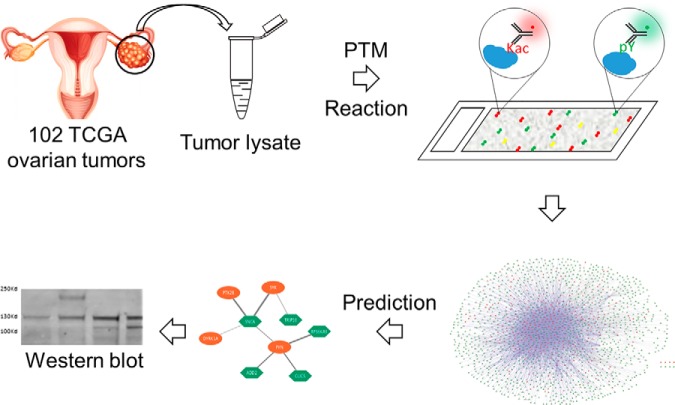
A systematic, multiplexed approach interrogating enzyme-substrate relationships in context of PTMs is fundamental in understanding the dynamics of these pathways in disease processes. Multiplexed PTM reactions on HuProt arrays were developed and applied to 102 TCGA ovarian tumor samples. Data integration and networks analysis led to the prediction that 19 tyrosine kinases were elevated and potentially responsible for the dysregulated pTyr signaling pathways in ovarian tumors. Elevated kinase activities of PTK2 and PTK2B were confirmed in several ovarian cancer cell lines.
Keywords: CPTAC, Protein array, Ovarian tumors, Kinase, Tyrosine phosphorylation
Keywords: Protein array, Ovarian cancer, Tyrosine Kinases*, Phosphorylation, Proteogenomics, CPTAC
Graphical Abstract
Highlights
Multiplexed PTM assays on HuProt array were developed using ovarian tumor lysates.
Proteome-wide Tyr phosphorylation with 102 ovarian tumors were performed and analyzed.
19 kinases were predicted to have elevated activities in ovarian tumor.
Elevated activities of PTK2 and PTK2B were confirmed in ovarian cancer cell lines.
Abstract
The recent accomplishment of comprehensive proteogenomic analysis of high-grade serous ovarian carcinoma (HGSOC) tissues reveals cancer associated molecular alterations were not limited to variations among DNA, and mRNA/protein expression, but are a result of complex reprogramming of signaling pathways/networks mediated by the protein and post-translational modification (PTM) interactomes. A systematic, multiplexed approach interrogating enzyme-substrate relationships in the context of PTMs is fundamental in understanding the dynamics of these pathways, regulation of cellular processes, and their roles in disease processes. Here, as part of Clinical Proteomic Tumor Analysis Consortium (CPTAC) project, we established a multiplexed PTM assay (tyrosine phosphorylation, and lysine acetylation, ubiquitylation and SUMOylation) method to identify protein probes' PTMs on the human proteome array. Further, we focused on the tyrosine phosphorylation and identified 19 kinases are potentially responsible for the dysregulated signaling pathways observed in HGSOC. Additionally, elevated kinase activity was observed when 14 ovarian cancer cell lines or tumor tissues were subjected to test the autophosphorylation status of PTK2 (pY397) and PTK2B (pY402) as a proxy for kinase activity. Taken together, this report demonstrates that PTM signatures based on lysate reactions on human proteome array is a powerful, unbiased approach to identify dysregulated PTM pathways in tumors.
The recently completed proteogenomic analysis of 174 TCGA ovarian high-grade serous ovarian carcinoma (HGSOC)1 tissues has demonstrated the ability of proteomic technologies to analyze large-scale, complex clinical specimens with broad protein coverage and high analytical precision to serve as an indispensable component of the comprehensive molecular characterization of cancer (1). Integrated analysis of the genome and corresponding proteome has revealed that the somatic genome drives the cancer proteome and the associated levels of protein post-translational modification (PTM) especially for proteins associated with chromosomal instability and protein acetylation associated with homologous recombination deficiency (1). These findings, together with recent reports in the literature (2–6), indicate that much of the cancer associated molecular alterations will not necessarily be limited to coordinated variations among DNA, copy number, and mRNA/protein expression, but is rather as complex rewiring of signaling pathways/networks mediated by the protein and post-translational modification (PTM) interactomes.
Ovarian cancer remains the most lethal gynecologic cancer and the fifth most common cause of cancer-related death for women in the United States (7). Nearly 22,000 women in the U.S. are diagnosed with ovarian cancer annually and ∼14,200 women die each year from this disease (7). HGSOC makes up ∼70% of all ovarian cancers and accounts for 90% of the deaths from this disease. HGSOC patients typically respond well to initial treatment. However, these individuals frequently develop chemoresistance and succumb to the disease. Interestingly, more and more literature reveals that proteins' PTMs, such as tyrosine phosphorylation and lysine acetylation, appear to have higher expressions and activities in cancer, which play an integral role in the development of ovarian cancer and its resistance to chemotherapy (1, 8–13). A systematic, multiplexed approach to identify dynamic PTMs would have an important impact on proteomic studies and be essential for gaining a comprehensive understanding of the PTMs involved in both normal and cancerous signaling networks. Further, analysis of disease-associated specific enzyme substrate relationships could provide the foundation for identifying enzymes responsible for the observed PTM-substrate modifications and allow for the construction of disease specific cellular signaling networks.
Protein PTM is essential for a host of vital mechanisms including protein maturation, regulation of protein functionality and cellular signaling pathways, protein localization, and protein degradation.
PTM refers to the additions of a chemical group covalently to proteins and is mostly achieved via enzymatic reactions after protein translation (14). The identification of the modified protein substrates and construction of the enzyme-substrate relationships are fundamental to understanding the regulation of cellular processes via PTMs and their roles in disease progression (5, 6).
Although developments in mass spectrometry (MS) and protein/peptide array technologies have tremendously improved our ability to profile the PTMs in a high-throughput fashion, these methods suffer from various shortcomings. For example, during MS detection, phosphopeptides are easily lost during the loading onto the reverse-phase columns because that the addition of anionic/acidic phosphate groups increases hydrophilicity resulting in reduced retention (15). Consequently, obtaining a global landscape of tyrosine phosphorylation in human tissues and/or tumors has remained challenging. Functional protein arrays on the other hand have proved to be well suited for determination of enzyme-substrate relationships as demonstrated by a study using 289 unique human kinases and human protein arrays to define >47,000 potential kinase-substrate relationships (KSRs) (16, 17). Through integration of this data set with known phosphorylation sites, mainly identified by MS/MS analyses, a high-resolution KSR network that connects 230 kinases with 2591 in vivo phosphorylation sites on 652 substrates (17). This network later served as the foundation to identify additional signaling components in the c-Met signaling via carrying out cell lysate phosphorylation on human protein arrays (18). Traditionally, isotope-labeling has been the primary method used to identify a modified substrate, but this can be challenging especially when dealing with a high-content array, such as the human proteome arrays (HuProt array), because of the high density of printed (100 μm of center-to-center distance). Alternately, antibody-based fluorescent detection allows for the interrogation of two PTM reactions simultaneously and has been successfully applied to identify proteins substrates of tyrosine phosphorylation, sulfation (19, 20), SUMOylation (21), and ubiquitylation (22).
As a part of the proteogenomic analysis of the TCGA HGSOC samples, we have established multiplexed PTM assays on HuProt arrays to identify candidate PTM substrates and constructed a corresponding kinase-substrate network in ovarian cancer tissues to identify dysregulated signaling pathways in ovarian tumors. Of the predict kinases, the autophosphorylation status of PTK2 (pY397) and PTK2B (pY402) was validated as a proxy for their activities in cell-based assays. These studies have demonstrated that this new approach can be widely applied to rapidly determine dysregulated signaling components in many other tumors and/or diseases.
