Abstract
Background
The concept of personalized medicine defines a promising approach in cancer care. High-throughput genomic profiling of tumor specimens allows the identification of actionable mutations that potentially lead to tailored treatment for individuals’ benefit. The aim of this study was to prove efficacy of a personalized treatment option in solid tumor patients after failure of standard treatment concepts.
Results
Final analysis demonstrates that 34 patients (62%) had a longer PFS upon experimental treatment (PFS1) when compared to previous therapy (PFS0); PFS ratio > 1.0 (p = 0.002). The median PFS under targeted therapy based on molecular profiling (PFS1) was 112 days (quartiles 66/201) and PFS0 = 61 days (quartiles 51/92; p = 0.002). Of the 55 patients, 31 (56%) showed disease control (DCR), consisting of 2 (4%) patients which achieved a complete remission, 14 (25%) patients with a partial remission and 15 (27%) patients who had a stabilization of disease. Median OS from start of experimental therapy was 348 days (quartiles 177/664).
Conclusion
The prospective trial EXACT suggests that treatment based on real-time molecular tumor profiling leads to superior clinical benefit.
Materials and Methods
In this prospective clinical phase II trial, 55 cancer patients, after failure of standard treatment options, aimed to achieve a longer progression-free survival on the experimental treatment based on the individual’s molecular profile (PFS1) when compared to the last treatment given before (PFS0). The personalized medicine approach was conceived to be clinical beneficial for patients who show a PFS ratio (PFS 1/PFS0) of > 1.0.
Keywords: precision medicine, molecular profile
INTRODUCTION
Etiological concepts on cancer development, malignant growth and tumor propagation have led to the discovery of various molecular driver mechanisms [1, 2]. Based on these advances, medical oncology has started to enter an era of individualized medicine where treatment selection is becoming tailored to druggable molecular targets [1–3]. This individualized treatment concept is mainly based on molecular and genetic characterization of the tumors including next generation sequencing (NGS), which allows to align the most appropriate treatment according to the patient’s disease [4]. Although there is a general acceptance towards such individualized approach, thereby stratifying and subgrouping patients to improve the quality of clinical care in oncology, molecular profiling has just started to assist prediction of the drug’s clinical benefit by identifying the most responsive patient subgroup. Specific molecules, such as EGFR, KRAS, BRAF, estrogen receptor, HER2, etc. are already associated with patient populations that respond more likely to target therapy [5–8]. However, most genes have been largely constrained to organ-specific analysis (e.g. estrogen receptor /HER2 for breast cancer, KRAS for colorectal cancer, BRAF for melanoma, etc.). Recently, molecular profile-based therapy has shown clinical benefit in different cancer types independent of their primary tumor site of origin [2, 9, 10]. Such a tailored treatment strategy revealed to be an effective approach to increase progression free survival (PFS), when compared to the patients’ most recent standard treatment regimen.
Here we aimed to verify the concept of individualized therapy by using molecular profiling of patient tumors in the era of immunotherapies. We could demonstrate that in certain patients an individualized treatment upon a real-time assessment of the tumor’s molecular profile might reflect an efficient strategy to control the disease.
RESULTS
Patient characteristics and study algorithm
Patients with metastatic solid tumors after failure of any standard treatment options were real-time biopsated and fresh tumor material was investigated for possible druggable targets via NGS, IHC and FISH. Results of the molecular profile were discussed by a multidisciplinary team for treatment decision. From 114 patients screened, 55 patients (48%) were eligible to start treatment upon the molecular profile derived from real-time biopsy. 35 (64%) men and 20 (36%) women with a mean age of 61 years (± 14) where included. At time of censoring (12/31/2016), 27 patients (48.2%) were still alive while 29 (51.8%) patients were deceased. Most common tumor types analyzed were colon cancer (n = 7; 12.3%), cholangiocellular cancer (n = 6; 10.5%), head and neck cancer (n = 5; 8.8%), thyroid cancer (n = 5; 8.8%) and lymphomas (n = 4; 7%) (Table 1).
Table 1. Patient characteristics and tumor types in EXACT.
| Gender | n | % |
|---|---|---|
| Male | 35 | 63.6 |
| Female | 20 | 36.4 |
| Age | years | Range |
| 61 | 23-84 | |
| n | % | |
| Alive | 27 | 49.1 |
| Dead | 28 | 50.9 |
| Tumor type | n | % |
| CRC | 7 | 12.3 |
| CCC | 6 | 10.5 |
| Head & neck cancer | 5 | 8.8 |
| Thyroid cancer | 5 | 8.8 |
| Lymphoma | 4 | 7.0 |
| Mesothelioma | 3 | 5.3 |
| CUP | 3 | 5.3 |
| HCC | 2 | 3.5 |
| Esophageal cancer | 2 | 3.5 |
| PEComa (renal) | 2 | 3.5 |
| Ovarian cancer | 2 | 3.5 |
| PDAC | 2 | 3.5 |
| NET | 2 | 3.5 |
| Adrenal cancer | 1 | 1.8 |
| Hepatoid Peritoneum | 1 | 1.8 |
| Vulvar carcinoma | 1 | 1.8 |
| Breast cancer | 1 | 1.8 |
| Prostate cancer | 1 | 1.8 |
| Urothelial cancer | 1 | 1.8 |
| Testicular cancer | 1 | 1.8 |
| Multiple Myeloma | 1 | 1.8 |
| Endometrial cancer | 1 | 1.8 |
| Melanoma | 1 | 1.8 |
| Total | 55 | 100.0% |
CRC: colorectal cancer; CCC: cholangiocarcinoma; CUP: cancer of unknown primary; HCC: hepatocellular carcinoma; PEComa: perivascular epithelioid cell tumor of the kidney; PDAC: pancreatic ductal adenocarcinoma; NET: neuroendocrine tumor.
