Abstract
The Ctp1 protein in Schizosaccharomyces pombe is essential for DNA double-strand break (DSB) repair by homologous recombination. Fission yeast Ctp1 and its budding yeast (Sae2) and human (CtIP) homologs control Mre11–Rad50–Nbs1 nuclease complex activity and harbor DNA-binding and -bridging activities. However, the molecular basis for Ctp1–DNA transactions remains undefined. Here, we report atomic force microscopy (AFM) imaging of S. pombe Ctp1–DNA complexes revealing that Ctp1 polymerizes on dsDNA molecules and forms synaptic filaments that bridge two dsDNA strands. We observed that Ctp1 DNA filaments are typified by an average filament length of ∼180 bp of dsDNA and a Ctp1 tetramer footprint of ∼15 bp. Biochemical results characterizing Ctp1 variants with impaired DNA-binding or -bridging properties were consistent with Ctp1-mediated DNA bridging requiring the intact and correctly folded Ctp1 tetramer. Furthermore, mutations altering Ctp1 oligomerization and DNA bridging in vitro conferred cell sensitivity to DSB-producing agents. Together, these results support an important role for Ctp1-regulated DNA strand coordination required for DNA DSB repair in S. pombe.
Keywords: DNA repair, homologous recombination, atomic force microscopy (AFM), Schizosaccharomyces pombe, structural biology, chromosome damage, CtIP, Ctp1, DNA bridging, Sae2, DNA-binding protein
Introduction
Cytotoxic DNA double-strand breaks (DSBs)4 are generated by exposure to exogenous clastogens, such as ionizing radiation and cancer chemotherapeutic topoisomerase 2 drugs, as well as during endogenous processes, including meiosis (1–3). The Ctp1 protein (Ctp1 in Schizosaccharomyces pombe, Sae2 in Saccharomyces cerevisiae, and CtIP in Homo sapiens) is an essential component of DNA DSB repair by homologous recombination (HR), where homologous DNA sequences, such as sister chromatids, are used to accurately repair the DSB. To initiate this repair, Ctp1 directly binds and regulates the Mre11–Rad50–Nbs1 (MRN) resection and nuclease complex (4–6). Mutations in either Ctp1 or MRN result in failed DSB processing and DNA damage sensitivity (7, 8), and deletion of CtIP in mice is embryonic lethal (9). Ctp1Sae2/CtIP directly interacts with MRN through a phosphorylation-dependent binding pocket on Nbs1 (4) and promotes the Mre11-endonuclease dependent removal of 5′-protein–DNA adducts (10–12), such as Rec12- and topoisomerase 2–DNA protein cross-links (13, 14). In addition, Ctp1 promotes resection of the DSB to produce ssDNA required for strand invasion of the homologous DNA template and subsequent ligation of the DSB (7, 15–17).
Although it remains unclear how DNA repair intermediates are coordinated during adduct removal and resection, the DNA-binding properties of Ctp1 suggest that it could play a role in DNA strand coordination (18–20). Specifically, S. pombe Ctp1 and its budding yeast counterpart Sae2 are oligomeric proteins that bind DNA and can bridge two dsDNA strands together (18, 21). As such, Ctp1 could protect DNA ends from nucleolytic degradation (22), facilitate association of the DSB with the sister chromatid for homologous repair (23), and/or ensure that coincident resection of DSBs occurs, a role ascribed to S. cerevisiae Sae2 (24). DNA-binding sites critical for DSB repair in S. pombe reside at the extreme termini of Ctp1. The conserved N terminus of Ctp1 forms a homotetramer composed of a helical domain that oligomerizes into a ∼120-Å-long parallel four-helical bundle called the tetrameric helical dimer of dimers (THDD), with DNA-binding sites located in the coiled-coil regions (18, 25). C-terminal to the THDD, Ctp1 contains a low complexity intrinsically disordered C-terminal region containing phosphorylated SXT motifs that tether Ctp1 to MRN through Nbs1 (4). Additional functionally critical DNA-binding interaction sites are found at the extreme C terminus in the conserved Sae2-like domain, which harbors a conserved RHR DNA-binding motif (18). In addition, alleles of the human CtIP homolog lacking the C-terminal DNA-binding domain are dominant-negative for Jawad–Seckel syndrome (26), suggesting that the DNA-binding properties of Ctp1Sae2/CtIP are important for maintaining genome stability. Despite the apparent importance of DNA binding by Ctp1, the molecular basis for Ctp1 DNA-binding and bridging activities remains undefined.
Here, we combine atomic force microscopy (AFM) with biochemical and in vivo studies to characterize WT and Ctp1 mutants that have impaired DNA-binding and/or -bridging properties. We report that Ctp1 polymerizes on dsDNA molecules, forming synaptic filaments that bridge two dsDNA strands, with extended filament lengths spanning ∼180 bp of DNA and an average Ctp1 tetramer footprint of ∼15 bp. Ctp1-mediated DNA bridging in vitro requires not only the previously reported DNA-binding motifs at N and C termini (18), but also the intact Ctp1 tetramer. Mutations altering Ctp1 oligomerization and DNA bridging in vitro also undermine Ctp1 DSB repair functions in S. pombe. Altogether, these results underline new functional roles for Ctp1 protein–DNA bridging filaments in the coordination of DNA repair intermediates during homologous recombination DSB repair, and they provide a mechanistic basis for the DNA repair defect that causes Jawad–Seckel syndrome in humans.
