Abstract
Captive-bred rhesus macaques of Indian origin represent one of the most important large animal models for infectious disease, solid organ transplantation and stem cell research. There is a dearth of information defining hematopoietic development, including neutrophil leukocyte differentiation in this species using multicolor flow cytometry. In the current study we sought to identify cell surface markers that delineate neutrophil progenitor populations with characteristic immunophenotypes. We defined four different post-mitotic populations based on their CD11b and CD87 expression pattern, and further refined their immunophenotypes using CD32, CD64, lactoferrin and myeloperoxidase as antigenic markers. The four subsets contained myelocyte, metamyelocyte, band, and segmented neutrophil populations. We compared our flow cytometry-based classification with the classical nuclear morphology-based classification. We found overlap of immunological phenotype between populations of different nuclear morphology and identified phenotypically different subsets within populations of similar nuclear morphology.
We assessed the responsiveness of these populations to stimulatory signals such as LPS, fMPL or PMA, and demonstrated significant differences between human and rhesus macaque neutrophil progenitors. In this study we provided evidence for species-specific features of granulopoiesis that ultimately manifested in the divergent immunophenotypes of the fully differentiated segmented neutrophils of humans and rhesus macaques. Additionally, we found functional markers that can be used to accurately quantify neutrophil progenitors by flow cytometry. While these markers do not coincide with the classical nuclear-morphology-based grading they enable us to perform functional studies monitoring immunophenotypic markers.
Keywords: immunophenotype, neutrophil progenitors, bone marrow, rhesus macaque
Summary sentence:
A flow cytometric protocol using species-specific characteristics to define rhesus macaque neutrophil progenitors in the bone marrow.
Introduction
Neutrophil leukocytes are the most abundant white blood cells in circulation. They represent first responders in any type of injury or inflammation (1, 2). Mature neutrophils are non-dividing cells with a lifespan of several days (3, 4). Therefore, their homeostasis requires continuous replenishment supported by an uninterrupted differentiation process in the bone marrow. Differentiation stages of human neutrophil progenitor cell populations are distinguished according to their nuclear morphology. In successive order these are myeloblasts, promyelocytes, myelocytes, metamyelocytes, bands, and segmented neutrophils (5). Accurate quantitation of these cell populations with Wright-Giemsa staining and microscopy is time consuming, inefficient, lacks any information about their functional capabilities, phenotypic heterogeneity, and provides no insights regarding through what steps neutrophil differentiation happens (6). Quantitation using flow cytometry-based methods might offer significant improvement provided that appropriate differentiation markers are identified (7, 8). In the case of human neutrophil progenitors these critical differentiating markers have been the CD11b (CR3), CD15 (Lewis-x antigen), CD16 (FcRγ III), CD33, and the CD49d (VLA-4) antigens (7, 9).
Although the primary benefit of using nonhuman primates in biomedical research is the similarity of their immune system to the human immune system, numerous species-specific differences exist that can affect study designs and outcomes (10–15). For example, circulating neutrophil leukocytes in humans express both low affinity immunoglobulin Fcγ receptors (FcγR II and III) CD32 and CD16 (16). Neutrophils of several nonhuman primate species including rhesus macaques, cynomolgus macaques, African green monkeys and baboons do not express CD16 (17, our unpublished data). However, the absence of CD16 on nonhuman primate neutrophils cannot be generalized since sooty mangabeys do express this receptor on their PMN (17). Additionally, while circulating neutrophils in rhesus macaques are CD32 positive, the same cells in African green monkeys are CD32 negative (our unpublished data).
Differences between humans and rhesus macaques exist in the expression patterns of CD11c (CR4) and CD64 (high affinity FcγR I) (our unpublished data). Disparities between the immunophenotypes of mature neutrophils raise the possibility of disparities between the differentiation phenotypes of neutrophil progenitors between the two species. Thus, defining characteristic immunophenotype profiles of the different neutrophil progenitor populations in rhesus macaques is necessary. Unfortunately, established differentiating markers used for human samples are of limited value for rhesus macaque samples. For example, an antibody recognizing the CD15 antigen in rhesus macaques is not available. The antibody specific for CD33 (clone AC104.3E3) does not cross-react in a large number of animals tested, and the CD49d antigen expression pattern does not allow reliable separation of the CD49d positive and negative populations.
In the current study we sought to establish a flow cytometric protocol with differentiation markers that unequivocally define distinct subsets, and provide a tool to accurately quantitate neutrophil progenitors in the bone marrow of rhesus macaques. We compared our panel of markers between healthy adult human and rhesus samples to determine similarity and species-specific differences.
Materials and Methods
Human bone marrow and blood samples
We purchased de-identified, freshly obtained human bone marrow and matching blood samples from 18–65 years old healthy individuals from AllCells LLC (Boston, USA), or Lonza Inc. (Mapleton, IL, USA). The donors signed a procedure-specific consent form. Pregnant women were not included in the study.