Molecular profile
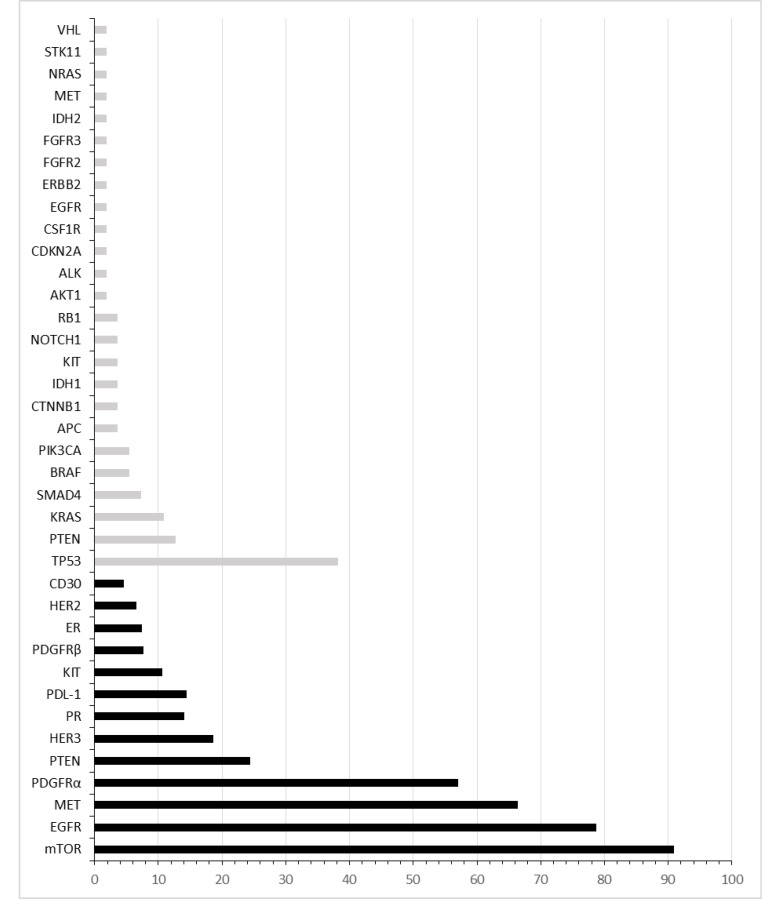
Tumor samples of 55 patients were real-time analyzed via NGS, IHC and FISH using a platform of a parallel study (unpublished data). The most frequent somatic mutations observed were in TP53 (38%), PTEN (13%) and KRAS (11%). Frequently observed increases in protein expression as assessed by IHC were EGFR (75%), phospho-mTOR (75%), MET (69%) and PDGFRa (55%) (Figure 1; for histochemistry examples refer to Figure S1). FISH was performed, but FISH results alone did not lead to treatment suggestions (data not shown).
Figure 1. Molecular profile assessed by immunohistochemistry and next generation sequencing.
All immunoreactivity positive cases (black bars) and patients with genetic mutations (grey bars) are represented as percentage of enrolled subjects.
Response rate
Patients were treated with kinase inhibitors, growth factor receptor antibodies in combination with hormone therapy or chemotherapy or were given immunotherapy (Table 2). The median PFS1/PFS0 ratio of all patient was 1.35 (quartile 0.7/2.9). From 55 patients, 34 (62%) showed a PFS1/PFS0 ratio >1.0 under the experimental therapy (PFS1) (p = 0.002) (Table 4). Thus, the primary study aim was met as the null hypothesis was rejected.
Table 2. Treatment of 34 patients (62%) based on molecular profiling with PFS1/PFS0 >1.0°.
