Bacterial endospores can remain dormant and highly resistant to environmental insults for long periods but can also rapidly germinate in response to a nutrient-rich environment. The persistence and subsequent germination of spores contribute to their colonization of new environments and to the spread of certain diseases. Proteins of Bacillus subtilis and Bacillus anthracis were identified that are associated with the spore membrane, a position that can allow them to contribute to germination. A set of identified proteins that are predicted to carry out ion transport were examined for their contributions to spore formation, stability, and germination. Greater knowledge of spore formation and germination can contribute to the development of better decontamination strategies.
KEYWORDS: Bacillus anthracis, germination, membrane, proteome, spore, subtilis
ABSTRACT
Bacterial endospores produced by Bacillus and Clostridium species can remain dormant and highly resistant to environmental insults for long periods, but they can also rapidly germinate in response to a nutrient-rich environment. Multiple proteins involved in sensing and responding to nutrient germinants, initiating solute and water transport, and accomplishing spore wall degradation are associated with the membrane surrounding the spore core. In order to more fully catalog proteins that may be involved in spore germination, as well as to identify protein changes taking place during germination, unbiased proteomic analyses of membrane preparations isolated from dormant and germinated spores of Bacillus anthracis and Bacillus subtilis were undertaken. Membrane-associated proteins were fractionated by SDS-PAGE, gel slices were trypsin digested, and extracted peptides were fractionated by liquid chromatography and analyzed by matrix-assisted laser desorption ionization–tandem time of flight mass spectrometry. More than 500 proteins were identified from each preparation. Bioinformatic methods were used to characterize proteins with regard to membrane association, cellular function, and conservation across species. Numerous proteins not previously known to be spore associated, 6 in B. subtilis and 68 in B. anthracis, were identified. Relative quantitation based on spectral counting indicated that the majority of spore membrane proteins decrease in abundance during the first 20 min of germination. The spore membranes contained several proteins thought to be involved in the transport of metal ions, a process that plays a major role in spore formation and germination. Analyses of mutant strains lacking these transport proteins implicated YloB in the accumulation of calcium within the developing forespore.
IMPORTANCE Bacterial endospores can remain dormant and highly resistant to environmental insults for long periods but can also rapidly germinate in response to a nutrient-rich environment. The persistence and subsequent germination of spores contribute to their colonization of new environments and to the spread of certain diseases. Proteins of Bacillus subtilis and Bacillus anthracis were identified that are associated with the spore membrane, a position that can allow them to contribute to germination. A set of identified proteins that are predicted to carry out ion transport were examined for their contributions to spore formation, stability, and germination. Greater knowledge of spore formation and germination can contribute to the development of better decontamination strategies.
INTRODUCTION
Bacterial endospores produced by Bacillus, Clostridium, and related genera can remain dormant for extended periods of time. In addition, these spores are highly resistant to many of the chemical and physical treatments commonly used to reduce bacterial contamination (1). Upon exposure to conditions conducive to resumption of vegetative growth these spores can rapidly germinate and resume metabolism (2, 3). These factors allow certain spore-forming species to act as significant human pathogens, including potential biological weapons (4), as agents of food poisoning and spoilage (5), and, on a positive note, as effective vehicles for delivery of antigens or metabolic and enzymatic activities of industrial, consumer, or patient benefit (6–8).
Spore dormancy and resistance properties are directly related to the relative dehydration of the spore core (cytoplasm) and high core concentrations of solutes such as calcium dipicolinate (Ca2+-DPA) (1). The dehydrated and metabolically inactive state of the core is maintained by the spore membrane, which exists in a novel nonfluid state (9), and the surrounding spore cortex peptidoglycan cell wall (10). Spore germination proceeds through the rapid release of the core Ca2+-DPA pool, a concomitant uptake of water (3), and the subsequent return of the spore membrane fluidity (9). This is immediately followed by degradation of most of the cortex wall (11), allowing full core rehydration, the resumption of metabolic activity, and spore outgrowth. Knowledge of the mechanisms driving spore germination will allow targeting of this process for the improvement of decontamination regimens, as well as regulating germination during antigen or activity delivery (12).
Genome sequences and transcriptome profiling have produced predictions of proteins present within spores (13–17). A number of proteome studies have examined spore fractions in several species (16, 18–26). Most of these studies, however, were biased toward soluble proteins, as opposed to membrane-associated proteins, even though many of the proteins currently known to function in the spore germination process are associated with the spore membrane. These include germinant receptor complexes (27, 28), the SpoVA proteins involved in Ca2+-DPA transport (29–32), the GerD lipoprotein (33, 34), and the YpeB-SleB proteins involved in cortex degradation (35). Some of these germination-associated proteins are known to decrease in amount or in their membrane associations during germination (34, 35).
The goals of the present study were to catalog the spore membrane proteomes of Bacillus subtilis and Bacillus anthracis and to examine changes in these proteomes during germination. More than 500 proteins were identified in each proteome, and approximately 100 of these were found to contain amino acid sequences predicted to result in integral or peripheral membrane association. The majority of previously identified membrane-associated germination proteins were identified, and many proteins were identified for the first time in the membrane proteomes of both species. Spectral counting methods for determining protein abundance changes during germination revealed that the levels of many spore membrane proteins decreased significantly during germination. The functions of several spore membrane proteins predicted to be involved in ion transport were subsequently examined.
RESULTS
Proteins identified in dormant spore membrane preparations.
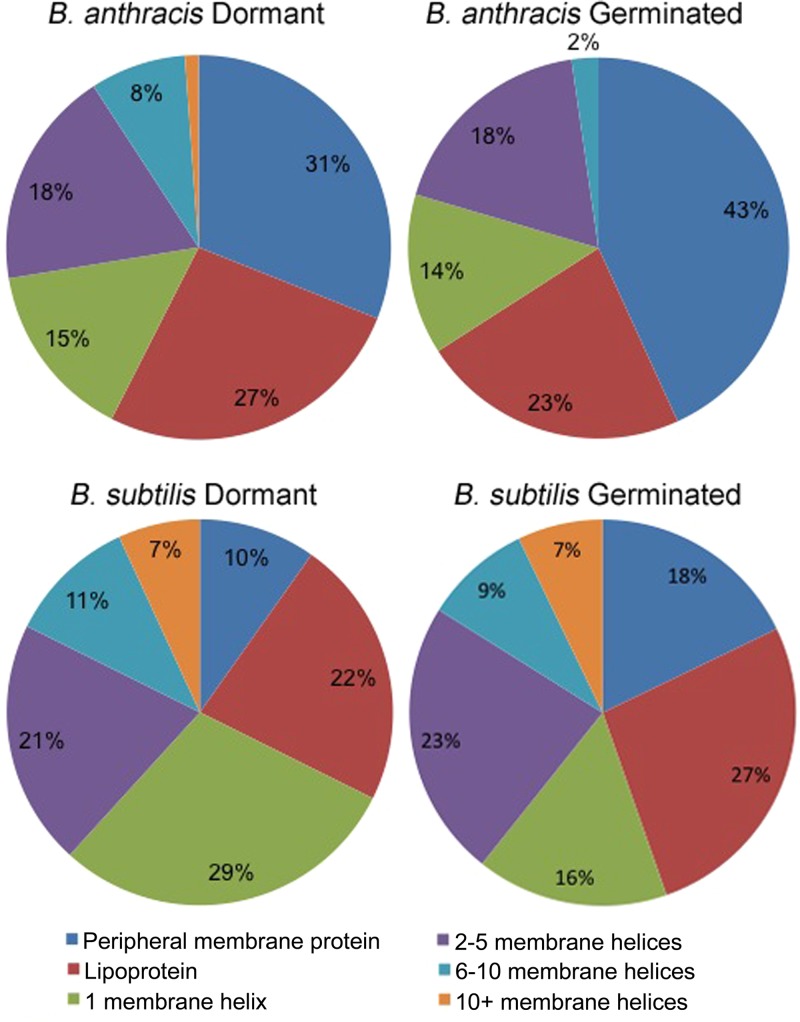
Membranes were isolated from three independent preparations of B. anthracis and B. subtilis dormant spores. Proteins were separated by SDS-PAGE (see Fig. S1 in the supplemental material), followed by gel slice preparation and processing to provide peptides that were submitted to liquid chromatography (LC) separation and matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) analysis. Totals of 603 and 592 proteins were identified in B. subtilis and in B. anthracis samples, respectively (see Tables S1 and S2 in the supplemental material). Bioinformatic predictions suggested that 104 (17%) and 87 (15%) of these proteins were membrane-associated proteins in B. subtilis and B. anthracis, respectively (Table S3). Many ribosomal proteins and proteins associated with macromolecular structures, such as spore coats and exosporium, were also identified, presumably due to pelleting of these structures during centrifugation of membranes or due to association of these structures with the membrane. Predicted membrane proteins were further categorized based on the type of membrane association and number of predicted membrane-spanning helices (Fig. 1). The presence of a wide variety of lipoprotein, peripheral, monotopic, and integral polytopic membrane proteins indicates that the method used was successful in recovering a broad membrane proteome. A total of 11 known germination-related membrane proteins were identified in B. subtilis samples (Table 1). Similarly, a total of five known germination related proteins were identified in B. anthracis samples (Table 1 ). The remaining identified membrane proteins have predicted functions in 14 COG (Clusters of Orthologous Groups) function categories.
FIG 1.
Predicted membrane-spanning domains of B. anthracis and B. subtilis spore membrane proteins. Predictions of membrane association mechanisms were made for proteins identified in membrane fractions as described in Materials and Methods. Proteins were further classified based upon their predicted number of membrane-spanning helices.
TABLE 1.
B. anthracis and B. subtilis spore germination proteins identified in spore membrane proteomes
| Gene | Function | UniProt no. |
Membrane predictiona | |
|---|---|---|---|---|
| B. subtilis | B. anthracis | |||
| gerAC | Germinant receptor | P07870 | Lipoprotein | |
| gerBC | Germinant receptor | P39571 | Lipoprotein | |
| gerD | Germinant response | P16450 | Q81VP4 | Lipoprotein |
| gerKC | Germinant receptor | P49941 | Lipoprotein | |
| prkC | Peptidoglycan receptor | O34507 | Integral | |
| spoVAC | DPA transport | P40868 | Q81X68 | Integral |
| spoVAD | DPA transport | P40869 | Q81X67 | Peripheral |
| spoVAF | DPA transport | P31845 | Q81MG2 | Integral |
| yfkR | Germinant receptor | O35028 | Lipoprotein | |
| yhcN | Outgrowth | P54598 | Lipoprotein | |
| ypeB | Cortex degradation | P38490 | Q81PQ4 | Integral |
The mechanism of membrane association was predicted as described in Materials and Methods.