EXPERIMENTAL PROCEDURES
Ovarian Tissue Collection
The tumor tissue samples were a subset of the TCGA high-grade serous ovarian carcinoma specimens, previously characterized from a genomic perspective (TCGA Research Network, 2011). analyzed in this study using HuProt arrays as a part of NCI/CPTAC funded research. All tumor samples analyzed (supplemental Table S1) were obtained through the TCGA Biospecimen Core Resource, as described previously (23). All specimens were first used for integrated genomic analysis as part of the TCGA program and residual materials from the same specimens were analyzed as part of this Clinical Proteomic Tumor Analysis Consortium (CPTAC) project. All the biospecimens were collected from newly diagnosed patients with HGSOC who underwent surgical tumor resection and did not receive prior treatment including chemotherapy. Each case was reviewed through TCGA by a board-certified pathologist to confirm that the frozen section was histologically consistent with HGSOC. The eligible tumors contained an average of 70% tumor cell nuclei with less than 20% necrosis (1).
HuProt Array Fabrication
As previously reported, the HuProt array was constructed from a protein library composed of over 17,000 human open reading frames using the pEGHA vector expressed and purified from yeast, each recombinant protein had a GST-His6 tag at the N-terminal (24). Together with negative and positive controls, all purified human proteins were printed in duplicate onto a Super epoxide 2 slides and stored at −80 °C until use.
Ovarian Cancer Tissue Lysate Preparation
To prepare the reactive lysate, tissue was homogenized into tiny pieces and then treated with sonication in 120 μl reaction buffer (20 mm HEPES/Tris pH7.5, 100 mm NaCl, 10 mm MgCl2, 10 mm MnCl2, 50 mm KCl, 1 mm NaF, 10 mm Na3VO4, 5 mm Nicotinamide, and 1 mm PMSF). After centrifugation at max speed (21,000 × g) for 10 min at 4 °C, the lysate was transferred to a new tube and assayed for protein concentration by Bradford assay according to the manufacturer's protocol (Thermo Fisher Scientific, Waltham, MA).
Multiplexed PTM Assay on HuProt Array
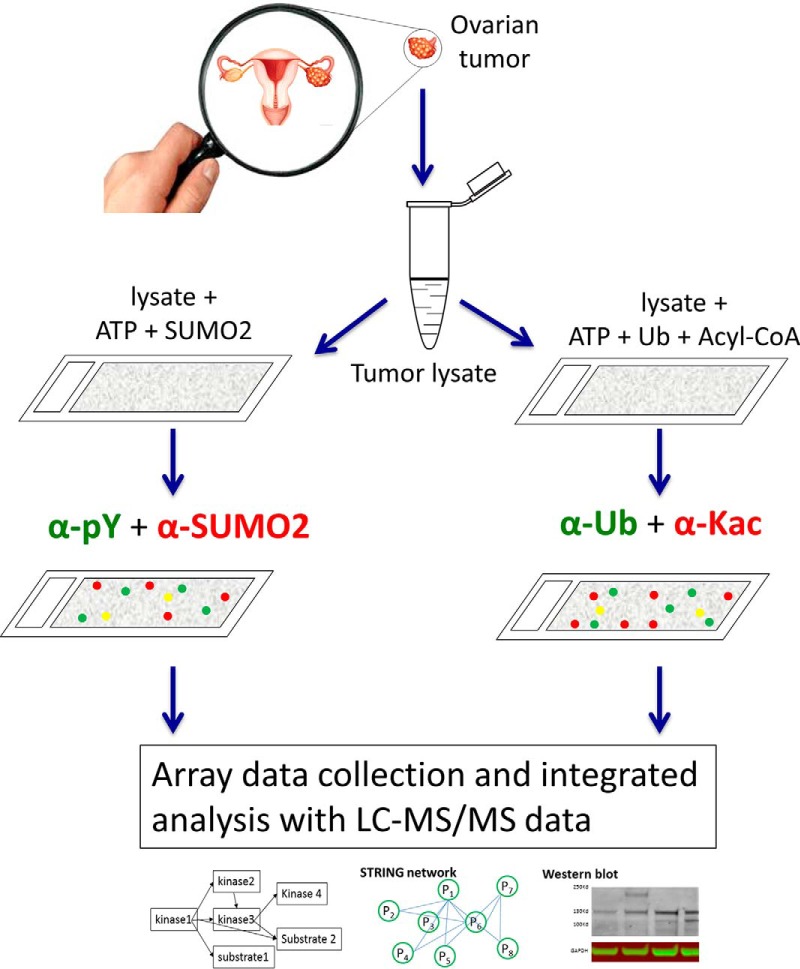
For the multiplexed PTM assay on the HuProt array, we focused on the following four types of PTM reaction assays, tyrosine phosphorylation (pY), and lysine acetylation, ubiquitylation and SUMOylation. A serial dilution of tissue lysate (0, 25 μg, 50 μg, 100 μg) in 120 μl reaction buffer (50 mm Tris-HCl and 25 mm HEPES-KOH at pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm MnCl2, 1 mm DTT, 1 mm EGTA, 2 mm Na3VO4, 2 mm NaF, and 1 mm ATP) were applied to four HuProt arrays to determine best condition for each PTM reaction. All the procedures were carried out as in our previous study (18). In brief, the HuProt array was immersed into 3% BSA in TBS buffer with 0.1% tween20 (TBST, pH 7.5) for 1 h at room temperature with gentle shaking. Then, tissue lysates were incubated on protein arrays, covered with a coverslip, and placed in a humidity chamber at 30 °C for 30 min. Following the termination of the reaction, the slides were subjected to three-10 min washes in 1X TBST, three-10 min washes in 1% SDS preheated to 65 °C, and one quick rinse with 1× TBST before PTM specific fluorescently labeled antibodies were added (Fig. 1). To simultaneously detect two PTM signals, we directly labeled anti-pY (i.e. pY100, Cell Signaling Technology, Danvers, MA) and anti-Ub monoclonal antibodies (Cell Signaling Technology) with Cy3, anti-lysine acetylation (Kac, PTM Biolabs Inc, Chicago, IL) and -SUMO2/3 monoclonal antibodies (Abcam, Cambridge, United Kingdom) with Cy5. Next, Cy3-labeled anti-pY and Cy5-labeled anti-SUMO2/3 antibodies were mixed in equal molar amount, whereas Cy3-labeled anti-Ub and Cy5-labeled anti-Kac antibodies were mixed the same way. After acquiring signals of each protein spot from the two HuProt arrays used for each duplicated lysate reaction, the scatter plot analysis was performed using the signals obtained from each duplicated reaction in Excel. The Trendline (i.e. the dotted blue lines) and the corresponding R2 value were next generated by Excel program. As a negative control, the pilot protein arrays were also separately incubated with the assay buffer without lysate following with the labeled two antibody mixtures and the positives scored on the arrays were removed from further analyses. Scanned images for each array were analyzed using GenePix Pro 6.0 (Molecular Devices, Sunnyvale, CA).
Fig. 1.
Scheme for profiling human multiplexed PTM assay on HuProt array. To establish multiplexed PTM reaction assays, lysates from each TCGA ovarian tumor samples were incubate on the HuProt array, which including over 17,000 purified recombinant human proteins as potential substrates, to obtain PTM signatures of protein kinases, acetyltransferases, and E3 ligases for ubiquitin or SUMO. As amount limitation of most available TCGA samples and the cost consideration of HuProt array, detection of Tyr-phosphorylation and SUMOylation were performed on one array simultaneously, whereas lysine acetylation and ubiquitylation reactions were multiplexed on a different HuProt array with their PTM-specific monoclonal antibodies respectively. Then, the identified PTM substrates were then used to predict corresponding enzymes for each type of PTMs to determine dysregulated pathways in the ovarian tumors.
Identification of Positive Substrates
Considering amount limitation of most available TCGA samples, the cost consideration of HuProt array and good replicability of the multiplexed PTM assay, we decide to detect Tyr-phosphorylation and lysine acetylation on one array simultaneously. Multiplexed pY and SUMO reactions were performed under the same conditions as the multiplexed acK and Ub reactions, except using different combinations of PTM-specific antibodies. Similarly, the multiplexed pY and acK reactions were performed under the same conditions as those described in the pilot assays except different combined primary antibodies.
For both pilot and regular multiplexed PTM assays performed on the HuProt arrays, positive hits were identified as in our previous studies (25). Briefly, the signal intensity (Rij) of each protein spot was defined as local signal value of foreground (Fij) minus that of local background (Bij). Because each protein is printed in duplicate on an array, Rij was averaged for each protein as Rp. A Z-score was determined for each protein on array by calculating the number of standard deviations above the mean value of all protein probes on the array. A stringent cutoff (Z ≥ 5) was used to determine the positives targets in this study.