| Tumor§ | Genetic profile | IHC | Therapy | Evidence supporting treatment decision | |
|---|---|---|---|---|---|
| CUP | Atypical ALK rearrangement | MET+, PR+ | Crizotinib Tamoxifen | [19, 20] | |
| CUP | KITmutation | KIT+ | Imatinib | [21] | |
| CUP | mTOR+ | Everolimus | [22, 23] | ||
| Thyroid | BRAF mutation (V600E) | Vemurafenib | [24] | ||
| Thyroid | mTOR+ EGFR+ |
Temsirolimus Cetuximab | [25, 26] | ||
| Thyroid | PDL-1+ | Pembrolizumab | [27, 28] | ||
| CCC | EGFR+ (RAS wildtype) | Irinotecan Cetuximab | [29] | ||
| CCC | EGFR+ (RAS wildtype) | Irinotecan Cetuximab | [29] | ||
| CCC | EGFR+ (RAS wildtype) | Irinotecan Cetuximab | [29] | ||
| CCC | FGFR2 mutation | MET+ | Regorafenib | [30, 31] | |
| HCC | EGFR+ | Irinotecan Cetuximab | [32, 33] | ||
| HCC | EGFR+ | Folfox Cetuximab | [34] | ||
| GEC | HER2+ | Trastuzumab Pertuzumab | [35] | ||
| GEC | HER2+ | Folfiri Trastuzumab | [35] | ||
| CRC | PDL1– MSHI high |
Pembrolizumab* | [36-38] | ||
| CRC | MSHI high | Pembrolizumab* | [36-38] | ||
| CRC | MET+ mTOR+ | Temsirolimus Bevacizumab beyond progression | [26] | ||
| CRC | KIT mutation | MET+ | Sunitinib | [39] | |
| RRC | EGFR+ HER2+ HER3+ |
Trastuzumab Lapatinib | [40] | ||
| Prostate | mTOR+ | Temsirolimus | [25, 41] | ||
| H&N | PDL-1+ | Pembrolizumab* | [42, 43] | ||
| H&N | mTOR+ | Carboplatin Everolimus | [44, 45] | ||
| H&N | PDGFRa+ | Docetaxel Sunitinib | [46] | ||
| NHL | mTOR+ | Everolimus | [22, 23] | ||
| NHL | CD30+ | Brentuximab | [47] | ||
| Ovarian | ER+ PR+ PDGFa+ mTOR+ |
Letrozol Bevacizumab | [48, 49] | ||
| Myeloma | PDL-1+ | Pembrolizumab | [50] | ||
| SCLC | EGFR mutation | EGFR + | Afatinib | [51] | |
| Pleuramesothelioma | PDL-1 + | Pembrolizumab | [52] | ||
| Pleuramesothelioma | PDGFRa+ PDGFRb+ |
Sunitinib | [39, 53] | ||
| Pleuramesothelioma | PDGFRb+ | Palbociclib | [54] | ||
| PEComa | PDGFRa+ PDGFRb+ EGFR+ |
Sunitinib | [39, 55] | ||
| Endometrial | ER+ mTOR+ | Exemestan Everolimus | [56] | ||
| Vulva | PDL-1+ | Pembrolizumab | [52] | ||
§CUP: cancer of unknown primary, CCC: cholangiocarcinoma, HCC: hepatocellular carcinoma, GEC: gastroesophageal cancer, CRC: colorectal cancer, RRC: renal cancer, H&N: head and neck cancer, SCLC: small cell lung cancer, NHL: non Hodgkin lymphoma, PEComa: perivascular epithelioid cell tumor of the kidney.
°The cut-offs values for the selection of putative druggable targets were determined as follows: PDL-1: presence of positive tumor cells, Tumor Proportion Score ≥1, mTOR: IHC score: 200–300, HER2: score ≥2 and confirmed amplification by FISH, KIT: IHC Score 100–300, PR: Allred Score ≥6, EGFR: IHC score 200–300, PDGFRα: IHC score 100–300, PDGFRβ: IHC score 200–300, ER: Allred Score ≥3, CD30: % of positive lymphoma cells, MET: IHC Score ≥2+ and HER3: IHC Score 100–300.
*At the time of treatment decision, pembrolizumab was not approved by neither the (U. S. Food and Drug Administration) FDA nor the European Medicines Agency (EMA).
Table 4. Survival data.
| Parameter | Median | Quartile |
|---|---|---|
| OS | 348 | 177/664 |
| PFS 0 | 61 | 51/92 |
| PFS 1 | 112 | 66/201 |
| Ratio PFS1/PFS0 | 1.35 | 0.7/2.9* |
Quartiles cover the 25% and 75% range; *for the ratio PFS1/PFS0, the range (min/max) is displayed.
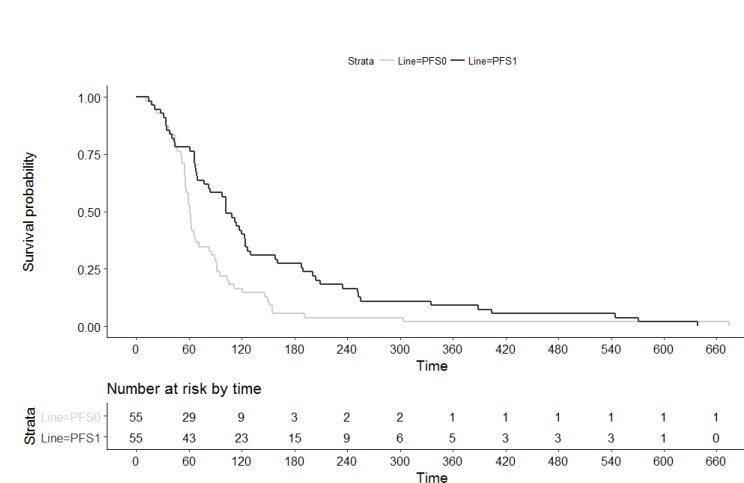
29% of patients (n = 16) showed an overall response according to RECIST. The disease control rate was 56% (n = 31). Out of 55 patients, 2 (4%) did show a complete remission and 14 patients (25%) had a partial remission while 15 patients (27%) had a stable disease according to RECIST 1.1 criteria (Table 3). 38% (n = 21) did not benefit from therapy and were progressive. Note that three patients were still under experimental therapy and were not evaluated for treatment response at the day of censoring (Figure 2).
Table 3. Treatment response rate upon experimental therapy.
| Treatment response | Number of patients | % of total |
|---|---|---|
| CR | 2 | 4 |
| PR | 14 | 26 |
| SD | 15 | 27 |
| PD | 21 | 38 |
| Ongoing | 3 | 5 |
| ORR | 16 | 29 |
| DCR | 31 | 56 |
| Total (n, %) | 55 | 100 |
CR: complete remission; PR: partial remission; SD: stable disease; PD: progressive disease; ORR: overall response rate (patients with complete or partial remissions); DCR: disease control rate (patients with complete remission, partial remission or stable disease).
Figure 2. Progression free survival upon last standard therapy (PFS0) and experimental individualized therapy (PFS1).
