Abstract
Drug abuse is a major public health problem in many countries in Europe and North America. Currently available platforms for drug abuse assessment are facing technical challenges of non-quantitation, inaccuracy, low throughput, incompatible with diverse complex specimens, long assay time and requirement of instrument and/or expertise for readout. Here, we report an integrated Competitive Volumetric-bar-chart Chip (CV-Chip) to assay multiple drug targets at the point-of-care (POC). To the best of our knowledge, it is the first time that a POC platform has been demonstrated to fully address the above-mentioned limitations. We applied this integrated CV-Chip platform to assay multiple drugs in 38 patient urine and serum samples and validated the on-chip results with an LC-MS/MS method, indicating a clinical sensitivity and specificity of 0.94 and 1.00, respectively. We further demonstrated that the combination of an on-chip blood separator with the CV-Chip enabled the platform to directly assay finger-prick whole blood samples, which has always been recognized as an ideal biospecimen for POC detections. In summary, this integrated CV-Chip is able to serve as a sensitive, accurate, fast, portable, readout visible, and minimally invasive platform for drug abuse assessment.
Keywords: drug abuse, point-of-care, microfluidics, finger-prick blood, blood separator
Graphical Abstract
INTRODUCTION
Drug abuse is a major public health concern, causing serious illness or injuries to millions of people.1 In 2014, 10% of the population aged 12 or older (~27.0 million) were found to be using illicit drugs in the United States.2 Fast, quantitative, non/minimally-invasive and cost-effective detection of abused drugs is of great importance, not only for clinical or forensic testing, but also for clinical trials to evaluate the efficacy of medications, which requires frequent analysis of patient samples to monitor changes in their illicit drug exposure over time.3–7
Currently, the most common methods for the measurement of illicit drugs are gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS).8–10 These instruments are sensitive, reliable and can provide accurate quantitative results, but they are commonly used in clinical lab11 though MS systems have the potential to be used for on-site analysis by combining with paper spray.12–14 Alternatively, low-cost technology platforms, such as commercially available point-of-care (POC) screening devices, facilitate multiplexed detection and give instant readout, but the results are qualitative/semi-quantitative and may suffer from false positives or false negatives.15–17 Other platforms, such as electrochemical immunosensors, offer advantages because they are sensitive, fast and easy to fabricate, but they still require accessory detector to measure the electrical or electrochemical signal.18–20
Microfluidics-based systems have become one potential approach for POC drug testing and personalized diagnostics, because of its potential capabilities of multiplexed and quantitative measurements, portability, low cost and high throughput.5,21–26 Over the last decade, there have been concerted efforts to develop microfluidic devices for drugs of abuse detection. Miyaguchi et al. embedded antibody-conjugated beads into the microdevice and measured methamphetamine in hair through a homemade apparatus.27 Andreou et al. presented a microfluidic device for the detection of methamphetamine in saliva using surface-enhanced Raman spectroscopy.5 These two promising methods can finish the measurement in 30 mins, but they can only detect one drug each time. Zhu et al. developed a microfluidic device for sample preparation and then coupled it with an HPLC-MS/MS system to analyze 14 drugs and metabolites in hair.28 Kirby et al. reported a method coupling microfluidics and a miniature MS platform to quantify multiple drugs of abuse in urine.29 These effective methods demonstrated high analytical sensitivity and multiplexed detection ability, but they still relied on the external MS systems for the quantitative readout.
As discussed above, currently available platforms for the detection of drugs of abuse are facing technical challenges of non-quantitation, inaccuracy, low-throughput, long assay time and requirement of instrument and/or expertise for readout. Here, we propose an integrated Competitive Volumetric-bar-chart Chip (CV-Chip) to assay multiple drug targets, which integrates the abilities to fully address the above-mentioned limitations. The CV-Chip platform was reported previously by us for qualitative analysis of biomarkers and drugs.30 It displayed visual positive or negative bar-chart results based on the direct competition of gas generated by the sample and the internal control. The current integrated CV-Chip is significantly improved over the previous platform in the following aspects: 1) it displays a quantitative readout, allowing it to be applied in more circumstances; 2) the assay time is significantly decreased from 1.5h to 10 min, making it more acceptable in on-site testing; 3) the ELISA procedure is simplified to one step, which may facilitate potential commercialization in future; 4) the ELISA probe is replaced with platinum nanoparticle (PtNP) from previous horseradish peroxidase (HRP), efficiently improving the analytical sensitivity over 10 times and also eliminating the incubation under 37°C; 5) it enables the detection of diverse complex specimens such as urine, serum and whole blood samples, thus greatly expanding its functions. We successfully applied the integrated CV-Chip to measure multiple drugs of abuse in urine and serum samples of 38 patients and further confirmed the results using an LC-MS/MS method. Furthermore, we combined an on-chip blood separator with the device to directly detect drugs in finger-prick whole blood. The results demonstrated that the platform could achieve good recovery rate for cocaine and amphetamine spiked in whole blood samples. In summary, the integrated CV-Chip provides a sensitive, accurate, fast, portable, readout visible, and minimal invasive platform for drugs of abuse assessment in urine, serum and whole blood.
EXPERIMENTAL
Materials and chemicals.
Glass slides (75×50×1 mm) were purchased from Corning Inc. (Corning, NY). SPR 220–7 and MF-CD26 was obtained from MicroChem Corp (Newton, MA) and Rohm and Haas Electronic Materials (Marlborough, MA), respectively. Hydrogen peroxide (H2O2, 35% wt in H2O), (3-glycidoxypropyl) trimethoxysilane (3-GPS), NH4F, HF, HNO3, toluene, ethanol, silicone oil, 3-Aminopropyl) triethoxysilane (APTES), glutaraldehyde, sodium cyanoborohydride, sulfuric acid, Tween 20, bovine serum albumin (BSA), Ethylenediaminetetraacetic acid dipotassium salt dehydrate (K2EDTA·2H2O) and the tested drugs were purchased from Sigma-Aldrich (St. Louis, MO). Phosphate buffered saline (PBS, 0.1 M, pH7.4) was obtained from Lonza (Allendale, NJ). Tridecafluoro-1, 1, 2, 2-tetrahydrooctyl-1-trichlorosilane was purchased from Pfaltz and Bauer (Waterbury, CT). Red ink was purchased from Fisher Scientific (Waltham, MA). Amorphous diamond-coated drill bits (0.031-inch cutter diameter) were purchased from Harvey Tool (Rowley, MA). Glass beads with various sizes were obtained from Corpuscular Inc. (Cold Spring, NY). All drug-BSA conjugates were obtained from Fitzgerald Industries International (Acton, MA). Drug standards were obtained from Sigma-Aldrich (Cocaine, C-008–1ML; Morphine, M-005–1mL; Amphetamine, A-007–1mL; Methadone, M-007–1mL; Methamphetamine, M-009–1mL) and Cerilliant (THC, T-005; Oxazepam, O-902). All anti-drug antibodies were purchased from Antibodies-Online (Atlanta, GA). FITC-conjugated, anti-mouse IgG secondary antibody was obtained from Abcam (Cambridge, MA). The abbreviations of COC, AMP, mAMP, BZO, OPI, THC and MTD were used for cocaine, amphetamine, methamphetamine, benzodiazepine, opiate, tetrahydrocannabinol and methadone, respectively. The abbreviation and cutoff values of the tested drugs are also shown in Table S1.
