Abstract
Bone morphogenetic protein (BMP) signaling is critical in renal development and disease. In animal models of chronic kidney disease (CKD), re-activation of BMP signaling is reported to be protective by promoting renal repair and regeneration. Clinical use of recombinant BMPs, however, requires harmful doses to achieve efficacy and is costly because of BMPs' complex synthesis. Therefore, alternative strategies are needed to harness the beneficial effects of BMP signaling in CKD. Key aspects of the BMP signaling pathway can be regulated by both extracellular and intracellular molecules. In particular, secreted proteins like noggin and chordin inhibit BMP activity, whereas kielin/chordin-like proteins (KCP) enhance it and attenuate kidney fibrosis or CKD. Clinical development of KCP, however, is precluded by its size and complexity. Therefore, we propose an alternative strategy to enhance BMP signaling by using small molecules, which are simpler to synthesize and more cost-effective. To address our objective, here we developed a small-molecule high-throughput screen (HTS) with human renal cells having an integrated luciferase construct highly responsive to BMPs. We demonstrate the activity of a potent benzoxazole compound, sb4, that rapidly stimulated BMP signaling in these cells. Activation of BMP signaling by sb4 increased the phosphorylation of key second messengers (SMAD-1/5/9) and also increased expression of direct target genes (inhibitors of DNA binding, Id1 and Id3) in canonical BMP signaling. Our results underscore the feasibility of utilizing HTS to identify compounds that mimic key downstream events of BMP signaling in renal cells and have yielded a lead BMP agonist.
Keywords: bone morphogenetic protein (BMP), BMP signaling, chemical biology, fibrosis, small molecule, SMAD transcription factor, TGF-beta signaling, acute kidney disease, chronic kidney disease, end-stage renal disease, high-throughput screening, sb4
Introduction
Bone morphogenetic proteins (BMPs)2 have essential roles in development, tissue homeostasis, and disease processes for a wide array of cell types. Genetically ablating key elements of the BMP signaling pathway causes embryonic lethality or leads to development defects across organs such as the kidneys (1), heart (2), lungs (3), and central nervous system (4). Tissue homeostasis of these organs and others is also heavily BMP-dependent, as disturbed BMP signaling is implicated in a multitude of diseases (5, 6). Modulating BMP signaling has proved beneficial in several disease contexts. Specifically, in animal models of liver (7), kidney (8, 9), cardiac (10), and pulmonary fibrosis and hypertension (11–13), restoration or enhancement of BMP signaling leads to reversal or attenuation of disease progression. In contrast, suppression of BMP signaling is beneficial in musculoskeletal and oncogenic diseases, such as fibrodysplasia ossificans progressiva and potentially in brainstem gliomas that have overactive BMP signaling (6, 14).
BMPs belong to the TGF-β superfamily of cytokines. In humans, a diverse set of BMP ligands have been identified that possess overlapping yet distinct functions. In particular, BMP4 and BMP7 ligands are critical in renal development (15, 16), whereas BMP2 and BMP7 ligands function in bone and cartilage formation (17). Several human type I BMP receptors (Alk1/2/3/6) and constitutively active type II BMP receptors (BMPR2, ACVR2A/B, and AMHR2) have been described (17). Cleaved and dimerized BMP ligands commence the BMP signaling cascade by engaging with a heterotetramer composed of two type I BMP receptors and two type II BMP receptors. Type II BMP receptors then trans-phosphorylate the glycine/serine-rich domains of the type I BMP receptors. In turn, the activated type I BMP receptors recruit and activate intracellular receptor-activated SMADs (R-SMADs). Canonical BMP signaling is transduced by SMAD-1/5/8/9, which form a transcriptional complex with common SMAD-4 (co-SMAD-4). Translocation of the SMAD heteromeric complex to the nucleus permits recruitment of tissue-specific transcription factors, which drive the expression of direct BMP gene targets. Specifically, bona fide canonical BMP signaling gene targets include the inhibitors of differentiation or inhibitors of DNA binding (ID) family of genes (18). Functionally, ID proteins sequester basic helix-loop-helix transcription factors to both positively regulate cell proliferation and negatively regulate cell differentiation (19).
To modulate the intensity and duration of BMP signaling, various steps of the pathway can be regulated at both intracellular and extracellular levels. For instance, inhibitory SMAD-6/7 (I-SMADs) compete for co-SMAD-4 and, thus, negatively regulate BMP signaling intracellularly, whereas noggin and chordin regulate extracellular levels of BMP ligands via binding sequestration and thereby suppress BMP signaling (20, 21). Secreted kielin/chordin-like proteins (KCPs), on the other hand, enhance BMP signaling via stabilizing BMP ligand–receptor interactions (22). Notably, KCP overexpression leads to increased p-SMAD-1/5/9 levels and lessens the severity of acute and chronic kidney disease, nonalcoholic fatty liver disease, and diet-induced obesity in mouse models (22–25).
Several laboratories have identified small molecules that stimulate both canonical and noncanonical BMP signaling. These agents have been characterized in a host of cell lines for potential applications toward bone repair (26–29), medulloblastoma (30, 31), pulmonary hypertension (12), and stem cell differentiation (32, 33). Immunosuppressants, sirolimus (rapamycin) and tacrolimus (FK506), promote activation of type I BMP receptors by liberating the glycine/serine-rich domain of type I BMP receptors from FKBP12 inhibition (12, 33). Potent and selective small-molecule inhibitors of BMP signaling have also been identified and include dorsomorphin (34), dorsomorphin homolog 1 (35), LDN-193189 (36), and LDN-212854 (37), all of which compete with ATP for binding to the type I BMP receptor kinase.
In this report, we developed a cell-based high-throughput screen for BMP agonists using human renal cells (HEK293s) carrying an integrated BMP reporter. After screening more than 63,000 small molecules, we identified sb4, a potent benzoxazole small molecule that activates a BMP reporter by stabilizing intracellular p-SMAD-1/5/9. The increased levels of phosphorylated SMAD-1/5/9 observed with sb4 result in activation of BMP target genes such as ID1 and ID3. Significantly, sb4-mediated activation of the BMP pathway is resistant to inhibition by noggin or type I BMP kinase inhibitors, which may prove beneficial in disease contexts where BMP inhibitors are also expressed.
Results
Creation of BMP agonist reporter cells for high-throughput screening (HTS)
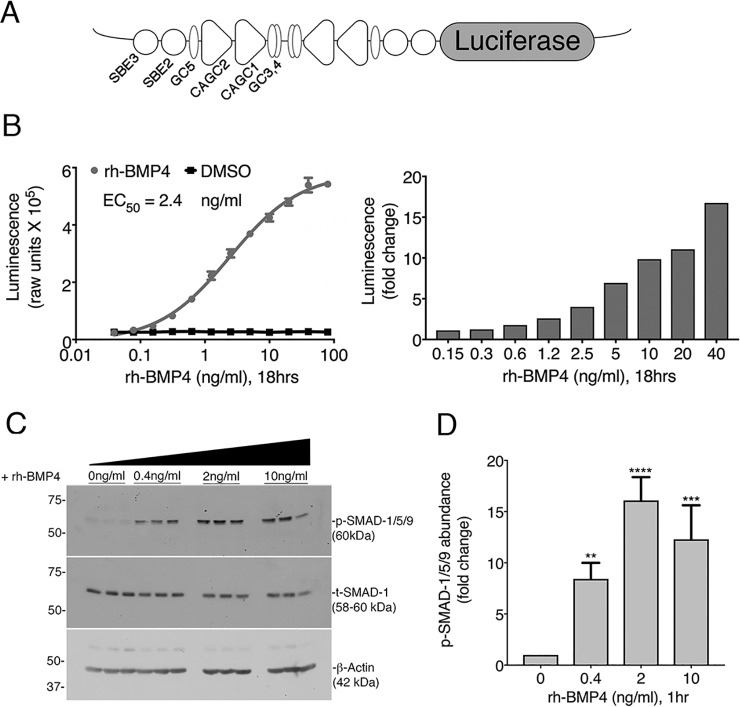
A modified BMP reporter plasmid (BRE-Luc) construct was designed by cloning an inverted repeat of conserved elements of the ID1 promoter that are necessary for p-SMAD-1/5/9 binding and transactivation (38) upstream of a minimal promoter and the firefly luciferase coding region (Fig. 1A). The BRE-Luc plasmid was linearized and stably integrated into the genome of human embryonic kidney cells (HEK293). Individual clonal isolates were then screened for responsiveness to both BMP4 and BMP7, and multiple responders were identified.
Figure 1.
Characterization and validation of BRE-Luc cells for HTS. A, the BRE-Luc reporter construct contains an inverted repeat of BMP-responsive elements driving luciferase. Elements of the ID1 promoter and SMAD-binding sites are marked as originally identified by Korchynskyi and ten Dijke (38). B, a dose response of BRE-Luc cells to increasing concentrations of rhBMP4 performed in triplicate. C, Western blotting of lysates from BRE-Luc cells treated with increasing concentrations of rhBMP4. Membranes were probed with p-SMAD-1/5/9 and total SMAD-1. β-Actin was used as an additional loading control. D, p-SMAD-1/5/9 protein levels were quantified by densitometry. The signal of p-SMAD-1/5/9 was normalized to total-SMAD-1 to control for loading variability. Data are expressed as -fold change relative to the medium alone, which was set to 1. Error bars, 1 S.D. from the mean of three independent biological replicates. ANOVA and Dunnett's post hoc multiple-comparison test generated p values as follows: p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****) relative to controls.