Survival data
The median PFS of the individualized treatment (PFS1) was 112 days (quartiles 66/201). The median PFS of the therapy given before (PFS0) was 61 days (quartiles 51/92). The overall survival at the day of censoring was 348 days (quartiles177/664). In the Wilcoxon Signed Rank Test PFS1 was significantly longer than PFS0 (p = 0.002). The 95% bootstrap confidence interval of the median of the ratio PFS1/PFS0 is [1.086; 2.034].
DISCUSSION
In this study we present a prospective clinical phase II trial to determine efficacy of individualized therapy. Tissue derived from real-time biopsies of 55 patients suffering from refractory cancer was characterized for their molecular profile and individualized treatment was suggested by a multidisciplinary team. From the 114 patients tested, 55 (48%) started treatment according to their tumor’s molecular profile. The median PFS under experimental treatment (PFS1) was 112 days and was significant longer than the median PFS upon the previous treatment. Notably and even more important, on the individual base, 34 (62%) patients achieved a longer PFS than upon the previous treatment, thus, the null hypothesis was clearly rejected. Thus, the majority of the patients had a clinical benefit from this individualized treatment concept in a late line setting. Furthermore, at time of censoring the median overall survival was 348 days (quartiles 177/664).
The ability to identify driver mutations in tumors had led to the introduction targeted treatments interfering with these tumor drivers. Although certain driver mutation are not tumor type specific, targeted treatments have so far been approved by authorities rather by origin of the tumor than by its mutation. In this context, the example of BRAF inhibitory drugs have been shown to be active in BRAF V600 mutated tumors [11], but when starting EXACT, approval of these drugs was limited to BRAF mutated melanoma. That BRAF inhibition might be effective in BRAF mutated tumors outside of melanomas is supported in this study. To address a potential biological activity of targeted drugs in rare mutations, so called basket trials are addressing a certain molecular mutation to assess whether a treatment concept is similarly amenable to identical targeted treatment. Other actionable biomarkers such as PD-L1 expression are thought to be less sensitive, but might be used as stratification factors in clinical trials. As somatic tumor mutations are amended to instability, real-time biopsy seems to be adequate for patients in treatment-refractory cancers [12–16].
In a meta-analysis, it was demonstrated that this concept has been achieved best results for efficacy of precision medicine in phase II trials [17]. This is consistent with our own observations of clonal selection during previous treatment and support the conclusion that targeted treatment should be exclusively suggested upon molecular profiling of real-time biopsies rather than from archival tissue.
In one of the first individualized treatment studies in cancer, von Hoff et al. reported beneficial effects of molecular profiling of patient`s tumors [16]. In this particular study 27% of the patients had a benefit from an individual target approach by resulting in a significant longer PFS when compared with the PFS of the previous regimen of the same patient (PFS ratio ≥ 1.3; 95% CI, 17% to 38%; one-sided, one-sample P = .007). Although a cross over between trials is not possible, our results of 58% of patients demonstrated an increased PFS under targeted treatment is stimulating. This might partially explained by the fact of progress in diagnostic techniques and therapeutic options within the last few years. In fact, we were considering beside other options checkpoint inhibitors as potential treatment options.
By recent advances of targeted treatment studies it becomes mandatory to evaluate potential effectiveness of marketed, targeted anticancer drugs when applied outside of their approved indications. In this context, ASCO is developing the Targeted Agent and Profiling Utilization Registry (TAPUR) study, which aims to facilitate patient access to marketed agents that are predicted to be beneficial based on analysis of patients’ tumor’s genomic profile. Furthermore, and most importantly, by capturing their outcomes in a prospective database this approach will improve the understanding which treatment concepts might lead to patient benefit. TAPUR is thereby conceptually similar to both the ongoing major initiative of the U.S. NCI, the Molecular Analysis for Therapy Choice (MATCH) trial as well as the AcSé program being conducted by the French National Cancer institute.
Although our study bears some weaknesses such as the fact that it was not randomized and the sample size was small, we feel it is important to emphasize that molecular profiling- based treatment decisions is feasible for a subgroup of patients to improve their prognosis when no standard treatment options are available. The use of a matched control arm to evaluate the benefit of molecular profiling in pretreated cancer patients is urged and the focus of a consecutive trial. With increasing number of new targeted treatment options available, the concept of an individualized therapy will likely be in focus of future treatment concepts in cancer.
MATERIALS AND METHODS
The study was conducted in accordance with the Declaration of Helsinki and was approved by the institutional ethic committee of the Medical University of Vienna (Nr.1541/2012).
Study design
The primary objective of the study was to prospectively validate the benefit of an individualized treatment concept based on molecular profiling from paraffin-embedded tumor tissue sections obtained before the start of treatment (real time biopsy).
Rejection of the null hypothesis was defined as follows: ≤40% of this patient population would have a PFS ratio of > 1.0. Thus, the individual patient served as his own control. The alternative proportion P1 (PFS ratio > 1.0) is set at least to 55% using a one-sided exact binomial test at a significance level of 0.0250. The null hypothesis can be rejected, if at least 30 out of 55 patients treated show a PFS ratio >1.0.
Furthermore, treatment response (ORR) and overall survival (OS) was assessed by by CT scans at 8-week intervals during therapy and judged according to the Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1 and evaluated as secondary end points.
Patients
Patient eligibility criteria included informed consent, any histologic type of metastatic cancer without further standard treatment option, tumor progression by RECIST criteria, age ≥ 18 years, ECOG performance status 0-1. Fresh tumor biopsy was obtained for pathologic analysis. Biopsies were performed by a heterogeneous group of different surgical techniques routinely used at the Department of Interventional Radiology. Patients were thought to be eligible if treatment could be initiated upon the molecular profile derived from real-time biopsy (see Supplementary Figure 2).