Proteins identified in germinated spore membrane preparations.
A portion of each spore preparation was exposed to nutrient germinants until the optical density (OD) of the suspension had decreased 50%, which resulted in the germination of ≥95% of the spores. The membrane fractions were then isolated, and the proteins were separated by SDS-PAGE (Fig. S1). Collection and processing of gel slices provided samples for LC separation and MALDI-TOF/TOF analysis. A total of 497 and 508 proteins were identified in B. subtilis and B. anthracis germinated spore membrane profiles, respectively (Tables S1 and S2). In each species, some proteins (82 and 88 proteins in B. subtilis and B. anthracis, respectively) were identified only in the germinated samples and not in the dormant spore samples, and these were predominantly predicted to be cytoplasmic proteins. Of the 52 (10.5%) and 38 (7.5%) proteins that were predicted to be membrane associated in germinated B. subtilis and B. anthracis spores, respectively, all but two in B. subtilis (P40780 and Q01464) were also present in the dormant spore samples.
Novel membrane proteins identified in spore membrane fractions.
The spore membranes are derived from the sporangium cytoplasmic membranes during the engulfment stage of sporulation, so it was no surprise to find that membrane proteins expected to be in vegetative cells were present in spore membrane fractions. Among the 104 predicted membrane proteins we detected in B. subtilis dormant spores, 98 and 78 had been identified in previous spore and vegetative proteome studies, respectively, and 75 of these were previously detected in both cell types (Table S3). Among the six predicted membrane proteins first identified in spores here (RbsA, YckB, YerB, YpmQ, YqaR, and YuaB), two have unknown functions, and others have predicted COG functions in amino acid transport and metabolism; carbohydrate transport and metabolism, colony structure, and energy production and conversion. GerAC, GerBC, GerKC, GerD, PrkC, SpoVAC, SpoVAD, SpoVAF, YfkR, YhcN, and YpeB are known germination-active proteins that were detected in spores in this study and previously (35, 36) (Table 1).
A similar analysis of the B. anthracis samples revealed that 12 predicted membrane proteins were reported previously in a vegetative cell proteome study, and 12 proteins were reported previously in spore proteome studies (Table S3), with one protein, YlaJ/BA2560, being found in both. Among the 75 predicted membrane proteins first identified in spores here, 16 are presently listed with unknown functions, 5 are known germination-active proteins (Table 1), and the rest sort into 12 different COG categories.
Membrane proteins under the control of sporulation-specific sigma factors.
Predicted membrane-associated proteins identified in B. subtilis samples were searched against the relatively well-characterized sporulation transcriptomes of that species (15, 17, 22, 37). Of the 104 predicted membrane-associated proteins, 29 have been shown to be controlled at the transcriptional level by the sporulation-specific sigma factors σE, σF, σG, and σK (Table S3). Only a small number of membrane proteins were identified from the mother cell-specific σE and σK regulons, four and two proteins, respectively. The remaining membrane proteins for which transcriptome data are available were in the forespore-specific σF (4 proteins) and σG (19 proteins) regulons, though the similarity between these two sigma factors results in some overlap of their regulons (17). The genes of 10 known germination-active proteins identified are all within the σG regulon. Eight σG-dependent proteins had not been previously detected in spore proteomes. Among these, YutC, YhcC, YrbG, and YqfX have unknown functions, while YveA, YwjE, YitG, and YthA were categorized into amino acid transport and metabolism, lipid biogenesis and metabolism, general transporter, and energy production and conversion, respectively.
Similarities between the spore membrane proteomes in the two Bacillus species.
Searches using the BLAST program were done to compare the predicted membrane proteins identified in samples from each species. Fifty-one B. anthracis spore membrane proteins showed high similarity to 48 spore membrane proteins identified in B. subtilis (Table 2). Forty-five of these proteins were considered to be orthologous, based on (i) a high level of amino acid sequence identity (>23%) and similarity (>42%) across an alignment of >62% of the protein sequences and (ii) synteny. Among these, GerD, MisCA, SpoVAC, SpoVAD, SpoVAF, YetF, YpeB, YthA, and YutC have been previously identified in spores under more defined studies and/or were expressed in the forespore-specific σF and σG regulons. Several of these are known germination-active proteins. MisCA (SpoIIIJ) is required for activation of σG after engulfment of the forespore is completed (38). YthA was suggested to compensate for the loss of cytochrome aa3, which is critical for sporulation as one of two heme copper terminal oxidases in B. subtilis (39). There is no known function reported for YutC and YetF. This leaves eight proteins that were detected in both species and had not been previously identified in spore proteomes or sporulation-specific regulons. ArtP, MetQ, and ZnuA are components of ABC transporters involved in amino acid or inorganic ion transport. YpmQ is involved in assembly of a copper center in cytochrome c oxidase, which has an important function in energy conversion and metabolism (40). YyxA was predicted to be a serine protease due to its sequence similarity to the S1B peptidase family. YndM, YugS and YkrK are proteins of unknown function.
TABLE 2.
Proteins detected in both Bacillus species spore membrane proteomesa
| Gene names (B. subtilis/B. anthracis) | UniProt no. |
Protein function | Sequence identity (%) | Sequence similarity (%) | Alignment lengthb (%) | |
|---|---|---|---|---|---|---|
| B. subtilis | B. anthracis | |||||
| ahpF/ahpF | P42974 | Q81ZC5 | NADH dehydrogenase | 84 | 91 | 100 |
| artP/GBAA_0367 | P54535 | Q6I447 | Arginine transport, ArtP | 37 | 55 | 95 |
| atpD/atpD | P37809 | Q81JZ5 | ATP synthase | 86 | 92 | 99 |
| atpF/atpF | P37814 | Q81JZ1 | ATP synthase | 57 | 76 | 98 |
| atpG/atpG | P37810 | Q81JZ4 | ATP synthase | 68 | 84 | 100 |
| bdbD/bdbD | O32218 | Q81YT8 | Disulfide bond formation | 42 | 64 | 85 |
| era/era | P42182 | Q81LT7 | GTPase Era | 75 | 89 | 100 |
| fhuD/GBAA_0351 | P37580 | Q81ZB8 | Iron-hydroxamate binding, FhuD | 38 | 61 | 93 |
| ftsH/ftsH | P37476 | Q81VX5 | Zinc metalloprotease, FtsH | 80 | 89 | 100 |
| gerD/gerD | P16450 | Q81VP4 | Germination, GerD | 43 | 69 | 89 |
| metQ/GBAA_5220 | O32167 | Q81XL5 | Methionine-binding lipoprotein | 57 | 76 | 100 |
| misCA/yidC2 | Q01625 | Q81JH1 | Membrane protein insertase | 68 | 81 | 100 |
| msmX/potA | P94360 | Q81TH8c | Maltodextrin import, MsmX | 51 | 71 | 76 |
| oppA/GBAA_0656 | P24141 | Q81V45 | Oligopeptide transport, OppA | 28 | 45 | 92 |
| oppB/GBAA_1192 | P24138 | Q81TS3 | Oligopeptide transport, OppB | 50 | 70 | 100 |
| oppC/GBAA_1193 | P24139 | Q81TS2 | Oligopeptide transport, OppC | 46 | 66 | 98 |
| oppD/GBAA_1194 | P24136 | Q81TS1 | Oligopeptide transport, OppD | 73 | 85 | 97 |
| oppF/GBAA_1195 | P24137 | Q81TS0 | Oligopeptide transport, OppF | 80 | 89 | 97 |
| pbpF/GBAA_1474 | P38050 | Q81T17 | Penicillin-binding protein 1F | 42 | 61 | 89 |
| plsY/plsY3 | Q45064 | Q81Y92 | Glycerol-3-PO4 acyltransferase | 63 | 75 | 94 |
| ponA/GBAA_2345 | P39793 | Q81QS3 | Penicillin-binding protein 1 | 46 | 65 | 89 |
| prsA/prsA1 | P24327 | Q81U45 | Foldase, PrsA | 47 | 66 | 96 |
| ptsG/ptsG | P20166 | Q81MH9 | PTS system | 61 | 79 | 100 |
| qcrA/qcrA | P46911 | Q81SV1 | Menaquinol-cytochrome c reductase | 70 | 80 | 95 |
| qcrB/qcrB | P46912 | Q81SV0 | Menaquinol-cytochrome c reductase | 95 | 97 | 100 |
| qoxA/ctaC | P34957 | Q81MT9c | Quinol oxidase | 36 | 56 | 62 |
| rbsA/ecsA | P36947 | Q81U40c | Ribose import, RbsA | 28 | 49 | 84 |
| resA/resA | P35160 | Q81SZ9 | Thiol-disulfide oxidoreductase | 52 | 69 | 99 |
| secDF/secDF | O32047 | Q81LH8 | Protein translocase, SecDF | 60 | 76 | 97 |
| secY/secY1 | P16336 | Q81VR0 | Protein translocase, SecY | 72 | 82 | 100 |
| spoVAC/GBAA_5375 | P40868 | Q81X68 | DPA transport, SpoVAC | 60 | 78 | 89 |
| spoVAD/GBAA_5376 | P40869 | Q81X67 | DPA transport, SpoVAD | 50 | 65 | 98 |
| spoVAF/spoVAF | P31845 | Q81MG2 | DPA transport SpoVAF | 63 | 80 | 96 |
| tagU/tagU | Q02115 | Q81K33 | Transcriptional regulator, LytR | 50 | 72 | 93 |
| tcyA/GBAA_0855 | P42199 | Q81UL3 | l-Cystine-binding protein TcyA | 53 | 69 | 98 |
| yetF/GBAA_5379 | O31533 | Q81X64 | Unknown | 35 | 59 | 63 |
| yfmC/GBAA_5330 | O34348 | Q81XB0c | Unknown | 25 | 46 | 93 |
| yfmC/fpuA | O34348 | Q81L65 | Fe-citrate binding, YfmC | 33 | 53 | 96 |
| yfmC/GBAA_0615 | O34348 | Q81V85c | Unknown | 34 | 49 | 99 |
| ykrK/GBAA_2841 | O31656 | Q81PG5 | Unknown | 37 | 59 | 94 |
| ylaJ/GBAA_2746 | O07634 | Q81PQ5 | Unknown, lipoprotein | 47 | 67 | 100 |
| yndM/BAS2909 | O31816 | Q6HWX1 | Unknown | 35 | 50 | 88 |
| yndM/GBAA_2961 | O31816 | Q81P56 | Unknown | 27 | 47 | 92 |
| ypeB/ypeB | P38490 | Q81PQ4 | Cortex degradation, YpeB | 57 | 77 | 99 |
| ypmQ/GBAA_2249 | P54178 | Q81R11 | SCO1 protein homolog | 49 | 70 | 96 |
| yqfX/BA_1410 | P54481 | Q81T77 | Unknown | 33 | 48 | 92 |
| ythA/cydA3 | C0SP90 | Q81WZ0 | Cytochrome bd menaquinol oxidase | 53 | 71 | 98 |
| yugS/GBAA_0608 | O05241 | Q81V91 | Unknown | 59 | 75 | 100 |
| yutC/GBAA_5199 | O32128 | Q81XN4 | Unknown, lipoprotein | 35 | 58 | 72 |
| yyxA/GBAA_5710 | P39668 | Q81JJ5 | Serine protease, YyxA | 50 | 68 | 97 |
| znuA/GBAA_2035 | O34966 | Q81RK9 | Zinc uptake, ZnuA | 37 | 55 | 97 |
Sequences were aligned using BLASTP (67). nt, nucleotides.