The inverted repeats of the p-SMAD-1/5/9 binding elements confer high sensitivity to rhBMP4 treatment and a wide dynamic range of detection of activated BMP signaling. In luciferase assays, we observed that rhBMP4 treatment caused a dose-dependent increase in luciferase activity in BRE-Lucs (EC50 = 2.4 ng/ml), whereas vehicle (DMSO) treatment had no effect (Fig. 1B). The BRE-Luc cell line displayed a highly specific response to BMP treatment, with 10 ng/ml rhBMP4 resulting in a 10-fold increase in luciferase activity over DMSO (Fig. 1B). Furthermore, immunoblotting assays revealed that stimulation of BRE-Lucs with 0.4, 2, or 10 ng/ml rhBMP4 yielded a dose-dependent induction of p-SMAD-1/5/9 (Fig. 1C), with 2 ng/ml rhBMP4 yielding a 16-fold induction of p-SMAD-1/5/9 compared with untreated cells (Fig. 1D). In the presence of increasing doses of rhBMP4, basal levels of the TGF-β effector, p-SMAD-2, remained constant (Fig. S1A). Thus, the BRE-Luc reporter cells displayed the sensitivity and specificity needed to detect activated BMP signaling in a cell-based HTS assay for small-molecule agonists of BMP signaling.
Identification of BMP signaling agonists via a cell-based HTS
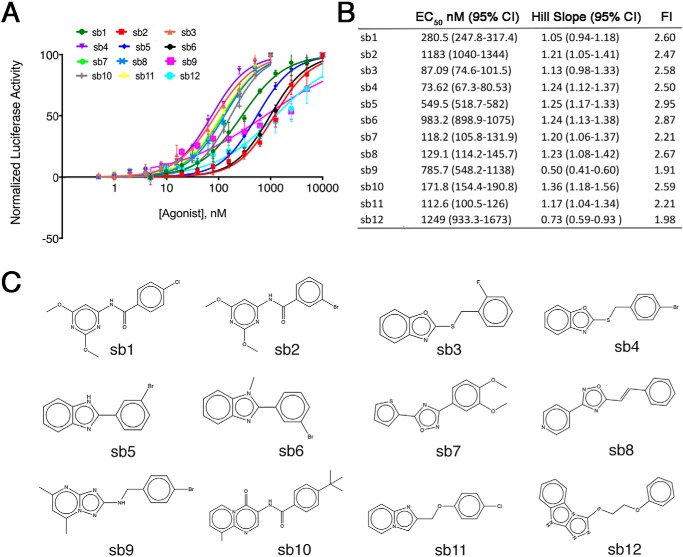
We next screened 63,608 small molecules from the Center for Chemical Genomics library at a single concentration (10 μm) in the BRE-Lucs in our primary HTS (Fig. 2, A and B). We identified small molecules whose activity was >18% the activity of the positive control (25 ng/ml rhBMP4) or >3 S.D. values above the negative control (0.1% DMSO) as a hit. To triage the primary screening hits, we applied various filters, such as pan-assay interference compounds (PAINS) (39), reactivity, aggregators, and solubility forecast index (40) implemented in MScreen (41). The initial 1,453 hits from our primary screen were retested in triplicate for confirmation. After applying MScreen filters, 211 were deemed dose–response candidates. Of these 211, 70 compounds showed specificity for the BMP response element in our counterscreen using a luciferase reporter cell line that contained a PAX protein response element unresponsive to BMPs (42). Fresh powders of the top 16 compounds were purchased to conduct dose response studies. Twelve of the 16 small molecules displayed dose–response activity with relatively potent EC50 values (Fig. 3). We determined that sb4 was the most promising compound because it displayed a sigmoidal dose–response curve with the lowest EC50 (74 nm) of the top 12 compounds. The Hill Slope of 1.24 indicates that sb4 binds noncooperatively (i.e. single ligand interaction) to its target. Luciferase activity after sb4 treatment was increased 2.5-fold compared with DMSO at the highest concentration of 1 μm.
Figure 2.
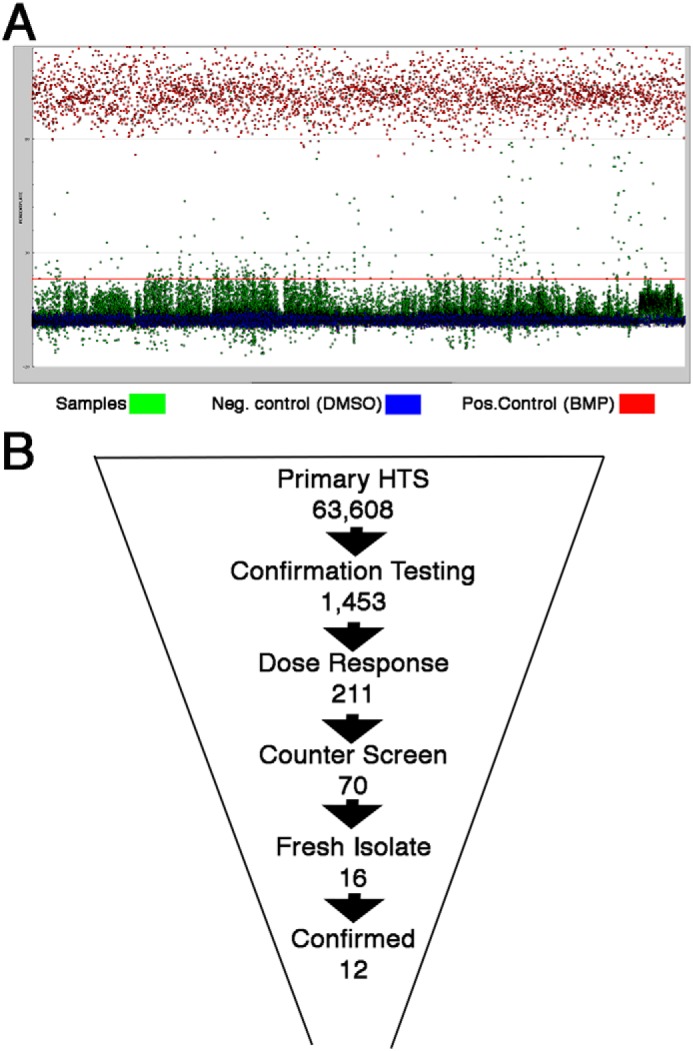
High-throughput screening strategy in BRE-Luc cells. A, a snapshot of the primary screen of 63,608 small molecules (green dots) at a single concentration (10 μm) in renal BRE-Luc cells. Positive controls were 25 ng/ml rhBMP4 (red dots), whereas negative controls were 0.1% DMSO (blue dots). The red line represents 3 S.D. values above the DMSO. The average luciferase induction for 25 ng/ml rhBMP4 was 355,000 ± 34,000; for DMSO, it was 44,000 ± 4,300; and for the small-molecule hits, it was 127,000. The average Z′ score for all of the plates screened was 0.71, indicating a high-quality screening assay. B, triage strategy used to winnow down the starting 63,608 small molecules from the primary HTS campaign, which allowed identification of the top 12 BMP signaling agonists based on dose–response curves and structures.
Figure 3.
Dose responses of the top 12 potential BMP agonists. A, 12 compounds were tested for BRE-Luc responses at 2-fold increasing concentrations from 0.5 nm to 10 μm in triplicate. B, effective concentrations at 50% maximum response (EC50), and the Hill slopes were calculated for each compound. Maximum -fold induction (FI) above DMSO controls is reported. C, chemical structures of the 12 candidate BMP agonists are shown schematically.
p-SMAD-1/5/9 is induced by BMP signaling agonists
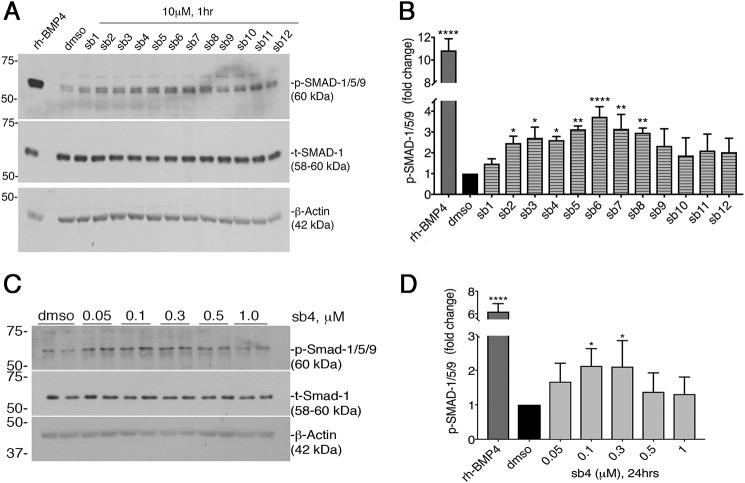
To determine whether phosphorylation of SMAD-1/5/9 was increased in the presence of our top 12 compounds, we treated HEK293 cells for 1 h with a 10 μm concentration of each compound and assayed for p-SMAD-1/5/9 induction by immunoblotting. 2 ng/ml rhBMP4 served as our positive control and induced p-SMAD-1/5/9 ∼11-fold over DMSO (Fig. 4, A and B). Additionally, we found that 1 h of treatment with several of our top 12 BMP signaling agonists significantly increased p-SMAD-1/5/9 levels at least 2-fold compared with DMSO treatment (Fig. 4, A and B). The increased levels of p-SMAD-1/5/9 within 1 h suggest a direct stimulation of canonical BMP signaling. Our data support the feasibility of identifying small molecules using high-throughput screening that can mimic key downstream effects of endogenous BMP ligands.