LC-MS/MS quantitation of drug concentrations.
Quantitation data of drug and metabolite concentrations in the urine and serum samples (#1- #20 and #29 - #38) were obtained from Associated Regional and University Pathologists, Inc. (ARUP laboratories, Salt Lake City, Utah) using its mass spectrometry methods. A LC-MS/MS method developed previously31 was used to quantify drug and metabolite concentrations in the clinical urine samples of #21- #28.
Fabrication of the integrated CV-Chip.
The pattern on the bottom and top plate of the integrated CV-Chip was fabricated following a standard photolithography process.25 Briefly, SPR220–7 molds (~10 μm) with the design were manufactured on glass slides (75×50×1 mm). Then, the glass slides were put into a glass etching solution (1:0.5:0.75 mol/L HF/NH4F/HNO3) to get the pattern (with a depth of ~ 50 μm). After that, sample inlets/outlets were obtained with a 0.03-inch diamond drill. Finally, the surface of the glass slides was treated with tridecafluoro-1, 1, 2, 2-tetrahydrooctyl-1-trichlorosilane for hydrophobic modification. Finally, the glass slides were cleaned with ethanol and dried with nitrogen gas.
Surface modification to the bottom plate.
Before the assay, the wells for ELISA (first lane) on the bottom plate required surface treatment.25,32 Previously, we used (3-glycidoxypropyl) trimethoxysilane (3-GPS) to introduce the epoxy group for antibody coating.25 Here, we changed the epoxy group to an aldehyde to increase the antibody coating efficacy.33,34 Briefly, wells of the first lane from the top and bottom ends were cleaned with piranha solution (H2SO4:H2O2=7:3) for 1 hour, rinsed with Millipore water, and dried with nitrogen gas. Next, 2 μL of 2.0% APTES in toluene was added into each well and incubated for 20 min and then heated at 120 °C for 15 min. Finally, the well was treated with 2 μL PBS solution containing 0.5% glutaraldehyde and 5 mM sodium cyanoborohydride for 1h and then washed with PBS.
Antibody coating in the ELISA wells.
The test for drugs of abuse is based on competitive ELISA principles. It requires only one antibody for the on-chip assay. Briefly, following surface modification, 2 μL capture antibody (~10μg/mL) was added to the ELISA well (the first lane from the top and bottom ends) and incubated 1h at room temperature. Then the wells were washed with PBS (containing 0.05% v/v Tween) three times and blocked with 5% w/v BSA for 1 h. Then 2 μL drug-BSA-PtNP solution was added and incubated for 1 h followed by washing.
Assembly of the device.
After the antibody coating (see Supplementary Information), the device was assembled as previously described 25,32,35. Briefly, 5 μL of silicone oil was carefully added to the non-pattern area of the top device plate (with the patterns facing up). Then, the top plate was assembled with the bottom plate. The silicone oil was evenly distributed on the two plates by sliding the plates against each other repeatedly, effectively sealing the two glass slides together and preventing solution leakage. This silicone oil-based sealing is also effective to prevent reagent evaporation during mediumtime storage (e.g., one-week) at room temperature.
Drug detection on the device.
The drug detection is divided into four steps. First, sample/control and buffer solutions are loaded into the device after the assembly. Second, a horizontal slide of the top plate will connect the sample/control and buffer solution. Red ink and H2O2 are also loaded at this step. In the third step, a pipette was used to generate a negative pressure at the inlet, pulling the sample/control and buffer solutions through the ELISA wells. In the final step, an oblique slide of the top plate will initiate the reaction between PtNPs and H2O2 to produce oxygen gas, which will finally push the red ink to generate the bar-chart readout.
Manufacture of the on-chip blood separator.
A big glass bead (~ 800 μm) was initially placed into a 0.1– 20 μL pipette tip using a tweezer. This bead was trapped in the tip. Then 10 μL of ~100 μm beads (dissolved in PBS in 107/mL) were added into the tip, followed by filling the tip with 10 μL of ~15 μm beads (dissolved in PBS in 108/mL). The small sized beads were stacked on the top of the big one, forming a microfilter. These glass beads were blocked with BSA before use to avoid non-specific binding. The manufactured microfilter tip was pretreated with K2EDTA (0.5 % wt in distilled water) to prevent potential coagulation of the whole blood sample. Then a 20-μL pipette with a sample-filled tip can be inserted into the microfilter tip. Push the pipette to let the blood sample go through the glass beads and the plasma can be obtained outside the microfilter tip.
RESULTS AND DISCUSSION
Working principle of the integrated CV-Chip.
CV-Chip is based on SlipChip technology.25,36 Design and performance of the CV-Chip has been demonstrated previously and the results proved that the device could assay drugs of abuse with good analytical sensitivity, wide dynamic range and others.30 But the assay time is relatively long (>1.5h). It is necessary to reduce the assay time to facilitate forensic drug analysis or drug assessment to monitor medication regimen or other POC applications.15,16 To efficiently decrease the assay time, the integrated CV-Chip combined a sample loading component to the original CV-Chip (Figure 1a, b), in which a series of sample loading and washing steps can be accomplished in one step. Sequential delivery of liquid segments containing samples and ELISA reagents has been reported to generate a rapid and simple ELISA reaction and was applied to detect protein biomarkers others.37–40However, no such integrated system focused on drug detection. Since drugs are detected based on competitive ELISA, it requires fewer steps than sandwich ELISA for protein markers, which makes the integrated CV-Chip much simpler than the reported platforms.