That is, the percentage of the B. subtilis protein sequence that was aligned with the B. anthracis sequence.
Three B. subtilis membrane proteins, YhcC, YveA, and YitG, that are expressed in the σG regulon (17, 37) and do not have similar proteins in the B. anthracis protein database, were detected. Three other σG regulon proteins—YhbJ, YrbG, and YwjE (37)—have highly similar proteins in B. anthracis (BAS4465, BAS4309, and BAS5195, respectively) but were detected only in B. subtilis.
Membrane protein changes during spore germination.
Spectral counting methods (41, 42) were applied to determine protein abundance information before and after spore germination. To test the validity of this method, we compared the ratio changes of proteins in these data to more precise, previously published quantification data for several germination proteins (43) (Table 3). The trends in terms of proteins increasing, decreasing, or not changing in membrane association during germination were generally consistent in the two data sets with only one protein, SpoVAC, exhibiting an inverse trend in the two studies. The decreased abundance of B. anthracis proteins BAS2560, BAS1302, and BAS4323 after spore germination was also consistent with a previous whole spore quantitative proteomic study (21). On the other hand, our results showed that BAS0812 and BAS3922 decreased 5.9- and 3.7-fold, respectively, and BAS0405 increased 2-fold after spore germination, but these proteins showed no change in quantity in the previous study (21). Preliminary protein quantity changes during germination were determined for 99 B. subtilis and 83 B. anthracis spore membrane proteins. Strikingly, most of the membrane proteins (>70%) in both species were either not detected (Table S4) or greatly reduced in quantity after spore germination (73 B. subtilis proteins and 65 B. anthracis proteins) (Table 4). In B. subtilis samples, two membrane proteins, YtxH and MinD, were present in germinated spore samples that were not detected in the dormant spore samples.
TABLE 3.
Validation of membrane protein quantification
| B. subtilis protein | Dormant spore/germinated spore ratio determined by: |
|
|---|---|---|
| Spectral counting | MRMa | |
| GerAC | 1.7 | 1.9 |
| GerBC | 0.8 | 1.5 |
| GerKC | 1.5 | 2.4 |
| GerD | 3.0 | 3.5 |
| PrkC | 1.9 | 3.3 |
| SpoVAC | 3.3 | 0.8 |
| SpoVAD | 0.5 | 0.6 |
| YpeB | 4.2 | 6.8 |
MRM data are from Chen et al. (43).
TABLE 4.
Changes in Bacillus spore membrane protein detection following germination
| Gene | UniProt no. |
Membrane prediction | Fold change in unique spectra (D/Gf ) | |
|---|---|---|---|---|
| B. subtilis | B. anthracis | |||
| fruA | P71012 | Integral | 5.4 | |
| qcrB | P46912 | Integral | 4.5 | |
| ypeB | P38490 | Integral | 4.2e | |
| yheB | O07543 | Integral | 3.9 | |
| znuA | O34966 | Lipoprotein | 3.6 | |
| ylaJ | O07634 | Lipoprotein | 3.3 | |
| oppC | P24139 | Integral | 3.3 | |
| atpF | P37814 | Integral | 3.3 | |
| spoVAC | P40868 | Integral | 3.3a | |
| fhuD | P37580 | Lipoprotein | 3.1 | |
| yugS | O05241 | Q81V91 | Integral | 3.0/3.3 |
| gerD | P16450 | Lipoprotein | 3.0e | |
| ythA | C0SP90 | Integral | 2.9 | |
| secDF | O32047 | Integral | 2.9 | |
| ypmQ | P54178 | Lipoprotein | 2.8 | |
| yitG | Q796Q1 | Integral | 2.7 | |
| yqfX | P54481 | Q81T77 | Integral | 2.6/INFb,c |
| oppA | P24141 | Q81V45 | Lipoprotein | 2.6/3.8 |
| qoxA | P34957 | Integral | 2.2 | |
| rbsA | P36947 | Peripheral | 2.2 | |
| yfmC | O34348 | Lipoprotein | 2.1 | |
| yhcN | P54598 | Lipoprotein | 2.0 | |
| yugP | O05248 | Integral | 2.0 | |
| spoVAD | P40869 | Q81 × 67 | Peripheral | 0.5e /1.4 |
| atpG | P37810 | Q81JZ4 | Peripheral | 0.5/3.9 |
| BAS4323 | Q6HSW8 | Lipoprotein | 8.9b | |
| GBAA_2961 | Q81P56 | Integral | 7.0 | |
| prsA1 | Q81U45 | Lipoprotein | 6.3 | |
| GBAA_0855 | Q81UL3 | Lipoprotein | 5.9c | |
| GBAA_0615 | Q81V85 | Lipoprotein | 4.8 | |
| GBAA_3048 | Q81NX4 | Peripheral | 4.1 | |
| GBAA_5684 | Q81JM0 | Integral | 3.9 | |
| GBAA_422 | Q81ML8 | Lipoprotein | 3.7d | |
| cccA | Q81LU6 | Integral | 3.3 | |
| psd | Q81LP7 | Peripheral | 3.3 | |
| GBAA_1195 | Q81TS0 | Peripheral | 3.0 | |
| GBAA_1523 | Q81SX1 | Integral | 2.2 | |
| GBAA_3927 | Q81WP4 | Lipoprotein | 13 | |
| GBAA_0418 | Q81Z55 | Peripheral | 0.5d | |
Result is not consistent that of Chen et al. (43).
Result is consistent with that of Jagtap et al. (21).
This protein was detected in B. anthracis dormant spores but not in germinated spores.
Result is not consistent with that of Jagtap et al. (21).
Result is consistent that of Chen et al. (43).
D/G, dormant/germinated.
Growth and sporulation of strains lacking putative ion transporters present in spore membranes.
Six proteins (ZnuA, YcnL, ChaA, YflS, YloB, and YugS) identified in one or both of the spore membrane proteomes are similar to components of known cation transport systems. These proteins may play distinct roles in the accumulation of ions during sporulation and/or the rapid release of ions during germination. Single and multiple mutants of the putative ion transporter genes in B. subtilis were obtained or generated, and the effects of these mutations were analyzed.
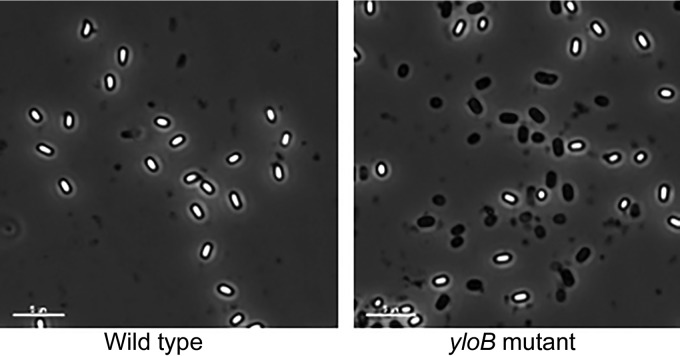
Cultures of B. subtilis strains lacking putative ion transporters were allowed to sporulate for 3 days, and the total and heat resistant viable counts were determined. At this time point, essentially no vegetative cells were visible by microscopy, and all of the remaining cells appeared to be spores (Fig. 2 and data not shown). All strains containing a yloB deletion showed a decrease in both total and heat-resistant CFU/ml in comparison to the wild-type strain (Table 5). This could potentially result from decreased vegetative growth, a low percentage of spore formation or completion of sporulation, or death of spores. When viewed under the microscope, 40 to 50% of spores from all the strains containing a yloB deletion were phase dark (Fig. 2). The phase-dark spore phenotype (loss of refractility) suggests the formation of unstable spores. Several of the multiple mutants lacking yloB also exhibited a relative decrease in heat resistance (a low ratio of heated to unheated CFU/ml). Strains without a yloB deletion, such as DPVB706, resembled the wild type phenotypically wherein >90% of the spores are phase bright. In order to more closely analyze the effect of mutations on sporulation and germination, four strains were chosen for further study: PS832 (wild type), DPVB693 (ΔyloB), DPVB706 (ΔznuA ΔyflS Δycnl [abbreviated “Δ3”]), and DPVB722 (ΔznuA ΔyflS Δycnl ΔyloB ΔyugS::mls ΔchaA::spec [abbreviated “Δ5ΔyloB”]).
FIG 2.
Mutant strains lacking YloB produce many phase-dark spores. B. subtilis strains were grown and sporulated in 2×SG medium, and spores were purified by water washing. Phase-contrast microscopy revealed that approximately 50% of the spores produced by all strains containing yloB deletion mutations were phase dark. Scale bars, 5 μm.
TABLE 5.