Figure 4.
Activation of p-SMAD-1/5/9 by small molecules. A, immunoblotting of whole cell protein lysates from BRE-Luc cells treated with either 2 ng/ml rhBMP4, 0.04% DMSO, or the top 12 BMP candidate agonists at 10 μm for 1 h in BRE-Luc cells. Membranes were probed with anti-p-SMAD-1/5/9 and anti-total-SMAD-1. β-Actin served as an additional loading control. B, quantitation of p-SMAD-1/5/9 protein levels as determined by densitometry of three independently derived Western blots. The p-SMAD-1/5/9 levels were normalized to total-SMAD-1. Final data are expressed as -fold change relative to the mean negative control signal, with error bars representing 1 S.D. (*, p < 0.05; **, p < 0.01; ****, p < 0.0001 relative to control). ANOVA, Dunnett's post hoc multiple comparisons test was used to determine significance. C, immunoblotting of lysates from serum-starved PRECs treated for 24 h with increasing concentrations of sb4 in serum-free medium. Total SMAD-1 and β-actin serve as loading controls. D, quantitation of p-SMAD-1/5/9 levels are shown. Statistical analyses of replicates were done as in B.
To determine whether sb4 can induce SMAD-1/5/9 phosphorylation in other cell types, we employed serum-starved primary mouse kidney epithelial cells (PRECs) for compound testing, which lack the BRE-Luc reporter construct. We observed induction of phosphorylated SMAD-1/5/9 in a dose-dependent manner (Fig. 4, C and D), with sb4 at 100 and 300 nm yielding a 2-fold induction over DMSO after 24 h of treatment. Although not as robust as rhBMP4, our results suggest that sb4 can significantly increase p-SMAD-1/5/9 abundance in PRECs under serum-starved conditions.
To ensure specificity, we examined p-SMAD-2 levels after treating cells with candidate BMP signaling agonists at 10 μm for 1 h. No significant effects were observed on the TGF-β effector p-SMAD-2 or p-SMAD-3 with any of the compounds tested (Fig. S1B). We also examined the activation of noncanonical BMP signaling by our top 12 compounds by immunoblotting for phosphorylation of mitogen-activated kinases p38, ERK1/2, and JNK. We found that our BMP signaling agonists (sb7–sb11) decreased levels of p-ERK1/2 after 1 h of treatment but had little effect on p-JNK or p-p38 (Fig. S2, C and D). In addition, BMPs can stimulate the phosphorylation of transforming growth factor β–activated kinase 1 (p-TAK-1). However, sb12 decreased levels of p-TAK-1, whereas there was no significant increase of p-TAK-1 by the other compounds (Fig. S2B).
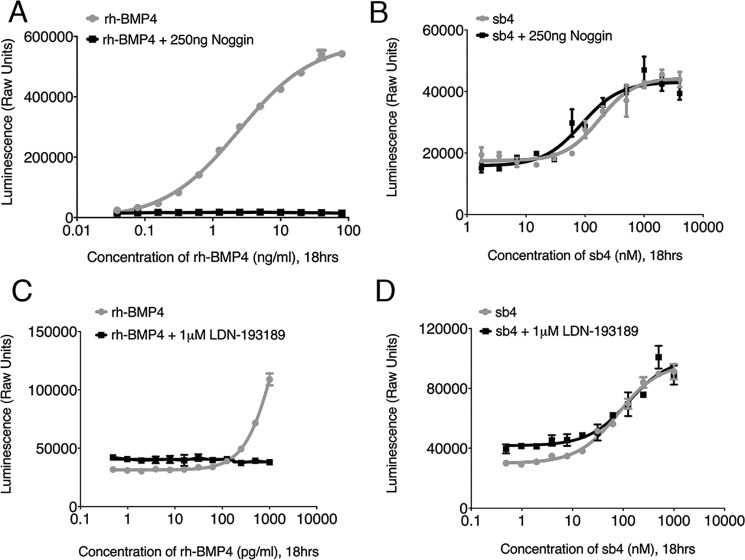
Noggin and type I BMP receptor inhibition is bypassed by sb4
To address the mechanism of sb4-mediated activation of BMP signaling, we conducted luciferase-based inhibition studies with both endogenous and chemical inhibitors of the BMP signaling pathway. Noggin is an endogenous inhibitor of BMP signaling that acts by sequestering dimerized BMP ligands away from BMP receptors and thus suppresses BMP signaling (21). The addition of 250 ng/ml of noggin completely suppressed the effects of rhBMP4 in the BRE-Luc cells (Fig. 5A). However, noggin had no effects on the sb4-mediated activation of BRE-Luc (Fig. 5B). Additionally, we tested the chemical inhibitor, LDN-193189, which selectively inactivates type I BMP receptors (36, 43). Inhibition of type I BMP receptors completely abolished the activation of luciferase by rhBMP4 (Fig. 5C). However, sb4 remained active even in the presence of 1 μm LDN-193189 (Fig. 5D). Taken together, our results indicate that sb4 can activate BMP signaling independent of noggin and type I BMP receptors, suggesting that it works through an alternate mechanism.
Figure 5.
Effects of BMP inhibitors on sb4 activity. A, BRE-Luc cells were subjected to increasing concentrations of rhBMP4 with (black) or without (red) 250 ng/ml rhNoggin. Maximum concentrations of 80 ng/ml for rhBMP4 were used, and cells were incubated for 18 h before measuring luminescence. B, a dose response for sb4 was measured in BRE-Luc cells alone (blue) or in the presence of 250 ng/ml rhNoggin (black). A maximum concentration of 4 μm sb4 was used. C, activity of increasing concentrations of rhBMP4 in BRE-Luc cells alone (red) or with a constant dose of 1 μm type I BMP receptor inhibitor (LDN-193189, black). For both curves, a maximum concentration of 1000 pg/ml rhBMP4 was used, and cells were incubated for 18 h before measuring luminescence. D, increasing concentrations of sb4 were added to BRE-Luc cells alone (blue) or in the presence of 1 μm LDN-193189 (black). A maximum of 1000 nm sb4 was used. Experiments were conducted in triplicate. Error bars, S.D.
BMP signaling efficacy is enhanced and maintained with sb4 treatment
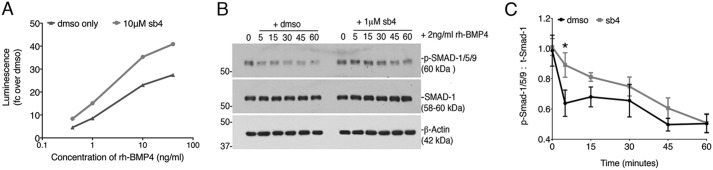
To determine whether the efficacy of BMP signaling is enhanced in the presence of sb4, we assayed varying concentrations of rhBMP4 in the presence of sb4. We found that sb4 enhanced the efficacy of signaling at each concentration of rhBMP4 tested (Fig. 6A). This effect was most pronounced at low concentrations of rhBMP4, whereby sb4 increased BRE-luc expression 2-fold at 0.4 ng/ml of rhBMP4. These data suggest that sb4 may act to stabilize p-SMAD-1/5/9 to enhance the transcriptional response. To test this more directly, we examined the decay of p-SMAD-1/5/9 after rhBMP4 stimulation in the presence or absence of sb4. The BRE-Luc cells were stimulated with 2 ng/ml rhBMP4 for 1 h, after which the media were removed and replaced with fresh media containing either DMSO or 1 μm sb4. p-SMAD-1/5/9 levels were then examined after 0, 5, 15, 30, 45, and 60 min of sb4 treatment and compared with DMSO controls (Fig. 6, C and D). At each time point, cells treated with sb4 had higher levels of p-SMAD-1/5/9. Furthermore, the decay of p-SMAD-1/5/9 was attenuated, suggesting that sb4 works primarily by increasing the half-life of p-SMAD-1/5/9.
Figure 6.
BMP signaling agonists enhance the efficacy of BMPs. A, BRE-Luc cells were treated with DMSO (black) or sb4 (gray) at a constant concentration of 10 μm in the presence of increasing concentrations of rhBMP4 (0.4, 1, 10, and 40 ng/ml) for 18 h. A representative experiment was conducted in triplicate, reported as -fold change over DMSO. B, Western blotting of protein lysates from BRE-luc cells treated for 1 h with rhBMP4 (2 ng/ml) followed by fresh media containing either vehicle (DMSO) or 1 μm sb4. Samples were harvested at 0, 5, 15, 30, 45, and 60 min after the addition of sb4. A representative Western blot for p-SMAD-1/5/9 from three independent biological experiments is shown. C, ratio of p-SMAD-1/5/9 to total SMAD-1 as quantified by densitometry is shown for experiments outlined in C. Ratio of p-SMAD-1/5/9 to total-SMAD-1 over time was compared by unpaired multiple t tests. *, p = 0.02. Note the increased stability of p-SMAD-1/5/9 in sb4-treated cells. Error bars, S.E.