Tissue samples
Tissues from metastatic cancer patients were formalin fixed and paraffin embedded at the Department of Pathology, Medical University Vienna. for histology and molecular diagnostics.
Cancer gene panel sequencing
The DNA library was generated by multiplex polymerase chain reaction (PCR) with the Ion AmpliSeq Cancer Hotspot Panel v2™ (Life Technologies, Carlsbad, CA). The panel covers mutation hotspots of 50 genes, mostly oncogenes and tumor suppressor genes that are frequently mutated in tumors (ABL, AKT, ALK, APC, ATM, BRAF, CDH, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAS, GNAQ, HNF1A, HRAS, IDH1, JAK2, JAK3, IDH2, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL). Sequencing was performed with an Ion Torrent PGM™ (Life Technologies). Nonsynonymous mutations detected with the Ion Torrent PGM™ were verified by capillary sequencing. The sequencing of PCR products was carried out with the BigDyeR Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA). The resulting DNA fragments were purified with the DyeEx 96 Kit (Qiagen) and sequenced with a 3500 Genetic Analyzer (Applied Biosystems). For sequence analysis we employed the SeqScape Version 2.7 software (Applied Biosystems).
Immunohistochemistry
Immunohistochemistry was performed with a Ventana Benchmark Ultra stainer (Ventana, Tucson, AZ). The following antibodies were employed: ALK (clone 1A4; Zytomed, Berlin, Germany), CD30 (clone BerH2; Dako, Vienna, Austria), CD20 (clone L26; Dako), EGFR (clone 3C6; Ventana), Estrogen-receptor (clone SP1; Ventana), HER2 (clone 4B5; Ventana), HER3 (clone SP71; Abcam), KIT (clone 9.7; Ventana), MET (clone SP44; Ventana), phospho-mTOR (clone 49F9; Cell Signalling, Danvers, MS), PDGFRA (rabbit polyclonal; Thermo Fisher Scientific), PDGFRB (clone 28E1, Cell Signalling), PD-L1 (clone E1L3N; Cell Signalling), Progesteron-receptor (clone 1E2; Ventana), PTEN (clone Y184; Abcam) and ROS1 (clone D4D6; Cell Signalling). For details refer to the Supplementary Files (Supplementary Table 1).
The diagnostic sensitivity and specifity of the antibodies has been validated at the Department of Pathology at the Medical University Vienna. For the validation appropriate positive and negative tissue controls were employed (see Supplementary Figure 1). Furthermore, the omission of primary antibodies and the replacement of primary antibodies by antibodies of the same species, isotype and concentration, having no known reactivity against human tissue, served as negative reagent controls. The antibodies employed in this study have been institutionally approved for the application in routine histopathological diagnostics. The antibodies to ALK, CD30, EGFR, HER2 and MET are additionally licensed in vitro diagnostics, the antibody to CD20 is CE marked.
For the evaluation of staining intensities with antibodies to EGFR, phospho-mTOR, PDGFRA, PDGFRB and PTEN an immunohistochemial score was determined by multiplying the percentage of positive cells by their respective staining intensity (0 = negative, 1 = weak, 2 = moderate, 3 = strong). Immunohistochemical score (maximum 300) = (% negative × 0) + (% weak × 1) + (% moderate × 2) + (% strong × 3).
ALK, CD30, CD20 and ROS1 stainings were categorised as positive or negative with the percentage of reactive neoplastic cells, but without scoring of staining intensities. ALK or ROS1 positive cases were consecutively interrogated for the presence of a respective gene translocation by FISH. HER2 staining was graduated according to the guidelines of the company Dako for the Dako HercepTestR with possible scores 0 (negative), 1+ (negative), 2+ (positive), 3+ (positive). HER2 2+ cases were further analysed by HER2 in-situ hybridization to verify a HER2 gene amplification. For PD-L1 the percentage of tumor cells with a membranous staining, irrespective of staining intensity, was determined (so-called “tumor proportion score”). MET staining was graduated according to a published scoring system that evaluated both staining intensity (negative, weak, moderate, or strong) and prevalence of these intensities in tumor cells [18].
Fluorescence in situ hybridization (FISH)
FISH was performed with 4 μm thick FFPE tissue sections. The following FISH probes were employed: ALK (2p23.1; Abbott, Abbott Park, IL), RET (10q11; Kreatech, Berlin, Germany), PTEN (10q23.31)/Centromer 10) and ROS1 (Zytovision, Bremerhaven, Germany). 200 cell nuclei per tumor were evaluated. To detect HER2, two diagnostic systems were applied: FISH (PathVysion II; Abbott) and CISH (Ventana Medical Systems by Roche Diagnostics).
Treatment algorithm
Patients with refractory metastatic cancer without any standard treatment options according to NCCN guidelines and/or local guidelines were included. Potential therapeutic targets in individual patient’s tumor sections were individualized by genomic tumor profiling (NGS and FISH) in combination with immunohistochemistry. The generated data were biostatistically combined with the actual data from clinical trials thus resulting in the identification of druggable targets (drivers) with the highest likelihood of response in each individual patient. Based on these recommendations, previous lines of treatment and patient performance status, final treatment decisions were made by at least two different medical oncologists in a molecular tumor board with participation of pathologists, radiologists and translational scientists. Only agents with marketing authorization and established safety profile for combinations were used in off-label settings in this trial.
