In the search for biomarkers of synapse loss associated with Alzheimer's disease (AD), we used shotgun proteomics to identify a panel of 9 synaptic proteins detectable in human cerebrospinal fluid (CSF). Expression at the human synapse was verified using super resolution microscopy. Using SRM, we monitored the panel in CSF from 3 independent clinical cohorts. We report 6 synaptic biomarkers that demonstrate changes in preclinical AD prior to markers of neurodegeneration, which could have clinical value for assessing disease progression.
Keywords: Selected reaction monitoring, Clinical proteomics, Alzheimer's disease, Biomarker: Prognostic, Cerebrospinal fluid
Graphical Abstract
Highlights
Proteomic analysis of cerebrospinal fluid and identification of synaptic component.
Use of super resolution microscopy to verify synapse-specificity in human tissue.
Selective reaction monitoring MS (SRM) of synaptic panel in 3 cohorts of Alzheimer's disease cerebrospinal fluid.
Synaptic protein changes precede tau in preclinical Alzheimer's disease.
Abstract
A biomarker of synapse loss, an early event in Alzheimer's disease (AD) pathophysiology that precedes neuronal death and symptom onset, would be a much-needed prognostic biomarker. With direct access to the brain interstitial fluid, the cerebrospinal fluid (CSF) is a potential source of synapse-derived proteins. In this study, we aimed to identify and validate novel CSF biomarkers of synapse loss in AD. Discovery: Combining shotgun proteomics of the CSF with an exhaustive search of the literature and public databases, we identified 251 synaptic proteins, from which we selected 22 for further study. Verification: Twelve proteins were discarded because of poor detection by Selected Reaction Monitoring (SRM). We confirmed the specific expression of 9 of the remaining proteins (Calsyntenin-1, GluR2, GluR4, Neurexin-2A, Neurexin-3A, Neuroligin-2, Syntaxin-1B, Thy-1, Vamp-2) at the human synapse using Array Tomography microscopy and biochemical fractionation methods. Exploration: Using SRM, we monitored these 9 synaptic proteins (20 peptides) in a cohort of CSF from cognitively normal controls and subjects in the pre-clinical and clinical AD stages (n = 80). Compared with controls, peptides from 8 proteins were elevated 1.3 to 1.6-fold (p < 0.04) in prodromal AD patients. Validation: Elevated levels of a GluR4 peptide at the prodromal stage were replicated (1.3-fold, p = 0.04) in an independent cohort (n = 60). Moreover, 7 proteins were reduced at preclinical stage 1 (0.6 to 0.8-fold, p < 0.04), a finding that was replicated (0.7 to 0.8-fold, p < 0.05) for 6 proteins in a third cohort (n = 38). In a cross-cohort meta-analysis, 6 synaptic proteins (Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, Syntaxin-1B and Thy-1) were reduced 0.8-fold (p < 0.05) in preclinical AD, changes that precede clinical symptoms and CSF markers of neurodegeneration. Therefore, these proteins could have clinical value for assessing disease progression, especially in preclinical stages of AD.
Synapse loss is a fundamental process underlying many neurological and psychiatric diseases including, but not limited to Alzheimers Disease (AD)1, Parkinson's disease, Lewy body diseases, schizophrenia and depression (1–3). Therefore, a biomarker capable of detecting synapse loss in living individuals has the potential to be a surrogate marker for disease severity, which would make an excellent addition to the biomarker arsenal for a wide range of neurological diseases. To search for novel synaptic biomarkers, we have selected AD as a disease model for synaptopathy. Synapse loss is an early event in AD, which precedes neuronal death (1) and evidence from animal models indicates that the synapse is the target of both AD pathological proteins, Aβ and tau (4). AD can be conceptualized as a continuum of preclinical and clinical phases, based on the clinical syndrome and biomarkers of brain amyloidosis and tau-mediated neurodegeneration. In this conceptualization, patients with mild cognitive impairment (5) or dementia (6) who are positive for AD biomarkers are labeled as patients with MCI because of AD (prodromal AD) or dementia because of AD (6). Likewise, the current guidelines from the National Institute on Aging-Alzheimer Association (NIA-AA) conceptualize three preclinical stages of AD (7) whereby cognitively normal subjects with signs of brain amyloidosis (preclinical Stage 1), amyloidosis and neurodegeneration (preclinical Stage 2) or both markers as well as subtle cognitive decline (preclinical Stage 3). Although markers of Aβ and tau pathology are excellent diagnostic biomarkers for AD, a marker of synapse degeneration would be invaluable for assessing disease progression in at-risk subjects. In this regard, many researchers have turned to biochemical markers in CSF, a biofluid with direct access to the central nervous system that can be extracted from living individuals by lumbar puncture.
Previous studies have reported elevated CSF levels of individual synaptic proteins such as neurogranin (8), SNAP-25 (9), synaptotagmin-1 (10) in AD dementia patients and neurexins 1, 2 and 3, and neurofascin (11) in prodromal AD patients. Although these findings support the idea that synaptic proteins in CSF may be informative in AD, the previously reported correlation between CSF levels of synaptic proteins with CSF levels of tau suggest that widespread neuronal loss could be a confounding factor when studying synaptic proteins in the CSF, particularly at clinical disease stages. The CSF profile of synaptic proteins in preclinical stages of AD, before widespread neurodegeneration has taken hold has not been explored in detail. This is an important aspect because a good CSF marker of underlying synaptic degeneration should demonstrate changes that precede those of neurodegeneration markers. The progressive staging of neurodegeneration markers in AD, make this disease an excellent model system to evaluate the potential relationship between CSF levels of proposed synaptic biomarkers and existing markers of neurodegeneration.
Here we report (1) a systematic proteomic study of the CSF with a thorough characterization of the synaptic composition (discovery), (2) development of Tier 2 SRM assays for a set of CSF proteins whose expression was confirmed at the human cortical synapse or in synapse-associated structures (verification), (3) assessment of the CSF profile of the synaptic panel in a clinical cohort that includes cognitively normal subjects and AD preclinical and clinical stages (exploration), and (4) confirmation in independent clinical AD cohorts (clinical validation).
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
This study is divided into the following stages: Discovery stage; peptide and protein identification in 7 pools of 60 CSF samples. Verification stage; Tier 2 SRM assay development for selected peptides in CSF and pathological study of selected proteins in human post-mortem tissue from 6 donors. Exploration stage; SRM of selected peptides in CSF from cognitively normal controls and pre-clinical and clinical stages of the AD continuum (n = 80) prospectively recruited from the Sant Pau Initiative in Neurodegeneration (SPIN) cohort at Hospital Sant Pau, Barcelona and by the CITA Foundation, Donostia (exploratory cohort). Validation stage; SRM of selected peptides in an independent collection of CSF (n = 60) prospectively recruited from the collection at Hospital Clinic, Barcelona (validation cohort-1) and an independent selection (validation cohort-2) of cognitively normal controls and preclinical stage 1 subjects (n = 38) from the SPIN cohort. Where possible, subjects included in each group were age and sex-matched. CSF samples were run on the mass spectrometer in a randomized order with respect to diagnostic group. No technical replicates were included in the SRM study. Biological controls for SRM included cognitively normal individuals (exploratory Cohort, n = 20, validation cohort-1, n = 18, validation cohort-2, n = 20). Biological replicates for SRM included patients and volunteers grouped according to established diagnostic criteria. As technical controls for SRM, BSA controls were run between each sample (shotgun and targeted LC-MS/MS).
Clinical CSF Cohorts
All participants gave their written consent. The study (IIBSP-BIO-2015–76) was approved by the local ethics committee following the ethical standards recommended by the Helsinki Declaration. All subjects were evaluated by neurologists with expertise in neurodegenerative diseases, by neuropsychologists using a previously published neuropsychological battery (12) and assessed for established AD biomarkers, namely brain amyloidosis (low CSF levels of Aβ1–42 or positive amyloid PET imaging) and neurodegeneration (high CSF levels of total tau or phosphorylated tau) based on local cut-offs. These cut-offs have high specificity and sensitivity to distinguish AD dementia patients from controls (13). Diagnoses of prodromal AD and AD dementia were made according to NIA-AA guidelines (5, 6). Subjects within the normal range following formal neuropsychological evaluation, when accounting for age and education (mostly recruited among patients' caregivers), were classified into preclinical AD stages in accordance with NIA-AA guidelines (7).
CSF Collection, Biomarker Assessment, and APOE Genotyping
CSF samples were collected following international consensus recommendations (14) as previously described (13). Samples had been previously stored at −80 °C and had not been thawed prior to analysis. Commercially available ELISA kits were used to determine levels of CSF Aβ1–42 (InnotestTM Aβ1–42, Fujirebio-Europe, Belgium), total Tau (InnotestTM hTAU Ag), Tau phosphorylated at threonine residue 181 (InnotestTM Phospho-Tau 181P). Our laboratory has extensive experience in CSF biomarker determination and participates in the Alzheimer's Association external quality control program for CSF biomarkers (15).
Post-mortem Human Brain Samples
All post-mortem brain tissue used in this study was collected by the Neurological Tissue Bank at Hospital Clínic (IDIBAPS, Barcelona). The study (IIBSP-ATM-2012–46) was approved by the local Ethics Committee of both the Tissue Bank and Sant Pau Research Institute. Fresh brain tissue used for array tomography was collected from the superior frontal cortex of a female donor who died at the age of 83 and showed low AD pathology (Braak stage II). Pre-collected frozen tissue blocks from 6 donors (4 male, 2 female, mean age-at-death 66 years) without AD pathology were used for synaptosome and PSD enrichment.
Shotgun Liquid Chromatography Mass Spectrometry (LC-MS/MS)
Where indicated, immunodepletion was performed using the ProteoPrep® Immunoaffinity Albumin & IgG Depletion Kit (Sigma-Aldrich, Missouri). Six pools each containing CSF from 10 individuals were precipitated with acetone and protein content was quantified by Bradford assay. Samples (50 ug) were reduced in 10 mm DTT, alkylated with 55 mm IAA, and, digested in-solution with trypsin and LysC overnight and desalted using a Desalted MicroSpin Column (GE healthcare, UK). Where indicated, samples were fractionated by strong cation exchange (SCX; Empore Disks Anion Exchange-SR, Sigma-Aldrich). An equivalent of 5 μl of each CSF Sample was analyzed using a LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to a nano-LC (Proxeon, Odense, Denmark) equipped with a reversed-phase chromatography 2-cm C18 pre-column (Acclaim PepMap-100, Thermo; 100 μm i.d., 5 μm), and a reversed-phase chromatography 25 cm C18 column (Nikkyo Technos, Japan; μm i.d., 1.9 μm) using a data-dependent acquisition mode. Acquired data were analyzed using the Proteome Discoverer software suite (v1.4.1.14, Thermo Fisher Scientific), and the Mascot search engine (v2.5.1, Matrix Science) was used for peptide identification. Data were searched against the Swiss Prot Human Protein database plus the most common contaminants (version 2014, 20884 entries). A precursor ion mass tolerance of 7 ppm at the MS1 level was used, and up to three missed cleavages for trypsin were allowed. The fragment ion mass tolerance was set to 0.5 Da. Oxidation of Methionine and N-terminal protein acetylation was defined as variable modification and carbamidomethylation of Cysteines was set as fixed modification. The identified peptides were filtered at 5% FDR calculated using a target-decoy database strategy. For each identified peptide, peptide peak areas were obtained as extracted ion chromatograms and protein abundances were estimated with the average peak area of the three most intense peptides per protein. For characterization of the CSF proteome, proteins without a reviewed Uniprot identifier were excluded. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (16) partner repository (https://www.ebi.ac.uk/pride/archive) with the data set identifier PXD010356.