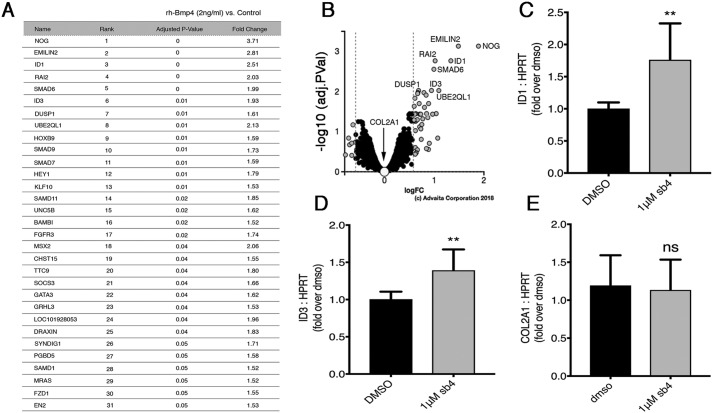
Enhancement of BMP signaling efficacy in our luciferase and Western blotting studies prompted us to ask whether endogenous BMP target genes are also up-regulated after sb4 dosing. Because our compounds mimic the activity of low-dose rhBMP4, we conducted a transcriptomic analysis in BRE-Luc cells treated for 4 h with 2 ng/ml rhBMP4. We identified 31 genes up-regulated between 1.5- and 3.7-fold above untreated cells (p < 0.05) (Fig. 7, A and B). We assayed two of the top rhBMP4 target genes by quantitative RT-PCR to determine whether sb4 could affect the expression of these genes. At 24 h, sb4 significantly increased the expression of Id1 and Id3 (Fig. 7, C and D) but had no effect on CoL2A1, which also did not respond to rhBMP4. These data demonstrate that at least some endogenous BMP target genes show increased expression in cells treated with sb4.
Figure 7.
sb4 can activate endogenous BMP4 target genes. A, the top 31 genes with increased transcriptional activation after 4 h of rhBMP4 (2 ng/ml) as determined by Affymetrix microarrays. Triplicate biological replicates were performed for treated and untreated groups. The p values ≤ 0.05 and -fold changes ≥ 1.5 are reported for treated versus untreated groups. B, volcano plot of 6,643 genes that were measured for expression. 51 genes were differentially expressed between treated and untreated groups. Significant differentially expressed genes are shown as gray dots; black and selected white dots represent nonsignificant genes. The volcano plot was generated by uploading transcriptomic data into iPathwayGuide, with cut-off -fold change and p values set to 0.58 and 1, respectively. C–E, quantitative RT-PCR analysis was used to determine the relative mRNA levels of endogenous rhBMP4 target genes after 1 μm sb4 treatment. The Id1 (C) and Id3 (D) genes show significant up-regulation, whereas CoL2A1 (E) remains unchanged. Data represent the mean and 1 S.D. from three independent biological experiments performed in triplicate (**, p < 0.01; ns, not significant: unpaired t test). Error bars, S.D.
Analogs of sb4 increase BMP signaling dose-dependently
Testing of structural analogs of sb4 could identify compounds with increased efficacy and also reveal structure–activity relationships. Thus, we compared 10 related compounds with sb4 and sb3 in our cell-based luciferase reporter assay and determined the EC50 values (Fig. 8). Of these 10 compounds, four were inactive and six were active compounds with EC50 values ranging from 16.2 to 274.4 nm. For these benzyl-thio-benzo-oxazole compounds, the substitution at the benzene ring of the benzyl group seems to be important. The data show very tight structure–activity relationships. For example, benzyl or pyridyl for R1 makes compounds inactive as in sb4.a1 and sb4.a4, but a methyl-substituted benzyl at the para position makes the most active compound with EC50 of 16.16 nm as in sb4.a2. Different substitutions of benzyl with fluorine, chlorine, and bromine at the para position as in sb4.a5, sb4.a3, and sb4, respectively, also make compounds active, but not as active as the methyl group. For para substitution, methyl is the best (sb4.a2: 16.16), followed by fluorine (sb4.a5: 60.14), bromine (sb4: 73.62), and then chlorine (sb4.a3: 77.05). Compared with substitution at the para position of the benzyl as in sb4.a5, substitution at the ortho position with fluorine as in sb3 reduces activity. Compared with substitution at the para position of the benzyl as in sb4.a3, substitution at the meta position with Cl as in sb4.a6 makes compound 5 times less active. Substitution at the ortho position of the benzyl with methoxy is 2 times less active than fluorine. The fluorine-substituted compound at the ortho position is 10% more active when chlorine is substituted at the other ortho position at the same time as in sb4.a10. The inactivity of sb4.a7 could be either due to the steric effect of the substitution of methyl at the benzo-oxazole or due to the lack of aromatic ring at the right-hand side, or both. The oxazolo-pyridine in place of the benzo-oxazole as in sb4.a9 kills the activity. Overall, the substitutions at the para or ortho position of benzyl group are preferred to that at the meta position. These data suggest that modification of benzo-oxazole is not tolerated.
Figure 8.
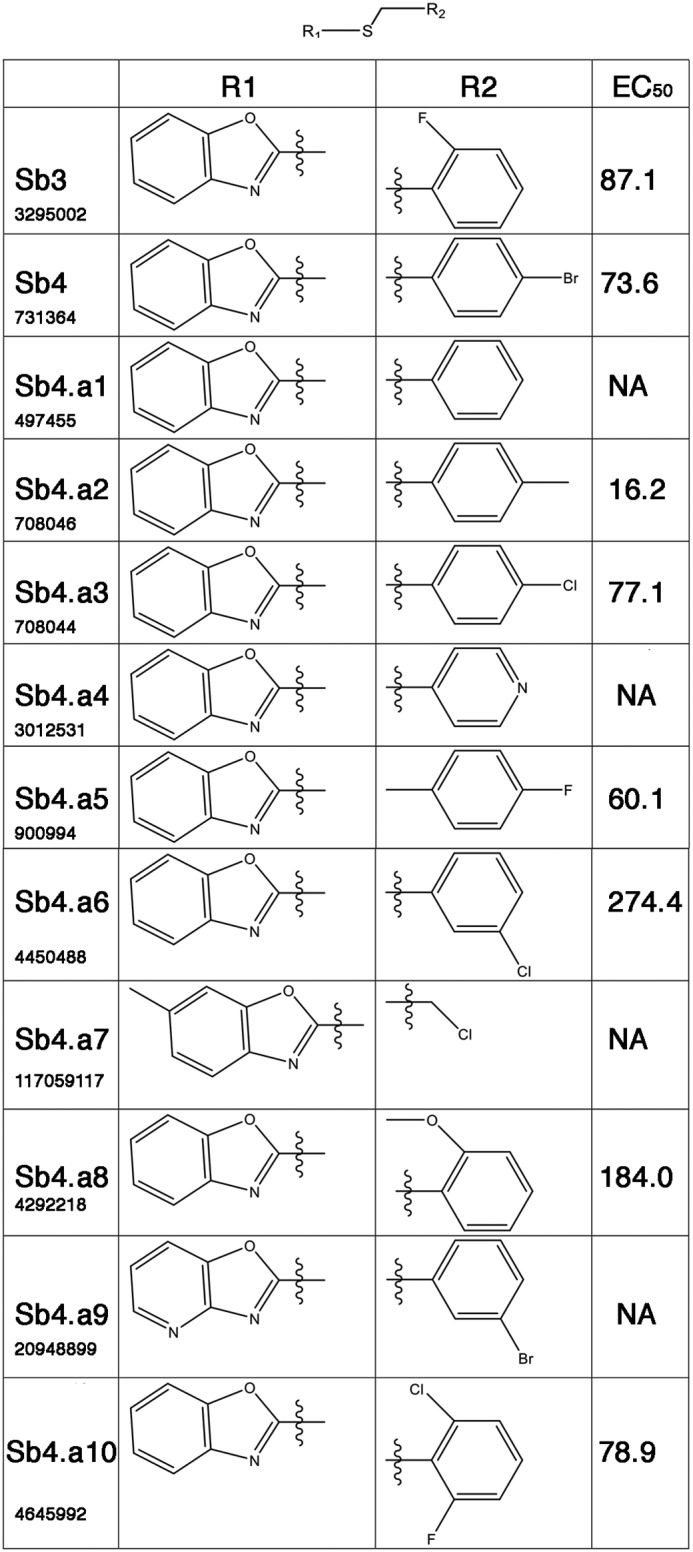
Structure–activity relationships among 11 sb4-like compounds. Dose–response curves for analog structures of sb4 were tested in BRE-luc cells. The effective concentration at 50% maximum activity (EC50) was calculated for each structure as shown. The PubChem ID numbers are listed below the designated name. NA, not active at concentration tested.
