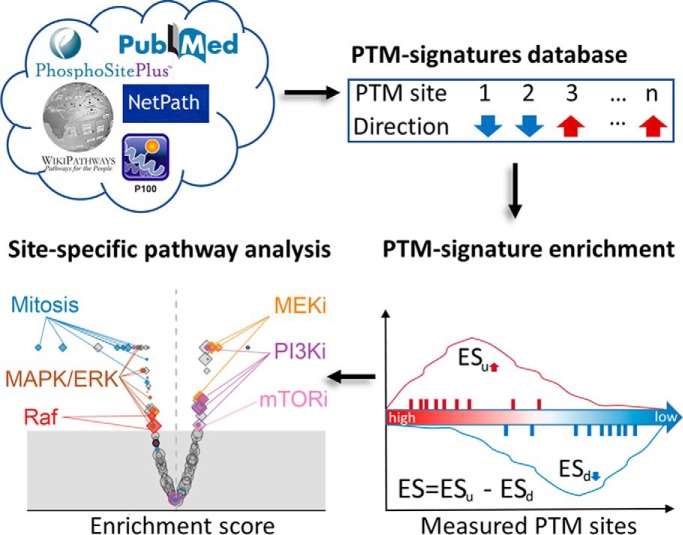
Pathway analysis of PTM data sets is typically performed at a gene-centric level because of the lack of appropriately curated PTM signature databases. We have developed a PTM signatures database (PTMsigDB) providing curated phosphorylation signatures of kinases, perturbations and signaling pathways to enable site-specific PTM signature enrichment analysis (PTM-SEA). Application of PTM-SEA to phosphoproteomes of several cell lines perturbed with growth factors, cell cycle inhibitors, or a specific PI3K inhibitor demonstrated the potential of our site centric approach to study dysregulated pathways in cancers.
Keywords: Phosphorylation, Pathway Analysis, Computational Biology, Database design, Post-translational modifications*
Graphical Abstract
Highlights
Database of PTM site-specific phosphorylation signatures of kinases, perturbations and signaling pathways (PTMsigDB).
PTM signature enrichment analysis (PTM-SEA) outperformed gene-centric analysis in detection of EGF induced phospho signaling events.
PI3K perturbation signatures were readily detected in PI3Ka inhibited human breast cancer cells.
PTMsigDB and PTM-SEA can be freely accessed at https://github.com/broadinstitute/ssGSEA2.0.
Abstract
Signaling pathways are orchestrated by post-translational modifications (PTMs) such as phosphorylation. However, pathway analysis of PTM data sets generated by mass spectrometry (MS)-based proteomics is typically performed at a gene-centric level because of the lack of appropriately curated PTM signature databases and bioinformatic tools that leverage PTM site-specific information. Here we present the first version of PTMsigDB, a database of modification site-specific signatures of perturbations, kinase activities and signaling pathways curated from more than 2,500 publications. We adapted the widely used single sample Gene Set Enrichment Analysis approach to utilize PTMsigDB, enabling PTM Signature Enrichment Analysis (PTM-SEA) of quantitative MS data. We used a well-characterized data set of epidermal growth factor (EGF)-perturbed cancer cells to evaluate our approach and demonstrated better representation of signaling events compared with gene-centric methods. We then applied PTM-SEA to analyze the phosphoproteomes of cancer cells treated with cell-cycle inhibitors and detected mechanism-of-action specific signatures of cell cycle kinases. We also applied our methods to analyze the phosphoproteomes of PI3K-inhibited human breast cancer cells and detected signatures of compounds inhibiting PI3K as well as targets downstream of PI3K (AKT, MAPK/ERK) covering a substantial fraction of the PI3K pathway. PTMsigDB and PTM-SEA can be freely accessed at https://github.com/broadinstitute/ssGSEA2.0.
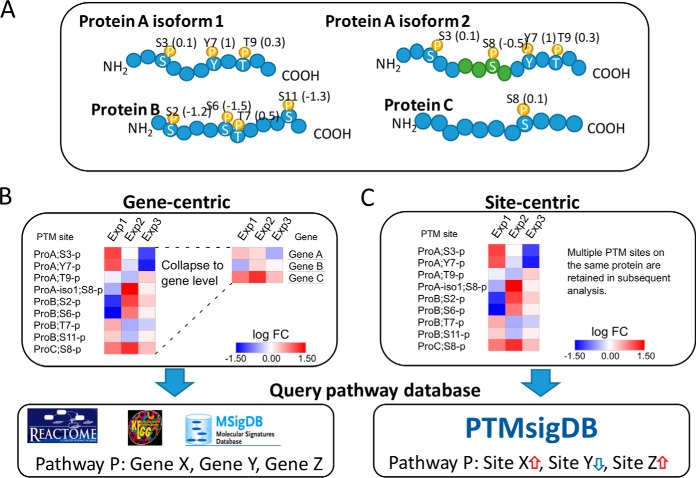
Identifying signaling pathways that are dysregulated in diseases such as cancer is crucial for understanding the molecular mechanisms underlying the disease and ultimately for developing better treatments. Post-translational modifications (PTMs)1 of proteins play a key role in practically every cellular process by regulating activity, localization and interaction of proteins. Mass spectrometry (MS)-based proteomics facilitates profiling of tens of thousands of PTM sites in a single experiment, most importantly phosphorylation, acetylation and ubiquitylation. Because of tremendous improvements in mass spectrometry technologies as well as sample processing workflows, large-scale analyses of PTMs have now become feasible for entire patient cohorts. This is exemplified by recent Clinical Proteomics Tumor Analysis Consortium (CPTAC) landmark studies of breast and ovarian cancer, in which pivotal findings were derived from phosphoproteomic analyses of up to 174 patient tumor samples (1, 2). For example, regulation of specific phosphorylation pathways revealed a novel breast cancer subtype that was unique to the phosphoproteomes of the tumors and could not be observed in RNA, DNA or protein space (1). Such analyses require well-annotated molecular signaling pathways and ideally would incorporate the role of specific modified sites rather than gene products alone. However, databases annotating biological pathways such as KEGG (3), Reactome (4) or MSigDB (5) are gene-centric and do not capture information for individual PTM sites. Therefore, pathway analysis of acquired phosphoproteomes typically involves collapsing discrete measurements of PTM sites on proteins into a single measurement represented by e.g. the mean or median of corresponding sites, which are mapped to their respective gene symbols. This procedure allows the resulting data set to be used for gene-centric pathway analysis, sacrificing the additional information provided by individual PTM sites (Fig. 1) and likely diluting the signal represented in the phosphoproteome data. The information loss is especially profound if multiple phosphosites on the same protein are differentially abundant. The lack of appropriately curated databases has impeded development of bioinformatics analysis tools that leverage site-centric PTM signatures. Further, existing tools that can be directly applied to site-centric data sets, such as PHOXTRACK (6), KSEA (7–9), KEA (10), IKAP (11) or PSEA (12), are solely intended to detect signatures of kinase activity in phosphorylation data by screening for known kinase substrate sites, and do not encompass analysis of molecular signaling pathways or signatures of kinase inhibition.
Fig. 1.
Pathway analysis of phosphoproteome data sets. A, The majority of proteins have multiple phospho-acceptor sites that can be differentially occupied and vary in abundance levels measured by MS. The four examples illustrate three proteoforms with varying number of phosphorylation sites (represented by amino acid residue and position in protein sequence) which are quantified with different fold changes (represented as numbers in parenthesis). Protein A exemplifies the presence of two protein isoforms carrying an isoform-specific phosphorylation site. B, Gene-centric pathway analysis typically involves combining fold changes of multiple sites mapping to the same gene symbol by calculating a center value of abundance (mean or median) or by choosing a single site characterized by high degree of variance (thus information) across a sample cohort. Additional information provided by multiple phosphorylation sites on a single protein as well as different sites on protein isoforms are not considered. Resulting gene-centric expression matrix can then be queried against gene-centric pathway databases such as Reactome, KEGG or MSigDB, in which each pathway is represented as a collection of gene symbols. C, Site-centric pathway analysis takes all quantified phosphorylation sites into account and requires a database of pathways annotated using individual phosphorylation sites. For that purpose, we developed PTMsigDB, annotating signatures of pathways, kinases and perturbations at the level of sites. Each site is annotated with the direction of regulation in a signature, i.e. whether its abundance is decreased or elevated, exemplified by blue and red arrows, respectively. Quantitative, site-centric phosphoproteomic data can be directly queried against PTMsigDB to identify signatures of phosphosites that correlate with annotated signature sets in PTMsigDB.
Here we present the first iteration of PTMsigDB, a collection of PTM site-specific signatures that have been assembled and curated from published data sets (Fig. 1). The current version is comprised exclusively of phosphorylation signatures but serves as a foundation for signatures of other PTMs like ubiquitination or acetylation to be added in the future. Although PTMsigDB enables kinase signature analysis similar to published tools described above, it additionally includes many curated signature sets of PTM sites mined from PhosphoSitePlus (13), Netpath (14) and WikiPathways (15) that have been i) quantitatively characterized in specified perturbation studies; ii) are known substrates of kinases; or iii) are known to be activated/deactivated in signaling pathways. A unique feature of PTMsigDB is the annotation of each PTM site with the direction of regulation, i.e. increased or decreased abundance upon a specific perturbation or in the context of signaling through a canonical pathway. We further present an extension of the widely used single sample Gene Set Enrichment Analysis (ssGSEA) (16, 17) that enables PTM Signature Enrichment Analysis (PTM-SEA) illustrated here for MS-based phosphoproteomics data sets.
We first assessed the utility of our approach using a well-characterized, literature-derived phosphoproteome data set of epidermal growth factor (EGF)-perturbed HeLa cells (18) and compared PTM-SEA to regular gene-centric ssGSEA. We next applied PTM-SEA to phosphoproteomes of human cell lines treated with different cell cycle inhibitors that have been profiled in the Library of Integrated Network-Based Cellular Signatures (LINCS) (19, 20). Finally, to demonstrate the value of PTM-SEA in perturbation studies targeting clinically relevant cancer pathways, we applied our tools to analyze a deep phosphoproteome of a PI3Ka-inhibited breast cancer cell line.
EXPERIMENTAL PROCEDURES
Assembly of PTMsigDB
The foundation of PTMsigDB is PTM sites curated by PhosphoSitePlus (PSP). All sites present in PTMsigDB were either directly derived from, or subsequently mapped to PSP. Each site in PTMsigDB is annotated with the PSP site group id, flanking amino acid sequence (± 7 amino acids) and UniProt accession number (Table I, supplemental Table S1). Each site in our database is annotated with the reported direction of regulation upon a specific perturbation or in a molecular pathway. We note that we do not capture functional annotations of PTM sites in PTMsigDB, such as activating or inactivating effect on the modified protein.
Table I. Representation of PTM sites in PTMsigDB.
| Database format | Site accession | Example |
|---|---|---|
| Uniprot-centric | Uniprot_acc;site-type;direction | Q06609;Y315-p;u |
| Flanking sequence | +/−7aa flanking seq-type;direction | ETRICKIYDSPCLPE-p;u |
| PSP site group id | site_grp_id-type;direction | 448324-p;u |
PTMsigDB is stored in Gene Matrix Transposed (GMT) format, a simple flat file format utilized by the gene-centric molecular signature database MSigDB (16, 21). Signature sets were assembled using three different modes of curation: i) entirely manual specification by domain experts (NetPath, WikiPathways, PSP kinase-substrate signatures); ii) semi-automatic specification (PSP perturbation signatures); and iii) fully automatic specification (LINCS perturbation signatures). Each signature is a unique collection of confidently localized PTM sites. Because sites are single amino acid residues, PTMsigDB requires single site-centric data matrices, and multiply phosphorylated peptides must be resolved into single site-centric data.
Although in this manuscript we focus on human signature sets, we have also started to derive signatures for mouse and rat (Table II, supplemental Table S2). Below we describe in more detail how signature sets of different categories have been assembled.
