs. S9 and S10). These data confirm that NAD+ binding to the BF-FAD site changes the electronic state of the EtfAB core, whereby the usual absorption spectrum of the ASQ is not observed even though a radical can be seen by EPR analysis (Fig. 5). Thus, it appears that a 3 e− reduced state of EtfAB accumulates upon reduction with NADH and by chemical reductant, based on both optical and EPR spectroscopy.
Reductive titrations of EtfABCX
The response of EtfABCX to chemical reductants and to NAD(H) was similar to that of EtfAB (Fig. 4 and Figs. S11 and S12) but more complex due to the additional flavin (QR-FAD) in EtfC and the two [4Fe-4S] clusters in EtfX. Titration of EtfABCX with NADH forms an intermediate ASQ that disappears and is replaced by absorption from an NSQ near 620 nm, mixed with the broad absorption of the CT complex, which we assigned to the interaction of NAD+ with the EtfAB core (Fig. S12). The additional flavin radical (the NSQ) we assigned to the QR-FAD in EtfC. A strong semiquinone radical signal (0.5 spins mol−1) at g = 2 was observed upon reduction of EtfABCX with excess NADH, in addition to several overlapping broad signals spanning the g = 2.08–1.80 region indicative of reduced iron–sulfur clusters (Fig. 5). The semiquinone EPR signal centered at g = 2.006 was resolved at 77 K. The line width (20 G) is most consistent with it originating from an NSQ radical (45) (Fig. 5C), possibly from the QR-FAD in EtfC (Fig. 1). The broad EPR signals were most intense near 10 K and disappeared above 30 K (Fig. 5G), consistent with the relaxation properties of reduced [4Fe-4S]1+ clusters. The reduction potentials of the two [4Fe-4S] clusters in P. furiosus Nfn (see Fig. S13) were estimated at −513 and −718 mV by direct electrochemistry (31), but unfortunately, P. aerophilum EtfABCX was not responsive to this approach.
The reduction by NADH of the lower-potential [4Fe-4S] clusters of EtfX is further evidence of the bifurcating activity of P. aerophilum EtfABCX, as is formation of the QR-FAD of EtfC to generate an NSQ seen by visible absorption. In addition, NAD+ also interacts with chemically reduced P. aerophilum EtfABCX, yielding CT bands, but, as with EtfAB, the spectrum is slightly different (lacking some of the 680-nm features) from that induced by NADH treatment alone, indicating that the CT bands are an inherent property of the interaction of NAD+ with the EtfAB core.
Discussion
The data presented herein show that the P. aerophilum EtfAB core possesses several crucial prerequisites for bifurcating activity: 1) a bifurcating flavin that accepts electron pairs from NADH, 2) a high potential electron transfer pathway consisting of a high-potential flavin (Em = −94 mV for the ASQ/HQ couple of ET-FAD) linking the bifurcating flavin to the quinone pool via QR-FAD, and 3) a low potential pathway by which the BF-FAD reduces Fd (via the two 4Fe clusters in EtfX). Our measured Em (−285 mV) for the OX/HQ couple of BF-FAD is more positive than that of NADH (E0′ −320 mV), indicating favorable electron transfer but with minimal dissipation of energy. However, the BF-FAD does not display a stable SQ state in our mediated equilibrium titrations, which suggests that it exhibits crossed potentials where its Em ASQ/HQ is more positive than its Em OX/ASQ, although their average must correspond to the measured Em OX/HQ of −285 mV (ASQ is proposed based on its observation by TAS). Thus, we can calculate a lower limit for the Em of the BF-FAD ASQ/HQ couple. Assuming that the ASQ/HQ couple of the ET-FAD is operative in turnover (27, 30), electron transfer to ASQ-ET-FAD at −94 mV is exergonic by 191 mV. It follows that electron transfer to Fd can be endergonic at a similar level, placing a lower boundary of −476 mV on the Em of the BF-FAD OX/ASQ, which is presumably sufficient to reduce Fd (3, 47). For the respiratory bifurcating bc1 complex, a stability constant for the low potential SQ state of the bifurcating quinone was estimated at ∼10−15, from which a potential span of ∼900 mV between the first high-potential electron and the second low-potential electron was derived (48). This concept has been adapted to calculate the potentials of other bifurcating centers, including several EtfAB-containing complexes (reviewed in Ref. 8). Although the quinone value does not apply to a flavin system, which has very different chemistry, it puts our value in perspective. When it is applied to P. aerophilum EtfABCX, we calculate a ASQ/HQ couple of +165 mV (−285 + (900 mV/2)) and an OX/ASQ couple of −735 mV (−285 − (900 mV/2)). Hence, based on these two approaches, we estimate the Em of ASQ/HQ to be between −94 and +165 mV and that of OX/ASQ to be between −735 and −476 mV, with an average corresponding to the measured value of −285 mV. These values are well within the range proposed previously for other Etf-type complexes and are consistent with the thermodynamically favorable reduction of Fd with an Em of approximately −500 mV (8, 30, 48).
As shown in Fig. 6, we propose that, based on the equilibrium titration data, the 1 e− reduced state of EtfAB within the EtfABCX complex is the resting form of the enzyme. This accepts a hydride from NADH to generate the HQ-BF-FAD in close proximity to ASQ-ET-FAD, in which the NAD+ stays bound and forms a NAD+·BF-FADHQ CT complex. The BF- and ET-FAD could share an electron switching between HQ-BF-FAD/ASQ-ET-FAD and ASQ-BF-FAD/HQ-ET-FAD, and the stable CT complex is formed by the strong interaction of NAD+ with BF-FAD, as shown in Fig. 6. In this state, the EtfAB subcomplex is “hot” and can transfer the low-potential electron from the BF-FAD to Fd through the proximal and distal iron–sulfur clusters in EtfX, leaving the EtfAB subcomplex as OX-BF-FAD/HQ-ET-FAD. The distance between the ET-FAD and the QR-FAD is too far for efficient electron transfer (Fig. 2 and Refs. 16, 21, 29, 30, and 49), but it has been proposed that ETF-type complexes undergo a large conformational change as part of the catalytic cycle (16, 21, 29, 30). We suggest that this is triggered by oxidation of the ASQ-BF-FAD and brings the ET-FAD close enough to the QR-FAD for electron transfer into the high-potential active site (30). The high-potential electron on HQ-ET-FAD cannot reduce the highly crossed BF-FAD, so it can only be transferred to EtfC, thereby returning the EtfAB core to its bifurcation-ready 1 e− reduced state (Fig. 6). It is not clear when NAD+ leaves the EtfAB core, but we assume that it is after or in conjunction with transfer of the low-potential electron from the BF-FAD.