Production of heat-resistant spores by B. subtilis strains lacking putative ion transportersa
| Strain | Genotype | Total CFU/ml | Heated CFU/ml | Heated/total | Heated/WT heated |
|---|---|---|---|---|---|
| PS832 | Wild type | 1.2 × 109 | 0.9 × 109 | 0.8 | 1 |
| DPVB689 | ΔznuA::mls | 1.4 × 109 | 1.3 × 109 | 0.9 | 1.4 |
| DPVB690 | Δycnl::mls | 1.3 × 109 | 1.4 × 109 | 1.1 | 1.6 |
| DPVB691 | ΔyflS::mls | 1.2 × 109 | 1.0 × 109 | 0.8 | 1.1 |
| DPVB693 | ΔyloB::mls | 4.3 × 109 | 3.4 × 108 | 0.8 | 0.4 |
| DPVB706b | ΔznuA ΔyflS Δycnl | 1.1 × 109 | 1.0 × 109 | 0.9 | 1.1 |
| DPVB715 | ΔyugS::mls | 0.9 × 109 | 6.9 × 108 | 0.8 | 0.8 |
| DPVB716 | ΔchaA::spec | 1.1 × 109 | 1.3 × 109 | 1.2 | 1.4 |
| DPVB717 | ΔznuA ΔyflS Δycnl ΔyloB::mls | 4.7 × 108 | 2.5 × 108 | 0.5 | 0.3 |
| DPVB718 | ΔyugS::mls ΔchaA::spec | 4.4 × 108 | 4.8 × 108 | 1.1 | 0.5 |
| DPVB719 | ΔznuA ΔyflS Δycnl ΔyloB | 6.0 × 108 | 2.2 × 108 | 0.3 | 0.2 |
| DPVB720 | ΔznuA ΔyflS Δycnl ΔyloB ΔchaA::spec | 4.4 × 108 | 2.4 × 108 | 0.5 | 0.3 |
| DPVB721 | ΔznuA ΔyflS Δycnl ΔyloB ΔyugS::mls | 5.0 × 108 | 2.0 × 108 | 0.4 | 0.2 |
| DPVB722c | ΔznuA ΔyflS Δycnl ΔyloB ΔyugS::mls ΔchaA::spec | 3.9 × 108 | 1.6 × 108 | 0.4 | 0.2 |
Values are averages of three independent experiments.
The genotype for DPVB706 is abbreviated as “Δ3” throughout the text.
The genotype for DPVB722 is abbreviated to “Δ5ΔyloB” throughout the text.
The growth and sporulation of strains were examined following inoculation into 2xSG medium at 37°C. All four strains grew and reached the initiation of sporulation (T0) at similar rates (Fig. S2A). Samples were collected at various time points for assay of glucose dehydrogenase (GDH) activity, an early sporulation marker, and dipicolinic acid (DPA) accumulation, a late sporulation marker. The timing and quantity of GDH activity were reasonably similar for all four strains, suggesting that the early stages of sporulation were not affected in B. subtilis strains with a yloB deletion (Fig. S2B). The four strains accumulated similar amounts of DPA (Fig. S2C), though for DPVB693 (ΔyloB) and DPVB722 (Δ5ΔyloB) DPA accumulation may have been slightly reduced in comparison PS832 (wild type) and DPVB706 (Δ3). This suggests that the mutant strains with a yloB deletion may be partially defective in the uptake or maintenance of DPA during late-stage sporulation.
Spores were generated from the wild-type and three mutant strains in chemically defined sporulation medium (CDSM) to which defined concentrations of Ca2+ were added. (The various components used to produce the CDSM contain some residual Ca2+ that allows growth and spore formation. The concentrations indicated are the Ca2+ added in addition to those residual amounts.) With 1 mM added Ca2+, all four strains produced similar numbers of heat-resistant spores (Table 6). As added Ca2+ was decreased from 1 mM to 0, the numbers of heat-resistant spores produced by the wild-type and DPVB706 (Δ3) strains decreased 18- and 19-fold, respectively, whereas the numbers of heat-resistant spores produced by DPVB693 (ΔyloB) and DPVB722 (Δ5ΔyloB) decreased 43- and 181-fold, respectively (Table 6). Similar dilution experiments with Mn2+ and Fe3+, in the presence of 1 mM Ca2+, showed no differences between the spore formation rates among these strains (data not shown). This suggests that there may be a high-affinity transporter for Ca2+ that is nonfunctional in the yloB mutant strains, which is consistent with the observation that yloB deletion strains appear to produce unstable spores.
TABLE 6.
Production of heat-resistant spores by B. subtilis ion transporter mutants with different Ca2+ concentrationsa
| Concn (mM) of added CaCl2 | Strain | Total CFU/ml | Heat-resistant CFU/ml |
|---|---|---|---|
| 1 | PS832 | 3.9 × 107 | 4.5 × 107 |
| DPVB693 | 5.8 × 107 | 4.0 × 107 | |
| DPVB706 | 4.2 × 107 | 5.0 × 107 | |
| DPVB722 | 2.9 × 107 | 6.7 × 107 | |
| 0.2 | PS832 | 3.6 × 107 | 3.1 × 107 |
| DPVB693 | 2.8 × 108 | 3.5 × 107 | |
| DPVB706 | 2.7 × 108 | 3.2 × 107 | |
| DPVB722 | 6.3 × 107 | 3.2 × 107 | |
| 0.04 | PS832 | 3.9 × 107 | 4.2 × 106 |
| DPVB693 | 1.4 × 108 | 5.7 × 106 | |
| DPVB706 | 6.8 × 107 | 4.5 × 106 | |
| DPVB722 | 1.4 × 108 | 3.5 × 106 | |
| 0 | PS832 | 3.4 × 107 | 2.5 × 106 |
| DPVB693 | 3.9 × 107 | 9.3 × 105 | |
| DPVB706 | 2.9 × 107 | 2.6 × 106 | |
| DPVB722 | 8.1 × 107 | 3.7 × 105 |
Values are averages of two independent experiments.
Quantification of metal ions in cellular compartments during sporulation.
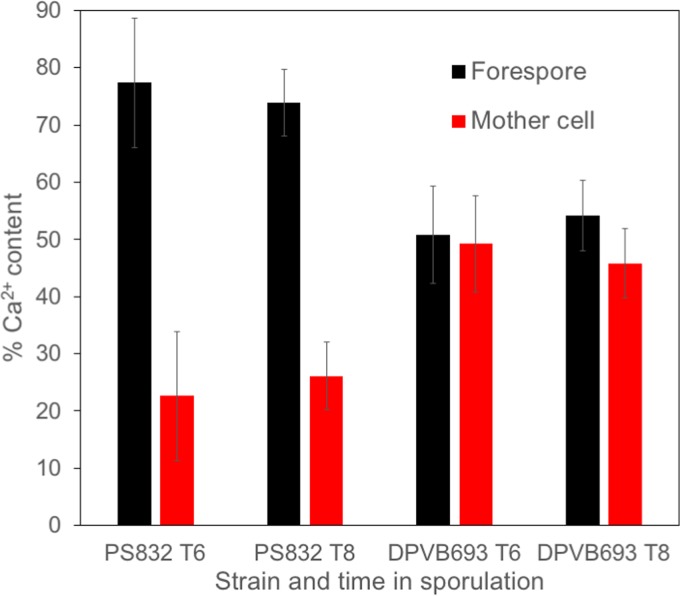
PS832 (wild type) and DPVB693 (ΔyloB) were grown to sporulation in 2×SG medium at 37°C and were harvested at 6 h (T6) or T8. The separation of mother cell and forespore contents was achieved through lysozyme treatment and differential centrifugation, followed by the quantification of Ca2+ using atomic emission spectroscopy. During sporulation, the Ca2+ content in the wild type was far greater in the forespore than in the mother cell, while in the yloB deletion mutant the Ca2+ content was nearly equal in the forespore and mother cell compartments (Fig. 3). This suggests that the transport of Ca2+ from the mother cell to the forespore may be affected in the yloB deletion mutant. In addition, at both time points, the yloB mutant sporangia contained ≤65% of the total Ca2+ found in the wild-type sporangia, suggesting that the failure to concentrate Ca2+ in the forespore impaired the overall Ca2+ accumulation.
FIG 3.
Ca2+ content of forespore and mother cell in sporulating cells. Strains were grown with shaking in 2×SG medium at 37°C and OD600 was measured. Samples were removed at T6 or T8 and treated with lysozyme and several rounds of centrifugation to separate forespore and mother cell compartments. The samples were extracted with HCl and Ca2+ was quantified using atomic emission spectroscopy. Values give the averages from three assays, and error bars indicate standard deviations.
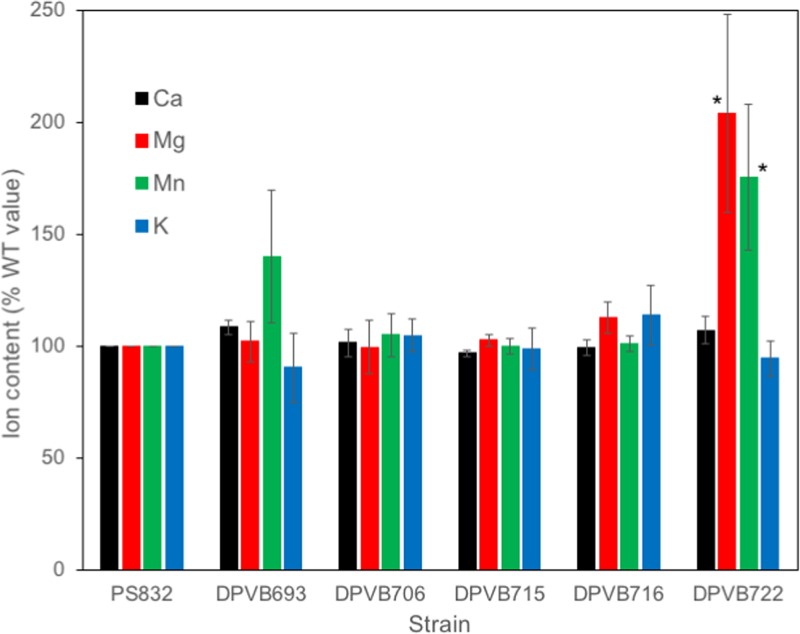
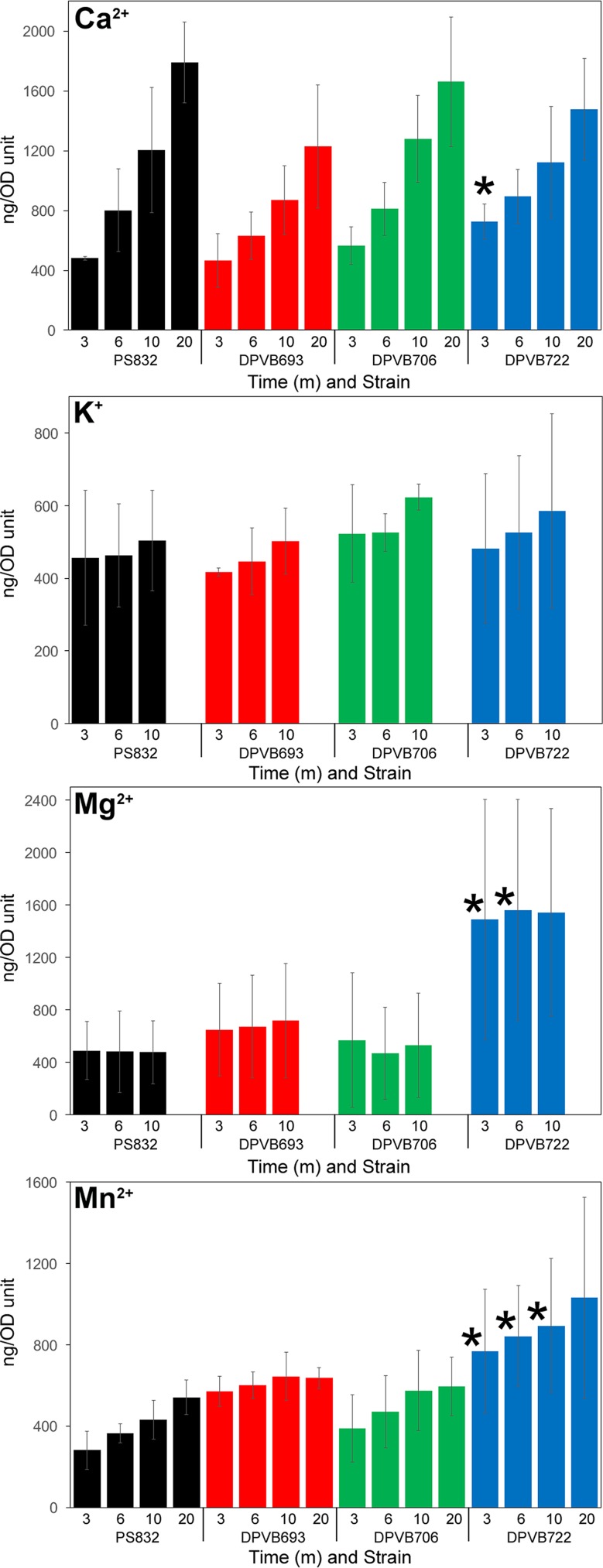
Assays of yloB mutant spore characteristics were complicated by the presence of a mixed population of phase-bright and phase-dark spores, which made various contributions to normalizing measurements such as the CFU and OD. Phase-bright spores were therefore further purified using density gradients. Purified phase-bright spores were extracted with HCl, and the amounts of several metal ions were quantified using atomic emission spectroscopy (Fig. 4). Phase-bright spores of the yloB deletion strain (DPVB693) contained amounts of all ions similar to those of wild-type spores. A surprising observation was a significant increase (P ≤ 0.05) in the levels of Mg2+ and Mn2+ in the sextuple transporter mutant in comparison to the wild type. This difference in ion content between strain DPVB722 (Δ5ΔyloB) and strains DPVB693 (ΔyloB) and DPVB706 (Δ3) suggested to us that the additional mutations in yugS or chaA that are present in DPVB722 (Δ5ΔyloB) might be responsible. We therefore assayed ion contents of spores produced by strains DPVB715 (ΔyugS::mls) and DPVB716 (ΔchaA::spec). These spores were indistinguishable from those of the wild type (Fig. 4).