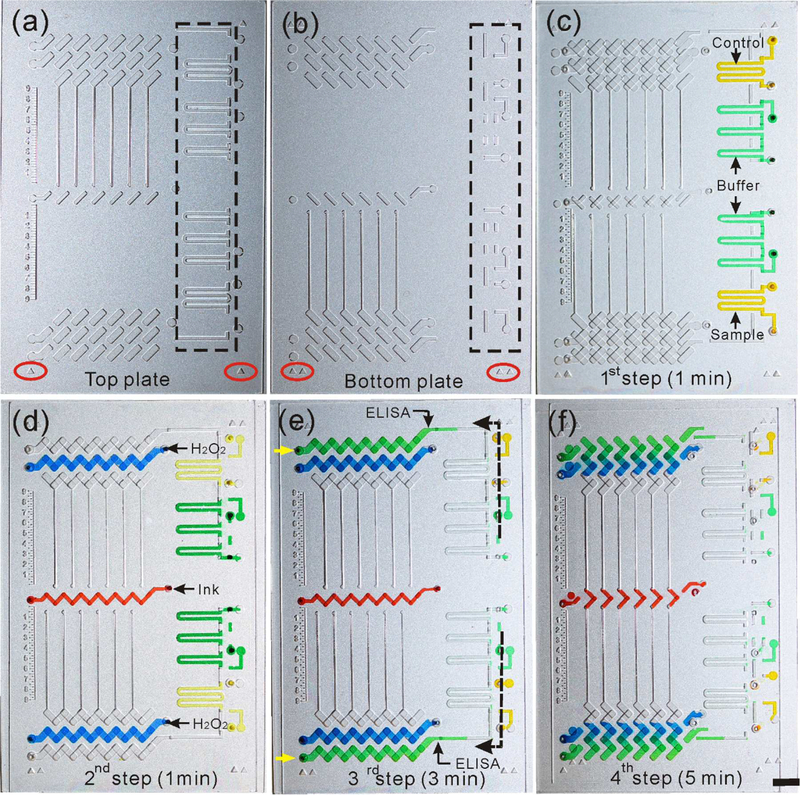
Figure 1.
Working principle of the integrated CV-Chip. (a) Top plate and (b) bottom plate of the device. The black dotted rectangle showed the channels for sample loading. The red ellipse showed the triangles for device alignment during the operation. (c) The first step of the operation. Sample/control (yellow dye) and washing buffer (green dye) were loaded into the device. It takes 1 min for this step. (d) The second step of the operation. Horizontally move the top plate to the left connected the sample/control and buffer channels. The buffer was separated to three segment by air, which was used to mimic three times of washing. Red ink (red dye) and H2O2 (blue dye) were loaded in this step. This step takes 1 min. (e) The third step of the operation. Sample/control and buffer flowed through the ELISA wells by applying a pipette at the inlet (pointed by the yellow arrow) to generate a negative pressure. It takes 3 min to finish the ELISA reaction and washing steps. (f) The fourth step of the operation. An oblique sliding of the top plate against the bottom plate connected the ELISA well with the H2O2, which initiated the reaction between PtNPs and H2O2 to generate oxygen and further produce the bar-chart readout. It takes 5 min to get the readout. Scale bar, 0.5 cm for (a-f).
As shown in Figure 1 and Figure S1, the integrated CV-Chip is composed of one bottom plate and one top plate and the operation has four steps. Firstly, sample and control (solution with the cutoff value of the target drug) and washing buffer were loaded to the relevant channels (Figure 1c). This step takes about 1 min. Here, the sample channel requires minimal solution (~ 2 μL), which makes it suitable to do minimally-invasive blood sampling. As a second step, the top plate was slid horizontally to make the wells and channels connect as shown in Figure 1d. Sample/control and washing buffer segments were formed in the side channels. There were three buffer segments separated by air, mimicking triplicate washing steps. Red ink and H2O2 were also loaded at this step. The second step takes about 1 min. Subsequently, a negative pressure, generated using a 20 μL pipette at the left inlet, drew the solutions through the ELISA lane (Figure 1e). In this step, target drugs in the sample competed with the preloaded drug-BSA-PtNPs to bind to the surface immobilized antibodies, followed by the washing buffer. It takes 3 min to finish this step. In the last step, an oblique slide of the top glass plate connected the ELISA well with the H2O2 well and initiated the reaction between PtNPs and H2O2, which generates oxygen gas (Figure 1f). The oxygen generated at the two ends will compete to push the red ink to produce a bar-chart in the center part of the device. The displacement of the ink bars can be quantified using the on-chip rulers etched beside the reading channel. The whole assay was finished in 10 min. We designed some markers on the top and bottom plate of the device to facilitate the alignment during the operation (Figure 1a-c). To further improve the user interface and ease the operation, we can design on-chip guide tracks on the device, as shown in Figure S2, to assist the sliding.
Sensitivity improvement to the integrated CV-Chip.
The ELISA reaction takes about 3 min on the integrated CV-Chip, which may raise concerns about the sensitivity of the platform, but similar integrated platforms have demonstrated sufficient analytical sensitivity for detecting protein biomarkers.37–40 Here, to make sure the analytical sensitivity can meet the requirements for testing drugs of abuse, we improved the assay in two aspects, one was the surface modification for antibody immobilization and the other was the ELISA probe.