Results
Molecular basis for Ctp1 DNA binding and bridging
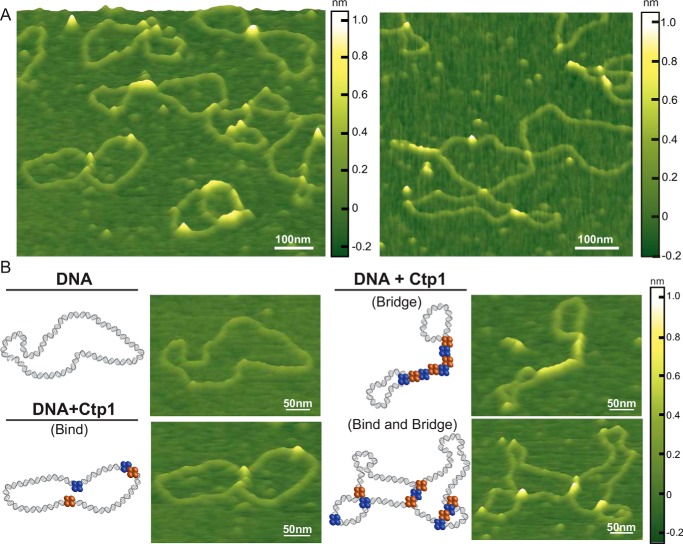
We employed AFM to capture images of unliganded Ctp1 and Ctp1–DNA complexes (Fig. 1, A and B). In the absence of DNA, WT Ctp1 molecules appear as short, diffuse peaks, coupled to regions of lower height, consistent with a predicted intrinsically disordered C-terminal region (Fig. S1A) (18, 27). The volume of proteins in AFM images depend linearly on their molecular weight (28–30); however, the low height of disordered regions of Ctp1 makes it difficult to determine an accurate volume of the proteins. Consequently, we cross-linked Ctp1 with glutaraldehyde prior to deposition, and the cross-linked Ctp1 exhibits taller, defined peaks (Fig. S1B). Based on a standard curve of AFM volume versus molecular weight, this volume corresponds to a molecular weight of 124 kDa (4 Ctp1 molecules) (28–30), which is consistent with Ctp1 (Mr ∼132 kDa) being predominantly tetrameric (Fig. S1C), consistent with multi-angle light scattering experiments (18).
Figure 1.
Ctp1 tetramers form bridging filaments on DNA. A, representative atomic force microscopy images of 800 nm Ctp1 with 1 nm pHOT1 relaxed plasmid DNA. White scale bar, 100 nm. B, selected examples of relaxed plasmid DNA, Ctp1–DNA binding, and Ctp1–DNA bridging complexes by AFM. Both intra- and intermolecular bridging events are shown. Blue and orange spheres, Ctp1 protomers involved in DNA interactions. White scale bar, 50 nm.
To define global properties of Ctp1–DNA interactions, we imaged Ctp1 bound to relaxed plasmid DNA. Given the decrease in background noise caused by the intrinsically disordered region of Ctp1 in the presence of glutaraldehyde, we attempted to image Ctp1–DNA complexes in the presence of glutaraldehyde; however, we did not observe any complexes. Further analysis of the Ctp1–DNA interaction by electrophoretic mobility shift assays (EMSAs) demonstrated that glutaraldehyde impaired the interaction of Ctp1 with DNA (data not shown). As a result, we conducted AFM imaging of the Ctp1–DNA interactions without chemical cross-linkers. In the absence of Ctp1, the DNA appears as an open circular structure with no high features, typical of relaxed plasmid DNA (31, 32). In contrast, in the presence of Ctp1, we observe tracts of protein bridging two dsDNA molecules together (DNA bridging; 60% Ctp1–DNA complexes) as well as tracts binding along a single dsDNA (DNA binding) (Fig. 1, A and B), similar to that seen in AFM images of H-NS–DNA and MutL-α–DNA complexes (31, 32). Most bridging events (75%) are intramolecular (“zippered” plasmids), but intermolecular plasmid-to-plasmid bridging events are also observed (Fig. S2A).
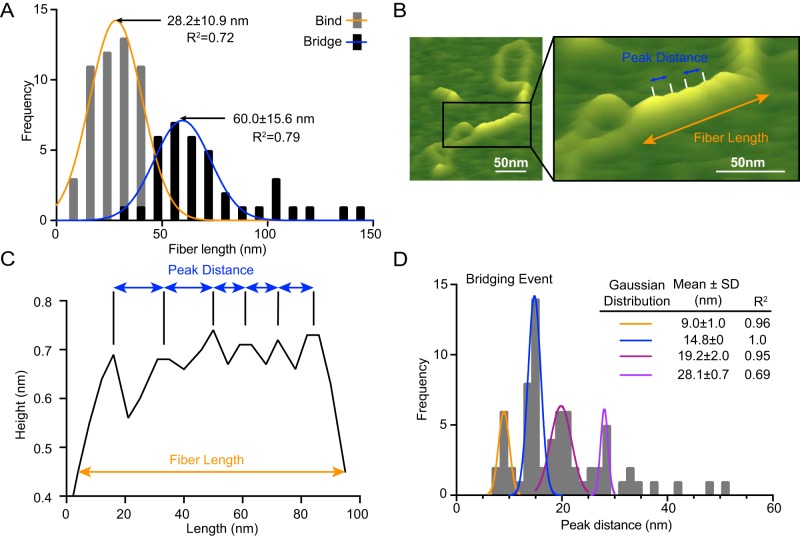
Analysis of the distribution of Ctp1 polymer lengths on DNA reveals that the average length of the Ctp1 protein–DNA filament tracts for the DNA-bridging events is approximately twice as long as that of the DNA-binding events (Fig. 2A). This result indicates that DNA bridging by Ctp1 promotes its cooperative assembly to “zipper up” two dsDNA chains, resulting in formation of elongated protein–DNA filaments. Notably, although the tracts along one dsDNA show no regular repeating features, the Ctp1–DNA bridging filaments show recurrent striations (Fig. 2, B and C). Quantitative analysis of the interpeak distances within these DNA-bridging complexes show quantized distributions of ∼10, 15, 20, and 30 nm (Fig. 2D). Given that AFM volume analysis shows that free Ctp1 exists primarily as a tetramer (Fig. S1C), these results suggest that the 5-nm periodicity represents individual tetramers of Ctp1. Interestingly, the volume/length ratio for Ctp1–DNA tracts on one dsDNA is identical to that for bridging tracts (Fig. S2B), suggesting that Ctp1 also has a 5-nm footprint when binding one dsDNA molecule. Furthermore, these results suggest that Ctp1 binding to one dsDNA may provide a nucleation site for bridging. Consistent with this idea, we also observe a greater percentage of Ctp1-bridging events relative to binding to one dsDNA throughout the AFM images (Fig. S2A). Ctp1 bridging does not significantly change the DNA contour length (Fig. S2, C and D), indicating that the Ctp1–DNA interactions do not induce DNA compression or wrapping.
