Abstract
Regulated intramembrane proteolysis (RIP) is a key mechanism for activating transmembrane proteins such as epithelial cell adhesion molecule (EpCAM) for cellular signaling and degradation. EpCAM is highly expressed in carcinomas and progenitor and embryonic stem cells and is involved in the regulation of cell adhesion, proliferation, and differentiation. Strictly sequential cleavage of EpCAM through RIP involves initial shedding of the extracellular domain by α-secretase (ADAM) and β-secretase (BACE) sheddases, generating a membrane-tethered C-terminal fragment EpCTF. Subsequently, the rate-limiting γ-secretase complex catalyzes intramembrane cleavage of EpCTF, generating an extracellular EpCAM-Aβ–like fragment and an intracellular EpICD fragment involved in nuclear signaling. Here, we have combined biochemical approaches with live-cell imaging of fluorescent protein tags to investigate the kinetics of γ-secretase–mediated intramembrane cleavage of EpCTF. We demonstrate that γ-secretase–mediated proteolysis of exogenously and endogenously expressed EpCTF is a slow process with a 50% protein turnover in cells ranging from 45 min to 5.5 h. The slow cleavage was dictated by γ-secretase activity and not by EpCTF species, as indicated by cross-species swapping experiments. Furthermore, both human and murine EpICDs generated from EpCTF by γ-secretase were degraded efficiently (94–99%) by the proteasome. Hence, proteolytic cleavage of EpCTF is a comparably slow process, and EpICD generation does not appear to be suited for rapidly transducing extracellular cues into nuclear signaling, but appears to provide steady signals that can be further controlled through efficient proteasomal degradation. Our approach provides an unbiased bioassay to investigate proteolytic processing of EpCTF in single living cells.
Keywords: epithelial cell adhesion molecule (EpCAM), gamma-secretase, proteasome degradation, regulated intramembrane proteolysis, protein processing, live-cell imaging, sheddase, cell adhesion, proteolytic processing, transmembrane protein, cell signaling
Introduction
Regulated intramembrane proteolysis is an important means of post-translational regulation for a growing number of transmembrane proteins (1, 2). This regulation comprises the initiation of signaling by receptors such as the NOTCH family members (3) and the tumor-associated antigen epithelial cell adhesion molecule (EpCAM)5 (4, 5), as well as the removal and subsequent degradation of membrane proteins to dispose of their function (6). The latter has, for example, been shown for the major cell adhesion molecule E-cadherin and its function in migration and segregation processes in the mouse intestine (7).
Central enzymes involved in RIP have been identified and studied to unravel the underlying molecular mechanisms (8). RIP is a sequential process initiated by ectodomain shedding, a mandatory initial cleavage of the extracellular domain of substrates by α- and β-secretases and other proteases as the rate-limiting step (9). The resulting truncated protein termed the C-terminal fragment (CTF) is a substrate for the intramembrane-cleaving γ-secretase protease complex that is composed of the four subunits presenilin 1 or 2 (PS), nicastrin (NCT), presenilin enhancer (PEN-2), and anterior pharynx-defective 1 (APH-1). Release of ectodomains from RIP substrates is a prerequisite for NCT to bind CTFs and thus facilitate cleavage by PS, the enzymatically active component of the γ-secretase complex (10–14). Cleavage by the γ-secretase complex results in the formation of commonly small soluble extracellular domains and cytoplasmic intracellular domains (ICDs). Hence, the cleavage kinetics of RIP substrates is dictated by two major sequential events: first, an initial and mandatory generation of CTFs by sheddases through ectodomain shedding and, second, the generation of ICDs by the γ-secretase complex.
EpCAM is a single transmembrane protein that is subject to RIP in carcinoma, teratocarcinoma, and embryonic stem cells (5, 15, 16), where it is highly and frequently expressed (17, 18). EpCAM was initially shown to provide epithelial cells with a weak cell adhesion through homophilic interactions (19, 20). However, the postulated homophilic nature of EpCAM-mediated cell adhesion has been questioned recently and is a matter of debate (16, 21). An involvement of EpCAM in cancer cell proliferation and stem cell differentiation was revealed when it was shown to induce the proto-oncogene c-myc (22) and to be generally involved in regulation of genes associated with proliferation and differentiation (23). Similar to Notch, amyloid precursor protein, and other substrates, RIP of EpCAM generates a soluble ectodomain termed EpEX through the action of ADAM10/17 and BACE1 proteases, a soluble extracellular Aβ-like fragment identified via MS analysis, and intracellular EpICD fragments following cleavage by γ-secretase (15, 16). EpEX is a ligand for intact EpCAM molecules that induces their further cleavage (5), as well as a novel ligand for EGFR in colon and head and neck carcinomas, as was described recently (24, 25). As such, EpEX induces EGFR-dependent signaling pathways, including ERK1/2 and AKT, and induces a moderate proliferation of carcinoma cells, but restricts EGF/EGFR/pERK1/2-dependent regulation of epithelial–mesenchymal transition (EMT) (25).
EpICD represents the functional intracellular signaling fragment that plays a crucial role in cancer cell proliferation and in the maintenance of stem cell phenotype (4, 5, 26–28). Despite these important roles of EpCAM fragments in physiological and malignant conditions, the cleavage pace and efficiency of EpCTF by γ-secretase to release EpICD, as well as EpICD stability, remain largely unclear.
In this study, we combined biochemical and live-cell imaging approaches using fluorescence-tagged EpCTF variants to characterize and quantify cleavage by the γ-secretase complex in living cells. Kinetics of EpCTF cleavage by γ-secretase and subsequent degradation of EpICD by the proteasome were determined in multiple cell lines. All data demonstrated that EpCTF cleavage is a very slow process, whereas the subsequent degradation of EpICD was highly efficient in both murine and human cells. Slow processing by γ-secretase was further confirmed with endogenous EpCAM in carcinoma cells. Cross-species swapping experiments further demonstrated that γ-secretase activity, not EpCTF-related features, defines the slow pace of cleavage. Hence, RIP of EpCAM probably provides cells with a comparably slow response signal and a means of disposal of EpCAM.
Results
EpCTF-YFP is correctly targeted to the plasma membrane
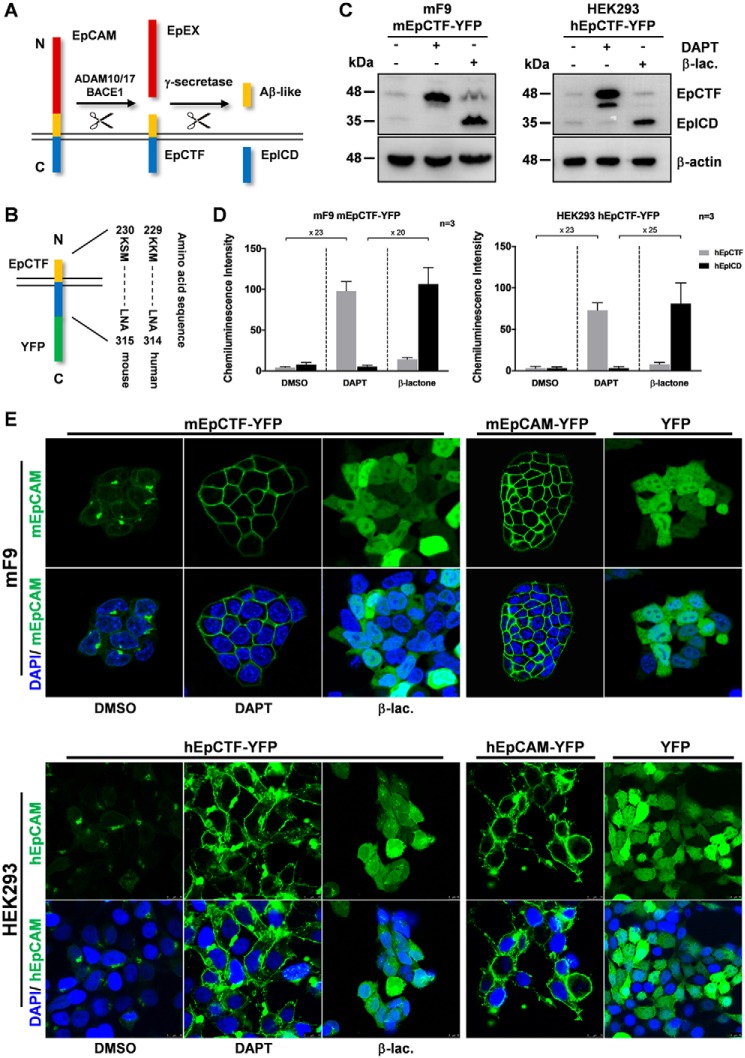
EpCAM is cleaved by ADAM10/17 or BACE1 and γ-secretase to sequentially generate EpEX, EpCTF, Aβ-like, and EpICD fragments (Fig. 1A). To study the pace and efficiency of EpCTF cleavage by γ-secretase (i.e. the second cleavage of EpCAM during RIP), human and murine variants of EpCAM composed of the signal peptide of EpCAM, a c-Myc tag, 35 membrane-proximal amino acids (aa) of the extracellular domain of EpCAM, and the transmembrane and intracellular domains of EpCAM were fused to yellow fluorescent protein (YFP) (Fig. 1B). These tagged mimics of EpCTF (in the following EpCTF-YFP) allow assessment of the proteolytic processing of EpCTF by γ-secretase in living cells by monitoring YFP fluorescence.