Previously, we used (3-glycidoxypropyl) trimethoxysilane (3-GPS) to introduce the epoxy group on the surface, which can react with the amine group of antibodies.25,30,32 This modification method is simple, but the coating density is not high enough. To make the antibody coating more efficient, we functionalized the glass surface with aldehyde groups instead of the epoxy functional group (Figure S3). This results in a more stable and efficient bioconjugation scheme.33,34 Our results showed that the aldehyde-modified surface was more homogeneous and demonstrated a higher efficiency for antibody immobilization than the epoxy-modified surface (Figure S4). In addition to the surface modification, we also changed the ELISA probe to PtNP from HRP. The readout of previous CV-Chip is based on nitrogen gas, which is produced by an HRP mediated reaction between luminol and H2O2.30 Though drug-HRP derivatives are commercially available and convenient, this reaction is not highly sensitive and it requires incubation at 37°C to achieve sufficient analytical sensitivity. Alternatively, PtNPs are reported to be very sensitive catalysts for the generation of oxygen gas from H2O2 at room temperature.41 We compared the activity of the two probes, PtNPs and HRP, for gas generation, which demonstrated that the analytical sensitivity was increased over 10 times by using PtNPs rather than HRP (Movie S1 and S2). Drug-PtNP conjugates were prepared by immobilizing drug-BSA derivatives onto the PtNPs. Successful conjugation was verified using dynamic light scattering measurements before and after immobilizing the drug-BSA conjugates on the PtNPs (Figure S5)
Test of spiked cocaine samples on the device.
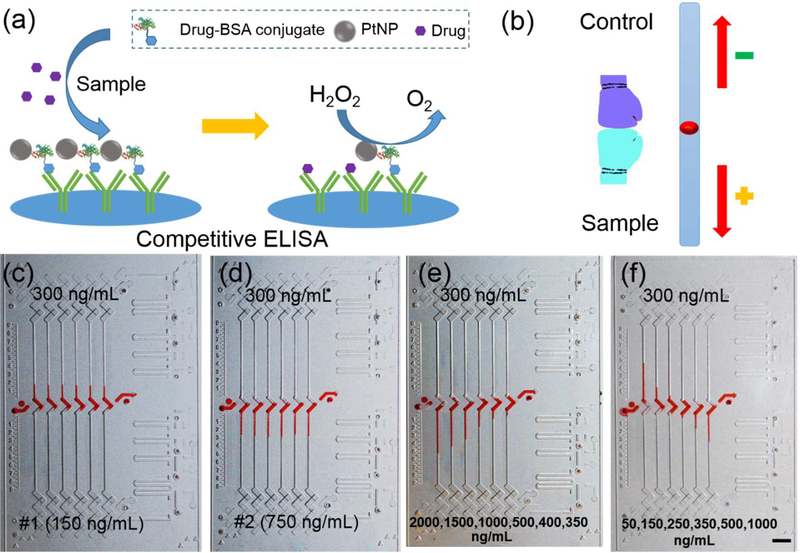
Figure 2a shows the competitive ELISA performed on the device. Higher drug concentrations in the sample will compete to bind to more antibodies, rendering less PtNP-probes left on the surface and resulting in less oxygen generated. Therefore, a positive sample will generate a downward bar on the device and a negative sample will generate an upward bar (Figure 2b). A Comparison of the assay time on the previous CV-Chip and the integrated CV-Chip was shown in Table S2. We first tested two cocaine samples spiked in PBS. The control with a cutoff value of 300 ng/mL was loaded at the top end; a negative sample (150 ng/mL) and a positive sample (750 ng/mL) were loaded at the bottom end in the two tests, respectively. As shown in Figure 2c and d, the negative sample showed upward homogeneous bars in the six channels, while the positive sample displayed six downward homogeneous bars. The intra-assay %CV for the two samples were 3.6 % and 4.2%, respectively. We then tested the two samples spiked in blank urine and serum substrates. The results in Figure S6 suggested that the complex matrices in human urine or serum provide little or no interference with the performance of the device, demonstrating the compatibility of our method to diverse complex specimens. Furthermore, we tested a series of positive samples (spiked in urine) with gradient concentrations, which produced a gradient bar-chart as shown in Figure 2e. Then three negative and three positive samples spiked in urine were tested simultaneously and the results provided in Figure 2f. The inter-assay %CV for all these tested samples were within 8.5%. These results demonstrated that the integrated CV-Chip had sufficient analytical sensitivity to distinguish the minimum differences between the control and samples.
Figure 2.
Test of spiked cocaine samples. (a) Scheme of competitive ELISA on the device. Antibodies have been coated on the glass surface and drug-BSA-PtNPs have bound to the antibodies. When a sample containing the target drugs is added, the drugs will compete with the drug-BSA-PtNPs to bind to the antibodies. Hence, more drugs will render less drug-BSA-PtNPs left, and result in less oxygen gas produced. (b) Principle of readout. The control with the cutoff value of the target drug is loaded at the top end, while the sample is loaded at the bottom end. The readout is based on the competition of oxygen gas generated by the control and sample; negative samples generate more gas than the control and produce an upward ink bar; conversely, positive samples produce a downward inks bar. (c, d) Testing results of a negative sample (c) and a positive sample (d) spiked in PBS. A solution with 300 ng/mL of cocaine was loaded at the top to serve as the control. Both of them generated a uniform bar-chart but with different directions. (e) Testing results of a series of positive samples spiked in urine. These samples with gradient concentrations generated a gradient bar-chart. (f) Testing results of three negative and three positive samples spiked in urine. Scale bar, 0.5 cm for (c-f).
Test of patient urine samples.