Figure 2.
Analysis of Ctp1–DNA interactions by AFM. A, frequency distribution of fiber lengths from both bridging and binding events. Data are represented as mean ± S.D. calculated from nonlinear Gaussian fit. n = 37 bridging events and 53 binding events. B, representative illustration highlighting measurements analyzed in AFM images. The AFM image is the same as in Fig. 1B but is enlarged and labeled to illustrate how fiber length and peak distance metrics were assessed for calculations documented in A and C. Black box, enlarged region. C, representative section trace from our custom analysis software ImageMetrics, illustrating peak distance and fiber length measurements. D, frequency distribution of peak distances in bridging events. Data are represented as mean ± S.D. calculated from nonlinear Gaussian fit. n = 37 bridging events.
Ctp1 oligomerization and N-terminal domain folding facilitates DNA bridging
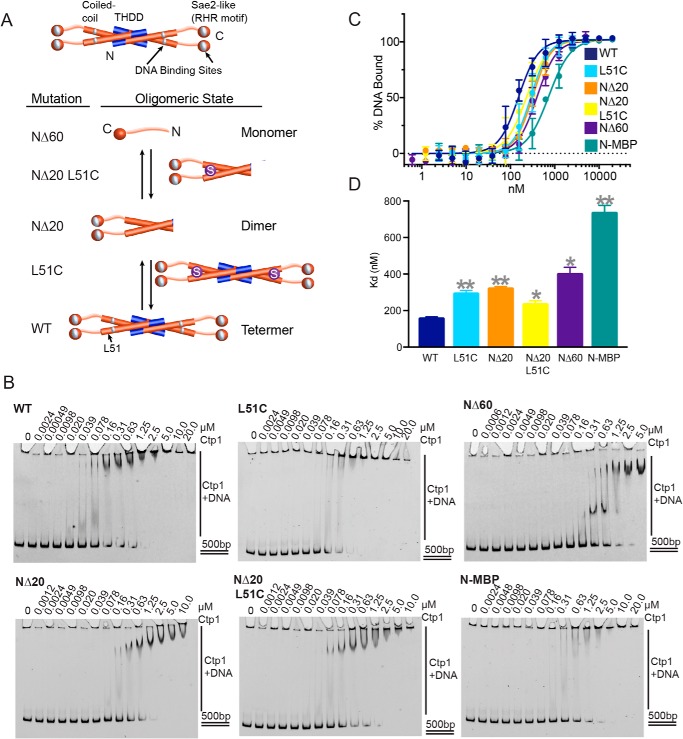
Based on our previous work, DNA binding and bridging requires conserved residues both in the THDD domain and C-terminal Sae2-like domain bearing an RHR motif of Ctp1 (18) (Fig. 3A); however, the precise roles of Ctp1 oligomerization in DNA-binding and -bridging activities are unclear. Therefore, to probe the roles of Ctp1 oligomerization on Ctp1 DNA binding and function, we engineered a series of THDD variants that alter Ctp1 oligomerization. Two N-terminal deletions convert the native Ctp1 tetramer into dimeric (Ctp1-NΔ20) or monomeric forms (Ctp1-NΔ60), as assessed by analytical gel-filtration chromatography (Fig. S3, A and B). Based on the crystal structure of the Ctp1 helical bundle (18), we further engineered a disulfide bond to replace an “a” heptad position of the Ctp1 leucine zipper repeat (Ctp1 variant L51C). This strategy covalently cross-links full-length tetrameric (L51C) and dimeric proteins (NΔ20-L51C) with a stable disulfide (Fig. 3A), visible as dimeric forms on an SDS-polyacrylamide gel even under denaturing and reducing conditions (Fig. S3A). An additional THDD variant, containing an N-terminal maltose-binding protein (MBP) fusion with a fixed-arm linker that does not inhibit tetramerization (18) but potentially blocks DNA access to the THDD, was used to assess the overall necessity of the THDD for Ctp1–DNA interactions. All tetrameric and dimeric forms of Ctp1 retain both the N-terminal and C-terminal DNA-binding motifs.
Figure 3.
Altering Ctp1 tetramerization decreases affinity for DNA binding in vitro. A, Ctp1 oligomerization mutants constructed by N-terminal deletions and point mutations. Blue, N-terminal 20 amino acids; orange cylinder, N-terminal 60 amino acids; purple sphere with S, L51C mutation. N-terminal domain length and protein length in solution are indicated (18). B, representative images of electrophoretic mobility shift assays of Ctp1 and Ctp1 oligomerization mutants with 500-bp dsDNA to calculate DNA-binding affinity. n = 3 independent experiments. C, Ctp1–DNA binding activity. n = 3 independent experiments; error bars, S.D. D, Ctp1 DNA-binding dissociation constants on a 500-bp dsDNA substrate, calculated from n = 3 independent experiments in Fig. 3C. Error bars, S.E. **, p < 0.01; *, p < 0.05 (two-tailed t test).
We evaluated the DNA-binding activity of the Ctp1 oligomerization variants by EMSAs (Fig. 3, B–D). WT Ctp1 shows a Kd = 150 ± 12 nm (Hill coefficient (h) = 2.0 ± 0.3) for a 500-bp duplex DNA substrate. By comparison, dimeric (NΔ20, Kd = 320 ± 13 nm, h = 2.1 ± 0.2) and monomeric forms (NΔ60, Kd = 400 ± 39 nm, h = 2.0 ± 0.3) bind ∼2- and ∼2.7-fold less tightly. Cross-linking of the coiled-coil via cysteines (L51C and NΔ20 L51C) also reduces DNA-binding affinity ∼1.5–2-fold (Kd = 290 ± 19 nm, h = 2.0 ± 0.3 and 230 ± 22 nm, h = 1.8 ± 0.3) compared with WT Ctp1, indicating that the leucine zipper conformation is important for native DNA binding. Furthermore, the fusion of the bulky MBP protein to the Ctp1 N terminus (N-MBP) causes a ∼5-fold decrease in affinity of Ctp1 on duplex DNA (Kd = 730 ± 44 nm, h = 1.8 ± 0.2) (Fig. 3D). As this MBP fusion construct is tetrameric (18), the globular MBP protein may sterically block the THDD DNA-binding site rather than impair oligomerization. Taken together, these results demonstrate that access to the Ctp1 THDD and THDD oligomerization are important for stable DNA binding, whereas covalent cross-links that restrict the conformational flexibility of the Ctp1 N-terminal coiled-coil region modulate DNA interactions.
