Abstract
Transforming growth factor β (TGFβ) potently activates hepatic stellate cells (HSCs), which promotes production and secretion of extracellular matrix (ECM) proteins and hepatic fibrogenesis. Increased ECM synthesis and secretion in response to TGFβ is associated with endoplasmic reticulum (ER) stress and the unfolded protein response (UPR). TGFβ and UPR signaling pathways are tightly intertwined during HSC activation, but the regulatory mechanism that connects these two pathways is poorly understood. Here, we found that TGFβ treatment of immortalized HSCs (i.e. LX-2 cells) induces phosphorylation of the UPR sensor inositol-requiring enzyme 1α (IRE1α) in a SMAD2/3-procollagen I–dependent manner. We further show that IRE1α mediates HSC activation downstream of TGFβ and that its role depends on activation of a signaling cascade involving apoptosis signaling kinase 1 (ASK1) and c-Jun N-terminal kinase (JNK). ASK1–JNK signaling promoted phosphorylation of the UPR-associated transcription factor CCAAT/enhancer binding protein β (C/EBPβ), which is crucial for TGFβ- or IRE1α-mediated LX-2 activation. Pharmacological inhibition of C/EBPβ expression with the antiviral drug adefovir dipivoxil attenuated TGFβ-mediated activation of LX-2 or primary rat HSCs in vitro and hepatic fibrogenesis in vivo. Finally, we identified a critical relationship between C/EBPβ and the transcriptional regulator p300 during HSC activation. p300 knockdown disrupted TGFβ- or UPR-induced HSC activation, and pharmacological inhibition of the C/EBPβ–p300 complex decreased TGFβ-induced HSC activation. These results indicate that TGFβ-induced IRE1α signaling is critical for HSC activation through a C/EBPβ-p300–dependent mechanism and suggest C/EBPβ as a druggable target for managing fibrosis.
Keywords: fibrosis, endoplasmic reticulum stress (ER stress), collagen, fibronectin, apoptosis signal-regulating kinase 1 (ASK1), c-Jun N-terminal kinase (JNK), cell signaling, adefovir dipivoxil, C/EBPβ, IRE1α, p300
Introduction
Hepatic stellate cell (HSC)4 activation increases production of extracellular matrix (ECM) proteins, leading to fibrogenesis (1, 2). ECM proteins undergo folding and processing within the endoplasmic reticulum (ER) prior to trafficking through the secretory pathway. Canonically, increased production of ECM proteins leads to ER stress and activation of the unfolded protein response (UPR), which facilitates their folding and trafficking out of the cell (3, 4). Based on this, several studies show that the UPR is critical for HSC activation. Indeed, stimuli that promote HSC activation, such as TGFβ, are associated with ER stress and UPR induction (5–7). Furthermore, chemical induction of the UPR in HSCs in vitro increased expression of ECM proteins in HSCs, whereas pharmacological inhibition of the UPR attenuated HSC activation (8–11). Despite these observations, the mechanisms by which the UPR regulates HSC activation, and in turn how HSC activation modulates UPR signaling, are unclear.
One of the major ER stress sensors that initiates the UPR is inositol-requiring enzyme 1α (IRE1α). IRE1α is critical for UPR signaling to accommodate increased protein folding and trafficking (12–14). Upon sensing increased levels of unfolded proteins in the ER, IRE1α dimerizes and undergoes autotransphosphorylation. This kinase activity is necessary for activation of its endonuclease domain as well as mediating downstream signaling cascades involving apoptosis-signaling kinase 1 (ASK1) (15–19). Both the kinase and endonuclease domains of IRE1α are associated with HSC activation (5, 8, 11), but the mechanisms that regulate and propagate IRE1α signaling during HSC activation and fibrogenesis are not completely understood.
We sought to better understand the mechanisms of IRE1α activation and signaling during fibrogenesis. First, we established that TGFβ induces IRE1α activation downstream of SMAD2/3 signaling and procollagen I expression. Mutational analysis revealed that IRE1α signaling through its kinase domain, but not its endonuclease domain, is sufficient to promote HSC activation. IRE1α signaling through ASK1 and JNK is critical for TGFβ up-regulation and phosphorylation of the UPR-responsive transcription factor CCAAT/enhancer binding protein β (C/EBPβ), which in turn is critical for HSC activation. Pharmacological antagonism of C/EBPβ through adefovir dipivoxil disrupted HSC activation in vitro and fibrogenesis in vivo. Finally, we found that the transcriptional regulator p300 is critical for both TGFβ- and UPR-mediated HSC activation and may act through a mechanism involving C/EBPβ. Together, our work shows that TGFβ induction of the UPR leads to a feed-forward signaling pathway that acts through C/EBPβ–p300 to promote fibrogenesis and that C/EBPβ can be therapeutically targeted to limit fibrosis in vivo.
Results
TGFβ induction of IRE1α signaling is critical for HSC activation
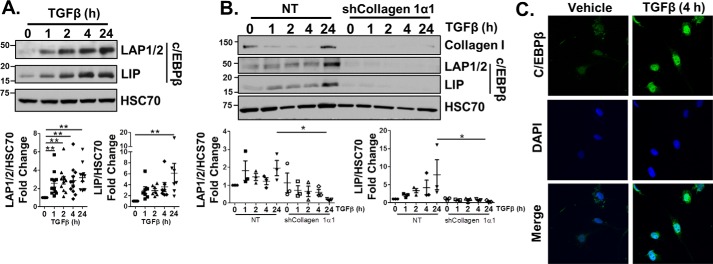
To better understand the relationship between UPR signaling and TGFβ, we first established a model to characterize and understand IRE1α signaling in the presence of TGFβ. Immortalized HSCs (LX-2 cells) treated with TGFβ (5 ng/ml) showed increased phosphorylation of IRE1α, indicative of its activation (Fig. 1A). TGFβ canonically signals through a pathway involving the transcription factors SMAD2/3, which up-regulates transcription of several genes involved in HSC activation and fibrogenesis, including procollagen 1α1 and 1α2 and fibronectin (20). To test whether TGFβ induced IRE1α phosphorylation downstream of SMAD2/3, we disrupted either SMAD2 or procollagen Iα1 expression using RNAi (siSMAD2 or shCollagen 1α1). IRE1α phosphorylation in response to TGFβ was attenuated in shCollagen 1α1 or siSMAD2 cells compared with nontargeting (NT) controls (Fig. 1 (A and B), quantification adjacent). Furthermore, loss of procollagen Iα1 reduced TGFβ up-regulation of fibronectin, as well as αSMA, a marker of activated HSCs. These observations led us to examine whether IRE1α signaling was critical for TGFβ-induced HSC activation. HSCs were isolated from mice harboring loxP sites within the gene encoding IRE1α and were treated with an adenovirus expressing Cre recombinase (AdCre) to disrupt IRE1α expression or with LacZ as a control (21, 22). Following TGFβ treatment, protein levels of fibronectin, collagen I, and αSMA were reduced in cells with Cre-mediated IRE1α knockdown (Fig. 1C, quantification adjacent). Similarly, treatment of LX-2 cells with the IRE1α inhibitor 4μ8C (15 μm) effectively blocked TGFβ induction of collagen I and fibronectin protein (Fig. 1D, quantification below). 4μ8C also reduced gene transcription of fibronectin, procollagen 1α1 and 1α2, and αSMA (Fig. 1E). Increased mRNA expression of peroxisome proliferator–activated receptor γ (PPARγ), which is associated with quiescent HSCs, was also observed. Thus, inhibition of IRE1α signaling limits TGFβ activation of HSCs.
Figure 1.
TGFβ induction of IRE1α is critical for HSC activation. A, LX-2 cells were stably infected with an shRNA targeting procollagen 1α1 (shCollagen 1α1) or an NT control. Cells were treated with 5 ng/ml TGFβ for 0, 1, 2, 4, or 24 h, followed by assessment of phosphorylation and protein levels of IRE1α, fibronectin (FN), αSMA, and collagen I. HSC70 served as a loading control. Quantification is shown adjacent. B, LX-2 cells were transfected with an siRNA targeting SMAD2 (siSMAD2) or a nontargeting siRNA (siControl). 24 h post-transfection, cells were serum-starved and treated with TGFβ for 0, 1, 2, 4, or 24 h. Cell lysates were harvested, and levels of phosphorylated and total IRE1α, SMAD2/3, and HSC70 (loading control) were assessed by immunoblotting. Quantification is shown adjacent. C, primary HSCs were isolated from IRE1αfl/fl mice and infected with adenovirus expressing Cre recombinase (AdCre) or LacZ as a control. 48 h postinfection, cells were treated with TGFβ or vehicle. Cell lysates were harvested and analyzed by immunoblotting for FN, collagen I, αSMA, and HSC70 (loading control). Quantification is shown adjacent. D and E, LX-2 cells were pretreated with the IRE1α inhibitor 4μ8C (15 μm) for 1 h, followed by TGFβ or vehicle for 24 h. Cell lysates or mRNA were harvested and analyzed by either immunoblotting (D) for FN, collagen I, and HSC70 (loading control) or qPCR (E) to assess fibronectin, procollagen 1α1 and 1α2, αSMA, or PPARγ expression. Quantification for D is shown below the blots. Statistics were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars, S.E.