Animals
We used freshly obtained whole blood and bone marrow from 1.5–9 years old healthy Indian rhesus macaques (Macaca mulatta). The animals in this study were assigned to various protocols approved by the University of Wisconsin Institutional Animal Care and Use Committee. They were cared for according to the NIH “Guide to the Care and Use of Laboratory Animals.”
Flow cytometric staining, data acquisition, and analysis
We dispersed the bone marrow cells in RPMI containing 10% FCS, and filtered them through a 100 μm cell strainer. We stained 106 cells in 100 μl tissue culture medium or 100 μl EDTA anticoagulated whole blood at RT with CD3 Alexa488 (clone SP34–2), HLA-DR FITC (clone L243), CD123 FITC (clone 7G3), CD66a-e PerCP-Vio700 (clone TET2, Miltenyi), CD11b PE-Cy7 (clone ICRF44), CD64 BV510 (clone 10.1 BioLegend), CD34 BV650 ((clone 563, Fisher Scientific), CD32 BV711 (clone FLI8.26), CD45 BV786 (clone D058–1283), CD23 Alexa700 (clone M-L233), and CD87 APC (clone VIM5, Fisher Scientific). Antibodies were obtained from BD Biosciences unless indicated otherwise. We used Near IR ARD (Life Technologies) to label dead cells. Each reagent was titered for optimal performance. After 15 minutes of incubation we added 1 ml of 1X BD Pharm Lysing Solution (BD Biosciences) for 5 minutes. After two washing steps, we enhanced the CD87 APC signal using APC Faser Kit (Miltenyi 130–091-762). We fixed the cells with 125 μl of 2% PFA for 15 minutes, pelleted them at 530 rcf for 5 minutes, and replaced the PFA, with 100 μl of Bulk Permeabilization reagent (Life Technologies GAS002S100). We stained for the intracellular antigens with anti-lactoferrin PE antibody (clone 4C5, Fisher Scientific), and anti-MPO eFluor450 antibody (clone MPO455–8E6, eBioscience) for 15 minutes. Following two washing steps we acquired the data on a BD LSR-II flowcytometer (Becton Dickinson, San Jose, CA) using FACSDiva™ 8.0.1. software. We analyzed the data with FlowJo™ 10.2 (Tree Star Inc., Ashland, OR).
Cell Sorting
We stained 107 bone marrow cells with CD3 FITC (clone SP34–2), CD123 FITC (clone 7G3), HLA-DR FITC (clone L243), Near IR ARD live/dead discriminator dye, CD11b PE-Cy7, and CD87 APC in 200 μl of RPMI- 10% FCS. Following a 15-minute incubation period, we removed the red blood cells with Pharm Lysing Solution. We enhanced the CD87 staining with the APC Faser kit. Sorting was performed with a FACSJazz cell sorter (Becton Dickinson, San Jose, CA) under biosafety level-3 conditions. Cell aggregates, dead cells, T cells, basophil granulocytes, macrophages, and B cells were excluded from the sort strategy. We collected between 17,000–93,000 cells for nuclear morphology analysis. Cells were spun onto a 7mm diameter area of a glass microscope slide using 530 rcf for 5 minutes, then stained with Wright-Giemsa stain (Wescor Aerospray® Hematology Slide Stainer/Cytocentrifuge, model 7150, Logan, UT).
Identification of neutrophil maturational stage.
Neutrophil maturational stage was based on generally accepted criteria of nuclear shape for veterinary species (18). Segmented nuclei contained two or more lobes connected by nuclear segments that were less than one-half the width of the widest lobe. Band nuclei were S- or U-shaped with parallel sides lacking distinct segmentation; any narrowing of the nucleus had to maintain a width greater than one-half the width the thickest part of the nucleus. Metamyelocyte nuclei were elongate ovals with an indentation (reniform). Myelocyte nuclei were round or oval without an indentation. Percentages of each maturational stage and leukocyte type were based on three hundred cell differential counts.
In vitro stimulation assays.
100 μl whole blood, or 106 freshly processed bone marrow cells were stimulated with 1 μg/ml LPS, PMA, or 1 μM fMLP for 15 minutes at 37oC in a 5% CO2 incubator. Following the stimulation, we stained the cells for surface antigens CD3, HLA-DR, CD123, CD66a-e, CD11b, CD64, CD32, CD45, CD34, CD23, CD87, and stimulation-induced lactoferrin and MPO release. We differentiated live cells from dead ones again with the Near IR ARD. After 15 minutes of staining we removed the red blood cells with 1 ml of 1X BD Pharm Lysing Solution. Following two washing steps, the CD87 APC signal was enhanced with the APC Faser Kit. Cells were fixed with 125 μl of 2% PFA for 15 minutes, pelleted, and permeabilized with 100 μl of Bulk Permeabilization reagent. We removed the permeabilization buffer with two washing steps than acquired the flow cytometric data.