Discussion
To identity chemical compounds that could be the basis for developing therapeutic BMP agonists, we designed a cell-based HTS and identified multiple specific small molecules that enhance and/or stabilize the BMP effectors p-SMAD-1/5/9. Activation of BMP signaling is reported to be protective in a variety of disease contexts, including animal models of acute and chronic kidney disease. Our unbiased HTS identified a novel compound, sb4, that rapidly activates BMP signaling in renal cell cultures. We found that sb4 increased levels of p-SMAD-1/5/9 within 1 h and increased the expression of direct key BMP4 target genes (Id1 and Id3). Additionally, our experiments revealed that sb4 acts downstream of type I BMP receptors and bypasses negative regulation by noggin, an extracellular inhibitor of BMP signaling. Moreover, in the presence of rhBMP4, sb4 enhances BMP signaling efficacy by slowing the turnover or decay of p-SMAD-1/5/9.
In contrast to animal models of CKD, human CKD patient samples have elevated levels of BMP expression (44) along with elevated levels of negative regulators of BMPs similar to noggin, such as gremlin (45, 46) and connective tissue growth factor (47). Hence, a mechanism such as that demonstrated by sb4, which bypasses negative regulation by noggin and similar negative regulators of BMP signaling, could be beneficial in specific clinical situations where these negative regulators are elevated. Further, the ability to activate BMP signaling when type I BMP receptors are inhibited could also be a useful mechanism in the event that receptor status is low due to diseased states (48, 49). Moreover, compounds like sb4 could synergize with endogenous levels of BMPs and potentially abrogate the need for harmful doses of rhBMPs that lead to unwanted side effects (50–52).
Several other laboratories have also reported the identification of activators/sensitizers of BMP signaling in other cell types. Of note, some of these previously published BMP activators, such as isoliquiritigenin (31), FK506 (12), and PD-407824 (32), were tested in our primary HTS screen. However, they did not meet our cut-off for further analysis due to either having low activity in our primary screen or not meeting the criteria to pass our counterscreen. This may underscore the potential of diverse compound structures to induce cell type–specific responses. In a broader sense, it stresses the importance of identifying direct targets and determining the mechanism of action of these diverse compounds, because each molecule may target different levels/aspects of canonical or noncanonical BMP signaling and, therefore, induce varying intensities of activation of BMP signaling culminating in diverse biological effects.
Canonical R-SMAD-1/5/8/9 activation occurs via phosphorylation by type I BMP receptors. Phosphorylation at the C-terminal transcriptional domain of SMAD-1/5/8/9 facilitates co-SMAD-4 docking and heteromeric SMAD transcriptional complex formation. In addition, the linker interdomain of SMAD-1/5/8/9 undergoes phosphorylation by cyclin-dependent kinases (CDK8/9) and glycogen synthase kinase 3 (GSK3), which renders R-SMADs fully functional (53). To control activated BMP/SMAD-1/5/8/9 signaling duration and intensity, fundamental turnover mechanisms, such as dephosphorylation and ubiquitination, are necessary. In particular, small C-terminal domain Ser/Thr phosphatases (SCPs) have been identified that dephosphorylate the C-terminal transcriptional domain of SMAD-1 after type I BMP receptor phosphorylation (54, 55) and the linker interdomain after CDK8/9 phosphorylation (55). Experimental siRNA-mediated knockdown of SCP-1/2 conferred increased BMP signaling and Id1 expression in human keratinocytes and osteosarcoma cell lines (54). Thus, small molecules such as sb4 could potentially inhibit such phosphatases and prove beneficial in disease states such as CKD.
The Smurf proteins (SMAD ubiquitination regulatory factors) also negatively regulate BMP/SMAD activity (56). GSK3 phosphorylation recruits Smurfs to the BMP/SMAD linker interdomain to ubiquitinate activated BMP/SMADs for proteasomal degradation (57, 58). Increased BMP signaling is also observed following siRNA-mediated knockdown of Smurf (59). Yes-associated proteins (YAPs) are also recruited to the linker region of SMAD-1 but, instead, function to enhance BMP/SMAD transcriptional activity and therefore signaling (59). Because sb4 activates BMP signaling independent of noggin or type I BMP receptor inhibition, it is plausible that the increased levels of p-SMAD-1/5/9 observed in this study can be potentially attributed to inhibiting SCP or Smurf activity or promotion of YAP function. Further studies are needed to identify the direct targets of sb4 and its analogs to unravel the precise mechanism of action.
Clinical trials have been conducted to assess the clinical use of peptide BMP agonist (THR-184) in acute kidney injury (60) as well as the use of a humanized, neutralizing TGF-β1 mAb in CKD (61). Both clinical trials yielded futile results due to lack of efficacy. This may suggest the need for combination treatments that not only repress TGF-β activity but simultaneously activate BMP signaling while bypassing negative regulators such as noggin. Compounds with a benzoxazole moiety similar to sb4 have been reported to have important clinical applications. Pharmacological activities include working as anti-inflammatory (62) and anti-diabetic agents (63). Our studies suggest that these agents could also possess anti-fibrotic pharmacological activity due to their ability (sb4 and its analogs) to biologically activate and enhance BMP signaling. Therefore, and in conclusion, identification of the renal BMP signaling agonists reported here provides a novel tool compound that can be used to interrogate BMP signaling preclinically in animal models of kidney disease.
Experimental procedures
Construction of BMP reporter cells and cell culture
Human embryonic kidney cells (HEK293) were transfected with a linearized plasmid containing two inverted repeats of a BMP-responsive element containing SMAD-binding sites and parts of the ID1 promoter as described (38). Transformants were selected in 800 mg/ml Geneticin (G418), and clones were derived by single-cell dilution. Individual BRE-Luc–containing HEK293 clones were tested for responsiveness to BMP4 and BMP7. For testing of small molecules, BRE-Lucs were maintained in complete medium (Dulbecco's modified Eagle's medium containing 4.5 g/liter d-glucose and 584 mg/liter l-glutamine supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin) and cultured to subconfluence at 37 °C and 5% CO2. PRECs were immortalized by using temperature-sensitive T-antigen transformation and cultured to subconfluence in complete medium containing 800 μg/ml Geneticin (G418) at 33 °C and 5% CO2.
Small-molecule screening library
63,608 drug/lead-like structures were screened in the Center for Chemical Genomics small-molecule library housed at the Life Science Institute at the University of Michigan. Small molecules were selected for their drug/lead-like physicochemical properties, which fell within the Lipinski's space for bioavailable/orally active drugs having molecular weights <500, hydrogen bond donors <5, hydrogen acceptors <10, and cLogPs <5 (64). The libraries utilized at the Center for Chemical Genomics consisted of compounds from the ChemDiv 100K diversity set in which 61,120 structures were screened, returning a hit ratio of 17.32%. 1,280 compounds from the Library of Pharmacologically Active Compounds (Sigma) returned a hit ratio of 9.06%. 1,280 compounds from the Prestwick library returned a hit ratio of 8.2%, and screening 320 compounds from the National Institutes of Health Clinical Compound library returned a 7.81% hit ratio. Instant JChem was used for structure database management, search, and prediction (Instant JChem 5.9.0, ChemAxon).
sb4 analog similarity search
Analogs similar to sb4 were obtained as fresh powders after doing a structure similarity search at the Aldrich Market Select website. A 70% similarity search type was used that included product filters to select for compounds with molecular weights <500 and cLogP <5. Purity of compounds ranged from 85 to 90%, and they were synthesized by AMS suppliers including AMS Private Supplier I (sb4.A7), ChemBridge (sb4, 85% purity), ChemDiv (sb4.A9, 90% purity), Labotest (sb3, sb4.A2-A6, sb4.A8, sb4.A10, 90% purity), and Pharmeks (sb4.A1, 90% purity).