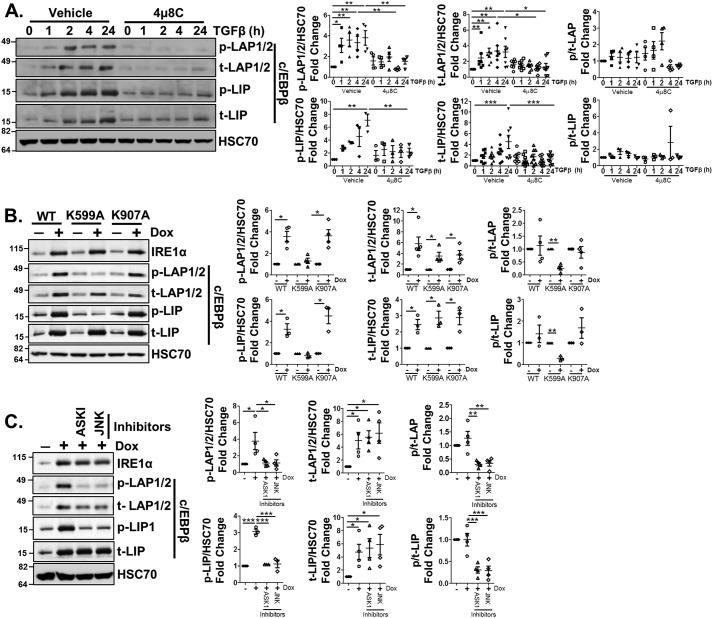
We next asked whether IRE1α signaling was sufficient to mediate profibrotic gene expression. A doxycycline-inducible IRE1α construct was stably expressed in LX-2 cells (doxIRE1α), and upon doxycycline treatment (5 μg/ml), increased protein levels of IRE1α, as well as fibronectin and collagen I, were observed (Fig. 2A, quantification adjacent). To elucidate the mechanisms downstream of IRE1α signaling responsible for this effect, we utilized doxycycline-inducible IRE1α mutants that specifically inactivate either the IRE1α kinase domain (K599A) or the endonuclease domain (K907A) (16). Overexpression of the K907A mutant recapitulated the effects of the WT IRE1α construct; however, overexpression of IRE1α K599A failed to up-regulate fibronectin or collagen I (Fig. 2A). This implicates the IRE1α kinase domain in HSC activation independent of IRE1α endonuclease activity. A major downstream target of the IRE1α kinase domain is ASK1, which in turn phosphorylates and activates signaling cascades through Jun N-terminal kinase (JNK) or p38 MAPK. To explore whether one or both of these pathways was critical for HSC activation downstream of IRE1α, doxIRE1α cells were treated with doxycycline in the presence of ASK1, JNK, or p38 inhibitors (GS-444217 (2 μm), SP600125 (10 μm), and SB203580 (0.5 μm) for ASK1, JNK, and p38, respectively) (23, 24). Inhibition of ASK1 or JNK attenuated the effects of IRE1α overexpression on fibronectin and collagen I expression (Fig. 2B, quantification adjacent), but inhibition of p38 had no effect. Inhibition of ERK1/2 (U0126, 5 μm), which acts downstream of TGFβ signaling, also showed no effect on IRE1α-mediated expression of fibronectin or collagen I (Fig. 2C, quantification adjacent). Together, these data suggest that TGFβ-induced IRE1α signaling drives HSC activation in an ASK1-JNK–dependent manner.
Figure 2.
IRE1α kinase activity promotes HSC activation. A, LX-2 cells were stably infected with doxycycline-inducible IRE1α constructs encoding for WT IRE1α (doxIRE1α WT), a kinase-dead mutant (K599A), or an endonuclease-dead mutant (K907A). Cells were treated with doxycycline (5 μg/ml) or vehicle for 24 h, cells were lysed, and protein levels of IRE1α, fibronectin, and collagen I levels were assessed. HSC70 served as a loading control. Quantification is shown adjacent. B, doxIRE1α cells were treated with GS-444217 (2 μm), SP600125 (10 μm), or SB203580 (0.5 μm) for 1 h to inhibit ASK1, JNK, or p38, respectively, followed by doxycycline treatment for 24 h. Cell lysates were harvested, and IRE1α, fibronectin, collagen I, phosphorylated and total JNK, and phosphorylated and total p38 were assessed by immunoblotting. HSC70 served as a loading control. Quantification is shown adjacent. C, doxIRE1α cells were pretreated with U0126 (5 μm) to inhibit ERK1/2 phosphorylation, followed by doxycycline treatment for 24 h. Cells were lysed and assessed by immunoblotting for IRE1α, fibronectin, collagen I, phosphorylated and total ERK1/2, and HSC70 as a loading control. Quantification is shown adjacent. Statistics were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01). n ≥ 3 biological replicates for each experiment. Error bars, S.E.
TGFβ induces expression and phosphorylation of C/EBPβ in an IRE1α-dependent mechanism
The role of IRE1α signaling in regulating expression of TGFβ target genes led us to explore the mechanisms responsible for this effect. The ENCODE database showed binding sites for the UPR-associated transcription factor C/EBPβ along several gene promoters, including procollagen 1α1 and fibronectin. C/EBPβ is a b-ZIP domain–containing protein transcribed from a single-exon gene as three different isoforms: liver-activating protein 1 and 2 (LAP1 and LAP2), which are transcriptional co-activators, and liver inhibitory protein (LIP), which acts as a co-repressor (25, 26). To establish a role for C/EBPβ in HSC activation, C/EBPβ protein levels were examined in response to TGFβ and found to be up-regulated; however, this up-regulation was disrupted by loss of collagen I (Fig. 3, A and B, quantification below). We also observed translocation of C/EBPβ into the nucleus after TGFβ treatment (4 h) by immunofluorescence (Fig. 3C). We next assessed whether TGFβ-induced C/EBPβ expression required IRE1α signaling. Pretreatment with 4μ8C attenuated TGFβ-induction of C/EBPβ protein expression (Fig. 4A, quantification adjacent). Phosphorylation of C/EBPβ at site Thr-235 (corresponding to Thr-223 on LAP2 and Thr-37 on LIP) is known to mediate its activation; thus, we also assessed whether C/EBPβ phosphorylation was modulated by a TGFβ/IRE1α mechanism (27). Indeed, C/EBPβ phosphorylation increased with TGFβ treatment but was attenuated in the presence of 4μ8C. Due to the increase in both total expression and phosphorylation of C/EBPβ in response to TGFβ, the ratio of phosphorylated/total C/EBPβ was unchanged. We further assessed C/EBPβ expression and phosphorylation in the doxIRE1α cells. Increased expression of C/EBPβ (total LAP1/2 and LIP) was observed with overexpression of WT, K599A, or K907A IRE1α; however, C/EBPβ phosphorylation (p-LAP1/2 or p-LIP) failed to be induced with the K599A mutant, which suggests dependence on an IRE1α-dependent kinase cascade (Fig. 4B, quantification adjacent). Similarly, both the ASK1 and JNK inhibitors blocked C/EBPβ phosphorylation downstream of IRE1α expression (Fig. 4C, quantification adjacent). Interestingly, whereas doxycycline treatment did not alter the ratio of phosphorylated/total C/EBPβ in doxIRE1α WT or K907A cells, phosphorylated/total C/EBPβ was significantly reduced following doxycycline treatment in doxIRE1α K599A cells or with inhibition of ASK1 or JNK. Thus, ASK1/JNK signaling promotes C/EBPβ activity downstream of IRE1α. Together, these data suggest that IRE1α signaling promotes TGFβ-mediated HSC activation through up-regulation of both C/EBPβ protein levels and phosphorylation.
Figure 3.
TGFβ promotes C/EBPβ expression. A, LX-2 cells were treated with 5 ng/ml TGFβ for 0, 1, 2, 4, or 24 h, harvested, and assessed by immunoblotting for expression of C/EBPβ isoforms (LAP1/2 and LIP). HSC70 served as a loading control. Quantification is shown below. B, shCollagen 1α1 or NT cells were treated with TGFβ for 0, 1, 2, 4, or 24 h, and cell lysates were analyzed by immunoblotting for collagen I or C/EBPβ isoforms. HSC70 served as a loading control. Quantification is shown below. C, LX-2 cells were treated with 5 ng/ml TGFβ for 4 h, fixed, permeabilized, and stained for C/EBPβ (green) and DAPI (blue). Representative images are shown. Statistics were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01). n ≥ 3 biological replicates. Error bars, S.E.
Figure 4.