Database and Literature Curation
A systematic search of PubMed (www.pubmed.com) for proteomic studies was performed using the search terms “Cerebrospinal fluid”/“CSF,” & “proteome”/“proteomics” & “human,” for the CSF proteome (April 2013) and “synaptosome”/”synapse”/“post-synaptic density” & “proteome”/“proteomics” & and “human”/”mouse”/”rat,” for the synapse proteome (April 2014). Only publications in English were reviewed. All identified proteins were extracted either directly from the published material or where available, from the PRIDE repository. All proteins with a known function related to the synapse were retrieved from 4 publicly available databases. Specifically, all proteins annotated with a GO related to the synapse were retrieved from AmiGO2 version 2.3.2 (http://amigo2.geneontology.org/amigo) using the search terms “dendritic spine,” “synapse,” “synaptic,” transmission of nerve impulse” and “neurotransmitter”. All proteins annotated to “Glutamatergic synapse,” “Cholinergic Synapse,” “Serotonergic synapse,” “GABergic synapse,” “Dopaminergic synapse,” “Synaptic vesicle cycle,” ”Long-term potentiation,” ”Long-term depression,” “Retrograde endocannabinoid signaling” according to the Kyoto Encyclopedia of Genes and Genomes were retrieved from http://www.kegg.jp. All proteins with “neurotransmitter release,” “synapse,” “synaptic,” or “synaptogenesis” as an annotated function were retrieved from Uniprot (www.uniprot.org). Because each data source used distinct protein identifiers that, in some cases, had been retired or updated, a unique, reviewed Uniprot identifier was assigned to each protein using the bioinformatic gene identification conversion tools, PIR (http://pir.georgetown.edu) and Bio-Mart (http://www.ensembl.org/biomart) to avoid duplication across studies. Proteins without a reviewed Uniprot identifier were removed from the study.
Synaptosome Enrichment
All steps were performed at 4 °C. 200 mg chunks were cut from frozen frontal cortex tissue blocks and homogenized in cold Buffer A (0.32 m sucrose, 1 mm NaHCO3, 1 mm MgCl2, 0.5 mm CaCl2, 1:2500 phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin). Homogenates were centrifuged (1400 × g, 10 min) and the supernatant transferred to a new tube. The pellet was resuspended in cold Buffer A and the previous step repeated with centrifugation at 710 × g. The two supernatants were combined and centrifuged (710 × g). The supernatant was subjected to a final centrifugation (30,000 × g, 15 min). The pellet was resuspended in Buffer B (0.32 m sucrose, 1 mm NaHCO3), layered over a sucrose gradient (0.85 m, 1 m, 1.2 m) and centrifuged (82,500 × g, 2 h). The synaptosomal fraction (a thick white band at the 1–1.2 M interface) was collected, diluted in 4x volume of Buffer B. An aliquot was centrifuged (48,200 × g, 20 min) and the pellet (synaptosome) resuspended in Buffer C (50 mm Tris pH 7.4, 1% SDS) and stored at −80 °C. The remaining aliquot was diluted in equal volume Buffer D (50 mm Tris pH 7.4) and 1× volume of 2% Triton-X, incubated for 10 min and centrifuged (maximum velocity, 30 min). The pellet (post-synaptic density; PSD) was re-suspended in Buffer C and stored at −80 °C.
Array Tomography
The array tomography protocol was applied using previously described methods (17, 18). Briefly, a 1 cm3 section was taken from the superior frontal cortex was fixed, dehydrated and polymerized in 100% LR-white resin. The embedded samples were sectioned using a diamond knife (Diatome, UK) creating 20 serial 70 nm thick sections, which were mounted onto coverslips. The ultrathin ribbons were washed with Tris buffer and blocked for 5 min. Primary antibodies used were as mentioned above with the following exceptions; anti-Tenascin-R (Abcam, UK; ab121916) anti-GluR2, anti-Neuroligin-2, anti-Neurexin2 (Merck Millipore, Massachusetts; MAB397, AB15510, ABN97), anti-synaptophysin (Osenses, Australia; oss00029w) and anti-PSD95 (Synaptic Systems, Germany; 124014), alexa-tagged secondary antibodies (Thermo Fisher Scientific). Coverslips were mounted onto the slides using Slowfade Gold with DAPI (Thermo Fisher Scientific). Images were captured using a fully automated epifluorescence upright microscope (custom adapted BX51, Olympus, Pennsylvania) with a 64 × 1.2 NA Plan Apochromat objective. Image analysis was performed using Matlab (Mathworks). The script has been deposited at https://github.com/MemoryUnitSantPau with the name SynSeg. Images from serial sections were stacked, aligned, thresholded and the nonspecific staining (not present in at least 2 consecutive sections) removed using a local threshold-based algorithm. 3-D reconstructions of representative synapses were generated using the ImageJ Volume Viewer plug-in with tricubic smooth interpolation.
SDS-Page and Western Blotting
The total protein content of homogenate, synaptosome and PSD enriched fractions was quantified by bicinchoninic acid assay. Aliquots containing 20 μg total protein were boiled, diluted in loading buffer (100 mm Tris-HCL, 4% SDS, 20% glycerol, 200 mm DTT and 200 mm β-mercaptoethanol) and loaded onto a 12% Tris-Tricine gel and electrophoresed. Proteins were transferred to a nitrocellulose membrane, which was immunostained using the following antibodies; anti-CLSTN1, anti-Synaptophysin, anti-Thy1, (Abcam; ab134130, ab8049, ab133350), anti-GluR2, anti-GluR4, anti-Vamp2, anti-PSD95 (Cell Signaling; 13607, 8070, 13508, 3450), Anti-neuroligin-2, anti-Syntaxin-1B (Synaptic Systems; 129203, 110403), anti-Neurexin3, anti-Tenascin R (Thermo Fisher; PA5–47714, PA5–47546), fluorescent dye-conjugated secondary antibodies (Li-COR Biosciences, Nebraska).
Targeted Liquid Chromatography Mass Spectrometry (LC-SRM)
Fifty-four proteotypic peptides (7 to 20 amino acids long) with tryptic terminals corresponding to 22 proteins brought forward from the shotgun data were selected based on previous LC-MS/MS data and database searches (Peptide Atlas). For each targeted peptide, corresponding crude heavy peptides were synthesized with 13C6 15N4 (Arg), or 13C6 15N2 (Lys) isotopes (Peprotech SRM custom peptides, grade 2, Thermo Fisher Scientific) for use as reference internal standards in the CSF samples (Thermo Fisher), and to generate a library of MS/MS spectra for the selection of interference-free transitions for the peptides or interest. Individual CSF samples were precipitated with acetone and redissolved in 6 m urea before reduction (10 mm, DTT), alkylation (55 mm, IAA) and in-solution digestion with LysC and Trypsin (1:10, 37 °C, overnight). Isotopically-labeled peptides were spiked in each sample, and an equivalent of 5 μl of each sample was analyzed in a 120-min gradient (0–35% ACN+0.1%FA) in SRM mode using a triple quadrupole-Qtrap mass spectrometer (5500 QTrap, Sciex, Massachussetts) coupled to a nano-LC chromatography column (300 μl/min, 25-cm C18 column, 75 μm I.d., 2 μm particle size). BSA control samples were analyzed between runs for instrument quality control. Transitions were visualized and analyzed using Skyline 3.5 and peak picking was manually reviewed based on the co-elution of the reference and endogenous peptides, peak shape and correlation of transition rank intensities between the reference and endogenous transitions. Of the initial 54 targeted peptides, 32 were eliminated from the study because of poor detection in a pilot detection experiment with 5 CSF samples. The remaining 22 peptides were taken forward for further monitoring in the exploratory and validation cohorts. Peptide stability was assessed by injecting a pool of all the samples over the duration of the mass spectrometric measurements and monitoring the peak area of the standard peptides. SRM transitions were processed using the dataProcess function of MSstats v3.5 package in R (19). Specifically, the EqualizeMedians function was used to normalize the data using the transition intensities of the isotope-labeled standard peptides. This method was chosen as it assumes that the samples of a data set are separated by a constant and scales the samples so that they have the same median. The results were not significantly altered when compared with normalization using the alternative Quantile method in MSstats. The normalized data were summarized (TMP: Tukey's Median Polish) according to either peptide or protein. Transitions with between run interference were identified using the “betweenRunInterferenceScore” (cut-off = <0.8). Samples with log base-2 endogenous intensities under the cut-off designated by the MSstats package for each cohort (9.5211, 8.4337, and 8.5219, respectively) were considered as censored missing values. The mass spectrometry-based targeted proteomics data have been deposited to the Panorama repository (https://panoramaweb.org/5Jugvs.url) and the PRIDE repository with the identifier PXD012138.
Statistical Analysis
To determine enrichment of synaptic prteins in synaptosome and PSD-enriched fractions isolated from post-mortem brain tissue, the intensity of bands corresponding to the synaptic proteins on SDS-PAGE membranes were quantified using Odyssey software (Li-COR Biosciences). The ratio of each fraction relative to that of the homogenate (enrichment) was calculated. Because enrichment ratios for the 10 synaptic proteins did not clearly deviate from a Gaussian distribution (Kolmogorov-Smirnov p > 0.1), mean enrichment ratios were compared in synaptosome and PSD fractions versus homogenates by One-Way Analysis of Variance (with Dunnett's post-hoc test). For SRM data, log2 fold-changes for each preclinical and clinical AD stage relative to controls were calculated using a mixed effect linear regression model (GroupComparison function in MSstats). For meta-analyses of the SRM data from the three cohorts, p values were calculated according to Fisher's method using the “metap” package in R. Where stated, adj.p values refers to p values adjusted using the Benjamini-Hochberg method to account for multiple testing.
RESULTS
Discovery Stage - Characterizing the Synaptic Component of the CSF
To characterize the CSF proteome, we performed a discovery proteomics screen of 7 CSF pools each containing samples from 10 individuals extracted from either cognitively normal controls (5 pools), AD patients (1 pool) or a mixture of both (1 pool), varying the LC-MS/MS conditions to optimize protein yield. Further details of the sample pools, LC-MS/MS conditions and identified contaminants can be found in supplemental Table S1. We found that depletion of abundant IgGs and albumins increased the protein yield by 115% and that SCX fractionation had a greater effect on protein yield (186% increase compared with runs without SCX, 6 h LC) than extended LC times (141% increase following SCX + 6 h LC compared with SCX + 2 h LC). Overall, IgG and albumin depletion, SCX fractionation with 6 h LC resulted in the greatest protein yield (mean yield = 1,259 proteins, range = 702–1,615). Across all LC-MS/MS runs, we identified 18,785 peptides (supplemental Table S2) corresponding to 2742 unique proteins (supplemental Table S3).
To gain a more complete picture of the CSF proteome, we performed a complementary search of published proteomic studies of CSF extracted from cognitively or neurologically normal individuals. The search identified 10 independent studies (20–29) listed in Table IA. We extracted 3662 unique proteins supplemental Table S3) from the 9 studies for which data were publicly available. By combining our dataset with the datasets from the literature, we report 4315 proteins that have been identified in cognitively/neurologically normal CSF and a further 296 proteins that we detected only in pools from AD patients and/or mixed pools.