After the test of spiked samples, we moved to test patient urine samples by using the integrated CV-Chip. Urine drug quantitation, though difficult due to the inter- and intra-individual variability in urine volume and drug excretion/renal function, is still an attractive approach worthy of further development.42 The rationale is: 1) Urine is usually the preferred matrix in the clinic for determining the presence or absence of drugs because it has a 1- to 3-day window of detection for most drugs and/or their metabolites and is currently the most extensively validated biological specimen for drug testing.7 Therefore, while drug testing may be performed by either testing urine, serum, oral fluids, sweat or hair, urine drug test (UDT) is predominantly used in the clinic. 2) UDT is also the simplest, most non-invasive approach for biological sample screening and widely accepted as the gold standard.43–45
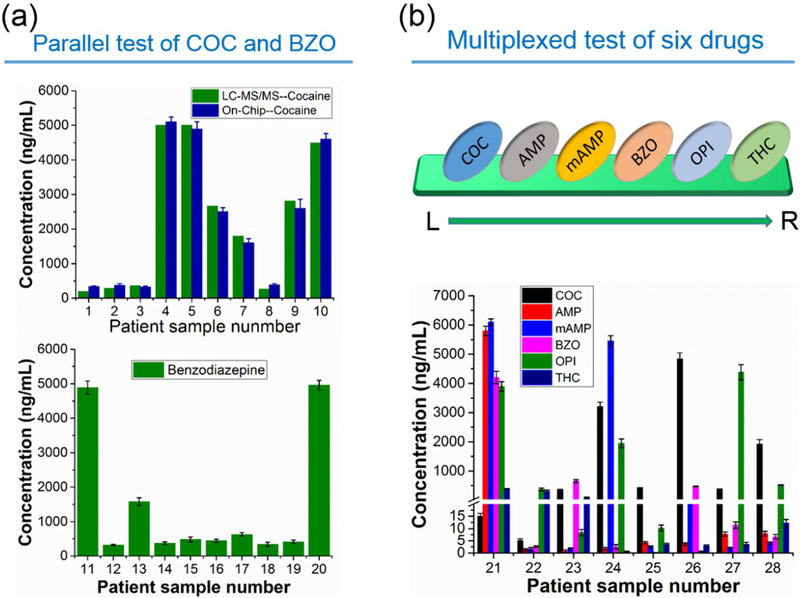
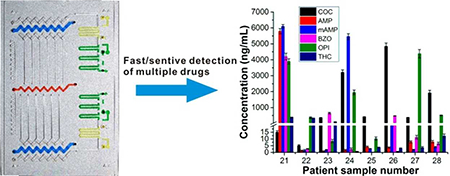
Before the test of patient samples, we first generated the calibration curves on the device by using spiked urine samples for six abused drugs. Since negative and positive samples produce upward and downward bars respectively, the calibration curves were plotted separately for these two groups of samples (Figure S7, S8). These response curves were pseudo-exponential rather than linear, which may be attributed to consumption of H2O2 during the reactions, thermodynamic effects and others.25,32 Then we tested COC in 10 patient urine samples (#1- #10) and BZO in another 10 patient samples (#11 – #20), one sample per test. The results were shown in Figure 3a. We further tested another eight patient samples (#21 – #28) in a multiplex assay to detect six drugs (COC, AMP, mAMP, BZO, OPI, and THC) per chip (Figure 3b). All patient samples were tested three times and the inter-assay %CV for all the samples were within 9.5%. Though the six drugs have different cutoff values and various concentration distributions, the device was still able to obtain quantitative readouts for all the drugs in these samples, demonstrating the advantages of wide dynamic range and good analytical specificity and sensitivity. The on-chip data were further confirmed by LC-MS analyses (Table S3, S4 and S5) and we found that results obtained from the two methods were in good agreement with each other (Figure 3b).
Figure 3.
Assay of patient urine samples. (a) Parallel test of patient urine samples for COC (#1– #10, top image) and BZO (#11–#20, bottom image). Patient sample was measured for each single drug in the six channels in each test. LC-MS/MS data were also plotted side by side with the on-chip data for cocaine (5000 ng/mL was used for #4 and #5). Data shown as mean of three independent measurements ± S.D. (b) Results of multiplexed detection of six drugs for samples #21–#28. Six drugs of COC, AMP, mAMP, BZO, OPI and THC were tested simultaneously in the six channels from left to right. Data shown as mean of three independent measurements ± S.D.
Test of MTD in patient serum samples.
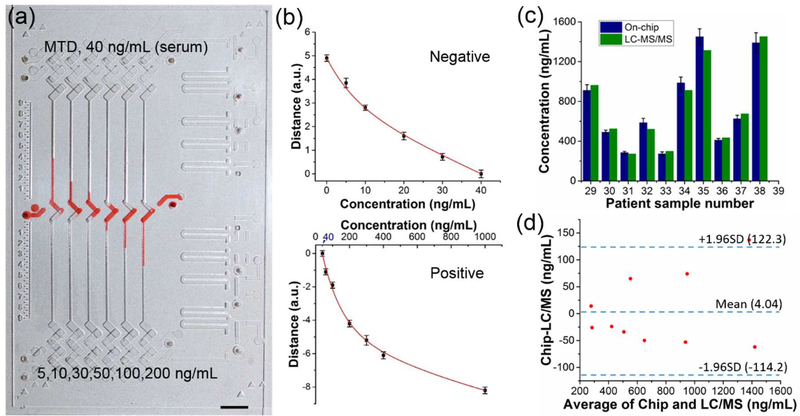
In general, the detection window is longest in hair, followed by urine, sweat, oral fluid, and blood.21 In blood, most drugs of abuse can be detected at the low nanogram per milliliter level for 1 or 2 days. Though blood drug detection windows are not as long as urine, blood drug concentration most accurately reflects the frequency and amount of drug of abuse.44,46 For this reason, we also validated the capability of the integrated CV-Chip in serum drug tests, using MTD as an example. The cutoff value of MTD in serum is 40 ng/mL. A series of spiked samples were first tested. As shown in Figure 4a, small differences in drug concentration among the samples could be effectively visualized on the device. Then, the calibration curves for the negative and positive MTD samples were generated by using spiked serum samples (Figure 4b). We tested 10 serum samples (#29 – #38) on the device. The on-chip results (with inter-assay CV% less than 10%) and the LC-MS/MS data were shown in Figure 4c and Table S6, which demonstrated that the results of both types of tests were correlated. We further used Bland–Altman analysis to examine the correlation between the two methods. Figure 4d shows that they agree well with each other, indicating the accuracy and reliability of our platform.
Figure 4.
Test of MTD in patient serum samples. (a) Bar-chart result of the test of MTD in spiked urine samples. These negative and positive samples showed proper inks bars on the device. Scale bar, 0.5 cm. (b) Calibration curves for negative (top) and positive (bottom) samples. (c) Bar graphs of the MTD concentration in the patient serum samples measured by the device (navy blue) and LC-MS/MS (olive), side-by-side. The value of methadone (not its metabolite) was used in the plot for LC-MS/MS data. On-chip data shown as mean of three independent measurements ± S.D. (d) Bland−Altman analysis for assessing agreement between results from the integrated CV-Chip and LC-MS/MS. The 95% confidence interval on the mean value is displayed in the graph. The results showed good correlation between the two methods.
Taken together all the on-chip results for the 38 samples, the 95% confidence interval (CI) for the clinical sensitivity and specificity were calculated to be 0.94 (CI: 0.8378– 0.9794) and 1.00 (CI: 0.8794– 1) respectively (Table S7), which is comparable to that of commercial drug test kits (such as Triage TOX Drug screen).15,17
Combination of an on-chip blood separator with the device for finger-prick whole blood assay.