C/EBPβ expression and phosphorylation is increased by TGFβ in an IRE1α-dependent manner. A, LX-2 cells were pretreated for 1 h with 15 μm 4μ8C, followed by TGFβ (5 ng/ml) treatment for 0, 1, 2, 4, or 24 h. Cell lysates were harvested, and protein expression or phosphorylation of C/EBPβ isoforms on Thr-235 (corresponding to Thr-223 on LAP2 and Thr-37 on LIP) was examined by immunoblotting. HSC70 served as a loading control. Quantification is shown in adjacent graphs. B, doxIREα WT, K599A, or K907A cells were treated with doxycycline (5 μg/ml) or vehicle for 24 h. Cells were lysed, and C/EBPβ phosphorylation, total C/EBPβ, and IRE1α expression were analyzed by immunoblotting. HSC70 served as a loading control. Quantification is shown in adjacent graphs. C, doxIRE1α cells were treated with GS-444217 (2 μm) or SP600125 (10 μm) for 1 h to inhibit ASK1 or JNK, respectively, followed by doxycycline treatment for 24 h. Cells were lysed, and phosphorylation of C/EBPβ isoforms and total C/EBPβ expression, as well as IRE1α expression, was analyzed. HSC70 served as a loading control. Quantification is shown in adjacent graphs. Statistics for A and C were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). For B, paired t tests were performed. n ≥ 3 biological replicates for each experiment. Error bars, S.E.
Loss of C/EBPβ disrupts TGFβ-induced HSC activation
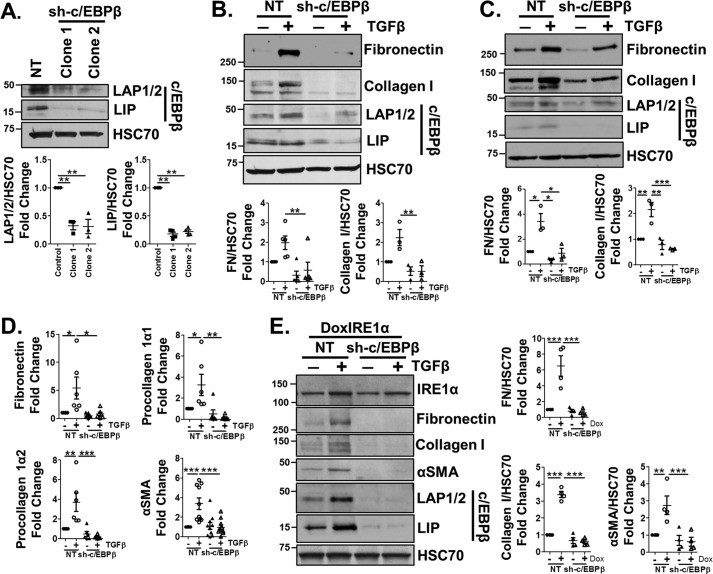
To determine whether C/EBPβ is critical for HSC activation, LX-2 cells were infected with a lentivirus expressing an shRNA against C/EBPβ (sh-C/EBPβ), or an NT shRNA. Two clones were selected that exhibited reduced expression of the activating and inhibitory isoforms of C/EBPβ, LAP1/LAP2 (69 and 68% reduction) and LIP (82 and 78% reduction) (Fig. 5A, quantification below). Reduced C/EBPβ expression attenuated the effects of TGFβ on fibronectin and collagen I protein levels compared with controls in both clones (Fig. 5 (B and C), quantification below). Loss of C/EBPβ expression also attenuated fibronectin, procollagen 1α1, procollagen 1α2, and αSMA mRNA expression in response to TGFβ, indicative of a crucial role for C/EBPβ in TGFβ-mediated gene transcription (Fig. 5D, clone 1). To determine whether C/EBPβ acted downstream of IRE1α, doxIRE1α cells were stably infected with the shRNA targeting C/EBPβ and treated with doxycycline. Despite induction of IRE1α, the associated up-regulation of fibronectin, collagen I, and αSMA was lost in doxIRE1α sh-C/EBPβ cells (Fig. 5E, quantification adjacent). Together, these data highlight a crucial role for C/EBPβ in mediating mRNA and protein expression of ECM proteins and αSMA downstream of TGFβ and IRE1α.
Figure 5.
C/EBPβ is critical for HSC activation in response to TGFβ or IRE1α signaling. A, LX-2 cells were infected with a lentivirus expressing an shRNA against C/EBPβ (sh-C/EBPβ) or an NT control. Two clonal cell lines were engineered and were assessed by immunoblotting for expression of C/EBPβ isoforms (LAP1/2 and LIP). HSC70 served as a loading control. Quantification is shown below. B and C, sh-C/EBPβ cells (clones 1 and 2) or NT cells were treated with TGFβ (5 ng/ml) for 24 h. Cell lysates were harvested and assessed by immunoblotting for collagen I, FN, C/EBPβ, and HSC70 (loading control). Quantification is shown below. D, sh-C/EBPβ or NT cells were treated with TGFβ (5 ng/ml), after which mRNA was harvested and qPCR was performed to assess fibronectin, procollagen 1α1 and 1α2, or αSMA expression. E, DoxIRE1α cells were infected with lentivirus expressing shRNA targeting C/EBPβ (sh-C/EBPβ) or an NT control, and clonal cell populations were selected. Cells were treated with doxycycline for 24 h, after which cell lysates were harvested and assessed by immunoblotting for IRE1α, fibronectin, collagen I, αSMA, C/EBPβ, and HSC70 (loading control). Quantification is shown adjacent. Statistics were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). n ≥ 3 biological replicates for each experiment. Error bars, S.E.
Adefovir dipivoxil inhibits C/EBPβ expression and blocks HSC activation in vitro and in vivo
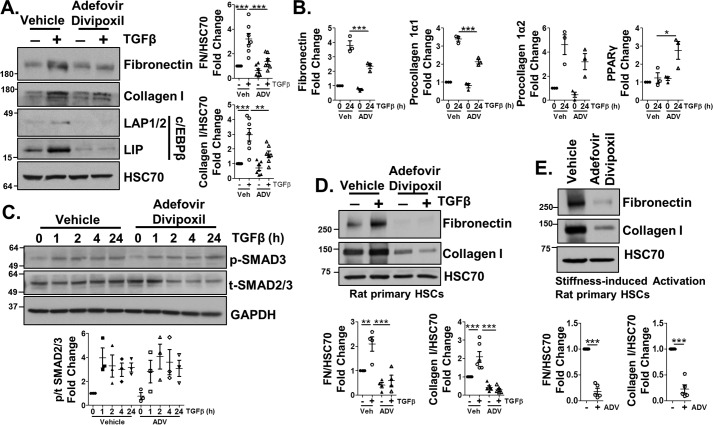
The crucial role of C/EBPβ in HSC activation led us to ask whether C/EBPβ could serve as an antifibrotic target. Recently, the hepatitis B antiviral drug adefovir dipivoxil was shown to antagonize C/EBPβ expression in vitro (28). Indeed, adefovir dipivoxil treatment (10 μm) of LX-2 cells attenuated TGFβ induction of C/EBPβ in response to TGFβ and blocked the effects of TGFβ on fibronectin and procollagen 1α1 at the mRNA and protein level (Fig. 6 (A and B), quantification adjacent) despite having no effect on TGFβ induction of SMAD2/3 phosphorylation (Fig. 6C, quantification below). The conserved SMAD2/3 phosphorylation indicates that adefovir dipivoxil is acting downstream of SMAD2/3. We also observed a significant increase in PPARγ mRNA expression. Adefovir dipivoxil similarly reduced collagen I and fibronectin protein levels in isolated rat primary HSCs in response to TGFβ or stiffness-induced activation (Fig. 6 (D and E), quantification below).
Figure 6.
Adefovir dipivoxil limits HSC activation through decreasing C/EBPβ expression. A and B, LX-2 cells were pretreated with 10 μm adefovir dipivoxil (ADV) for 1 h, followed by TGFβ (5 ng/ml) for 24 h. Cell lysates or mRNA were harvested and assessed by immunoblotting (A) for FN, collagen I, C/EBPβ isoforms LAP1/2 and LIP, and HSC70 (loading control) or qPCR (B) for fibronectin, procollagen 1α1 and 1α2, or PPARγ expression. Quantification for A is shown in adjacent graphs. C, cells were treated with TGFβ in the presence of adefovir dipivoxil or vehicle for 0, 1, 2, 4, or 24 h. Cell lysates were harvested and assessed for phosphorylated SMAD3 or total SMAD2/3. HSC70 served as a loading control. D, primary HSCs were harvested from rats and pretreated with adefovir dipivoxil for 1 h, followed by either vehicle or TGFβ for 24 h. Lysates were harvested and analyzed by immunoblotting for fibronectin or collagen I. HSC70 served as a loading control. Quantification is shown in the graphs below. E, primary HSCs were harvested from rats, followed by adefovir dipivoxil treatment on days 2, 4, and 6 post-isolation. Cell lysates were harvested on day 7 and analyzed by immunoblotting for fibronectin, collagen I expression, or HSC70 (loading control). Quantification is shown in adjacent graphs. Statistics were performed using ANOVA followed by Tukey post hoc analysis for A–D and paired t test for E (*, p < 0.05; **, p < 0.01; ***, p < 0.001). n ≥ 3 biological replicates for each experiment. Error bars, S.E.