Figure 6.
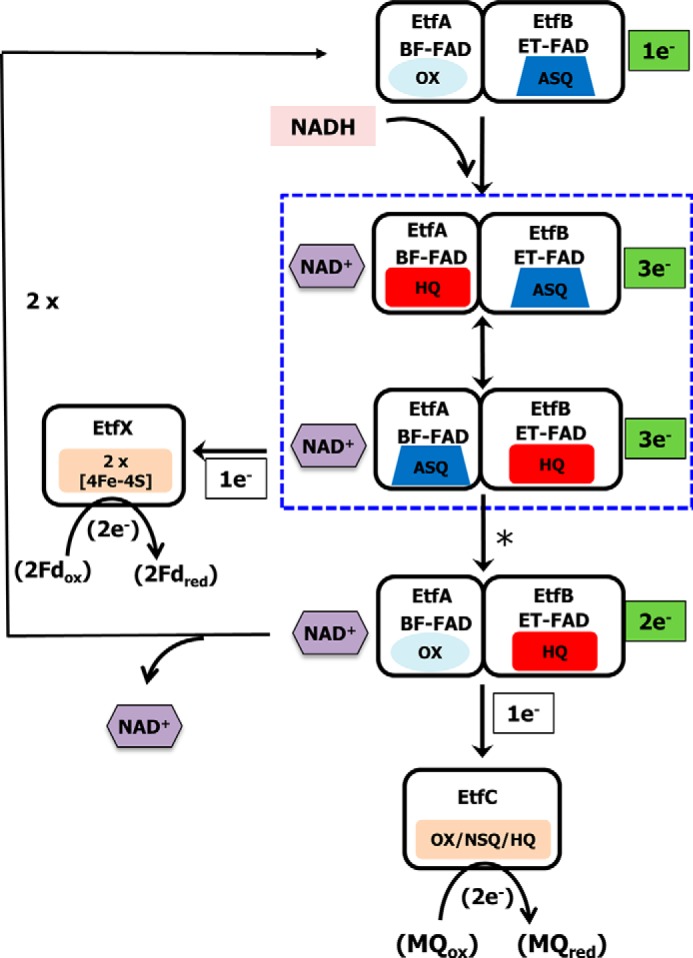
Proposed catalytic cycle of EtfABCX. The complex is depicted in its bifurcation-ready state in which the ET-FAD is in the ASQ state. In the first round of NADH oxidation, the BF-FAD is reduced to the HQ state and forms a CT complex with NAD+. The first electron is transferred to the ASQ-ET-FAD to generate the HQ state, leaving the ASQ-BF-FAD. The transfer of an electron to the low-potential branch [4Fe-4S] cluster triggers a protein conformational change (indicated by the asterisk) that then enables HQ-ET-FAD to reduce the FAD of EtfC with 1 e− converting it to the NSQ state. The catalytic cycle continues with the oxidation of a second NADH and the transfer of electrons down the low- and high-potential branches leading to the reduction of Fd and MQ, respectively. Boxed with a blue dotted line are the complexes with NAD+ that could form CT complexes.
Once the 1 e− reduced EtfAB core of EtfABCX is generated by loss of the high-potential electron, the enzyme then undergoes a second round of reduction by NADH. Note that only the first round of NADH oxidation is shown in Fig. 6. Hence, to complete the overall 4 e− reaction (Reaction 1), a second NADH binds, a second molecule of Fd is reduced via EtfX, and a second electron is transferred to NSQ-QR-FAD, which in its fully reduced HQ form reduces one molecule of MQ. Thus, the two rounds of bifurcation that constitute the catalytic cycle of EtfABCX (Reaction 1) are in essence the same in terms of the BF- and ET-FAD, where the second NADH interacts with the same bifurcation-ready 1 e− reduced form of the EtfAB subcomplex as the first NADH.
It is important to point out that our proposed mechanism (Fig. 6) eliminates the possibility of the second electron from BF-FAD passing to the ET-FAD, despite the favorable thermodynamics, because the ET-FAD is fully reduced to its HQ state upon accepting the first electron derived from NADH and therefore is unable to accept a second electron (30). Thus, the strongly reducing electron of the ASQ-BF-FAD would be constrained to use the low-potential branch formed by the [4Fe-4S] clusters in EtfX. In contrast, our in vitro reductions of fully oxidized EtfAB enable NADH to provide two electrons and reduce the ET-FAD from the OX to the HQ state, whereupon electron transfer between HQ-ET-FAD-EtfAB and OX-ET-FAD-EtfAB generates two ASQ-ET-FAD-EtfABs, thereby explaining the formation of the ASQ state during NADH titrations (Fig. 4A and Fig. S9). This was also proposed for the related EtfAB–Bcd complex (16). This ASQ state is also formed in the P. aerophilum EtfABCX complex when all partners are present (Fig. 4B and Fig. S12).
Reduction of P. aerophilum EtfAB and EtfABCX by NADH generates a striking CT band spanning 600 to 1000 nm (Fig. 4, A and B) with little absorbance remaining at 454 nm. Chemical reduction of EtfAB and EtfABCX both generate a flavin ASQ that does not undergo further reduction with excess reductant (Figs. S6 and S11), and upon NAD+ binding, the optical ASQ signal disappears and a CT signal appears instead (Fig. 4C and Fig. S11). Previous reports on related ETF systems from mesophilic organisms either do not collect data for the full 600–1000-nm region or report much weaker absorption (16, 21). We propose that the much stronger signal seen from the P. aerophilum proteins is a consequence of the hyperthermophilic organism that we have used as the gene donor to produce EtfABCX. The optical spectra were collected at room temperature, which is 80 °C lower than the optimum growth temperature of P. aerophilum (25). In contrast, the related Etf systems caffeyl-CoA reductase (CarCDE), butyryl-CoA dehydrogenase-Etf, and FixABCX derive from mesophilic microorganisms (4, 19, 24). The enhanced stability of hyperthermophilic enzyme might intensify the effect of NAD(H) binding, leading to more distinct spectral features for the CT complexes.