Integration of Kinase-Substrates Relationship and HuProt Array-based pY Profiling to Predict the Dysregulated Kinases in Ovarian Tumors
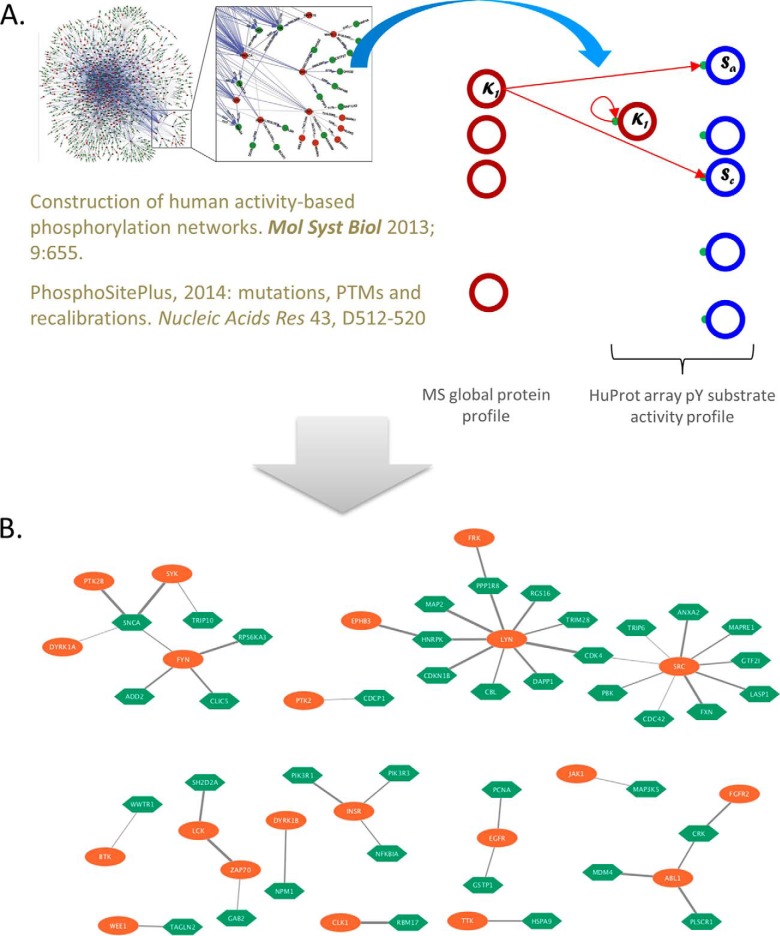
Through performing large-scale pY analysis profiling using HuProt arrays, a cohort of substrates modified with tyrosine phosphorylation were identified. In order to predict the kinases in ovarian tumor lysates responsible for phosphorylating these proteins, we integrated the pY profiling data and the LC-MS/MS global protein abundance data of the same samples generated by the CPTAC and generate the dysregulated KSR network through integrated activity-based analysis (1) (Fig. 4A). Briefly, pairs of proteins with high correlations between MS-based global protein abundance and pY activities were selected to test against the two KSR database, PhosphoNetworks (17, 26) and PhosphoSitePlus (27), to identify the potential active kinase-substrate pairs. PhosphoSitePlus is an open, comprehensive resource for studying experimentally observed PTMs. PhosphoNetworks database represents a high-resolution map of the human phosphorylation networks constructed by integration between a large dataset obtained by performing individual kinase reactions on human protein microarrays and phosphosites identified using MS/MS approaches (17). Active kinase substrate network in ovarian cancer was plotted using Cytoscape (28). Functional annotation and pathway analysis were performed by Ingenuity Pathway Analysis (IPA) software (29). (Qiagen, Hilden, Germany).
Fig. 4.
Integrated activity-based analysis identified active tyrosine kinases among ovarian high-grade serious carcinoma. A, Systemic schema for integrated activity-based analysis. An Integrated tyrosine kinase substrate relationship database was constructed from two data sources; and then correlation of kinase-substrate pairs were assessed between the LC-MS/MS quantitation of kinases and HuProt array-based activity quantitation of substrates; B, Identified correlated KSR network with 19 kinase and 40 substrates. Orange ovals represent the kinases; green hexagons represent their substrates with known kinases substrates relationships based on KSR database; and the edges represent correlations between the LC-MS/MS quantitation of kinases and HuProt array-based activity quantitation of substrates (width representing correlation strength). See also supplemental Table S2.
Cell Culture
Ovarian carcinoma cell lines SKOV3, ES2, and OVCAR3 were obtained from the American Tissue Culture Center (Rockville, MD). MPSC1 cell line was established from a low-grade serous carcinoma (30). OVISE and OVTOKO were obtained from the Japanese Health Science Research Resources Bank (Osaka, Japan). EFO21, KK, KOC7C, OVCAR5 and TYKNU were kind gifts from Dr. B. Karlan, Cedars Sinai Medical Center, Los Angeles, California, Dr. K. Nakayama, Shimane University, Japan and Dr. S. Baylin, Johns Hopkins Medicine, Baltimore, MD. Of these 11 ovarian cancer cells, EFO21, KK and SKOV3 were derived from cystadenocarcinoma, a malignant form of a cystadenoma and a cancer from glandular epithelium, in which cystic accumulations of retained secretions are formed; OVISE, OVTOKO, KOC7C, and ES-2 were derived from ovarian clear cell adenocarcinoma, a subtype of epithelial ovarian cancer; TYKNU and OVCAR3 were derived from high grade ovarian serous adenocarcinoma; MPSC1 was from low grade ovarian serous adenocarcinoma. Normal counterparts of ovarian cancer cells including ovarian surface epithelial cell lines (OSE4 and OSE10) as well as an immortalized fallopian tube epithelial cell line FT2821were included as controls. Cell lines were maintained in RPMI 1640 or Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin.
Western Blotting
Protein lysates were prepared by lysing cells with RIPA buffer supplemented with protease and phosphatase inhibitors (Roche, Basel, Switzerland). Aliquots of 15 μg total proteins from each cell lysate were separated on 4–12% NuPAGE Bis-Tris Gels (Thermo Fisher Scientific, Waltham, MA), and transferred onto PVDF membranes using Trans-Blot Turbo rapid Western blotting transfer system (Bio-Rad Laboratories, Hercules, CA). The membrane was incubated with the appropriate antibodies and ECL developing solution (General Electric, Boston, MA). The antibodies used include Anti-Lyn antibody (phospho Y396, Abcam), Anti-BTK antibody (phospho Y551, Abcam), Anti-PTK2 antibody (phospho Y397, Abcam), anti-GAPDH antibody (Sigma-Aldrich, St. Louis, MO), anti-mouse HRP and anti-rabbit HRP (Jackson ImmunoResearch, West Grove, PA). GAPDH was used as a loading control.
RESULTS
Overall Research Strategy
Our goal was to identify dysregulated PTM signaling pathways via summarization of PTM signatures generated with each TCGA ovarian tumor samples as a part of CPTAC (31). Because the amount in most available TCGA samples was limited, we decided to develop a multiplexed PTM assays to obtain PTM signatures for each ovarian tumor sample with minimum consumption of the samples. To establish multiplexed PTM reaction assays on HuProt array with tumor lysates we used over 17,000 purified recombinant human proteins spotted on the HuProt array as potential substrates to obtain PTM signatures of protein kinases, acetyltransferases, and E3 ligases for ubiquitin or SUMO. Comparison of these PTM signatures was used to predict PTMs status and dysregulated signal pathways in ovarian tumors (Fig. 1). Here, Tyr-phosphorylation and SUMOylation were performed on one array simultaneously, whereas lysine acetylation and ubiquitylation reactions were multiplexed on a different HuProt array. After the PTM reactions, modified substrate proteins were identified with PTM-specific monoclonal antibodies. The identified PTM substrates were then used to predict corresponding enzymes for each type of PTM to determine dysregulated pathways in the ovarian tumors.