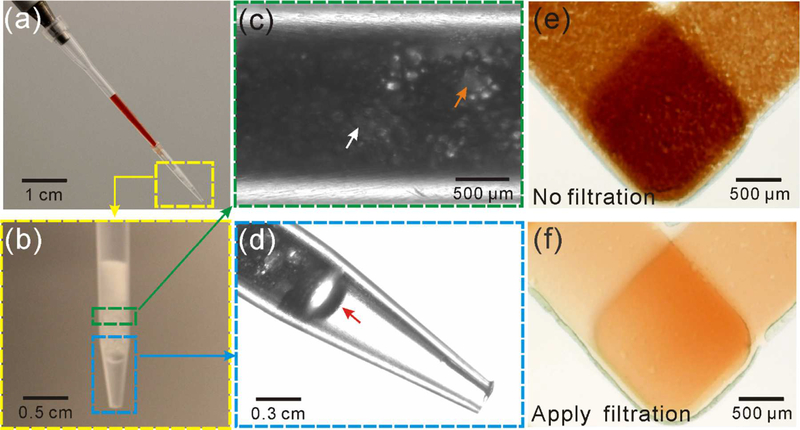
When it comes to POC detection, an ideal sample of choice is finger-prick whole blood,47–50 while it still remains a great challenge for current platforms. The integrated CV-Chip was validated with serum samples. To expand the function of our platform in POC tests, we move to test whole blood sample directly. To assay whole blood, it is necessary to separate blood cells before the on-chip assay. Centrifugation can be applied to get serum or plasma from blood samples. But for POC testing in resource limited settings, access to centrifuges is not so convenient though there have been some portable centrifuges.51,52 There are some commercial blood separators based on filter paper (GE Healthcare Life Science and Pall Life Science), which are used to get plasma from whole blood in a simple way. But this kind of separator is only ideal for lateral flow assays where the filter paper itself is the substrate for the assay. To retrieve serum from the separator, it requires combination with other complex platforms and it takes a relatively long time (>10 min) to get sufficient volume of serum.53 In addition to the filter paper-based blood separator, microbeads are integrated into microfluidic chip to separate blood cells.54 Here, to get plasma simply, fast and easily, we developed a blood separator based on pipette tip and glass beads, which served as a microfilter. This microfilter-based blood separator can separate blood cells based on size: plasma can go through the gap between the beads, while blood cells are trapped on the top or in the gap. Figure 5 showed the details for the glass beads-based microfilter tip and the results before and after applying the filtration. More than 90% of blood cells were removed. The plasma separation efficiency of the microfilter tip is about 80% compared to conventional centrifugation method (800g, 10 min). We also evaluated the effect of potential hemolysis on the assay during the microfiltration and no significant influence was found on the detection results (Supplementary Information). This home-made tip makes it easy, fast and efficient to directly assay whole blood (such as finger-prick blood) on POC platforms, which also minimizes handling and cryo-preservation artifacts that may hinder assay reliability. To further increase the ease of operation, we can integrate the beads-based microfilter on the device by adding a specific area at the sample inlet for preloaded microbeads. Briefly, a trap area at the inlet with a different height from the channels/wells can be designed to stack the microbeads, which enables plasma to go through and blood cells to be trapped.
Figure 5.
Microfilter-based blood separator. (a) A pipette tip with blood inserted into a filter tip with glass beads. Push the pipette to let the serum go through the glass beads, while the blood cells were trapped in the gaps between the beads. (b) Filter tip filled with three kinds of glass beads (600 μm, 100 μm and 15 μm). (c) Glass beads inside the tip. The yellow and white arrow indicates one of the 100 μm and 15 μm beads, respectively. (d) An amplified view of the filter tip end. The yellow and red arrow indicates the 100 μm and 600 μm bead, respectively. (e, f)Blood samples loaded into the device without (e) or with the filter (f). The graphs confirmed the performance of this home-made microfilter tip.
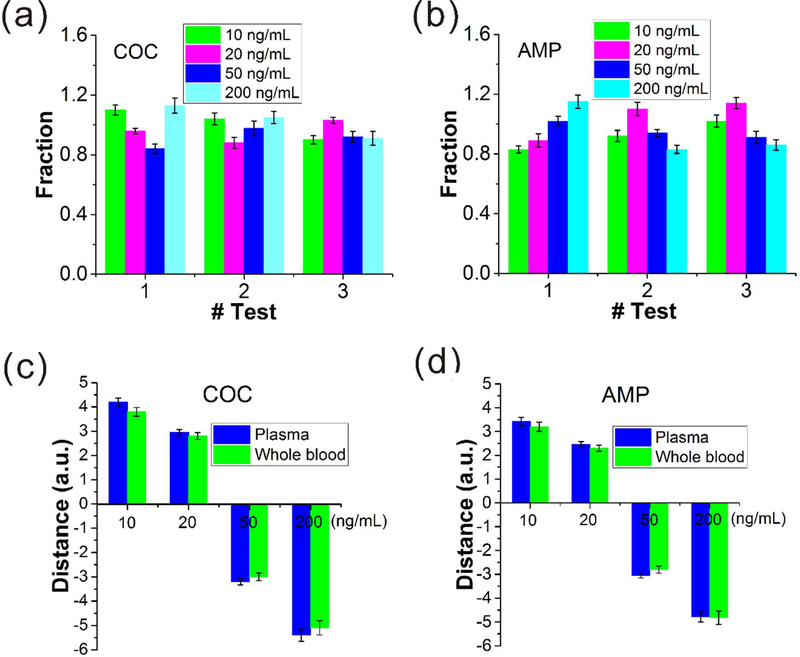
We then applied the glass beads-based microfilter tip to test spiked blood samples. Since it requires ~ 2 μL of sample each time, a single finger-prick can provide enough blood for obtaining 3–5 samples (Figure S9). The cutoff values of COC and AMP in serum are 30 ng/mL. We prepared 4 COC samples and 4 AMP samples with concentrations of 10, 20, 50 and 200 ng/mL by spiking the drugs into finger-prick whole blood collected from drug-free volunteers. All these samples were tested three times. Then the tests were examined by the analysis of the recovery rate (amount of drug measured on the device/amount of drug spiked), a factor used to determine assay accuracy, with an acceptable range between 0.8 and 1.2.32,55 As shown in Figure 6a and b, the fraction of recovery for the drugs in all the tests were within the desired range. Furthermore, we prepared 8 plasma samples (plasma was obtained by centrifuging the blood drawn from drug-free volunteers) by spiking COC and AMP with the same concentrations as the blood samples and compared the detection results with those obtained from the blood samples. Figure 6c and d revealed that the results were in good correlation, proving that the integration of microfilter tip enabled the integrated CV-Chip to efficiently assay finger-prick blood samples for drug testing. For most commercial drug test kits (e.g., At Home Drug Test kit), they are able to test one or multiple drugs in urine and give out fast qualitative results. But our CV-Chip device is able to quantitate illicit drugs in urine, serum and whole blood for multiple drugs. For other POC platforms used for drug test, most of them require external instruments/devices to assist the readout but our device can get the quantitation data based on naked-eyes, making it a portable and low-cost method.