Table II. Extending PTMsigDB to other model organisms.
| Organism | No. signatures total | No. perturbation signatures | No. kinase signatures | No. pathway signatures |
|---|---|---|---|---|
| Human | 491 | 223 (1045 sites) | 220 (7,104 sites) | 48 (1164 sites) |
| Mouse | 119 | 48 (254 sites) | 71 (1133 sites) | - |
| Rat | 82 | 39 (203 sites) | 42 (614 sites) | 1 (17 sites) |
Signature Extraction from PSP Perturbation Data
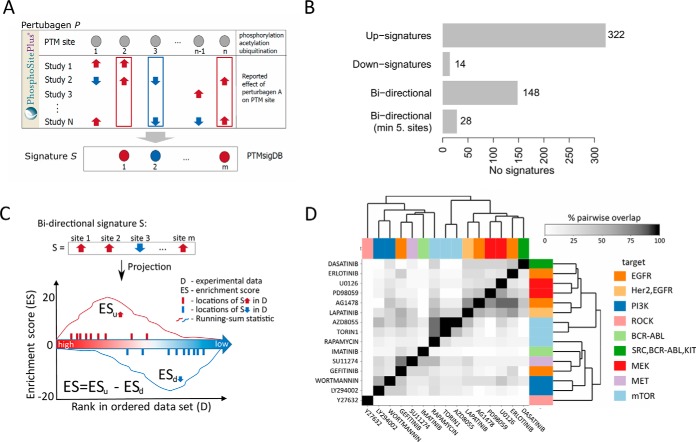
Perturbation signatures were derived from PTM sites manually curated from literature describing the phosphoproteomic impact of perturbations such as small molecules and growth factors. The initial database represented results from 2,483 perturbation studies. Many studies employed common perturbations (for instance, the same small molecule) with different experimental conditions (such as different cell lines, dosages or time points). To manage the bias inherent in such differential representation of perturbations we assembled “consensus-signatures” that combined results consistently reported across different studies of a common perturbation. For each perturbation P we extracted all PTM sites that were increased or decreased in abundance by P. For each PTM site S belonging to the P-altered set we then required a minimum of two independent studies that reported S to be regulated by P in the same direction (Fig 2A). Requiring consistency between two independent studies balanced the competing interests of retaining large numbers of signatures and PTM sites per signature and maximizing the likely relevance of such signatures to novel perturbation data sets. Each site in this category is annotated by the Pubmed identifier (PMID) of the corresponding publication in the Description field of the GMT file.
Fig. 2.
PTMsigDB signature sets and scoring scheme. Signatures sets in PTMsigDB are divided into three categories (perturbations, kinases and signatures of molecular pathways) derived from four major sources (PhosphoSitePlus, NetPath, WikiPathways, LINCS). A, Perturbation signatures from PhosphoSitePlus were assembled from a total of 2483 publications. To reduce experimental noise we require that each site be consistently reported by at least two independent studies (consensus signatures). Experimental noise can be introduced by use of different cell systems (e.g. in vivo versus in vitro), different protocols (e.g. dosage of perturbation) or different technologies (e.g. low-throughput versus high throughput). B, Bar chart depicting the distribution of unidirectional (all sites of a signature are annotated with decreased OR increased abundance levels) and bidirectional (sites of a signature include both decreased AND increased abundance levels) signature sets. C, Modified scoring scheme of the single sample Gene Set Enrichment Analysis (ssGSEA) algorithm enables scoring of directional signature sets. For each direction a running-sum statistic (y axis) is calculated by walking down the ranked list of PTM sites in the data set (x axis). An enrichment score (ES) reflected by the area under the resulting curve is calculated for each direction (ESu for “up” sites and ESd for “down” sites) separately and combined by subtracting ESd from ESu. D, Heatmap depicting the overlap between phosphorylation sites commonly affected by kinase-inhibitors. The similarity matrix is based on the number of shared phosphosites between signatures. The upper and lower triangular matrices are normalized by the total number of sites of signatures listed in rows and columns, respectively.
Kinase-Substrate Signatures from PSP
The file Kinase_Substrate_Data set contains a list of experimentally-determined kinase substrates and metadata curated from the literature by PSP and was downloaded from their website (https://www.phosphosite.org/staticDownloads) on March 21, 2018. Kinase-substrate signatures were parsed from the downloaded file using an in-house developed R-script. Briefly, all annotated substrate sites for each kinase were extracted and a minimum of five distinct sites was required to form a kinase signature. The script was used to parse kinase signatures for human, mouse and rat.
Signatures of Molecular Signaling Pathways from NetPath
Annotation of pathways with phosphorylation was performed manually by domain experts and was based on 909 publications. All sites were first mapped to PSP and were annotated by flanking amino acid sequence and site group id. Only sites that could be mapped to PSP and that were consistently annotated with either increased or decreased levels were retained. Each site in this category is annotated by either the Pubmed identifier (PMID) or by a string concatenating first author and publication year of the corresponding manuscript. This information can be found the Description field of the GMT file.
Signatures of Molecular Signaling Pathways from WikiPathways
All human, mouse and rat specific pathways in gpml format were downloaded from (https://www.wikipathways.org/index.php/Download_Pathways) on March 20, 2018. An in-house developed R-script was used to parse each file representing a single pathway and to extract information about annotated phosphorylation sites. A minimum of five distinct sites was required to form a signature. All signature sets were exported in GMT format.
Perturbation Signatures from LINCS Database
Perturbation data derived from the P100 assay (19) were downloaded from PanoramaWeb (https://panoramaweb.org/project/LINCS/P100/begin.view) (22) on August 30, 2017. We retrieved level 4 data (L4) which were sample-normalized (columns), QC'd and plate-normalized (rows) comprising a total of 189 perturbations and 12 human cell lines. Data for the P100 assay were acquired in PRM or in DIA mode and analyzed using Skyline (23). A more detailed description of the different data processing steps, including the analysis of DIA data, are described in (24). We first normalized P100 probes to the DMSO control experiment, which was present in each 96 well plate, by subtracting the average log2 DMSO ratio followed by Z-score transformation (per sample column). To derive signatures for a specific perturbation we considered probes with Z-scores more extreme than ± 2 per sample column observed in at least two samples (replicates or cell lines) measured on the same 96 well plate. Perturbations measured on multiple plates were combined into a single perturbation signature set. All metadata (plate number, cell line, dose, etc.) for each site of a signature can be found in the Description field. All sites were mapped to PSP and were annotated by flanking amino acid sequence and site group id. A minimum of five distinct sites was required to form a signature and signature sets were exported in GMT format.
Single Sample Gene Set Enrichment Analysis (ssGSEA)
Gene Set Enrichment Analysis (GSEA) (16) is a tool for determining enrichment of gene sets using genes differentially expressed between two phenotypes measured in a sufficient number of replicates. The approach to apply GSEA on a single, pre-ranked data vector representing gene abundance, especially from a single sample, has been introduced in (17) and is typically referred to as the signature projection method or single sample GSEA (ssGSEA). Briefly, expression values of genes in one sample are rank-ordered and an enrichment score for a signature S is calculated based on the difference in the Empirical Cumulative Distribution Functions (ECDFs) of genes in S and the set of all genes. This ECDF difference between signature S and remaining genes (ΔECDF) is calculated by walking down a rank-sorted list of genes, increasing a running-sum statistic when a gene belongs to S, and decreasing the running-sum statistic if the gene does not belong to S. The enrichment score ES reflects the degree to which genes in S are overrepresented at the top or bottom of the ranked-ordered list. Two methods have been proposed to calculate ES: the original GSEA publication (16) described a weighted Kolmogorov-Smirnov-like statistic defined as maximal distance between both ECDFs:
Alternatively, the ES can be calculated as the area under the delta ECDF
with p being the maximal rank of genes in the sample.
To account for different signature sizes, and correlations between gene set and the data set, a normalized enrichment score (NES) is calculated by permuting the genes in the sample using a user-defined number of permutations n (typically n = 1000):
Permutations are also used to calculate a p value for a given NES of a signature S:
Calculated p values can be further corrected for multiple hypothesis testing.
ssGSEA 2.0 and PTM-SEA Specific Modifications
We obtained the R program for ssGSEA analysis developed by the original authors and re-implemented parts of the software to optimize execution speed (support of multiple CPU cores). We further modified the handling of missing values in the data set more robust by discarding them before calculating pathway enrichment scores and implemented support for GCT (Gene Cluster Text) v1.3 files which enable storing data and metadata in a single text file. Import, export and manipulation of GCT files was implemented using the cmapR R-package (https://github.com/cmap/cmapR). We further extended the generated output of the original implementation of ssGSEA by utilizing GCT v1.3 metadata fields. For instance, for each scored gene set information on the number of scored genes and total percentage of genes scored set are included in the results files.
We extended the regular ssGSEA algorithm to enable site-specific, directional (Fig 2B) PTM signature enrichment analysis by incorporating directionality into the scoring scheme (Fig 2C). Each part of a directional signature set is scored separately by first calculating enrichment scores (ES) of PTM sites annotated with increased abundance (ESu). If the signature set contains PTM sites that were annotated with decreased abundance, a separate enrichment score (ESd) for those sites is calculated. Both scores are combined into a single enrichment score by calculating: ES = ESu − ESd. The resulting combined score reflects a measure of correlation between annotated sites in a signature and the abundance ranks of the signature in the actual data: positive values indicate positive correlation; negative values indicate anti-correlation. Normalized enrichment scores and p values are calculated on the combined score based on a user-defined number of permutations applied to a single sample column.
PTM-SEA is Available on GenePattern (25), a powerful platform to deploy and run software and entire analysis pipelines in a web browser, and can be accessed using the following URL: https://tinyurl.com/PTM-SEA-GP. Source code, documentation and instructions to run ssGSEA2.0/PTM-SEA on a local computer and on GenePattern are available on GitHub and can be accessed using the following URL: https://github.com/broadinstitute/ssGSEA2.0.
Preprocessing of Data Sets for Site-specific PTM-SEA
The input to the PTM-SEA R program is a single site-centric data matrix, m, stored in GCT format and a signature set database in GMT (Gene Matrix Transposed) format. Each row in m represents a single phosphorylation site confidently localized to a specific amino acid residue, with measured abundances across samples specified in columns in m. Multiple phosphorylation sites detected on the same peptide must be converted into separate site-specific entities for every site. Although some proteomics software packages, such as MaxQuant (26), readily produce single site-centric PTM reports, the use of other software packages might require additional preprocessing steps. To resolve the abundance of phosposites that have been measured on different phospho-proteoform peptides we used the abundance measured on the monophosphorylated version. If no monophosphorylated peptide version has been observed, we reported the abundance of the peptide carrying the least number of phosphate groups. Because we cannot disambiguate the abundance of multiple phosphosites measured on the same peptide in general, each site was assigned the measured abundance of the corresponding peptide and reported as separate rows in m. Lastly, if a site was measured on two or more equivalent peptides (e.g. one site exclusively detected on two doubly phosphorylated peptides), we reported the average abundance of corresponding peptides.
The general PTM-SEA workflow, including necessary pre-processing steps and description of data file formats is depicted in supplemental Fig. S1.
Parameters for PTM-SEA and ssGSEA2.0
Abundance levels of proteins/genes or PTM sites were first rank-normalized (sample.norm.type = “rank”). Rank-normalized profiles were further Z-scored (correl.type = “z.score”) and weighted (weight = 0.75). To obtain signature enrichment scores we calculated the difference of the ECDFs of the sites part of the signature and of all sites (statistic = “area.under.RES”) as described in (17). Normalized enrichment scores (NES) and p values were calculated using 10,000 permutations (nperm = 10000). We note that 1,000 permutations were typically enough to obtain stable p values. Although p values were calculated on a sample-by-sample basis, we further corrected permutation-derived p values for multiple hypothesis testing using the method proposed by Benjamini and Hochberg (27) using the entire data set (global.fdr = 'TRUE').
Sharma et al. EGF Data Set
We retrieved supplemental Table S2 from the manuscript by Sharma et al. (18). The authors used the MaxQuant software suite (26, 28) to obtain label-free intensity estimates of phosphosite abundances. We converted absolute intensities into relative ratios by dividing each phosphosite by its average intensity measured in six replicates of DMSO-treated control samples and subsequently applied log2 transformation. We performed the conversion to ratio data to intrinsically compare the effect of EGF and nocodazole treatment to the DMSO control experiment. We note that this conversion was not required to apply PTM-SEA, as PTM-SEA can also be directly applied to non-ratio data. All phosphosites with localization probability > 0.75 and a DMSO ratio observed in at least one sample were considered in further analyses (n = 27,307).