Immunoblotting
BRE-Lucs were passaged in Dulbecco's modified Eagle's medium supplemented with 5% FBS (v/v) for both HTS and 6-well plate studies. Compounds were added in the presence of 5% FBS. PRECs were serum-starved for 1 h before the addition of factors in serum-free medium. Subconfluent BRE-Luc or PREC cultures were seeded into 6-well plates at 3.0 × 105 or 2.0 × 105 cells/well respectively, using an automated cell counter (CountessTM II). Cells were treated with rh-BMP4 (R&D Systems; 314-BP), vehicle (DMSO-d6), or compound (10 μm) for 1 or 24 h. After the indicated time points, treatment medium was removed, and adhered cells were washed once in 1× PBS and subsequently harvested in 1× PBS and pelleted by low centrifugation. The cell pellet was then suspended in PK lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 10% glycerol, 1% Triton X-100, 1 mm Na3VO4, 50 mm NaF) containing both phosphatase and protease inhibitors (PhosSTOP and cOmplete Mini; Roche Applied Science) and allowed to lyse on ice for 30 min with intermittent vortexing. To pellet the insoluble fraction, the suspended pellet was centrifuged at 15,000 rpm for 15 min at 4 °C. Supernatant was transferred to clean Eppendorf tubes, and 6× SDS was added 1:5 and heated for 5 min at 95 °C. Equal amounts of total protein lysates were resolved on hand-casted 8% SDS-polyacrylamide gels. Separated proteins were then electrophoretically transferred onto Immobilon®-FL polyvinylidene difluoride membranes. Membranes were dried and reactivated in methanol rinsed in 1× TBS and blocked for 1 h in 5% BSA or 5% milk dissolved in 1× TBS-T (1× TBS + 0.1% Tween® 20) and then incubated overnight with primary antibodies consisting of one of the following: anti-Rb phospho-SMAD-1/5/9 1:1,000 (CST®; 13820), anti-Rb total SMAD-1 1:2,000 (CST®; 9734), anti-Rb phospho-SMAD-2 1:1000 (CST®; 3104), anti-Rb phospho-SMAD-3 1:1000 (CST®; 9520), anti-Rb total SMAD-2/3 1:1000 (CST®; 3102), anti-Rb phospho-TAK1 (CST®; 9339), anti-Rb phospho-MAPK family sampler kit (CST®; 9910T), and anti-β-actin 1:10,000 (Proteintech®; 60008-1-Ig). The next day, membranes were washed in 1% nonfat dry milk prepared in 1× TBS-T and incubated for 1 h at room temperature in anti-Rb HRP or anti-Ms-HRP diluted in 1% nonfat dry milk (1× TBS-T). Membranes were washed in TBS-T and rinsed twice with 1× TBS and incubated with ECL Western blotting substrate (PierceTM; 32106). Finally, membranes were incubated with HyBlot ESTM autoradiography film (Denville Scientific) and developed on the Kodak X-OMAT 2000A processor and quantified using ImageJ (version 2.0.0).
Luciferase assays
Compounds were dissolved in MagniSolvTM DMSO-d6 (DMSO-d6, EMD Millipore; 2206-27-1) and then diluted in 5% FBS medium and added to 96-well plates. Recombinant proteins (rhBMP4 or rhNoggin, R&D Systems; 314-BP or 6057-NG) were reconstituted per the manufacturer's instructions and diluted in 5% FBS medium. BRE-Lucs were passaged in 5% FBS medium and seeded into factor-containing 96-well plates at 3.0 × 103 cells/well using an automated cell counter (CountessTM II). BRE-Lucs were incubated overnight at 37 °C and 5% CO2. The next day, 85% of the culture medium was aspirated using a microplate washer (BioTek® ELx405TM), and an equal volume of luciferase reagent (Steady-Glo® Promega) was added to the remaining culture medium in each well using a MultidropTM dispenser. Following Steady-Glo® cell lysis, luminescence activity was measured on a luminometer (PHEARstar® BMG Labtech).
BRE-Luc cell transcriptome profiling
To identify direct gene targets of low-dose rhBMP4 (2 ng/ml) specifically in BRE-Lucs, cells were seeded at 3.0 × 105 cells in 6-well plates in triplicate for each sample. Cells were cultured in 5% FBS medium overnight and treated the next day with or without 2 ng/ml rhBMP4 for 4 h. After treatment, BRE-Lucs were harvested to isolate total RNA using the RNeasy® minikit (Qiagen) by following the manufacturer's protocol. Transcriptome profiles of low-dose rhBMP4 (2 ng/ml) (treated) and 5% FBS medium-only (untreated) cells were assessed via AffymetrixTM microarray analysis performed by the Microarray Core housed within University of Michigan DNA Sequencing Core. Concisely, the integrity and yield of total RNA were assessed by using the RNA 6000 Nano kit on the 2100 Bioanalyzer (Agilent Technologies). Reported RNA integrity numbers were above 9 for the two samples submitted in triplicate. 400 ng of total RNA was used to generate fragmented and biotin-conjugated single-stranded cDNA using the GeneChipTM Whole Transcript PLUS reagent kit (Applied Biosystems). Single-stranded cDNA was then hybridized to the Affymetrix® GeneAtlas Human Gene version 2.1 Sense Target microarray. Following hybridization, wash, stain, and imaging procedures were carried out according to the manufacturer's protocol. The robust multiarray average method (65) was used to correct for variations between microarrays. Oligo and Limma packages of Bioconductor/R were used to identify differentially expressed genes by fitting normalized gene expression data to weighted linear models (66). Probe sets with a variance >0.025 and -fold changes >1.5 were selected. Multiplicity assessment was carried out by using the false discovery rate method to adjust p values (67). Final data were loaded into iPathwayGuide to generate a volcano plot (Advaita Bioinformatics).
Quantitative RT-PCR
To assess the capacity of HTS compounds to induce the expression of endogenous direct gene targets of BMP signaling, BRE-Lucs were seeded in 6-well plates at 3.0 × 105 cells/well and cultured in 5% FBS medium overnight. The following day, BRE-Lucs were treated with low-dose rhBMP4 (2 ng/ml), 0.04% DMSO-d6, or increasing concentrations of compounds (all with 0.04% DMSO-d6). After 24 h of treatment, BRE-Lucs were harvested for total RNA isolation using the RNeasy® minikit (Qiagen) by following the manufacturer's protocol. Synthesis of first-strand cDNA was prepared by using 1 μg of total RNA, SuperscriptTM III (Invitrogen) reverse transcriptase, and 100 ng of random primers. Amplified cDNA templates were diluted 1:50, and SYBR® Green Master Mix was used to monitor cDNA template amplification on the Applied Biosystems® 7500 real-time PCR system. Hprt was used to normalize gene expression. For each sample, -fold change of gene expression over 0.04% DMSO-d6 was calculated. Human primer pair sequences used were obtained from PrimerBank (68) (except for Hprt) and include the following: Id1, CTGCTCTACGACATGAACGG (forward) and GAAGGTCCCTGATGTAGTCGAT (reverse); Id3, GAGAGGCACTCAGCTTAGCC (forward) and TCCTTTTGTCGTTGGAGATGAC (reverse); CoL2A1, CCAGATGACCTTCCTACGCC (forward) and TTCAGGGCAGTGTACGTGAAC (reverse); Hprt, ATGGACAGGACTGAACGTCTT (forward) and TCCAGCAGGTCAGCAAAGAA (reverse).
Statistical analysis
The following statistical tests were used to assess significance and derive p values; for each test, α = 0.05%: one-way ANOVA with post hoc Dunnett's multiple-comparison test, unpaired two-tailed t test, and multiple t tests (one unpaired t test per row) by using GraphPad Prism version 7.0d.
Author contributions
G. R. D. conceptualized and designed the research project. S. T. J. B., E. J. R., E. G., and G. R. D. performed experiments. S. T. J. B., P. H. L., and G. R. D. analyzed and interpreted data. S. T. J. B. and G. R. D. drafted and edited original manuscript and prepared figures.
Supplementary Material
Acknowledgments
We are grateful to the Center for Chemical Genomics at the University of Michigan Life Science Institute for providing technical expertise and support. Specifically, we thank M. Larsen for technical assistance with the high-throughput screening campaign and triage of active compounds as well as A. White (Vahlteich Medicinal Chemistry Core) for also assisting with active compound triage and selection. We also thank the University of Michigan DNA Sequencing Core for assistance performing the Affymetrix® microarray gene expression analysis and C. Johnson for performing the statistical analysis of Affymetrix® GeneChip data. We are also thankful for helpful discussions and advice from A. Soofi, A. Higashi, S. Patel, and Z. Nikolovska-Coleska.
This work was supported by NIDDK, National Institutes of Health, Grants 5R01DK054740-16 and 3R01DK054740-16S1. Funding was also provided by the University of Michigan Center for the Discovery of New Medicines directed by V. Groppi. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
- BMP
- bone morphogenetic protein
- R-SMAD
- receptor-activated SMAD
- co-SMAD4
- common SMAD-4
- HTS
- high-throughput screen
- p-
- phosphorylated
- PREC
- primary mouse kidney epithelial cell
- ERK
- extracellular signal–regulated kinase
- JNK
- c-Jun N-terminal kinase
- TGF-β
- transforming growth factor
- TAK-1
- TGF-β–activated kinase 1
- CDK
- cyclin-dependent kinase
- GSK3
- glycogen synthase kinase 3
- YAP
- Yes-associated protein
- HRP
- horseradish peroxidase.