With P. aerophilum EtfABCX, we experimentally distinguished two different CT optical signatures. Both have a broad CT feature with a maximum around 780 nm, but if NADH is used as the reductant, a second feature is seen near 680 nm (Fig. 4 (A and B) and Figs. S9 and S12). From our data, the CT complex is formed by NAD+ binding to the BF-FAD, and a flavin radical is present. We propose that both BF-FAD and ET-FAD are electronically coupled and influence the nature of the NAD+-bound complex. It was previously suggested for a related EtfAB complex that a tyrosine residue situated between the BF-FAD and ET-FAD mediates an electronic coupling between the two flavin cofactors (27). In P. aerophilum EtfAB, a tryptophan residue occupies this position and would also be competent to mediate such a coupling. Although a charge transfer between NADH and the OX-BF-FAD could also be possible (50, 51), this is inconsistent with both the radical EPR signal and the absence of an optical signature expected from the ET-FAD ASQ. The exact nature of the NAD+-bound complexes in P. aerophilum EtfABCX is therefore unclear at present.
The reaction mechanism of P. aerophilum EtfABCX involves a bifurcating flavin with crossed potentials, where removing one electron from the HQ state generates a high-energy intermediate (ASQ) with a potential lower than that of the donor electron pair (from NADH) that is capable of ultimately reducing Fd (1, 31), although it should be noted that crossed potentials may not be an absolute requirement for bifurcating activity (52). It is generally accepted that this ASQ derived from a highly crossed BF-FAD is “unstable” (53). In our new NAD+-dependent mechanism, we propose that the formation of a NAD+ BF-FAD complex might protect this high-energy low-potential site from potentially harmful chemical reactions, especially when the EtfAB core is not complexed with its partners (Fig. 6). For example, it was previously shown that molecular oxygen can be reduced by the analogous EtfAB–Bcd complex, and this leads to formation of superoxide with potentially disastrous consequences for the cell (22). Based on our reaction scheme (Fig. 6), we propose that the addition of NADH to the EtfAB–Bcd complex also forms a NAD+ BF-FAD complex and that, in the absence of the high-potential substrate (crotonyl-CoA), this minimizes reaction of the BF-FAD with oxygen. Protection of a flavin site from oxygen through formation of a NAD+ complex to prevent the formation of damaging superoxide has been shown before with enzymes such as toluene dioxygenase and NADH quinone oxidoreductase (54, 55). The formation of a NAD+ charge transfer complex might therefore be an integral feature of EtfAB complexes and play an important role not only in its mechanism but also in protecting the cell from reactive oxygen species, a particularly important feature in (micro)aerophilic microorganisms, such as P. aerophilum (8, 29, 30, 48). Unexpectedly, we did not observe the complete 4 e− reduced form of EtfAB (or the EtfABCX equivalent) with either the physiological electron donor NADH or with an artificial reductant. This interesting characteristic of the EtfAB core is presumably related to the formation of the NAD+ BF-FAD complex, although the reason for this and the detailed nature of the different electronic states will be a topic of future studies.
The only other type of bifurcating enzyme that has been studied in detail mechanistically is the phylogenetically unrelated NfnI of P. furiosus (31). At first glance, this catalyzes a similar reaction to that of P. aerophilum EtfABCX, except that the high-potential acceptor is NAD rather than quinone (Fig. 1). However, other than a common site (BF-FAD) to bifurcate the two electrons from the hydride transfer from NAD(P)H, the proposed catalytic cycle of P. aerophilum EtfABCX (Fig. 6) is very different from that of NfnI (Fig. S13). In both enzymes, reduction of BF-FAD by NAD(P)H provides electrons for the high- and low-potential pathways, but the stable nicotinamide-dependent CT complex observed with EtfABCX is not evident with NfnI. In further contrast, the 1 e− reduced form of EtfABCX with the ASQ state of the ET-FAD is the proposed physiological resting form, which is ready to accept a hydride from NADH to form 3 e− reduced intermediates. However, the resting state of NfnI is fully oxidized, and its reduction by NADPH generates a stable 2 e− reduced state (Fig. S13). Moreover, in Nfn, the subsequent oxidation of ASQ-BF-FAD by reduction of ET-FAD is gated by an intervening [2Fe-2S] cluster, which mediates electron flow via the ET-FAD to the high-potential acceptor (in this case, NAD+) in the absence of any significant conformational change (56). In further contrast, electron flow to the high-potential pathway in EtfABCX is gated by a flavin 1 e− acceptor and a rather dramatic conformational change that enables oxidation of ET-FAD (31).
The results presented herein therefore show that, despite a common bifurcating flavin, P. aerophilum EtfABCX and P. furiosus NfnI differ by formation of a stable nicotinamide-dependent CT complex in the former and not the latter, in the nature of the resting and bifurcating-ready states of the enzymes, in how electron flow down the exergonic path is gated, and in the two 2 e− catalytic cycles that constitute the overall 4 e− bifurcation reaction. They also differ in the overall energetics of their reactions. Whereas both reduce Fd (Em ∼−500 mV), the driving force for EtfABCX (ΔE = 250 mV) is derived from NADH oxidation (E0′ = −320 mV) coupled to MQ reduction (E0′ = −70 mV). This provides more than twice the driving force for bifurcation than in NfnI (ΔE = 100 mV), which couples the oxidation of NADPH (Em ∼−380 mV, under physiological conditions) to the reduction of NAD+ (Em ∼−280 mV). It is not clear whether the overall energetics are related to any differences in the overall catalytic cycles for the two enzymes. This emphasizes that there might not be a general flavin-based bifurcating mechanism; rather, each type of enzyme harbors a unique reaction scheme (3, 8, 11, 12). Nevertheless, the reaction scheme shown in Fig. 6 is proposed as a framework to guide future work in elucidating more mechanistic details of the bifurcating reaction of P. aerophilum EtfABCX and of bifurcating ETFs in general.