FIG 4.
Ion contents of purified phase-bright spores. Purified spores were extracted with HCl, and the amounts of various ions were quantified using atomic emission spectroscopy. Values are averages from three to nine assays, depending on the strain, and are expressed as percentages of the values found in PS832 (wild type) spores prepared on the same day. The strains examined were DPVB693 (ΔyloB), DPVB706 (Δ3), DPVB715 (ΔyugS::mls), DPVB716 (ΔchaA::spec), and DPVB722 (Δ5ΔyloB). Error bars indicate standard errors. *, significant difference from PS832 (P ≤ 0.05).
Germination rate and release of ions during germination.
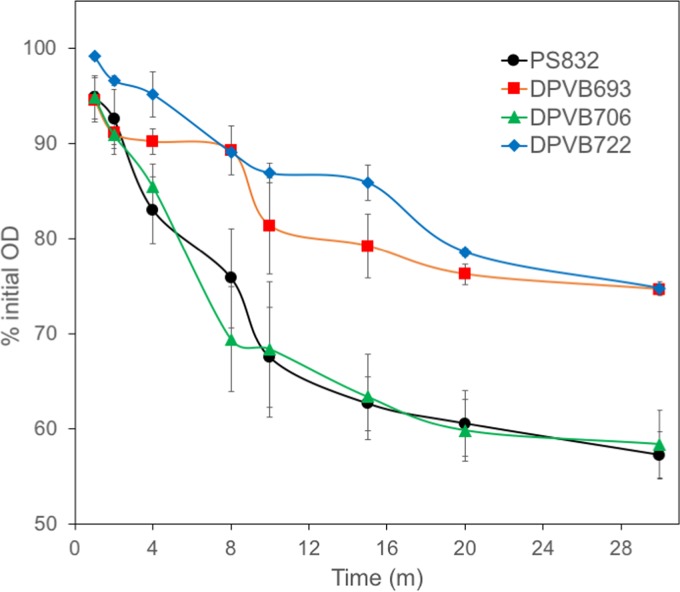
Purified phase-bright spores were heat activated and stimulated to germinate by addition of 10 mM l-alanine. The rate of germination as assayed by OD decrease was slower in the strains with a yloB deletion than for the wild type (Fig. 5). Levels of release of Ca2+ and K+ during spore germination were similar for all strains (Fig. 6). Levels of release of Mg2+ and Mn2+ were also similar for strains except for the sextuple mutant, DPVB722 (Δ5ΔyloB), which released significantly larger amounts than the wild type. The release of K+ and Mg2+ appeared to be essentially complete within 3 min of the start of germination, whereas the release of Ca2+ and Mn2+ continued to increase gradually for 10 to 20 min.
FIG 5.
Germination of purified phase-bright spores. Purified spores of B. subtilis wild-type and mutant strains were heat activated and stimulated to germinate by the addition of 10 mM l-alanine. Values are averages from three assays, and error bars indicate the standard deviations. For DPVB693 (ΔyloB) all points after 2 min and for DPVB722 (Δ5ΔyloB) all points are significantly different from those of PS832 (wild type) and DPVB706 (Δ3) (P ≤ 0.05).
FIG 6.
Release of ions by germinating spores. Spores were germinated as described in Materials and Methods. Samples were collected at the indicated times and centrifuged briefly to pellet spores. Ions in the germination exudate were quantified using atomic emission spectroscopy. Values are averages from three assays, and error bars indicate standard deviations. *, points significantly different from those of PS832 (wild type) and DPVB693 (ΔyloB) (P ≤ 0.05).
DISCUSSION
Proteins associated with the membranes separating the mother cell and forespore play crucial roles in communication between the cells during sporulation, allowing the two cells to coordinate the timing of gene expression changes and morphological development. Membrane proteins localized at the inner spore membrane have been demonstrated to play key roles in multiple stages of spore germination. In addition, germination processes such as ion efflux and water influx are likely to involve membrane proteins, yet few proteins involved in these processes have been identified. The goal of this study was to compare and contrast spore membrane-associated proteins present in two Bacillus species in an effort to further our understanding of cell differentiation processes at the molecular level and to identify new germination-active proteins. A gel-based approach was used to provide a set of integral membrane proteins, lipoproteins, and peripheral membrane proteins. This study identified 65 membrane-associated spore proteins that had not been previously reported in any B. subtilis or B. anthracis vegetative or spore proteomic study. The percentages of proteins identified that are predicted to be membrane associated were high, 15 to 17% of all proteins identified, which is a significant improvement over some previous spore proteomic studies (<5%) (19, 21, 22) and similar to more recent studies (25, 26). Identification of these proteins may provide a pathway to better understand the role of membrane proteins during sporulation and germination.
To provide preliminary information about the fate of spore membrane proteins during germination, relative quantities were derived using label-free spectral counting methods. This strategy is based on the empirical observation that the amount of unique tandem mass spectrometry (MS/MS) counts of a protein has strong linear correlation with relative protein abundance (41, 42). Although this method is not as accurate as targeted MRM-based methods, it has higher dynamic range than other mass spectrometry quantification methods (44) and therefore is a popular method to detect global protein changes between experimental groups. Note that this relative quantification method was established using data generated by electrospray ionization mass spectrometry. The limitations of applying this method on data generated by MALDI mass spectrometry have not been previously examined. However, the relative quantity values for multiple proteins acquired in this work were consistent with those acquired in our previous MRM study (43) and a previous stable isotope-labeled quantitative study (21), suggesting that this method is applicable to MALDI-generated data. Results published while this work was in progress indicate that membrane fractions produced by methods such as those used here recover only a portion of the spore membrane proteins, and extensive chemical extraction is required to recover the full amount of several spore membrane proteins (45). However, such a chemical extraction precludes the separation of membrane and soluble proteins. In the absence of a spore membrane extraction process that provides high recovery, protein quantitation will have to be done using whole-spore extracts or will require evidence that the partial membrane extract is representative of the entire membrane.
A notable trend in our study is that the number of identified membrane proteins is greatly reduced after spore germination. Several membrane proteins, mostly lipoproteins, have been previously suggested to dissociate from the membrane during spore germination (21, 34, 43). Spectral counting showed similar results with most lipoproteins diminished or undetectable following germination. The decreased recovery of lipoproteins may be due to proteolysis or may indicate a change in their ability to maintain membrane association during fractionation, which in turn could be associated with the membrane reorganization process. Surprisingly, over half of the membrane proteins that decreased to below detectable levels during germination of both Bacillus species spores were integral membrane proteins. Although previous efforts to quantify spore integral membrane proteins are rare, decreased levels of BAS1302 after spore germination was consistent with previous work (21). Since the membrane proteins identified that were observed to decrease are involved in multiple COG function categories, this decrease appears to be a general spore germination phenomenon. The shift from a relatively nonfluid impermeable membrane to a fluid membrane with solute transfer may require a rather complex rearrangement of the membrane and membrane-associated proteome.
Several proteins detected in spore membranes are involved in protein degradation. B. subtilis HtrC (YyxA, YycK), a predicted Htr-type serine protease, was suggested to be transcribed from its own σG promoter (46). Loss of HtrC produced no obvious phenotypic change in B. subtilis vegetative cells (46, 47); however, the presence of HtrC, and its apparent B. anthracis ortholog, which is also expressed during sporulation (16), in the spore membrane suggested that it may play a role in spore formation or germination. Disruption of htrC in both B. anthracis and B. subtilis caused a reduction in proteolysis of YpeB during spore germination, but this did not alter the progress of germination of these spores (48). B. subtilis HtpX and B. anthracis RasP are two predicted zinc proteases detected in the spore membranes. Their functions have not been characterized, but they may be involved in membrane protein quality control based on the function of the well-known homolog FtsH (49). Further characterization of the roles these proteases play in the spore may shed light on the observed decrease in membrane proteins during spore germination.