Statistical analysis
For statistical analysis we employed Students’ paired t test using Microsoft Excel software version 16.16.1 for Macintosh.
Results and Discussion
Our goal was to establish a flow cytometric method for the quantification of neutrophil progenitor populations in the bone marrow of rhesus macaques. To that end, we had to find immunologic markers that defined separate differentiation stages reproducibly and unequivocally. We designed a multicolor flow cytometry panel selecting the CD11b, CD32, CD64, CD87, lactoferrin and MPO antigens due to their established timeline in human neutrophil maturation (4). CD11b mediates complement-coated pathogen uptake and regulates leukocyte adhesion as part of the CD11b/CD18 heterodimer (19, 20). In humans, the absence of CD11b separates myeloblasts and promyelocytes from later neutrophil progenitor populations, as this antigen first appears at the myelocyte stage (21). The low affinity γ immunoglobulin receptor CD32 appears as early as the myeloblast stage in humans, and its expression increases in the segmented neutrophil stage (8, 22). The high affinity γ immunoglobulin receptor CD64 expression is maintained at a very low level until it is entirely downregulated at the band stage in humans (8, 22, 23). CD87 (uPAR) has been described as a marker to identify the band and segmented neutrophil populations in human bone marrow (24). MPO can be detected in the primary azurophilic granules from the promyelocyte phase to the fully matured segmented neutrophils (7, 25–27). Lactoferrin, a multifunctional protein with antimicrobial activity, appears first in the secondary granules at the metamyelocyte stage of human granulopoiesis (7, 28–30).
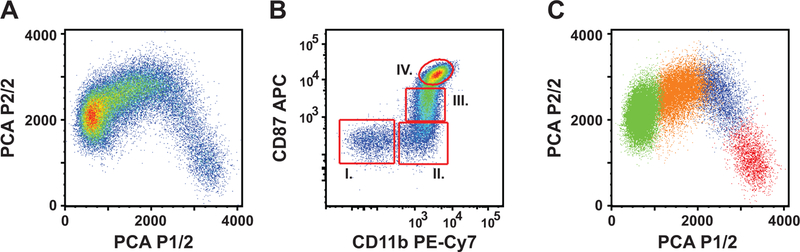
As the initial step of our analysis, we excluded lymphocytes, basophil and eosinophil granulocytes, stem cells, macrophages, dead cells, and cell aggregates (SFigure 1). Then we used principal component analysis to two components to identify those markers that accounted for the most variance within the CD66a-e+CD23− gate, and thus defined distinct cell subsets robustly (Figure 1A). CD87 (−0.66), CD32 (−0.59) and CD11b (−0.38) possessed the highest Eigenvalues in the Eigenvector of the first component, which accounted for 70% of the variance. CD11b (0.55), and CD64 (0.48) had the highest Eigenvalues in the Eigenvector of the second component, which accounted for additional 9.4% of the variance. Based on these results we selected CD11b and CD87 in a bivariate dot plot display to define four well-separated populations (designated I. through IV.) (Figure 1B). These populations covered the PCA dot plots entirely and on mutually exclusive manner (Figure 1C). The relative frequency of the populations from I through IV were: 10.1±7.6%, 13.6±7.3%, 21.6±7.5%, and 49.4±8.5% of the parent population respectively (n=5).
Figure 1: Principal component analysis of neutrophil progenitor immunophenotype markers in the bone marrow of rhesus macaques.
A: PCA for two components. Plot is based on the expression of eight (CD10, CD11b, CD32, CD64, CD87, lactoferrin, and MPO) different parameters. B: CD11b/CD87 defined population I-IV. Single, live, CD66a-e positive cells, negative for CD3, CD123, HLA-DR, CD34, and CD23 antigens were gated as shown in supplemental figure 1. C: relative location of populations I-IV on the PCA plot. Red = population I, blue = population II, orange = population III, and green = population IV.