Statistics
For statistical analysis whether PFS0 and PFS1 are different, a Wilcoxon Signed Rank Test was performed. To find possible patient characteristics that influence the difference between PFS0 and PFS1 a linear model was performed with the difference (PFS1 - PFS0) as dependent, and age as well as gender as independent variables. Furthermore a 95% confidence interval for the median of the ratio PFS1/PFS0 was calculated through bootstrap with 1000 samples. Note that three of the 55 patients had still an ongoing therapy, but were treated as if the therapy has already ended. Analysis was performed using statistical Software R Version 3.3.0.
SUPPLEMENTARY MATERIALS FIGURES AND TABLE
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 2.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, Shyr Y, Camidge DR, Sequist LV, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elez E, Argiles G, Tabernero J. First-Line Treatment of Metastatic Colorectal Cancer: Interpreting FIRE-3, PEAK, and CALGB/SWOG 80405. Curr Treat Options Oncol. 2015;16:52. doi: 10.1007/s11864-015-0369-x. [DOI] [PubMed] [Google Scholar]
- 4.Wong DJ, Ribas A. Targeted Therapy for Melanoma. Cancer Treat Res. 2016;167:251–262. doi: 10.1007/978-3-319-22539-5_10. [DOI] [PubMed] [Google Scholar]
- 5.Andre F, Bachelot T, Commo F, Campone M, Arnedos M, Dieras V, Lacroix-Triki M, Lacroix L, Cohen P, Gentien D, Adelaide J, Dalenc F, Goncalves A, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15:267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 6.Meric-Bernstam F, Brusco L, Shaw K, Horombe C, Kopetz S, Davies MA, Routbort M, Piha-Paul SA, Janku F, Ueno N, Hong D, De Groot J, Ravi V, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol. 2015;33:2753–2762. doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohal DP, Rini BI, Khorana AA, Dreicer R, Abraham J, Procop GW, Saunthararajah Y, Pennell NA, Stevenson JP, Pelley R, Estfan B, Shepard D, Funchain P, et al. Prospective Clinical Study of Precision Oncology in Solid Tumors. J Natl Cancer Inst. 2015;108 doi: 10.1093/jnci/djv332. [DOI] [PubMed] [Google Scholar]
- 8.Stockley TL, Oza AM, Berman HK, Leighl NB, Knox JJ, Shepherd FA, Chen EX, Krzyzanowska MK, Dhani N, Joshua AM, Tsao MS, Serra S, Clarke B, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8:109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jabbour E. Chronic myeloid leukemia: First-line drug of choice. Am J Hematol. 2016;91:59–66. doi: 10.1002/ajh.24249. [DOI] [PubMed] [Google Scholar]
- 10.Rocco G, Morabito A, Leone A, Muto P, Fiore F, Budillon A. Management of non-small cell lung cancer in the era of personalized medicine. Int J Biochem Cell Biol. 2016;78:173–179. doi: 10.1016/j.biocel.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, Wolf J, Raje NS, Diamond EL, Hollebecque A, Gervais R, Elez-Fernandez ME, Italiano A, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373:726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Jr, Tsao A, Stewart DJ, Hicks ME, Erasmus J, Jr, Gupta S, Alden CM, Liu S, Tang X, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Tourneau C, Delord JP, Goncalves A, Gavoille C, Dubot C, Isambert N, Campone M, Tredan O, Massiani MA, Mauborgne C, Armanet S, Servant N, Bieche I, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 14.Papadimitrakopoulou V, Lee JJ, Wistuba II, Tsao AS, Fossella FV, Kalhor N, Gupta S, Byers LA, Izzo JG, Gettinger SN, Goldberg SB, Tang X, Miller VA, et al. The BATTLE-2 Study: A Biomarker-Integrated Targeted Therapy Study in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2016;34:3638–3647. doi: 10.1200/JCO.2015.66.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsimberidou AM, Wen S, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Aldape K, Ye Y, Kurzrock R, Berry D. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res. 2014;20:4827–4836. doi: 10.1158/1078-0432.CCR-14-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Von Hoff DD, Stephenson JJ, Jr, Rosen P, Loesch DM, Borad MJ, Anthony S, Jameson G, Brown S, Cantafio N, Richards DA, Fitch TR, Wasserman E, Fernandez C, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 17.Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, Lazar V, Kurzrock R. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33:3817–3825. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spigel DR, Ervin TJ, Ramlau RA, Daniel DB, Goldschmidt JH, Jr, Blumenschein GR, Jr, Krzakowski MJ, Robinet G, Godbert B, Barlesi F, Govindan R, Patel T, Orlov SV, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2013;31:4105–4114. doi: 10.1200/JCO.2012.47.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan VC. Tamoxifen as the first targeted long-term adjuvant therapy for breast cancer. Endocr Relat Cancer. 2014;21:R235–246. doi: 10.1530/ERC-14-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazandjian D, Blumenthal GM, Chen HY, He K, Patel M, Justice R, Keegan P, Pazdur R. FDA approval summary: crizotinib for the treatment of metastatic non-small cell lung cancer with anaplastic lymphoma kinase rearrangements. Oncologist. 2014;19:e5–11. doi: 10.1634/theoncologist.2014-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Hadiya BM, Bakheit AH, Abd-Elgalil AA. Imatinib mesylate. Profiles Drug Subst Excip Relat Methodol. 2014;39:265–297. doi: 10.1016/B978-0-12-800173-8.00006-4. [DOI] [PubMed] [Google Scholar]