Experimental procedures
Expression and purification of P. aerophilum EtfABCX
The plasmid encoding P. aerophilum EtfABCX was constructed with the following fragments via Gibson assembly (New England Biolabs): an 8.8-kb backbone BAC vector containing the pyrF genetic marker, the flanking homologous recombination region targeting the interspace between convergent genes PF1232 and PF1233, the slp promoter region (PF1399), and the 5.04-kb EtfBAXC genes PAE0721–0723, with a His9-Ala tag at the N terminus of the EtfA subunit (PAE0725). The order of genes in this cluster was rearranged to EtfABCX (PAE0722, PAE0721, PAE0725, and PAE0723) with a P. furious ribosomal binding site inserted in front of each Etf subunit. This plasmid, termed pGL098, was sequence-verified and linearized before transformation of P. furiosus COM1 strain as described previously (57). Positive transformants were cultured and purified twice on liquid and solid defined cellobiose medium. The genomic DNA was isolated using the ZymoBeadTM genomic DNA kit (Zymo Research), and PCR was used for isolate screening, using a primer pair that targeted outside the homologous flanking region mentioned above. The strain containing the EtfABCX overexpression construct was designated as MW393. All primers used in plasmid and strain construction are listed in Table S2.
Large-scale fermentation of recombinant P. furiosus was performed as described previously (58). Frozen cells (∼100 g wet weight) were lysed in 50 mm phosphate buffer, pH 7.5, with 1 mm DTT under strict anaerobic conditions. Cell debris and membranes were removed by centrifugation at 100,000 × g. The supernatant was loaded on a 5-ml Histrap column (GE Healthcare) using the recommended phosphate buffer with the addition of 1 mm DTT. The green-colored EtfABCX started eluting when ∼50 mm imidazole was applied. Based on the chromatogram, the green/yellow fractions were pooled and concentrated to ∼5–10 ml using ultrafiltration under anaerobic conditions. The concentrated sample was applied to a Superdex S200 column (60/600; GE Healthcare) and eluted with 25 mm Tris-HCl, pH 8, containing 300 mm NaCl (but lacking DTT). Fractions corresponding to EtfAB and EtfABCX were selected on the basis of flavin content and gel electrophoresis (Miniprotean TGX stainfree gels; Bio-Rad).
Enzyme assays
NADH-linked activities of EtfAB and ABCX were measured anaerobically at 80 °C in 3-ml glass cuvettes sealed with rubber stoppers containing 50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm NADH, and 200 μm iodonitrotetrazolium chloride. The formation of the red formazan was followed at 500 nm (ϵ = 19.3 mm−1 cm−1) in a Cary 100 UV-visible spectrophotometer with a Peltier-based temperature controller (Agilent). When menadione (100 μm) was used as electron acceptor, the activity was followed by the oxidation of NADH (200 μm, ϵ = 6.2 mm−1 cm−1 at 340 nm). Bifurcating assays were performed anaerobically in quartz 4-ml fluorescence cuvettes sealed with silicon stoppers using a RF-5301-PC fluorimeter (Shimadzu). P. furiosus Fd (50 μm) and menadione (100 μm) were reduced with 400 μm titanium citrate (reduction of Fd was monitored by its UV-visible spectrum). NAD+ (1 mm) was added as electron acceptor, and the formation of NADH was followed by fluorescence (excitation 340 nm/emission 460 nm).
Redox titrations
Titrations were performed at 25 °C in an upcycled Agilent HP 8453 operating under OlisWorks (Olis) using 600-μl masked quartz cuvettes (Starna) anaerobically sealed with silicon stoppers. Reductant was anaerobically added in small quantities with a 10-μl gas-tight syringe (Hamilton) to samples prepared in 50 mm MOPS, pH 7.5, with 200 mm NaCl. UV-visible spectra, 200–1000 nm, were recorded after each addition. All solutions were prepared under strict anaerobic conditions, and stock solutions of titanium citrate were prepared as described previously (59).
To determine reduction midpoint potentials at pH 7.0 (E0′), protein samples were prepared in 25 mm Bistris propane, pH 7.0, containing 200 mm NaCl. E0′ values were measured using the xanthine/xanthine oxidase system (27, 60, 61). Attainment of equilibrium was confirmed by comparing E0′ values obtained in slower and faster titrations; 40 min was typical (62), and all titrations were performed in duplicate or more. The dyes used were thionin acetate (nDye = 2, E0′Dye = +56 mV, λmax = 598 nm), Nile blue chloride (nDye = 2, E0′Dye = −116 mV, λmax = 636 nm), and safranin-O (nDye = 2, E0′Dye = −289 mV, λmax = 520 nm, and the dye isosbestic at 412 nm was used for monitoring the flavin). Data were analyzed using Equation 1 derived by equating the Nernst equation for the dye with that of the enzyme, at equilibrium with the dye, for the example of the OX/ASQ couple. At each point in the titrations,
| (Eq. 1) |
the concentrations of oxidized and reduced dye and reacting flavin were determined from the absorbance values of each at the appropriate wavelength. To focus on only the one flavin undergoing reduction in each phase of reduction, difference spectra were used. In these, the final spectrum of each phase was subtracted from each other spectrum in the phase, and the three phases were identified by noting the different changes in A374 characterizing initial reduction of OX to ASQ, ASQ to HQ, and OX to HQ (Fig. S6). Thus, we calculated the concentration of FAD or dye remaining in the oxidized state of the reaction under consideration ([OxFAD] or [Oxdye]), and from these and the total concentrations, we obtained the concentrations of the corresponding reduced states ([RedFAD] or [Reddye]). Plots of log10([OxFAD]/[RedFAD˙̄]) versus log10([OxDye]/[RedDye]) then yielded nFAD, the number of electrons taken up by the FAD revealing the nature of each reaction, from the magnitude of the slope, m = nFAD/ndye. The intercept with the vertical axis at log([Oxdye]/[Reddye]) = 0, b, then yielded the flavin E0′ of the reaction from b = nFAD(E0′Dye − E0′FAD)/25.7 mV, where 25.7 mV replaces the physical constants and temperature RT/F (T = 25 °C), and all E0′ values are quoted versus the normal hydrogen electrode.