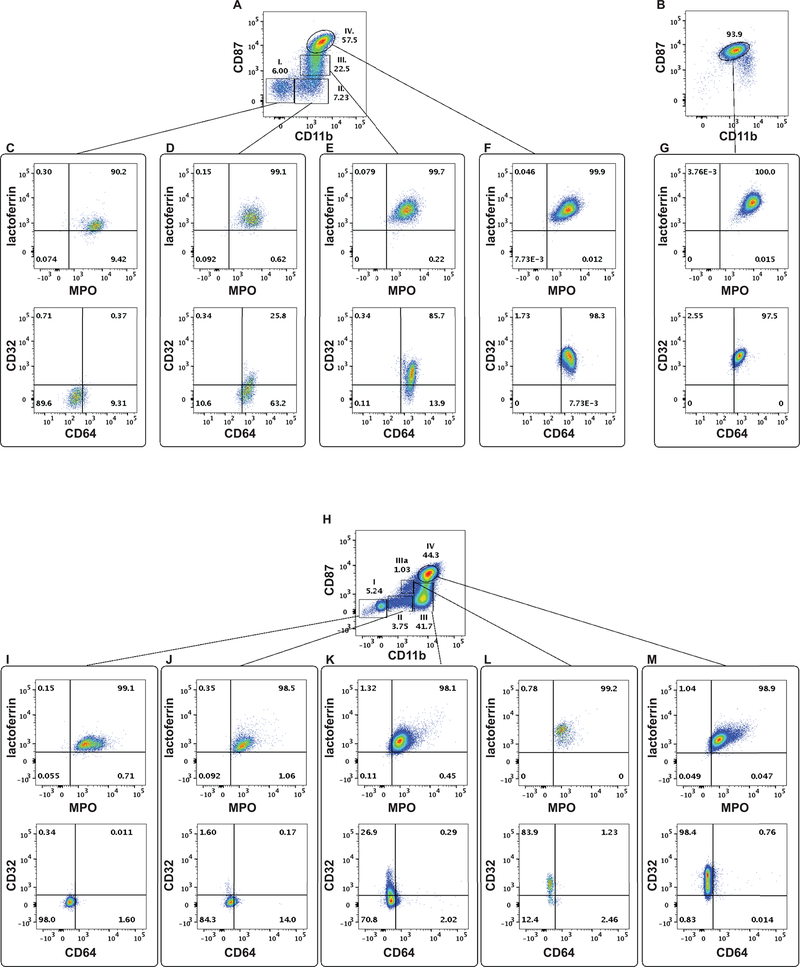
Next, we determined the expression patterns of MPO, lactoferrin, CD32 and CD64 in these populations. MPO and lactoferrin were present in all four populations. While the MPO content/cell decreased, the intracellular level of lactoferrin increased in the consecutive populations (Table 1). We detected CD64 on the cell surface at the CD11b+CD87− phase (population II), and cells remained positive for this protein throughout the final stages of granulopoiesis (Figure 2 D-F). Thus, emergence of CD64 coincided with the appearance of CD11b, but preceded CD87 expression. On the other hand, CD32 first appeared in population III (Figure 1 D, E) with increased expression in population IV (Figure 2 C-F, Table 1). To summarize the data above we can refine the immunophenotype of the four populations as follows:
Table 1.
The neutrophil progenitor populations defined by the CD11b and CD87 markers display different expression level of the CD64, CD32, lactoferrin, MPO, and CD49d antigens.
| Population | CD64 | CD32 | lactoferrin | MPO | CD11b | CD87 | |
|---|---|---|---|---|---|---|---|
| Rhesus macaques | I. | 338±92 | 95± 101 | 946±267 | 6320±1916 | 46±17 | 327±59 |
| II. | 1019±254** | 235±156** | 2138±742* | 4110±946* | 1397±670** | 333±76 | |
| III. | 1477±390* | 479±176** | 4155±1166** | 3254±707* | 1766±944 | 1384±633* | |
| IV. | 1278±281 | 1654±529** | 5201±1490 | 3823±1205 | 2821±1671* | 6989±4425* | |
| Neut in blood | 1125±507 | 2753±1085* | 5920±1114 | 5582±2840 | 2571±1449 | 5428±1532 | |
| Human samples | I. | 515±94 | 181±45 | 862±134 | 7478±5236 | 73±47 | 333±90 |
| II. | 516±109 | 193±34 | 1013±158 | 3972±2510* | 1519±573** | 586±107** | |
| III. | 430±85 | 345±71 | 1762±660 | 2532±1561 | 9530±4353* | 1247±309** | |
| IIIa. | 325±43 | 1107±418* | 3142±959* | 3076±2182 | 2754±1215* | 1988±426** | |
| IV. | 302±53 | 1484±316** | 2330±1250 | 2247±1370 | 13039±5215** | 4364±2371** | |
| Neut in blood | 307±25 | 3339±576* | 11234±5514 | 11688±2340* | 9986±2404 | 5774±1756 |
Numbers are the average of Geometric Mean of Fluorescence Intensity ± SD (n=5),
indicates p<0.05
indicates p<0.005 difference from the previous population listed in the table.
Figure 2: Neutrophil progenitor populations defined by the CD11b and CD87 expression pattern possess distinct immunophenotypes.
A: Populations I-IV according to their CD11b-CD87 expression pattern in rhesus macaque bone marrow. B. CD11b/CD87 expression by neutrophil leukocytes in rhesus macaque peripheral blood. C-F. Lactoferrin, MPO, CD64 and CD32 expression of populations I, II, III and IV in rhesus macaque bone marrow. G. Lactoferrin, MPO, CD64 and CD32 expression by neutrophil granulocytes in rhesus macaque blood. H: Populations I-IV according to their CD11b-CD87 expression pattern in human bone marrow. I-M. Lactoferrin, MPO, CD64 and CD32 expression of populations I, II, IIIa, III, and IV in human bone marrow.