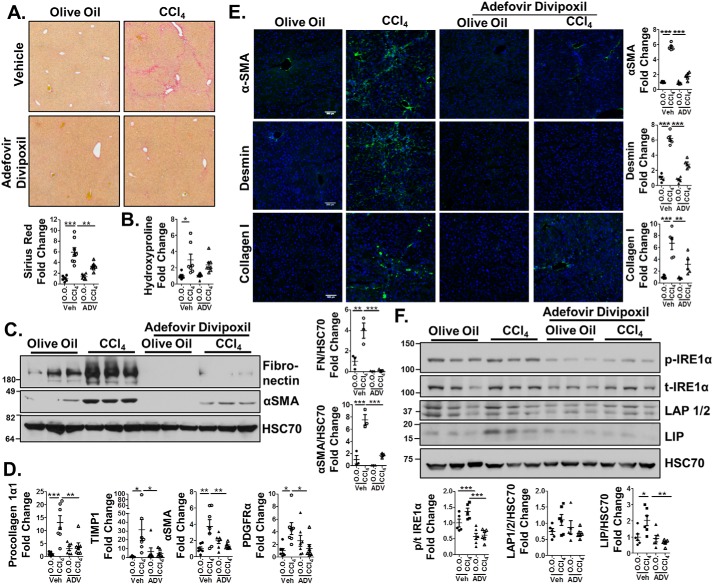
We next tested adefovir dipivoxil as an antifibrotic agent in vivo, injecting 10-week-old, sex-matched C57Bl/6J mice for 6 weeks with CCl4 or vehicle (2 days/week) and 10 mg/kg adefovir dipivoxil (or vehicle) on the remaining 5 days/week. Livers were then harvested and assessed for fibrosis. Mice receiving adefovir dipivoxil in conjunction with CCl4 exhibited significantly less collagen deposition as assessed by sirius red staining (Fig. 7A, quantified below). Hydroxyproline analysis showed a significant increase in collagen content in CCl4-treated mice compared with controls, but no significant difference between Adefovir-CCl4 and Adefovir-olive oil groups (Fig. 7B). Furthermore, adefovir dipivoxil reduced CCl4-induced protein expression of fibronectin and αSMA (Fig. 7C, quantification adjacent) and gene expression of procollagen 1α1, TIMP1, αSMA, and PDGFRα (Fig. 7D). Finally, immunofluorescence was performed on liver sections and showed reduced αSMA, desmin (a marker of HSCs), and collagen I in adefovir-CCl4–treated mice compared with CCl4 alone (Fig. 7E). Assessment of C/EBPβ protein levels in whole-liver tissue revealed that LIP increased in response to CCl4 but was significantly decreased with adefovir dipivoxil treatment (Fig. 7F, quantification below). Alternatively, LAP1/2 protein levels were unaffected. We also analyzed IRE1α phosphorylation in whole liver and observed increased phosphorylation of IRE1α with CCl4 treatment, whereas treatment with adefovir dipivoxil blocked this effect (Fig. 7F). Finally, analysis of the data revealed no differential effect of CCl4 or adefovir dipivoxil between male and female mice (data not shown). Thus, adefovir dipivoxil reduces HSC activation in vitro and fibrogenesis in vivo.
Figure 7.
Adefovir dipivoxil–treated mice display reduced fibrogenesis. Age- and sex-matched C57Bl/6 mice (n = 7–8 mice/group) were treated with CCl4 or olive oil twice a week for 6 weeks, in conjunction with injections of 10 mg/kg adefovir dipivoxil or 0.05 m citric acid (vehicle) 5 times a week during the same time period. Whole liver was harvested and analyzed for fibrosis. A, liver sections underwent sirius red staining, and quantification was performed using ImageJ (shown below). B, hydroxyproline analysis for collagen content. C, whole liver lysates were harvested and immunoblotted for fibronectin, αSMA, and HSC70 (loading control). Quantification is shown adjacent. D, qPCR was performed on mRNA harvested from whole liver to analyze expression of procollagen 1α1, TIMP1, αSMA, and PDGFRα. E, immunofluorescence was performed on frozen liver sections to assess αSMA, desmin (a marker of HSCs), and collagen I (green). DAPI was used as a nuclear stain (blue). Representative images are shown. Staining was quantified using ImageJ and is shown in the adjacent graphs. F, whole-liver lysates were harvested and immunoblotted for phospho-IRE1α, total IRE1α, C/EBPβ, and HSC70 (loading control). Statistics were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars, S.E.
C/EBPβ mediates HSC activation through a mechanism involving p300
With a link between C/EBPβ and HSC activation identified, we questioned the mechanism of C/EBPβ regulation of profibrotic genes. C/EBPβ canonically acts as a homo- or heterodimer to mediate transcription. Analysis of the ENCODE database revealed C/EBPβ-binding sites in close proximity to sites bound by the transcriptional regulator p300 along the promoters of TGFβ-responsive genes, including procollagen 1α1 and fibronectin. p300 has been implicated in HSC activation and is a known C/EBPβ-binding partner (29, 30). In vivo analysis of p300 protein levels showed increased p300 in mice treated with CCl4, but this effect was blocked in mice receiving both CCl4 and adefovir dipivoxil (Fig. 8A, quantification below). This prompted us to study p300 in both TGFβ- and UPR-induced HSC activation. LX-2 cells were infected with a lentivirus expressing an shRNA that targets p300 (sh-p300). sh-p300 or NT cells were treated with TGFβ or tunicamycin (Tm; 1 μg/ml), a potent inducer of ER stress, for 24 h or vehicle alone. TGFβ or Tm treatment increased protein levels of fibronectin, collagen I, and αSMA, but p300 knockdown attenuated this effect (Fig. 8, B and C). Additionally, p300 knockdown in doxIRE1α cells (doxIRE1α-sh-p300) attenuated IRE1α-mediated up-regulation of fibronectin, collagen I, and αSMA protein levels (Fig. 8D, quantification below).
Figure 8.
p300 is critical for TGFβ- or UPR-mediated HSC activation and may involve a C/EBPβ-dependent mechanism. A, whole-liver lysates harvested from mice treated with olive oil, CCl4, adefovir dipivoxil, or CCl4 + adefovir dipivoxil and assessed by immunoblotting for protein levels of p300. HSC70 served as a loading control. B and C, LX-2 cells were infected with a lentivirus encoding an shRNA targeting p300 (sh-p300) or NT control and were selected to yield a clonal cell population. NT or sh-p300 cells were treated with 5 ng/ml TGFβ (B) or 1 μg/ml Tm (C) for 24 h, and cell lysates were harvested and analyzed by immunoblotting for p300, fibronectin, collagen I, αSMA, and HSC70 (loading control). Quantification is adjacent. D, doxIRE1α cells were stably infected with a lentivirus encoding an shRNA targeting p300 (doxIRE1α sh-p300) or NT, treated with doxycycline, and assessed by immunoblotting for IRE1α, p300, fibronectin, collagen I, αSMA, and HSC70 (loading control). Quantification is adjacent. Statistics were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). n ≥ 3 biological replicates for each experiment. Error bars, S.E.
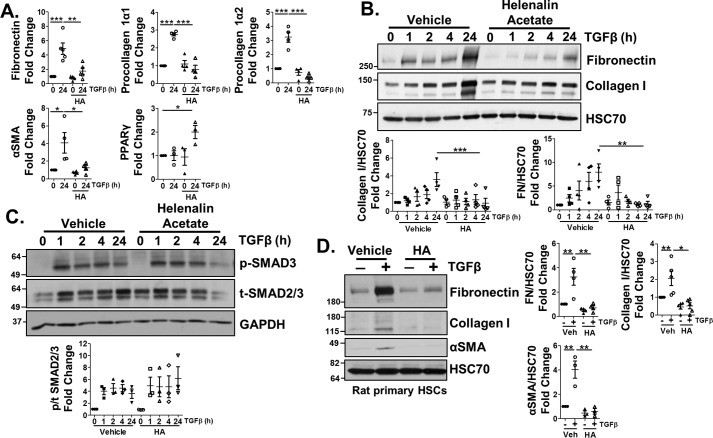
The similar effects of p300 and C/EBPβ knockdown on TGFβ- and IRE1α-mediated HSC activation suggested that these proteins may act together. Previous studies showed that p300 binding to C/EBPβ led to transcriptional activation in a mechanism that involved C/EBPβ phosphorylation at Thr-235. Thus, we asked whether p300 is responsible for the effects of C/EBPβ during TGFβ-mediated HSC activation (29). To this end, we utilized helenalin acetate, a sesquiterpene lactone reported to disrupt the interaction between C/EBPβ and p300 (31). Co-treatment of LX-2 cells with TGFβ and helenalin acetate (1 μm) disrupted TGFβ induction of fibronectin, procollagen 1α1, procollagen 1α2, and αSMA mRNA levels, as well as increased PPARγ mRNA (Fig. 9A). Helenalin acetate also attenuated TGFβ induction of fibronectin and collagen I protein levels, despite no effect on TGFβ-mediated SMAD2/3 phosphorylation (Fig. 9 (B and C), quantification below). Helenalin acetate similarly reduced TGFβ induction of collagen I and fibronectin in primary HSCs isolated from rats (Fig. 9D, quantification adjacent). Together, these data show that p300 is critical for mediating HSC activation downstream of TGFβ-IRE1α signaling and may act through a mechanism involving C/EBPβ.