Ablelin et al. Drug Perturbation Data Set
The results of the discovery data described in (19) were downloaded from MassIVE using accession number MSV000079524. We downloaded the datafile Supp_DataDet1_P100_Discovery.gct and re-formatted the GCT id column to match the UniProt-centric format of PTMsigDB, as described above (supplemental Fig. S1). The resulting GCT file could be readily used for PTM-SEA using parameters described above. Because this discovery data set was used to derive the targeted P100 probes (from which we curated LINCS signature sets) we removed all P100 signatures from PTMsigDB prior to analysis to avoid a potential contamination of training and test data sets.
Cluster Analysis
All cluster analyses were based on hierarchical clustering using average linkage and Euclidean distance metric. Heatmaps were generated in R using the pretty heatmap package (pheatmap) or in Morpheus (https://software.broadinstitute.org/morpheus).
CCLE PI3K Perturbation Experiment—
Cell Culture and Treatment
T47D cells were cultured in RPMI medium with 10% FBS and 1% Penicillin/streptomycin. Cells were plated in 15 cm plates at a final density of 3.25e6 and 2e6, respectively, in a total volume of 29 ml, and incubated at 37 °C under standard culture conditions. Forty-five hours (approximately) after plating, 1 ml of BYL719 diluted in RPMI+FBS+P/S medium was added to the treated plates to achieve a final concentration of 1uM, with 1 ml of DMSO-dilution in RPMI+FBS+P/S medium added to the control-plates. Plates were then placed back in the 37 °C incubator. Plates were o harvested at 6-h and 24-h time points. For each time point, BYL719-treated and DMSO-treated plates were removed from the incubator in batches of 2, placed on ice, and washed once with ice-cold PBS. Cells were then scraped off the plate in fresh ice-cold PBS and placed in 15 ml conical tubes on ice. The process was repeated in batches of 2 until all plates for that specific time point and treatment were collected into a single 15 ml conical tube. Cells were then spun at 1200 RPM in a 4 °C centrifuge for 10 min. PBS was removed by aspiration, and the cell pellet was resuspended in ice cold PBS for a third wash. The cell suspension was then distributed, proportionally, into individual Eppendorf tubes intended for proteomics profiling, RNAseq and immunoblotting analysis. Eppendorfs were spun at 3000rpm in a 4 °C table top centrifuge, the PBS supernatant was manually removed, and cell pellets were snap frozen in a 70% ethanol/dry ice bath. Frozen pellets were stored at −80C° until processing for proteomics experiments. Time course treatments were performed in technical triplicates, each performed on different days.
Lysis and Digestion
Cell pellets were lysed in 50 mm Tris-HCl buffer (pH 8.0) containing 8 m Urea, 75 mm NaCl, 1 mm EDTA, protease and phosphatase inhibitors as previously described (29). Protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Pierce, Thermo Fisher Scientific, Bremen, Germany). Following reduction with 5 mm dythiothreitol (DTT) for 30min at room temperature (RT) and alkylation with 10 mm iodoacetamide for 45min at RT in the dark, urea concentration was reduced to 2 m and samples were digested with Endoproteinase LysC (Wako Chemicals, Richmond, VA) at an enzyme-to-substrate ratio of 1:50 for 2 h at 30 °C followed by sequencing grade trypsin (Promega, Madison, WI) at an enzyme-to-substrate ratio of 1:50 overnight at 30 °C. Reactions were quenched the next day with formic acid added to a final concentration of 1%. The samples were desalted using tC18 SepPak 200 mg cartridges (Waters, Milford, MA). Briefly, the cartridges were conditioned with 3 ml of Acetonitrile (ACN) followed by 3 ml of 50% ACN containing 0.1% formic acid (FA). After equilibrating cartridges with 3 × 4 ml of 0.1% trifluoroacetic acid (TFA)/water, samples were loaded and washed with 3 × 3 ml of 0.1% TFA/water followed by 1 × 3 ml of 1%FA/water. Samples were eluted with 2 × 1.5 ml 0.1% FA/50% ACN, frozen at −80 °C and dried down by vacuum centrifugation.
Labeling
For the quantitative phosphoproteomics experiment, 10 samples were labeled with TMT 10-plex reagent (Lot Number: PE201534A) (Thermo Fisher Scientific) following the manufacturer's protocol. Briefly, 1 mg of each sample was reconstituted in 1 ml of HEPES buffer at pH 8.5. Eight milligram of each of the TMT10 reagents was reconstituted in 410μl of ACN and added to the corresponding sample (Table III). After incubation at RT with shaking for 1 h the reactions were quenched by adding 80μl of 5% hydroxylamine to each and incubating at RT with shaking for 15min. The samples were combined and dried down by vacuum centrifugation. The combined sample was reconstituted in 0.1% FA/3% ACN and desalted using tC18 SepPack 500 mg cartridge as described above. Eluate was dried down completely.
Table III. TMT-10 reporter ion channel layout for PI3Ka inhibitor experiment.
| Channel | 126 | 127N | 127C | 128N | 128C | 129N | 129C | 130N | 130C | 131 |
|---|---|---|---|---|---|---|---|---|---|---|
| Pert | DMSO | DMSO | DMSO | DMSO | BYL719 | BYL719 | BYL719 | BYL719 | BYL719 | BYL719 |
| Time | 6h | 24h | 6h | 24h | 6h | 24h | 6h | 24h | 6h | 24h |
| Replicate | R1 | R1 | R2 | R2 | R1 | R1 | R2 | R2 | R3 | R3 |
Phosphoptyrosine (pY) Antibody Enrichment
The sample was reconstituted in 3 ml of immunoaffinity purification (IAP) buffer consisting of 50 mm MOPS at pH 7.2, 10 mm sodium phosphate, and 50 mm NaCl. One aliquot of pY1000 antibody beads (Cell Signaling Technologies, Danvers, MA) was washed 3 times with 1.5 ml IAP buffer. Washed antibody beads were added to the sample and incubated at 4 °C with end-over-end rotation for 1 h. After spinning down the antibody beads, the flow-through was collected to be desalted for the deep phospho-profiling. The beads were further washed 4x in 1.5 ml of ice-cold PBS, and pY-containing peptides eluted with 2 × 50μl of 0.15% TFA, desalted using C18 stage tips as described before (29) and dried down.
Flow-through was desalted using tC18 SepPack 500 mg cartridge as described above and eluate was dried down.
Basic Reversed Phase (bRP) Fractionation of Peptides
Desalted flow-through sample was fractionated on Agilent 1100 HPLC system using Zorbax 300 Extend-C18 4.6 mm ID x 250 mm length column with buffers A and B of 5 mm ammonium formate pH 10.0, in 2 and 90% ACN, respectively. Sample was reconstituted in 900μl of buffer A and 2 injections of 425 μl were performed. Sample was fractionated at a flow rate of 1 ml/min with the following gradient: 0 –16% buffer B for 6min, 16 - 40% buffer B for 60min, 40 - 44% buffer B for 4min, 44 - 60% buffer B for 5min and isocratic hold at 60% B for 14min. One-minute fractions were collected throughout the entire gradient for both injections into the same 96-deep well plate (GE Healthcare, Chicago, IL). Collected fractions were pooled into 24 fractions using a concatenation strategy as described before (29). Fractions were acidified with FA for a final concentration of 0.1%. 150 μl (5%) from each of the fractions was transferred into an autosampler vial and dried down for LC-MS/MS analysis of the proteome. The remaining 95% of the 24 fractions was dried down for detection and quantification of phosphopeptides.
Immobilized Metal Affinity Chromatography (IMAC) of the Fractions
Ni-NTA Superflow Agarose beads (Qiagen, Venlo, The Netherlands) were used for IMAC enrichment of the peptides as described previously (29). Briefly, an appropriate amount of bead slurry for 24 fractions was transferred to an Eppendorf tube and washed with HPLC water. Beads were stripped of Ni by incubating with 100 mm EDTA for 30 min at RT with end-over-end rotation and iron was added by incubating with 10 mm iron chloride (FeCl3) for 30min at RT with end-over-end rotation. Beads were washed 3× in 1 ml HPLC water and resuspended in an appropriate volume of 1:1:1 (v/v/vol) ratio ACN/Methanol/0.01% Acetic acid for aliquoting of 30 μl of slurry (corresponding to 10μl of beads) for each fraction. Dried-down fractions were reconstituted in 1 ml of binding buffer (0.1% TFA/80% ACN) and added to the beads. Samples were incubated for 30 min at RT with gentle shaking. After spinning down the beads, the supernatant consisting of unbound peptides was removed and beads reconstituted with phosphopeptides in 200 μl of binding buffer. Beads were transferred to stage tips for elution and desalting of the phosphopeptides as previously described (29). Eluted phosphopeptides were dried down completely.
LC-MS/MS Analysis of pY and IMAC Samples
pY and IMAC samples were resuspended in 9 μl of 0.1% FA/3% ACN and 4 μl of each was analyzed on a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) coupled to Easy-nLC 1000 liquid chromatography system (Thermo Fisher Scientific) with buffers A and B of 0.1% FA in 3 and 90% ACN, respectively. Peptides were separated at a flow rate of 200 nl/min on 75 μm ID picofrit columns (New Objective, Woburn, MA) packed in-house to 22–25 cm length with Reprosil-Pur C18-AQ 1.9 μm beads (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) and heated to 50 °C. IMAC samples were separated with a solvent B ramp of 2 - 6% in 1min followed by a linear gradient of 6–30% in 84min for a total run time of 110 min. Eluted peptides were analyzed in data-dependent mode acquiring MS1 scan at 70,000 resolution with automatic gain control (AGC) set to 3e6 followed by MS/MS on the 12 most abundant ions with resolution of 35,000, AGC of 5e4 with maximum inject time of 100ms, isolation width of 1.6Da with 0.3Da offset, normalized collision energy (NCE) set to 31(optimized for the specific QE Plus), peptide match function set to preferred, isotope exclusion function enabled, and dynamic exclusion set to 30 s. pY samples were separated in 2 × 4 μl injections of the sample with a 154min LC-MS/MS method where linear gradient of 6–35% solvent B was performed in 120min. MS parameters were as described above other than the maximum inject time for MS/MS being set to 120ms and dynamic exclusion set to 5 s.
CCLE Perturbation Data Analysis
Raw mass spectrometry data was analyzed using Spectrum Mill Workbench v5.1 (pre-release) (Agilent Technologies, Santa Clara, CA) as described before (29). Briefly, extracted MS/MS spectra were searched against human UniProt protein sequence database (downloaded on 12/28/2017) complemented with 264 commonly observed lab contaminants (65,068 database entries). Enzyme definition was defined as “trypsin allow P” for full tryptic specificity and up to five missed cleavages were allowed. Carbamidomethylation of cysteines was set as a fixed modification together with TMT10 isobaric labels at lysine residues (N termini would be considered regardless if it was TMT-labeled). Acetylation of protein N termini, oxidation of methionine and phosphorylation of serine, threonine and tyrosine were set as variable modifications with a precursor MH+ shift range of 0 to 272 Da. Database spectral matching was performed by Spectrum Mill search engine using ±20 ppm mass tolerance for precursor ions and product ions. Peptide-spectrum matches (PSMs) were automatically designated as confidently assigned using the Spectrum Mill autovalidation module employing a target-decoy search strategy to control false discovery rate (FDR). Individual PSMs were rolled up to phosphorylation sites that were separately FDR-controlled resulting in an estimated FDR <1.3% at the level of individual phosphorylation sites. Phosphorylation sites that could be unambiguously assigned to specific serine, threonine and tyrosine residues as automatically determined by Spectrum Mill software (1) were considered for further analyses (n = 24,035). TMT reporter ion intensities were corrected for isotope impurities using a previously described strategy (30) integrated into Spectrum Mill. TMT reporter ion ratios for BYL719/DMSO treatments of the first two replicates were calculated by matched replicates (e.g. BYL719R1/DMSOR1) (Table III). The third replicate did not have a paired control experiment and the average reporter ion intensity of both DMSO replicates was used as denominator. TMT reporter ion ratios were log2 transformed and median-centered for subsequent analysis.