Development of HuProt Array-based Lysate Profiling of Multiplexed PTM Signatures
Recent studies have revealed that several types of PTMs are involved in the development and progression of ovarian cancer, such as Tyr phosphorylation (32), SUMOylation (33), ubiquitylation (34), and acetylation (35), presumably caused by dysregulation of the corresponding enzymes. Therefore, it is informative to identify dysregulated signaling pathways governed by these enzymes.
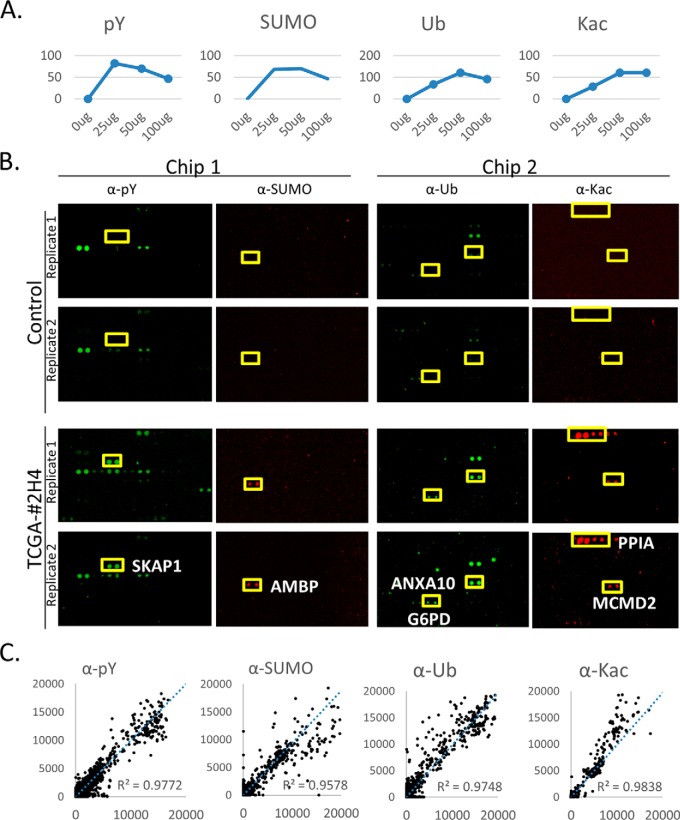
To develop the multiplexed reactions, we first used two frozen ovarian tumor samples of relatively high amounts for optimization on a pilot HuProt array. Each tumor sample was first lysed and concentration of total protein was measured. The whole tumor lysates were serially diluted to various concentrations, ranging from 0.21 to 0.83 μg/μl. By plotting numbers of positives obtained at different lysate concentrations, 0.42 μg/μl of the total protein was determined as the optimal concentration for the multiplexed lysate reactions (Fig. 2A).
Fig. 2.
Determination of optimal lysate concentrations and test of reproducibility of the HuProt array-based lysate profiling of multiplexed PTM signatures. A, the number of positive hits at increased amount of total tumor lysates. Consideration of the reaction of all four type of PTMs, 50 μg/120 μl lysate each reaction appeared relative high hit numbers. B, Representative cropped images of HuProt array for each PTM assay. For example, Src kinase-associated phosphoprotein 1(SKAP) was found to be readily Tyr-phosphorylated and Protein AMBP was SUMOylated, whereas Annexin A10 (ANXA10) and Glucose-6-phosphate 1-dehydrogenase (G6PD) were strongly ubiquitylated, Peptidyl-prolyl cis-trans isomerase A (PPIA) and Minichromosome maintenance domain-containing protein 2 (MCMD2) were acetylated, respectively, by the lysates tested. C, Four types of PTM reactions on different HuProt arrays showed high reproducibility with TCGA sample #2H4).
To evaluate the reproducibility of the multiplexed PTM reactions, four TCGA ovarian tumor samples of relatively large size were lysed and incubated on the HuProt arrays under the optimized conditions (i.e. 0.42 μg/μl) in duplicate (Fig. 2B). As illustrated in Fig. 2C, all the four PTM reactions of showed high reproducibility (e.g. TCGA sample #2H4), with the correlation coefficient R2 values ranging from 0.9578 to 0.9838. Several representative examples are shown in Fig. 2B. For example, SKAP was found to be readily Tyr-phosphorylated and Protein AMBP was SUMOylated; whereas ANXA10 and G6PD were strongly ubiquitylated; PPIA and MCMD2 were acetylated, respectively, by the lysates tested. As a comparison, none of them showed any detectable signals in the negative control reactions, indicating that the observed PTM signals were specific to TCGA samples.
Large-scale, HuProt Array-based Lysate Profiling of TCGA Ovarian Tumors
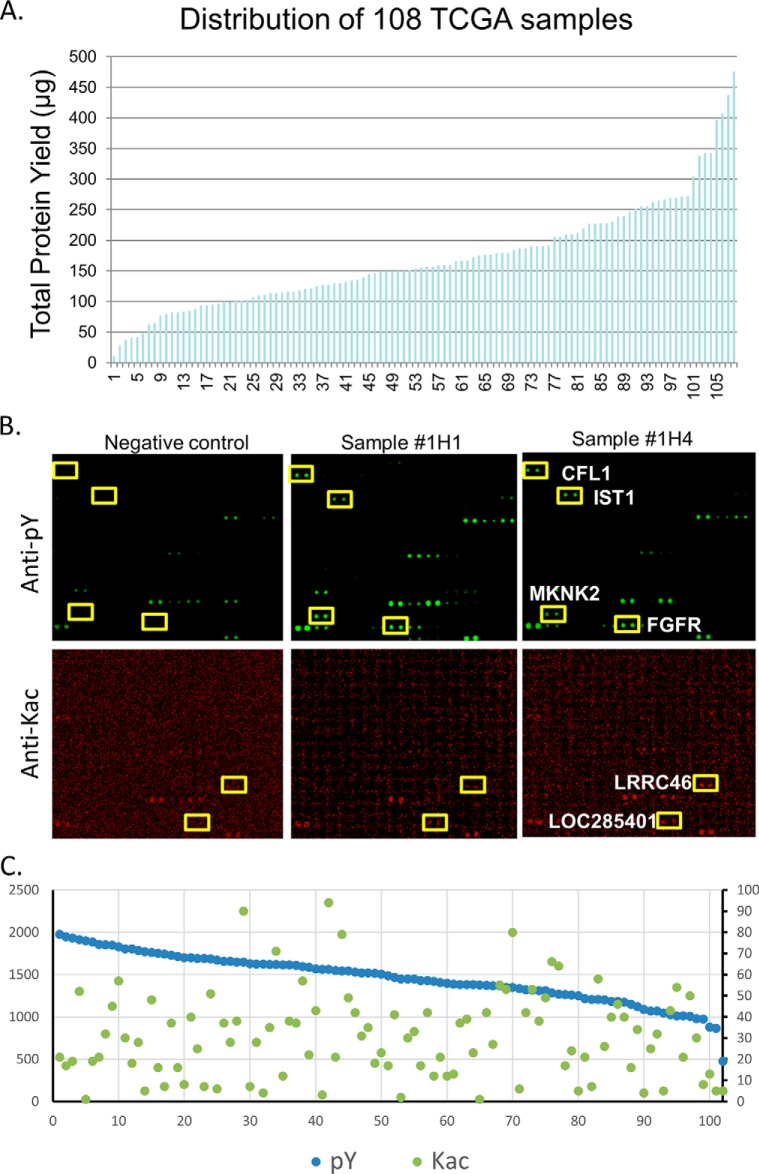
We next carried out a large-scale PTM signature profiling on HuProt arrays. All the 108 available ovarian tumor samples, ranging from 1.1 to 21 mg wet weights, were lysed and the concentration quantified using Bradford method. The total lysate proteins ranged from 10.5 to 560 μg per sample with a total protein concentration ranging from 0.3 to 3.5 μg/μl (Fig. 3A). Based on the optimized PTM assay conditions, 102 of the 108 samples produced enough protein for a single multiplex PTM assay on HuProt arrays, whereas fewer than 80 produced adequate protein for performing two multiplex PTM assays. Considering the high reproducibility of the multiplex PTM reactions and the importance of surveying a large number of tumor samples on HuProt arrays, this study focused on multiplexed Tyr phosphorylation and acetylation reactions for a total of 102 ovarian tumor samples.