Figure 9.
Helenalin acetate limits TGFβ-induced HSC activation. LX-2 cells were pretreated with 1 μm helenalin acetate (HA) for 1 h, followed by TGFβ treatment for 24 h. mRNA (A) or cell lysates (B and C) were harvested and analyzed by qPCR for fibronectin, procollagen 1α1 and 1α2, αSMA, or PPARγ or by immunoblotting for fibronectin, collagen I, p-SMAD3, total SMAD2/3, and HSC70 (loading control). Quantification for B and C is shown below the blots. D, primary rat HSCs (rHSCs) were pretreated with helenalin acetate for 1 h, followed by TGFβ for 48 h. Cells were lysed and assessed for protein expression of fibronectin, collagen I, and αSMA. HSC70 served as a loading control. Quantification is shown in adjacent graphs. Statistics were performed using ANOVA followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). n ≥ 3 biological replicates for each experiment. Error bars, S.E.
Discussion
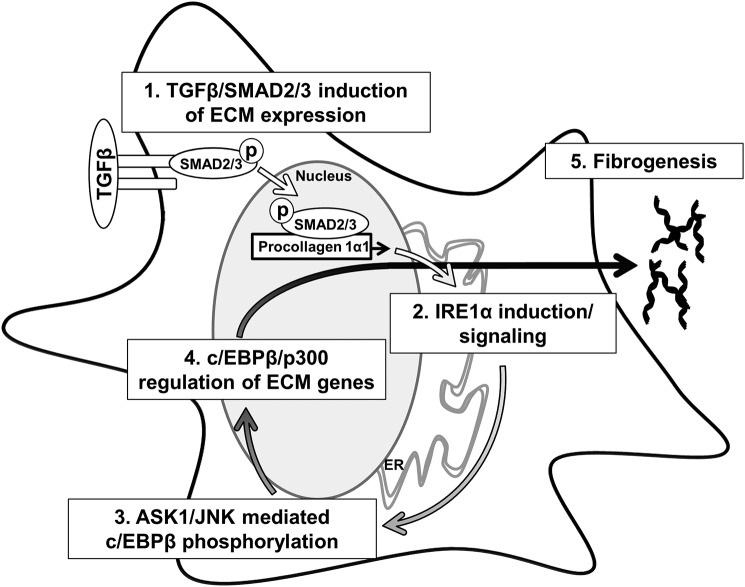
UPR signaling plays an important role during HSC activation, although the mechanisms that govern the relationship between HSC activation, UPR signaling, and fibrogenesis are not fully understood. Here, we make four key findings that provide mechanistic insight into how IRE1α drives fibrogenesis in response to TGFβ (Fig 10). We show that TGFβ induces the UPR and IRE1α signaling through a SMAD2/3-collagen I–dependent mechanism. Next, we identify C/EBPβ as a crucial mediator of HSC activation that is regulated by IRE1α in an ASK1/JNK-dependent mechanism. Furthermore, C/EBPβ can be pharmacologically targeted by adefovir dipivoxil to block HSC activation in vitro and fibrogenesis in vivo. Finally, we show that p300 is a crucial mediator of HSC activation downstream of the UPR, which may act together with C/EBPβ in this role. Together, these data map out a UPR-dependent feed-forward mechanism that drives fibrogenesis through C/EBPβ–p300.
Figure 10.
Cross-talk between TGFβ and IRE1α promotes HSC activation. 1, TGFβ-mediated phosphorylation of SMAD2/3 leads to increased expression of ECM proteins leading to ER stress. 2, ER stress leads to IRE1α autotransphosphorylation. 3, IRE1α signaling leads to phosphorylation of C/EBPβ downstream of ASK1 and JNK. 4, activation of C/EBPβ facilitates ECM production through a mechanism that involves p300. 5, together, this signaling cascade is critical for fibrogenesis.
UPR signaling in activated HSCs is thought to accommodate increased protein translation, trafficking, and secretion. Chemical induction of the UPR can also drive fibrogenesis independently of canonical activation signals. The relationship between canonical HSC activation mechanisms and the UPR, however, is still unclear. We show here that IRE1α signaling through its kinase domain is crucial for HSC activation. IRE1α kinase activity leads to a signaling cascade involving ASK1 and JNK, which are critical for phosphorylation of C/EBPβ. Phosphorylation of the Thr-235/Thr-223/Thr-37 on C/EBPβ is associated with increased transcriptional activity and is a known target of the MAPK family of kinases, consistent with our data (27). This role of IRE1α is interesting for a few reasons. First, IRE1α signaling has been implicated in HSC fibrogenic gene expression through both its endonuclease domain and through p38 signaling; however, we find that neither of these mechanisms are critical for HSC activation in our inducible IRE1α system or necessary for C/EBPβ phosphorylation. p38 activation was previously associated with SMAD3 phosphorylation and collagen I expression in response to chemically induced ER stress (11), whereas activation of the transcription factor XBP1 downstream of IRE1α endonuclease activity is associated with ER dilation and up-regulation of genes involved in protein trafficking and secretion (5, 6, 8). Furthermore, we show that overexpression of either the K907A or K599A IRE1α mutant still led to up-regulation of total C/EBPβ expression, implicating a kinase- and endonuclease-independent mechanism of C/EBPβ regulation. Thus, IRE1α acts through several mechanisms to contribute to HSC activation. The role of ASK1–JNK signaling in this process is also interesting, as a separate ASK1 inhibitor, selonsertib, is associated with reduced collagen content and liver stiffness when used in a clinical trial to treat patients with non-alcoholic steatohepatitis (32, 33). Our work suggests that this reduction may be due to inhibition of C/EBPβ phosphorylation, subsequently limiting HSC activation. Finally, the loss of procollagen and fibronectin expression in the absence of IRE1α signaling provides insight into HSC activation mechanisms. We propose here that initial SMAD2/3-dependent up-regulation of ECM proteins leads to UPR signaling and that the UPR is critical for driving and maintaining the fibrogenic phenotype, thus placing procollagen I as both a cause and a consequence of UPR signaling during HSC activation. Alternatively, UPR induction prior to increased protein load has been observed in immune cells (34). Further assessment of UPR induction in response to different activating stimuli may provide insight into the role of the UPR in HSCs.
Our data indicating that C/EBPβ is critical for HSC activation in a UPR-dependent manner provide some context for the regulation of C/EBPβ and ECM production in HSCs. Previous studies show that C/EBPβ can positively or negatively regulate procollagen 1α1 expression, and these effects are dependent on the stimulus, isoform, and coregulators (35–38). C/EBPβ transcription is unique, in that three isoforms are transcribed from the same gene. The activating isoforms of C/EBPβ (LAP1 and LAP2) bind DNA as well as interact with several transcriptional co-activators, whereas the inhibitory LIP isoform serves as a dominant negative. Thus, the ratio of LAP/LIP greatly influences transcriptional programming. This is evident during the UPR, where LAP1/LAP2 are typically up-regulated during early adaptive UPR, and LIP is up-regulated later and associated with proapoptotic UPR signaling (39). LIP overexpression also inhibits collagen I expression (38). Thus, targeting C/EBPβ or a specific isoform, such as the full-length LAP1, which can bind to p300, could reduce the activated HSC population in vivo (31).
UPR-mediated phosphorylation of C/EBPβ may influence recruitment of transcriptional coactivators during HSC activation. Mutation of mouse C/EBPβ at Thr-188 to alanine (corresponding to Thr-235 in human C/EBPβ) blocks C/EBPβ induction of the c-fos promoter as well as p300 recruitment to the same site (40, 41). A similar mechanism may be responsible during HSC activation, as loss of Thr-235 phosphorylation is associated with attenuated ECM production in response to IRE1α overexpression, and helenalin acetate also limits the effects of TGFβ. The mechanisms and the transcriptional complexes involved in TGFβ- and IRE1α-mediated gene transcription through C/EBPβ merit further study, as both C/EBPβ and p300 can interact with several transcription factors, including SMADs. Of further interest is the requirement for p300 in UPR-induced HSC activation. There is limited evidence of p300 involvement downstream of the UPR, aside from acetylation of the transcription factor XBP1 (34). p300 may complex with additional UPR-responsive transcription factors to regulate transcriptional activity.
Whereas we have focused here on UPR signaling through IRE1α, signaling through the other two UPR sensors, PERK and ATF6α, also likely plays a role in HSC activation (4). In vivo studies show a positive relationship between PERK signaling and HSC activation, and subsequent studies showed that PERK increases SMAD2 expression through a mechanism involving phosphorylation of heterogeneous nuclear riboprotein A1 and inhibiting the degradation of SMAD2 mRNA (9). Additionally, C/EBPβ is canonically downstream of the PERK pathway and thus may provide mechanisms for further regulation of C/EBPβ in HSCs (39). Less is known regarding ATF6α in HSCs. Inhibition of ATF6α reduced HSC activation in response to chemically induced ER stress, but no direct mechanisms for ATF6α signaling during HSC activation have been identified (11). Further studies are needed to fully understand the integration of IRE1α, PERK, and ATF6α signaling during HSC activation and how cross-talk between activation signals (such as TGFβ) and UPR signaling influences HSC activation and survival.