Population I. CD66+CD11b−CD87−MPO+lactoferrin+CD64−CD32−,
Population II. CD66+CD11b+CD87−MPO+lactoferrin+CD64+CD32−,
Population III. CD66+CD11b+CD87+MPO+lactoferrin+CD64+CD32+, and
Population IV. CD66+CD11b+CD87++MPO+lactoferrin+CD64+CD32++.
Immunophenotype of the neutrophil leukocytes in the peripheral blood shared the characteristics of population IV (Figure 2 F and G, Table 1).
In humans, CD11b can be detected at the myelocyte stage first (21), and the appearance of CD87 on the cell surface indicates the final steps (bands and segmented neutrophils) of maturation (24). However, the expression pattern of CD11b and CD87 antigens together on neutrophil progenitors has not been published. To validate our staining panel for rhesus macaques, we stained human bone marrow samples. While we found similarities between the two species, we also identified one major difference originating from three different levels of CD11b expression. In human samples, CD87 and CD11b defined five separate sub-populations that we designated, I, II, III, IIIa, and IV (Figure 2 H). The phenotype of these populations can be defined as (Figure 2 I-M):
Population I. CD66+CD11b−CD87−MPO+lactoferrin+CD64−CD32−,
Population II. CD66+CD11b+CD87−MPO+lactoferrin+CD64+/−CD32+/−,
Population III. CD66+CD11b++CD87−MPO+lactoferrin+CD64+/−CD32+/−,
Population IIIa. CD66+CD11b+CD87+MPO+lactoferrin+CD64−CD32+/−, and
Population IV. CD66+CD11b++CD87++MPO+lactoferrin+CD64+/−CD32++.
The relative frequency of the populations from I through IV within the CD66a-e+CD23− gate were: 4.3±0.7%, 4.0±0.5%, 1.1±0.9%, 30.7±6.3%, and 56.1±7.3% respectively (n=5).
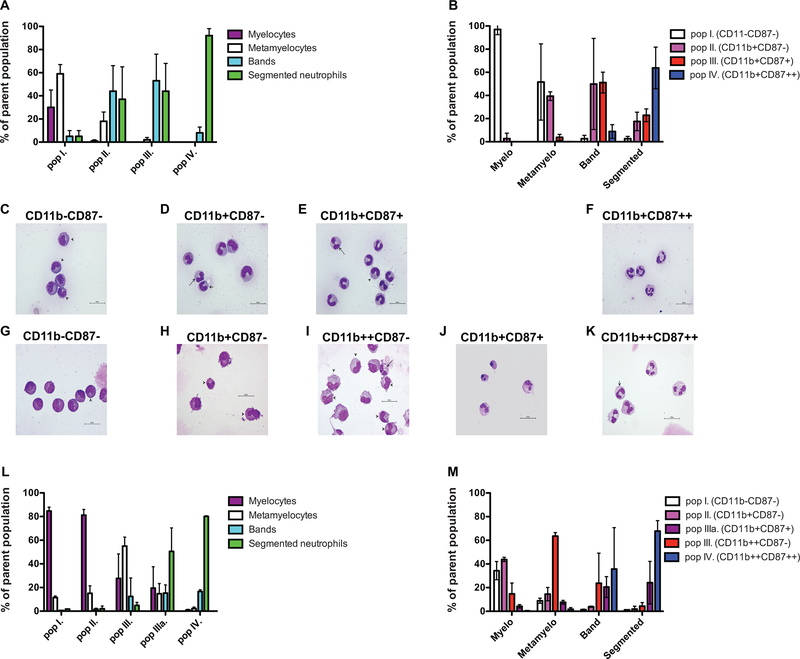
Since the phenotype of >97% of the circulating neutrophils resembled the phenotype of population IV (Table 1, Figure 2 B), we proposed that population I represents the most immature, and population IV is the most mature subset. To confirm this hypothesis, we sorted the CD11b/CD87 defined subsets and determined their cellular composition by nuclear morphology-based staging. We found overlap between the morphologically identified stages and the flow cytometry-based immunophenotypic categorization. In rhesus myelocytes and metamyelocytes represented the majority of population I: 30±15% and 59±8% respectively with 5±5% presence of each of the band and segmented neutrophils. Population II contained negligible numbers of myelocytes 1±1%, 18±8% metamyelocytes, 44±22% band, and 37±28% segmented neutrophils. Population III consisted of 2±2% of metamyelocytes, 53±23% band and 44±24% segmented neutrophils, while the overwhelming majority 92±6% of population IV was segmented neutrophils with 8±5% of bands (Figure 3 A, C-F). These results indeed support our hypothesis that population I represents cells at the less mature stage and the differentiation pathway leads towards population IV. We quantitated the heterogeneity of the populations classified by their nuclear morphology (Figure 3 B). We found that in rhesus myelocytes display almost exclusively the population I phenotype (97±4.5%), metamyelocytes are a combination of populations I and II (51.6±32.9 and 49.9±39.4% respectively), bands are mainly a mix of populations II and III (39.4±3.7 and 51.1±9.0%), and the segmented neutrophils are a combination of III and IV (22.9±5.5 and 63.8±18%). A more complex picture emerges in humans (Figure 3 G-M). Most myelocytes display either population I or II phenotype (34.3±7.7 and 43.8±1.8 respectively). This is primarily due to the earlier expression of CD11b, compared to the differentiation in rhesus macaques. Interestingly, in mice CD11b and CD32 are expressed on the cell surface throughout the entire granulopoiesis (31). While metamyelocytes can be found in populations I, II, and IIIa (Figure 3 L), the majority of them (63.5±3%) display III phenotype (Figure 3 M). Bands may express population IIIa, III, or IV phenotype (20.5±8.8%, 23.7±25.4%, and 35.7±35%), showing high variability between different individuals. The majority of segmented neutrophils were found in populations IIIa and IV (24.2±18% and 67.8±8.8%). The data summarized in Figure 3 provide evidence that the induction of nuclear segmentation and the changes in these immunophenotypic markers are not strongly connected.