Several proteins involved in amino acid transport were identified in the spore membranes. YveA was previously characterized to be the primary l-aspartate transporter in B. subtilis vegetative cells (50), and transcriptome analysis showed that this gene was expressed under the control of the forespore-specific σG (17). MetQ and ArtP were previously characterized in B. subtilis vegetative cells as involved in methionine and arginine transport, respectively (51, 52). These proteins, as well as their apparent B. anthracis orthologs, were present in the spore membranes. The roles of these amino acid transporters in either sporulation or germination processes are unknown at present.
Cations such as Ca2+, Mn2+, and Zn2+ accumulate in spores during the sporulation of Bacillus species (53), and the rapid release of cations is a distinct early germination step (54). Divalent metal ions, predominantly Ca2+, conjugated with DPA contribute to spore resistance to heat (55, 56) and ionizing radiation (57). Six membrane proteins with predicted functions involved in inorganic ion transport were identified in the spores of both species. These proteins may play roles in accumulation of ions during sporulation and/or the rapid release of ions during germination.
A mutant lacking yloB appeared to have a defect in accumulation of Ca2+ into developing forespores, and a subpopulation of these spores were unstable and did not retain refractility and heat resistance. A previous study of a yloB mutant did not detect a reproducible defect in Ca2+ uptake by sporulating cells; however, that study did not differentiate Ca2+ uptake into the mother cell versus the developing spore (58). In that study, the yloB mutant was found to have defects in spore germination and spore heat resistance (58), and we found similar defects in this mutant. A potential explanation for this suite of phenotypic properties is that Ca2+ transport from the mother cell into the forespore is slowed in the yloB mutant, and this results in the spores not achieving the normal Ca2+ content. A subpopulation of developing spores that does not reach some threshold Ca2+ content is unstable, losing refractility and viability relatively rapidly. The subpopulation that remains phase bright includes those spores that reached a threshold level of Ca2+-DPA content that we could not differentiate from that of the wild-type spores. These stable ΔyloB spores are still not entirely normal and exhibited reduced heat resistance and germination rate.
An interesting parallel to Ca2+ transport into the spore may exist for transport of DPA, which complexes with Ca2+ in the spore core. DPA is synthesized in the mother cell (59), transported across the outer forespore membrane by mother cell-expressed SpoVV (60) and then across the inner forespore membrane by the SpoVA complex (30). Mother cell-expressed YloB may play the (partially redundant) role of Ca2+ transport across the outer membrane, with another system responsible for transport across the inner membrane.
The sextuple mutant lacking six putative ion transport components, including YloB, exhibits essentially the same phenotype as the yloB mutant but in addition accumulates larger amounts of Mn2+ and Mg2+ into the dormant spores. This might be due to a failure to export these ions out of the forespore, or perhaps they accumulate to higher levels in the developing spore, along with DPA, when Ca2+ transport is diminished. We note that during germination, Mn2+ is released slowly, like Ca2+ and DPA, whereas Mg2+ is released more rapidly, similar to K2+. This suggests that the excess Mn2+ may be associated with DPA, while the excess Mg2+ is not bound to DPA.
In both Bacillus species, one quarter of the identified spore membrane proteins have no assigned COG functional category and most have no significant sequence similarity to known genes or functional domains. More interestingly, most of these proteins were not identified in vegetative cell membranes previously, and some were present in both Bacillus spores, suggesting that these proteins are likely expressed for particular purposes in spores. Further characterization of these proteins may reveal roles in the sporulation, germination, or outgrowth processes. For example, B. subtilis YetF and YutC were previously characterized to be expressed under the control of spore-specific σF and σG, respectively (37, 61), and their apparent B. anthracis orthologs were also identified in this study. Five other B. subtilis proteins together with YetF are grouped in an uncharacterized protein family (UPF0702), in which a σG-regulated protein YrbG (37) was also identified in B. subtilis spore membrane. Homologs of these proteins with no known function are not found in other bacteria, indicating that they might be very specific for Bacillus species. Considering their expression is under the control of forespore sigma factors, these proteins may play yet uncharacterized roles in spore formation, stabilization, and/or germination processes.
MATERIALS AND METHODS
Spore preparation.
Spores of B. anthracis Sterne strain 34F2, an attenuated vaccine strain, were prepared in modified G broth (62). Spores of B. subtilis strain PS832 and derivatives were prepared in 2×SG broth (63). Spores were harvested after 3 to 4 days incubation at 37°C, washed in water for several days, and purified by centrifugation through a 50% sodium diatrizoate (Sigma) layer as described previously (64). All spores used in proteome analyses were 99% free of vegetative cells and were stored in deionized water at 4°C until analysis.
To prepare germinated spores, a 10-ml suspension of dormant spores at an OD at 600 nm (OD600) of 20 in water was heat activated at 70°C for 30 min (B. anthracis) or 75°C for 30 min (B. subtilis) and cooled on ice for 10 min. The spores were then germinated at 37°C and at an OD600 of 2 with 50 mM l-alanine plus 1 mM inosine (B. anthracis) or 10 mM l-valine (B. subtilis) in 25 mM Tris-HCl buffer (pH 7.4). The germination of spores was terminated after the OD600 dropped to 50% of the initial value (within 10 and 35 min after germinant addition for B. anthracis and B. subtilis, respectively). Germinated spores were collected by centrifugation at 12,000 × g for 5 min at 4°C, quickly washed with cold deionized water, centrifuged again, and frozen at –80°C. Examination by phase-contrast microscopy indicated that >95% of the spores in each preparation had germinated.
Preparation of spore membrane fractions.
Spore membrane fractions were prepared by a modification of a previously described method (43). Dormant and germinated spores were lyophilized, and the dry spores (∼19 mg for germinated spores and ∼24 mg for dormant spores) were pulverized with 100 mg of glass beads in a dental amalgamator (Wig-L-Bug) at 4,600 rpm for pulses of 30 s each, with 30-s pauses on ice between pulses. Spore disruption was monitored by suspending an aliquot of spore material in H2O and observing under phase-contrast microscopy. After >80% of spores were disrupted, the dry powder was suspended in 0.5 ml of 4°C extraction buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 2 mg/ml RNase A, 2 mg/ml DNase I, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The suspension was centrifuged (6,000 × g, 10 min, 4°C), and the resultant supernatant was centrifuged again (13,000 × g, 10 min, 4°C) to remove insoluble material. The remaining supernatant was centrifuged at 100,000 × g for 60 min at 4°C, and the resulting pellet was designated the crude spore membrane fraction. This membrane fraction was homogenized in 1 ml of alkaline buffer (100 mM Na2CO3-HCl [pH 11], 10 mM EDTA, 100 mM NaCl, 1 mM PMSF) and gently shaken for 60 min at 4°C. The homogenate was subjected to ultracentrifugation as described above. The resulting pellet was homogenized in 1 ml high-salt buffer (20 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1 M NaCl, 1 mM PMSF) and again subjected to ultracentrifugation. After a final wash with 1 ml of TE buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1 mM PMSF), the resulting pellet was homogenized in 200 μl of TE buffer, flash frozen, and stored at –80°C until analysis. Protein concentrations were determined by acid hydrolysis and amino acid analysis (65) with comparison to a standard set of amino acids (Sigma).
SDS-PAGE, trypsin digestion, and peptide fractionation.
Membrane fractions were dried and resuspended in SDS-PAGE sample loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.05% bromophenol blue) to a final protein concentration of 2 μg/μl. SDS-PAGE was used to separate 30 μg of protein of each spore membrane sample in both 10% polyacrylamide and 12.5 to 14% gradient polyacrylamide gels for B. subtilis samples and in a 12.5 to 14% gradient polyacrylamide gel for B. anthracis samples. Gels were stained with ProtoBlue Safe (National Diagnostics). Each gel lane was cut into 10 to 12 slices, consistently for all samples within each species, in an effort to isolate regions containing many lower abundance proteins away from higher abundance proteins that could dominate MS profiles. Gel slices were ground and destained with 50% LC/MS-grade acetonitrile supplemented with 25 mM NH4HCO3. The gel slices were then dehydrated with 100% acetonitrile and vacuum dried. Gel slices were soaked in 25 mM NH4HCO3 containing 10 μg/ml trypsin (Sigma), and digestion was carried out at 37°C for at least 16 h. Tryptic peptides were extracted from gel slices into 50% LC/MS-grade acetonitrile–0.1% trifluoroacetic acid (TFA) using a sonication bath. Peptides were vacuum dried and resuspended in 40 μl of 2% LC/MS-grade acetonitrile–0.1% TFA.
An Eksigent Nano2D-LC unit, flowing at 0.7 μl/min, was used to inject 10 μl of each peptide sample through a Captrap cartridge (Michrom Bioresources) and a self-packed New Objective Integrafrit column (50 by 0.1 mm), both packed with Magic C18 AQ (200 Å, 3 μm; Michrom Bioresources). Elution was performed using 5% acetonitrile for 25 min, a 5-min linear increase to 14% acetonitrile, and a 95-min linear increase to 34% acetonitrile. An Ekspot plate spotter (Eksigent) was used to spot the eluate onto MALDI target plates at a rate of 10 to 15 s per spot depending on the band intensity and the size of a gel slice, and the spots were air dried.
Mass spectrometry and protein identification.
Matrix was prepared by suspending approximately 200 mg of αCHCA (Aldrich) in 1 ml of 100 mM acetic acid, followed by vigorous mixing. After centrifugation for 2 min at 1,000 × g, the supernatant was removed, and the wash procedure was repeated. Two additional washes were performed with 100% acetonitrile, and the αCHCA was vacuum dried and stored at 4°C. The matrix solution was prepared by dissolving 4 mg of washed αCHCA in 1 ml of 1:1:0.001 (vol/vol/vol) water-acetonitrile-TFA; 2 M NH4Cl was then added to a final concentration of 10 mM. Each dried sample spot on the MALDI plates was overlaid with 1 μl of matrix solution and air dried.
For calibrating and tuning the MALDI-TOF/TOF 4800 analyzer (AB Sciex), 200 μl of the matrix solution was added to an aliquot of a mix of six standard peptides (Anaspec, catalog no. 60882). This mix was spotted onto all calibration spots when analyzing a sample MALDI plate. For each peptide sample spot, a scan for the m/z range from 800 to 4,000 was acquired with averaged data from 1,000 laser shots in reflector positive-ion mode. The 10 most abundant ions for each spot, above a minimum signal-to-noise ratio (>50), were then automatically selected for subsequent MS/MS analysis. Parent ions chosen for one spot were excluded from analyses in subsequent spots. MS/MS scans were the averages of 3,000 laser shots (1 kV, positive-ion mode).