Fig. 3.
Profiling of multiplexed Tyr phosphorylation and Lys acetylation for 102 TCGA ovarian tumor samples. A, Preparation of total lysates for 102 TCGA ovarian tumor samples. B, Representative cropped image of HuProt array for multiplexed Tyr phosphorylation and Lys acetylation assay. For example, the Tyr residues of Cofilin-1(CFL1), Putative MAPK-activating protein PM28 (IST1), MAP kinase-interacting serine/threonine-protein kinase 2(MKNK2), and Fibroblast growth factor receptor 1 (FGFR) were heavily phosphorylated by both TCGA samples #1H1 and #1H4. Although, Leucine-rich repeat-containing protein 46 (LRRC46) and LOC285401 were both acetylated by the same two TCGA tumor samples as detected with a pan anti-Kac antibody. C, Number of significantly Tyr-phosphorylated and Lys-acetylated positive hits for each of the 102 TCGA ovarian tumor samples.
In addition to the successful profiling of Tyr phosphorylation and acetylation signatures for all 102 ovarian tumor samples, negative control reactions with no lysate added were carried out in duplicate. Many proteins were found strongly phosphorylated on their Tyr residues by tumor lysates only (36, 37). For example, CFL1, IST1, MKNK2, and FGFR were heavily phosphorylated by both TCGA samples #1H1 and #1H4 (upper panel; Fig. 3B). None showed any detectable signal in the negative control reactions. Similarly, many proteins were also acetylated only by the tumor lysates. For example, LRRC46 and LOC285401 were both acetylated by the same two TCGA tumor samples as detected with a pan anti-Kac antibody (see Methods) (lower panel; Fig. 3B).
To identify those proteins that demonstrated robust Tyr phosphorylation and/or acetylation signals, we employed a similar protocol as described previously (24). We observed that, with a few exceptions, the overall distributions for both Tyr phosphorylation and Lys acetylation were very similar from sample to sample, indicating the high quality of the assays (supplemental Fig. S1). Based on histogram analysis of the signal intensity, we calculated the standard deviation (S.D.) value for each PTM reaction on the HuProt arrays. Using a very stringent cut off value of ≥ 5 S.D., positives were determined for each reaction. After removal of false positives based on negative control reactions, the number of significantly Tyr-phosphorylated proteins ranged from 448 to 1854 among the 102 samples, whereas the number of significantly acetylated proteins was from 1 to 94 (Fig. 3C). There was no apparent correlation between Tyr phosphorylation and Lys acetylation in our assays.
Data Integration to Predict Signaling Pathways Dysregulated in Ovarian Cancer
Because the relationship between acetyltransferases and their specific substrates remained largely unknown, we decided to focus on the Tyr phosphorylation analysis using an integrated kinase-substrate interaction (KSR) database based on PhosphoSitePlus (27) and PhosphoNetworks (17). Integration of these two databases allowed us to construct a new phosphotyrosine-specific KSR database, containing a total of 989 KSRs involving 83 kinase and 561 substrates. After integrated analysis of the tyrosine phosphorylation signals on HuProt array using ovarian tumor lysates, 54 kinases were predicted as the kinases responsible for the observed Tyr phosphorylation; together with the corresponding 118 substrates, they form 245 active KSRs in ovarian tumor (supplemental Table S2). Through integrated activity-based analysis, a network with19 kinases and 40 substrates having high correlation (Spearman correlation, p < 0.2) between MS-based global protein abundance and pY activities was shown in Fig. 4B. Interestingly, integrated activity-based analysis identified SYK as a highly active tyrosine kinase among ovarian high-grade serious carcinoma (Fig. 4B and supplemental Table S2). The Ingenuity® Pathway Analysis (IPA) analysis of active KSRs showed that the top enriched molecular and cellular functions include cell movement (p value = 1.38E-64), migration of cells (p value = 1.26E-62), and apoptosis (p value = 1.20E-56).
Validation of Dysregulated Tyrosine Kinase Signaling Pathways
The above IPA analysis predicted that members in the Src kinase family were significantly enriched in dysregulated Tyr kinases in ovarian tumors. Src kinases are non-receptor tyrosine kinases, including nine members: Src, Yes, Fyn, Fgr, Lck, Hck, Blk, Lyn, and Frk, many of which are known to play an important role in cancer biology (37–42). Except for Blk, these Src kinases were predicted as dysregulated in ovarian tumor based on our analyses. Importantly, polyclonal antibodies against seven of these eight Src kinases (Src, Yes, Fyn, Hck, Lyn, Fgr, and Frk) showed strong immunohistochemistry (IHC) staining in tissue sections of ovarian tumor samples according to Human Protein Atlas (HPA), whereas little or no detectable IHC signals were seen in normal ovarian tissues. The HPA data are in close agreement with our prediction, indirectly validated our prediction.
Spleen tyrosine kinase (SYK) is another interesting kinase predicted to be dysregulated in ovarian tumors (Fig. 4). SYK is known to play an essential role in various autoimmune diseases, and malignancies (43). The reason we highlighted it here is that SYK has become one of the major tyrosine kinases in ovarian cancer (10, 44, 45), the biochemistry and biology of SYK in ovarian cancer pathogenesis have been recently reported and an NIH registered clinical trial using a SYK inhibitor (fostamatinib) is ongoing. As a modulator of tumorigenesis, however, SYK plays a dual role as a tumor promoter and tumor suppressor, depending on tissue types (46). In HGSOCs, SYK expression and phospho-SYK (the active form of SYK) were elevated in recurrent post-chemotherapy ovarian carcinomas as compared with treatment naïve tumors before paclitaxel/carboplatin treatment (10, 45). Pharmacological or genetic inactivation of SYK could enhances paclitaxel cytotoxicity in vitro and in vivo and reduces ovarian cancer cell motility and invasion through modulating SYK downstream proteins including tubulins and microtubule-associated proteins (10, 47).
We also found that two focal adhesion kinases, PTK2 and PTK2B, were both predicted to be activated in ovarian tumors. Focal adhesion kinases are known to regulate the cell reorganization of the actin cytoskeleton, cell polarization, cell migration, adhesion, spreading, and bone remodeling. Further, recent studies have demonstrated that both PTK2 and PTK2B are involved in various cancers (48–51). Directly relevant to this study was the report that PTK2 was found overexpressed in most invasive ovarian cancers and plays a functionally significant role in ovarian tumor cell migration and invasion (52). To initiate activation this important non-receptor Tyr kinase, PTK2 autophosphorylates at Tyr397 and Src., another non-receptor kinase, is recruited via its interaction through the SH2 domain. PTK2 is fully activated through phosphorylation of Tyr576/577 residues by Src (53).
Similarly, PTK2B has been identified as a prognostic factor and a critical downstream signaling pathway for ascites-induced ovarian cancer cell migration and the expression of phosphorylated PTK2B in serous ovarian tumors was found to be associated with shorter progression-free survival (54). Similarly to PTK2, after PTK2B autophosphorylation at Tyr402, Src is recruited and activated by PTK2B; the activated Src fully activates PTK2B by phosphorylating Tyr579/580/881 (55, 56). Importantly, IHC studies in the HPA database have shown that protein levels of both PTK2B and PTK2 were found significantly elevated in ovarian tumors (57). Therefore, the activation status of the two kinases could be readily examined using phosphosite-specific antibodies.