References
- 1. Luo G., Hofmann C., Bronckers A. L., Sohocki M., Bradley A., and Karsenty G. (1995) BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev. 9, 2808–2820 10.1101/gad.9.22.2808 [DOI] [PubMed] [Google Scholar]
- 2. Chen H., Shi S., Acosta L., Li W., Lu J., Bao S., Chen Z., Yang Z., Schneider M. D., Chien K. R., Conway S. J., Yoder M. C., Haneline L. S., Franco D., and Shou W. (2004) BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131, 2219–2231 10.1242/dev.01094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu B., Chen C., Chen H., Zheng S. G., Bringas P. Jr, Xu M., Zhou X., Chen D., Umans L., Zwijsen A., and Shi W. (2011) Smad1 and its target gene Wif1 coordinate BMP and Wnt signaling activities to regulate fetal lung development. Development 138, 925–935 10.1242/dev.062687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meyers E. A., and Kessler J. A. (2017) TGF-β family signaling in neural and neuronal differentiation, development, and function. Cold Spring Harb. Perspect Biol. 9, a022244 10.1101/cshperspect.a022244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang R. N., Green J., Wang Z., Deng Y., Qiao M., Peabody M., Zhang Q., Ye J., Yan Z., Denduluri S., Idowu O., Li M., Shen C., Hu A., Haydon R. C., et al. (2014) Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105 10.1016/j.gendis.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomez-Puerto M. C., Iyengar P. V., García de Vinuesa A., Ten Dijke P., and Sanchez-Duffhues G. (2019) Bone morphogenetic protein receptor signal transduction in human diseases. J. Pathol. 247, 9–20 10.1002/path.5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung Y. H., Huang Y. H., Chu T. H., Chen C. L., Lin P. R., Huang S. C., Wu D. C., Huang C. C., Hu T. H., Kao Y. H., and Tai M. H. (2018) BMP-2 restoration aids in recovery from liver fibrosis by attenuating TGF-β1 signaling. Lab. Invest. 98, 999–1013 10.1038/s41374-018-0069-9 [DOI] [PubMed] [Google Scholar]
- 8. Morrissey J., Hruska K., Guo G., Wang S., Chen Q., and Klahr S. (2002) Bone morphogenetic protein-7 improves renal fibrosis and accelerates the return of renal function. J. Am. Soc. Nephrol. 13, S14–S21 [PubMed] [Google Scholar]
- 9. Zeisberg M., Bottiglio C., Kumar N., Maeshima Y., Strutz F., Müller G. A., and Kalluri R. (2003) Bone morphogenic protein-7 inhibits progression of chronic renal fibrosis associated with two genetic mouse models. Am. J. Physiol. Renal Physiol. 285, F1060–F1067 10.1152/ajprenal.00191.2002 [DOI] [PubMed] [Google Scholar]
- 10. Zeisberg E. M., Tarnavski O., Zeisberg M., Dorfman A. L., McMullen J. R., Gustafsson E., Chandraker A., Yuan X., Pu W. T., Roberts A. B., Neilson E. G., Sayegh M. H., Izumo S., and Kalluri R. (2007) Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 13, 952–961 10.1038/nm1613 [DOI] [PubMed] [Google Scholar]
- 11. Myllärniemi M., Lindholm P., Ryynänen M. J., Kliment C. R., Salmenkivi K., Keski-Oja J., Kinnula V. L., Oury T. D., and Koli K. (2008) Gremlin-mediated decrease in bone morphogenetic protein signaling promotes pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 177, 321–329 10.1164/rccm.200706-945OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spiekerkoetter E., Tian X., Cai J., Hopper R. K., Sudheendra D., Li C. G., El-Bizri N., Sawada H., Haghighat R., Chan R., Haghighat L., de Jesus Perez V., Wang L., Reddy S., Zhao M., et al. (2013) FK506 activates BMPR2, rescues endothelial dysfunction, and reverses pulmonary hypertension. J. Clin. Invest. 123, 3600–3613 10.1172/JCI65592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Long L., Ormiston M. L., Yang X., Southwood M., Gräf S., Machado R. D., Mueller M., Kinzel B., Yung L. M., Wilkinson J. M., Moore S. D., Drake K. M., Aldred M. A., Yu P. B., Upton P. D., and Morrell N. W. (2015) Selective enhancement of endothelial BMPR-II with BMP9 reverses pulmonary arterial hypertension. Nat. Med. 21, 777–785 10.1038/nm.3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taylor K. R., Vinci M., Bullock A. N., and Jones C. (2014) ACVR1 mutations in DIPG: lessons learned from FOP. Cancer Res. 74, 4565–4570 10.1158/0008-5472.CAN-14-1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manson S. R., Austin P. F., Guo Q., and Moore K. H. (2015) BMP-7 signaling and its critical roles in kidney development, the responses to renal injury, and chronic kidney disease. Vitam. Horm. 99, 91–144 10.1016/bs.vh.2015.05.003 [DOI] [PubMed] [Google Scholar]
- 16. Oxburgh L., Dudley A. T., Godin R. E., Koonce C. H., Islam A., Anderson D. C., Bikoff E. K., and Robertson E. J. (2005) BMP4 substitutes for loss of BMP7 during kidney development. Dev. Biol. 286, 637–646 10.1016/j.ydbio.2005.08.024 [DOI] [PubMed] [Google Scholar]
- 17. Salazar V. S., Gamer L. W., and Rosen V. (2016) BMP signalling in skeletal development, disease and repair. Nat. Rev. Endocrinol 12, 203–221 10.1038/nrendo.2016.12 [DOI] [PubMed] [Google Scholar]
- 18. Hollnagel A., Oehlmann V., Heymer J., Rüther U., and Nordheim A. (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem. 274, 19838–19845 10.1074/jbc.274.28.19838 [DOI] [PubMed] [Google Scholar]
- 19. Miyazono K., and Miyazawa K. (2002) Id: a target of BMP signaling. Sci. STKE 2002, pe40 [DOI] [PubMed] [Google Scholar]
- 20. Piccolo S., Sasai Y., Lu B., and De Robertis E. M. (1996) Dorsoventral patterning in Xenopus: inhibition of ventral signals by direct binding of chordin to BMP-4. Cell 86, 589–598 10.1016/S0092-8674(00)80132-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zimmerman L. B., De Jesús-Escobar J. M., and Harland R. M. (1996) The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86, 599–606 10.1016/S0092-8674(00)80133-6 [DOI] [PubMed] [Google Scholar]
- 22. Lin J., Patel S. R., Cheng X., Cho E. A., Levitan I., Ullenbruch M., Phan S. H., Park J. M., and Dressler G. R. (2005) Kielin/chordin-like protein, a novel enhancer of BMP signaling, attenuates renal fibrotic disease. Nat. Med. 11, 387–393 10.1038/nm1217 [DOI] [PubMed] [Google Scholar]
- 23. Soofi A., Zhang P., and Dressler G. R. (2013) Kielin/chordin-like protein attenuates both acute and chronic renal injury. J. Am. Soc. Nephrol. 24, 897–905 10.1681/ASN.2012070759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soofi A., Wolf K. I., Ranghini E. J., Amin M. A., and Dressler G. R. (2016) The kielin/chordin-like protein KCP attenuates nonalcoholic fatty liver disease in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G587–G598 10.1152/ajpgi.00165.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soofi A., Wolf K. I., Emont M. P., Qi N., Martinez-Santibanez G., Grimley E., Ostwani W., and Dressler G. R. (2017) The kielin/chordin-like protein (KCP) attenuates high-fat diet-induced obesity and metabolic syndrome in mice. J. Biol. Chem. 292, 9051–9062 10.1074/jbc.M116.771428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okada M., Sangadala S., Liu Y., Yoshida M., Reddy B. V., Titus L., and Boden S. D. (2009) Development and optimization of a cell-based assay for the selection of synthetic compounds that potentiate bone morphogenetic protein-2 activity. Cell Biochem. Funct. 27, 526–534 10.1002/cbf.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kato S., Sangadala S., Tomita K., Titus L., and Boden S. D. (2011) A synthetic compound that potentiates bone morphogenetic protein-2-induced transdifferentiation of myoblasts into the osteoblastic phenotype. Mol. Cell Biochem. 349, 97–106 10.1007/s11010-010-0664-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao Y., Wang C., Zhang X., Xing G., Lu K., Gu Y., He F., and Zhang L. (2014) Selective small molecule compounds increase BMP-2 responsiveness by inhibiting Smurf1-mediated Smad1/5 degradation. Sci. Rep. 4, 4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baek S. H., Choi S. W., Park S. J., Lee S. H., Chun H. S., and Kim S. H. (2015) Quinoline compound KM11073 enhances BMP-2-dependent osteogenic differentiation of C2C12 cells via activation of p38 signaling and exhibits in vivo bone forming activity. PLoS One 10, e0120150 10.1371/journal.pone.0120150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Genthe J. R., Min J., Farmer D. M., Shelat A. A., Grenet J. A., Lin W., Finkelstein D., Vrijens K., Chen T., Guy R. K., Clements W. K., and Roussel M. F. (2017) Ventromorphins: a new class of small molecule activators of the canonical BMP signaling pathway. ACS Chem. Biol. 12, 2436–2447 10.1021/acschembio.7b00527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vrijens K., Lin W., Cui J., Farmer D., Low J., Pronier E., Zeng F. Y., Shelat A. A., Guy K., Taylor M. R., Chen T., and Roussel M. F. (2013) Identification of small molecule activators of BMP signaling. PLoS One 8, e59045 10.1371/journal.pone.0059045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feng L., Cook B., Tsai S. Y., Zhou T., LaFlamme B., Evans T., and Chen S. (2016) Discovery of a small-molecule BMP sensitizer for human embryonic stem cell differentiation. Cell Rep. 15, 2063–2075 10.1016/j.celrep.2016.04.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee K. W., Yook J. Y., Son M. Y., Kim M. J., Koo D. B., Han Y. M., and Cho Y. S. (2010) Rapamycin promotes the osteoblastic differentiation of human embryonic stem cells by blocking the mTOR pathway and stimulating the BMP/Smad pathway. Stem Cells Dev. 19, 557–568 10.1089/scd.2009.0147 [DOI] [PubMed] [Google Scholar]