In our EMSA binding experiments, we reproducibly observe progressive supershifting of DNA–protein complexes with increasing addition of Ctp1, consistent with multiple Ctp1 proteins binding or bridging DNA and/or multiple DNA-binding sites in the Ctp1 tetramer mediating tethering of DNA molecules (18) (Fig. 3A). To examine the basis for multisite DNA binding, we evaluated the influence of Ctp1 oligomerization on its DNA-bridging activity with an in vitro DNA affinity pulldown assay (Fig. 4A) (18). Robust DNA bridging was observed for full-length Ctp1, but DNA-bridging activity was severely impaired for the NΔ20-L51C, NΔ60, and N-MBP variants. Unexpectedly, we found that although the NΔ20 construct contains both N- and C-terminal DNA-binding sites and thus retains residual DNA-bridging activity, the disulfide-bonded form (NΔ20-L51C) of this deletion that displays moderate DNA-binding activity (Fig. 4, B and C) has no detectable bridging activity (Fig. 4, C and D).
Figure 4.
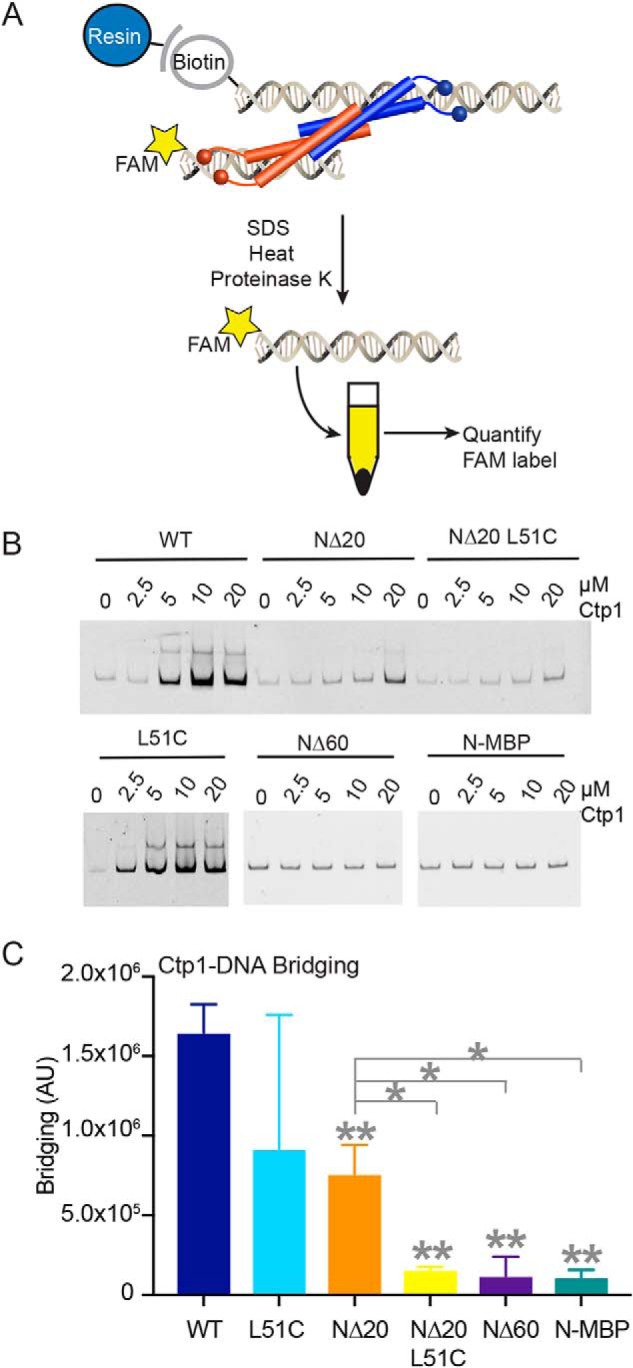
Altering Ctp1 tetramerization impacts DNA bridging in vitro. A, illustration of DNA-bridging assay. B, representative gels of bridging activity for Ctp1 and Ctp1 oligomerization mutants used to calculate bridging activity. C, quantification of DNA-bridging assays. n = 3 independent experiments; error bars, S.D. **, p < 0.01; *, p < 0.05 (two-tailed t test). p values comparing WT and mutants are denoted on the tops of columns. p values comparing NΔ20 and other mutants are indicated by gray lines.
AFM imaging of mutant Ctp1 proteins in the presence of DNA was consistent with DNA-binding and -bridging in vitro assays (Fig. S4, A and B). NΔ20-L51C binds circular plasmid DNA (Fig. S4A) but did not display filamentous bridging architectures typical of the WT protein. By comparison, NΔ60 did not appreciably interact with plasmid DNA under the conditions of AFM imaging (Fig. S4B), consistent with the decreased DNA-binding affinity of this mutant. Analysis of peak volumes in the AFM images indicates that Ctp1 NΔ20-L51C and NΔ60 have an approximate molecular weight of 78 and 34 kDa, consistent with primarily dimeric (NΔ20-L51C, calculated molecular weight = 61 kDa) and monomeric species (NΔ60, calculated molecular weight = 26 kDa), under AFM experimental conditions (Fig. S4, C and D). Together, these results indicate that both oligomerization and mobility of the Ctp1 N-terminal coiled-coiled domain are critical for efficient DNA bridging. Moreover, the NΔ20-L51C mutant has unique characteristics in that it retains moderate DNA binding (Fig. 3D) but ablates DNA bridging (Fig. 4C) in vitro.