The MS and MS/MS data were analyzed using Proteinpilot software version 4.0 (AB Sciex) and Scaffold version 3.0 software (Proteome Software, Inc.). The parameters used in Proteinpilot were as follows: identification as sample type, no cysteine alkylation, trypsin as digestion enzyme, gel-based identification as special factors, biological modifications and amino acid substitutions as ID focus, thorough ID as search effort, and detected protein threshold (unused prot score) > 0.05 (10%) as result quality. The B. subtilis 168 protein database (2/10/2012) and B. anthracis (BACAN) protein database (12/18/2012) were downloaded from Universal Protein Resources (http://www.uniprot.org/). Based on a Proteinpilot false discovery rate analysis of the acquired protein list, a cutoff line at a 5% false discovery rate and a minimum of one peptide at 95% identification confidence was applied to each biological sample protein profile. The raw data are available from the PRIDE database under access number PXD002136 (66). Proteins identified in at least two biological replicates were considered a valid identification. All proteins in dormant and germinated protein profiles were then annotated according to their accession number using two databases: the National Center for Biological Information (https://www.ncbi.nlm.nih.gov/) and Universal Protein Resources. Other online resources used for predicting membrane associations included the Brinkman Laboratory at Simon Fraser University (http://www.psort.org/psortb/index.html) and the PRED-LIPO server at University of Athens (http://bioinformatics.biol.uoa.gr/PRED-LIPO/input.jsp). Protein NCBI Clusters of Orthologous Groups (COG) were predicted using the COMBREX database (http://combrex.bu.edu/).
To acquire preliminary relative protein quantification, the Mascot data files of three biological replicates were merged, and onsite Mascot searches were performed for each peptide fragmentation mass spectrum against the B. subtilis and B. anthracis protein databases. Mascot parameters were as follows: trypsin specificity with one missed cleavage, deamidation, and pyro-cmC and oxidations were considered variable modifications. The peptide mass tolerance and fragment mass tolerance were set to ±500 ppm and ±0.2 Da, respectively. Search results were analyzed using Scaffold version 3.4.9. The Scaffold parameters were 95% protein threshold and one minimum identified peptide at 95% of threshold, and the normalized unique spectrum count of a protein was used to relatively quantify the protein changes between two samples (41).
All identified B. anthracis membrane proteins were searched against the whole B. subtilis protein database and the proteins identified in B. subtilis spore membrane profile using the NCBI protein BLAST tool (67). Only sequences with an alignment coverage of ≥60% were considered possible orthologs in the two species. If the protein in the full B. subtilis protein database that had the highest identity percentage against a B. anthracis membrane query protein was also identified in the B. subtilis spore membrane profile, the two proteins were considered to be the most likely orthologs. If a protein in B. subtilis spore membrane proteome had high similarity to a B. anthracis membrane query protein, but it was not the highest identity match in the full B. subtilis protein database, then the two proteins were considered to be likely paralogs.
Generation of mutants lacking putative ion transporter genes.
Single mutants lacking four genes (znuA, ycnL, yflS, and yloB) were obtained from the Bacillus subtilis Genetic Stock Center. Each mutation was a deletion, with the gene of interest replaced by an erythromycin resistance gene flanked by two loxP sites (68). The mutations were introduced into B. subtilis strain PS832 by natural transformation with selection for erythromycin (2.5 μg/ml) and lincomycin (12.5 μg/ml) (MLS) resistance. The Cre recombinase was expressed from plasmid pDR244 (68) to stimulate deletion of the resistance gene, leaving an unmarked in-frame deletion mutation. This process was repeated to produce multiple mutants. All mutations were verified by PCR and agarose gel electrophoresis.
Two additional mutations were generated using the long-flanking homology PCR (LFH-PCR) method (69, 70). PCR products were prepared containing ∼1 kb of sequence upstream of the gene, the first 10 to 50 bp of the coding sequence, a spectinomycin or MLS resistance cassette, the last 10 to 50 bp of the coding sequence, and ∼1 kb downstream. The PCR products were used to transform PS832 competent cells to generate the single mutants ΔchaA::spec and ΔyugS::mls, respectively. The mutations were verified by PCR and agarose gel electrophoresis.
Analysis of sporulation properties.
Spores were prepared in 2×SG broth and in CDSM minimal medium (71) to which different concentrations of calcium ions were added. The number of heat-resistant spores was estimated by heat treatment at 80°C for 10 min and serial dilution onto Luria-Bertani plates. The rate of progression through sporulation in 2×SG medium 37°C was analyzed by collecting samples for assay of GDH activity and DPA accumulation as previously described (64).
Separation of mother cell and forespore fractions.
Strains were grown with shaking in 2×SG medium at 37°C. Samples (30 ml) of culture were collected at T6 and T8, and centrifuged at 7,741 × g for 5 min. The supernatant was removed and the pellet was suspended in 5 ml of SMM (72). This was followed by the addition of 25 mg of lysozyme (Sigma-Aldrich) and incubation at 37°C for 15 min (73). The protoplasts were centrifuged at 7,741 × g for 10 min, and the supernatant was discarded. The pellet was suspended in 5 ml of cold, sterile deionized water, vortexed, and centrifuged at 7,741 × g for 10 min. The supernatant contained the lysed mother cell fraction, and the pellet contained the purified forespores. Ca2+ was quantified in each fraction using inductively coupled plasma atomic emission spectroscopy at the Virginia Tech Soil Testing Laboratory (CirOS VISION ICP-AES; Spectro Analytical Instruments) (74).
Quantification of ions using atomic emission spectroscopy.
Phase-bright spores were purified away from phase-dark spores by centrifugation through a 50% sodium diatrizoate layer as described previously (64) and were suspended at an OD600 of 10 in 1 ml of 200 mM Tris-HCl (pH 8.0) to remove coat-associated ions. The spore suspension was rocked at room temperature for 20 min and washed three times with fresh deionized water by centrifugation at 15,800 × g for 2 min. Pellets were suspended in 1 ml of 6 M ultrapure HCl (Fisher Chemicals) and heated at 100°C for 10 min, followed by centrifugation at 15,800 × g for 10 min. The supernatant was collected, and the amounts of individual ions were quantified by using atomic emission spectroscopy.
Purified phase-bright spores were heat activated (70°C for 20 min and cooling on ice) and stimulated to germinate at a starting OD600 of 10 by addition of 10 mM l-alanine in 50 mM NaPO4 buffer at pH 7.0. At different time intervals, samples were removed and centrifuged for 2 min at 15,800 × g. The ion contents of germination exudate (supernatant) samples were analyzed using atomic emission spectroscopy. Significant differences between values were determined by using unpaired two-tailed Student t tests.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of Allergy and Infectious Disease of the National Institutes of Health under award R21AI088298. The mass spectrometry resources used in this work are maintained in part through funding by the Fralin Life Science Institute at Virginia Tech and the Agricultural Experiment Station Hatch Program at Virginia Tech (CRIS project VA-135981).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00662-18.
REFERENCES
- 1.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat, and chemicals. J Appl Microbiol 101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 2.Moir A. 2006. How do spores germinate? J Appl Microbiol 101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 3.Setlow P. 2003. Spore germination. Curr Opin Microbiol 6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Mallozzi M, Viswanathan VK, Vedantam G. 2010. Spore-forming Bacilli and Clostridia in human disease. Future Microbiol 5:1109–1123. doi: 10.2217/fmb.10.60. [DOI] [PubMed] [Google Scholar]
- 5.Setlow P, Johnson EA. 2007. Spores and their significance, p 35–67. In Doyle MP, Beuchat LR (ed), Food microbiology: fundamentals and frontiers, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 6.Amuguni H, Tzipori S. 2012. Bacillus subtilis: a temperature-resistant and needle-free delivery system of immunogens. Hum Vaccin Immunother 8:979–986. doi: 10.4161/hv.20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutting SM, Hong HA, Baccigalupi L, Ricca E. 2009. Oral vaccine delivery by recombinant spore probiotics. Int Rev Immunol 28:487–505. doi: 10.3109/08830180903215605. [DOI] [PubMed] [Google Scholar]
- 8.Knecht LD, Pasini P, Daunert S. 2011. Bacterial spores as platforms for bioanalytical and biomedical applications. Anal Bioanal Chem 400:977–989. doi: 10.1007/s00216-011-4835-4. [DOI] [PubMed] [Google Scholar]
- 9.Cowan AE, Olivastro EM, Koppel DE, Loshon CA, Setlow B, Setlow P. 2004. Lipids in the inner membrane of dormant spores of Bacillus species are largely immobile. Proc Natl Acad Sci U S A 101:7733–7738. doi: 10.1073/pnas.0306859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koshikawa T, Beaman TC, Pankratz HS, Nakashio S, Corner TR, Gerhardt P. 1984. Resistance, germination, and permeability correlates of Bacillus megaterium spores successively divested of integument layers. J Bacteriol 159:624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popham DL, Heffron JD, Lambert EA. 2012. Degradation of spore peptidoglycan during germination, p 121–142. In Abel-Santos E. (ed), Bacterial spores: current research and applications. Caister Academic Press, Norwich, UK. [Google Scholar]
- 12.Indest KJ, Buchholz WG, Faeder JR, Setlow P. 2009. Workshop report: modeling the molecular mechanism of bacterial spore germination and elucidating reasons for germination heterogeneity. J Food Sci 74:R73–R78. doi: 10.1111/j.1750-3841.2009.01245.x. [DOI] [PubMed] [Google Scholar]
- 13.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. 2006. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol 188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327:945–972. doi: 10.1016/S0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Bergman NH, Thomason B, Shallom S, Hazen A, Crossno J, Rasko DA, Ravel J, Read TD, Peterson SN, Yates J, Hanna PC. 2004. Formation and composition of the Bacillus anthracis endospore. J Bacteriol 186:164–178. doi: 10.1128/JB.186.1.164-178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. 2006. The forespore line of gene expression in Bacillus subtilis. J Mol Biol 358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 18.Abhyankar W, Beek AT, Dekker H, Kort R, Brul S, de Koster CG. 2011. Gel-free proteomic identification of the Bacillus subtilis insoluble spore coat protein fraction. Proteomics 11:4541–4550. doi: 10.1002/pmic.201100003. [DOI] [PubMed] [Google Scholar]