- 22.Hasskarl J. Everolimus. Recent Results Cancer Res. 2014;201:373–392. doi: 10.1007/978-3-642-54490-3_23. [DOI] [PubMed] [Google Scholar]
- 23.Rocha T, Fortier SC, Fischer T, Perini GF, Gaiolla RD, Fogliatto L, Delamain MT, Costa AFD, Castro NS, Barretos WG, Souza CA, Buccheri V, Chiattone CS. Everolimus as a single agent in refractory or relapsed Hodgkin’s lymphoma: the Brazilian Named Patient Program Experience. Rev Bras Hematol Hemoter. 2017;39:216–222. doi: 10.1016/j.bjhh.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brose MS, Cabanillas ME, Cohen EE, Wirth LJ, Riehl T, Yue H, Sherman SI, Sherman EJ. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: a non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:1272–1282. doi: 10.1016/S1470-2045(16)30166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malizzia LJ, Hsu A. Temsirolimus, an mTOR inhibitor for treatment of patients with advanced renal cell carcinoma. Clin J Oncol Nurs. 2008;12:639–646. doi: 10.1188/08.CJON.639-646. [DOI] [PubMed] [Google Scholar]
- 26.Merchan JR, Qin R, Pitot H, Picus J, Liu G, Fitch T, Maples WJ, Flynn PJ, Fruth BF, Erlichman C. Safety and activity of temsirolimus and bevacizumab in patients with advanced renal cell carcinoma previously treated with tyrosine kinase inhibitors: a phase 2 consortium study. Cancer Chemother Pharmacol. 2015;75:485–493. doi: 10.1007/s00280-014-2668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandini S, Massi D, Mandala M. PD-L1 expression in cancer patients receiving anti PD-1/PD-L1 antibodies: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2016;100:88–98. doi: 10.1016/j.critrevonc.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Chowdhury S, Veyhl J, Jessa F, Polyakova O, Alenzi A, MacMillan C, Ralhan R, Walfish PG. Programmed death-ligand 1 overexpression is a prognostic marker for aggressive papillary thyroid cancer and its variants. Oncotarget. 2016;7:32318–32328. doi: 10.18632/oncotarget.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim R, Sun Y, Im SA, Hsieh RK, Yau TK, Bonaventura A, Cheirsilpa A, Esser R, Mueser M, Advani S. Cetuximab plus irinotecan in pretreated metastatic colorectal cancer patients: the ELSIE study. World J Gastroenterol. 2011;17:1879–1888. doi: 10.3748/wjg.v17.i14.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, Humblet Y, Bouche O, Mineur L, Barone C, Adenis A, Tabernero J, Yoshino T, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303–312. doi: 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 31.Sun W, Patel A, Normolle D, Patel K, Ohr J, Lee JJ, Bahary N, Chu E, Streeter N, Drummond S. A phase 2 trial of regorafenib as a single agent in patients with chemotherapy refractory advanced and metastatic biliary adenocarcinoma/cholangiocarcinoma. Cancer. 2018 Dec 18 doi: 10.1002/cncr.31872. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faugeras L, Dili A, Druez A, Krug B, Decoster C, D’Hondt L. Treatment options for metastatic colorectal cancer in patients with liver dysfunction due to malignancy. Crit Rev Oncol Hematol. 2017;115:59–66. doi: 10.1016/j.critrevonc.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 33.Bardelli A, Janne PA. The road to resistance: EGFR mutation and cetuximab. Nat Med. 2012;18:199–200. doi: 10.1038/nm.2646. [DOI] [PubMed] [Google Scholar]
- 34.Utsunomiya S, Okumura A, Watanabe K, Kunii S, Ishikawa D, Hirosaki T, Yamada K, Kaga A. A case of liver metastases with hyperbilirubinemia that was safely treated with chemotherapy (FOLFOX plus Cetuximab) Annals of Oncology. 2017;28 [Google Scholar]
- 35.Palle J, Tougeron D, Pozet A, Soularue E, Artru P, Leroy F, Dubreuil O, Sarabi M, Williet N, Manfredi S, Martin-Babau J, Rebischung C, Abdelghani MB, et al. Trastuzumab beyond progression in patients with HER2-positive advanced gastric adenocarcinoma: a multicenter AGEO study. Oncotarget. 2017;8:101383–101393. doi: 10.18632/oncotarget.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.U.S. Food, Drug Administration FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication. Available at https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm560040.htm.