Table I. Details of published proteomic studies of the CSF and synapse. The number of proteins identified by at least 1 proteotypic peptide in each study of (A) the CSF, (B) whole synapse, (C) active zone and synaptic vesicles and (D) PSD/MAGUK-associated signalling complexes (MASC) is shown. *Data were not publicly available for retrieval. The species, tissue, clinical status (of CSF donors), synaptic fraction (synapse), protein identification and number of proteins are shown for each study.
| Study | Species (n) | Tissue | Clinical status (A)/Fraction analyzed (B–D) | Protein identification method | #Proteins published |
|---|---|---|---|---|---|
| (A) Proteomic studies of the CSF | |||||
| (20) Begcevic | Human (6) | CSF | Non-pathological | SCX+LC-MS/MS | 2615* |
| (21) Zhang 2015 | Human (14) | CSF | Non-neurological Surgery | HPLC+LC-MS/MS | 2513 |
| (22) Guldbrandsen | Human (21) | CSF | Neurologically healthy | RP-AX LC-MS/MS | 3081 |
| (23) Holtta | Human (3) | CSF | Neurologically healthy | LC-MALDI TOF/TOF | 104 |
| (24) Schutzer | Human (11) | CSF | Non-neurologic | SCX + LC-MS/MS | 2630 |
| (25) Stoop | Human (40) | CSF | Non-neurological Surgery | LC-MS & MALDI-FT-ICR-MS | 178 |
| (26) Pan 2006 | Human (19) | CSF | Cognitively normal | LC-MS/MS | 216 |
| (27) Xu | Human (22) | CSF | Cognitively normal | SCX + LC-MS/MS | 784 |
| (28) Zhang 2005 | Human (22) | CSF | Cognitively normal | SCX + LC-MS/MS | 312 |
| (29) Wenner | Human (10) | Postmortem CSF | Cognitively normal | SCX + LC-MS/MS | 249 |
| (B) Proteomic studies of the whole synapse | |||||
| (30) DiGiorgis | Human (3) | cortex | Synaptosome (phosphoproteins) | MS/MS | 24 |
| (31) Wilhelm | Rat (4) | cortex &cerebellum | Synaptosome | iBAQ | 1113 |
| (32) Abdul-Husn | Mouse (10) | hippocampus | Presynapse | In-gel(26)/In-solution: LC-MS/MS | 131 |
| Rat (10) | striatum | Presynapse | In-gel(26)/In-solution: LC-MS/MS | 113 | |
| (C) Proteomic studies of the active zone/synaptic vesicles (SV) | |||||
| (33) Weingarten | Mouse (12) | whole brain | Presynaptic active zone, SV | microOTOF-Q II/MALDI-TOF/TOF | 482 |
| (34) Blondeau | Rat | whole brain | SV (Ficoll) | In-gel(61): LCMS/MS | 209 |
| (35) Boyken | Rat | whole brain | SV (fraction 4–7, 19–21, vGLUT1/VGAT+) | iTRAQ AP+LC-MS/MS | 491 |
| (36) Coughenour | Rat | forebrain | SV (fraction 13–21) | In-gel NanosprayMS/MS QTOF | 35 |
| (37) Morciano 2005 | Rat (3) | whole brain | SV (fraction 5–11, 28–34/SV2-complex) | In-gel(36) MALDI-TOF | 93 |
| (38) Morciano 2009 | Rat | whole brain | SV (fraction 5–11, 28–34/SV2-complex) | In-gel MALDI-TOF-MS/WB | 218 |
| (39) Takamori | Rat | whole brain | SV | 2D-GE Q-TOF MS | 400 |
| (D) Proteomic studies of the post-synaptic density (PSD)/ MASC | |||||
| (40) Bayés 2011 | Human (9) | neocortex | PSD | LC-MS | 1451 |
| (41) Fernandez | Mouse | forebrain | PSD (PSD-95+complexes) | AP + LC-MS/MS LTQ-FT | 117 |
| (42) Cheng | Rat | Forebrain/cerebellum | PSD | ICAT-LC-MS/MS | 238 |
| (44) Li 2004 | Rat | forebrain | PSD | 2D-GE+MALDI-TOF/TOF | 92 |
| (43) Li 2005 | Rat | forebrain | PSD | ICAT LC-MS TOF/TOF - ICAT | 113 |
| (45) Peng | Rat | forebrain | PSD | LC-MS/MS LCQ-DECA XP Ion trap | 365 |
| (46) Walikonis | Rat | forebrain | PSD | 2D-GE, MALDI-TOF MS | 26 |
| (47) Bayés 2012 | Mouse (3) | cortex | PSD | LC-MS | 1546 |
| (48) Collins 2006 | Mouse | whole brain | PSD | SDS-PAGE-LC-MS/MS Q-TOF | 617 |
| (49) Trinidad | Mouse | whole brain | PSD | SCX Nano-LC-ESI-QTOF MS/MS | 242 |
| (50) Bayés 2014 | Human | neocortex | MASC | AP-Orbitrap LCMS/MS | 289 |
| (48) Collins 2006 | Mouse | whole brain | MASC | SDS-PAGE-LC-MS/MS Q-TOF | 186 |
To identify the synaptic component of the CSF, we next sought to characterize the human synaptic proteome also using publicly available data, but in this case related to the characterization of the synapse. Here we have defined the synaptic proteome as proteins that (1) have an annotated synaptic function according to 4 selected public databases (see Experimental Procedures), and (2) have been detected in at least 1 of 23 published proteomic studies (30–50) of synapse-enriched fractions from rodent or human brain tissue (Table IB–ID). supplemental Table S4 shows the 537 proteins that satisfy these criteria. Cross-referencing the CSF with this shortlist, we identified 251 synaptic proteins that are detectable in the CSF by shotgun mass spectrometry (supplemental Table S5). Using these criteria, we report that ∼6% of CSF proteins are of synaptic origin.
Verification Stage - Selecting a Panel of Synapse-specific Proteins Suitable for SRM
The 251 synaptic proteins that were detectable in the CSF were manually curated and an initial list of 22 proteins that participate in one or more core synaptic process (Calsyntenin-1 (51), GABAB receptor 1 (52), GABAA receptor A2 (53), GABAA receptor A3, GluR2 (54), GluR4 (55), mGluR1 (56), Munc18 (57), Neurexin-2A (58) Neurexin-3A (59), Neuroligin-1 (60), Neuroligin-2 (61), Neuroplastin (62), Synapsin-1 (63), Synaptic Ras-GAP 1 (64), Synaptic vesicle glycoprotein 2B (65), Synaptotagmin-1 (66), Syntaxin-1A (67), Syntaxin-1B, Tenascin-R (68), Thy-1 and Vamp-2 (69)) were selected with the goal of defining a subset of proteins that is amenable for multiplex quantification. To confirm detection of the 22 proteins in CSF by targeted mass spectrometry, we developed SRM assays using 54 isotopically-labeled peptides corresponding to the 22 selected proteins to be used as internal references. We monitored the peptides in a preliminary set of 5 CSF samples and eliminated 32 peptides from the study because of poor detection (supplemental Table S6).
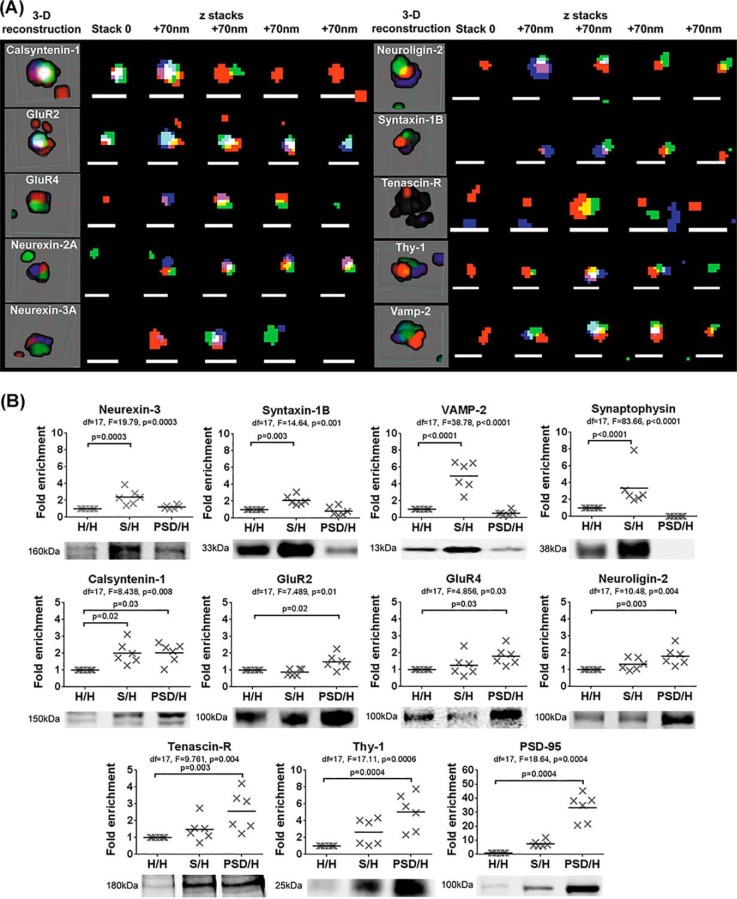
To investigate the specificity of the expression of the remaining 10 proteins in human synapses, we applied array tomography (AT) microscopy to our panel of proteins in human cortical tissue. AT is particularly suited to the study of synapses as it provides improved spatial resolution in the axial plane compared with other light microscopy techniques (i.e. confocal microscopy). By obtaining ultrathin (70 nm) tissue sections, single synapse terminals may be identified. Representative 3D reconstructions of single synapse terminals clearly show the expression of 9 of the panel proteins directly at the synapse, marked by pre (synaptophysin) and post (PSD-95) synaptic markers (Fig 1A). In contrast, Tenascin-R was found surrounding the synapse without making direct contact. This is consistent with the literature where Tenascin-R has been reported to reside in extracellular perineuronal nets that surround the synapse (70).
Fig. 1.
Expression of the panel proteins at the human cortical synapse. A, Using AT microscopy, ultrathin tissue slices (70 nm) from human post-mortem cortical tissue from one brain donor were immunostained for pre- (synaptophysin, red) and post- (PSD95, green) synaptic markers and the synaptic panel proteins (blue). A representative 3-D reconstruction of a representative synapse is shown for each panel protein. The segmented immunofluorescence of the 3 proteins in each individual stack (at 70 nm increments) is shown to the right of the reconstruction. Scale bars representing 1 μm are shown at the bottom of each z stack. B, Mean fold-enrichment plotted for each panel protein in homogenate (H), synaptosome-enriched (S) and PSD-enriched (PSD) fractions taken from post-mortem human cortex (n = 6). S/H and PSD/H; intensity in S or PSD fractions relative to H for the same sample. H/H; intensity in the H fraction for each sample relative to the mean intensity in the H fraction across all samples. Enrichment of the pre- (synaptophysin) and post (PSD-95) markers are also shown. Degrees of freedom (df), F statistic and p values for the ANOVA are shown at the top of each plot and Dunnett's p values are shown for significant pair-wise comparisons (α = 0.05).