Figure 1.
Generation of YFP-tagged murine and human EpCTF variants. A, schematic representation of RIP of EpCAM by ADAM10/17, BACE1, and γ-secretase, including the resulting protein fragments (EpCAM extracellular domain (EpEX), EpCAM CTF (EpCTF), and EpCAM intracellular domain (EpICD)). B, EpCTF-YFP variants consist of the signal peptide of murine or human EpCAM (residues 1–23), a short linker peptide consisting of two amino acids (KL), and the CTF sequence of murine EpCAM (residues 251–315) and human EpCAM (residues 250–314) followed by YFP. C, expression of EpCTF-YFP and EpICD-YFP was analyzed by immunoblotting with YFP-specific antibodies in stable mF9 and HEK293 transfectants following the indicated treatments (γ-secretase inhibitor (DAPT) and β-lactone proteasome inhibitor (β-lac.)). Equal protein loading was verified with staining against β-actin. Shown are representative results from n = 3 independent experiments. D, immunoblotting results presented in C were quantified from n = 3 independent experiments. Shown are mean values ± S.E. (error bars). E, mF9 and HEK293 cells with murine and human EpCTF-YFP were incubated with DMSO, DAPT, and β-lactone, respectively. The green YFP signal was assessed by laser-scanning confocal microscopy. As controls, cells expressing a YFP-tagged full-length version of murine and human EpCAM or YFP only were assessed in parallel. Each image was adjusted independently to achieve the highest quality and contrast. Shown are representative results from n = 3 independent experiments.
To confirm EpCTF-YFP regulation via a γ-secretase/proteasome–dependent pathway like endogenous EpCAM, immunoblotting was performed with whole-cell lysates of stable transfectants of murine EpCTF-YFP in murine F9 teratoma cells and of human EpCTF-YFP in human embryonic kidney HEK293 cells. Both cell lines were treated with the γ-secretase inhibitor DAPT to block EpCTF cleavage and with the proteasome inhibitor β-lactone to block subsequent degradation of EpICD. Untreated cells expressed only minute amounts of murine and human EpCTF-YFP and EpICD-YFP fragments (Fig. 1C). After 24 h of DAPT treatment, EpCTF-YFP was detected as a prominent band of 45 kDa and a minor band of 42 kDa, whereas EpICD was not or barely detectable (Fig. 1C). As expected, proteasome inhibition after 12 h of treatment with β-lactone led to an accumulation of EpICD-YFP at 32 kDa after (Fig. 1C). Protein quantification of human and murine EpCTF and EpICD fragments was conducted from three independent immunoblotting experiments. Stabilization of mEpCTF and hEpCTF through treatment of cells with the γ-secretase inhibitor DAPT resulted in a 23-fold increase of both EpCTFs as compared with DMSO-treated control cells (Fig. 1D). Stabilization of mEpICD and hEpICD through treatment of cells with the proteasome inhibitor β-lactone resulted in a 20- and 25-fold increased amount of EpICD, respectively, as compared with conditions of DAPT treatment (i.e. inhibition of EpICD generation) (Fig. 1D). Hence, murine and human EpCTF-YFP are processed by γ-secretase, and, thereafter, the resulting EpICD fragments are degraded by the proteasome.
Next, the correct subcellular localization of EpCTF fusions was assessed by laser-scanning confocal microscopy. mF9 and HEK293 cells with murine and human EpCTF-YFP were treated with DMSO, DAPT, or β-lactone. The YFP signal was barely visualized at the plasma membrane following DMSO treatment and only displayed faint perinuclear staining and intracellular aggregates (Fig. 1E). In DAPT-treated cells, murine and human EpCTF-YFP localized on the plasma membrane. After β-lactone incubation in the absence of DAPT, murine and human EpICD-YFP accumulated homogeneously in the cytoplasm (Fig. 1E). Control cell lines expressing either murine or human full-length EpCAM-YFP demonstrated the expected plasma membrane localization, whereas in YFP-expressing cells, the signal was evenly distributed in cells (Fig. 1E).
Taken together, the results showed that murine and human EpCTF-YFP variants expressed in mF9 and HEK293 cells, respectively, were correctly localized and responded to inhibitors comparably with endogenous EpCTF (5).
Live-cell imaging of EpCTF proteolysis by γ-secretase
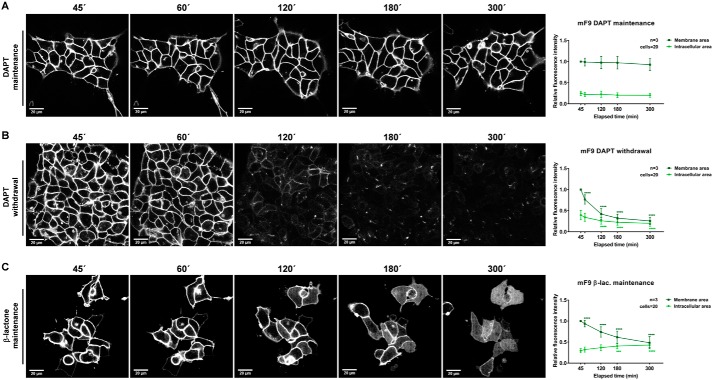
To analyze and quantify EpCTF cleavage by γ-secretase, we monitored EpCTF-YFP with live-cell confocal time-lapse microscopy. mF9 and HEK293 cells stably expressing either murine and human EpCTF-YFP variants, respectively, were pretreated with DAPT for 24 h to stabilize EpCTF-YFP at the cell surface. Thereafter, either cells were further kept in DAPT-containing medium or DAPT was withdrawn by wash-out. In a third group, β-lactone was added 12 and 2 h prior to DAPT withdrawal in murine mF9 and human HEK293 cell lines, respectively, and was maintained throughout the imaging period. Frames were acquired every 5 or 7.5 min in mF9 or HEK293 cells, respectively (Videos S1–S6). At selected time points, the mean fluorescence intensity from membrane and intracellular areas was quantified. The results were normalized to the intensity at the first time point of membrane fluorescence recording.
The presence of the γ-secretase inhibitor DAPT resulted in stable localization of mEpCTF-YFP at the cell surface over a period of observation of 5 h (Fig. 2A (left panels) and Video S1). Quantification of fluorescence at the plasma membrane and in intracellular areas in n = 20 cells confirmed a steady intensity over time (Fig. 2A, right panel). Following DAPT withdrawal, fluorescence intensity of mEpCTF-YFP gradually decreased over 5 h. Compared with the initial time point, we observed a ≥70% loss of fluorescence intensity at the plasma membrane (Fig. 2B and Video S2). Inhibition of the proteasome with β-lactone in combination with a release of γ-secretase inhibition allowed for the cleavage and intracellular accumulation of mEpICD-YFP (Fig. 2C and Video S3). Under these conditions, EpCTF-YFP fluorescence at the membrane decreased by 50%, and fluorescence intensity of mEpICD-YFP in the intracellular area increased from 25 to 50% (Fig. 2C). Taken together, these data support the view that mEpICD-YFP is slowly cleaved from EpCTF and thereafter efficiently degraded by proteasome.
Figure 2.
Live-cell imaging of murine EpCTF-YFP cleavage. Murine EpCTF-YFP was stably expressed in murine F9 teratoma cells. Following pretreatment with DAPT (see “Experimental procedures”), cells were maintained in DAPT-containing medium (A), washed and maintained in normal medium (B), or washed and maintained in β-lactone–containing medium (C) for a further 5 h. Cells were monitored every 5 min at five positions. Shown are representative images at the indicated time points from n = 3 independent experiments (left panels). Quantification of YFP fluorescence at the plasma membrane and in the intracellular area was performed on n = 20 cells from n = 3 independent experiments. Shown are mean values ± S.E. (error bars) (right panels). p values were calculated with one-way ANOVA. ****, p < 0.0001.