- 34. Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., and Peterson R. T. (2008) Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33–41 10.1038/nchembio.2007.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hao J., Ho J. N., Lewis J. A., Karim K. A., Daniels R. N., Gentry P. R., Hopkins C. R., Lindsley C. W., and Hong C. C. (2010) In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem. Biol. 5, 245–253 10.1021/cb9002865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cuny G. D., Yu P. B., Laha J. K., Xing X., Liu J. F., Lai C. S., Deng D. Y., Sachidanandan C., Bloch K. D., and Peterson R. T. (2008) Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg. Med. Chem. Lett. 18, 4388–4392 10.1016/j.bmcl.2008.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Williams E., and Bullock A. N. (2018) Structural basis for the potent and selective binding of LDN-212854 to the BMP receptor kinase ALK2. Bone 109, 251–258 10.1016/j.bone.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Korchynskyi O., and ten Dijke P. (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277, 4883–4891 10.1074/jbc.M111023200 [DOI] [PubMed] [Google Scholar]
- 39. Baell J. B., and Holloway G. A. (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 10.1021/jm901137j [DOI] [PubMed] [Google Scholar]
- 40. Hill A. P., and Young R. J. (2010) Getting physical in drug discovery: a contemporary perspective on solubility and hydrophobicity. Drug. Discov. Today 15, 648–655 10.1016/j.drudis.2010.05.016 [DOI] [PubMed] [Google Scholar]
- 41. Jacob R. T., Larsen M. J., Larsen S. D., Kirchhoff P. D., Sherman D. H., and Neubig R. R. (2012) MScreen: an integrated compound management and high-throughput screening data storage and analysis system. J. Biomol. Screen. 17, 1080–1087 10.1177/1087057112450186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grimley E., Liao C., Ranghini E. J., Nikolovska-Coleska Z., and Dressler G. R. (2017) Inhibition of Pax2 transcription activation with a small molecule that targets the DNA binding domain. ACS Chem. Biol. 12, 724–734 10.1021/acschembio.6b00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu P. B., Deng D. Y., Lai C. S., Hong C. C., Cuny G. D., Bouxsein M. L., Hong D. W., McManus P. M., Katagiri T., Sachidanandan C., Kamiya N., Fukuda T., Mishina Y., Peterson R. T., and Bloch K. D. (2008) BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 14, 1363–1369 10.1038/nm.1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rudnicki M., Eder S., Perco P., Enrich J., Scheiber K., Koppelstätter C., Schratzberger G., Mayer B., Oberbauer R., Meyer T. W., and Mayer G. (2007) Gene expression profiles of human proximal tubular epithelial cells in proteinuric nephropathies. Kidney Int. 71, 325–335 10.1038/sj.ki.5002043 [DOI] [PubMed] [Google Scholar]
- 45. Mezzano S., Droguett A., Burgos M. E., Aros C., Ardiles L., Flores C., Carpio D., Carvajal G., Ruiz-Ortega M., and Egido J. (2007) Expression of gremlin, a bone morphogenetic protein antagonist, in glomerular crescents of pauci-immune glomerulonephritis. Nephrol. Dial. Transplant. 22, 1882–1890 10.1093/ndt/gfm145 [DOI] [PubMed] [Google Scholar]
- 46. Dolan V., Murphy M., Sadlier D., Lappin D., Doran P., Godson C., Martin F., O'Meara Y., Schmid H., Henger A., Kretzler M., Droguett A., Mezzano S., and Brady H. R. (2005) Expression of gremlin, a bone morphogenetic protein antagonist, in human diabetic nephropathy. Am. J. Kidney Dis. 45, 1034–1039 10.1053/j.ajkd.2005.03.014 [DOI] [PubMed] [Google Scholar]
- 47. Turk T., Leeuwis J. W., Gray J., Torti S. V., Lyons K. M., Nguyen T. Q., and Goldschmeding R. (2009) BMP signaling and podocyte markers are decreased in human diabetic nephropathy in association with CTGF overexpression. J. Histochem. Cytochem. 57, 623–631 10.1369/jhc.2009.953224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeisberg M., and Kalluri R. (2008) Reversal of experimental renal fibrosis by BMP7 provides insights into novel therapeutic strategies for chronic kidney disease. Pediatr. Nephrol. 23, 1395–1398 10.1007/s00467-008-0818-x [DOI] [PubMed] [Google Scholar]
- 49. Bosukonda D., Shih M. S., Sampath K. T., and Vukicevic S. (2000) Characterization of receptors for osteogenic protein-1/bone morphogenetic protein-7 (OP-1/BMP-7) in rat kidneys. Kidney Int. 58, 1902–1911 10.1111/j.1523-1755.2000.00362.x [DOI] [PubMed] [Google Scholar]
- 50. Carragee E. J., Hurwitz E. L., and Weiner B. K. (2011) A critical review of recombinant human bone morphogenetic protein-2 trials in spinal surgery: emerging safety concerns and lessons learned. Spine J. 11, 471–491 10.1016/j.spinee.2011.04.023 [DOI] [PubMed] [Google Scholar]
- 51. Muchow R. D., Hsu W. K., and Anderson P. A. (2010) Histopathologic inflammatory response induced by recombinant bone morphogenetic protein-2 causing radiculopathy after transforaminal lumbar interbody fusion. Spine J. 10, e1–e6 10.1016/j.spinee.2010.06.020 [DOI] [PubMed] [Google Scholar]
- 52. Chen N. F., Smith Z. A., Stiner E., Armin S., Sheikh H., and Khoo L. T. (2010) Symptomatic ectopic bone formation after off-label use of recombinant human bone morphogenetic protein-2 in transforaminal lumbar interbody fusion Report of 4 cases. J. Neurosurg. Spine 12, 40–46 [DOI] [PubMed] [Google Scholar]
- 53. Aragón E., Goerner N., Zaromytidou A. I., Xi Q., Escobedo A., Massagué J., and Macias M. J. (2011) A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 25, 1275–1288 10.1101/gad.2060811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Knockaert M., Sapkota G., Alarcón C., Massagué J., and Brivanlou A. H. (2006) Unique players in the BMP pathway: small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 11940–11945 10.1073/pnas.0605133103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sapkota G., Knockaert M., Alarcón C., Montalvo E., Brivanlou A. H., and Massagué J. (2006) Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-β pathways. J. Biol. Chem. 281, 40412–40419 10.1074/jbc.M610172200 [DOI] [PubMed] [Google Scholar]
- 56. Zhu H., Kavsak P., Abdollah S., Wrana J. L., and Thomsen G. H. (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400, 687–693 10.1038/23293 [DOI] [PubMed] [Google Scholar]
- 57. Bruce D. L., and Sapkota G. P. (2012) Phosphatases in SMAD regulation. FEBS Lett. 586, 1897–1905 10.1016/j.febslet.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 58. Sapkota G., Alarcón C., Spagnoli F. M., Brivanlou A. H., and Massagué J. (2007) Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell 25, 441–454 10.1016/j.molcel.2007.01.006 [DOI] [PubMed] [Google Scholar]
- 59. Alarcón C., Zaromytidou A. I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A. N., Manova-Todorova K., Macias M. J., Sapkota G., Pan D., and Massagué J. (2009) Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139, 757–769 10.1016/j.cell.2009.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Himmelfarb J., Chertow G. M., McCullough P. A., Mesana T., Shaw A. D., Sundt T. M., Brown C., Cortville D., Dagenais F., de Varennes B., Fontes M., Rossert J., and Tardif J. C. (2018) Perioperative THR-184 and AKI after cardiac surgery. J. Am. Soc. Nephrol 29, 670–679 10.1681/ASN.2017020217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Voelker J., Berg P. H., Sheetz M., Duffin K., Shen T., Moser B., Greene T., Blumenthal S. S., Rychlik I., Yagil Y., Zaoui P., and Lewis J. B. (2017) Anti-TGF-β1 antibody therapy in patients with diabetic nephropathy. J. Am. Soc. Nephrol. 28, 953–962 10.1681/ASN.2015111230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Seth K., Garg S. K., Kumar R., Purohit P., Meena V. S., Goyal R., Banerjee U. C., and Chakraborti A. K. (2014) 2-(2-Arylphenyl)benzoxazole as a novel anti-inflammatory scaffold: synthesis and biological evaluation. ACS Med. Chem. Lett. 5, 512–516 10.1021/ml400500e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim M. J., An H. J., Kim D. H., Lee B., Lee H. J., Ullah S., Kim S. J., Jeong H. O., Moon K. M., Lee E. K., Yang J., Akter J., Chun P., Moon H. R., and Chung H. Y. (2018) Novel SIRT1 activator MHY2233 improves glucose tolerance and reduces hepatic lipid accumulation in db/db mice. Bioorg. Med. Chem. Lett. 28, 684–688 10.1016/j.bmcl.2018.01.021 [DOI] [PubMed] [Google Scholar]
- 64. Lipinski C. A., Lombardo F., Dominy B. W., and Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- 65. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., and Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 66. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 67. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 68. Wang X., Spandidos A., Wang H., and Seed B. (2012) PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40, D1144–D1149 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.