Figure 3. Cellular composition of the CD11b/CD87 defined populations.
Cells were sorted as detailed in the Materials and Methods section. Panels A-F display data from rhesus macaque samples. Panels G-M display data from human samples. A: Relative frequency of the neutrophil progenitors within the four CD11b/CD87 subsets (mean+/−SD; n=3). B: Relative frequency of the CD11b/CD87 subsets within the neutrophil progenitor populations defined by nuclear morphology classification. Representative photomicrographs of subsets described as C: population I−: three metamyelocytes and two myelocytes; D: population II−: one segmented, one band, and three metamyelocyte neutrophils; E: population III: one segmented, seven band, and one metamyelocyte neutrophils; and F: population IV: four segmented neutrophils in a rhesus macaque sample. Representative photomicrographs of subsets described as G: population I−: one metamyelocyte and seven myelocytes; H: population II−: two metamyelocytes and two myelocytes; I: population III: six metamyelocytes, three myelocytes, six metamyelocytes, and one segmented neutrophil; J: population IIIa: two segmented neutrophils and two pyknotic cells; and K: population IV: four segmented and one band neutrophil in a human bone marrow sample. Segmented neutrophils - long arrow, band - medium arrow, and metamyelocytes -short arrow (X1000 magnification). L: Relative frequency of the neutrophil progenitors within the five CD11b/CD87 subsets. M: Relative frequency of the CD11b/CD87 subsets within the neutrophil progenitor populations defined by nuclear morphology classification.
It is important to emphasize here that while CD87 is expressed at the two terminal stages of neutrophil differentiation in rhesus, it is only found on 53.9+/−3.1% of band, and 83+/−7.5% (n=3) of segmented neutrophils at detectable levels. (None of the myelocytes, and less than five percent of the metamyelocytes (4.3+/−3.5) are CD87 positive). Therefore, this marker alone cannot be used to quantitate all band and segmented neutrophils in the bone marrow of rhesus macaques.
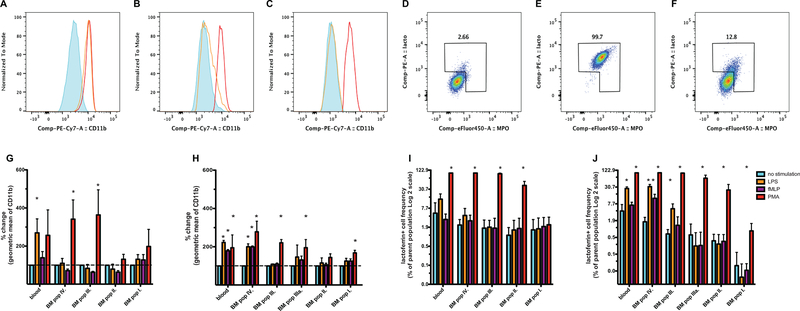
Next, to assess the functional capabilities of each neutrophil progenitor population subset, we stimulated matching blood and bone marrow samples in vitro with either LPS, fMLP, or PMA. We monitored the level of CD11b expression (Figure 4. A-C), and the release of lactoferrin and MPO to the cell surface (Figure 4. D-F) as indicators of activation (32, 33). Interestingly, while PMA induced significant changes in the phenotype of populations II-IV, neither LPS, nor fMLP elicited activation in these populations. We detected LPS-induced CD11b increases only in the blood-derived neutrophil population of rhesus macaques (Figure 4 G, I). In the human samples, however, we measured significant increase of CD11b expression and lactoferrin-positive cell frequency after LPS and PMA treatments in bone marrow populations III and IV which matched changes in blood samples (Figure 4 H, J). We found no measurable response to LPS and fMLP in populations I, II, and IIIa. These data suggest that while the human neutrophil progenitors complete their entire maturation in the bone marrow, the situation in rhesus macaques could be different. Perhaps neutrophils in rhesus macaques reach full maturation at another anatomical site. Several studies have provided evidence for this to occur in mice (34, 35).