Specificity of synaptic expression was further evaluated by quantifying the enrichment of the panel proteins in synaptosome and PSD fractions extracted from 6 human cortical tissue samples without pathology (supplemental Table S7). The 2A isoform of Neurexin-2 could not be analyzed because of the lack of a commercially-available specific antibody suitable for Western blotting. Fig 1B shows that all 9 proteins tested were enriched in synaptic fractions compared with the homogenate (p < 0.03). Specifically, Calsyntenin-1, Neurexin-3A, Syntaxin-1B and Vamp-2 were enriched 2.0 to 5.0-fold in synaptosomes (p < 0.02) with Vamp-2 in particular, showing greater enrichment (5-fold, p < 0.0001) than the widely-used pre-synaptic marker, synaptophysin (3-fold, p < 0.0001). Calsyntenin-1, GluR2, GluR4, Neuroligin-2, Tenascin-R and Thy-1 were enriched 1.8 to 5.0-fold in the PSD fraction (p < 0.03). Therefore all 10 proteins are expressed specifically at the human synapse or in the surrounding area (Tenascin R).
Exploration Phase - Characterizing the CSF Profile of the Synaptic Panel in AD
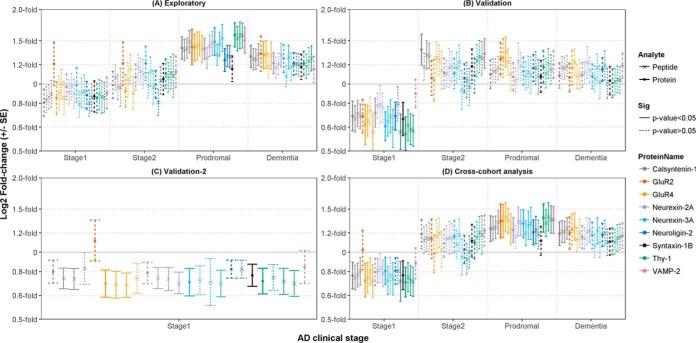
Because Tenascin-R, was not directly expressed at the synapse, this protein was not brought forward for evaluation. Table II provides further information regarding the 9 synaptic proteins and their corresponding peptides that were monitored by SRM in the exploratory cohort comprising 80 CSF samples from controls and all preclinical and clinical stages of the AD continuum (Table IIIA). Supplemental Table S8A lists the transitions included and excluded from the analysis. Fig 2A shows the relative fold-change in CSF levels of the 20 individual peptides and summarized protein levels at each AD stage compared with the cognitively normal, AD biomarker negative, control group (n = 20). supplemental Table S9A shows the raw values. Peptides were not significantly altered in individuals at preclinical AD stage 1 (n = 10) compared with controls, albeit that 17 peptides from 8 proteins were subtly reduced 0.8 to 0.9-fold (p > 0.1). At preclinical AD stage 2 (n = 10), no differences were observed (0.9 to 1.1-fold, p > 0.2). In prodromal AD patients (n = 20), 19 peptides from 8 proteins were elevated 1.3 to 1.6-fold (p < 0.04, adj.p < 0.04). In patients at the dementia stage (n = 20), Calsyntenin-1 (1.2 to 1.3-fold, p < 0.02, adj.p = 0.1), GluR4 (1.3-fold, p < 0.04, adj.p = 0.1), Neurexins-2A and 3A and Thy-1 (1.2 to 1.3-fold, p < 0.04, adj.p = 0.1) peptides remained elevated. Overall, levels of peptides from the same protein were highly correlated across all samples (ρ = 0.77 to 0.98). Excepting the GluR2 peptide, which showed relatively elevated levels (p > 0.05) at preclinical stage 1, the fold-changes of all peptides and proteins were comparable to each other at all disease stages, differing only in significance level, indicating very little peptide or protein-specific differences. In fact, the levels of all proteins across all disease stages were positively correlated (n = 80, all ρ>0.37, all p < 7 × 10−4), Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, Neuroligin-2 and Thy-1 in particular (ρ = 0.66 to 0.90).
Table II. Synaptic proteins and corresponding peptides monitored by SRM in CSF samples. The proteins (A) are listed with a summary of the reported synaptic function of the protein and coverage achieved by quantifying the peptides listed in (B).
| (A) Protein |
(B) Peptides |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name; UniprotID | Dendritic spine assembly, maintenance and maturation | Postsynaptic Ca2+ signalling | AMPA receptor endocytosis and trafficking | Synaptic transmission and plasticity | Synaptogenesis | Synaptic vesicle exocytosis | Presynaptic differentiation | Length; Coverage | Sequence | Position; Domain; Protein Isoform; Protein products where the peptide resides |
| Calsyntenin-1; O94985 | (51) | (87) | 981aa; 3% | LTVTAYDCGK | 235–244; extracellular;1,2; Mature, sAlc-α | |||||
| GNLAGLTLR | 537–545; extracellular;1,2; Mature, sAlc-α | |||||||||
| IISTITR | 684–690; extracellular;1,2; Mature, sAlc-α | |||||||||
| GluR2; P42262 | (88) | (89) | (90) | 883aa; 2% | YTSALTYDAVQVMTEAFR | 295–312; extracellular; Flip,Flop,3 4; Mature | ||||
| GluR4; P48058 | (90) | 902aa; 5% | LQNILEQIVSVGK | 218–230; extracellular;1,2; Mature | ||||||
| GYHYIIANLGFK | 234–245; extracellular;1,2; Mature | |||||||||
| IQGLTGNVQFDHYGR | 354–368; extracellular;1,2; Mature | |||||||||
| Neurexin-2A; Q9P2S2 | (58) | (91) | 1712aa; 2% | LSALTLSTVK | 161–170; extracellular;1A,2A; Mature, sNRX2A | |||||
| LGERPPALLGSQGLR | 184–198; extracellular;1A,2A; Mature, sNRX2A | |||||||||
| LQGDLSFR | 478–485; extracellular;1A,2A; Mature, sNRX2A | |||||||||
| Neurexin-3A; Q9Y4C0 | (58) | (59) | (91) | 1643aa; 3% | SDLSFQFK | 49–56; extracellular;1A,4A; Mature, sNRXN3 | ||||
| NGLILHTGK | 293–301; extracellular;1A,4A; Mature, sNRXN3 | |||||||||
| ANDGEWYHVDIQR | 537–549; extracellular;1A,3A,4A; Mature, sNRXN3 | |||||||||
| Neuroligin-2; Q8NFZ4 | (92) | (61) | (93) | 835aa; 4% | ELVDQDVQPAR | 336–346; extracellular;1; Mature, sNLGN2 | ||||
| TLLALFTDHQWVAPAVATAK | 450–469; extracellular;1; Mature, sNLGN2 | |||||||||
| Syntaxin-1B; P61266 | (94) | (95) | 288aa; 5% | AIEQSIEQEEGLNR | 94–107; cytoplasmic;1,2; Mature | |||||
| Thy-1; P04216 | (96) | (97) | (98) | 161aa; 24% | VTSLTACLVDQSLR | 22–35; polypeptide chain;1; Mature | ||||
| HVLFGTVGVPEHTYR | 61–75; polypeptide chain;1; Mature | |||||||||
| VLYLSAFTSK | 88–97; polypeptide chain;1; Mature | |||||||||
| Vamp-2; P63027 | (69) | 116aa; 14% | LQQTQAQVDEVVDIMR | 32–47; cytoplasmic, v-SNARE motif;1; Mature | ||||||
Table III. Demographic and biomarker levels for CSF cohorts used for Targeted SRM. The number of samples, percentage female and APOE ϵ4 (E4+) carriers as well as the mean, standard deviation (S.D.) and range of the age-at-lumbar puncture, mini-mental stage examination (MMSE) score, and CSF levels of Aβ42, total tau and phosphorylated tau (p-tau) are shown for each diagnostic group of the exploratory (A) and validation cohorts 1 (B) and 2 (C). Diagnostic groups were classified according to AD biomarkers for amyloidosis (+/−) and neurodegeneration (+/−) and degree of cognitive impairment (−; cognitively normal, +; amnestic mild cognitive impairment, ++; Dementia). Local biomarker cut-offs for positivity were as follows; Aβ42. <550 ng/ml (Exploratory), <500 ng/ml (Validation). PET positive, total tau >350 ng/ml (Exploratory), >450 ng/ml (age50–70) />500 ng/ml (age>70) (Validation), phosphorylated tau (p-tau) >61 ng/ml (Exploratory), >75 ng/ml (Validation).
| Clinical stage | Amyloid- osis | Neuro-degeneration | Cognitive decline | N (% female, %E4+) | Age at collection | Mean +/− S.D. (range) |
|||
|---|---|---|---|---|---|---|---|---|---|
| MMSE score | Aβ42 (ng/ml) | total tau (ng/ml) | p-tau (ng/ml) | ||||||
| (A) | |||||||||
| Controls | − | − | − | 20 (75,0) | 67+/−5 (60–77) | 29+/−1 (28–30) | 899+/−175 (618–1147) | 205+/−35 (144–274) | 38+/−6 (30–49) |
| Preclinical 1 | + | − | − | 10 (60,86) | 69+/−4 (64–75) | 28+/−2 (26–30) | 406+/−92 (245–509) | 232+/−67 (155–318) | 43+/−7 (30–52) |
| Preclinical 2/3 | + | + | − | 10 (50,75) | 68+/−5 (63–76) | 29+/−2 (26–30) | 412+/−43 (361–485) | 601+/−591 (323–2270) | 85+/−42 (56–201) |
| Prodromal AD | + | + | + | 20 (65,83) | 70+/−4 (60–77) | 28+/−2 (24–30) | 449+/−62 (300–548) | 716+/−355 (352–1771) | 108+/−45 (66–230) |
| AD dementia | + | + | ++ | 20 (65,70) | 72 (62–81)+/−6 | 23+/−2 (19–28) | 400+/−87 (178–530) | 708+/−398 (353–1784) | 91+/−36 (45–206) |
| (B) | |||||||||
| Controls | − | − | − | 18 (61,17) | 60+/−5 (52–68) | 29+/−1 (26–30) | 788+/−227 (528–1351) | 235+/−79 (111–429) | 54+/−13 (37–71) |
| Preclinical 1 | + | − | − | 9 (78,44) | 64+/−7 (55–73) | 28+/−1 (26–30) | 375+/−84 (263–466) | 182+/−60 (103–258) | 43+/−14 (26–61) |
| Preclinical 2/3 | + | + | − | 8 (38,38) | 76+/−6 (68–85) | 28+/−1 (26–29) | 362+/−94 (229–486) | 677+/−539 (389–2093) | 100+/−45 (64–213) |
| Prodromal AD | + | + | + | 10 (50,100) | 67+/−9 (53–77) | 25+/−3 (18–29) | 357+/−73 (243–456) | 773+/−217 (451–1253) | 111+/−20 (75–144) |
| AD dementia | + | + | ++ | 15 (60,60) | 66+/−9 (51–83) | 23 (18–26) | 322+/−85 (206–441) | 1039+/−484 (430–2000) | 142+/−49 (76–222) |
| (C) | |||||||||
| Controls | − | − | − | 20 (55,25) | 57+/−8 (45–76) | 29+/−1 (28–30) | 835+/−137 (614–1151) | 151+/−58 (79–290) | 31+/−11 (4–59) |
| Preclinical 1 | + | − | − | 18 (56,39) | 57+/−8 (40–74) | 29+/−1 (27–30) | 516+/−81 (384–734) | 136+/− (61–292) | 29+/−10 (18–57) |
Fig. 2.