Adefovir dipivoxil was identified as a pharmacological inhibitor of C/EBPβ during a screen for compounds that preferentially targeted the LIP isoform of C/EBPβ. In the drug screen, adefovir dipivoxil reduced the ratio of LIP/LAP but also impacted expression of the activating isoforms, prompting us to test whether adefovir dipivoxil could limit HSC activation through targeting C/EBPβ. Adefovir dipivoxil effectively reduced protein levels of all C/EBPβ isoforms in LX-2 cells and attenuated TGFβ-stimulated HSC activation, as well as fibrogenesis in vivo. Interestingly, LIP, but not LAP1/2, was significantly reduced in whole liver in vivo. These data suggest a crucial and targetable role for C/EBPβ, and the ratio of LIP to LAP1/2 in fibrosis, further supported by previous findings where mesenchymal C/EBPβ deletion attenuated pulmonary fibrosis (27). Based on the known role of C/EBPβ in both proliferation and apoptosis, we anticipate that adefovir dipivoxil may also limit HSC proliferation or promote HSC apoptosis, both of which are favorable for fibrosis regression and resolution (42–45). Finally, whereas we anticipate that C/EBPβ is one of the major targets of adefovir dipivoxil in HSCs, off-target effects may contribute to the antifibrotic nature of the drug. Another acyclic nucleotide analog used in hepatitis B patients, tenofovir, was associated with reduced sirius red staining in a thioacetamide model of liver injury, but its effects on C/EBPβ or UPR signaling were not assessed (46).
In conclusion, we show that TGFβ induces UPR signaling in HSCs, which acts in a feed-forward mechanism through IRE1α to promote fibrotic gene expression through C/EBPβ–p300.
Experimental procedures
Cell culture
Immortalized HSCs (LX-2, ATCC) and primary HSCs were cultured in Dulbecco's modified Eagle's medium (Gibco) + 10% fetal bovine serum + 1% penicillin/streptomycin. Compounds used include TGFβ (5 ng/ml; R&D Biosystems), doxycycline (5 μg/ml; Clontech NC0424034), 4μ8C (15 μm; Selleck-chem S7272), GS-444217 (2 μm; Gilead Sciences), SB203580 (0.5 μm; Selleckchem S1076), SP600125 (10 μm; Selleckchem S1460), U0126 (5 μm; InvivoGen (San Diego, CA)), adefovir dipivoxil (10 μm in vitro and 10 mg/kg in vivo; Sigma A9730), tunicamycin (1 μg/ml; Sigma 654380), and helenalin acetate (1 μm; Cayman Chemical 17050). For all experiments using compounds in conjunction with TGFβ, cells were serum-starved for 4 h and pretreated with the indicated compound for 1 h, followed by treatment with TGFβ or vehicle. Doxycycline-inducible cell lines were cultured in Dulbecco's modified Eagle's medium plus 10% tetracycline-free fetal bovine serum (Clontech) and 1% penicillin/streptomycin. For experiments using ASK1, JNK, or p38 inhibitors in conjunction with doxycycline, the inhibitors were added to the cells for 1 h followed by the addition of doxycycline for 24 h. Transient transfection of LX-2 cells with siRNA against SMAD2 (Qiagen) or a nontargeting control was achieved using Oligofectamine (Invitrogen), and experiments were conducted 48 h post-transfection.
Plasmids and generation of stable cell lines
DNA encoding WT, K599A, or K907A IRE1α was cloned in a pLVX-tetone vector (Clontech) with a hygromycin selection cassette (pLVX-tetone-hygro-IRE1α, pLVX-tetone-hygro-IRE1αK599A, or pLVX-tetone-hygro-IRE1αK907A). shRNA against procollagen 1α1, C/EBPβ, p300, or a nontargeting shRNA control was purchased from Sigma. To generate lentivirus encoding the aforementioned plasmids, the plasmids were transfected into HEK293T cells as described previously (7). The virus-containing medium was harvested and used to infect LX-2 cells, followed by selection with puromycin (2 μg/ml) for shRNA constructs or hygromycin (2 μg/ml) for pLVX-tetone-hygro constructs.
Primary HSC isolation and infection
HSCs were harvested from IRE1αfl/fl mice (21, 47, 48) or adult female Lewis rats (Charles River Laboratories) (49) as described previously. HSCs isolated from IRE1αfl/fl mice were infected with adenovirus encoding for Ad-Cre-eGFP to induce gene recombination or with an Ad-LacZ control, as described previously (50). All procedures were reviewed and approved by the Mayo Clinic Institutional Animal Care and Use Committee.
In vivo treatment and analysis
Eight-week-old C57Bl/6J mice were purchased from Envigo. Following a 1-week acclimation period, age- and sex-matched mice were separated into four treatment groups: olive oil + vehicle (n = 7), CCl4 + vehicle (n = 8), olive oil + adefovir dipivoxil (n = 8), and CCl4 + adefovir dipivoxil (n = 8). CCl4 (0.5 μl/mg, 0.08 ml/mouse) was administered by intraperitoneal injection twice a week for 6 weeks. Each group consisted of four males and four females, which were housed together according to their treatment group. Mice were housed in transparent polycarbonate cages subjected to 12-h light/dark cycles under a temperature of 21 °C and a relative humidity of 50%. A standard chow diet and freshwater were provided ad libitum. Adefovir dipivoxil (Sigma-Aldrich, SML0240) was dissolved in sterile in citric acid (0.05 m, pH 2.0) and administered at a concentration of 10 mg/kg/day 5 days a week for 6 weeks. Mice were observed daily for distress and tolerated the experimental protocol without adverse effects, aside from one mouse unexpectedly reaching end point criteria (olive oil + vehicle group). After 6 weeks, mice were sacrificed, and livers were harvested for analysis of fibrosis. Assessment of collagen content by sirius red staining or hydroxyproline was performed as described previously (7). For analysis of gene expression, mRNA was harvested using an RNeasy kit (Qiagen), followed by reverse transcription into cDNA and assessment by qPCR. For protein assessment, livers were homogenized in lysis buffer as described previously and analyzed by Western blotting (7). For immunofluorescence, liver samples were snap-frozen in OCT embedding compound, and 7-μm frozen sections were obtained using a Leica cryostat. Sections were fixed with 4% paraformaldehyde, followed by incubation with primary antibodies against collagen I, desmin, and αSMA. Anti-goat IgG and anti-rabbit IgG conjugated with Alexa fluorochromes were used as secondary antibodies (Molecular Probes, Inc., Eugene, OR). DAPI was used at 1:2000 for counterstaining. Microscopy was performed by Axio Observer (Carl Zeiss, Thornwood, NY), in which appropriate light and filter combinations were selected according to excitation and emission spectrum features of the Alexa fluorochromes. All procedures were reviewed and approved by the Mayo Clinic Institutional Animal Care and Use Committee.
Immunoblotting
Following the indicated treatment, cells were lysed in a modified radioimmune precipitation buffer and spun down to remove cell debris (7). Lysates were denatured using 6× sample buffer (Boston Bioproducts BP-111R), resolved using SDS-PAGE, transferred to nitrocellulose membrane, and blocked in 5% BSA. Membranes were incubated in the indicated primary antibody, followed by the appropriate secondary antibody, and developed. All experiments were performed in triplicate biological replicates at a minimum, with quantification and statistics performed using ImageJ and PRISM, respectively. HSC70 or GAPDH were used as loading control for all immunoblots.
Antibodies
The following antibodies were used: HSC70 (Santa Cruz Biotechnology, Inc., 7298), Collagen I (Southern Biotech 1310-01), IRE1α (Cell Signaling 3294), phospho-IRE1α (Abcam ab124945), SMAD2/3 (Cell Signaling 3102S), phospho-SMAD3 (Cell Signaling 9520S), fibronectin (for LX-2 cells, BD Biosciences 611012), αSMA (Sigma A5228), phospho-p38 (Cell Signaling 9211S), total p-38 (Cell Signaling 9212S), phospho-JNK (Cell Signaling 9255S), total JNK (Cell Signaling 9252S), C/EBPβ (Developmental Studies Hybridoma Bank at the University of Iowa, PCRP-CEBPB-3D10), phospho-C/EBPβ (Thr-235 on LAP1, Thr-223 on LAP, and Thr-37 on LIP; Cell Signaling 3084S), fibronectin (for mouse and rat HSCs, mouse tissue, Abcam ab2413), PDGFRβ (Cell Signaling 28E1), desmin (Abcam 15200), and P300 (Santa Cruz Biotechnology sc-8981).
qPCR analysis
Following treatment, cells were harvested using an RNeasy kit (Qiagen). Equal amounts of mRNA were reverse-transcribed, and qPCR was performed. Human GAPDH or mouse 18S primers were used for normalization in experiments using human cells or mouse liver lysates, respectively. Primer sequences used for qPCR analysis are listed in Table 1.
Table 1.