Figure 4. Stimulation induced phenotype changes of neutrophil lineage cells in rhesus macaque or human samples.
A: CD11b expression by neutrophils in blood; B: population IV in bone marrow; C: and population III in the bone marrow of a healthy rhesus macaque. Red histogram: PMA stimulation; yellow histogram: LPS stimulation; blue histogram: unstimulated sample. D: surface lactoferrin/MPO in unstimulated; E: PMA stimulated, or F: LPS stimulated neutrophils in the blood of a healthy rhesus macaque. G: Stimulation induced increase of the geometric mean fluorescence of CD11b in rhesus macaque blood and bone marrow (n=3), or H: in human blood and bone marrow (n=3). I: Stimulation induced increase of the frequency of lactoferrin/MPO surface positive cells in rhesus macaque blood and bone marrow (n=3), or J: in human blood and bone marrow (n=3).
Herein we have provided a flow cytometry-based method to characterize four immunophenotypically different subsets of neutrophil progenitors in rhesus macaques. With this protocol we have introduced a technique that offers improved sensitivity and more dimensionality than the traditional nuclear morphology-based classification. Most importantly this method enables us to monitor the functional aspect of neutrophil maturation. It can be easily combined with applications addressing scientific questions at the genome or transcriptome level. We filled in an important knowledge gap that allows correct data interpretation of studies conducted in rhesus macaque model systems. It will be particularly beneficial for solid organ transplantation research, where extensive surgery induces emergency granulopoiesis, and infectious disease studies, where control of pathogenesis might depend on the homeostasis of neutrophil leukocytes (36, 37).
Supplementary Material
Acknowledgments
This work was supported by NIH grant #5P51OD011106 to the Wisconsin National Primate Research Center at the University of Wisconsin-Madison. Animals were handled in accordance with the standards of the American Association for the Accreditation of Laboratory Animal Care (AAALAC). We are grateful to Dr. Eileen Maher for editing our manuscript and Ms. Jessica Furlott for her technical assistance.
Abbreviations
- APC
Allophycocyanin
- ARD
Amino Reactive Dye
- BV
Brilliant Violet
- CR3
Complement Receptor 3
- CR4
Complement Receptor 4
- fMLP
N-Formylmethionyl-leucyl-phenylalanine
- FCS
Fetal Calf Serum
- LPS
Lipopolysaccharide
- Near IR ARD
Near Infrared amino reactive d
- MPO
Myeloperoxidase
- PCA
Principal Component Analysis
- PE
Phycoerythrin
- PerCP
Peridinin Chlorophyll
- PFA
paraformaldehyde
- PMA
Phorbol 12-myristate 13-acetate
- PMN
Polymorphonuclear Neutrophilic Granulocyte
- RT
Room Temperature
- uPAR
urokinase Plasminogen Activator Receptor
Footnotes
Conflict of Interest Disclosure
The authors declare no conflict of interest.