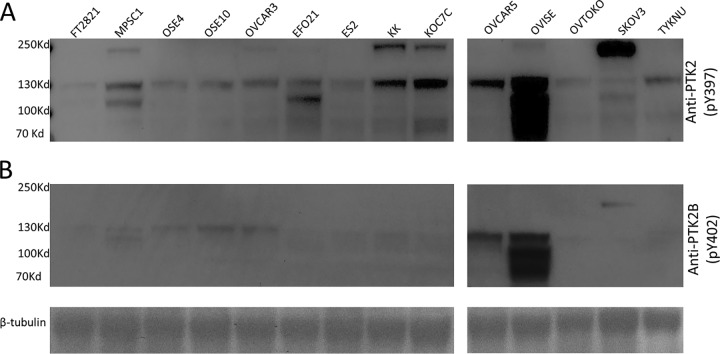
To test our hypothesis, the autophosphorylation status of PTK2 (pY397) and PTK2B (pY402) was examined as a proxy for their activities in 14 cell lines derived from either ovarian associated normal or tumor tissues, including three negative controls derived from a normal-appearing fallopian tube epithelial sample (FT2821) and two ovarian surface epithelial cell lines immortalized by SV40 large T antigen (OSE4 and OSE10) and other 11 varies of human ovarian carcinoma cells (MPSC1, OVCAR3, EFO21, ES2, KK, KOC7C, OVCAR5, OVISE, OVTOKO, SKOV3, TYKNU) (Fig. 5A). As predicted based on the HPA results, strong anti-PTK2 (pY397) immunoblot signals were detected in most ovarian tumor cell lines, including MPSC1, KK, KOC7C, OVCAR5, OVISE SKOV3, whereas a few ovarian tumor cell lines, OVCAR3, EFO21, ES2, OVTOKO, and TYKNU, showed somewhat lower signals. Anti-PTK2 (pY397) showed very low or undetectable immunoblot signals in three negative control cell lines, namely FT2821, OSE4, and OSE10. In addition, ovarian tumor cell SKOV3 displayed very low immunoblot signal at the expected molecular mass of PTK2 monomer; however, a very strong band was observed at twice molecular weight of PTK2, suggesting that a highly activated homodimer had formed. This is consistent with previous reports where PTK2 was observed to dimerize via the C-terminal region (FAT domain) swapping in helix 1 to stabilize a FAT: FERM interaction and play a dominant role in Y397 phosphorylation (58, 59) (60). Further, PTK2 dimerization was also observed in cell of KK and KOC7C, both of which had strong immunoblot signals of mono-PTK2 activity based on the autophosphorylation status of pY397 (Fig. 5A).
Fig. 5.
Validation of the dysregulated tyrosine kinases FAK and PTK2B. To test the dysregulated of PTK2 and PTK2B in ovarian cancer, 14 cell lines derived from either ovarian associated normal or tumor tissues, including three negative controls (FT2821, OSE4 and OSE10) and 11 varies of human ovarian carcinoma cells (MPSC1, OVCAR3, EFO21, ES2, KK, KOC7C, OVCAR5, OVISE, OVTOKO, SKOV3, TYKNU), were subject to Western blotting test for autophosphorylation status of PTK2 (pY397) and PTK2B (pY402) as a proxy for their activities. A, Activity status of PTK2 (pY397) in 14 cell lines; B, Activity status of PTK2B (pY402) in 14 cell lines.
Similarly, highly activated PTK2B signals, as detected by anti-PTK2B (pY 402), were only observed in ovarian tumor cell lines, OVCAR5 and OVISE (Fig. 5B). Further, a weak band double the size of PTK2B was detected in SKOV3, suggesting PTK2B dimer formation presumably via Ca2+-dependent regulation of PTK2B autophosphorylation (61). Similarly, anti-PTK2B (pY402) immunoblot analysis showed very low signals in all the three negative control cell lines Finally, we performed immunoblot analysis with both anti-PTK2 (pY397) and PTK2B (pY402) against a variety of immortalized cell lines, including Vero cell from African green monkey kidney epithelial, HEK293 cell from human embryonic kidney cell and GM12878 from human B-lymphocyte, and six tumor cells, K562 and Jurkat from leukemia, HeLa from cervical cancer, NCI-H929 from plasmacytoma and A172 from brain glioblastoma. As expected, no detectable immunoblot signals were observed in these cell lines, suggesting that PTK2 and PTK2B activation is highly specific in ovarian tumors (Data not shown).
DISCUSSION
Rapid development of new technologies in genomics, especially the use of next generation sequencing, has greatly advanced understanding of the connections between molecular alternations at the DNA level and tumorigenesis. However, the recent accomplishment of complete proteogenomic analysis by CPTAC consortium provided strong evidence that much of the cancer associated molecular alterations will not necessarily be limited to the form of coordinated variations among DNA, copy number, and mRNA/protein expression, but is rather a result of complex rewiring of signaling pathways or networks mediated by protein and PTM interactomes (62–64). To achieve an efficient and cost-effective approach to identify dysregulated PTM pathways in tumor tissues, we proposed to generate a kinase activity signature using total cell lysates to perform phosphorylation reactions on the human protein array; then, based on phosphorylated substrates, upstream activated protein kinases could be identified (18). Although our previous study using similar method have successfully identified and confirmed new signaling components downstream of the c-Myc activation through comparison of phosphorylation signatures generated by lysates treated with hepatocyte growth factor (HGF) with the untreated cells (18). This new principle has not been tested in real tumor tissues.
In this study, we applied this principle with several improvements to obtain PTM signatures of four different types with a large number of human ovarian tumor samples as part of the CPTAC consortium (1). First, we employed the currently largest HuProt array as a substrate identification platform to ensure comprehensive coverage of the human proteome. Second, we developed and optimized a multiplexed PTM assay on HuProt to simultaneously obtain Tyr phosphorylation and acetylation signatures using pan antibodies to detect the modification. Third, we developed and optimized another multiplexed PTM reaction to detect ubiquitylation and SUMOylation signatures. During optimization, we observed that p-Tyr-100, of the three widely used pan anti-phosphotyrosine monoclonal antibodies (mAbs) (4G10, p-Tyr-100 and pY20), produced better sensitivity with lower background (65) and was used for the large-scale pY profiling. On the other hand, the two pan anti-acetyl-Lys mAbs performed similarly and were kept for later anti-AceK studies.
We also noticed that some HuProt proteins could be readily recognized by the above pan antibodies in mock reaction controls without adding any tissue lysates. One possible explanation was that some human proteins were already phosphorylated and/or acetylated during induction in the budding yeast by conserved protein kinases or acetyltransferases. Another possibility was because of autophosphorylation or autoacetylation of the HuProt proteins during the incubation with the reaction mixture. Indeed, we observed strong pY or acLys signals for some known kinases and acetyltransferases (data not shown). To simplify data analysis, these proteins were considered false-positives and were removed from downstream analysis.
As a part of CPTAC consortium, all the ovarian tumor samples were provided by TCGA Biospecimen Core Resource. Because of the limited sample volume and because of high reproducibility of the PTM reactions as observed during our optimization (Fig. 2), we decided to focus on generation of p-Tyr and ac-Lys signatures using a single reaction on HuProt for 108 of the 120 available ovarian tumor samples. Using stringent cutoff criteria, we identified many significant pY substrates, ranging from 448 to 1854, and significant ac-Lys substrates ranging from 1 to 94 across the 102 individual ovarian tumors. Further bioinformatics analyses allowed us to identify a subset of enriched p-Tyr substrates, although no significant counterparts were identified among ac-Lys substrates.
This integrative approach allows to predict Tyr kinases with elevated expression or activity based on previously established kinase-substrate relationships (i.e. KSR and PhosphoSitePlus) and literature (17, 18). Among the predicted kinases, members in the non-receptor Tyr kinase Src family were found significantly enriched. Although no healthy ovarian tissues were available for comparison in this study because of a lack of such sample in the TCGA consortium rich IHC data sets that are publicly available through the Human Protein Atlas served as a reference (57). Significantly, the majority of the Src family kinases were found highly expressed in the ovarian tumor sections compared with the healthy tissues used by HPA (see above). Further, we could correctly predict the activation of SYK in ovarian tumors, a finding consistent with a recent study (10). Finally, our validation with phosphosite-specific mAbs confirmed our prediction in cell lines derived from ovarian tumors. Taken together, these findings demonstrate the power of identifying PTM signatures on HuProt arrays identify dysregulated signaling pathways in tumors and potentially many disease states.
This new approach, however, is not without limitation. Because we used pan antibodies to detect PTM signals on HuProt arrays, the quality in terms of specificity and sensitivity of the mAbs could dictate the outcomes of this approach. Moreover, prediction of dysregulated upstream enzymes is heavily reliant on availability of the existing enzyme-substrate relationships. In this case, Tyr kinases were successfully predicted and validated as a result of the long history of studying phosphorylation events both in vitro and in vivo. Unfortunately, such relationships between the ubiquitin and SUMO substrates and their respective E3 ligases are largely uncharacterized, making it difficult to predict E3 activities based solely on substrate modification state. Therefore, the unbiased, systematic interrogation of enzyme-substrate relationships will serve as a foundation to further understand signaling pathways especially those related to disease.