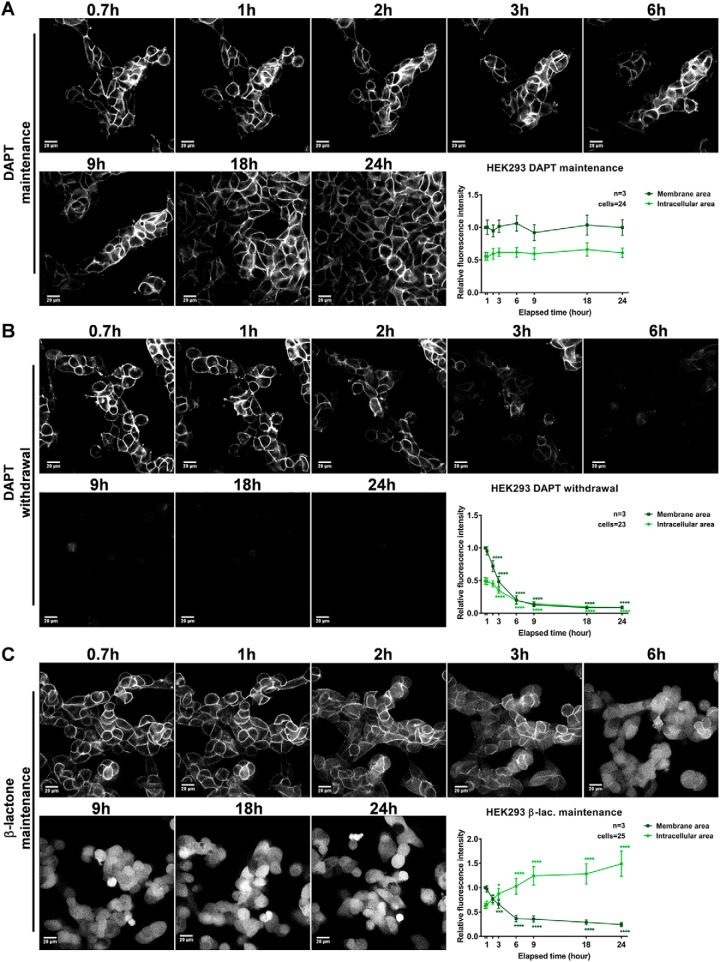
Next, the efficiency and pace of cleavage of hEpCTF-YFP were addressed in stable HEK293 transfectants. Similar to mEpCTF-YFP in mF9 cells, hEpCTF-YFP remained tethered to the membrane in cells constantly treated with DAPT over 24 h (Fig. 3A and Video S4). No significant decline of fluorescence could be measured (Fig. 3A, bottom right panel). Withdrawal of DAPT allowed for the cleavage of hEpCTF-YFP to an ∼80% reduction of fluorescence after 24 h, which was slower than that of mEpCTF-YFP in mF9 cells (Fig. 3B and Video S5). Upon release from DAPT and concomitant treatment with proteasome inhibitor, we observed accumulation of hEpICD-YFP in the cytoplasm and nucleus as soon as 2 h after the start of treatment (Fig. 3C and Video S6). After 24 h, accumulation in the intracellular area resulted in a >3-fold increase in fluorescence intensity as compared with the initial time point (Fig. 3, bottom right panel).
Figure 3.
Live-cell imaging of human EpCTF-YFP cleavage. Human EpCTF-YFP was stably expressed in human HEK293 cells. Following pretreatment with DAPT (see “Experimental procedures”), cells were maintained in DAPT-containing medium (A), washed and maintained in normal medium (B), or washed and maintained in β-lactone–containing medium (C) for a further 24 h. Cells were monitored every 7.5 min at five positions. Shown are representative images at the indicated time points from n = 3 independent experiments (left panels). Quantification of YFP fluorescence at the plasma membrane and in the intracellular area was performed on n = 20 cells from n = 3 independent experiments. Shown are mean values ± S.E. (error bars) (right panels). p values were calculated with one-way ANOVA. ****, p < 0.0001.
To exclude potential effects of the inhibitors on gene expression, mRNA levels of the catalytic component of γ-secretase (i.e. expression of murine and human presenilin 1 and 2) were assessed by qPCR. In both mF9 and HEK293 cells, we observed no significant differences in presenilin 1 and 2 mRNA levels at time points equivalent to the start and end of the time-lapse interval (Fig. S1). Taken together, our results support the view that proteolysis of murine and human EpCTF by γ-secretase is a slow process that is followed by an efficient degradation of the newly generated EpICD by the proteasome.
Biochemical assessment of EpCTF cleavage by γ-secretase
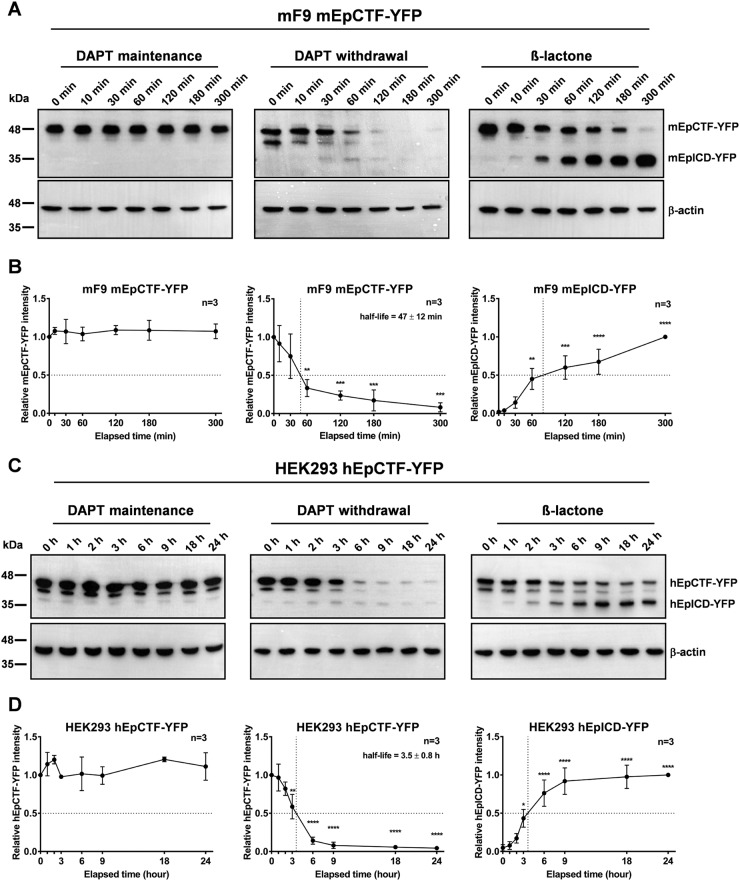
To calculate the 50% protein turnover of EpCTF and quantify EpICD degradation efficiency, time course experiments were performed and combined with immunoblotting detection. In analogy to live-cell imaging experiments, cells were either maintained in DAPT throughout the detection period, released from DAPT, or released from DAPT and concomitantly treated with β-lactone. Protein levels of mEpCTF-YFP and mEpICD-YFP in mF9 cells were analyzed at 0, 10, 30, 60, 120, 180, and 300 min. In the presence of DAPT, immunoblotting for mEpCTF-YFP did not reveal any significant changes over 5 h (Fig. 4, A and B, left panels). Upon DAPT withdrawal, we observed 50% cleavage of mEpCTF-YFP at 47 ± 12 min (Fig. 4, A and B, middle panels). Upon additional β-lactone treatment, quantification of immunoblots for mEpCTF-YFP and mEpICD-YFP showed 50% of mEpICD-YFP accumulation after ∼70 min (Fig. 4, A and B, right panels).
Figure 4.
Quantification of murine and human EpCTF-YFP cleavage. A, murine EpCTF-YFP was stably expressed in murine F9 teratoma cells. Following pretreatment with DAPT (see “Experimental procedures”), cells were maintained in DAPT-containing medium, washed and maintained in normal medium, or washed and maintained in β-lactone–containing medium for a further 5 h. mEpCTF-YFP and mEpICD-YFP expression was assessed by immunoblotting with YFP-specific antibodies at the indicated time points. Shown are representative results from n = 3 independent experiments. B, quantification of mEpCTF-YFP and mEpICD-YFP expression and calculation of mEpCTF 50% protein turnover were performed from n = 3 independent experiments. Shown are mean values ± S.E. (error bars). p values were calculated with one-way ANOVA. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C, human EpCTF-YFP was stably expressed in human HEK293 cells. Following pretreatment with DAPT (see “Experimental procedures”), cells were maintained in DAPT-containing medium, washed and maintained in normal medium, or washed and maintained in β-lactone–containing medium for a further 24 h. hEpCTF-YFP and hEpICD-YFP expression was assessed by immunoblotting with YFP-specific antibodies at the indicated time points. Shown are representative results from n = 3 independent experiments. D, quantification of hEpCTF-YFP and hEpICD-YFP expression and calculation of hEpCTF 50% protein turnover were performed from n = 3 independent experiments. Shown are mean values with S.E. p values were calculated with one-way ANOVA. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Similar to mEpCTF, hEpCTF-YFP levels of stable HEK293 transfectants were not significantly altered significantly over 24 h in DAPT (Fig. 4, C and D, left panels). Upon withdrawal of DAPT, we determined a 50% hEpCTF-YFP cleavage at 3.5 ± 0.8 h (Fig. 4, C and D, middle panels). Withdrawal of DAPT and concomitant treatment with β-lactone resulted in hEpCTF-YFP cleavage and stabilization of hEpICD-YFP (Fig. 4C, right panels). Quantification of immunoblots demonstrated an accumulation 50% of maximal hEpICD-YFP amounts at ∼3.5 h (Fig. 4D, right panel).