Figure 6.
Test of samples spiked in whole blood. (a, b) Recovery rate of four spiked samples for COC (a) and AMP (b) in three tests. The whole blood samples were applied the filtration before loaded into the device. All the tests were within the acceptable range. Error bar represents S.D. in the six parallel channels. (c, d) Comparison of ink bar distances between the plasma samples and whole blood samples (applied filtration). These two kinds of samples produced similar bar lengths, confirming the good performance of the on-chip blood separator and the whole detection system.
CONCLUSIONS
In this manuscript, we presented an integrated platform for assaying drugs of abuse. The integrated CV-Chip is very suitable for the detection of abused drugs because: 1) the cutoff value of the targeted drug can be directly put at one end of the device to serve as the internal control and it will generate clear negative or positive bar chart; 2) small differences between the sample and the cutoff can be distinguished based on the direct competition; 3) though different drugs have different cutoff values, we only need to put different controls at one end to do multiplexed detection. The device is able to complete the assay of multiple drugs in 10 min with less than 2 μL of sample solution. By using this platform, we tested multiple illicit drugs in 38 patient urine and serum samples and confirmed these on-chip results with an LC-MS/MS method. We also demonstrated that the combination of an on-chip blood separator with the device enabled the platform to assay finger-prick whole blood. In summary, the integrated CV-Chip enables the detection of drugs of abuse in urine, serum and whole blood to be fast, sensitive, quantitative, cost-effective and minimally invasive.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the funding support from NIH-R01 DA035868, R01 CA180083, R56 AG049714, and R21 CA191179.
REFERENCES
- (1).United Nations Office on Drugs and Crime, World Drug Report 2015. (United Nations publication, Sales No. E.15.XI.6).
- (2).Center for Behavioral Health Statistics and Quality.(2015). Behavioral health trends in the United States: Results from the 2014 National Survey on Drug Use and Health (HHS Publication No. SMA 15–4927, NSDUH Series H-50).
- (3).Marks V Ann. Clin. Biochem 1988, 25, 220–225. [DOI] [PubMed] [Google Scholar]
- (4).George S; Braithwaite RA Clin. Chem 2002, 48, 1639–1646. [PubMed] [Google Scholar]
- (5).Andreou C; Hoonejani MR; Barmi MR; Moskovits M; Meinhart CD Acs Nano 2013, 7, 7157–7164. [DOI] [PubMed] [Google Scholar]
- (6).Li GH; Brady JE; Chen QX Accident Anal. Prev 2013, 60, 205–210. [DOI] [PubMed] [Google Scholar]
- (7).Vindenes V; Lund HME; Andresen W; Gjerde H; Ikdahl SE; Christophersen AS; Oiestad EL Forensic Sci. Int 2012, 219. [DOI] [PubMed] [Google Scholar]
- (8).Eichhorst JC; Etter ML; Hall PL; Lehotay DC Methods Mol. Biol 2012, 902, 29–41. [DOI] [PubMed] [Google Scholar]
- (9).Lee Y; Lai KK; Sadrzadeh SM Clin, Biochem 2013, 46, 1118–1124. [DOI] [PubMed] [Google Scholar]
- (10).Tang MHY; Ching CK; Lee CYW; Lam YH; Mak TWL J. Chromatogr. B 2014, 969, 272–284. [DOI] [PubMed] [Google Scholar]
- (11).Singh RJ; Eisenhofer G Clin. Chem 2007, 53, 1565–1567. [DOI] [PubMed] [Google Scholar]
- (12).Espy RD; Manicke NE; Ouyang Z; Cooks RG Analyst 2012, 137, 2344–2349. [DOI] [PubMed] [Google Scholar]
- (13).Su Y; Wang H; Liu JJ; Wei P; Cooks RG; Ouyang Z Analyst 2013, 138, 4443–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Espy RD; Teunissen SF; Manicke NE; Ren Y; Ouyang Z; van Asten A; Cooks RG Analytical Chemistry 2014, 86, 7712–7718. [DOI] [PubMed] [Google Scholar]
- (15).Attema-de Jonge ME; Peeters SY; Franssen EJ J. Emerg. Med 2012, 42, 682–691. [DOI] [PubMed] [Google Scholar]
- (16).Beck O; Carlsson S; Tusic M; Olsson R; Franzen L; Hulten P Scand J. Clin. Lab Invest 2014, 74, 681–686. [DOI] [PubMed] [Google Scholar]
- (17).Lin CN; Nelson GJ; McMillin GA J Anal Toxicol 2013, 37, 30–36. [DOI] [PubMed] [Google Scholar]
- (18).Feng TT; Wang Y; Qiao XW Electroanalysis 2017, 29, 662–675. [Google Scholar]
- (19).Chikkaveeraiah BV; Bhirde AA; Morgan NY; Eden HS; Chen XY Acs Nano 2012, 6, 6546–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).de la Escosura-Muniz A; Parolo C; Merkoci A Mater. Today 2010, 13, 17–27. [Google Scholar]
- (21).Ng AHC; Wheeler AR Clin. Chem 2015, 61, 1233–1234. [DOI] [PubMed] [Google Scholar]
- (22).Sackmann EK; Fulton AL; Beebe DJ Nature 2014, 507, 181–189. [DOI] [PubMed] [Google Scholar]