Synaptic panel peptide levels in the CSF across the AD continuum. The log2 fold-change (± standard error; S.E.) in CSF levels of the synaptic panel peptides and summarized protein levels are plotted for each preclinical and clinical AD stage versus cognitively normal controls for (A) the exploratory cohort (Controls n = 20, Stage1 n = 10, Stage2 n = 10, Prodromal n = 20, AD dementia n = 20), (B) validation cohort-1 (Controls n = 18, Stage1 n = 9, Stage2 n = 8, Prodromal n = 10, AD dementia n = 15), (C) validation cohort-2 (Controls n = 20, Stage1 n = 18) and (D) the combined mean log2 fold-change across the 3 cohorts. For ease of interpretation, the natural values are labeled on the y axis on a log2 scale. The linestyle of the error bars were determined by p value cut-offs for pair-wise group comparisons using a mixed effect linear regression model (see legend). Stage1; preclinical AD stage 1, Stage2; preclinical stage 2,Prodromal; prodromal AD, Dementia; AD dementia.
Validation Phase - Validation of the Synaptic CSF Profile in an Independent AD Cohort
All 20 peptides were brought forward for validation in an independent clinical cohort comprising 60 CSF samples controls and preclinical and clinical stages of the AD continuum (Table 3B). Supplemental Table S8B lists the transitions included and excluded from the analysis. Fig 2B shows the relative fold-change in CSF levels of the individual peptides and summarized proteins at each AD stage compared with the cognitively normal, AD biomarker negative control group (n = 18). Supplemental Table S9B shows the raw values. Like the exploratory cohort, levels of peptides from the same protein were highly correlated across all samples (ρ = 0.52 to 0.98) and all 20 peptides showed comparable CSF profiles at all disease stages. All proteins were positively correlated (n = 60, all ρ>0.36 all p < 0.005), GluR4, Neurexin-2A, Neuroligin-2, and Thy-1 in particular (ρ = 0.61 to 0.80). The greatest fold-change was observed in individuals at preclinical stage 1 (n = 9) in whom Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, Neuroligin-2, Syntaxin-1, and Thy-1 peptides were reduced 0.6 to 0.8-fold (p < 0.05, adj.p = 0.03 to 0.09). In individuals at preclinical stage 2 (n = 8), Calsyntenin-1 and Vamp-2 peptides were elevated 1.3-fold (p < 0.05, adj.p > 0.1). Although all peptides showed positive fold-changes (1.1 to 1.3-fold) in prodromal AD patients (n = 10), the only peptide to validate the elevated levels observed in the exploratory analyses was for GluR4, which was elevated 1.3-fold (p = 0.04, adj.p = 0.5). None of the peptides were significantly elevated in patients at the dementia stage (1.0 to 1.2-fold, p > 0.09).
Although, all synaptic proteins moderately correlated with total tau levels, in both cohorts (exploratory; n = 80, ρ = 0.45 to ρ = 0.67, validation; n = 60, ρ = 0.36 to 0.56), by definition, total tau levels are unaltered in individuals at preclinical AD stage 1. Therefore, a reduction of synaptic proteins at preclinical stage 1 would be of increased clinical value because individuals at this early stage are asymptomatic and have yet to demonstrate widespread neuronal degeneration. We therefore sought to replicate this finding in a larger set of controls and preclinical AD stage 1 (n = 38) from the SPIN cohort. Supplemental Table S8C lists the transitions included and excluded from the analysis. Fig 2C shows that compared with control subjects, Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, Syntaxin-1 and Thy-1 were reduced 0.7 to 0.8-fold (p < 0.05). Supplemental Table S9C shows the raw values.
Finally, we performed a cross-cohort meta-analysis by combining the p values (Fisher's p value) at each disease stage for peptides that demonstrated fold-changes in the same direction across all cohorts. Supplemental Table S9D shows the raw values. The mean of fold-change for each peptide at each disease stage is plotted in Fig 2D. In this meta-analysis, Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, Syntaxin-1B, and Thy-1 peptides were reduced (0.8-fold, Fisher's p < 0.05) in individuals at preclinical AD stage 1, Calsyntenin-1, GluR2, GluR4, Neurexin-2A, Neurexin-3A, Neuroligin-2, Thy-1, and Vamp-2 peptides were elevated (1.2 to 1.4-fold, Fisher's p < 0.04) in prodromal AD patients and Calsyntenin-1, GluR4, Neurexin-2A, and Neurexin-3A were elevated (1.2 to 1.3-fold, p = 0.04) at the dementia stage. Therefore, these data support a biphasic profile for 5 proteins (Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, and Thy-1) whereby compared with controls, CSF peptide levels are reduced at the earliest preclinical stage 1 of AD when neurodegeneration has yet to take hold, as indicated by total tau levels, but elevated at the prodromal AD stage when neurodegeneration is widespread.
DISCUSSION
This is the first systematic study of the CSF proteome and thorough characterization of its synaptic component. Using this resource, we selected a panel of 10 synaptic proteins for evaluation as potentially novel biomarkers of synapse degeneration in neurological diseases characterized by synaptopathy. We confirmed the specific expression of 9 of the synaptic proteins at the human cortical neuronal synapse therefore increasing the probability that their CSF levels are directly related to synapse integrity. A thorough evaluation of the CSF profile of the 9 synaptic proteins in three independent clinical cohorts comprising controls and pre-clinical and clinical stages of AD revealed a set of six synaptic proteins (Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, Syntaxin-1B and Thy-1) that were reduced 0.8-fold (p < 0.05) at the earliest preclinical stage of AD (stage 1, cognitively normal with positive amyloid markers) compared with controls. Five of these proteins were also elevated in patients at the clinical stages (cognitive impairment and positive neurodegeneration markers). We propose that the reduced levels in preclinical stage 1 may reflect reduced synaptic density in these individuals who already show signs of brain amyloidosis, an effect that is confounded by widespread neurodegeneration at later disease stages.
In the discovery stage of this study, a thorough proteomic screen of 60 CSF samples combined with a literature search of proteomic studies of the CSF identified 4,315 proteins in cognitively normal individuals. To put this into context, the latest release of the Human Protein Atlas (v17) comprises RNA expression for 10,226 unique genes that were positively detected in the brain (cortex, cerebellum or hippocampus), of which we report that 26% are detectable in the CSF. According to the Human Protein Atlas data, we estimate that 79% of the brain-expressed proteins detected in the CSF are known to be expressed in the cortex, 65% in the hippocampus and 67% in the cerebellum.
We then searched the literature and public databases for proteins that are both physically and functionally related to the synapse, thereby prioritizing well-characterized synaptic proteins. The 537 proteins retrieved represent 4% of those with positive RNA brain expression data included in the Human Protein Atlas database, suggesting that this is a relatively stringent classification. Regional expression data from the Human Protein Atlas provide evidence for 95% of these synaptic transcripts in the cerebral cortex, 75% in the hippocampus and 87% in the cerebellum. We conclude that most of the proteins we have classified as synaptic are expressed across multiple brain regions. This lack of regional specificity offers the possibility that these proteins could be informative across the range of neurological and psychiatric diseases, which demonstrate distinct regional susceptibility. Notably, our CSF database included 47% of these synaptic proteins. When compared with the 26% yield of brain-expressed proteins in CSF, we conclude that the coverage of synaptic proteins in our CSF database was relatively high.
In the verification stage, we employed AT microscopy, a technique especially suited to study single synapses (17, 18) to verify the synaptic expression of a set of proteins detectable in the CSF by SRM and with a known synaptic function at the human cortical synapse. With improved antibody penetration compared with other super resolution techniques and improved resolution compared with traditional confocal microscopy, we report with high confidence that our panel of 9 proteins are expressed directly at or surrounding human cortical neuronal synapses. The synaptic panel includes proteins with a range of synaptic functions, including but not limited to, spine assembly and maturation, synaptic transmission and plasticity, synaptogenesis and synaptic vesicle exocytosis. No clear difference in CSF profile could be attributed to any function. The protein expression of 7 of these proteins has been assessed by the Human Protein Atlas, all of which were detected in the cortical neuropil. Calsyntenin-1 and Neuroligin-2 were the only proteins detected, at low levels, in glial cells. Consistent with the extracellular expression in perineuronal nets, Tenascin-R was detected in the neuropil but not in neurons or glia, further supporting its exclusion from the remainder of the study. In AD, reduced brain expression at the transcriptomic or proteomic level has been reported for GluR2, GluR4, Neurexin-2A, Syntaxin-1B, Tenascin-R, Thy-1, and VAMP-2, whereas increased expression has been reported for Neuroligin-2 (71–74). Pertinently, there is evidence in the literature that some of the panel proteins may be involved in mechanisms directly related to AD pathogenesis. For example, GluR2, (75) GluR4 (76), and Neurexin-2A (77) have been postulated as direct targets of Aβ oligomers. Additionally, there is evidence that Calsyntenin-1 (78, 79) and Vamp-2 (80) play a role in Aβ production, trafficking or secretion and a role for the Neurexin-Neuroligin complex in regulating Aβ metabolism and function has also been postulated (81). The potential involvement of these proteins in AD pathogenesis makes these proteins particularly attractive candidate biomarkers for AD. This does not exclude their potential as biomarkers of synapse loss in other neurological diseases. Defective GluR2 RNA editing may be relevant to amyotrophic lateral sclerosis (82), whereas variations in genes encoding Neuroligin-2, Neurexin-3 and Syntaxin-1B have been implicated in schizophrenia (83), autism spectrum disorder (84) and epilepsy (85), respectively.
In the exploration and validation stages of this study, we quantified the 9 synapse-verified proteins in three independent clinical cohorts of CSF samples from cognitively normal controls and individuals from the AD continuum. As has been reported for other synaptic proteins, such as neurogranin (86), synaptotagmin-1 (10) and SNAP-25 (9), we observed an increase in CSF levels of 8 of the panel proteins in prodromal AD patients compared with controls. The moderate correlation of the synaptic panel proteins with a marker of tau-mediated neurodegeneration suggests that synapse and neuronal loss are interrelated in AD and that the panel proteins partially reflect both processes. GluR2, Neurexin-2A, Neuroligin-2, Syntaxin-1B, and Vamp-2 showed a weaker correlation with CSF total tau than that reported for neurogranin, synaptotagmin and SNAP-25. It remains to be determined whether the greater independence of these proteins from CSF tau levels compared with other synaptic proteins provides additional clinical value when used as biomarkers. We also report that 12 peptides from Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, and Thy-1 were reduced at preclinical AD stage 1. Our data support that reduced levels of synaptic proteins in the CSF reflect a reduced synaptic density in these individuals, which is only apparent when not being masked by widespread neurodegeneration. These proteins could be valuable markers for monitoring disease progression in at-risk individuals before the appearance of cognitive symptoms.
In conclusion, we have identified and provided clinical validation for a set of synaptic proteins (Calsyntenin-1, GluR4, Neurexin-2A, Neurexin-3A, Syntaxin-1B, and Thy-1) that can be detected in CSF by targeted liquid chromatography mass spectrometry without depletion, immunoprecipitation or fractionation of the sample that could provide added value in the clinical setting to assess disease progression in individuals at-risk for AD and AD patients and could improve enrichment and monitoring of drug efficacy in pharmaceutical drug trials. To our knowledge, this is the first study to demonstrate a non-linear profile of a subset of synaptic proteins in AD with changes observed at the earliest preclinical AD stage before neurodegeneration is widespread. This study serves as a basis for future research on the dynamics of synaptic proteins in the CSF. Because the cross-sectional design of the current study does not allow assessment of the prognostic capacity of the synaptic panel proteins, future longitudinal studies in large cohorts are necessary to further explore the potential of the synaptic panel proteins as prognostic biomarkers that precede markers of neurodegeneration. The potential of the proteins described herein to be useful prognostic biomarkers across a range of neurological and psychiatric disease opens many avenues for further investigation.