Enzymatic cleavage with a 50% turnover of substrates at 47 min and 3.5 h of murine and human EpCTFs, respectively, is a relatively slow process. To confirm that these values are not cell type–specific, mEpCTF-YFP and hEpCTF-YFP were stably expressed in murine NIHT3T3 fibroblasts and in the human head and neck squamous cell carcinoma line FaDu, respectively. Gradual reduction of mEpCTF-YFP in NIH3T3 cells was observed by epifluorescence microscopy over 24 h (Fig. S2A). Quantification of fluorescence intensity over time in n = 10 cells of three independent experiments (n = 30 total cells) resulted in a calculated average half-life of 5.0 h (Fig. S2B). Biochemical analysis confirmed the cleavage of mEpCTF-YFP over time (Fig. S2C), and immunoblot quantification determined a 50% protein turnover of mEpCTF-YFP in NIH3T3 cells at 5.3 ± 1.1 h (Fig. S2D). Similar results were obtained in FaDu cells transfected with human EpCTF-YFP. Assessment of YFP fluorescence disclosed an average half-life of 4.7 h (Fig. S3, A and B). Biochemical assessment of hEpCTF-YFP cleavage confirmed a 50% protein turnover of hEpCTF-YFP at 5.1 ± 1.1 h (Fig. S3, C and D). Hence, EpCTF proteolysis by γ-secretase is a slow process in all cell lines tested.
The pace of proteolysis is dictated by γ-secretase, not EpCTF
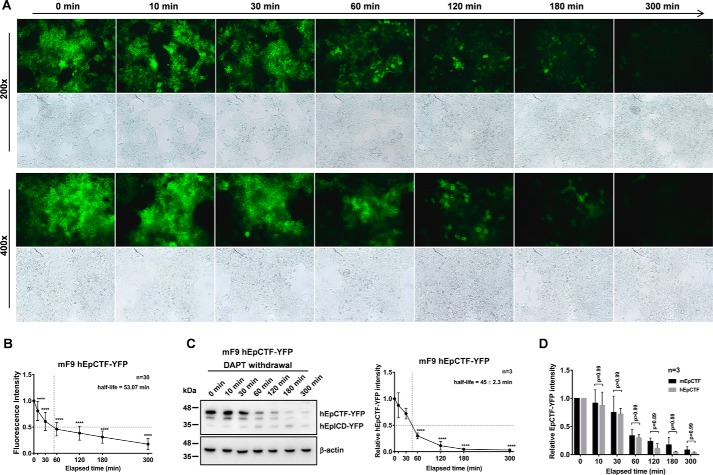
Although proteolysis of mEpCTF and hEpCTF was slow, 50% protein turnovers of EpCTFs still differed considerably across all cell lines tested (45 min to 5.3 h in mF9 and HEK293 cells, respectively). To address the limiting factors in EpCTF cleavage, cross-species swapping experiments were conducted, where hEpCTF-YFP was expressed in murine F9 cells and mEpCTF-YFP was expressed in HEK293 cells. Cleavage of hEpCTF-YFP in mF9 cells was recorded by epifluorescence microscopy. We observed a significant drop in fluorescence starting between 30 and 60 min (Fig. 5A). Quantification of YFP fluorescence intensity over time in n = 10 cells of three independent experiments (n = 30 total cells) resulted in a calculated average half-life of 53.07 min (Fig. 5B). Immunoblotting analysis of hEpCTF-YFP in murine F9 cells confirmed the timing observed in fluorescence imaging. We observed a 50% protein turnover at 45 ± 2.3 min (Fig. 5C). Hence, both microscopy and immunoblotting experiments suggest that murine and human EpCTF-YFP are cleaved at almost the same rate in mF9 cells, with 50% of hEpCTF-YFP being cleaved at 45 min in mF9 cells rather than 3.5 h in HEK293 cells. To confirm similar cleavage rates, Two-way ANOVA was performed at each time point between mEpCTF and hEpCTF in mF9 cells. With an overall p value of 0.96, ANOVA analysis demonstrates no significant difference in cleavage rate of either construct (Fig. 5D).
Figure 5.
Cleavage of hEpCTF in murine F9 teratoma cells. A, human EpCTF-YFP was stably expressed in murine F9 teratoma cells. Following pretreatment with DAPT (see “Experimental procedures”), cells were maintained in normal medium for a further 5 h. Cells were monitored at the indicated time points by immunofluorescence microscopy. Shown are representative pictures at the indicated time points from n = 3 independent experiments at ×200 and ×400 magnification. B, immunofluorescence microscopy results shown in A were quantified from n = 30 cells from n = 3 independent experiments. Shown are mean values ± S.E. (error bars). p values were calculated with one-way ANOVA. ****, p < 0.0001. C, visualization and quantification of hEpCTF-YFP and hEpICD-YFP expression and calculation of hEpCTF 50% protein turnover was performed from n = 3 independent immunoblotting experiments. Shown are mean values with S.E. p values were calculated with one-way ANOVA. ****, p < 0.0001. D, two-way ANOVA was used to calculate the difference at each time point between mEpCTF and hEpCTF in mF9 cells. The overall p value of the test is 0.96 with individual p values ranging from 0.88 to 0.99.
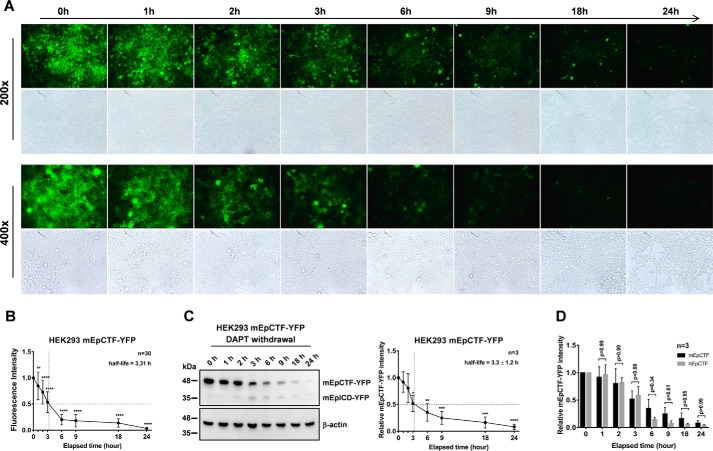
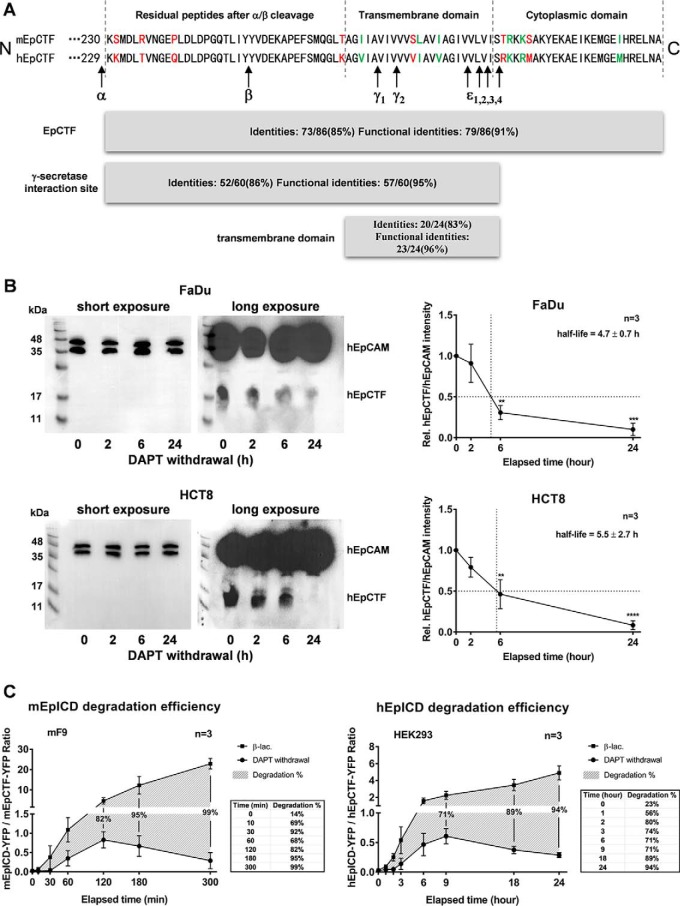
We then analyzed the cleavage pace of mEpCTF-YFP in HEK293 cells by epifluorescence microscopy and immunoblotting. Both assays confirmed a reduction of mEpCTF-YFP over 24 h, and we calculated an average 50% protein turnover at 3.31 h from fluorescence intensity kinetics of n = 30 total cells (Fig. 6, A and B). Biochemical evaluation of mEpCTF-YFP half-life in HEK293 cells disclosed a 50% decrease at 3.3 ± 1.2 h (Fig. 6C). No statistically significant difference between hEpCTF and mEpCTF cleavage pace was observed in HEK293 cells, with an overall p value of the two-way ANOVA test at 0.10 and individual p values at distinct time points ranging from 0.34 to 0.99 (Fig. 6D). Thus, cross-species swapping experiments demonstrated that the cleavage pace of EpCTF variants was associated with endogenous γ-secretase activity rather than the species of origin of the substrate EpCTF. This notion is further corroborated by sequence similarities between murine and human EpCTFs. aa sequences were analyzed for identity, similarity in function, and differences within the protein stretch from aa 230–315 of mEpCTF and 229–314 of hEpCTF. mEpCTF and hEpCTF are characterized by 91% functional identity and have even greater identity within γ-secretase cleavage sites and transmembrane domains, with 95 and 96%, respectively (Fig. 7A). We have previously identified cleavage sites of γ-secretase within the transmembrane domains of murine and human EpCAM have been identified in our previous work (15, 16). γ and ϵ cleavage sites of EpCTF are identical between mEpCTF and hEpCTF, except the ϵ4 site (Fig. 7A). The highly conserved structure of EpCTF from different species, especially in γ-secretase cleavage sites (15), further suggests that cleavage pace depends upon the γ-secretase complex rather than the substrate.