Experimental Design and Statistical Rationale
The PI3K perturbation experiment comprised a single cell line (T47D, n = 1), analyzed at 6h and 24h time points, of inhibitor (BYL719) and paired control (DMSO) treatments. The BYL719 treatments were measured in three biological replicates, whereas the control experiments were measured in two biological replicates to accommodate the entire experiment in a single TMT-10 plex (Table III). BYL719/DMSO ratios were calculated and median-normalized as described above. To combine the triplicate measurements in each time point into a single readout as input for PTM-SEA, we employed a moderated one-sample t test using the limma R-package. This appropriately accounted for the variance observed across replicates captured in a p value. The resulting vector of p values was log-transformed and multiplied by the sign of the average log2 reporter ion ratio:
The transformed p values were used as input for PTM-SEA. We did not rank-normalize the transformed p values (sample.norm.type = 'none') and set the weight parameter to 1 (weight = 1) to incorporate the magnitude of the transformed phosphosite p values into the calculation of enrichment scores and p values for each signature.
RESULTS
Signature Sets in PTMsigDB
We developed PTMsigDB, a curated phosphosite-centric resource of molecular signatures of perturbations, kinase activity and molecular pathways. The foundation of PTMsigDB is PhosphoSitePlus (PSP) (13), a comprehensive systems biology resource for PTMs, which provides high-quality curation and annotation of PTMs at the individual residue level. We define a collection of PTM sites, whose levels are collectively regulated in a curated pathway or upon a perturbation, as a signature set. To derive signature sets, and to build up PTMsigDB, we mined PhosphoSitePlus (PSP), NetPath (NP), WikiPathways (WP) and the LINCS databases. Depending on the source of a signature, the curation process was either entirely manual (WP, NP), semi-automated (PSP kinase signatures, PSP perturbation signatures) or fully automated (LINCS). Manually curated signature sets representing molecular pathways were assembled by human domain experts. For signatures sets derived in a semi-automated or fully automated manner, we required that consistent results (site and direction of change) be reported by more than one independent study (see Methods for details) (Fig 2A, supplemental Fig. S2A).
Signature sets in PTMsigDB can be separated into three categories: 1) Perturbation signatures derived from treatment of cells with perturbations such as small molecules or growth factors (PSP, LINCS); 2) Signature sets of molecular signaling pathways (NP, WP); and 3) Kinase-substrate signatures (PSP). The different categories contain different numbers of signature sets, associated proteins and PTM sites (Table IV, supplemental Fig. S2B). In total, PTMsigDB contains 491 signature sets comprising 8029 phosphorylation sites on 2650 proteins. To assess the proportion of sites of known functional relevance, we compared sites in PTMsigDB to regulatory PTM sites in PSP, a set of functionally annotated sites that regulate downstream cellular processes, molecular functions and protein-protein interactions that we refer to as a “gold standard” of annotated PTM sites. Of 6,685 human phosphosites in the PSP regulatory site collection, 70% were present in signatures of PTMsigDB, emphasizing the functional relevance of most phosphorylation sites in PTMsigDB (supplemental Fig. S2C).
Table IV. Categories of signatures in PTMsigDB Numbers refer to human signatures.
| Category | Source | Annotation type | No. of signatures | No. of sites | No. of regulatory sites | |
|---|---|---|---|---|---|---|
| Perturbation | PSP | Manual and Semi-automated | 112 | 223 | 977 | 756 (77.4%) |
| Perturbation | LINCS | Fully automated | 111 | 72 | 10 (13.9%) | |
| Pathway | NetPath | Manual curation | 35 | 48 | 1082 | 498 (46.0%) |
| Pathway | WikiPathways | Manual curation | 13 | 145 | 141 (97.2%) | |
| Kinase | PSP | Manual and Semi-automated | 220 | 7104 | 4,475 (63.0%) | |
| Total | 491 | 8029 | 4,676 (58.2%) | |||
Kinases are key players in molecular signaling and a single kinase can play an essential role in multiple different pathways. Depending on the essentiality of a kinase, its substrate site can be assigned to several molecular pathways or perturbation signatures in PTMsigDB. To assess the degree of essentiality of phosphosites in PTMsigDB, we determined for each site the number of signatures they contribute to. Whereas the most redundant, and hence essential, sites occur in about 20% of all signatures, the median redundancy across all phosphosites was 2.5 signatures. Interestingly, six of the 10 most redundant PTM-sites in our database were T-x-Y activation loops of several mitogen activated protein kinases (MAPKs) underlining their essential role in a large number of molecular signaling pathways (Table V). Although this observation is consistent with gene-centric databases (MAPK3 is the most redundant gene symbol in MSigDB 5.1), in PTMsigDB we capture the increased phosphorylation levels of the activation loop (MAPK3 Thr-202 and Tyr-204), and so provide a direct readout of enzyme activation rather than merely elevated gene expression levels of MAPK3 as represented in MSigDB.
Table V. Most essential phosphosites in PTMsigDB. Site annotations were derived from UniProt.
| Site | Gene symbol | Protein name | Site annotations (UniProt) | No. signatures |
|---|---|---|---|---|
|
P27361;Y204-p P27361;T202-p |
MAPK3 | Mitogen-activated protein kinase 3 | Dually phosphorylated on Thr-202 and Tyr-204, which activates the enzyme. | 93 90 |
| P31749;S473-p | AKT1 | RAC-alpha serine/threonine-protein kinase | In kinase domain, needed to activate enzyme. | 82 |
|
P28482;Y187-p P28482;T185-p |
MAPK1 | Mitogen-activated protein kinase 1 | Dually phosphorylated on Thr-185 and Tyr-187, which activates the enzyme. | 85 79 |
| P62753;S235-p | RPS6 | 40S ribosomal protein S6 | Phosphorylated at Ser-235 and Ser-236 by RPS6KA1 and RPS6KA3; phosphorylation at these sites facilitates the assembly of the preinitiation complex | 55 |
| P49841;S9-p | GSK3B | Glycogen synthase kinase-3 beta | Inactivated by phosphorylation at Ser-9 (Probable). | 54 |
|
Q16539;Y182-p Q16539;T180-p |
MAPK14 | Mitogen-activated protein kinase 14 | Dually phosphorylated on Thr-180 and Tyr-182 by the MAP2Ks MAP2K3/MKK3, MAP2K4/MKK4 and MAP2K6/MKK6 in response to inflammatory cytokines, environmental stress or growth factors, which activates the enzyme. | 49 46 |
| Q15149;T4030-p | PLEC | Plectin | P100 probe | 47 |
Tyrosine phosphorylation of proteins presents the key mechanism to propagate extracellular signals, mediated by growth factor ligands, into the cell via receptor tyrosine kinases (RTKs), a class of transmembrane proteins which induce intracellular signal cascades and ultimately initiate cell proliferation (31). Compared with its low frequency in the human proteome (2.6%), tyrosine sites were overrepresented in PTMsigDB (20.4%) (supplemental Fig. S2D), once again emphasizing the functional relevance of phosphorylation sites in PTMsigDB.
A unique advantage of PTMsigDB over other pathway databases is the annotation of each PTM site with its reported direction of change upon a specific perturbation or signaling event. For instance, stimulation with growth factors results in an activation of specific signaling cascades; thus, PTM sites involved in these cascades are typically up-regulated, whereas inhibitory perturbations such as kinase inhibitors result in down-regulation of affected PTM sites. Although most signature sets in PTMsigDB were unidirectional (i.e. solely consisted of either up- or downregulated sites (Fig 2B), about 30% of signature sets were bidirectional and contained sites with both increased and decreased levels. However, most bidirectional signature sets were dominated by sites demonstrating up-regulation. For instance, requiring an arbitrarily chosen minimum of five PTM sites per category (increased/decreased abundance) resulted in 28 signatures that were distinctly bidirectional (Fig 2B). With ongoing collection of high-quality quantitative MS-based PTM data and associated curation efforts, we expect to see more site-specific phosphorylation signatures of a bidirectional nature. To account for the additional information provided by annotated direction of abundance changes, we adapted the scoring scheme of the widely used single sample Gene Set Enrichment Analysis (ssGSEA) algorithm (17) (see Methods). The resulting signature score reflects the degree of correlation between signature and data set. Positive signature scores indicate positive correlation, scores around zero indicate no correlation and negative scores indicate anti-correlation between signature and data (Fig 2C).
PTMsigDB includes phosphosite signatures of 45 kinase inhibitors (KI), a class of small molecules that revolutionized cancer therapies by enabling targeted inhibition of pathways dysregulated in specific cancers. Whereas most KI signature sets were assembled automatically from the LINCS database (see Methods), fifteen of these KI signatures sets were assembled semi-automatically from a set of manually curated phosphorylation sites derived from PSP. These signature sets comprised inhibitors targeting ABL, EGFR, mTOR, cMET, MEK, PI3K, ROCK and SRC kinases and were assembled from 131 phosphorylation sites with only moderate overlap (Fig 2D). To group KIs according to their phosphosite signatures we constructed a distance matrix based on the number of phosphosites shared between KI signatures which we subsequently subjected to hierarchical clustering. Interestingly, KIs targeting PI3K (LY294002, wortmannin) and MEK (PD98059, U0126) clearly cluster together whereas signatures of KIs targeting other kinases such as EGFR and mTOR were not characterized by high overlap in phosphosites pointing to different downstream effects introduced upon kinase inhibition. Although the vast majority of phosphosites from KI signatures were annotated with decreased abundance levels upon KI treatment, 17 out of 131 sites were annotated with increased abundance levels including Y861 on PTK2 (erlotinib) andY977 on PLCG1 (imatinib). A list of all KI signature sets, including fully-automatically derived signatures, can be found in supplemental Table S3.
Modified Amino Acid Residues in PTMsigDB Are Robustly Represented Across Databases and Organisms
Gene-centric pathway databases such as KEGG, Reactome and MSigDB use gene symbols as standardized primary identifiers (32). However, there is no standardized nomenclature for protein accession numbers. Protein database resources such as UniProt (33), NCBI RefSeq (34) or Ensemble (35) have distinct accession number schemes, which makes it difficult to compare proteins across databases. Moreover, underlying gene models of a given protein, such as translation start sites (TSS) or exon-intron boundaries, can vary across databases and between different database releases. This directly affects the representation of modified protein residues in terms of protein accession and residue number. Approaches to circumvent this issue include the use of amino acids flanking the modified residue as a unique identifier. However, these flanking residues can vary between isoforms or can be altered by non-synonymous mutations when searching customized sequence databases (36). PSP has developed a robust classification scheme providing unique identifiers for modification sites (site group ID) combining homologous and orthologous sites within proteoforms and between species (13). Every site in PTMsigDB was either directly derived from, or first mapped to PSP, thereby ensuring an unambiguous identifier across proteoforms and species for every PTM site. To ensure a high degree of compatibility to phosphorylation data sets generated by different software packages and searched against different protein sequence databases, PTMsigDB represents signatures using three different identifiers for phosphorylation sites: i) PSP site group ID; ii) UniProt; iii) Flanking sequence (Table I). Although the PSP site group ID provides an unambiguous representation of PTM sites within protein families and across species (13), using this type of identifier restricts the analysis to PTM sites present in PSP. We generally recommend using the flanking sequence as site identifier, because these are more invariant to updates made to protein sequence databases. The database format was kept generic and can be readily used to incorporate signatures of other PTMs beyond phosphorylation.