DNA-bridging mutants compromise Ctp1 genome protective functions
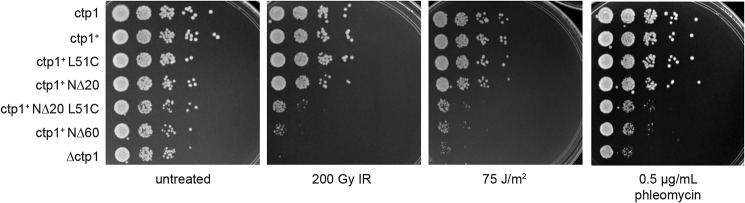
To probe the biological importance of DNA binding and bridging in DSB repair, we examined the effects of Ctp1 DNA-binding and -bridging mutations on clastogen sensitivity of S. pombe. Immunoblotting of the C-terminal 5x-FLAG epitope tag in ctp1+ strains showed that mutant proteins were present at similar levels to WT (Fig. S5). The ctp1+ L51C and ctp1+ NΔ20 display growth phenotypes after DNA damage comparable with WT. In contrast, the NΔ20-L51C mutation exhibits acute sensitivity to all DNA-damaging agents evaluated (Fig. 5 and Fig. S5). Given that the NΔ20-L51C mutant binds DNA (Fig. 3D) but does not efficiently bridge DNA (Fig. 4C), these data suggest that the ability of Ctp1 to form oligomeric tracts that bridge two dsDNAs together plays an important role in DSB repair.
Figure 5.
Altering Ctp1 tetramerization affects DNA repair in vivo. Ctp1 oligomerization mutants displayed sensitivity to acute ionizing radiation, acute UV irradiation, and chronic phleomycin treatment. Shown are representative images from n = 3 technical replicates.
Discussion
Ctp1 is a critical component of the DNA DSB repair machinery, engaging with the MRN complex to initiate resection of the DNA DSB (4–6); however, the mechanism of Ctp1 action in this role is poorly defined. For efficient repair to occur, Ctp1 must form oligomers capable of efficient DNA-binding and -bridging activity (Fig. 6) (18). Based on our AFM data (Figs. 1 and 2), this bridging action occurs through the formation of protein–DNA filaments. At the gross structural level defined by AFM, the Ctp1 filaments reported herein are reminiscent of bacterial nucleoid-associated protein H-NS and the mismatch repair protein Mlh1-Pms1 from yeast, which form bridging filaments (31, 32). Protein–DNA filaments are an architectural feature frequently found to regulate genomic structure and repair (1, 2). In DSB repair, HR enlists multiple RPA and Rad51 molecules to bind ssDNA and dsDNA, creating presynaptic filaments in preparation for homologous pairing (33). In nonhomologous end-joining, the XRCC4-XLF filament bridges two DNA molecules independent of DNA ends to maintain DNA DSBs in proximity for repair (34). Ctp1 may play an analogous role in HR and microhomology-mediated end-joining (MMEJ) repair, because in humans, HR and MMEJ employ CtIP tetramers and dimers, respectively (25). Our studies show that both dimeric and tetrameric Ctp1 are capable of DNA bridging (Fig. 4C) and that mutations altering DNA bridging exhibit marked DNA damage sensitivity (Fig. 5), implicating DNA bridging as an important function of Ctp1.
Figure 6.
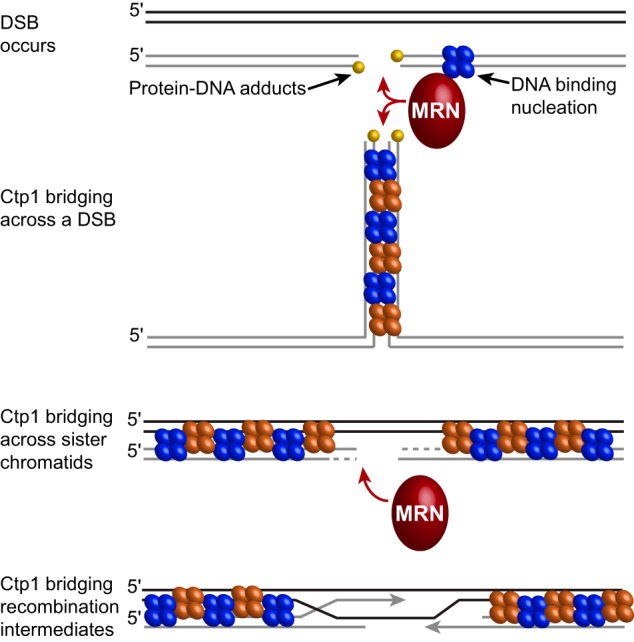
Possible roles for Ctp1 bridging in homologous recombination repair. When a DSB occurs, Ctp1 associates with MRN at the break site and may initiate nucleation. Ctp1 filaments can then form and may bridge across the DSB (intramolecular bridging), adjacent to the break site or may bridge between damaged DNA and the sister chromatid (intermolecular bridging), which would stabilize the repair complex and stimulate resection by Mre11. Ctp1 filaments may also bridge across sister chromatids to stabilize recombination intermediates.