- 19.Delvecchio VG, Connolly JP, Alefantis TG, Walz A, Quan MA, Patra G, Ashton JM, Whittington JT, Chafin RD, Liang X, Grewal P, Khan AS, Mujer CV. 2006. Proteomic profiling and identification of immunodominant spore antigens of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Appl Environ Microbiol 72:6355–6363. doi: 10.1128/AEM.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CM, Foster KW, DeSilva TS, Van Kampen KR, Elmets CA, Tang DC. 2004. Identification of Bacillus anthracis proteins associated with germination and early outgrowth by proteomic profiling of anthrax spores. Proteomics 4:2653–2661. doi: 10.1002/pmic.200400831. [DOI] [PubMed] [Google Scholar]
- 21.Jagtap P, Michailidis G, Zielke R, Walker AK, Patel N, Strahler JR, Driks A, Andrews PC, Maddock JR. 2006. Early events of Bacillus anthracis germination identified by time-course quantitative proteomics. Proteomics 6:5199–5211. doi: 10.1002/pmic.200600314. [DOI] [PubMed] [Google Scholar]
- 22.Kuwana R, Kasahara Y, Fujibayashi M, Takamatsu H, Ogasawara N, Watabe K. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971–3982. doi: 10.1099/00221287-148-12-3971. [DOI] [PubMed] [Google Scholar]
- 23.Lai EM, Phadke ND, Kachman MT, Giorno R, Vazquez S, Vazquez JA, Maddock JR, Driks A. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J Bacteriol 185:1443–1454. doi: 10.1128/JB.185.4.1443-1454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao L, Jiang S, Wang B, Chen L, Yao Q, Chen K. 2011. Protein profile of Bacillus subtilis spore. Curr Microbiol 63:198–205. doi: 10.1007/s00284-011-9967-4. [DOI] [PubMed] [Google Scholar]
- 25.Swarge BN, Roseboom W, Zheng L, Abhyankar WR, Brul S, de Koster CG, de Koning LJ. 2018. “One-Pot” sample processing method for proteome-wide analysis of microbial cells and spores. Prot Clin Appl 12:1700169. doi: 10.1002/prca.201700169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng L, Abhyankar W, Ouwerling N, Dekker HL, van Veen H, van der Wel NN, Roseboom W, de Koning LJ, Brul S, de Koster CG. 2016. Bacillus subtilis spore inner membrane proteome. J Proteome Res 15:585–594. doi: 10.1021/acs.jproteome.5b00976. [DOI] [PubMed] [Google Scholar]
- 27.Korza G, Setlow P. 2013. Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J Bacteriol 195:1484–1491. doi: 10.1128/JB.02262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paidhungat M, Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J Bacteriol 183:3982–3990. doi: 10.1128/JB.183.13.3982-3990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fort P, Errington J. 1985. Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis. J Gen Microbiol 131:1091–1105. doi: 10.1099/00221287-131-5-1091. [DOI] [PubMed] [Google Scholar]
- 30.Tovar-Rojo F, Chander M, Setlow B, Setlow P. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J Bacteriol 184:584–587. doi: 10.1128/JB.184.2.584-587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J Bacteriol 187:5677–5682. doi: 10.1128/JB.187.16.5677-5682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vepachedu VR, Setlow P. 2007. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J Bacteriol 189:1565–1572. doi: 10.1128/JB.01613-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igarashi T, Setlow B, Paidhungat M, Setlow P. 2004. Effects of a gerF (lgt) mutation on the germination of spores of Bacillus subtilis. J Bacteriol 186:2984–2991. doi: 10.1128/JB.186.10.2984-2991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelczar PL, Setlow P. 2008. Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores. J Bacteriol 190:5635–5641. doi: 10.1128/JB.00670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atrih A, Foster SJ, Moir A, Chirakkal H, O’Rourke M. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383–2392. doi: 10.1099/00221287-148-8-2383. [DOI] [PubMed] [Google Scholar]
- 36.Stewart KA, Yi X, Ghosh S, Setlow P. 2012. Germination protein levels and rates of germination of spores of Bacillus subtilis with overexpressed or deleted genes encoding germination proteins. J Bacteriol 194:3156–3164. doi: 10.1128/JB.00405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steil L, Serrano M, Henriques AO, Volker U. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiol 151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- 38.Serrano M, Corte L, Opdyke J, Moran CP Jr, Henriques AO. 2003. Expression of spoIIIJ in the prespore is sufficient for activation of sigma G and for sporulation in Bacillus subtilis. J Bacteriol 185:3905–3917. doi: 10.1128/JB.185.13.3905-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winstedt L, von Wachenfeldt C. 2000. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J Bacteriol 182:6557–6564. doi: 10.1128/JB.182.23.6557-6564.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattatall NR, Jazairi J, Hill BC. 2000. Characterization of YpmQ, an accessory protein required for the expression of cytochrome c oxidase in Bacillus subtilis. J Biol Chem 275:28802–28809. doi: 10.1074/jbc.M002741200. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, Sadygov RG, Yates JR. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 42.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ. 2006. Quantitative proteomics analysis of the secretory pathway. Cell 127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Ray WK, Helm RF, Melville SB, Popham DL. 2014. Levels of germination proteins in Bacillus subtilis dormant, superdormant, and germinating spores. PLoS One 9:e95781. doi: 10.1371/journal.pone.0095781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, Scrivens JH. 2009. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J Proteome Res 8:3752–3759. doi: 10.1021/pr900080y. [DOI] [PubMed] [Google Scholar]
- 45.Stewart KA, Setlow P. 2013. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J Bacteriol 195:3575–3582. doi: 10.1128/JB.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabret C, Hoch JA. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol 180:6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573–1583. doi: 10.1099/00221287-146-7-1573. [DOI] [PubMed] [Google Scholar]
- 48.Bernhards CB, Chen Y, Toutkoushian H, Popham DL. 2015. HtrC is involved in proteolysis of YpeB during germination of Bacillus anthracis and Bacillus subtilis spores. J Bacteriol 197:326–336. doi: 10.1128/JB.02344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalbey RE, Wang P, van Dijl JM. 2012. Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev 76:311–330. doi: 10.1128/MMBR.05019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorca G, Winnen B, Saier MH. Jr, 2003. Identification of the l-aspartate transporter in Bacillus subtilis. J Bacteriol 185:3218–3222. doi: 10.1128/JB.185.10.3218-3222.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekowska A, Robin S, Daudin JJ, Henaut A, Danchin A. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol 2:RESEARCH0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hullo MF, Auger S, Dassa E, Danchin A, Martin-Verstraete I. 2004. The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, d- and l-methionine. Res Microbiol 155:80–86. doi: 10.1016/j.resmic.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Charney J, Fisher WP, Hegarty CP. 1951. Manganese as an essential element for sporulation in the genus Bacillus. J Bacteriol 62:145–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swerdlow BM, Setlow B, Setlow P. 1981. Levels of H+ and other monovalent cations in dormant and germinating spore of Bacillus megaterium. J Bacteriol 148:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerhardt P, Marquis RE. 1989. Spore thermoresistance mechanisms, p 43–63. In Smith I, Slepecky RA, Setlow P (ed), Regulation of prokaryotic development. American Society for Microbiology, Washington, DC. [Google Scholar]
- 56.Paidhungat M, Setlow B, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J Bacteriol 182:5505–5512. doi: 10.1128/JB.182.19.5505-5512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Granger AC, Gaidamakova EK, Matrosova VY, Daly MJ, Setlow P. 2011. Effects of Mn and Fe levels on Bacillus subtilis spore resistance and effects of Mn2+, other divalent cations, orthophosphate, and dipicolinic acid on protein resistance to ionizing radiation. Appl Environ Microbiol 77:32–40. doi: 10.1128/AEM.01965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raeymaekers L, Wuytack E, Willems I, Michiels CW, Wuytack F. 2002. Expression of a P-type Ca2+-transport ATPase in Bacillus subtilis during sporulation. Cell Calcium 32:93. doi: 10.1016/S0143-4160(02)00125-2. [DOI] [PubMed] [Google Scholar]
- 59.Daniel RA, Errington J. 1993. Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J Mol Biol 232:468–483. doi: 10.1006/jmbi.1993.1403. [DOI] [PubMed] [Google Scholar]
- 60.Ramirez-Guadiana FH, Meeske AJ, Rodrigues CDA, Barajas-Ornelas RDC, Kruse AC, Rudner DZ. 2017. A two-step transport pathway allows the mother cell to nurture the developing spore in Bacillus subtilis. PLoS Genet 13:e1007015. doi: 10.1371/journal.pgen.1007015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arrieta-Ortiz ML, Hafemeister C, Bate AR, Chu T, Greenfield A, Shuster B, Barry SN, Gallitto M, Liu B, Kacmarczyk T, Santoriello F, Chen J, Rodrigues CD, Sato T, Rudner DZ, Driks A, Bonneau R, Eichenberger P. 2015. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol Syst Biol 11:839. doi: 10.15252/msb.20156236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim HU, Goepfert JM. 1974. A sporulation medium for Bacillus anthracis. J Appl Bacteriol 37:265–267. doi: 10.1111/j.1365-2672.1974.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 63.Leighton TJ, Doi RH. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem 254:3189–3195. [PubMed] [Google Scholar]
- 64.Nicholson WL, Setlow P. 1990. Sporulation, germination, and outgrowth, p 391–450. In Harwood CR, Cutting SM (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Ltd, Chichester, England. [Google Scholar]
- 65.González-Castro MJ, López-Hernández J, Simal-Lozano J, Oruña-Concha MJ. 1997. Determination of amino acids in green beans by derivitization with phenylisothiocyanate and high-performance liquid chromatography with ultraviolet detection. J Chrom Sci 35:181–185. doi: 10.1093/chromsci/35.4.181. [DOI] [Google Scholar]
- 66.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. 2014. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 68.Koo BM, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann AB, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291–305.e297. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 70.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast 12:259–265. doi:. [DOI] [PubMed] [Google Scholar]
- 71.Hageman JH, Shankweiler GW, Wall PR, Franich K, McCowan GW, Cauble SM, Grajeda J, Quinones C. 1984. Single, chemically defined sporulation medium for Bacillus subtilis: growth, sporulation, and extracellular protease production. J Bacteriol 160:438–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wyrick PB, Rogers HJ. 1973. Isolation and characterization of cell wall-defective variants of Bacillus subtilis and Bacillus licheniformis. J Bacteriol 116:456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meador-Parton J, Popham DL. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J Bacteriol 182:4491–4499. doi: 10.1128/JB.182.16.4491-4499.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soil and Plant Analysis Council I. 1999. Methods of instrumental analysis, p 207–224. In Jones J., Jr (ed), Soil analysis handbook of reference methods. CRC Press, Inc, Boca Raton, FL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.