PTM-SEA Captured Underlying Signaling Cascades in EGF- and Nocodazole-treated HeLa Cells
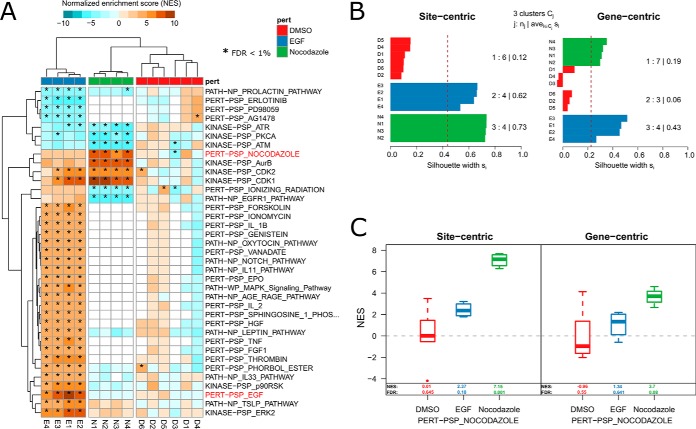
To assess our approach in the context of known biology we applied PTM-SEA to a recent high quality study of the well-characterized phosphoproteome of HeLa cells (18). In this study the authors evaluated the phosphoproteome dynamics of HeLa S3 cells stimulated with epidermal growth factor (EGF) and arrested in mitosis by nocodazole treatment. Because signatures of both EGF and nocodazole perturbation are part of PTMsigDB, this study provided a useful benchmark data set to assess the utility of our approach. The authors report >50K phosphorylation sites, of which >36K sites could be confidently localized to a specific residue and so were used in subsequent analysis. We first converted the label-free intensity values into ratios to the control experiment (DMSO) and subsequently applied PTM-SEA to project phosphosites to signature sets. PTM-SEA showed strongest enrichment of the EGF and nocodazole signatures in PTMsigDB in replicate samples of corresponding treatments (FDR < 1%) (Fig 3A). In addition, we detected cell cycle-related signatures in the nocodazole treated samples, which was used to mitotically arrest cells. For instance, signatures of cell cycle kinases CDK1, CDK2 and Aurora B were enriched. Among depleted signatures were kinases involved in DNA damage response (ATR, ATM) and the EGFR pathway.
Fig. 3.
Phosphoproteome signatures of EGF and nocodazole treatment in HeLa cells. We applied PTM-SEA to a phosphoproteome data set from the literature. Sharma et al. (18) studied phosphoproteome dynamics of EGF treatment in HeLa S3 cells. To mitotically arrest the cells, the antineoplastic agent nocodazole was used. Because PTMsigDB contained signatures for both perturbations, this data set was used as a benchmark to evaluate PTM-SEA. A, Heatmap depicting normalized enrichment scores (NES) of signatures (rows) in PTMsigDB that were consistently enriched or depleted at FDR < 0.01 in replicate measurements of EGF/nocodazole treatments (marked by asterisks). Hierarchical clustering of enrichment scores separated samples (columns) by experimental condition (EGF, nocodazole and control). Enrichment scores of nocodazole and EGF signature showed highest magnitude in the respective experiments and are highlighted in red. B, Silhouette analysis comparing hierarchical clustering of site-centric (left panel) and gene-centric (right panel) signature enrichment scores into three clusters. The bar chart depicts silhouette scores (x axis) of each sample (y axis) colored according to the assigned cluster. Average silhouette scores for each cluster are depicted at the right side of the bar charts. C, Signature enrichment scores of the nocodazole perturbation signature calculated using site-centric (left panel) and gene-centric (right panel) approach. Numbers indicate the median NES and FDR in replicate measurements of respective perturbations (x axis).
Samples treated with EGF showed enrichment of signatures of other growth factors (fibroblast growth factor, FGF1; hepatocyte growth factor, HGF), signatures of different cytokines (tumor necrosis factor, TNF; erythropoietin; EPO, and interleukin IL1B, IL2 IL33 pathway, IL11 pathway), signatures of signaling pathways (e.g. MAPK signaling pathway; thymic stromal lymphopoietin pathway, TSLP; AGE/RAGE pathway) and kinases acting downstream of these pathways (ERK2, p90RSK). No consistent enrichment or depletion of signatures was observed in the DMSO-treated control samples. Focusing solely on kinase enrichment scores, we compared PTM-SEA to KSEA (KSEA App) (7, 9) which demonstrated good agreement between both scoring approaches (supplemental Fig. S3). Taken together, these results demonstrated that PTM-SEA captured underlying signaling cascades stimulated by EGF or nocodazole treatment, beyond kinase activity signatures.
PTM-SEA Increased Enrichment of Expected Signaling Signatures
We then used the same data set to compare the performance of PTM-SEA's site-centric approach (in which individual sites were considered) to a gene-centric pathway analysis (in which multiple sites on the same gene were combined), and assessed how well each approach captured the underlying signaling events upon EGF and nocodazole treatment. To enable a direct comparison of both approaches we generated a gene-centric version of PTMsigDB in which signatures were represented as non-redundant sets of gene symbols without annotated regulatory direction, mimicking conventional gene-centric pathway databases like MSigDB. Additionally, in the HeLa data set described above we collapsed abundance of multiple sites mapping to the same gene symbol to their average DMSO normalized abundance. The resulting gene-centric expression matrix was projected to gene set enrichment scores by application of ssGSEA using the same parameters as described above. We used hierarchical clustering (supplemental Fig. S4A, S4B) and subsequent silhouette analysis to assess how well site-centric phospho signatures and gene-centric pathways captured signaling events introduced upon stimulation with EGF and cell cycle arrest by nocodazole. Given a distance matrix D and number of clusters K, silhouette coefficients represent a measure of intra-cluster similarity and inter-cluster distance for each clustered item i, and range between −1 and 1. A value close to 1 indicates similarity of item i with all other items in the same cluster and low similarity to items assigned to neighboring clusters, whereas negative values indicate closer similarity of item i to items in neighboring clusters. Silhouette scores were calculated for each sample (replicate measurements of EGF, nocodazole and DMSO treatments) using K = 3 as the cluster number to match the number of different treatments. Importantly, silhouette scores from site-centric clustering were consistently higher than those from gene-centric analysis (Fig 3B). In addition, hierarchical clustering of gene-centric enrichment scores into three cluster did not consistently group samples of the same treatments together (Fig 3B, supplemental Fig. S4B), whereas the site-centric analysis clearly separated different treatments and clustered together samples of EGF, nocodazole and control treatment (supplemental Fig. S4A). Although enrichment scores of the nocodazole signature were specifically enriched in the nocodazole-treated samples for both approaches, the extent of enrichment was more significant in the site-centric approach (FDR = 0.001) compared with the gene-centric approach (FDR = 0.08) (Fig 3C). Similar results were observed for the EGF signature (supplemental Fig. S4C, S4D) demonstrating better representation of the signature using individual phospho sites rather than collapsed gene-centric measures.
Site-specific Approach Outperforms Gene-specific Scoring
We next wanted to assess whether scoring each site-localized modified amino acid residue in a protein provides any benefit over simply scoring multiple phosphorylation events on single proteins (non-site-specific). The latter approach would enable a pseudo site-centric analysis of PTM-data sets using gene-centric databases and ssGSEA for which a substantial number of well-curated, gene-centric pathways provided by MSigDB, KEGG, Biocarta or Reactome are available. Instead of matching exact residue positions between data and database, gene symbols of multiply phosphorylated proteins were represented multiple times in calculation of enrichment scores corresponding to the number of observed phosphorylation events. The resulting bias toward higher weights of longer proteins that carry more phosphorylation sites was intrinsically addressed by calculation of normalized enrichment scores (NES). Each phosphosite was represented by the gene symbol mapping to the corresponding protein, which enabled the use of gene-centric pathway databases. Because of the resulting redundancy in repeated gene symbols we refer to this approach as gene-centric-redundant. To enable the scoring of redundant lists of gene symbols, we extended the ssGSEA algorithm accordingly and applied gene-centric-redundant ssGSEA to the EGF/nocodazole treated HeLa data set described above. To enable a direct comparison to the site-centric and gene-centric approaches, we used the gene-centric version of PTMsigDB and performed the same silhouette analysis based on hierarchical clustering as described above (supplemental Fig. S5). Like the gene-centric analysis, hierarchical clustering of gene-centric-redundant enrichment scores did not clearly separate the different treatments and resulted in a mixed cluster containing DMSO- and nocodazole-treated samples (supplemental Fig. S5A). As a result, silhouette scores were consistently lower compared with site-centric analysis (supplemental Fig. S5B) even for the correctly assigned EGF cluster pointing to a less stable clustering.
Focusing on nocodazole and EGF signatures alone, which were used as positive controls (see previous section), the gene-centric-redundant approach showed better enrichment of the EGF signature (FDR: 0.001) (supplemental Fig. S5C) and nocodazole signature (FDR: 0.001) (supplemental Fig. S5D) compared with the gene-centric EGF (FDR: 0.013) (Fig 3C) and nocodazole signature (FDR: 0.083) (supplemental Fig. S4D). Like in the site-centric analysis, the gene-centric-redundant approach detected an enrichment of the EGF and nocodazole signatures at an FDR of 0.001 in the corresponding treatments, therefore achieving comparable performance.
In summary, results obtained by the gene-centric-redundant approach outperformed the gene-centric approach but did not result in a stable clustering of samples according to treatments compared with site-centric enrichment analysis. We therefore conclude that scoring the exact residue number to derive signature enrichment scores best represented the underlying signaling events introduced by EGF and nocodazole stimulation. Because availability of site-specific signature sets in PTMsigDB is still limited compared with gene centric database such as MSigDB we note that the gene-centric-redundant approach might be employed to score gene-centric signatures, in particular for modifications other than phosphorylation. A systematic assessment of this approach however is beyond the scope of this manuscript.
A table of all scored signatures derived from the site-centric, gene-centric and gene-centric-redundant approach can be found in supplemental Table S4.
PTM-SEA Facilitates Detection of Differential Kinase Activity Signatures of Cell Cycle Inhibitors
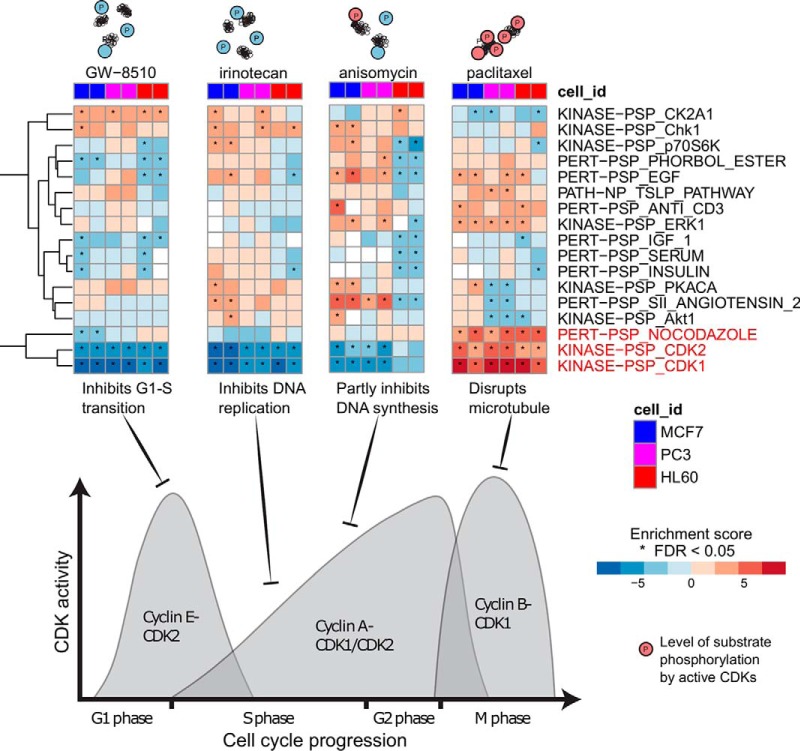
We applied PTM-SEA to perturbed phosphoproteomes of three human cancer cell lines representing breast cancer (MCF7), prostate cancer (PC3) and leukemia (HL60). Cell lines were treated with 23 different chemically active compounds as part of the Library of Integrated Network-Based Cellular Signatures (LINCS). These data were collected to derive a reduced representation of phosphorylation signatures (19) and were therefore acquired in single-shot LC-MS/MS mode without sample fractionation, resulting in relatively shallow phosphoproteome coverage (1.3K p-sites per LC-MS/MS run). Despite the resulting bias toward detection of highly abundant phosphorylated peptides, several signatures present in PTMsigDB were readily observed, demonstrating the feasibility of our approach in data sets comprising ∼1K phosphosites (Fig. 4, supplemental Table S5). To avoid potential contamination of the test data set (single shot data used to design the P100 assay) with training data (PRM/DIA data from application of the P100 assay), signatures derived from the P100 assay were removed from PTMsigDB prior to this analysis. The strongest signatures consistently observed across all three cell lines were detected in perturbation experiments involving cell cycle inhibitors and were predominantly comprised of cyclin kinases (CDK1, CDK2). The large number of substrate sites known to be phosphorylated by CDK1 (n = 487) and CDK2 (n = 341) facilitated the scoring of these signatures in relatively shallow phosphoproteome data sets. Kinase activity of CDKs is largely driven by binding to small regulatory proteins called cyclins to form a CDK-cyclin complex representing the active kinase. Different CDKs bind to different cyclins whose concentration varies across the cell cycle. Depending on the cell cycle phase in which the inhibitor is active, PTM-SEA detected distinct signatures of CDK activity that clearly correlated with concentration of Cyclin A and B (Fig. 4). For instance, treatment with Irinotecan, which inhibits DNA replication specifically during S phase when the concentration of Cyclin A is low, led to negative enrichment of the CDK1 and CDK2 signatures, indicating inhibition. Positive enrichment of the CDK1 signature was detected after treatment with Paclitaxel, which disrupts microtubules resulting in a cell cycle arrest in prometaphase during mitosis. During this phase the concentration of cyclin B has already increased, thereby, leading to high activity of CDK1. We also observed increased activity of CDK2, which regulates events in prophase right before prometaphase. Other inhibitors resulted in decreased CDK activity either by inhibiting CDK2 directly (GW8510) or by inhibiting DNA synthesis (anisomycin). These results demonstrated the ability of our approach to detect and quantify signatures of kinase activities in perturbed cell systems that clearly correlated with the mode of action (MoA) of the respective compounds.