Figure 6.
Cleavage of mEpCTF in human HEK293 cells. A, murine EpCTF-YFP was stably expressed in human HEK293 cells. Following pretreatment with DAPT (see “Experimental procedures”), cells were maintained in normal medium for a further 24 h. Cells were monitored at the indicated time points by immunofluorescence microscopy. Shown are representative pictures at the indicated time points from n = 3 independent experiments at ×200 and ×400 magnification. B, immunofluorescence microscopy results shown in A were quantified from n = 30 cells from n = 3 independent experiments. Shown are mean values ± S.E. (error bars). p values were calculated with one-way ANOVA. ****, p < 0.0001. C, visualization and quantification of mEpCTF-YFP and mEpICD-YFP expression, and calculation of mEpCTF 50% protein turnover was performed from n = 3 independent immunoblotting experiments. Shown are mean values with S.E. p values were calculated with one-way ANOVA. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. D, two-way ANOVA was used to calculate the difference at each time point between mEpCTF and hEpCTF in mF9 cells. The overall p value of the test is 0.10 with individual p values ranging from 0.34 to 0.99.
Figure 7.
Amino acid similarities of EpCTF variants and EpICD degradation efficiencies. A, amino acid sequences of murine and human EpCTF are compared. Amino acids in red type represent differences between murine and human sequence. Amino acids in green type represent different but functionally equivalent residues. Functional identities between mEpCTF and hEpCTF are 91%, suggesting a highly conserved amino acid sequence. Functional identities of γ-secretase cleavage sites and transmembrane domains are 95 and 96%, respectively. The majority of γ and ϵ cleavage sites of EpCTF variants are identical except for the ϵ4 site. B, cleavage of endogenous EpCTF was assessed in FaDu and HCT8 carcinoma cells. FaDu and HCT8 cells were treated with γ-secretase inhibitor DAPT for 24 h. Thereafter, cell inhibition was released upon withdrawal of DAPT, and EpCTF expression was monitored by immunoblotting with EpICD-specific antibodies. Shown are representative results from n = 3 independent immunoblotting experiments (left panels) and mean values with S.E. (error bars) (right panels). p values were calculated with one-way ANOVA. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C, ratios of EpICD-YFP to EpCTF-YFP were calculated from immunoblotting results to quantify EpICD degradation efficiency. Percentages of mEpICD degradation are represented by the Δ value between two groups. 99% of mEpICD was degraded in the absence of β-lactone at 5 h (left panel shadow area and table). 94% hEpICD was degraded at 24 h in HEK293 cells (right panel). Shown are mean values with S.E. (error bars) from n = 3 independent experiments.
In the following, we aimed at validating the findings of exogenously expressed EpCTF-YFP proteins in the context of cleavage of endogenous EpCTF in membrane fractions of carcinoma cells. Head and neck squamous carcinoma cell line FaDu and ileocecal adenocarcinoma cell line HCT8 were treated for 24 h with DAPT to block cleavage of membrane-tethered endogenous EpCTF. Kinetics of EpCTF cleavage were then monitored after withdrawal of DAPT in immunoblotting experiments. Treatment of FaDu and HCT8 cells with DAPT resulted in the accumulation of endogenous EpCTF, which was slowly cleaved after DAPT withdrawal (Fig. 7B, left panels). Quantification of endogenous EpCTF relative to full-length EpCAM revealed EpCTF half-lives of 4.7 ± 0.7 and 5.5 ± 2.7 h for FaDu and HCT8 cells, respectively (Fig. 7B, right panel). Thus, cleavage of endogenous human EpCTF by γ-secretase was confirmed to be slow in carcinoma cells.
Degradation of EpICD by proteasome is highly efficient
Following the generation of EpICD through cleavage of EpCTF by γ-secretase, EpICD can translocate into the nucleus (4, 5). Generally, the stability of EpICD appears rather low, with small amounts detectable in whole-cell lysates and in the nucleus (5, 25), where EpICD deploys its major signaling function (5, 27–31). To dispose of EpICD functions, the cell requires a degradation modality that is assumed by the proteasome. Therefore, to quantify the efficiency of proteasomal degradation of murine and human EpICD, ratios of EpICD to EpCTF were calculated from immunoblots in the presence and absence of β-lactone. In β-lactone–treated mF9 cells, ratios gradually increased from 0.02 to 22.88, demonstrating an accumulation of mEpICD. In the absence of β-lactone after DAPT withdrawal, the ratio was initially increased to 0.83 and dropped to 0.29 at later time points, suggesting that mEpICD sequentially accumulated and was later degraded (Fig. 7C, left panel). Percentages of mEpICD degradation are represented by the differences in ratios between the two treatment groups. After 5 h, 99% of mEpICD was degraded in absence of β-lactone (Fig. 7C, left panel). Similarly, 94% of hEpICD was degraded after 24 h in HEK293 cells (Fig. 7C, right panel). Hence, degradation of murine and human EpICD fragments by the proteasome is a highly efficient process, which takes place following EpCTF cleavage by γ-secretase.
Discussion
The expression of EpCAM is a tightly regulated process throughout differentiation, under physiological and pathological conditions. Early in embryonic development during gastrulation and in vitro in model systems, EpCAM expression is retained in endodermally derived cells, whereas it is repressed in cells differentiating into mesodermal lineages (32). Spatiotemporally selective expression of EpCAM is important for proper maturation of embryonic stem cells (ESCs), as a balanced combination of EpCAM+ and EpCAM− cells is required for full differentiation (32). Similarly, EpCAM is characterized by a dynamic expression during cancer progression and is frequently lost during EMT (33–35). Loss of EpCAM expression in carcinoma cells during EMT is primarily associated with enhanced migration and invasion properties (33, 36–39). A direct involvement of the soluble extracellular domain of EpCAM (EpEX) in the regulation of EMT by EGFR has been revealed recently and represents a molecular basis for the strongly differing clinical performance of patients with head and neck cancers (HNSCC) based on their EpCAM/EGFR expression profile (25). These findings are in line with the report on the expression of EpCAM in single HNSCC cells with a primarily epithelial phenotype, as opposed to cells with an EMT signature. The latter ones were characterized by metastases formation and poorer survival (40).
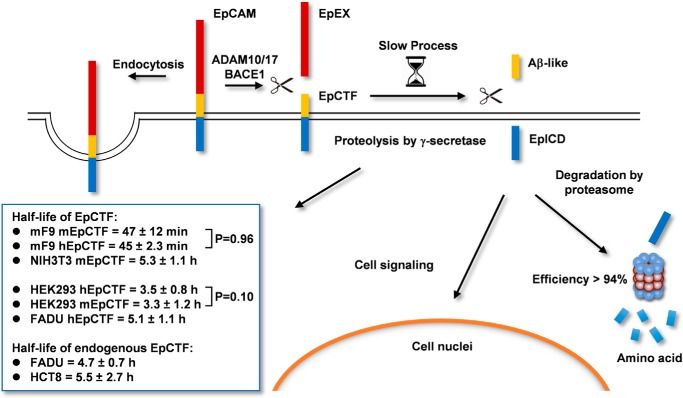
In our previous study, we have addressed the precise timing for the strict differential regulation of EpCAM in an in vitro three-dimensional model of ESC differentiation. Complete loss of EpCAM in subsets of cells occurs within ∼12 h of spontaneous differentiation (32). However, down-regulation of the EPCAM transcription levels ensued with a slight delay. Together with a 50% protein turnover of the EpCAM protein of 21 h (41), these results suggested a post-translational mechanism to actively down-regulate EpCAM protein within 12 h. RIP and endocytosis are two major mechanisms by which cells dispose of EpCAM from the cell surface (Fig. 8) (15, 16). Furthermore, RIP is central to the signaling functions of EpCAM in cancer and stem cells, where it was described to regulate the expression of pluripotency-associated genes (4, 5, 27, 30, 42, 43). Therefore, understanding the pace of RIP is a central point to further interpret EpCAM functions in the cellular context of differentiation and signaling in normal and malignant cells.