EPR spectroscopy
Samples of P. aerophilum EtfAB and EtfABCX were prepared for analysis by EPR spectroscopy under strictly anaerobic conditions in an N2 atmosphere glove box (MBraun). Both EtfAB (150 μm) and EtfABCX (120 μm) were prepared in 25 mm Tris buffer containing 300 mm NaCl and 5% (v/v) glycerol. EtfAB was treated with either NADH (1 mm), sodium dithionite (10 mm), or sequential treatments of sodium dithionite (10 mm) and NAD+ (10 mm). For the latter, NAD+ was added to a thawed sample that had been initially treated and frozen with sodium dithionite. All samples were prepared in sealed tubes and slowly frozen in liquid N2. EPR spectra were recorded with a Bruker E-500 spectrometer (X-band, 9.38 GHz) equipped with an SHQ resonator, an in-cavity cryogen-free VT system (ColdEdge Technologies), and a MercuryiTC temperature controller (Oxford). EPR parameters for measuring radical signals were as follows: modulation frequency, 100 kHz; modulation amplitude, 2 G; microwave power, 1 milliwatt; temperature, 77 K. EPR parameters for measuring [Fe-S] signals were as follows: modulation frequency, 100 kHz; modulation amplitude, 10 G; microwave power, 1 milliwatt; temperature, 10 K. Spin quantification of radical signals was carried out by comparison of signal intensities with standards using HO-TEMPO (4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxy, 5–100 μm concentration) and double integration of the spectra using the OriginPro software package.
TAS
The ultrafast (100 fs to 5.1 ns) TAS spectrometer employed here was operated as described previously (31). The pump wavelength was 400 nm for sample excitation, and the experiment was conducted at room temperature. For all measurements, the sample was prepared in an Mbraun glove box (N2 atmosphere), sealed in a 2-mm quartz cuvette, and constantly stirred to prevent photodegradation. The concentrations of P. aerophilum EtfAB and EtfABCX were ∼150 and 100 μm, respectively. The data collection (350 pump shots per time point) was carried out four consecutive times and then averaged. The experiment was repeated two times. Data were corrected for spectral chirp using SurfaceXplorer (Ultrafast Systems). Fitting was performed in Igor Pro using single- or double-exponential fit functions.
MS analysis
Protein identification (63), intact protein analysis (64), and protein–protein interaction analysis (24, 65) were performed as described previously. The P. aerophilum EtfABCX complex (7.9 μm) was surface-labeled with 0.3 mm DnsCl (Acros) in a 100 mm sodium phosphate buffer, pH 8.0, at 25 °C. The reaction was quenched after 5, 15, and 30 min with 2 μl of 100 mm ammonium acetate, pH 8.0. Samples were digested with trypsin followed by LC-MS/MS analysis (63). Native MS experiments were conducted on a SYNAPT G2-Si instrument (Waters) (66). Samples were exchanged to 500 mm ammonium acetate, pH 7.0 (Sigma), and infused from in-house prepared gold-coated borosilicate glass capillaries at a protein concentration of 1.5 μm and a flow rate of 90 nl min−1. Data analysis was performed in MassLynx version 4.1 (Waters). The EtfABCX model was produced using Swiss Modeler (67), followed by manual adjustment of a few side chains and unstructured strands and then energy minimization in Chimera (68). These were tested on the basis of the fit of the cofactors from the template structures within the modeled P. aerophilum components.
Author contributions
G. J. S., N. M.-R., M. T.-L., D. W. M., D. M. N. N., G. L. L., J. P. H., C. E. L., P. W. K., B. B., A.-F. M., and M. W. W. A. data curation; G. J. S., N. M.-R., M. T.-L., D. W. M., D. M. N. N., G. L. L., J. P. H., A. P., C. E. L., P. W. K., B. B., A.-F. M., and M. W. W. A. formal analysis; G. J. S., N. M.-R., M. T.-L., D. W. M., D. M. N. N., G. L. L., J. P. H., A. P., C. E. L., P. W. K., and B. B. investigation; G. J. S., N. M.-R., M. T.-L., D. W. M., D. M. N. N., G. L. L., J. P. H., A. P., C. E. L., P. W. K., and B. B. methodology; G. J. S. and A.-F. M. writing-original draft; G. J. S., N. M.-R., M. T.-L., D. W. M., D. M. N. N., G. L. L., J. P. H., A. P., C. E. L., P. W. K., J. W. P., B. B., A.-F. M., and M. W. W. A. writing-review and editing; J. W. P. and M. W. W. A. funding acquisition; J .W. P. and M. W. W. A. project administration; M. W. W. A. conceptualization; M. W. W. A. resources; M. W. W. A. supervision.
Supplementary Material
Acknowledgments
The Mass Spectrometry Facility at Montana State University is supported in part by the Murdock Charitable Trust and National Institutes of Health IDEA Program Grant P20GM103474. We thank the Microfabrication Facility of Montana State University for help in preparation of gold-coated borosilica capillaries for noncovalent MS and Zachary Wood (University of Georgia) for use of the upcycled Agilent HP 8453.
This work and all authors were supported as part of the Biological and Electron Transfer and Catalysis (BETCy) EFRC, an Energy Frontier Research Center funded by the United States Department of Energy, Office of Science, Basic Energy Sciences under Award DE-SC0012518. Additional support was provided by the Research Challenge Trust fund of Kentucky (NM-R) and by the “Unifying Concepts in Catalysis” cluster of excellence (UNICAT) and the Technische Universität-Berlin (to A.-F. M.). This work was authored in part by the National Renewable Energy Laboratory, operated by Alliance for Sustainable Energy, LLC, for the U.S. Department of Energy (DOE) under Contract No. DE-AC36–08GO28308 (to D. W. M., C. E. L., and P. W. K.). Funding for TAS and EPR measurements provided by the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences (D. W. M., C. E. L., and P. W. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The views expressed in the article do not necessarily represent the views of the DOE or the U.S. Government.
This article contains Tables S1 and S2 and Figs. S1–S13.
- ETF
- electron transfer flavoprotein
- BF-FAD
- bifurcating flavin
- ET-FAD
- electron transfer flavin
- QR-FAD
- quinone reductase flavin
- Fd
- ferredoxin
- BS3
- bis(sulfosuccinimidyl)suberate
- GEE
- glycine ethyl ester
- DnsCl
- dansyl chloride
- MQ
- menaquinone
- ASQ
- anionic semiquinone
- TAS
- transient absorption spectroscopy
- CT
- charge transfer
- NSQ
- neutral semiquinone
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane.