Fig. 4.
MoA-specific kinase signatures of cell cycle inhibitors. Heatmaps depicting perturbation and kinase signature enrichment scores of three human cancer cell lines (MCF7, HL60, PC3) treated with four different cell cycle inhibitors measured in duplicates. Asterisks indicate signature scores at FDR < 0.05. Cartoons above the heatmap graphically illustrate the increasing levels (blue to red) of CDK1/2 substrate phosphorylations. The density plot illustrates cyclin A/B/E concentrations across cell cycle stages that correlate with CDK1/2 activity levels. Black T-arrows indicate stage of cell cycle the corresponding inhibitor is active. Signatures of CDK1 and CDK2 activity strongly correlate with the mode of action of each compound.
Signatures of PI3Ka Inhibition in Human Breast Cancer Cells Readily Detected by PTM-SEA
As part of the cancer cell line encyclopedia (CCLE) project (37), which aims to provide genetic and pharmacological characterization of >1K human cancer cell lines, our laboratory has started to generate deep phosphoproteomes of selected cell lines treated with clinically relevant kinase inhibitors targeting dysregulated pathways in the respective cancers. Here we applied PTM-SEA to the PI3K-inhibited (PI3Ki) phosphoproteome of T47D human breast cancer cells to characterize signaling events effected upon kinase inhibition. To that end, T47D cells were treated with the PI3Ka inhibitor BYL719 for 6 h and 24 h, using DMSO as control treatment in both time points. Experiments were performed in triplicate employing chemical peptide labeling (TMT-10) for sample multiplexing. Off-line basic pH reversed phase fractionation of phospho-enriched peptides prior to analysis by liquid chromatography coupled to high performance tandem mass spectrometry (1, 38) resulted in the identification of 38,587 phosphorylation events in a single TMT-10 plex. 23,937 phosphorylation sites could be unambiguously assigned to a specific amino acid residue and were used in subsequent analyses (supplemental Table S6). Notably, phosphosite ratios normalized for measured protein expression levels showed high correlation to unnormalized phosphosite ratios demonstrating negligible effects of protein expression changes on phosphosite abundances (supplemental Fig. S6).
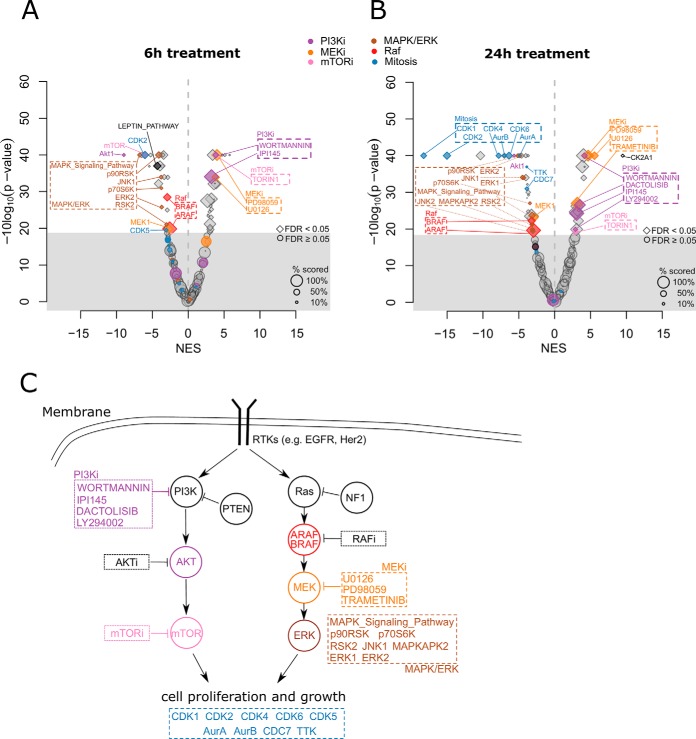
We applied PTM-SEA to phosphosite ratios (BYL719/DMSO treatment) to calculate enrichment scores for signature sets in PTMsigDB. Hierarchical clustering of signature scores calculated in individual experiments closely grouped triplicate measurements of both time point treatments together demonstrating high reproducibility of signature scores within replicate measurements of each time point (supplemental Fig. S7). To obtain a single enrichment score for each time point while appropriately accounting for biological variance across replicate measurements, we applied a one sample moderated t test to replicates of each time point, and used the signed, log-transformed p values as input to PTM-SEA (see Methods for details). In total we detected 52 significant signatures (FDR < 0.05) after 6 h and 56 signatures after 24 h of PI3K inhibition (Fig. 5). Although a BYL719 signature was not represented in PTMsigDB we detected signatures of several PI3K inhibitors that were positively enriched upon drug treatment. In the 6 h time point wortmannin, a nonspecific covalent inhibitor of PI3K, and IPI145 (duvelisib), a PI3K delta and gamma inhibitor, were significantly enriched (Fig 5A). In the 24 h time point two additional signatures of PI3K inhibitors, dactolisib and LY294002, were significantly enriched (Fig 5B) because of longer inhibition of PI3K. These signatures were semi-automatically (wortmannin, LY294002) and fully-automatically (IPI145, dactolisib) curated, validating the parameters used in the automatic curation pipelines to extract perturbation signatures from the LINCS and PSP databases (see Methods). We further observed signatures of MEK and mTOR inhibitors downstream of PI3K signaling positively enriched in both time points. Besides positively enriched KI signatures we observed an enrichment of the protein kinase CK2 signature (CK2A1), after 24 h treatment, pointing to an elevated activity of the kinase which was found to be dysregulated in many cancers (39, 40) (supplemental Fig. S8A).
Fig. 5.
Signatures of PI3K inhibition in breast cancer cells. T47D cells were treated with the PI3Ka inhibitor BYL719 and DMSO as control for 6h and 24h and deep phosphoproteomes were acquired resulting in ∼24K phosphosites localized to a specific Ser/Thr/Tyr residue. A, Volcano plot depicting enrichment of phosphoproteome signatures in the 6h time point. The x axis represents the normalized enrichment score (NES) between DMSO (left side) and drug treatment (right side). The size of the dots scale with the relative number of scored phosphorylation sites in a signature. The gray area contains signatures that did not significantly change upon drug treatment (permutation-based FDR ≥ 5%). PTM signature sets of inhibitors are annotated with an “i” after the drug target. B, Volcano plot depicting phospho signatures after treatment for 24 h. C, Schematic representation of the PI3K-AKT-mTOR and Ras-Raf-MEK-ERK pathways. Circles indicate kinases, boxes indicate specific kinase inhibitors. Highlighted in colors are significant phospho signatures shown in the volcano plots in A and B. The pathway representation is based on Fig. 1 in (44).
Among signatures that showed negative enrichment upon PI3K inhibition were AKT1, MEK1, mTOR, two isoforms of Raf kinase (ARAF, BRAF) and MAPK/ERK pathways downstream of PI3K, and associated kinases such as ERK1, ERK2, JNK1, p90RSK and p70S6K. Further downstream of PI3K we observed decreased activity of CDK (CDK1, CDK2, CDK4, CDK5, CDK6) and aurora (AurA, AurB) kinases which are essential regulators of the cell cycle, thereby highlighting the effect of PI3K inhibition on the inactivation of the cell proliferation machinery. These cell cycle-related signatures were more pronounced in the 24h time point (Fig 5B). We also observed decreased activity of the Leptin-induced pathway (supplemental Fig. S8B) which has been reported as a potential prognostic marker for breast cancer (41). PTMsigDB contains a PI3K-AKT pathway signature obtained from WikiPathways that we expected to observe with negative enrichment levels upon BYL7109 treatment. We detected the lowest enrichment of this pathway in the 24 h time point and the signature score did not pass statistical significance (FDR = 0.13, supplemental Table S7). However, further inspection of the phosphorylation sites in the PI3K-AKT pathway signature demonstrated a clear trend toward the inactivation of the pathway (supplemental Fig. S8C). A complete list of detected signature sets, together with signature enrichment scores and FDR-corrected p values can be found in (supplemental Table S7).
In summary, these findings were concordant with known cancer biology and covered a substantial part of the canonical PI3K-AKT-mTOR and Ras-Raf-MEK-ERK pathways that are affected by PI3K inhibition (Fig 5C). Our results demonstrated the potential of site-specific PTM signature enrichment analysis in interpreting perturbation studies that attempt to decipher the molecular mechanism of clinically relevant kinase inhibitors.
DISCUSSION
We describe an approach to perform phosphosite-specific signature analysis (PTM-SEA) based on a curated database of phosphosite-specific signatures (PTMsigDB). Curation and maintenance of such a database requires knowledge from domain experts, which in this study are represented by curators from PhosphoSitePlus, NetPath and WikiPathways. We consider PTMsigDB to be the foundation of a more comprehensive resource for PTM site-specific signatures of pathways and perturbations, rather than a mature and completely developed database. We aim to further extend PTMsigDB when more curated, site-specific molecular pathways are made available by repositories such as PSP. Moreover, we envision that this curation process will become a community effort in which researchers studying PTMs in particular pathways will contribute to curating these pathways at the level of PTM sites. Resources like WikiPathways that are maintained by the scientific community recently started to include PTM site-specific pathway annotations and provide a promising source for future expansion of PTMsigDB.
We have demonstrated the potential of PTMsigDB in analyzing MS-based phosphoproteomic data sets derived from perturbation studies involving EGF, cell cycle inhibitors and a specific PI3K inhibitor. The relatively small total number of signature sets (∼490) and unequal representation of perturbations, kinases and signaling pathways still limits the true potential of a PTM site-centric database. Our approach performs best when applied to perturbation data sets in which we expect a clear molecular phenotype. Subtle abundance differences in unperturbed baseline phosphoproteomes are much more challenging to detect with the current selection of available signature sets in PTMsigDB, especially because of the underrepresentation of molecular signaling pathways. Curation of the latter requires domain experts as well as the availability of detailed metadata accurately describing the biochemical experiments that were conducted to study signaling events in a pathway. However, these metadata (such as dosage of stimulant, cell system and time points of treatment) are often buried in respective subsections of manuscripts rather than stored together with the data itself, making curation of pathways prohibitively and unnecessarily difficult.
About 40% of signatures in PTMsigDB were derived from perturbation studies in which pathways were specifically activated (e.g. via growth factors) or inhibited (e.g. via kinase inhibitors). The cellular responses upon perturbation treatment are represented in perturbation signatures in PTMsigDB. Many of the respective phosphosites are of low stoichiometry and even after enrichment for modified peptides are of too low abundance to be sequenced by single-shot LC-MS/MS analysis. Although kinase-substrate signatures in PTMsigDB consist of 46 sites on average, this number is substantially lower for perturbation signatures (16 sites on average). Therefore, deeper phosphoproteome coverage is required to detect signatures beyond kinase activity, which typically involves extensive sample fractionation.