- 39.Jeong W, Doroshow JH, Kummar S. United States Food and Drug Administration approved oral kinase inhibitors for the treatment of malignancies. Curr Probl Cancer. 2013;37:110–144. doi: 10.1016/j.currproblcancer.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu ZQ, Zhang Y, Li N, Liu PJ, Gao L, Gao X, Tie XJ. Efficacy and safety of lapatinib and trastuzumab for HER2-positive breast cancer: a systematic review and meta-analysis of randomised controlled trials. BMJ Open. 2017;7:e013053. doi: 10.1136/bmjopen-2016-013053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armstrong AJ, Shen T, Halabi S, Kemeny G, Bitting RL, Kartcheske P, Embree E, Morris K, Winters C, Jaffe T, Fleming M, George DJ. A phase II trial of temsirolimus in men with castration-resistant metastatic prostate cancer. Clin Genitourin Cancer. 2013;11:397–406. doi: 10.1016/j.clgc.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, Cheng JD, Chow LQ. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 43.Mehra R, Seiwert TY, Gupta S, Weiss J, Gluck I, Eder JP, Burtness B, Tahara M, Keam B, Kang H, Muro K, Geva R, Chung HC, et al. Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma: pooled analyses after long-term follow-up in KEYNOTE-012. Br J Cancer. 2018;119:153–159. doi: 10.1038/s41416-018-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaishampayan U, Shevrin D, Stein M, Heilbrun L, Land S, Stark K, Li J, Dickow B, Heath E, Smith D, Fontana J. Phase II Trial of Carboplatin, Everolimus, and Prednisone in Metastatic Castration-resistant Prostate Cancer Pretreated With Docetaxel Chemotherapy: A Prostate Cancer Clinical Trial Consortium Study. Urology. 2015;86:1206–1211. doi: 10.1016/j.urology.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh J, Novik Y, Stein S, Volm M, Meyers M, Smith J, Omene C, Speyer J, Schneider R, Jhaveri K, Formenti S, Kyriakou V, Joseph B, et al. Phase 2 trial of everolimus and carboplatin combination in patients with triple negative metastatic breast cancer. Breast Cancer Res. 2014;16:R32. doi: 10.1186/bcr3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergh J, Mariani G, Cardoso F, Liljegren A, Awada A, Vigano L, Huang X, Verkh L, Kern KA, Giorgetti C, Gianni L. Clinical and pharmacokinetic study of sunitinib and docetaxel in women with advanced breast cancer. Breast. 2012;21:507–513. doi: 10.1016/j.breast.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Donato EM, Fernández-Zarzoso M, Hueso JA, de la Rubia J. Brentuximab vedotin in Hodgkin lymphoma and anaplastic large-cell lymphoma: an evidence-based review. Onco Targets Ther. 2018;11:4583–4590. doi: 10.2147/OTT.S141053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinzelmann-Schwarz V, Knipprath Meszaros A, Stadlmann S, Jacob F, Schoetzau A, Russell K, Friedlander M, Singer G, Vetter M. Letrozole may be a valuable maintenance treatment in high-grade serous ovarian cancer patients. Gynecol Oncol. 2018;148:79–85. doi: 10.1016/j.ygyno.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 49.Martin M, Loibl S, von Minckwitz G, Morales S, Martinez N, Guerrero A, Anton A, Aktas B, Schoenegg W, Munoz M, Garcia-Saenz JA, Gil M, Ramos M, et al. Phase III trial evaluating the addition of bevacizumab to endocrine therapy as first-line treatment for advanced breast cancer: the letrozole/fulvestrant and avastin (LEA) study. J Clin Oncol. 2015;33:1045–1052. doi: 10.1200/JCO.2014.57.2388. [DOI] [PubMed] [Google Scholar]
- 50.Badros A, Hyjek E, Ma N, Lesokhin A, Dogan A, Rapoport AP, Kocoglu M, Lederer E, Philip S, Milliron T, Dell C, Goloubeva O, Singh Z. Pembrolizumab, pomalidomide, and low-dose dexamethasone for relapsed/refractory multiple myeloma. Blood. 2017;130:1189–1197. doi: 10.1182/blood-2017-03-775122. [DOI] [PubMed] [Google Scholar]
- 51. (January 12, 2018) FDA broadens afatinib indication to previously untreated, metastatic NSCLC with other non-resistant EGFR mutations. https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm592558.htm. [Google Scholar]
- 52.Alley EW, Lopez J, Santoro A, Morosky A, Saraf S, Piperdi B, van Brummelen E. Clinical safety and activity of pembrolizumab in patients with malignant pleural mesothelioma (KEYNOTE-028): preliminary results from a non-randomised, open-label, phase 1b trial. Lancet Oncol. 2017;18:623–630. doi: 10.1016/S1470-2045(17)30169-9. [DOI] [PubMed] [Google Scholar]
- 53.Porta R, Borea R, Coelho A, Khan S, Araujo A, Reclusa P, Franchina T, Van Der Steen N, Van Dam P, Ferri J, Sirera R, Naing A, Hong D, et al. FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit Rev Oncol Hematol. 2017;113:256–267. doi: 10.1016/j.critrevonc.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Beaver JA, Amiri-Kordestani L, Charlab R, Chen W, Palmby T, Tilley A, Zirkelbach JF, Yu J, Liu Q, Zhao L, Crich J, Chen XH, Hughes M, et al. FDA Approval: Palbociclib for the Treatment of Postmenopausal Patients with Estrogen Receptor-Positive, HER2-Negative Metastatic Breast Cancer. Clin Cancer Res. 2015;21:4760–4766. doi: 10.1158/1078-0432.CCR-15-1185. [DOI] [PubMed] [Google Scholar]
- 55.Mejean A, Ravaud A, Thezenas S, Colas S, Beauval JB, Bensalah K, Geoffrois L, Thiery-Vuillemin A, Cormier L, Lang H, Guy L, Gravis G, Rolland F, et al. Sunitinib Alone or after Nephrectomy in Metastatic Renal-Cell Carcinoma. N Engl J Med. 2018;379:417–427. doi: 10.1056/NEJMoa1803675. [DOI] [PubMed] [Google Scholar]
- 56.Generali D, Montemurro F, Bordonaro R, Mafodda A, Romito S, Michelotti A, Piovano P, Ionta MT, Bighin C, Sartori D, Frassoldati A, Cazzaniga ME, Riccardi F, et al. Everolimus Plus Exemestane in Advanced Breast Cancer: Safety Results of the BALLET Study on Patients Previously Treated Without and with Chemotherapy in the Metastatic Setting. Oncologist. 2017;22:648–654. doi: 10.1634/theoncologist.2016-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.