References
- 1. Peters J. W., Miller A. F., Jones A. K., King P. W., and Adams M. W. (2016) Electron bifurcation. Curr. Opin. Chem. Biol. 31, 146–152 10.1016/j.cbpa.2016.03.007 [DOI] [PubMed] [Google Scholar]
- 2. Brandt U. (1996) Bifurcated ubihydroquinone oxidation in the cytochrome bc1 complex by proton-gated charge transfer. FEBS Lett. 387, 1–6 10.1016/0014-5793(96)00436-X [DOI] [PubMed] [Google Scholar]
- 3. Buckel W., and Thauer R. K. (2013) Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113 10.1016/j.bbabio.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 4. Li F., Hinderberger J., Seedorf H., Zhang J., Buckel W., and Thauer R. K. (2008) Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190, 843–850 10.1128/JB.01417-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Metcalf W. W. (2016) Classic spotlight: electron bifurcation, a unifying concept for energy conservation in anaerobes. J. Bacteriol. 198, 1358 10.1128/JB.00185-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitchell P. (1976) Possible molecular mechanisms of the protonmotive function of cytochrome systems. J. Theor. Biol. 62, 327–367 10.1016/0022-5193(76)90124-7 [DOI] [PubMed] [Google Scholar]
- 7. Schut G. J., and Adams M. W. (2009) The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457 10.1128/JB.01582-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buckel W., and Thauer R. K. (2018) Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 118, 3862–3886 10.1021/acs.chemrev.7b00707 [DOI] [PubMed] [Google Scholar]
- 9. Herrmann G., Jayamani E., Mai G., and Buckel W. (2008) Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190, 784–791 10.1128/JB.01422-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buckel W., and Thauer R. K. (2018) Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: a historical review. Front. Microbiol. 9, 401 10.3389/fmicb.2018.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peters J. W., Beratan D. N., Schut G. J., and Adams M. W. W. (2018) On the nature of organic and inorganic centers that bifurcate electrons, coupling exergonic and endergonic oxidation-reduction reactions. Chem. Commun. 54, 4091–4099 10.1039/C8CC01530A [DOI] [PubMed] [Google Scholar]
- 12. Peters J. W., Beratan D. N., Bothner B., Dyer R. B., Harwood C. S., Heiden Z. M., Hille R., Jones A. K., King P. W., Lu Y., Lubner C. E., Minteer S. D., Mulder D. W., Raugei S., Schut G. J., et al. (2018) A new era for electron bifurcation. Curr. Opin. Chem. Biol. 47, 32–38 10.1016/j.cbpa.2018.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia Costas A. M., Poudel S., Miller A. F., Schut G. J., Ledbetter R. N., Fixen K. R., Seefeldt L. C., Adams M. W. W., Harwood C. S., Boyd E. S., and Peters J. W. (2017) Defining electron bifurcation in the electron-transferring flavoprotein family. J. Bacteriol. 199, e00440–e00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toogood H. S., Leys D., and Scrutton N. S. (2007) Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes. FEBS J. 274, 5481–5504 10.1111/j.1742-4658.2007.06107.x [DOI] [PubMed] [Google Scholar]
- 15. Tsai M. H., and Saier M. H. Jr. (1995) Phylogenetic characterization of the ubiquitous electron transfer flavoprotein families ETF-α and ETF-β. Res. Microbiol. 146, 397–404 10.1016/0923-2508(96)80285-3 [DOI] [PubMed] [Google Scholar]
- 16. Chowdhury N. P., Mowafy A. M., Demmer J. K., Upadhyay V., Koelzer S., Jayamani E., Kahnt J., Hornung M., Demmer U., Ermler U., and Buckel W. (2014) Studies of the mechanism of electron bifurcation catalyzed by electron transferring flavoprotein (Etf) and butyryl-CoA dehydrogenase (Bcd) of Acidaminococcus fermentans. J. Biol. Chem. 289, 5145–5157 10.1074/jbc.M113.521013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sato K., Nishina Y., and Shiga K. (2003) Purification of electron-transferring flavoprotein from Megasphaera elsdenii and binding of additional FAD with an unusual absorption spectrum. J. Biochem. 134, 719–729 10.1093/jb/mvg199 [DOI] [PubMed] [Google Scholar]
- 18. Weghoff M. C., Bertsch J., and Müller V. (2015) A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ. Microbiol. 17, 670–677 10.1111/1462-2920.12493 [DOI] [PubMed] [Google Scholar]
- 19. Bertsch J., Parthasarathy A., Buckel W., and Müller V. (2013) An electron-bifurcating caffeyl-CoA reductase. J. Biol. Chem. 288, 11304–11311 10.1074/jbc.M112.444919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brockman H. L., and Wood W. A. (1975) d-Lactate dehydrogenase of Peptostreptococcus elsdenii. J. Bacteriol. 124, 1454–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato K., Nishina Y., and Shiga K. (2013) Interaction between NADH and electron-transferring flavoprotein from Megasphaera elsdenii. J. Biochem. 153, 565–572 10.1093/jb/mvt026 [DOI] [PubMed] [Google Scholar]
- 22. Chowdhury N. P., Kahnt J., and Buckel W. (2015) Reduction of ferredoxin or oxygen by flavin-based electron bifurcation in Megasphaera elsdenii. FEBS J. 282, 3149–3160 10.1111/febs.13308 [DOI] [PubMed] [Google Scholar]
- 23. Edgren T., and Nordlund S. (2004) The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J. Bacteriol. 186, 2052–2060 10.1128/JB.186.7.2052-2060.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ledbetter R. N., Garcia Costas A. M., Lubner C. E., Mulder D. W., Tokmina-Lukaszewska M., Artz J. H., Patterson A., Magnuson T. S., Jay Z. J., Duan H. D., Miller J., Plunkett M. H., Hoben J. P., Barney B. M., Carlson R. P., et al. (2017) The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 56, 4177–4190 10.1021/acs.biochem.7b00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Völkl P., Huber R., Drobner E., Rachel R., Burggraf S., Trincone A., and Stetter K. O. (1993) Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl. Environ. Microbiol. 