Supplementary Material
Acknowledgments
We thank the CPTAC project for making their proteomics data sets publicly available and the CPTAC consortium for the constructive suggestions.
Footnotes
* This work was funded by NCI's Clinical Proteomic Tumor Analysis Consortium initiative (NIH Grants U24CA160036 and U24CA210985), NIH/NCI grants U01CA200469, CA200469, RO1CA215483, P50CA228991, Ovarian Cancer Research Foundation Alliance grant OCRFA#458972, and Richard W. TeLinde Endowment, Department of Gynecology and Obstetrics, Johns Hopkins Medical Institutions.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
Conflict of Interest: The authors declare that they have no conflicts of interest.
1 The abbreviations used are:
- HGSOC
- high-grade serous ovarian carcinoma tumor
- PTM
- post-translational modification
- CPTAC
- Clinical Proteomic Tumor Analysis Consortium
- MS
- mass spectrometry
- KSR
- kinase-substrate relationship
- HPA
- Human Protein Atlas
- IPA
- Ingenuity Pathway Analysis
- IHC
- immunohistochemistry
- SYK
- spleen tyrosine kinase.
REFERENCES
- 1. Zhang H., Liu T., Zhang Z., Payne S. H., Zhang B., McDermott J. E., Zhou J. Y., Petyuk V. A., Chen L., Ray D., Sun S., Yang F., Chen L., Wang J., Shah P., Cha S. W., Aiyetan P., Woo S., Tian Y., Gritsenko M. A., Clauss T. R., Choi C., Monroe M. E., Thomas S., Nie S., Wu C., Moore R. J., Yu K. H., Tabb D. L., Fenyo D., Bafna V., Wang Y., Rodriguez H., Boja E. S., Hiltke T., Rivers R. C., Sokoll L., Zhu H., Shih I. M., Cope L., Pandey A., Zhang B., Snyder M. P., Levine D. A., Smith R. D., Chan D. W., Rodland K. D., and Investigators C. (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giguere S. S., Guise A. J., Jean Beltran P. M., Joshi P. M., Greco T. M., Quach O. L., Kong J., and Cristea I. M. (2016) The proteomic profile of deleted in breast cancer 1 (DBC1) interactions points to a multifaceted regulation of gene expression. Mol. Cell. Proteomics 15, 791–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boja E. S., and Rodriguez H. (2014) Proteogenomic convergence for understanding cancer pathways and networks. Clin. Proteomics 11, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ardito F., Giuliani M., Perrone D., Troiano G., and Lo Muzio L. (2017) The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 40, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kolch W., and Pitt A. (2010) Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat. Rev. Cancer 10, 618–629 [DOI] [PubMed] [Google Scholar]
- 6. Krueger K. E., and Srivastava S. (2006) Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol. Cell. Proteomics 5, 1799–1810 [DOI] [PubMed] [Google Scholar]
- 7. Siegel R., Ma J., Zou Z., and Jemal A. (2014) Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 [DOI] [PubMed] [Google Scholar]
- 8. Marsh D. J., Shah J. S., and Cole A. J. (2014) Histones and their modifications in ovarian cancer - drivers of disease and therapeutic targets. Front. Oncol. 4, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galdieri L., Gatla H., Vancurova I., and Vancura A. (2016) Activation of AMP-activated protein kinase by metformin induces protein acetylation in prostate and ovarian cancer cells. J. Biol. Chem. 291, 25154–25166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu Y., Gaillard S., Phillip J. M., Huang T. C., Pinto S. M., Tessarollo N. G., Zhang Z., Pandey A., Wirtz D., Ayhan A., Davidson B., Wang T. L., and Shih Ie M. (2015) Inhibition of spleen tyrosine kinase potentiates paclitaxel-induced cytotoxicity in ovarian cancer cells by stabilizing microtubules. Cancer Cell 28, 82–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ntanasis-Stathopoulos I., Fotopoulos G., Tzanninis I. G., and Kotteas E. A. (2016) The emerging role of tyrosine kinase inhibitors in ovarian cancer treatment: a systematic review. Cancer Invest. 34, 313–339 [DOI] [PubMed] [Google Scholar]
- 12. Morotti M., Becker C. M., Menada M. V., and Ferrero S. (2013) Targeting tyrosine-kinases in ovarian cancer. Expert Opin. Investig. Drugs 22, 1265–1279 [DOI] [PubMed] [Google Scholar]
- 13. Wilken J. A., Badri T., Cross S., Raji R., Santin A. D., Schwartz P., Branscum A. J., Baron A. T., Sakhitab A. I., and Maihle N. J. (2012) EGFR/HER-targeted therapeutics in ovarian cancer. Future Med. Chem. 4, 447–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Theillet F. X., Smet-Nocca C., Liokatis S., Thongwichian R., Kosten J., Yoon M. K., Kriwacki R. W., Landrieu I., Lippens G., and Selenko P. (2012) Cell signaling, post-translational protein modifications and NMR spectroscopy. J. Biomol. NMR 54, 217–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steen H., Jebanathirajah J. A., Rush J., Morrice N., and Kirschner M. W. (2006) Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics 5, 172–181 [DOI] [PubMed] [Google Scholar]
- 16. Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., Wan J., Tao S. C., Qian J., Zhao Y., Boeke J. D., Berger S. L., and Zhu H. (2009) Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136, 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Newman R. H., Hu J., Rho H. S., Xie Z., Woodard C., Neiswinger J., Cooper C., Shirley M., Clark H. M., Hu S., Hwang W., Jeong J. S., Wu G., Lin J., Gao X., Ni Q., Goel R., Xia S., Ji H., Dalby K. N., Birnbaum M. J., Cole P. A., Knapp S., Ryazanov A. G., Zack D. J., Blackshaw S., Pawson T., Gingras A. C., Desiderio S., Pandey A., Turk B. E., Zhang J., Zhu H., and Qian J. (2013) Construction of human activity-based phosphorylation networks. Mol. Syst. Biol. 9, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woodard C. L., Goodwin C. R., Wan J., Xia S., Newman R., Hu J., Zhang J., Hayward S. D., Qian J., Laterra J., and Zhu H. (2013) Profiling the dynamics of a human phosphorylome reveals new components in HGF/c-Met signaling. PLoS ONE 8, e72671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labots M., Gotink K. J., Dekker H., Azijli K., van der Mijn J. C., Huijts C. M., Piersma S. R., Jimenez C. R., and Verheul H. M. (2016) Evaluation of a tyrosine kinase peptide microarray for tyrosine kinase inhibitor therapy selection in cancer. Exp. Mol. Med. 48, e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang B. Y., Chen P. C., Chen B. H., Wang C. C., Liu H. F., Chen Y. Z., Chen C. S., and Yang Y. S. (2017) High-throughput screening of sulfated proteins by using a genome-wide proteome microarray and protein tyrosine sulfation system. Anal. Chem. 89, 3278–3284 [DOI] [PubMed] [Google Scholar]
- 21. Cox E., Uzoma I., Guzzo C., Jeong J. S., Matunis M., Blackshaw S., and Zhu H. (2015) Identification of SUMO E3 ligase-specific substrates using the HuProt human proteome microarray. Methods Mol. Biol. 1295, 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu J. Y., Lin Y. Y., Qian J., Tao S. C., Zhu J., Pickart C., and Zhu H. (2008) Functional dissection of a HECT ubiquitin E3 ligase. Mol. Cell. Proteomics 7, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cancer Genome Atlas Research, N. (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu C. J., Song G., Huang W., Liu G. Z., Deng C. W., Zeng H. P., Wang L., Zhang F. C., Zhang X., Jeong J. S., Blackshaw S., Jiang L. Z., Zhu H., Wu L., and Li Y. Z. (2012) Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Mol. Cell. Proteomics 11, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H. S., Woodard C., Wang H., Jeong J. S., Long S., He X., Wade H., Blackshaw S., Qian J., and Zhu H. (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139, 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu J., Rho H. S., Newman R. H., Zhang J., Zhu H., and Qian J. (2014) PhosphoNetworks: a database for human phosphorylation networks. Bioinformatics 30, 141–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jimenez-Marin A., Collado-Romero M., Ramirez-Boo M., Arce C., and Garrido J. J. (2009) Biological pathway analysis by ArrayUnlock and Ingenuity Pathway Analysis. BMC Proc. 3, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pohl G., Ho C. L., Kurman R. J., Bristow R., Wang T. L., and Shih Ie M. (2005) Inactivation of the mitogen-activated protein kinase pathway as a potential target-based therapy in ovarian serous tumors with KRAS or BRAF mutations. Cancer Res. 65, 1994–2000 [DOI] [PubMed] [Google Scholar]