Ctp1–DNA fibers typically extended to much longer tracts of regularly repeated peaks when Ctp1 interacted with two dsDNA molecules in a bridging event, as compared with a binding event (Fig. 2), suggesting a cooperative mechanism of binding. The shortest repeat of bridging filaments observed by AFM is 10 nm or ∼30 bp (10-nm length of DNA at 34 Å/10 bp for B-DNA). The 5-nm Ctp1–DNA footprint indicates that DNA-bridging events involve on average ∼12 Ctp1 tetramers, whereas DNA-binding events involve ∼6 Ctp1 tetramers (Fig. 2, A and D). In addition, the overall DNA length is unchanged by bridging (Fig. S2, C and D). Because the maximum particle dimension of free Ctp1 based on small-angle X-ray scattering analysis is 26 nm (Fig. S6A) (18), these observations imply that Ctp1 undergoes a conformational compaction during DNA binding. Structural compaction likely takes place within the flexible, intrinsically disordered C terminus of Ctp1, which could take on a variety of conformations when interacting with DNA (Fig. S6B). This change in structure of the Ctp1 C-terminal tails might facilitate a spatial search during DNA bridging. The Ctp1 N-terminal coiled-coil domain may also have important structural rearrangements upon DNA bridging, as locking the Ctp1 coiled-coil in place with a disulfide bond (L51C mutation) abolishes DNA bridging in Ctp1 NΔ20 (Fig. 4C). A critical role for the conserved C-terminal RHR motifs in the Sae2-like domain in mediating DNA binding (18, 25) suggests that this region is likely also important for filament formation. Understanding the precise nature of Ctp1–DNA interactions will require more detailed high-resolution structural analysis of Ctp1–DNA complexes by X-ray crystallography or cryo-EM.
Biologically, during coordination of the DNA repair intermediates in both HR and MMEJ, the short tracts of Ctp1 along a single dsDNA molecule may serve to nucleate the extended tracts of Ctp1 filaments through cooperative DNA binding (Fig. 6). Nucleation of Ctp1–DNA filaments may be controlled by MRN complex binding at the DSB (4). Ctp1 does not require free DNA ends for bridging (18), and DNA bridging occurs with relaxed circular plasmid DNA (Fig. 1). Ctp1 can furthermore mediate both intra- and intermolecular bridging (Fig. 1B and Fig. S2A). These observations provide possible explanations for results showing that the Ctp1 homolog Sae2 is required for in vivo intrachromosomal DNA bridging of HO-induced DSBs (21). Ctp1 filaments may bridge across sister chromatids to coordinate homologous templates or within recombination intermediates in HR (Fig. 6). Tethering DNA repair intermediates adjacent to the break site by Ctp1 filaments could protect DNA from unregulated nucleolytic degradation while also regulating MRN activity near the DSB ends to facilitate coincident (coordinated) DSB resection, as observed for Sae2 (24). The tethering of DNA by Ctp1 may also be important for restricting mobility of broken DNA ends during repair, effectively stitching DNA ends while repair proteins heal the break, and to possibly regulate subsequent strand invasion steps. That the MRN complex can also bridge DNA ends (7, 35–37) indicates that there are distinct functional requirements to coordinate DSB intermediates before, during, and after DNA end processing and that multiple strand coordination activities control the initiation of HR.
The precise roles of Ctp1–DNA filament architectures in regulating the removal of protein-blocked DNA termini by MRN remain to be determined. We posit that Ctp1 protein–DNA filament formation may limit an otherwise distributive MRN endonuclease activity to cut only near protein-blocked termini where filaments cannot extend. Ctp1, Sae2, and CtIP also exhibit structure-specific DNA end-binding properties (10, 12, 18, 19), so the specific MRN interaction with Ctp1/Sae2/CtIP on DNA ends may alternatively dictate site-specific cleavage near protein-blocked DNA ends. Future structural work investigating the complex interplay between Ctp1, DNA, and the MRN complex will be required to more clearly define the role of Ctp1-bridging filaments in regulating MRN activity.
Experimental procedures
Protein expression and purification
Ctp1 WT and NΔ60 were expressed and purified as described previously (18) with a final purification on a 16/60 S200 size-exclusion chromatography column (GE Healthcare) in 20 mm Tris, pH 7.5, 500 mm NaCl, and 0.1% β-mercaptoethanol. Ctp1 NΔ20 was expressed and purified as a His6-tagged fusion from pMCSG7 (39) identical to Ctp1 NΔ60. L51C point mutations were introduced with QuikChange mutagenesis (Agilent Technologies, Santa Clara, CA) into both Ctp1 WT and NΔ20. Point mutations were expressed and purified identical to their WT counterparts. MBP-Ctp1 was expressed and purified as described previously for MBP-Ctp1 fusions generated in the pMALX vector (18, 38).
Atomic force microscopy
Protein–DNA complexes were made by incubating 800 nm Ctp1 with 1 nm pHOT1-relaxed plasmid (TopoGEN Inc., Buena Vista, CA) for 30 min at room temperature in binding buffer (40 mm Hepes, pH 7, 5 mm MgCl2, 25 mm NaCl, 0.1 mm tris(2-carboxyethyl)phosphine) in a total volume of 20 μl. The entire reaction was deposited on freshly cleaved mica (Ted Pella, Inc., Redding, CA) for less than 1 min and then rinsed with 800 μl of molecular biology grade water. The mica surface was then blotted dry and fully dried by a nitrogen stream. Images were captured in air with a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA) microscope in tapping mode using PointProbe®Plus-NCL silicon probes (Nanosensors, Neuchatel, Switzerland). Images were collected at a size of 2 × 2 μm.
Image processing and analysis
Image Metrics version 1.13 (in-house designed MatLab (MathWorks Inc., Natick, MA) program) was used for image processing and analysis. Standard image processing utilized plane subtraction and flattening. Analysis included measuring DNA contour lengths and protein–DNA complex lengths, heights, and volumes. Bridging events were defined as protein interacting with two molecules of dsDNA, whereas binding events were defined as protein interacting with one molecule of dsDNA. Particle analysis to determine bridging versus binding events included protein–DNA complexes with a maximum height of ≥0.6 nm, as this is greater than 3 times the average DNA height of 0.20 ± 0.0008 nm determined by image analysis of DNA particles only; n values are indicated in the figure legends. Additional analysis for Fig. S2B used images that had defined interpeak distances as shown in Fig. 2, B and C. Statistical analyses were carried out using GraphPad Prism version 7.0 (GraphPad Software Inc., La Jolla, CA). Molecular weights of Ctp1–DNA complexes were derived from volumes as determined by ImageMetrics, using a published standard curve (28).