Figure 8.
Schematic representation of RIP, degradation, and endocytosis of EpCAM. Sequential cleavage of EpCAM via RIP is depicted, including enzymes involved and resulting cleavage products with half-lives.
RIP encompasses an initial cleavage of EpCAM by α- and β-secretases to generate the soluble extracellular domain EpEX and an EpCTF fragment that is still resident in the plasma membrane and is further cleaved at γ and ϵ sites by the γ-secretase complex. Only very limited knowledge is available so far with respect to the frequency of α- and β-cleavage of EpCAM (e.g. a requirement for cell–cell contact and a capacity of EpEX to serve as a ligand for intact EpCAM molecules) (5, 44). Furthermore, regulation of EpCAM cleavage by EGF/EGFR during phenotypic changes along the EMT have been described for endometrial cancer cells (45). However, we could not reproduce such EGF/EGFR-dependent RIP of EpCAM in an array of carcinoma cell lines of various entities, including endometrial cells, thus suggesting that it does not represent a common mechanism in cancer cells (25). Generally, the pace of EpCTF proteolysis by γ-secretase will further determine the pace of signaling through EpICD and of EpCAM disposal from the plasma membrane. In addition, the stability of EpICD in cells will control the availability for and duration of nuclear signaling by EpCAM.
In the present study, we demonstrate that the pace of murine and human EpCTF proteolysis by γ-secretase is slow and that the subsequent degradation of EpICD by the proteasome is efficient. Tracking of EpCTF cleavage by live-cell imaging and biochemical approaches in combination with fluorescent protein-tagged variants of EpCAM revealed 50% protein turnover of EpCTF ranging from 45 min to 5.3 h in different cell lines. These findings are in line with the reported slow and inefficient proteolysis of amyloid precursor protein CTF by γ-secretase (46) and with cleavage by rhomboid proteases (47). Swapping experiments demonstrated that the species of provenance of EpCTF had no impact on the pace of proteolysis. In fact, the pace of proteolysis was transferred to EpCTF variants through the choice of cell lines, which in turn displayed a steady pace of cleavage independently of the species origin of the substrate. The notion that γ-secretase rather than its substrate CTF dictates the pace of catalysis is corroborated by high identities in aa of murine and human EpCTFs.
The cleavage pace of endogenous EpCTF was demonstrated to be similarly slow in carcinoma cells of the head and neck area and the ileocecum as compared with exogenously expressed variants of EpCTF, with 4.7 and 5.5 h, respectively. Based on the slow and inefficient cleavage of EpCTF by γ-secretase, rapid signaling by EpCAM through RIP appears unlikely. We therefore suggest that RIP of EpCAM is responsible for more steady signaling through EpICD in tumor cells (4, 5) and in stem cells (27, 28), rather than for rapid transmission of extracellular cues into de novo gene transcription activities. Additionally, EpICD-mediated signaling is tightly controlled by a highly ≥94% efficient degradation of EpICD by the proteasome. Furthermore, differences in EpICD nuclear localization between malignant and normal cells, as observed in colon tissue (5), might impact EpCAM signaling through RIP and result in a lack of signals in normal tissue, as has been recently reported for primary human liver cells (48). However, degradation of EpCAM through RIP and proteasomal activity (e.g. during ESC differentiation and in cells undergoing EMT during the metastatic cascade) appears as a potentially central means of disposal of EpCAM.
Taken together, we conclude that proteolysis of EpCTF by γ-secretase is a slow process that is subsequently followed by highly efficient degradation of the cleavage product EpICD by the proteasome.
Experimental procedures
Cell lines
Murine F9 teratoma (kind gift from Dr. Marcus Conrad, Munich) cells and murine embryonic fibroblasts (NIH3T3) were cultured in Dulbecco's modified Eagle's medium (high-glucose) supplemented with 20% FCS (Biochrom AG, Heidelberg, Germany) and 1% penicillin/streptomycin. Human embryonic kidney 293 cells (HEK293) and human squamous cell carcinoma cells (FaDu and HCT8) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. All cell lines were grown in a 5% CO2 atmosphere at 37 °C. Human cell lines were confirmed by STR-typing at the Helmholtz Center (Munich, Germany).
Transfections and expression vectors
Transfections were performed with the MATra reagent (Iba, Goettingen, Germany) following the manufacturer's recommendations. Full-length murine and human EpCAM (315 and 314 aa, respectively) were cloned in fusion with a YFP tag to generate EpCAM-YFP constructs. EpCTF-YFP constructs consist of the signal peptide of murine or human EpCAM (residues 1–23), a short linker peptide consisting of two amino acids (KL), and the CTF sequence of murine (residues 251–315) or human EpCAM (residues 250–314) followed by YFP. All constructs mentioned above were cloned into the 141 pCAG-3SIP expression vector by using EcoRI and NheI restriction enzyme sites. Stable selection of transfectants was performed with puromycin (1 μg/ml) in the appropriate culture medium starting 24 h after transfection.
Inhibitors
Inhibition of γ-secretase and proteasome activity was achieved by treating cells with DAPT (10 μm; Sigma, Munich, Germany) or lactacystin β-lactone (10 μm; Cayman Chemical), respectively. Inhibitors were supplemented when medium was changed.
Laser-scanning confocal microscopy
For fluorescence staining, cells were plated on glass slides and grown to confluence followed by incubation with different inhibitors. Cells were washed with PBS, fixed in 3% paraformaldehyde, and covered with VectaShield containing 4′,6-diamidino-2-phenylindole to stain nuclei (Vector Laboratories, Burlingame, CA). Images were analyzed with a TCS-SP5 system, ×63 oil immersion objective, and LAS AF software (Leica Microsystems, Wetzlar, Germany).
Epifluorescence microscopy
Cells were cultured on 10-cm tissue culture dishes and grown to 50% confluence. Cells were pretreated with DAPT for 24 h and washed five times with PBS, and new medium without DAPT was added. Images were acquired with an Olympus motorized inverted research microscope IX81. The system was run by MMI Cellcut Plus software (Molecular Machines and Industries, Eching, Germany). Fluorescence intensity across the plasma membrane areas was quantified with Fiji software (48). Exposure time for all cell lines was set to 959 ms in ×200 magnification and 1180 ms in ×400 magnification, respectively. Gain value was set to 0.5 dB, and camera offset value was subtracted from all images before quantification. Mean intensity value of fluorescence at the first time point of measurement was set to 100% as a reference, and all other values were normalized to the reference.
Live-cell imaging
Cells were cultured on 35-mm glass or plastic bottom (Ibidi, Munich, Germany) dishes and grown to 50% confluence. Cells were pretreated with DAPT for 24 h, washed five times with PBS, and separated into three treatment groups: DAPT maintenance, DAPT withdrawal, and DAPT withdrawal with β-lactone maintenance. Images were acquired on a Nikon Ti-E microscope equipped with a Opterra-II swept scan confocal head (Bruker) and automated stage (ASI) with a ×60 numerical aperture 1.4 Plan Apo oil immersion objective or a ×40 numerical aperture 0.95 Plan Apo air objective, respectively (both from Nikon), onto a Photometrics Evolve 512 Delta EMCCD camera. The system was run by PrairieView software (Bruker). Temperature was maintained by a full enclosure incubator (In Vivo Scientific) with a feedback-controlled heating unit (World Precision Instruments), with the temperature set such that 37 °C was maintained at the sample. An atmosphere of 5% CO2 was maintained by mixing pressured air with 100% CO2 by 190/10 flow rates through connected flow meters (Voegtlin, Germany). Air was humidified through a semipermeable membrane tube immersed in water. A 488-nm excitation wavelength and GFP filter set (500LP or 520/40BP, Chroma, Semrock) was used to visualize YFP. Fluorescence intensity across the plasma membrane and intracellular areas was quantified with Fiji (49). Camera offset value was subtracted from all images before quantification. Mean intensity value of fluorescence at the first time point of measurement was set as a reference to 100%, and all other values were normalized to the reference.