PTMsigDB contains signature sets for 45 kinase inhibitors (KI) and, to our knowledge, presents the first set of KI signatures curated at the level of phosphorylation sites. Although it is well known that KIs often target more than one protein, a systematic characterization of clinically relevant KIs and their targets was released only recently (42). The availability of molecular signatures of KIs curated at the PTM-site level enables further systematic screening for potential off-target effects of certain KIs. Though the number of KI signatures is still relatively small in the current version of PTMsigDB, this category of signatures is expected to be especially valuable for the scientific community.
Phosphosite-specific analysis of kinase substrate relationships is not an entirely new concept and it has been successfully employed by several groups (6, 7, 10–12). Instead of using the kinase expression as proxy for its activity, the abundance profiles of phosphorylation sites that are known substrates of the respective kinase are monitored. In this study we detected differential signatures of cyclin-dependent kinase (CDK) 1 and 2 in three human cancer cell lines upon treatment with different cell cycle inhibitors. Most importantly, treatment with paclitaxel resulted in increased activity of CDK1/2 whereas other inhibitors showed decreased kinase activity. Paclitaxel is an antineoplastic chemotherapy medication that targets tubulin by stabilizing the microtubule polymer preventing it from disassembly. The resulting inhibition of the mitotic spindle function blocks the progression of mitosis in which the concentration of cyclin A is high. Cyclin A specifically binds to CDK1, and the resulting complex is in an active state phosphorylating a multitude of different substrates that were identified by PTM-SEA.
Application of PTM-SEA to the deeply fractionated phosphoproteome of PI3K inhibited human breast cancer cells detected signatures that covered a substantial part of the canonical PI3K-AKT-mTOR and Ras-Raf-MEK-ERK pathways inhibitory signatures of kinases downstream (MEK, AKT, ERK) of PI3K. We also detected increased activity of the catalytic subunit α of casein kinase 2 (CK2a), a ubiquitous protein kinase known to be dysregulated in multiple cancers (39). Attenuation of PI3K/Akt signaling upon selective inhibition of CK has been reported previously (43). Our detection of higher activity levels of this kinase after inhibition of the PI3K pathway is an interesting observation for which the rationale remain unclear. Importantly, signatures of several PI3K inhibitors were enriched upon treatment and demonstrated decreased phosphorylation levels of sites that are part of these signatures, thereby supporting the selection of phosphosites for these inhibitors in PTMsigDB. Not surprisingly, the strongest effects of PI3K inhibition were observed in the cell proliferation machinery, exemplified by significantly reduced activity of mitotic kinases (CDKs and Aurora kinases).
With ongoing curation efforts to create PTM site-specific signatures spanning more molecular signaling pathways in PTMsigDB, we expect to achieve a more fine-grained view on signaling events disrupted by targeted inhibition of cancer-relevant pathways. We present here a uniform method and guidelines to the community to create these PTM signatures. In cases where no site-specific information is available yet, our gene-centric-redundant ssGSEA approach provides an alternative to standard gene centric approaches.
DATA AVAILABILITY
The original mass spectra for the PI3K perturbation experiment may be downloaded from MassIVE (http://massive.ucsd.edu) using the identifier: MSV000083128. The data is directly accessible via ftp://massive.ucsd.edu/MSV000083128.
Mass spectra of identified phosphopeptides were uploaded to MS-Viewer (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) and can be accessed with search key gxo6ksgdby or using the link below: http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=gxo6ksgdby&search_name=msviewer, PTMsigDB can be downloaded from https://github.com/broadinstitute/ssGSEA2.0.
Supplementary Material
Acknowledgments
We thank Alexander R. Pico from the Gladstone Institutes and cofounder of WikiPathways for constructive discussions around PTMsigDB.
Footnotes
* This work was supported by grants from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium (CPTAC) to the Broad Institute of MIT and Harvard: to S.A.C. and M.A.G. (U24CA210986); to D. R. Mani (U24CA210979); and to S.A.C. (U01CA214125). This work was further supported by NCI grants U01CA217885 and R01CA121941 to P.T.
 This article contains supplemental Figures and Tables. The authors do not declare any conflict of interest.
This article contains supplemental Figures and Tables. The authors do not declare any conflict of interest.
1 The abbreviations used are:
- PTM
- post translational modification
- PTM-SEA
- PTM signature enrichment analysis
- GSEA
- gene set enrichment analysis
- ssGSEA
- single sample gene set enrichment analysis
- PTMsigDB
- PTM signatures database
- MSigDB
- molecular signatures database
- NP
- NetPath
- WP
- WikiPathways
- FDR
- false discovery rate
- EGF
- epidermal growth factor
- KEGG
- kyoto encyclopedia of genes and genomes
- PSP
- PhosphoSitePlus
- LINCS
- library of integrated network-based cellular signatures
- NaCl
- sodium chloride
- EDTA
- ethylenediaminetetraacetic acid
- Ni
- nickel
- HPLC
- high pressure liquid chromatography
- MS
- mass spectrometry
- LC-MS/MS
- liquid chromatography-MS/MS
- GCT
- gene cluster text
- GMT
- gene matrix transposed.
REFERENCES
- 1. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., Gatza M. L., Wilkerson M., Perou C. M., Yellapantula V., Huang K.-L., Lin C., McLellan M. D., Yan P., Davies S. R., Townsend R. R., Skates S. J., Wang J., Zhang B., Kinsinger C. R., Mesri M., Rodriguez H., Ding L., Paulovich A. G., Fenyö D., Ellis M. J., Carr S. A., and NCI CPTAC. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang H., Liu T., Zhang Z., Payne S. H., Zhang B., McDermott J. E., Zhou J. Y., Petyuk V. A., Chen L., Ray D., Sun S., Yang F., Chen L., Wang J., Shah P., Cha S. W., Aiyetan P., Woo S., Tian Y., Gritsenko M. A., Clauss T. R., Choi C., Monroe M. E., Thomas S., Nie S., Wu C., Moore R. J., Yu K. H., Tabb D. L., Fenyö D., Vineet V., Wang Y., Rodriguez H., Boja E. S., Hiltke T., Rivers R. C., Sokoll L., Zhu H., Shih I. M., Cope L., Pandey A., Zhang B., Snyder M. P., Levine D. A., Smith R. D., Chan D. W., Rodland K. D., Carr S. A., Gillette M. A., Klauser K. R., Kuhn E., Mani D. R., Mertins P., Ketchum K. A., Thangudu R., Cai S., Oberti M., Paulovich A. G., Whiteaker J. R., Edwards N. J., McGarvey P. B., Madhavan S., Wang P., Chan D. W., Pandey A., Shih I. M., Zhang H., Zhang Z., Zhu H., Cope L., Whiteley G. A., Skates S. J., White F. M., Levine D. A., Boja E. S., Kinsinger C. R., Hiltke T., Mesri M., Rivers R. C., Rodriguez H., Shaw K. M., Stein S. E., Fenyo D., Liu T., McDermott J. E., Payne S. H., Rodland K. D., Smith R. D., Rudnick P., Snyder M., Zhao Y., Chen X., Ransohoff D. F., Hoofnagle A. N., Liebler D. C., Sanders M. E., Shi Z., Slebos R. J. C., Tabb D. L., Zhang B., Zimmerman L. J., Wang Y., Davies S. R., Ding L., Ellis M. J. C., and Townsend R. R. (2016) Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 166, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kanehisa M., and Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Croft D., Mundo A. F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M. R., Jassal B., Jupe S., Matthews L., May B., Palatnik S., Rothfels K., Shamovsky V., Song H., Williams M., Birney E., Hermjakob H., Stein L., and D'Eustachio P. (2014) The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., and Mesirov J. P. (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weidnery C., Fischery C., and Sauer S. (2014) PHOXTRACK-a tool for interpreting comprehensive data sets of post-translational modifications of proteins. Bioinformatics 30, 3410–3411 [DOI] [PubMed] [Google Scholar]
- 7. Hernandez-Armenta C., Ochoa D., Goncalves E., Saez-Rodriguez J., and Beltrao P. (2017) Benchmarking substrate-based kinase activity inference using phosphoproteomic data. Bioinformatics 33, 1845–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zagorac I., Fernandez-Gaitero S., Penning R., Post H., Bueno M. J., Mouron S., Manso L., Morente M. M., Alonso S., Serra V., Muñoz J., Gómez-López G., Lopez-Acosta J. F., Jimenez-Renard V., Gris-Oliver A., Al-Shahrour F., Piñeiro-Yañez E., Montoya-Suarez J. L., Apala J. V., Moreno-Torres A., Colomer R., Dopazo A., Heck A. J. R., Altelaar M., and Quintela-Fandino M. (2018) In vivo phosphoproteomics reveals kinase activity profiles that predict treatment outcome in triple-negative breast cancer. Nat. Commun. 9, 3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wiredja D. D., Koyutürk M., and Chance M. R. (2017) The KSEA App: a web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics 33, 3489–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lachmann A., and Ma'ayan A. (2009) KEA: kinase enrichment analysis. Bioinformatics 25, 684–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mischnik M., Sacco F., Cox J., Schneider H.-C., Schäfer M., Hendlich M., Crowther D., Mann M., and Klabunde T. (2016) IKAP: A heuristic framework for inference of kinase activities from Phosphoproteomics data. Bioinformatics 32, 424–431 [DOI] [PubMed] [Google Scholar]
- 12. Suo S.-B., Qiu J.-D., Shi S.-P., Chen X., and Liang R.-P. (2015) PSEA: Kinase-specific prediction and analysis of human phosphorylation substrates. Sci. Rep. 4, 4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kandasamy K., Mohan S. S., Raju R., Keerthikumar S., Kumar G. S. S., Venugopal A. K., Telikicherla D., Navarro J. D., Mathivanan S., Pecquet C., Gollapudi S. K., Tattikota S. G., Mohan S., Padhukasahasram H., Subbannayya Y., Goel R., Jacob H. K. C., Zhong J., Sekhar R., Nanjappa V., Balakrishnan L., Subbaiah R., Ramachandra Y. L., Rahiman B. A., Prasad T. S. K., Lin J.-X., Houtman J. C. D., Desiderio S., Renauld J.-C., Constantinescu S. N., Ohara O., Hirano T., Kubo M., Singh S., Khatri P., Draghici S., Bader G. D., Sander C., Leonard W. J., and Pandey A. (2010) NetPath: a Public Resource of Curated Signal Transduction Pathways. Genome Biol. 11, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E. L., Bohler A., Mélius J., Waagmeester A., Sinha S. R., Miller R., Coort S. L., Cirillo E., Smeets B., Evelo C. T., and Pico A. R. (2016) WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 44, D488–D494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. a., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barbie D. A., Tamayo P., Boehm J. S., Kim S. Y., Susan E., Dunn I. F., Schinzel A. C., Sandy P., Meylan E., Fröhling S., Chan E. M., Sos M. L., Michel K., Silver S. J., Weir B. A., Reiling J. H., Sheng Q., Piyush B., Wadlow R. C., Le H., Hoersch S., Wittner B. S., Ramaswamy S., Livingston D. M., Sabatini D. M., Thomas R. K., Lander E. S., Mesirov J. P., David E., Gilliland D. G., Jacks T., and Hahn W. C. (2010) Systematic RNA interference reveals that oncogenic KRAS- driven cancers require TBK1. Nature 462, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma K., D'Souza R. C. C. J., Tyanova S., Schaab C., Wiśniewski J. J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 19. Abelin J. G., Patel J., Lu X., Feeney C. M., Fagbami L., Creech A. L., Hu R., Lam D., Davison D., Pino L., Qiao J. W., Kuhn E., Officer A., Li J., Abbatiello S., Subramanian A., Sidman R., Snyder E., Carr S. A., and Jaffe J. D. (2016) Reduced-representation phosphosignatures measured by quantitative targeted MS capture cellular states and enable large-scale comparison of drug-induced phenotypes. Mol. Cell. Proteomics 15, 1622–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keenan A. B., Jenkins S. L., Jagodnik K. M., Koplev S., He E., Torre D., Wang Z., Dohlman A. B., Silverstein M. C., Lachmann A., Kuleshov M. V., Ma'ayan A., Stathias V., Terryn R., Cooper D., Forlin M., Koleti A., Vidovic D., Chung C., Schürer S. C., Vasiliauskas J., Pilarczyk M., Shamsaei B., Fazel M., Ren Y., Niu W., Clark N. A., White S., Mahi N., Zhang L., Kouril M., Reichard J. F., Sivaganesan S., Medvedovic M., Meller J., Koch R. J., Birtwistle M. R., Iyengar R., Sobie E. A., Azeloglu E. U., Kaye J., Osterloh J., Haston K., Kalra J., Finkbiener S., Li J., Milani P., Adam M., Escalante-Chong R., Sachs K., Lenail A., Ramamoorthy D., Fraenkel E., Daigle G., Hussain U., Coye A., Rothstein J., Sareen D., Ornelas L., Banuelos M., Mandefro B., Ho R., Svendsen C. N., Lim R. G., Stocksdale J., Casale M. S., Thompson T. G., Wu J., Thompson L. M., Dardov V., Venkatraman V., Matlock A., Van Eyk J. E., Jaffe J. D., Papanastasiou M., Subramanian A., Golub T. R., Erickson S. D., Fallahi-Sichani M., Hafner M., Gray N. S., Lin J.