DNA-binding assays
500-bp dsDNA was generated by PCR using a phiX174 Virion DNA template (New England BioLabs Inc., Ipswich, MA) and primers 5′-FAM-AGTTTTATCGCTTCCATGAC-3′ (where FAM represents fluorescein amidite) and 5′-TCAGAAAATCGAAATCATCTTC-3′ (Integrated DNA Technologies, Coralville, IA). The 500-bp dsDNA was gel-purified using a QiaQuick gel extraction kit (Qiagen, Hilden, Germany). Ctp1 protein concentration was calculated based on a functional oligomeric unit of that mutant (i.e. tetramer, dimer, or monomer). Serial dilutions of Ctp1, diluted in 20 mm Tris, pH 7.5, 0.1 mm tris(2-carboxyethyl)phosphine, 50 mm potassium acetate, and 10% glycerol, were incubated with 15 nm DNA substrate, 5% glycerol, and 1× reaction buffer (20 mm Tris, pH 7.5, 250 mm potassium acetate, 0.1 mm DTT, 10 μg μl−1 BSA, 0.5% glycerol) for 20 min at 20 °C. Reactions were resolved on 4–20% Novex TBE gels (Invitrogen) run at 4 °C. Experiments (n = 3 for each Ctp1 protein) were imaged with a Typhoon FLA 9500 (GE Healthcare) and analyzed with ImageJ. Binding curves were calculated based on the amount of unbound DNA remaining compared with the DNA-only control. Dissociation constants were calculated using specific binding with Hill slope in GraphPad Prism version 7.0a (GraphPad Software Inc.).
DNA-bridging assays
DNA-bridging assays were carried out as described previously (18), with the following changes. The 500-bp dsDNA substrate used was that described in the DNA-binding assays. 10 μl of Streptavidin Sepharose High Performance resin (GE Healthcare) was used in place of magnetic beads, and beads were collected by 2-min spins at 1000 × g in a microcentrifuge. Reactions were resolved on 4–20% Novex TBE gels (Invitrogen). Experiments (n = 3 for each Ctp1 protein) were imaged and analyzed as described for the DNA-binding assay. Total intensity was determined as intensity in reactions of Ctp1 + DNA minus the intensity in DNA-only reactions. Statistical analyses (t test, two-tailed) were performed by GraphPad Prism version 7.0a (GraphPad Software Inc.).
Strain construction
Methods and growth media for S. pombe genetics followed standard protocols (39). All strains used in this study are listed in Table S1. Point mutations were inserted by QuikChange mutagenesis (Agilent Technologies). Strains were constructed as described previously (18), and sequencing verified the presence of correct truncations and point mutations.
Immunoblotting
Immunoblotting was performed as described previously (18), except Western blotting processing was carried out with the iBind system (Thermo Fisher Scientific) using fluorescent detection and probing first with a rabbit anti-FLAG antibody at 1:400 dilution (Sigma-Aldrich, F7425, lot 085M4774V; validation on the manufacturer's website), followed by probing with IRDye 680LT goat anti-rabbit IgG at 1:1000 dilution (LI-COR Biosciences (Lincoln, NE), 925-68021, lot C51007-05; validation on the manufacturer's website). Imaging was performed with a LI-COR Odyssey FC (LI-COR Biosciences). The membrane was then stripped before being reprobed with monoclonal anti-PSTAIR (mouse) antibody at 1:1000 dilution (Sigma-Aldrich, P7962; lot 010M4766; validation on the manufacturer's website and in Tournier et al. (40)) followed by probing with IRDye 800CW goat anti-mouse IgG at 1:1000 dilution (LI-COR Biosciences 925-32210, lot C60107-06; validation on the manufacturer's website).
Clastogen sensitivity assays
Assays were performed as described previously (18) and treated with clastogens as labeled in Fig. 6 and Fig. S4.
Author contributions
S. N. A. and R. S. W. conceptualization; S. N. A., Z. M. L., and D. A. E. formal analysis; S. N. A. investigation; S. N. A., Z. M. L., and D. A. E. methodology; S. N. A. writing-original draft; S. N. A., D. A. E., and R. S. W. writing-review and editing; D. A. E. and R. S. W. supervision; D. A. E. and R. S. W. funding acquisition; R. S. W. project administration.
Supplementary Material
Acknowledgments
We thank M. Resnick and M. Schellenberg for comments.
This work was supported by the intramural research program of the National Institutes of Health; NIEHS, National Institutes of Health, Grant 1Z01ES102765 (to R. S. W.); and NIGMS, National Institutes of Health, Grants GM080294 and GM109832 (to D. A. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Table S1 and Figs. S1–S6.
- DSB
- double-strand break
- HR
- homologous recombination
- MRN
- Mre11–Rad50–Nbs1
- ssDNA
- single-stranded DNA
- THDD
- tetrameric helical dimer of dimers
- AFM
- atomic force microscopy
- EMSA
- electrophoretic mobility shift assay
- MBP
- maltose-binding protein
- MMEJ
- microhomology-mediated end-joining.