Quantitative RT-PCR
Total mRNA was isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany) and reverse-transcribed with the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). cDNA was amplified using SYBR Green PCR mastermix (Qiagen, Hilden, Germany) and gene-specific primers in a LightCycler® 480 (Roche Diagnostics). Normalizations across samples were performed using an average of constitutive gene expression of GUSB or GAPDH. Gene expression levels were calculated according the expression, 2−ΔΔCT, where ΔCT is defined as CTgene of interest − CTendogenous control, and ΔΔCT is defined as ΔCTcontrol − ΔCTtreatment. Primers were as follows: GusB, 5′-CAACCTCTGGTGGCCTTACC-3′ (forward) and 5′-GGGTGTAGTAGTCAGTCACAGAC-3′ (reverse); mouse presenilin 1, 5′-CAGTGAGGAGTGGGAGGCCCA-3′ (forward) and 5′-GCACAGGCCGATCAGTATGGCTACA-3′ (reverse); GAPDH, 5′-TGCACCACCAACTGCTTAGC (forward) and 5′-GGCATGGACTGTGGTCATGAG-3′ (reverse); human presenilin 1, 5′-TACCTGCACCGTTGTCCTAC-3′ (forward) and 5′-CTCATCTTGCTCCACCACCT-3′ (reverse); mouse presenilin 2, 5′-TCCTCAACTCCGTGCTTAAC-3′ (forward) and 5′-TGATGCTCCTCTTCTTGTTCAC-3′ (reverse); human presenilin 2, 5′-AGCCTCCCTTGACTGGCTAC-3′ (forward) and 5′-TTCTACAGTGTGTCTGGTGGG-3′ (backward).
Immunoblotting
Cells were lysed in PBS containing 1% Triton X-100 and protease inhibitors (Roche Complete, Roche Diagnostics). Protein concentrations were determined by a BCA assay (Thermo Scientific, Schwerte, Germany). 10–40 μg of proteins were separated by 10–15% SDS-PAGE, followed by immunoblotting with β-actin (Santa Cruz Biotechnology, Inc. (Dallas, TX), sc-47778; 1:5000) and GFP antibodies able to bind YFP (Abcam, Berlin, Germany), horseradish peroxidase–conjugated secondary antibodies, and detection with ECL reagent (Millipore, Darmstadt, Germany) in a Chemidoc XRS imaging system (Bio-Rad, Munich, Germany). Membrane extraction was performed as follows. Cells were resuspended in homogenizing buffer (10 nm MOPS, pH 7.0, 10 mm KCl (4 °C), 1× Roche Complete protease inhibitor), Dounce homogenized in 23-gauge needles (10 strokes), and centrifuged at 1000 × g at 4 °C for 15 min to generate a post-nuclear supernatant. Post-nuclear supernatant was then centrifuged at 16,000 × g at 4 °C for 20 min, and the resulting pellet was washed in 500 μl of homogenizing buffer before centrifugation at 16,000 × g at 4 °C for 3 min. The resulting pellet was resolved in lysis buffer (TBS-T, 1% Triton X-100).
Quantification of protein expression was conducted as follows. Chemiluminescence intensity of protein bands from each time point was measured using the volume tool implemented in the Image Lab software (Bio-Rad). The first time point of EpCTF expression was labeled as a reference band within the DAPT withdrawal/maintenance group, whereas the EpICD band from the last time point was set as a reference band within the β-lactone maintenance group. All remaining protein expression values represent the ratio of the antigen intensity divided by the reference band and were displayed as relative quantity.
Statistics
Results are presented as mean ± S.E. of at least three independent experiments. Significance of differences from the first time point was calculated using one-way ANOVA with Bonferroni's multiple-comparison test; the significance of differences between two groups at each time point was calculated using two-way ANOVA with Sidak's multiple-comparison test. The level of significance was displayed as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Author contributions
Y. H., A. C., M. P., V. K., and A. E. data curation; Y. H., A. C., G. K., M. P., V. K., and A. E. formal analysis; Y. H. and A. C. investigation; Y. H., A. C., G. K., M. P., V. K., and A. E. methodology; Y. H. and O. G. writing-review and editing; A. C. validation; A. E. and O. G. supervision; O. G. conceptualization; O. G. funding acquisition; O. G. writing-original draft; O. G. project administration.
Supplementary Material
This work was funded in part by Deutsche Forschungsgemeinschaft Grants GI 540/3-1 and GI 540/3-2 (to O. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Videos S1–S6.
- EpCAM
- epithelial cell adhesion molecule
- HNSCC
- head and neck squamous cell carcinoma
- RIP
- regulated intramembrane proteolysis
- CTF
- C-terminal fragment
- PS
- presenilin 1 or 2
- NCT
- nicastrin
- ICD
- intracellular domain
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- EMT
- epithelial–mesenchymal transition
- YFP
- yellow fluorescent protein
- aa
- amino acid(s)
- ANOVA
- analysis of variance
- ESC
- embryonic stem cell
- STR
- short tandem repeat
- BACE
- β-secretase
- DAPT
- N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester.
References
- 1. Lal M., and Caplan M. (2011) Regulated intramembrane proteolysis: signaling pathways and biological functions. Physiology 26, 34–44 10.1152/physiol.00028.2010 [DOI] [PubMed] [Google Scholar]
- 2. Medina M., and Dotti C. G. (2003) RIPped out by presenilin-dependent γ-secretase. Cell. Signal. 15, 829–841 10.1016/S0898-6568(03)00041-X [DOI] [PubMed] [Google Scholar]
- 3. Schroeter E. H., Kisslinger J. A., and Kopan R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 10.1038/30756 [DOI] [PubMed] [Google Scholar]
- 4. Chaves-Pérez A., Mack B., Maetzel D., Kremling H., Eggert C., Harréus U., and Gires O. (2013) EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene 32, 641–650 10.1038/onc.2012.75 [DOI] [PubMed] [Google Scholar]
- 5. Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P. A., Munz M., and Gires O. (2009) Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 11, 162–171 10.1038/ncb1824 [DOI] [PubMed] [Google Scholar]
- 6. Kopan R., and Ilagan M. X. (2004) γ-Secretase: proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 5, 499–504 10.1038/nrm1406 [DOI] [PubMed] [Google Scholar]
- 7. Solanas G., Cortina C., Sevillano M., and Batlle E. (2011) Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nat. Cell Biol. 13, 1100–1107 10.1038/ncb2298 [DOI] [PubMed] [Google Scholar]
- 8. Lichtenthaler S. F., Haass C., and Steiner H. (2011) Regulated intramembrane proteolysis–lessons from amyloid precursor protein processing. J. Neurochem. 117, 779–796 10.1111/j.1471-4159.2011.07248.x [DOI] [PubMed] [Google Scholar]
- 9. Lichtenthaler S. F., Lemberg M. K., and Fluhrer R. (2018) Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 37, e99456 10.15252/embj.201899456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Strooper B. (2003) Aph-1, Pen-2, and nicastrin with presenilin generate an active γ-secretase complex. Neuron 38, 9–12 10.1016/S0896-6273(03)00205-8 [DOI] [PubMed] [Google Scholar]
- 11. Edbauer D., Winkler E., Regula J. T., Pesold B., Steiner H., and Haass C. (2003) Reconstitution of γ-secretase activity. Nat. Cell Biol. 5, 486–488 10.1038/ncb960 [DOI] [PubMed] [Google Scholar]
- 12. Sato T., Diehl T. S., Narayanan S., Funamoto S., Ihara Y., De Strooper B., Steiner H., Haass C., and Wolfe M. S. (2007) Active γ-secretase complexes contain only one of each component. J. Biol. Chem. 282, 33985–33993 10.1074/jbc.M705248200 [DOI] [PubMed] [Google Scholar]
- 13. Bolduc D. M., and Wolfe M. S. (2014) Structure of nicastrin unveils secrets of γ-secretase. Proc. Natl. Acad. Sci. U.S.A. 111, 14643–14644 10.1073/pnas.1416637111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu P., Bai X. C., Ma D., Xie T., Yan C., Sun L., Yang G., Zhao Y., Zhou R., Scheres S. H. W., and Shi Y. (2014) Three-dimensional structure of human γ-secretase. Nature 512, 166–170 10.1038/nature13567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hachmeister M., Bobowski K. D., Hogl S., Dislich B., Fukumori A., Eggert C., Mack B., Kremling H., Sarrach S., Coscia F., Zimmermann W., Steiner H., Lichtenthaler S. F., and Gires O. (2013) Regulated intramembrane proteolysis and degradation of murine epithelial cell adhesion molecule mEpCAM. PLoS One 8, e71836 10.1371/journal.pone.0071836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsaktanis T., Kremling H., Pavšič M., von Stackelberg R., Mack B., Fukumori A., Steiner H., Vielmuth F., Spindler V., Huang Z., Jakubowski J., Stoecklein N. H., Luxenburger E., Lauber K., Lenarčič B., and Gires O. (2015) Cleavage and cell adhesion properties of human epithelial cell adhesion molecule (HEPCAM). J. Biol. Chem. 290, 24574–24591 10.1074/jbc.M115.662700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baeuerle P. A., and Gires O. (2007) EpCAM (CD326) finding its role in cancer. Br. J. Cancer 96, 417–423 10.1038/sj.bjc.6603494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balzar M., Winter M. J., de Boer C. J., and Litvinov S. V. (1999) The biology of the 17-1A antigen (Ep-CAM). J. Mol. Med. 77, 699–712 10.1007/s001099900038 [DOI] [PubMed] [Google Scholar]