59, 2918–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fitz-Gibbon S. T., Ladner H., Kim U. J., Stetter K. O., Simon M. I., and Miller J. H. (2002) Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. U.S.A. 99, 984–989 10.1073/pnas.241636498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duan H. D., Lubner C. E., Tokmina-Lukaszewska M., Gauss G. H., Bothner B., King P. W., Peters J. W., and Miller A. F. (2018) Distinct properties underlie flavin-based electron bifurcation in a novel electron transfer flavoprotein FixAB from Rhodopseudomonas palustris. J. Biol. Chem. 293, 4688–4701 10.1074/jbc.RA117.000707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watmough N. J., and Frerman F. E. (2010) The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim. Biophys. Acta 1797, 1910–1916 10.1016/j.bbabio.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 29. Demmer J. K., Bertsch J., Öppinger C., Wohlers H., Kayastha K., Demmer U., Ermler U., and Müller V. (2018) Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction. FEBS Lett. 592, 332–342 10.1002/1873-3468.12971 [DOI] [PubMed] [Google Scholar]
- 30. Demmer J. K., Pal Chowdhury N., Selmer T., Ermler U., and Buckel W. (2017) The semiquinone swing in the bifurcating electron transferring flavoprotein/butyryl-CoA dehydrogenase complex from Clostridium difficile. Nat. Commun. 8, 1577 10.1038/s41467-017-01746-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lubner C. E., Jennings D. P., Mulder D. W., Schut G. J., Zadvornyy O. A., Hoben J. P., Tokmina-Lukaszewska M., Berry L., Nguyen D. M., Lipscomb G. L., Bothner B., Jones A. K., Miller A. F., King P. W., Adams M. W. W., and Peters J. W. (2017) Mechanistic insights into energy conservation by flavin-based electron bifurcation. Nat. Chem. Biol. 13, 655–659 10.1038/nchembio.2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song Y., Nelp M. T., Bandarian V., and Wysocki V. H. (2015) Refining the structural model of a heterohexameric protein complex: surface induced dissociation and ion mobility provide key connectivity and topology information. ACS Cent. Sci. 1, 477–487 10.1021/acscentsci.5b00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baud A., Gonnet F., Salard I., Le Mignon M., Giuliani A., Mercère P., Sclavi B., and Daniel R. (2016) Probing the solution structure of Factor H using hydroxyl radical protein footprinting and cross-linking. Biochem. J. 473, 1805–1819 10.1042/BCJ20160225 [DOI] [PubMed] [Google Scholar]
- 35. Fernandez-Martinez J., Kim S. J., Shi Y., Upla P., Pellarin R., Gagnon M., Chemmama I. E., Wang J., Nudelman I., Zhang W., Williams R., Rice W. J., Stokes D. L., Zenklusen D., Chait B. T., Sali A., and Rout M. P. (2016) Structure and function of the nuclear pore complex cytoplasmic mRNA export platform. Cell 167, 1215–1228.e25 10.1016/j.cell.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu W. H., Roemer S. C., Zhou Y., Shen Z. J., Dennehey B. K., Balsbaugh J. L., Liddle J. C., Nemkov T., Ahn N. G., Hansen K. C., Tyler J. K., and Churchill M. E. (2016) The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones. Elife 5, e18023 10.7554/eLife.18023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mummadisetti M. P., Frankel L. K., Bellamy H. D., Sallans L., Goettert J. S., Brylinski M., Limbach P. A., and Bricker T. M. (2014) Use of protein cross-linking and radiolytic footprinting to elucidate PsbP and PsbQ interactions within higher plant Photosystem II. Proc. Natl. Acad. Sci. U.S.A. 111, 16178–16183 10.1073/pnas.1415165111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmidt C., Zhou M., Marriott H., Morgner N., Politis A., and Robinson C. V. (2013) Comparative cross-linking and mass spectrometry of an intact F-type ATPase suggest a role for phosphorylation. Nat. Commun. 4, 1985 10.1038/ncomms2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zanphorlin L. M., Lima T. B., Wong M. J., Balbuena T. S., Minetti C. A., Remeta D. P., Young J. C., Barbosa L. R., Gozzo F. C., and Ramos C. H. (2016) Heat shock protein 90 kDa (Hsp90) has a second functional interaction site with the mitochondrial import receptor Tom70. J. Biol. Chem. 291, 18620–18631 10.1074/jbc.M115.710137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merkley E. D., Rysavy S., Kahraman A., Hafen R. P., Daggett V., and Adkins J. N. (2014) Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 10.1002/pro.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hagedoorn P. L., Chen T., Schröder I., Piersma S. R., de Vries S., and Hagen W. R. (2005) Purification and characterization of the tungsten enzyme aldehyde:ferredoxin oxidoreductase from the hyperthermophilic denitrifier Pyrobaculum aerophilum. J. Biol. Inorg. Chem. 10, 259–269 10.1007/s00775-005-0637-5 [DOI] [PubMed] [Google Scholar]
- 42. Elling F. J., Becker K. W., Könneke M., Schröder J. M., Kellermann M. Y., Thomm M., and Hinrichs K. U. (2016) Respiratory quinones in Archaea: phylogenetic distribution and application as biomarkers in the marine environment. Environ. Microbiol. 18, 692–707 10.1111/1462-2920.13086 [DOI] [PubMed] [Google Scholar]
- 43. Nguyen D. M. N., Schut G. J., Zadvornyy O. A., Tokmina-Lukaszewska M., Poudel S., Lipscomb G. L., Adams L. A., Dinsmore J. T., Nixon W. J., Boyd E. S., Bothner B., Peters J. W., and Adams M. W. W. (2017) Two functionally distinct NADP+-dependent ferredoxin oxidoreductases maintain the primary redox balance of Pyrococcus furiosus. J. Biol. Chem. 292, 14603–14616 10.1074/jbc.M117.794172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Iyanagi T. (1977) Redox properties of microsomal reduced nicotinamide adenine dinucleotide-cytochrome b5 reductase and cytochrome b5. Biochemistry 16, 2725–2730 10.1021/bi00631a021 [DOI] [PubMed] [Google Scholar]
- 45. Medina M., Gomez-Moreno C., and Cammack R. (1995) Electron spin resonance and electron nuclear double resonance studies of flavoproteins involved in the photosynthetic electron transport in the cyanobacterium Anabaena sp. PCC 7119. Eur. J. Biochem. 227, 529–536 10.1111/j.1432-1033.1995.tb20420.x [DOI] [PubMed] [Google Scholar]