References
- 1. Andres S. N., Schellenberg M. J., Wallace B. D., Tumbale P., and Williams R. S. (2015) Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutagen. 56, 1–21 10.1002/em.21892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciccia A., and Elledge S. J. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell. 40, 179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andres S. N., and Williams R. S. (2017) CtIP/Ctp1/Sae2, molecular form fit for function. DNA Repair (Amst.) 56, 109–117 10.1016/j.dnarep.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams R. S., Dodson G. E., Limbo O., Yamada Y., Williams J. S., Guenther G., Classen S., Glover J. N., Iwasaki H., Russell P., and Tainer J. A. (2009) Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139, 87–99 10.1016/j.cell.2009.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Limbo O., Chahwan C., Yamada Y., de Bruin R. A., Wittenberg C., and Russell P. (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 28, 134–146 10.1016/j.molcel.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., and Bartek J., Baer R., Lukas J., and Jackson S. P. (2007) Human CtIP promotes DNA end resection. Nature 450, 509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams R. S., Williams J. S., and Tainer J. A. (2007) Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520 10.1139/O07-069 [DOI] [PubMed] [Google Scholar]
- 8. Stracker T. H., and Petrini J. H. (2011) The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen P. L., Liu F., Cai S., Lin X., Li A., Chen Y., Gu B., Lee E. Y., and Lee W. H. (2005) Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25, 3535–3542 10.1128/MCB.25.9.3535-3542.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anand R., Ranjha L., Cannavo E., and Cejka P. (2016) Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 64, 940–950 10.1016/j.molcel.2016.10.017 [DOI] [PubMed] [Google Scholar]
- 11. Cannavo E., and Cejka P. (2014) Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature 514, 122–125 10.1038/nature13771 [DOI] [PubMed] [Google Scholar]
- 12. Wang W., Daley J. M., Kwon Y., Krasner D. S., and Sung P. (2017) Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 31, 2331–2336 10.1101/gad.307900.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartsuiker E., Neale M. J., and Carr A. M. (2009) Distinct requirements for the Rad32 Mre11 nuclease and Ctp1 CtIP in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell. 33, 117–123 10.1016/j.molcel.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rothenberg M., Kohli J., and Ludin K. (2009) Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 5, e1000722 10.1371/journal.pgen.1000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Langerak P., Mejia-Ramirez E., Limbo O., and Russell P. (2011) Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271 10.1371/journal.pgen.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mimitou E. P., and Symington L. S. (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou Y., Caron P., Legube G., and Paull T. T. (2014) Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 42, e19 10.1093/nar/gkt1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andres S. N., Appel C. D., Westmoreland J. W., Williams J. S., Nguyen Y., Robertson P. D., Resnick M. A., and Williams R. S. (2015) Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat. Struct. Mol. Biol. 22, 158–166 10.1038/nsmb.2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., and Paull T. T. (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell. 28, 638–651 10.1016/j.molcel.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. You Z., Shi L. Z., Zhu Q., Wu P., Zhang Y. W., Basilio A., Tonnu N., Verma I. M., Berns M. W., and Hunter T. (2009) CtIP links DNA double-strand break sensing to resection. Mol. Cell 36, 954–969 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clerici M., Mantiero D., Lucchini G., and Longhese M. P. (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 280, 38631–38638 10.1074/jbc.M508339200 [DOI] [PubMed] [Google Scholar]
- 22. Eid W., Steger M., El-Shemerly M., Ferretti L. P., Peña-Diaz J., König C., Valtorta E., Sartori A. A., and Ferrari S. (2010) DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 11, 962–968 10.1038/embor.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu M., Zhao H., Limbo O., and Russell P. (2018) Mre11 complex links sister chromatids to promote repair of a collapsed replication fork. Proc. Natl. Acad. Sci. U.S.A. 115, 8793–8798 10.1073/pnas.1808189115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Westmoreland J. W., and Resnick M. A. (2013) Coincident resection at both ends of random, γ-induced double-strand breaks requires MRX (MRN), Sae2 (Ctp1), and Mre11-nuclease. PLoS Genet. 9, e1003420 10.1371/journal.pgen.1003420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davies O. R., Forment J. V., Sun M., Belotserkovskaya R., Coates J., Galanty Y., Demir M., Morton C. R., Rzechorzek N. J., Jackson S. P., and Pellegrini L. (2015) CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 22, 150–157 10.1038/nsmb.2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qvist P., Huertas P., Jimeno S., Nyegaard M., Hassan M. J., Jackson S. P., and Børglum A. D. (2011) CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet. 7, e1002310 10.1371/journal.pgen.1002310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sacho E. J., Kadyrov F. A., Modrich P., Kunkel T. A., and Erie D. A. (2008) Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL α. Mol. Cell. 29, 112–121 10.1016/j.molcel.2007.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang H., Yang Y., and Erie D. (2006) Characterization of protein–protein interactions using atomic force microscopy. In Protein Interactions: Biophysical Approaches for the Study of Complex Reversible Systems (Schuck P., ed) pp. 39–77, Springer, New York [Google Scholar]
- 29. Ratcliff G. C., and Erie D. A. (2001) A novel single-molecule study to determine protein-protein association constants. J. Am. Chem. Soc. 123, 5632–5635 10.1021/ja005750n [DOI] [PubMed] [Google Scholar]
- 30. Yang Y., Wang H., and Erie D. A. (2003) Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods 29, 175–187 10.1016/S1046-2023(02)00308-0 [DOI] [PubMed] [Google Scholar]
- 31. Dame R. T., Wyman C., and Goosen N. (2000) H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 28, 3504–3510 10.1093/nar/28.18.3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hall M. C., Wang H., Erie D. A., and Kunkel T. A. (2001) High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J. Mol. Biol. 312, 637–647 10.1006/jmbi.2001.4958 [DOI] [PubMed] [Google Scholar]
- 33. Morrical S. W. (2015) DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol. 7, a016444 10.1101/cshperspect.a016444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brouwer I., Sitters G., Candelli A., Heerema S. J., Heller I., de Melo A. J., Zhang H., Normanno D., Modesti M., Peterman E. J. G., and Wuite G. J. L. (2016) Sliding sleeves of XRCC4–XLF bridge DNA and connect fragments of broken DNA. Nature 535, 566–569 10.1038/nature18643 [DOI] [PubMed] [Google Scholar]
- 35. Chen L., Trujillo K., Ramos W., Sung P., and Tomkinson A. E. (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell. 8, 1105–1115 10.1016/S1097-2765(01)00388-4 [DOI] [PubMed] [Google Scholar]
- 36. Williams R. S., Moncalian G., Williams J. S., Yamada Y., Limbo O., Shin D. S., Groocock L. M., Cahill D., Hitomi C., Guenther G., Moiani D., Carney J. P., Russell P., and Tainer J. A. (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135, 97–109 10.1016/j.cell.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., and Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 10.1007/978-1-59745-196-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moon A. F., Mueller G. A., Zhong X., and Pedersen L. C. (2010) A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 19, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moreno S., Klar A., and Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- 40. Tournier S., Gachet Y., and Hyams J. (1997) Identification and preliminary characterization of p31, a new PSTAIRE-related protein in fission yeast. Yeast 13, 727–734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.