-R., Mills C. E., Muhlich J. L., Niepel M., Shamu C. E., Williams E. H., Wrobel D., Sorger P. K., Heiser L. M., Gray J. W., Korkola J. E., Mills G. B., LaBarge M., Feiler H. S., Dane M. A., Bucher E., Nederlof M., Sudar D., Gross S., Kilburn D. F., Smith R., Devlin K., Margolis R., Derr L., Lee A., and Pillai A. (2018) The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Systems 6, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liberzon A., Birger C., Thorvaldsdó H., Ghandi M., Mesirov J. P., Tamayo P., and Thorvaldsdó Ttir H. (2015) The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems 1, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sharma V., Eckels J., Schilling B., Ludwig C., Jaffe J. D., MacCoss M. J., and MacLean B. (2018) Panorama Public: A public repository for quantitative data sets processed in Skyline. Mol. Cell. Proteomics, mcp.RA117.000543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Litichevskiy L., Peckner R., Abelin J. G., Asiedu J. K., Creech A. L., Davis J. F., Davison D., Dunning C. M., Egertson J. D., Egri S., Gould J., Ko T., Johnson S. A., Lahr D. L., Lam D., Liu Z., Lyons N. J., Lu X., MacLean B. X., Mungenast A. E., Officer A., Natoli T. E., Papanastasiou M., Patel J., Sharma V., Toder C., Tubelli A. A., Young J. Z., Carr S. A., Golub T. R., Subramanian A., MacCoss M. J., Tsai L.-H., and Jaffe J. D. (2018) A Library of Phosphoproteomic and Chromatin Signatures for Characterizing Cellular Responses to Drug Perturbations. Cell Systems 6, 424–443.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., and Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 [DOI] [PubMed] [Google Scholar]
- 26. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. Benjamini Y., and Hochberg Y. (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Roy. Statistical Soc. 57, 289–300 [Google Scholar]
- 28. Cox J., Neuhauser N., Michalski A., Scheltema R. a., Olsen J. V., and Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 29. Mertins P., Tang L. C., Krug K., Clark D. J., Gritsenko M. A., Chen L., Clauser K. R., Clauss T. R., Shah P., Gillette M. A., Petyuk V. A., Thomas S. N., Mani D. R., Mundt F., Moore R. J., Hu Y., Zhao R., Schnaubelt M., Keshishian H., Monroe M. E., Zhang Z., Udeshi N. D., Mani D., Davies S. R., Townsend R. R., Chan D. W., Smith R. D., Zhang H., Liu T., and Carr S. A. (2018) Reproducible workflow for multiplexed deep-scale analysis of the proteome and phosphoproteome of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 13, 1632–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shadforth I. P., Dunkley T. P. J., Lilley K. S., and Bessant C. (2005) i-Tracker: For quantitative proteomics using iTRAQTM. BMC Genomics 6, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hunter T. (2014) The genesis of tyrosine phosphorylation. Cold Spring Harb. Perspect. Biol. 6, a020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gray K. A., Yates B., Seal R. L., Wright M. W., and Bruford E. A. (2015) Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 43, D1079–D1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bateman A., Martin M. J., O'Donovan C., Magrane M., Apweiler R., Alpi E., Antunes R., Arganiska J., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Chavali G., Cibrian-Uhalte E., Da Silva A., De Giorgi M., Dogan T., Fazzini F., Gane P., Castro L. G., Garmiri P., Hatton-Ellis E., Hieta R., Huntley R., Legge D., Liu W., Luo J., Macdougall A., Mutowo P., Nightingale A., Orchard S., Pichler K., Poggioli D., Pundir S., Pureza L., Qi G., Rosanoff S., Saidi R., Sawford T., Shypitsyna A., Turner E., Volynkin V., Wardell T., Watkins X., Zellner H., Cowley A., Figueira L., Li W., McWilliam H., Lopez R., Xenarios I., Bougueleret L., Bridge A., Poux S., Redaschi N., Aimo L., Argoud-Puy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M. C., Boeckmann B., Bolleman J., Boutet E., Breuza L., Casal-Casas C., De Castro E., Coudert E., Cuche B., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Jungo F., Keller G., Lara V., Lemercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T., Nouspikel N., Paesano S., Pedruzzi I., Pilbout S., Pozzato M., Pruess M., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A. L., Wu C. H., Arighi C. N., Arminski L., Chen C., Chen Y., Garavelli J. S., Huang H., Laiho K., McGarvey P., Natale D. A., Suzek B. E., Vinayaka C. R., Wang Q., Wang Y., Yeh L. S., Yerramalla M. S., and Zhang J. (2015) UniProt: A hub for protein information. Nucleic Acids Res. 43, D204–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C. M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V. S., Kodali V. K., Li W., Maglott D., Masterson P., McGarvey K. M., Murphy M. R., O'Neill K., Pujar S., Rangwala S. H., Rausch D., Riddick L. D., Schoch C., Shkeda A., Storz S. S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R. E., Vatsan A. R., Wallin C., Webb D., Wu W., Landrum M. J., Kimchi A., Tatusova T., DiCuccio M., Kitts P., Murphy T. D., and Pruitt K. D. (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zerbino D. R., Achuthan P., Akanni W., Amode M. R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C. G., Gil L., Gordon L., Haggerty L., Haskell E., Hourlier T., Izuogu O. G., Janacek S. H., Juettemann T., To J. K., Laird M. R., Lavidas I., Liu Z., Loveland J. E., Maurel T., McLaren W., Moore B., Mudge J., Murphy D. N., Newman V., Nuhn M., Ogeh D., Ong C. K., Parker A., Patricio M., Riat H. S., Schuilenburg H., Sheppard D., Sparrow H., Taylor K., Thormann A., Vullo A., Walts B., Zadissa A., Frankish A., Hunt S. E., Kostadima M., Langridge N., Martin F. J., Muffato M., Perry E., Ruffier M., Staines D. M., Trevanion S. J., Aken B. L., Cunningham F., Yates A., and Flicek P. (2018) Ensembl 2018. Nucleic Acids Res. 46, D754–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruggles K. V., Tang Z., Wang X., Grover H., Askenazi M., Teubl J., Cao S., McLellan M. D., Clauser K. R., Tabb D. L., Mertins P., Slebos R., Erdmann-Gilmore P., Li S., Gunawardena H. P., Xie L., Liu T., Zhou J.-Y., Sun S., Hoadley K. A., Perou C. M., Chen X., Davies S. R., Maher C. A., Kinsinger C. R., Rodland K. D., Zhang H., Zhang Z., Ding L., Townsend R. R., Rodriguez H., Chan D., Smith R. D., Liebler D. C., Carr S. A., Payne S., Ellis M. J., and Fenyo D. (2015) An analysis of the sensitivity of proteogenomic mapping of somatic mutations and novel splicing events in cancer. Mol. Cell. Proteomics, mcp.M115.056226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., Morais P., Meltzer J., Korejwa A., Jané-Valbuena J., Mapa F. A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I. H., Cheng J., Yu G. K., Yu J., Aspesi P., de Silva M., Jagtap K., Jones M. D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R. C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J. P., Gabriel S. B., Getz G., Ardlie K., Chan V., Myer V. E., Weber B. L., Porter J., Warmuth M., Finan P., Harris J. L., Meyerson M., Golub T. R., Morrissey M. P., Sellers W. R., Schlegel R., and Garraway L. A. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mundt F., Rajput S., Li S., Ruggles K. V., Mooradian A. D., Mertins P., Gillette M. A., Krug K., Guo Z., Hoog J., Erdmann-Gilmore P., Primeau T., Huang S., Edwards D. P., Wang X., Wang X., Kawaler E., Mani D. R., Clauser K. R., Gao F., Luo J., Davies S. R., Johnson G. L., Huang K.-L., Yoon C. J., Ding L., Fenyö D., Ellis M. J., Townsend R. R., Held J. M., Carr S. A., and Ma C. X. (2018) Mass spectrometry-based proteomics reveals potential roles of NEK9 and MAP2K4 in resistance to PI3K inhibitors in triple negative breast cancers. Cancer Res. canres.1990.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Unger G. M., Davis A. T., Slaton J. W., and Ahmed K. (2004) Protein kinase CK2 as regulator of cell survival: implications for cancer therapy. Curr. Cancer Drug Targets 4, 77–84 [DOI] [PubMed] [Google Scholar]
- 40. Hou Z., Nakanishi I., Kinoshita T., Takei Y., Yasue M., Misu R., Suzuki Y., Nakamura S., Kure T., Ohno H., Murata K., Kitaura K., Hirasawa A., Tsujimoto G., Oishi S., and Fujii N. (2012) Structure-Based Design of Novel Potent Protein Kinase CK2 (CK2) Inhibitors with Phenyl-azole Scaffolds. J. Med. Chem. 55, 2899–2903 [DOI] [PubMed] [Google Scholar]
- 41. Ando S., Barone I., Giordano C., Bonofiglio D., and Catalano S. (2014) The Multifaceted Mechanism of Leptin Signaling within Tumor Microenvironment in Driving Breast Cancer Growth and Progression. Front. Oncol. 4, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.-A., Reinecke M., Ruprecht B., Petzoldt S., Meng C., Zecha J., Reiter K., Qiao H., Helm D., Koch H., Schoof M., Canevari G., Casale E., Depaolini S. R., Feuchtinger A., Wu Z., Schmidt T., Rueckert L., Becker W., Huenges J., Garz A.-K., Gohlke B.-O., Zolg D. P., Kayser G., Vooder T., Preissner R., Hahne H., Tõnisson N., Kramer K., Götze K., Bassermann F., Schlegl J., Ehrlich H.-C., Aiche S., Walch A., Greif P. A., Schneider S., Felder E. R., Ruland J., Médard G., Jeremias I., Spiekermann K., and Kuster B. (2017) The target landscape of clinical kinase drugs. Science 358, eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siddiqui-Jain A., Drygin D., Streiner N., Chua P., Pierre F., O'Brien S. E., Bliesath J., Omori M., Huser N., Ho C., Proffitt C., Schwaebe M. K., Ryckman D. M., Rice W. G., and Anderes K. (2010) CX-4945, an Orally Bioavailable Selective Inhibitor of Protein Kinase CK2, Inhibits Prosurvival and Angiogenic Signaling and Exhibits Antitumor Efficacy. Cancer Res. 70, 10288–10298 [DOI] [PubMed] [Google Scholar]
- 44. Jokinen E., and Koivunen J. P. (2015) MEK and PI3K inhibition in solid tumors: rationale and evidence to date. Ther. Adv. Med. Oncol. 7, 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original mass spectra for the PI3K perturbation experiment may be downloaded from MassIVE (http://massive.ucsd.edu) using the identifier: MSV000083128. The data is directly accessible via ftp://massive.ucsd.edu/MSV000083128.
Mass spectra of identified phosphopeptides were uploaded to MS-Viewer (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) and can be accessed with search key gxo6ksgdby or using the link below: http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=gxo6ksgdby&search_name=msviewer, PTMsigDB can be downloaded from https://github.com/broadinstitute/ssGSEA2.0.