- 31. Ellis M. J., Gillette M., Carr S. A., Paulovich A. G., Smith R. D., Rodland K. K., Townsend R. R., Kinsinger C., Mesri M., Rodriguez H., Liebler D. C., and Clinical Proteomic Tumor Analysis, C. (2013) Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 3, 1108–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davies S., Holmes A., Lomo L., Steinkamp M. P., Kang H., Muller C. Y., and Wilson B. S. (2014) High incidence of ErbB3, ErbB4, and MET expression in ovarian cancer. Int. J. Gynecol. Pathol. 33, 402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eifler K., and Vertegaal A. C. (2015) SUMOylation-Mediated Regulation of Cell Cycle Progression and Cancer. Trends Biochem. Sci. 40, 779–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sane S., and Rezvani K. (2017) Essential Roles of E3 Ubiquitin Ligases in p53 Regulation. Int. J. Mol. Sci. 18, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bryson B. D., and White F. M. (2015) Quantitative profiling of lysine acetylation reveals dynamic crosstalk between receptor tyrosine kinases and lysine acetylation. PLoS ONE 10, e0126242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hunter T. (2009) Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21, 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sen B., and Johnson F. M. (2011) Regulation of SRC family kinases in human cancers. J. Signal Transduct. 2011, 865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Summy J. M., and Gallick G. E. (2003) Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 22, 337–358 [DOI] [PubMed] [Google Scholar]
- 39. Zhang S., and Yu D. (2012) Targeting Src family kinases in anti-cancer therapies: turning promise into triumph. Trends Pharmacol. Sci. 33, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parsons S. J., and Parsons J. T. (2004) Src family kinases, key regulators of signal transduction. Oncogene 23, 7906–7909 [DOI] [PubMed] [Google Scholar]
- 41. Emaduddin M., Bicknell D. C., Bodmer W. F., and Feller S. M. (2008) Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 105, 2358–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Belsches-Jablonski A. P., Biscardi J. S., Peavy D. R., Tice D. A., Romney D. A., and Parsons S. J. (2001) Src family kinases and HER2 interactions in human breast cancer cell growth and survival. Oncogene 20, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 43. Ma T. K., McAdoo S. P., and Tam F. W. (2016) Spleen tyrosine kinase: a crucial player and potential therapeutic target in renal disease. Nephron 133, 261–269 [DOI] [PubMed] [Google Scholar]
- 44. Yu Y., Suryo Rahmanto Y., Lee M. H., Wu P. H., Phillip J. M., Huang C. H., Vitolo M. I., Gaillard S., Martin S. S., Wirtz D., Shih I. M., and Wang T. L. (2018) Inhibition of ovarian tumor cell invasiveness by targeting SYK in the tyrosine kinase signaling pathway. Oncogene 37, 3778–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jinawath N., Vasoontara C., Jinawath A., Fang X., Zhao K., Yap K. L., Guo T., Lee C. S., Wang W., Balgley B. M., Davidson B., Wang T. L., and Shih Ie M. (2010) Oncoproteomic analysis reveals co-upregulation of RELA and STAT5 in carboplatin resistant ovarian carcinoma. PLoS ONE 5, e11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krisenko M. O., and Geahlen R. L. (2015) Calling in SYK: SYK's dual role as a tumor promoter and tumor suppressor in cancer. Biochim. Biophys. Acta 1853, 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu Y., Suryo Rahmanto Y., Lee M. H., Wu P. H., Phillip J. M., Huang C. H., Vitolo M. I., Gaillard S., Martin S. S., Wirtz D., Shih I. M., and Wang T. L. (2018) Inhibition of ovarian tumor cell invasiveness by targeting SYK in the tyrosine kinase signaling pathway. Oncogene 37, 3778–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sulzmaier F. J., Jean C., and Schlaepfer D. D. (2014) FAK in cancer: mechanistic findings and clinical applications. Nat. Rev. Cancer 14, 598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meads M. B., Fang B., Mathews L., Gemmer J., Nong L., Rosado-Lopez I., Nguyen T., Ring J. E., Matsui W., MacLeod A. R., Pachter J. A., Hazlehurst L. A., Koomen J. M., and Shain K. H. (2016) Targeting PYK2 mediates microenvironment-specific cell death in multiple myeloma. Oncogene 35, 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fan H., and Guan J. L. (2011) Compensatory function of Pyk2 protein in the promotion of focal adhesion kinase (FAK)-null mammary cancer stem cell tumorigenicity and metastatic activity. J. Biol. Chem. 286, 18573–18582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao C., Chen G., Kuan S. F., Zhang D. H., Schlaepfer D. D., and Hu J. (2015) FAK/PYK2 promotes the Wnt/beta-catenin pathway and intestinal tumorigenesis by phosphorylating GSK3beta. Elife 4, e10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sood A. K., Coffin J. E., Schneider G. B., Fletcher M. S., DeYoung B. R., Gruman L. M., Gershenson D. M., Schaller M. D., and Hendrix M. J. (2004) Biological significance of focal adhesion kinase in ovarian cancer: role in migration and invasion. Am. J. Pathol. 165, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitra S. K., Hanson D. A., and Schlaepfer D. D. (2005) Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 54. Lane D., Matte I., Laplante C., Garde-Granger P., Carignan A., Bessette P., Rancourt C., and Piche A. (2016) CCL18 from ascites promotes ovarian cancer cell migration through proline-rich tyrosine kinase 2 signaling. Mol. Cancer 15, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Park S. Y., Avraham H. K., and Avraham S. (2004) RAFTK/Pyk2 activation is mediated by trans-acting autophosphorylation in a Src-independent manner. J. Biol. Chem. 279, 33315–33322 [DOI] [PubMed] [Google Scholar]
- 56. Avraham H., Park S. Y., Schinkmann K., and Avraham S. (2000) RAFTK/Pyk2-mediated cellular signalling. Cell Signal 12, 123–133 [DOI] [PubMed] [Google Scholar]
- 57. Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P. H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Ponten F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 58. Fang X., Liu X., Yao L., Chen C., Lin J., Ni P., Zheng X., and Fan Q. (2014) New insights into FAK phosphorylation based on a FAT domain-defective mutation. PLoS ONE 9, e107134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arold S. T., Hoellerer M. K., and Noble M. E. (2002) The structural basis of localization and signaling by the focal adhesion targeting domain. Structure 10, 319–327 [DOI] [PubMed] [Google Scholar]
- 60. Brami-Cherrier K., Gervasi N., Arsenieva D., Walkiewicz K., Boutterin M. C., Ortega A., Leonard P. G., Seantier B., Gasmi L., Bouceba T., Kadare G., Girault J. A., and Arold S. T. (2014) FAK dimerization controls its kinase-dependent functions at focal adhesions. EMBO J. 33, 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lysechko T. L., Cheung S. M., and Ostergaard H. L. (2010) Regulation of the tyrosine kinase Pyk2 by calcium is through production of reactive oxygen species in cytotoxic T lymphocytes. J. Biol. Chem. 285, 31174–31184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duan G., and Walther D. (2015) The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 11, e1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karve T. M., and Cheema A. K. (2011) Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids 2011, 207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Merbl Y., and Kirschner M. W. (2011) Protein microarrays for genome-wide posttranslational modification analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 347–356 [DOI] [PubMed] [Google Scholar]
- 65. Tinti M., Nardozza A. P., Ferrari E., Sacco F., Corallino S., Castagnoli L., and Cesareni G. (2012) The 4G10, pY20 and p-TYR-100 antibody specificity: profiling by peptide microarrays. Nat. Biotechnol. 29, 571–577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.