Primer sequences used for qPCR analysis
| Gene name | Primers |
|---|---|
| Human fibronectin | |
| Forward | GTGGCTGCCTTCAATTCTC |
| Reverse | GTGGGTTGCAAACCTTCAAT |
| Human procollagen 1α1 | |
| Forward | TGTGAGGCCACGCATGAG |
| Reverse | CAGATCACGTCATCGCACAA |
| Human procollagen 1α2 | |
| Forward | GGCCCTCAAGGTTTCCAAGG |
| Reverse | CACCCTGTGGTCCAACAACTC |
| Human αSMA | |
| Forward | AATGCAGAAGGAGATCACGG |
| Reverse | TCCTGTTTGCTGATCCACATC |
| Human pPARγ | |
| Forward | ACCACTCCCACTCCTTTG |
| Reverse | GCAGGCTCCACTTTGATT |
| Human GAPDH | |
| Forward | CTCTGCTCCTCCTGTTCGAC |
| Reverse | TTAAAAGCAGCCCTGGTGAC |
| Mouse 18S | |
| Forward | GTACAAAGGGCAGGGACTTAAT |
| Reverse | AGGTCTGTGATGCCCTTAGA |
| Mouse procollagen 1α1 | |
| Forward | GAGCGGAGAGTACTGGATCG |
| Reverse | GCTTCTTTTCCTTGGGGTTC |
| Mouse TIMP1 | |
| Forward | CCTTGCAAACTGGAGAGTGACA |
| Reverse | AAGCAAAGTGACGGCTCTGGT |
| Mouse αSMA | |
| Forward | AAACAGGAATACGACGAAG |
| Reverse | GAATGATTTGGAAAGGA |
| Mouse PDGFRα | |
| Forward | GTTGCCTTACGACTCCAGATG |
| Reverse | TCACAGCCACCTTCATTACAG |
Statistical analysis
For all experiments with three or more groups, statistics were performed using analysis of variance (ANOVA) followed by Tukey post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). All statistical analysis comparing two groups utilized paired t tests. n ≥ 3 biological replicates for each experiment.
Author contributions
Z. L., C. L., and J. L. M. data curation; Z. L., C. L., and J. L. M. investigation; Z. L., C. L., N. K., H. M., V. H. S., and J. L. M. writing-review and editing; C. L. and J. L. M. formal analysis; C. L. and J. L. M. writing-original draft; N. K., H. M., V. H. S., and J. L. M. conceptualization; N. K. and H. M. resources; H. M., V. H. S., and J. L. M. funding acquisition; H. M. and J. L. M. methodology; V. H. S. and J. L. M. supervision; J. L. M. project administration.
Acknowledgments
We thank the Mayo Clinic Center for Cell Signaling for resources through the Optical Microscopy Core and the Gene Editing and Epigenomics Core as well as the Biomaterials and Histomorphometry Core Laboratory at Mayo Clinic. We also thank Gilead Sciences for providing GS-444217.
This work was supported by NIDDK, National Institutes of Health, Grants K01 DK112915, RO1 DK111378, R01 DK59615, and P30DK084567; NCI, National Institutes of Health, Grant R01 CA160069; and a research fellowship from the China Scholarship Council. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HSC
- hepatic stellate cell
- ECM
- extracellular matrix
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- IRE1α
- inositol-requiring enzyme 1α
- ASK1
- apoptosis signaling kinase 1
- C/EBP
- CCAAT/enhancer binding protein β
- PPARγ
- peroxisome proliferator–activated receptor γ
- JNK
- c-Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- αSMA
- smooth muscle actin
- p-
- phosphorylated
- TGFβ
- transforming growth factor β
- Tm
- tunicamycin
- LIP
- liver inhibitory protein
- LAP
- liver-activating protein
- qPCR
- quantitative PCR
- DAPI
- 4′,6-diamidino-2-phenylindole
- ANOVA
- analysis of variance
- NT
- nontargeting
- FN
- fibronectin.
References
- 1. Iwaisako K., Jiang C., Zhang M., Cong M., Moore-Morris T. J., Park T. J., Liu X., Xu J., Wang P., Paik Y. H., Meng F., Asagiri M., Murray L. A., Hofmann A. F., Iida T., et al. (2014) Origin of myofibroblasts in the fibrotic liver in mice. Proc. Natl. Acad. Sci. U.S.A. 111, E3297–E3305 10.1073/pnas.1400062111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiang J. X., and Török N. J. (2013) Liver injury and the activation of the hepatic myofibroblasts. Curr. Pathobiol. Rep. 1, 215–223 10.1007/s40139-013-0019-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schröder M., and Kaufman R. J. (2005) ER stress and the unfolded protein response. Mutat. Res. 569, 29–63 10.1016/j.mrfmmm.2004.06.056 [DOI] [PubMed] [Google Scholar]
- 4. Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- 5. Heindryckx F., Binet F., Ponticos M., Rombouts K., Lau J., Kreuger J., and Gerwins P. (2016) Endoplasmic reticulum stress enhances fibrosis through IRE1α-mediated degradation of miR-150 and XBP-1 splicing. EMBO Mol. Med. 8, 729–744 10.15252/emmm.201505925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hernández-Gea V., Hilscher M., Rozenfeld R., Lim M. P., Nieto N., Werner S., Devi L. A., and Friedman S. L. (2013) Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J. Hepatol. 59, 98–104 10.1016/j.jhep.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maiers J. L., Kostallari E., Mushref M., deAssuncao T. M., Li H., Jalan-Sakrikar N., Huebert R. C., Cao S., Malhi H., and Shah V. H. (2017) The unfolded protein response mediates fibrogenesis and collagen I secretion through regulating TANGO1 in mice. Hepatology 65, 983–998 10.1002/hep.28921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim R. S., Hasegawa D., Goossens N., Tsuchida T., Athwal V., Sun X., Robinson C. L., Bhattacharya D., Chou H. I., Zhang D. Y., Fuchs B. C., Lee Y., Hoshida Y., and Friedman S. L. (2016) The XBP1 arm of the unfolded protein response induces fibrogenic activity in hepatic stellate cells through autophagy. Sci. Rep. 6, 39342 10.1038/srep39342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koo J. H., Lee H. J., Kim W., and Kim S. G. (2016) Endoplasmic reticulum stress in hepatic stellate cells promotes liver fibrosis via PERK-mediated degradation of HNRNPA1 and up-regulation of SMAD2. Gastroenterology 150, 181–193.e8 10.1053/j.gastro.2015.09.039 [DOI] [PubMed] [Google Scholar]
- 10. Wang J. Q., Chen X., Zhang C., Tao L., Zhang Z. H., Liu X. Q., Xu Y. B., Wang H., Li J., and Xu D. X. (2013) Phenylbutyric acid protects against carbon tetrachloride-induced hepatic fibrogenesis in mice. Toxicol. Appl. Pharmacol. 266, 307–316 10.1016/j.taap.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 11. de Galarreta M. R., Navarro A., Ansorena E., Garzón A. G., Mòdol T., López-Zabalza M. J., Martínez-Irujo J. J., and Iraburu M. J. (2016) Unfolded protein response induced by Brefeldin A increases collagen type I levels in hepatic stellate cells through an IRE1α, p38 MAPK, and Smad-dependent pathway. Biochim. Biophys. Acta 1863, 2115–2123 10.1016/j.bbamcr.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 12. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., and Ron D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- 13. Lipson K. L., Fonseca S. G., Ishigaki S., Nguyen L. X., Foss E., Bortell R., Rossini A. A., and Urano F. (2006) Regulation of insulin biosynthesis in pancreatic β cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 4, 245–254 10.1016/j.cmet.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 14. Moore K. A., and Hollien J. (2012) The unfolded protein response in secretory cell function. Annu. Rev. Genet. 46, 165–183 10.1146/annurev-genet-110711-155644 [DOI] [PubMed] [Google Scholar]
- 15. Tirasophon W., Welihinda A. A., and Kaufman R. J. (1998) A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes. Dev. 12, 1812–1824 10.1101/gad.12.12.1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tirasophon W., Lee K., Callaghan B., Welihinda A., and Kaufman R. J. (2000) The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes. Dev. 14, 2725–2736 10.1101/gad.839400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang J., Liu H., Li L., Liu H., Shi W., Yuan X., and Wu L. (2016) Structural insights into IRE1 functions in the unfolded protein response. Curr. Med. Chem. 23, 4706–4716 10.2174/0929867323666160927142349 [DOI] [PubMed] [Google Scholar]
- 18. Hu P., Han Z., Couvillon A. D., Kaufman R. J., and Exton J. H. (2006) Autocrine tumor necrosis factor α links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1α-mediated NF-κB activation and down-regulation of TRAF2 expression. Mol. Cell Biol. 26, 3071–3084 10.1128/MCB.26.8.3071-3084.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uemura A., Oku M., Mori K., and Yoshida H. (2009) Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell Sci. 122, 2877–2886 10.1242/jcs.040584 [DOI] [PubMed] [Google Scholar]
- 20. Fabregat I., Moreno-Càceres J., Sánchez A., Dooley S., Dewidar B., Giannelli G., Ten Dijke P., and IT-LIVER Consortium (2016) TGF-β signalling and liver disease. FEBS J. 283, 2219–2232 10.1111/febs.13665 [DOI] [PubMed] [Google Scholar]