DATA AVAILABILITY
The shotgun proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (16) partner repository (https://www.ebi.ac.uk/pride/archive) with the dataset identifier PXD010356. The targeted proteomics data have been deposited to the Panorama repository (https://panoramaweb.org/5Jugvs.url) and the PRIDE repository with the identifier PXD012138. The script used for Matlab image analysis has been deposited at https://github.com/MemoryUnitSantPau with the name SynSeg.
Supplementary Material
Acknowledgments
We thank the patients and their families for their participation in this study.
Footnotes
* This work was supported by the following research grants: Fondos de Investigaciones Sanitarias (PI15/00058, PI14/01561, PI18/000327) from the “Fondo Europeo de Desarrollo Regional (FEDER)”, Unión Europea, “Una manera de hacer Europa” and the “Instituto de Salud Carlos III, Ministerio de Economia y Competitividad, Gobierno de España” (ISCIII). Additional funding came from the “Departament de Salut, Generalitat de Cataluña - Pla Estratègic de Recerca i Innovació en Salut (PERIS) 2016-2020, 2017-2019” (SLT002/16/00408, 2017SGR547), “Programa 1 Enfermedad de Alzheimer y otras demencias degenerativas” from the Centro de Investigación Biomédica en Red Enfermedades Neurodegenerativas (CIBERNED), the “Fundació Bancaria La Caixa” (4560/6393) and “La Marató” organized by the television channel, TV3 (201426 10). OB is supported by the Miguel Servet program (CP13/00091) from the ISCIII and FEDER. AB is supported by the Ministerio de Economia y Competitividad, Gobierno de España and FEDER (BFU2012-34398 and BFU2015-69717-P), by the Career Integration Grant from the European Union (ref. 304111, Marie Curie Actions), by the Ramón y Cajal Fellowship (ref. RYC-2011-08391) from the Ministerio de Economia y Competitividad, Gobierno de España and by the CERCA Programme from the Generalitat de Catalunya. RN-L is supported by the research grant from the Generalitat de Catalunya (PERIS SLT/2381/2016/00099). The proteomics analyses were performed in the CRG/UPF Proteomics Unit which is part of the Proteored, PRB3 and is supported by grant PT17/0019, of the PE I+D+i 2013–2016, funded by ISCIII and ERDF.
 This article contains supplemental material.
This article contains supplemental material.
The authors declare that the Biomedical Research Institute Sant Pau (IIB Sant Pau) has filed a patent application (pending) to the European Patent Office (EP18382175.0) to protect the intellectual property included in this manuscript. O Belbin, A Lleó, Á Bayés, J Fortea and D Alcolea are the named inventors.
1 The abbreviations used are:
- AD
- Alzheimer's disease
- AT
- array tomography
- CSF
- cerebrospinal fluid
- FDR
- false discovery rate
- GO
- gene ontology
- LC
- liquid chromatography
- MS/MS
- mass spectrometry
- NIH-AA
- National Institute of Health-Institute of Aging
- PSD-95
- post-synaptic density
- SCX
- strong cation exchange
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Selkoe D. J. (2002) Alzheimer's disease is a synaptic failure. Science 298, 789–791 [DOI] [PubMed] [Google Scholar]
- 2. Calabrese F., Riva M. A., and Molteni R. (2016) Synaptic alterations associated with depression and schizophrenia: potential as a therapeutic target. Expert. Opin. Ther. Targets 20, 1195–1207 [DOI] [PubMed] [Google Scholar]
- 3. Bellucci A., Mercuri N. B., Venneri A., Faustini G., Longhena F., Pizzi M., Missale C., and Spano P. (2016) Review: Parkinson's disease: from synaptic loss to connectome dysfunction. Neuropathol. Appl. Neurobiol. 42, 77–94 [DOI] [PubMed] [Google Scholar]
- 4. Spires-Jones T. L., and Hyman B. T. (2014) The intersection of amyloid beta and tau at synapses in Alzheimer's disease. Neuron 82, 756–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Albert M. S., DeKosky S. T., Dickson D., Dubois B., Feldman H. H., Fox N. C., Gamst A., Holtzman D. M., Jagust W. J., Petersen R. C., Snyder P. J., Carrillo M. C., Thies B., and Phelps C. H. (2011) The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKhann G. M., Knopman D. S., Chertkow H., Hyman B. T., Jack C. R. Jr, Kawas C. H., Klunk W. E., Koroshetz W. J., Manly J. J., Mayeux R., Mohs R. C., Morris J. C., Rossor M. N., Scheltens P., Carrillo M. C., Thies B., Weintraub S., and Phelps C. H. (2011) The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sperling R. A., Aisen P. S., Beckett L. A., Bennett D. A., Craft S., Fagan A. M., Iwatsubo T., Jack C. R. Jr, Kaye J., Montine T. J., Park D. C., Reiman E. M., Rowe C. C., Siemers E., Stern Y., Yaffe K., Carrillo M. C., Thies B., Morrison-Bogorad M., Wagster M. V., and Phelps C. H. (2011) Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7, 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thorsell A., Bjerke M., Gobom J., Brunhage E., Vanmechelen E., Andreasen N., Hansson O., Minthon L., Zetterberg H., and Blennow K. (2010) Neurogranin in cerebrospinal fluid as a marker of synaptic degeneration in Alzheimer's disease. Brain Res. 1362, 13–22 [DOI] [PubMed] [Google Scholar]
- 9. Brinkmalm A., Brinkmalm G., Honer W. G., Frolich L., Hausner L., Minthon L., Hansson O., Wallin A., Zetterberg H., Blennow K., and Ohrfelt A. (2014) SNAP-25 is a promising novel cerebrospinal fluid biomarker for synapse degeneration in Alzheimer's disease. Mol. Neurodegener 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ohrfelt A., Brinkmalm A., Dumurgier J., Brinkmalm G., Hansson O., Zetterberg H., Bouaziz-Amar E., Hugon J., Paquet C., and Blennow K. (2016) The pre-synaptic vesicle protein synaptotagmin is a novel biomarker for Alzheimer's disease. Alzheimers Res. Ther. 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duits F. H., Brinkmalm G., Teunissen C. E., Brinkmalm A., Scheltens P., Van der Flier W. M., Zetterberg H., and Blennow K. (2018) Synaptic proteins in CSF as potential novel biomarkers for prognosis in prodromal Alzheimer's disease. Alzheimers Res. Ther. 10, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sala I., Illan-Gala I., Alcolea D., Sanchez-Saudinos M. B., Salgado S. A., Morenas-Rodriguez E., Subirana A., Videla L., Clarimon J., Carmona-Iragui M., Ribosa-Nogue R., Blesa R., Fortea J., and Lleo A. (2017) Diagnostic and Prognostic Value of the Combination of Two Measures of Verbal Memory in Mild Cognitive Impairment due to Alzheimer's Disease. J. Alzheimers Dis. 58, 909–918 [DOI] [PubMed] [Google Scholar]
- 13. Alcolea D., Martinez-Lage P., Sanchez-Juan P., Olazaran J., Antunez C., Izagirre A., Ecay-Torres M., Estanga A., Clerigue M., Guisasola M. C., Sanchez Ruiz D., Marin Munoz J., Calero M., Blesa R., Clarimon J., Carmona-Iragui M., Morenas-Rodriguez E., Rodriguez-Rodriguez E., Vazquez Higuera J. L., Fortea J., and Lleo A. (2015) Amyloid precursor protein metabolism and inflammation markers in preclinical Alzheimer disease. Neurology 85, 626–633 [DOI] [PubMed] [Google Scholar]
- 14. Teunissen C. E., Tumani H., Bennett J. L., Berven F. S., Brundin L., Comabella M., Franciotta D., Federiksen J. L., Fleming J. O., Furlan R., Hintzen R. Q., Hughes S. G., Jimenez C. R., Johnson M. H., Killestein J., Krasulova E., Kuhle J., Magnone M. C., Petzold A., Rajda C., Rejdak K., Schmidt H. K., van Pesch V., Waubant E., Wolf C., Deisenhammer F., Giovannoni G., and Hemmer B. (2011) Consensus Guidelines for CSF and Blood Biobanking for CNS Biomarker Studies. Mult. Scler. Int. 2011, 246412. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Mattsson N., Andreasson U., Persson S., Carrillo M. C., Collins S., Chalbot S., Cutler N., Dufour-Rainfray D., Fagan A. M., Heegaard N. H., Robin Hsiung G. Y., Hyman B., Iqbal K., Kaeser S. A., Lachno D. R., Lleo A., Lewczuk P., Molinuevo J. L., Parchi P., Regeniter A., Rissman R. A., Rosenmann H., Sancesario G., Schroder J., Shaw L. M., Teunissen C. E., Trojanowski J. Q., Vanderstichele H., Vandijck M., Verbeek M. M., Zetterberg H., and Blennow K. (2013) CSF biomarker variability in the Alzheimer's Association quality control program. Alzheimers Dement 9, 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Colom-Cadena M., Pegueroles J., Herrmann A. G., Henstridge C. M., Munoz L., Querol-Vilaseca M., Martin-Paniello C. S., Luque-Cabecerans J., Clarimon J., Belbin O., Nunez-Llaves R., Blesa R., Smith C., McKenzie C. A., Frosch M. P., Roe A., Fortea J., Andilla J., Loza-Alvarez P., Gelpi E., Hyman B. T., Spires-Jones T. L., and Lleo A. (2017) Synaptic phosphorylated alpha-synuclein in dementia with Lewy bodies. Brain 140, 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pickett E. K., Henstridge C. M., Allison E., Pitstick R., Pooler A., Wegmann S., Carlson G., Hyman B. T., and Spires-Jones T. L. (2017) Spread of tau down neural circuits precedes synapse and neuronal loss in the rTgTauEC mouse model of early Alzheimer's disease. Synapse 71, e21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choi M., Chang C. Y., Clough T., Broudy D., Killeen T., MacLean B., and Vitek O. (2014) MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 [DOI] [PubMed] [Google Scholar]
- 20. Begcevic I., Brinc D., Drabovich A. P., Batruch I., and Diamandis E. P. (2016) Identification of brain-enriched proteins in the cerebrospinal fluid proteome by LC-MS/MS profiling and mining of the Human Protein Atlas. Clin. Proteomics 13, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Guo Z., Zou L., Yang Y., Zhang L., Ji N., Shao C., Sun W., and Wang Y. (2015) A comprehensive map and functional annotation of the normal human cerebrospinal fluid proteome. J. Proteomics 119, 90–99 [DOI] [PubMed] [Google Scholar]
- 22. Guldbrandsen A., Vethe H., Farag Y., Oveland E., Garberg H., Berle M., Myhr K. M., Opsahl J. A., Barsnes H., and Berven F. S. (2014) In-depth characterization of the cerebrospinal fluid (CSF) proteome displayed through the CSF proteome resource (CSF-PR). Mol. Cell Proteomics 13, 3152–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holtta M., Zetterberg H., Mirgorodskaya E., Mattsson N., Blennow K., and Gobom J. (2012) Peptidome analysis of cerebrospinal fluid by LC-MALDI MS. PLoS ONE 7, e42555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schutzer S. E., Liu T., Natelson B. H., Angel T. E., Schepmoes A. A., Purvine S. O., Hixson K. K., Lipton M. S., Camp D. G., Coyle P. K., Smith R. D., and Bergquist J. (2010) Establishing the proteome of normal human cerebrospinal fluid. PLoS ONE 5, e10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stoop M. P., Coulier L., Rosenling T., Shi S., Smolinska A. M., Buydens L., Ampt K., Stingl C., Dane A., Muilwijk B., Luitwieler R. L., Sillevis Smitt P. A., Hintzen R. Q., Bischoff R., Wijmenga S. S., Hankemeier T., van Gool A. J., and Luider T. M. (2010) Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples. Mol. Cell Proteomics 9, 2063–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan S., Wang Y., Quinn J. F., Peskind E. R., Waichunas D., Wimberger J. T., Jin J., Li J. G., Zhu D., Pan C., and Zhang J. (2006) Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J. Proteome Res. 5, 2769–2779 [DOI] [PubMed] [Google Scholar]