- 19. Litvinov S. V., Bakker H. A., Gourevitch M. M., Velders M. P., and Warnaar S. O. (1994) Evidence for a role of the epithelial glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell Adhes. Commun. 2, 417–428 10.3109/15419069409004452 [DOI] [PubMed] [Google Scholar]
- 20. Litvinov S. V., Velders M. P., Bakker H. A., Fleuren G. J., and Warnaar S. O. (1994) Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 125, 437–446 10.1083/jcb.125.2.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaber A., Kim S. J., Kaake R. M., Benčina M., Krogan N., Šali A., Pavšič M., and Lenarčič B. (2018) EpCAM homo-oligomerization is not the basis for its role in cell-cell adhesion. Sci. Rep. 8, 13269 10.1038/s41598-018-31482-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Münz M., Kieu C., Mack B., Schmitt B., Zeidler R., and Gires O. (2004) The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene 23, 5748–5758 10.1038/sj.onc.1207610 [DOI] [PubMed] [Google Scholar]
- 23. Maaser K., and Borlak J. (2008) A genome-wide expression analysis identifies a network of EpCAM-induced cell cycle regulators. Br. J. Cancer 99, 1635–1643 10.1038/sj.bjc.6604725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang K. H., Tso H. C., Hung S. H., Kuan I. I., Lai J. K., Ke F. Y., Chuang Y. T., Liu I. J., Wang Y. P., Chen R. H., and Wu H. C. (2018) Extracellular domain of EpCAM enhances tumor progression through EGFR signaling in colon cancer cells. Cancer Lett. 433, 165–175 10.1016/j.canlet.2018.06.040 [DOI] [PubMed] [Google Scholar]
- 25. Pan M., Schinke H., Luxenburger E., Kranz G., Shakhtour J., Libl D., Huang Y., Gaber A., Pavšič M., Lenarčič B., Kitz J., Jakob M., Schwenk-Zieger S., Canis M., Hess J., et al. (2018) EpCAM ectodomain EpEX is a ligand of EGFR that counteracts EGF-mediated epithelial-mesenchymal transition through modulation of phospho-ERK1/2 in head and neck cancers. PLoS Biol. 16, e2006624 10.1371/journal.pbio.2006624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. González B., Denzel S., Mack B., Conrad M., and Gires O. (2009) EpCAM is involved in maintenance of the murine embryonic stem cell phenotype. Stem Cells 27, 1782–1791 10.1002/stem.97 [DOI] [PubMed] [Google Scholar]
- 27. Lu T. Y., Lu R. M., Liao M. Y., Yu J., Chung C. H., Kao C. F., and Wu H. C. (2010) Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J. Biol. Chem. 285, 8719–8732 10.1074/jbc.M109.077081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang H. P., Chen P. H., Yu C. Y., Chuang C. Y., Stone L., Hsiao W. C., Li C. L., Tsai S. C., Chen K. Y., Chen H. F., Ho H. N., and Kuo H. C. (2011) Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J. Biol. Chem. 286, 33520–33532 10.1074/jbc.M111.256164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denzel S., Mack B., Eggert C., Massoner P., Stöcklein N., Kemming D., Harréus U., and Gires O. (2012) MMP7 is a target of the tumour-associated antigen EpCAM. Int. J. Exp. Pathol. 93, 341–353 10.1111/j.1365-2613.2012.00826.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin C. W., Liao M. Y., Lin W. W., Wang Y. P., Lu T. Y., and Wu H. C. (2012) Epithelial cell adhesion molecule regulates tumor initiation and tumorigenesis via activating reprogramming factors and epithelial-mesenchymal transition gene expression in colon cancer. J. Biol. Chem. 287, 39449–39459 10.1074/jbc.M112.386235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu T., Ma Y., and Wang H. (2017) EpCAM intracellular domain promotes porcine cell reprogramming by upregulation of pluripotent gene expression via β-catenin signaling. Sci. Rep. 7, 46315 10.1038/srep46315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarrach S., Huang Y., Niedermeyer S., Hachmeister M., Fischer L., Gille S., Pan M., Mack B., Kranz G., Libl D., Merl-Pham J., Hauck S. M., Paoluzzi Tomada E., Kieslinger M., Jeremias I., Scialdone A., and Gires O. (2018) Spatiotemporal patterning of EpCAM is important for murine embryonic endo- and mesodermal differentiation. Sci. Rep. 8, 1801 10.1038/s41598-018-20131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gires O., and Stoecklein N. H. (2014) Dynamic EpCAM expression on circulating and disseminating tumor cells: causes and consequences. Cell Mol. Life Sci. 71, 4393–4402 10.1007/s00018-014-1693-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang H., Stoecklein N. H., Lin P. P., and Gires O. (2017) Circulating and disseminated tumor cells: diagnostic tools and therapeutic targets in motion. Oncotarget 8, 1884–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gorges T. M., Tinhofer I., Drosch M., Röse L., Zollner T. M., Krahn T., and von Ahsen O. (2012) Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 12, 178 10.1186/1471-2407-12-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Driemel C., Kremling H., Schumacher S., Will D., Wolters J., Lindenlauf N., Mack B., Baldus S. A., Hoya V., Pietsch J. M., Panagiotidou P., Raba K., Vay C., Vallböhmer D., Harréus U., et al. (2014) Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene 33, 4904–4915 10.1038/onc.2013.441 [DOI] [PubMed] [Google Scholar]
- 37. Sankpal N. V., Fleming T. P., Sharma P. K., Wiedner H. J., and Gillanders W. E. (2017) A double-negative feedback loop between EpCAM and ERK contributes to the regulation of epithelial-mesenchymal transition in cancer. Oncogene 36, 3706–3717 10.1038/onc.2016.504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sankpal N. V., Mayfield J. D., Willman M. W., Fleming T. P., and Gillanders W. E. (2011) Activator protein 1 (AP-1) contributes to EpCAM-dependent breast cancer invasion. Breast Cancer Res. 13, R124 10.1186/bcr3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sankpal N. V., Willman M. W., Fleming T. P., Mayfield J. D., and Gillanders W. E. (2009) Transcriptional repression of epithelial cell adhesion molecule contributes to p53 control of breast cancer invasion. Cancer Res. 69, 753–757 10.1158/0008-5472.CAN-08-2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Puram S. V., Tirosh I., Parikh A. S., Patel A. P., Yizhak K., Gillespie S., Rodman C., Luo C. L., Mroz E. A., Emerick K. S., Deschler D. G., Varvares M. A., Mylvaganam R., Rozenblatt-Rosen O., Rocco J. W., et al. (2017) Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Munz M., Fellinger K., Hofmann T., Schmitt B., and Gires O. (2008) Glycosylation is crucial for stability of tumour and cancer stem cell antigen EpCAM. Front. Biosci. 13, 5195–5201 [DOI] [PubMed] [Google Scholar]
- 42. Kuan I. I., Liang K. H., Wang Y. P., Kuo T. W., Meir Y. J., Wu S. C., Yang S. C., Lu J., and Wu H. C. (2017) EpEX/EpCAM and Oct4 or Klf4 alone are sufficient to generate induced pluripotent stem cells through STAT3 and HIF2α. Sci. Rep. 7, 41852 10.1038/srep41852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Munz M., Baeuerle P. A., and Gires O. (2009) The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 69, 5627–5629 10.1158/0008-5472.CAN-09-0654 [DOI] [PubMed] [Google Scholar]
- 44. Denzel S., Maetzel D., Mack B., Eggert C., Bärr G., and Gires O. (2009) Initial activation of EpCAM cleavage via cell-to-cell contact. BMC Cancer 9, 402 10.1186/1471-2407-9-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hsu Y. T., Osmulski P., Wang Y., Huang Y. W., Liu L., Ruan J., Jin V. X., Kirma N. B., Gaczynska M. E., and Huang T. H. (2016) EpCAM-regulated transcription exerts influences on nanomechanical properties of endometrial cancer cells that promote epithelial-to-mesenchymal transition. Cancer Res. 76, 6171–6182 10.1158/0008-5472.CAN-16-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kamp F., Winkler E., Trambauer J., Ebke A., Fluhrer R., and Steiner H. (2015) Intramembrane proteolysis of beta-amyloid precursor protein by γ-secretase is an unusually slow process. Biophys. J. 108, 1229–1237 10.1016/j.bpj.2014.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dickey S. W., Baker R. P., Cho S., and Urban S. (2013) Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell 155, 1270–1281 10.1016/j.cell.2013.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gerlach J. C., Foka H. G., Thompson R. L., Gridelli B., and Schmelzer E. (2018) Epithelial cell adhesion molecule fragments and signaling in primary human liver cells. J. Cell. Physiol. 233, 4841–4851 10.1002/jcp.26286 [DOI] [PubMed] [Google Scholar]
- 49. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.