- (23).Song YJ; Huang YY; Liu XW; Zhang XJ; Ferrari M; Qin LD Trends Biotechnol. 2014, 32, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Teerinen T; Lappalainen T; Erho T Anal Bioanal Chem 2014, 406, 5955–5965. [DOI] [PubMed] [Google Scholar]
- (25).Song Y; Zhang Y; Bernard PE; Reuben JM; Ueno NT; Arlinghaus RB; Zu Y; Qin L Nat. Commun 2012, 3, 1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhu BW; Niu ZQ; Wang H; Leow WR; Wang H; Li YG; Zheng LY; Wei J; Huo FW; Chen XD Small 2014, 10, 3625–3631. [DOI] [PubMed] [Google Scholar]
- (27).Miyaguchi H; Takahashi H; Ohashi T; Mawatari K; Iwata YT; Inoue H; Kitamori T Forensic Sci. Int 2009, 184, 1–5. [DOI] [PubMed] [Google Scholar]
- (28).Zhu KY; Leung KW; Ting AKL; Wong ZCF; Ng WYY; Choi RCY; Dong TTX; Wang TJ; Lau DTW; Tsim KWK Anal. Bioanal. Chem 2012, 402, 2805–2815. [DOI] [PubMed] [Google Scholar]
- (29).Kirby AE; Lafreniere NM; Seale B; Hendricks PI; Cooks RG; Wheeler AR Anal. Chem 2014, 86, 6121–6129. [DOI] [PubMed] [Google Scholar]
- (30).Li Y; Xuan J; Xia T; Han X; Song Y; Cao Z; Jiang X; Guo Y; Wang P; Qin L Anal. Chem 2015, 87, 3771–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Cao Z; Kaleta E; Wang PJ Anal. Toxicol. 2015, 39, 335–346. [DOI] [PubMed] [Google Scholar]
- (32).Li Y; Xuan J; Song YJ; Qi WJ; He BS; Wang P; Qin LD Acs Nano 2016, 10, 1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Goddard JM; Erickson D Anal. Bioanal. Chem 2009, 394, 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lin MY; Ho FH; Yang CY; Yeh JA; Yang YS Chem Commun 2012, 48, 4902–4904. [DOI] [PubMed] [Google Scholar]
- (35).Li Y; Xuan J; Song Y; Wang P; Qin L Lab Chip 2015, 15, 3300–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Du WB; Li L; Nichols KP; Ismagilov RF Lab Chip 2009, 9, 2286–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chin CD; Laksanasopin T; Cheung YK; Steinmiller D; Linder V; Parsa H; Wang J; Moore H; Rouse R; Umviligihozo G; Karita E; Mwambarangwe L; Braunstein SL; van de Wijgert J; Sahabo R; Justman JE; El-Sadr W; Sia SK Nat. Med 2011, 17, 1015–U1138. [DOI] [PubMed] [Google Scholar]
- (38).Yang F; Zuo XL; Li ZH; Deng WP; Shi JY; Zhang GJ; Huang Q; Song SP; Fan CH Adv. Mater 2014, 26, 4671–4676. [DOI] [PubMed] [Google Scholar]
- (39).Laksanasopin T; Guo TW; Nayak S; Sridhara AA; Xie S; Olowookere OO; Cadinu P; Meng FX; Chee NH; Kim J; Chin CD; Munyazesa E; Mugwaneza P; Rai AJ; Mugisha V; Castro AR; Steinmiller D; Linder V; Justman JE; Nsanzimana S; Sia SK Sci. Transl. Med 2015, 7. [DOI] [PubMed] [Google Scholar]
- (40).Song Y; Wang Y; Qi W; Li Y; Xuan J; Wang P; Qin L Lab Chip 2016, 16, 2955–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Song Y; Xia X; Wu X; Wang P; Qin L Angew. Chem. Int. Ed. Engl 2014, 53, 12451–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Xiong LJ; Wang R; Liang C; Teng XM; Jiang FL; Zeng LB; Ye HY; Ni CF; Yuan XL; Rao YL; Zhang YR J. Chromatogr. A 2015, 1395, 99–108. [DOI] [PubMed] [Google Scholar]
- (43).Mehta N; Kunkel F; Shaparin N; Stripp R; Borg D; Fey EJ Pain 2015, 16, S7–S7. [Google Scholar]
- (44).Wille SMR; Baumgartner MR; Di Fazio V; Samyn N; Kraemer T Bioanalysis 2014, 6, 2193–2209. [DOI] [PubMed] [Google Scholar]
- (45).Zorec-Karlovsek M; Niedbala S; Fritch D; Steinmeyer S; Manns A Forensic Sci. Int 2003, 136, 310–310. [Google Scholar]
- (46).Vidal JC; Bertolín JR; Bonel L; Asturias L; Arcos-Martínez MJ; Castillo JR J. Phamaceut. Biomed 2016, 125, 54–61. [DOI] [PubMed] [Google Scholar]
- (47).St John A; Price CP Clin. Biochem. Rev 2014, 35, 155–167. [PMC free article] [PubMed] [Google Scholar]
- (48).Gaieski DF; Drumheller BC; Goyal M; Fuchs BD; Shofer FS; Zogby K West J. Emerg. Med 2013, 14, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Haleyur Giri Setty MK; Hewlett IK AIDS Res. Treat 2014, 2014, 497046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Steinmetzer K; Seidel T; Stallmach A; Ermantraut EJ Clin. Microbiol 2010, 48, 2786–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Travassos MA; Beyene B; Adam Z; Campbell JD; Mulholland N; Diarra SS; Kassa T; Oot L; Sequeira J; Reymann M; Blackwelder WC; Pasetti MF; Sow SO; Steinglass R; Kebede A; Levine MM Am. J. Trop. Med. Hyg 2015, 93, 416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Bhamla MS; Benson B; Chai C; Katsikis G; Johri A; Prakash M Nat. Biomed. Engin 2017, 1, 0009. [Google Scholar]
- (53).Liu C; Liao SC; Song J; Mauk MG; Li X; Wu G; Ge D; Greenberg RM; Yang S; Bau HH Lab Chip 2016, 16, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Shim JS; Ahn CH Lab Chip 2012, 12, 863–866. [DOI] [PubMed] [Google Scholar]
- (55).Castanheira AP; Barbosa AI; Edwardsa AD; Reis NM Analyst 2015, 140, 5609–5618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.