- 27. Xu J., Chen J., Peskind E. R., Jin J., Eng J., Pan C., Montine T. J., Goodlett D. R., and Zhang J. (2006) Characterization of proteome of human cerebrospinal fluid. Int. Rev. Neurobiol. 73, 29–98 [DOI] [PubMed] [Google Scholar]
- 28. Zhang J., Goodlett D. R., Peskind E. R., Quinn J. F., Zhou Y., Wang Q., Pan C., Yi E., Eng J., Aebersold R. H., and Montine T. J. (2005) Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiol. Aging 26, 207–227 [DOI] [PubMed] [Google Scholar]
- 29. Wenner B. R., Lovell M. A., and Lynn B. C. (2004) Proteomic analysis of human ventricular cerebrospinal fluid from neurologically normal, elderly subjects using two-dimensional LC-MS/MS. J. Proteome Res. 3, 97–103 [DOI] [PubMed] [Google Scholar]
- 30. DeGiorgis J. A., Jaffe H., Moreira J. E., Carlotti C. G. Jr, Leite J. P., Pant H. C., and Dosemeci A. (2005) Phosphoproteomic analysis of synaptosomes from human cerebral cortex. J. Proteome Res. 4, 306–315 [DOI] [PubMed] [Google Scholar]
- 31. Wilhelm B. G., Mandad S., Truckenbrodt S., Krohnert K., Schafer C., Rammner B., Koo S. J., Classen G. A., Krauss M., Haucke V., Urlaub H., and Rizzoli S. O. (2014) Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 [DOI] [PubMed] [Google Scholar]
- 32. Abul-Husn N. S., Bushlin I., Moron J. A., Jenkins S. L., Dolios G., Wang R., Iyengar R., Ma'ayan A., and Devi L. A. (2009) Systems approach to explore components and interactions in the presynapse. Proteomics 9, 3303–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weingarten J., Lassek M., Mueller B. F., Rohmer M., Lunger I., Baeumlisberger D., Dudek S., Gogesch P., Karas M., and Volknandt W. (2014) The proteome of the presynaptic active zone from mouse brain. Mol. Cell Neurosci. 59, 106–118 [DOI] [PubMed] [Google Scholar]
- 34. Blondeau F., Ritter B., Allaire P. D., Wasiak S., Girard M., Hussain N. K., Angers A., Legendre-Guillemin V., Roy L., Boismenu D., Kearney R. E., Bell A. W., Bergeron J. J., and McPherson P. S. (2004) Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. U.S.A. 101, 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boyken J., Gronborg M., Riedel D., Urlaub H., Jahn R., and Chua J. J. (2013) Molecular profiling of synaptic vesicle docking sites reveals novel proteins but few differences between glutamatergic and GABAergic synapses. Neuron 78, 285–297 [DOI] [PubMed] [Google Scholar]
- 36. Coughenour H. D., Spaulding R. S., and Thompson C. M. (2004) The synaptic vesicle proteome: a comparative study in membrane protein identification. Proteomics 4, 3141–3155 [DOI] [PubMed] [Google Scholar]
- 37. Morciano M., Burre J., Corvey C., Karas M., Zimmermann H., and Volknandt W. (2005) Immunoisolation of two synaptic vesicle pools from synaptosomes: a proteomics analysis. J. Neurochem. 95, 1732–1745 [DOI] [PubMed] [Google Scholar]
- 38. Morciano M., Beckhaus T., Karas M., Zimmermann H., and Volknandt W. (2009) The proteome of the presynaptic active zone: from docked synaptic vesicles to adhesion molecules and maxi-channels. J. Neurochem. 108, 662–675 [DOI] [PubMed] [Google Scholar]
- 39. Takamori S., Holt M., Stenius K., Lemke E. A., Gronborg M., Riedel D., Urlaub H., Schenck S., Brugger B., Ringler P., Muller S. A., Rammner B., Grater F., Hub J. S., De Groot B. L., Mieskes G., Moriyama Y., Klingauf J., Grubmuller H., Heuser J., Wieland F., and Jahn R. (2006) Molecular anatomy of a trafficking organelle. Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 40. Bayés À., van de Lagemaat L. N., Collins M. O., Croning M. D., Whittle I. R., Choudhary J. S., and Grant S. G. (2011) Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 14, 19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fernandez E., Collins M. O., Uren R. T., Kopanitsa M. V., Komiyama N. H., Croning M. D., Zografos L., Armstrong J. D., Choudhary J. S., and Grant S. G. (2009) Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Syst. Biol. 5, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng D., Hoogenraad C. C., Rush J., Ramm E., Schlager M. A., Duong D. M., Xu P., Wijayawardana S. R., Hanfelt J., Nakagawa T., Sheng M., and Peng J. (2006) Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell Proteomics 5, 1158–1170 [DOI] [PubMed] [Google Scholar]
- 43. Li K., Hornshaw M. P., van Minnen J., Smalla K. H., Gundelfinger E. D., and Smit A. B. (2005) Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J. Proteome Res. 4, 725–733 [DOI] [PubMed] [Google Scholar]
- 44. Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K. H., and Smit A. B. (2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 279, 987–1002 [DOI] [PubMed] [Google Scholar]
- 45. Peng J., Kim M. J., Cheng D., Duong D. M., Gygi S. P., and Sheng M. (2004) Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 279, 21003–21011 [DOI] [PubMed] [Google Scholar]
- 46. Walikonis R. S., Jensen O. N., Mann M., Provance D. W. Jr, Mercer J. A., and Kennedy M. B. (2000) Identification of proteins in the postsynaptic density fraction by mass spectrometry. J. Neurosci. 20, 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bayés À., Collins M. O., Croning M. D., van de Lagemaat L. N., Choudhary J. S., and Grant S. G. (2012) Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS ONE 7, e46683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Collins M. O., Husi H., Yu L., Brandon J. M., Anderson C. N., Blackstock W. P., Choudhary J. S., and Grant S. G. (2006) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, 16–23 [DOI] [PubMed] [Google Scholar]
- 49. Trinidad J. C., Thalhammer A., Specht C. G., Schoepfer R., and Burlingame A. L. (2005) Phosphorylation state of postsynaptic density proteins. J. Neurochem. 92, 1306–1316 [DOI] [PubMed] [Google Scholar]
- 50. Bayés À., Collins M. O., Galtrey C. M., Simonnet C., Roy M., Croning M. D., Gou G., van de Lagemaat L. N., Milward D., Whittle I. R., Smith C., Choudhary J. S., and Grant S. G. (2014) Human post-mortem synapse proteome integrity screening for proteomic studies of postsynaptic complexes. Mol. Brain 7, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ster J., Steuble M., Orlando C., Diep T. M., Akhmedov A., Raineteau O., Pernet V., Sonderegger P., and Gerber U. (2014) Calsyntenin-1 regulates targeting of dendritic NMDA receptors and dendritic spine maturation in CA1 hippocampal pyramidal cells during postnatal development. J. Neurosci. 34, 8716–8727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Terunuma M., Revilla-Sanchez R., Quadros I. M., Deng Q., Deeb T. Z., Lumb M., Sicinski P., Haydon P. G., Pangalos M. N., and Moss S. J. (2014) Postsynaptic GABAB receptor activity regulates excitatory neuronal architecture and spatial memory. J. Neurosci. 34, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koksma J. J., Fritschy J. M., Mack V., Van Kesteren R. E., and Brussaard A. B. (2005) Differential GABAA receptor clustering determines GABA synapse plasticity in rat oxytocin neurons around parturition and the onset of lactation. Mol. Cell Neurosci. 28, 128–140 [DOI] [PubMed] [Google Scholar]
- 54. Gainey M. A., Hurvitz-Wolff J. R., Lambo M. E., and Turrigiano G. G. (2009) Synaptic scaling requires the GluR2 subunit of the AMPA receptor. J. Neurosci. 29, 6479–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mokin M., and Keifer J. (2004) Targeting of GLUR4-containing AMPA receptors to synaptic sites during in vitro classical conditioning. Neuroscience 128, 219–228 [DOI] [PubMed] [Google Scholar]
- 56. Ohtani Y., Miyata M., Hashimoto K., Tabata T., Kishimoto Y., Fukaya M., Kase D., Kassai H., Nakao K., Hirata T., Watanabe M., Kano M., and Aiba A. (2014) The synaptic targeting of mGluR1 by its carboxyl-terminal domain is crucial for cerebellar function. J. Neurosci. 34, 2702–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meijer M., Cijsouw T., Toonen R. F., and Verhage M. (2015) Synaptic Effects of Munc18–1 Alternative Splicing in Excitatory Hippocampal Neurons. PLoS ONE 10, e0138950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Graf E. R., Zhang X., Jin S. X., Linhoff M. W., and Craig A. M. (2004) Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aoto J., Martinelli D. C., Malenka R. C., Tabuchi K., and Sudhof T. C. (2013) Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 154, 75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bemben M. A., Shipman S. L., Hirai T., Herring B. E., Li Y., Badger J. D. 2nd, Nicoll R. A., Diamond J. S., and Roche K. W. (2014) CaMKII phosphorylation of neuroligin-1 regulates excitatory synapses. Nat. Neurosci. 17, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim J. Y., Megat S., Moy J. K., Asiedu M. N., Mejia G. L., Vagner J., and Price T. J. (2016) Neuroligin 2 regulates spinal GABAergic plasticity in hyperalgesic priming, a model of the transition from acute to chronic pain. Pain 157, 1314–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Beesley P. W., Herrera-Molina R., Smalla K. H., and Seidenbecher C. (2014) The Neuroplastin adhesion molecules: key regulators of neuronal plasticity and synaptic function. J. Neurochem. 131, 268–283 [DOI] [PubMed] [Google Scholar]