- 21. Zhang K., Wang S., Malhotra J., Hassler J. R., Back S. H., Wang G., Chang L., Xu W., Miao H., Leonardi R., Chen Y. E., Jackowski S., and Kaufman R. J. (2011) The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J. 30, 1357–1375 10.1038/emboj.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drinane M. C., Yaqoob U., Yu H., Luo F., Greuter T., Arab J. P., Kostallari E., Verma V. K., Maiers J., De Assuncao T. M., Simons M., Mukhopadhyay D., Kisseleva T., Brenner D. A., Urrutia R., et al. (2017) Synectin promotes fibrogenesis by regulating PDGFR isoforms through distinct mechanisms. JCI Insight 2, 92821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darling N. J., and Cook S. J. (2014) The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim. Biophys. Acta 1843, 2150–2163 10.1016/j.bbamcr.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 24. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., and Ron D. (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666 10.1126/science.287.5453.664 [DOI] [PubMed] [Google Scholar]
- 25. Descombes P., and Schibler U. (1991) A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67, 569–579 10.1016/0092-8674(91)90531-3 [DOI] [PubMed] [Google Scholar]
- 26. Ramji D. P., and Foka P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 10.1042/bj20020508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakajima T., Kinoshita S., Sasagawa T., Sasaki K., Naruto M., Kishimoto T., and Akira S. (1993) Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. U.S.A. 90, 2207–2211 10.1073/pnas.90.6.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zaini M. A., Müller C., Ackermann T., Reinshagen J., Kortman G., Pless O., and Calkhoven C. F. (2017) A screening strategy for the discovery of drugs that reduce C/EBPβ-LIP translation with potential calorie restriction mimetic properties. Sci. Rep. 7, 42603 10.1038/srep42603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mink S., Haenig B., and Klempnauer K. H. (1997) Interaction and functional collaboration of p300 and C/EBPβ. Mol. Cell Biol. 17, 6609–6617 10.1128/MCB.17.11.6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dou C., Liu Z., Tu K., Zhang H., Chen C., Yaqoob U., Wang Y., Wen J., van Deursen J., Sicard D., Tschumperlin D., Zou H., Huang W. C., Urrutia R., Shah V. H., and Kang N. (2018) P300 acetyltransferase mediates stiffness-induced activation of hepatic stellate cells into tumor-promoting myofibroblasts. Gastroenterology 154, 2209–2221.e14 10.1053/j.gastro.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jakobs A., Steinmann S., Henrich S. M., Schmidt T. J., and Klempnauer K. H. (2016) Helenalin acetate, a natural sesquiterpene lactone with anti-inflammatory and anti-cancer activity, disrupts the cooperation of CCAAT box/enhancer-binding protein β (C/EBPβ) and co-activator p300. J. Biol. Chem. 291, 26098–26108 10.1074/jbc.M116.748129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Younossi Z. M., Stepanova M., Lawitz E., Charlton M., Loomba R., Myers R. P., Subramanian M., McHutchison J. G., and Goodman Z. (2018) Improvement of hepatic fibrosis and patient-reported outcomes in non-alcoholic steatohepatitis treated with selonsertib. Liver Int. 38, 1849–1859 10.1111/liv.13706 [DOI] [PubMed] [Google Scholar]
- 33. Loomba R., Lawitz E., Mantry P. S., Jayakumar S., Caldwell S. H., Arnold H., Diehl A. M., Djedjos C. S., Han L., Myers R. P., Subramanian G. M., McHutchison J. G., Goodman Z. D., Afdhal N. H., Charlton M. R., and GS-US-384–1497 Investigators (2017) The ASK1 inhibitor selonsertib in patients with nonalcoholic steatohepatitis: a randomized, phase 2 trial. Hepatology 10.1002/hep.29514 10.1002/hep.29514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang F. M., Chen Y. J., and Ouyang H. J. (2011) Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem. J. 433, 245–252 10.1042/BJ20101293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. García-Trevijano E. R., Iraburu M. J., Fontana L., Domínguez-Rosales J. A., Auster A., Covarrubias-Pinedo A., and Rojkind M. (1999) Transforming growth factor β1 induces the expression of α1(I) procollagen mRNA by a hydrogen peroxide-C/EBPβ-dependent mechanism in rat hepatic stellate cells. Hepatology 29, 960–970 10.1002/hep.510290346 [DOI] [PubMed] [Google Scholar]
- 36. Attard F. A., Wang L., Potter J. J., Rennie-Tankersley L., and Mezey E. (2000) CCAAT/enhancer binding protein β mediates the activation of the murine α1(I) collagen promoter by acetaldehyde. Arch. Biochem. Biophys. 378, 57–64 10.1006/abbi.2000.1803 [DOI] [PubMed] [Google Scholar]
- 37. Dodig M., Ogunwale B., Dasarathy S., Li M., Wang B., and McCullough A. J. (2007) Differences in regulation of type I collagen synthesis in primary and passaged hepatic stellate cell cultures: the role of α5β1-integrin. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G154–G164 10.1152/ajpgi.00432.2006 [DOI] [PubMed] [Google Scholar]
- 38. Iraburu M. J., Domínguez-Rosales J. A., Fontana L., Auster A., García-Trevijano E. R., Covarrubias-Pinedo A., Rivas-Estilla A. M., Greenwel P., and Rojkind M. (2000) Tumor necrosis factor α down-regulates expression of the α1(I) collagen gene in rat hepatic stellate cells through a p20C/EBPβ- and C/EBPδ-dependent mechanism. Hepatology 31, 1086–1093 10.1053/he.2000.5981 [DOI] [PubMed] [Google Scholar]
- 39. Li Y., Bevilacqua E., Chiribau C. B., Majumder M., Wang C., Croniger C. M., Snider M. D., Johnson P. F., and Hatzoglou M. (2008) Differential control of the CCAAT/enhancer-binding protein beta (C/EBPβ) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J. Biol. Chem. 283, 22443–22456 10.1074/jbc.M801046200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cui T. X., Piwien-Pilipuk G., Huo J. S., Kaplani J., Kwok R., and Schwartz J. (2005) Endogenous CCAAT/enhancer binding protein β and p300 are both regulated by growth hormone to mediate transcriptional activation. Mol. Endocrinol. 19, 2175–2186 10.1210/me.2004-0502 [DOI] [PubMed] [Google Scholar]
- 41. Cui T. X., Lin G., LaPensee C. R., Calinescu A. A., Rathore M., Streeter C., Piwien-Pilipuk G., Lanning N., Jin H., Carter-Su C., Qin Z. S., and Schwartz J. (2011) C/EBPβ mediates growth hormone-regulated expression of multiple target genes. Mol. Endocrinol. 25, 681–693 10.1210/me.2010-0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elsharkawy A. M., Oakley F., and Mann D. A. (2005) The role and regulation of hepatic stellate cell apoptosis in reversal of liver fibrosis. Apoptosis 10, 927–939 10.1007/s10495-005-1055-4 [DOI] [PubMed] [Google Scholar]
- 43. Friedman S. L., and Bansal M. B. (2006) Reversal of hepatic fibrosis—fact or fantasy? Hepatology 43, S82–S88 10.1002/hep.20974 [DOI] [PubMed] [Google Scholar]
- 44. Iredale J. P., Benyon R. C., Pickering J., McCullen M., Northrop M., Pawley S., Hovell C., and Arthur M. J. (1998) Mechanisms of spontaneous resolution of rat liver fibrosis: hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J. Clin. Invest. 102, 538–549 10.1172/JCI1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kisseleva T., and Brenner D. A. (2006) Hepatic stellate cells and the reversal of fibrosis. J. Gastroenterol. Hepatol. 21, S84–S87 10.1111/j.1440-1746.2006.04584.x [DOI] [PubMed] [Google Scholar]
- 46. Feig J. L., Mediero A., Corciulo C., Liu H., Zhang J., Perez-Aso M., Picard L., Wilder T., and Cronstein B. (2017) The antiviral drug tenofovir, an inhibitor of Pannexin-1-mediated ATP release, prevents liver and skin fibrosis by downregulating adenosine levels in the liver and skin. PLoS One 12, e0188135 10.1371/journal.pone.0188135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mederacke I., Dapito D. H., Affò S., Uchinami H., and Schwabe R. F. (2015) High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 10.1038/nprot.2015.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kakazu E., Mauer A. S., Yin M., and Malhi H. (2016) Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1α-dependent manner. J. Lipid Res. 57, 233–245 10.1194/jlr.M063412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weiskirchen R., and Gressner A. M. (2005) Isolation and culture of hepatic stellate cells. Methods Mol. Med. 117, 99–113 [DOI] [PubMed] [Google Scholar]
- 50. Yaqoob U., Cao S., Shergill U., Jagavelu K., Geng Z., Yin M., de Assuncao T. M., Cao Y., Szabolcs A., Thorgeirsson S., Schwartz M., Yang J. D., Ehman R., Roberts L., Mukhopadhyay D., and Shah V. H. (2012) Neuropilin-1 stimulates tumor growth by increasing fibronectin fibril assembly in the tumor microenvironment. Cancer Res. 72, 4047–4059 10.1158/0008-5472.CAN-11-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]