- 46. DeRose V. J., Woo J. C., Hawe W. P., Hoffman B. M., Silverman R. B., and Yelekci K. (1996) Observation of a flavin semiquinone in the resting state of monoamine oxidase B by electron paramagnetic resonance and electron nuclear double resonance spectroscopy. Biochemistry 35, 11085–11091 10.1021/bi960749f [DOI] [PubMed] [Google Scholar]
- 47. Deistung J., and Thorneley R. N. F. (1986) Electron transfer to nitrogenase. Characterization of flavodoxin from Azotobacter chroococcum and comparison of its redox potentials with those of flavodoxins from Azotobacter vinelandii and Klebsiella pneumoniae (nifF-gene product). Biochem. J. 239, 69–75 10.1042/bj2390069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bergdoll L., Ten Brink F., Nitschke W., Picot D., and Baymann F. (2016) From low- to high-potential bioenergetic chains: thermodynamic constraints of Q-cycle function. Biochim. Biophys. Acta 1857, 1569–1579 10.1016/j.bbabio.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 49. Page C. C., Moser C. C., Chen X., and Dutton P. L. (1999) Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52 10.1038/46972 [DOI] [PubMed] [Google Scholar]
- 50. Peregrina J. R., Sánchez-Azqueta A., Herguedas B., Martínez-Júlvez M., and Medina M. (2010) Role of specific residues in coenzyme binding, charge-transfer complex formation, and catalysis in Anabaena ferredoxin NADP+-reductase. Biochim. Biophys. Acta 1797, 1638–1646 10.1016/j.bbabio.2010.05.006 [DOI] [PubMed] [Google Scholar]
- 51. Batie C. J., and Kamin H. (1986) Association of ferredoxin-NADP+ reductase with NADP(H) specificity and oxidation-reduction properties. J. Biol. Chem. 261, 11214–11223 [PubMed] [Google Scholar]
- 52. Zhang P., Yuly J. L., Lubner C. E., Mulder D. W., King P. W., Peters J. W., and Beratan D. N. (2017) Electron bifurcation: thermodynamics and kinetics of two-electron brokering in biological redox chemistry. Acc. Chem. Res. 50, 2410–2417 10.1021/acs.accounts.7b00327 [DOI] [PubMed] [Google Scholar]
- 53. Nitschke W., and Russell M. J. (2012) Redox bifurcations: mechanisms and importance to life now, and at its origin: a widespread means of energy conversion in biology unfolds. Bioessays 34, 106–109 10.1002/bies.201100134 [DOI] [PubMed] [Google Scholar]
- 54. Lin T. Y., Werther T., Jeoung J. H., and Dobbek H. (2012) Suppression of electron transfer to dioxygen by charge transfer and electron transfer complexes in the FAD-dependent reductase component of toluene dioxygenase. J. Biol. Chem. 287, 38338–38346 10.1074/jbc.M112.374918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sena F. V., Sousa F. M., Oliveira A. S. F., Soares C. M., Catarino T., and Pereira M. M. (2018) Regulation of the mechanism of Type-II NADH: quinone oxidoreductase from S. aureus. Redox Biol. 16, 209–214 10.1016/j.redox.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Berry L., Poudel S., Tokmina-Lukaszewska M., Colman D. R., Nguyen D. M. N., Schut G. J., Adams M. W. W., Peters J. W., Boyd E. S., and Bothner B. (2018) H/D exchange mass spectrometry and statistical coupling analysis reveal a role for allostery in a ferredoxin-dependent bifurcating transhydrogenase catalytic cycle. Biochim. Biophys. Acta Gen. Subj. 1862, 9–17 10.1016/j.bbagen.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 57. Lipscomb G. L., Stirrett K., Schut G. J., Yang F., Jenney F. E. Jr, Scott R. A., Adams M. W., and Westpheling J. (2011) Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77, 2232–2238 10.1128/AEM.02624-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Verhagen M. F., Menon A. L., Schut G. J., and Adams M. W. (2001) Pyrococcus furiosus: large-scale cultivation and enzyme purification. Methods Enzymol. 330, 25–30 10.1016/S0076-6879(01)30368-3 [DOI] [PubMed] [Google Scholar]
- 59. Zehnder A. J., and Wuhrmann K. (1976) Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 194, 1165–1166 10.1126/science.793008 [DOI] [PubMed] [Google Scholar]
- 60. Efimov I., Parkin G., Millett E. S., Glenday J., Chan C. K., Weedon H., Randhawa H., Basran J., and Raven E. L. (2014) A simple method for the determination of reduction potentials in heme proteins. FEBS Lett. 588, 701–704 10.1016/j.febslet.2013.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Massey V. (1991) A simple method for determination of redox potentials. in Flavins and Flavoproteins (Curti B., Ronchi S., and Zanetti G., eds) pp. 59–66, Walter de Gruyer, Berlin [Google Scholar]
- 62. Trimmer E. E., Ballou D. P., Galloway L. J., Scannell S. A., Brinker D. R., and Casas K. R. (2005) Aspartate 120 of Escherichia coli methylenetetrahydrofolate reductase: evidence for major roles in folate binding and catalysis and a minor role in flavin reactivity. Biochemistry 44, 6809–6822 10.1021/bi0477236 [DOI] [PubMed] [Google Scholar]
- 63. Yang Z. Y., Ledbetter R., Shaw S., Pence N., Tokmina-Lukaszewska M., Eilers B., Guo Q., Pokhrel N., Cash V. L., Dean D. R., Antony E., Bothner B., Peters J. W., and Seefeldt L. C. (2016) Evidence that the Pi release event is the rate-limiting step in the nitrogenase catalytic cycle. Biochemistry 55, 3625–3635 10.1021/acs.biochem.6b00421 [DOI] [PubMed] [Google Scholar]
- 64. Poudel S., Tokmina-Lukaszewska M., Colman D. R., Refai M., Schut G. J., King P. W., Maness P. C., Adams M. W., Peters J. W., Bothner B., and Boyd E. S. (2016) Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim. Biophys. Acta 1860, 1910–1921 10.1016/j.bbagen.2016.05.034 [DOI] [PubMed] [Google Scholar]
- 65. Zhang H., Wen J., Huang R. Y., Blankenship R. E., and Gross M. L. (2012) Mass spectrometry-based carboxyl footprinting of proteins: method evaluation. Int. J. Mass Spectrom. 312, 78–86 10.1016/j.ijms.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Luo M. L., Jackson R. N., Denny S. R., Tokmina-Lukaszewska M., Maksimchuk K. R., Lin W., Bothner B., Wiedenheft B., and Beisel C. L. (2016) The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucleic Acids Res. 44, 7385–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