- 63. Verstegen A. M., Tagliatti E., Lignani G., Marte A., Stolero T., Atias M., Corradi A., Valtorta F., Gitler D., Onofri F., Fassio A., and Benfenati F. (2014) Phosphorylation of synapsin I by cyclin-dependent kinase-5 sets the ratio between the resting and recycling pools of synaptic vesicles at hippocampal synapses. J. Neurosci. 34, 7266–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Araki Y., Zeng M., Zhang M., and Huganir R. L. (2015) Rapid dispersion of SynGAP from synaptic spines triggers AMPA receptor insertion and spine enlargement during LTP. Neuron 85, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Morgans C. W., Kensel-Hammes P., Hurley J. B., Burton K., Idzerda R., McKnight G. S., and Bajjalieh S. M. (2009) Loss of the Synaptic Vesicle Protein SV2B results in reduced neurotransmission and altered synaptic vesicle protein expression in the retina. PLoS ONE 4, e5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bacaj T., Wu D., Burre J., Malenka R. C., Liu X., and Sudhof T. C. (2015) Synaptotagmin-1 and -7 Are Redundantly Essential for Maintaining the Capacity of the Readily-Releasable Pool of Synaptic Vesicles. PLos Biol. 13, e1002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Watanabe Y., Katayama N., Takeuchi K., Togano T., Itoh R., Sato M., Yamazaki M., Abe M., Sato T., Oda K., Yokoyama M., Takao K., Fukaya M., Miyakawa T., Watanabe M., Sakimura K., Manabe T., and Igarashi M. (2013) Point mutation in syntaxin-1A causes abnormal vesicle recycling, behaviors, and short term plasticity. J. Biol. Chem. 288, 34906–34919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saghatelyan A. K., Dityatev A., Schmidt S., Schuster T., Bartsch U., and Schachner M. (2001) Reduced perisomatic inhibition, increased excitatory transmission, and impaired long-term potentiation in mice deficient for the extracellular matrix glycoprotein tenascin-R. Mol Cell Neurosci. 17, 226–240 [DOI] [PubMed] [Google Scholar]
- 69. Hussain S., and Davanger S. (2015) Postsynaptic VAMP/synaptobrevin facilitates differential vesicle trafficking of GluA1 and GluA2 AMPA receptor subunits. PLoS ONE 10, e0140868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hagihara K., Miura R., Kosaki R., Berglund E., Ranscht B., and Yamaguchi Y. (1999) Immunohistochemical evidence for the brevican-tenascin-R interaction: colocalization in perineuronal nets suggests a physiological role for the interaction in the adult rat brain. J. Comp. Neurol. 410, 256–264 [PubMed] [Google Scholar]
- 71. Berchtold N. C., Coleman P. D., Cribbs D. H., Rogers J., Gillen D. L., and Cotman C. W. (2012) Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer's disease. Neurobiol. Aging 34, 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Counts S. E., Alldred M. J., Che S., Ginsberg S. D., and Mufson E. J. (2014) Synaptic gene dysregulation within hippocampal CA1 pyramidal neurons in mild cognitive impairment. Neuropharmacology 79, 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Manavalan A., Mishra M., Feng L., Sze S. K., Akatsu H., and Heese K. (2013) Brain site-specific proteome changes in aging-related dementia. Exp. Mol. Med. 45, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Leifer D., and Kowall N. W. (1992) Thy-1 in hippocampus: normal anatomy and neuritic growth in Alzheimer's disease. J. Neuropathol. Exp. Neurol. 51, 133–141 [PubMed] [Google Scholar]
- 75. Liu S. J., Gasperini R., Foa L., and Small D. H. (2010) Amyloid-beta decreases cell-surface AMPA receptors by increasing intracellular calcium and phosphorylation of GluR2. J. Alzheimers Dis. 21, 655–666 [DOI] [PubMed] [Google Scholar]
- 76. Chan S. L., Griffin W. S., and Mattson M. P. (1999) Evidence for caspase-mediated cleavage of AMPA receptor subunits in neuronal apoptosis and Alzheimer's disease. J. Neurosci. Res. 57, 315–323 [DOI] [PubMed] [Google Scholar]
- 77. Brito-Moreira J., Lourenco M. V., Oliveira M. M., Ribeiro F. C., Ledo J. H., Diniz L. P., Vital J. F. S., Magdesian M. H., Melo H. M., Barros-Aragao F., de Souza J. M., Alves-Leon S. V., Gomes F. C. A., Clarke J. R., Figueiredo C. P., De Felice F. G., and Ferreira S. T. (2017) Interaction of amyloid-beta (Abeta) oligomers with neurexin 2alpha and neuroligin 1 mediates synapse damage and memory loss in mice. J. Biol. Chem. 292, 7327–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Steuble M., Diep T. M., Schatzle P., Ludwig A., Tagaya M., Kunz B., and Sonderegger P. (2012) Calsyntenin-1 shelters APP from proteolytic processing during anterograde axonal transport. Biol. Open. 1, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vagnoni A., Perkinton M. S., Gray E. H., Francis P. T., Noble W., and Miller C. C. (2012) Calsyntenin-1 mediates axonal transport of the amyloid precursor protein and regulates Abeta production. Hum. Mol. Genet. 21, 2845–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sevlever D., Zou F., Ma L., Carrasquillo S., Crump M. G., Culley O. J., Hunter T. A., Bisceglio G. D., Younkin L., Allen M., Carrasquillo M. M., Sando S. B., Aasly J. O., Dickson D. W., Graff-Radford N. R., Petersen R. C., Deak F., and Belbin O. (2015) Genetically-controlled Vesicle-Associated Membrane Protein 1 expression may contribute to Alzheimer's pathophysiology and susceptibility. Mol. Neurodegener. 10, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sindi I. A., Tannenberg R. K., and Dodd P. R. (2014) Role for the neurexin-neuroligin complex in Alzheimer's disease. Neurobiol. Aging 35, 746–756 [DOI] [PubMed] [Google Scholar]
- 82. Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., and Kwak S. (2004) Glutamate receptors: RNA editing and death of motor neurons. Nature 427, 801. [DOI] [PubMed] [Google Scholar]
- 83. Sun C., Cheng M. C., Qin R., Liao D. L., Chen T. T., Koong F. J., Chen G., and Chen C. H. (2011) Identification and functional characterization of rare mutations of the neuroligin-2 gene (NLGN2) associated with schizophrenia. Hum. Mol. Genet. 20, 3042–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Vaags A. K., Lionel A. C., Sato D., Goodenberger M., Stein Q. P., Curran S., Ogilvie C., Ahn J. W., Drmic I., Senman L., Chrysler C., Thompson A., Russell C., Prasad A., Walker S., Pinto D., Marshall C. R., Stavropoulos D. J., Zwaigenbaum L., Fernandez B. A., Fombonne E., Bolton P. F., Collier D. A., Hodge J. C., Roberts W., Szatmari P., and Scherer S. W. (2012) Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am. J. Hum. Genet. 90, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schubert J., Siekierska A., Langlois M., May P., Huneau C., Becker F., Muhle H., Suls A., Lemke J. R., de Kovel C. G., Thiele H., Konrad K., Kawalia A., Toliat M. R., Sander T., Ruschendorf F., Caliebe A., Nagel I., Kohl B., Kecskes A., Jacmin M., Hardies K., Weckhuysen S., Riesch E., Dorn T., Brilstra E. H., Baulac S., Moller R. S., Hjalgrim H., Koeleman B. P., Jurkat-Rott K., Lehman-Horn F., Roach J. C., Glusman G., Hood L., Galas D. J., Martin B., de Witte P. A., Biskup S., De Jonghe P., Helbig I., Balling R., Nurnberg P., Crawford A. D., Esguerra C. V., Weber Y. G., and Lerche H. (2014) Mutations in STX1B, encoding a presynaptic protein, cause fever-associated epilepsy syndromes. Nat. Genet. 46, 1327–1332 [DOI] [PubMed] [Google Scholar]
- 86. Kvartsberg H., Duits F. H., Ingelsson M., Andreasen N., Ohrfelt A., Andersson K., Brinkmalm G., Lannfelt L., Minthon L., Hansson O., Andreasson U., Teunissen C. E., Scheltens P., Van der Flier W. M., Zetterberg H., Portelius E., and Blennow K. (2015) Cerebrospinal fluid levels of the synaptic protein neurogranin correlates with cognitive decline in prodromal Alzheimer's disease. Alzheimers Dement 11, 1180–1190 [DOI] [PubMed] [Google Scholar]
- 87. Vogt L., Schrimpf S. P., Meskenaite V., Frischknecht R., Kinter J., Leone D. P., Ziegler U., and Sonderegger P. (2001) Calsyntenin-1, a proteolytically processed postsynaptic membrane protein with a cytoplasmic calcium-binding domain. Mol. Cell Neurosci. 17, 151–166 [DOI] [PubMed] [Google Scholar]
- 88. Yang Y., Wang X. B., and Zhou Q. (2010) Perisynaptic GluR2-lacking AMPA receptors control the reversibility of synaptic and spines modifications. Proc. Natl. Acad. Sci. U.S.A. 107, 11999–12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Biou V., Bhattacharyya S., and Malenka R. C. (2008) Endocytosis and recycling of AMPA receptors lacking GluR2/3. Proc. Natl. Acad. Sci. U.S.A. 105, 1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang J. Q., Arora A., Yang L., Parelkar N. K., Zhang G., Liu X., Choe E. S., and Mao L. (2005) Phosphorylation of AMPA receptors: mechanisms and synaptic plasticity. Mol. Neurobiol. 32, 237–249 [DOI] [PubMed] [Google Scholar]
- 91. Biederer T., and Sudhof T. C. (2000) Mints as adaptors. Direct binding to neurexins and recruitment of munc18. J. Biol. Chem. 275, 39803–39806 [DOI] [PubMed] [Google Scholar]
- 92. Panzanelli P., Fruh S., and Fritschy J. M. (2017) Differential role of GABAA receptors and neuroligin 2 for perisomatic GABAergic synapse formation in the hippocampus. Brain Struct. Funct. 222, 4149–4161 [DOI] [PubMed] [Google Scholar]
- 93. Scheiffele P., Fan J., Choih J., Fetter R., and Serafini T. (2000) Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669 [DOI] [PubMed] [Google Scholar]
- 94. Davis S., Rodger J., Hicks A., Mallet J., and Laroche S. (1996) Brain structure and task-specific increase in expression of the gene encoding syntaxin 1B during learning in the rat: a potential molecular marker for learning-induced synaptic plasticity in neural networks. Eur. J. Neurosci. 8, 2068–2074 [DOI] [PubMed] [Google Scholar]
- 95. Bennett M. K., Calakos N., and Scheller R. H. (1992) Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 257, 255–259 [DOI] [PubMed] [Google Scholar]
- 96. Nosten-Bertrand M., Errington M. L., Murphy K. P., Tokugawa Y., Barboni E., Kozlova E., Michalovich D., Morris R. G., Silver J., Stewart C. L., Bliss T. V., and Morris R. J. (1996) Normal spatial learning despite regional inhibition of LTP in mice lacking Thy-1. Nature 379, 826–829 [DOI] [PubMed] [Google Scholar]
- 97. Liu C. J., Chaturvedi N., Barnstable C. J., and Dreyer E. B. (1996) Retinal Thy-1 expression during development. Invest. Ophthalmol. Vis. Sci. 37, 1469–1473 [PubMed] [Google Scholar]
- 98. Jeng C. J., McCarroll S. A., Martin T. F., Floor E., Adams J., Krantz D., Butz S., Edwards R., and Schweitzer E. S. (1998) Thy-1 is a component common to multiple populations of synaptic vesicles. J. Cell Biol. 140, 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The shotgun proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (16) partner repository (https://www.ebi.ac.uk/pride/archive) with the dataset identifier PXD010356. The targeted proteomics data have been deposited to the Panorama repository (https://panoramaweb.org/5Jugvs.url) and the PRIDE repository with the identifier PXD012138. The script used for Matlab image analysis has been deposited at https://github.com/MemoryUnitSantPau with the name SynSeg.