References
- 1.Nathan C (2006) Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol 6, 173–182. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Cassatella MA, Costantini C, Jaillon S (2011) Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol 11, 519–531. [DOI] [PubMed] [Google Scholar]
- 3.Lahoz-Beneytez J, Elemans M, Zhang Y, Ahmed R, Salam A, Block M, Niederalt C, Asquith B, Macallan D (2016) Human neutrophil kinetics: modeling of stable isotope labeling data supports short blood neutrophil half-lives. Blood 127, 3431–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pillay J, den Braber I, Vrisekoop N, Kwast LM, de Boer RJ, Borghans JA, Tesselaar K, Koenderman L (2010) In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood 116, 625–627. [DOI] [PubMed] [Google Scholar]
- 5.Kruger P, Saffarzadeh M, Weber AN, Rieber N, Radsak M, von Bernuth H, Benarafa C, Roos D, Skokowa J, Hartl D (2015) Neutrophils: Between host defence, immune modulation, and tissue injury. PLoS Pathog 11, e1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slukvin II (2013) Hematopoietic specification from human pluripotent stem cells: current advances and challenges toward de novo generation of hematopoietic stem cells. Blood 122, 4035–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mora-Jensen H, Jendholm J, Fossum A, Porse B, Borregaard N, Borregaad N, Theilgaard-Mönch K (2011) Technical advance: immunophenotypical characterization of human neutrophil differentiation. J Leukoc Biol 90, 629–634. [DOI] [PubMed] [Google Scholar]
- 8.Elghetany MT (2002) Surface antigen changes during normal neutrophilic development: a critical review. Blood Cells Mol Dis 28, 260–274. [DOI] [PubMed] [Google Scholar]
- 9.Pedersen CC, Borup R, Fischer-Nielsen A, Mora-Jensen H, Fossum A, Cowland JB, Borregaard N (2016) Changes in Gene Expression during G-CSF-Induced Emergency Granulopoiesis in Humans. J Immunol 197, 1989–1999. [DOI] [PubMed] [Google Scholar]
- 10.Bontrop RE, Watkins DI (2005) MHC polymorphism: AIDS susceptibility in non-human primates. Trends Immunol 26: 227–233. [DOI] [PubMed] [Google Scholar]
- 11.Carter DL, Shieh TM, Blosser RL, Chadwick KR, Margolick JB et al. (1999) CD56 identifies monocytes and not natural killer cells in rhesus macaques. Cytometry 37: 41–50. [PubMed] [Google Scholar]
- 12.Dykhuizen M, Ceman J, Mitchen J, Zayas M, MacDougall A et al. (2000) Importance of the CD3 marker for evaluating changes in rhesus macaque CD4/CD8 T-cell ratios. Cytometry 40: 69–75. [DOI] [PubMed] [Google Scholar]
- 13.LaBonte ML, Choi EI, Letvin NL (2004) Molecular determinants regulating the pairing of NKG2 molecules with CD94 for cell surface heterodimer expression. J Immunol 172: 6902–6912. [DOI] [PubMed] [Google Scholar]
- 14.Reeves RK, Gillis J, Wong FE, Yu Y, Connole M et al. (2010) CD16- natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood 115: 4439–4446. PMCID: PMC2881505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugimoto C, Hasegawa A, Saito Y, Fukuyo Y, Chiu KB et al. (2015) Differentiation Kinetics of Blood Monocytes and Dendritic Cells in Macaques: Insights to Understanding Human Myeloid Cell Development. J Immunol 195: 1774–1781. PMCID: PMC4530075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruhns P (2012) Properties of mouse and human IgG receptors and their contribution to disease models. Blood 119, 5640–5649. [DOI] [PubMed] [Google Scholar]
- 17.Rogers KA, Scinicariello F, Attanasio R (2006) IgG Fc receptor III homologues in nonhuman primate species: genetic characterization and ligand interactions. J Immunol 177, 3848–3856. [DOI] [PubMed] [Google Scholar]
- 18.Radin MJ, Wellman ML. Chapter 7 Granulopoiesis (pp. 43–49). In: Schalm’s Veterinary Hematology, 6th ed. Eds: Weiss DJ and Wardrop KJ, Wiley Blackwell, Ames, IA. [Google Scholar]
- 19.Jones DH, Anderson DC, Burr BL, Rudloff HE, Smith CW, Krater SS, Schmalstieg FC (1988) Quantitation of intracellular Mac-1 (CD11b/CD18) pools in human neutrophils. J Leukoc Biol 44, 535–544. [DOI] [PubMed] [Google Scholar]
- 20.Beller DI, Springer TA, Schreiber RD (1982) Anti-Mac-1 selectively inhibits the mouse and human type three complement receptor. J Exp Med 156, 1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terstappen LW, Safford M, Loken MR (1990) Flow cytometric analysis of human bone marrow. III. Neutrophil maturation. Leukemia 4, 657–663. [PubMed] [Google Scholar]
- 22.Fleit HB, Wright SD, Durie CJ, Valinsky JE, Unkeless JC (1984) Ontogeny of Fc receptors and complement receptor (CR3) during human myeloid differentiation. J Clin Invest 73, 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimmerjahn F, Ravetch JV (2006) Fcgamma receptors: old friends and new family members. Immunity 24, 19–28. [DOI] [PubMed] [Google Scholar]
- 24.Elghetany MT, Patel J, Martinez J, Schwab H (2003) CD87 as a marker for terminal granulocytic maturation: assessment of its expression during granulopoiesis. Cytometry B Clin Cytom 51, 9–13. [DOI] [PubMed] [Google Scholar]
- 25.Gadd SJ, Majdic O, Kasinrerk W, Stockinger H, Maurer D, Eher R, Knapp W (1990) M5, a phosphoinositol-linked human myelomonocytic activation-associated antigen. Clin Exp Immunol 80, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bainton DF (1981) Selective abnormalities of azurophil and specific granules of human neutrophilic leukocytes. Fed Proc 40, 1443–1450. [PubMed] [Google Scholar]
- 27.Nauseef WM, Olsson I, Arnljots K (1988) Biosynthesis and processing of myeloperoxidase--a marker for myeloid cell differentiation. Eur J Haematol 40, 97–110. [DOI] [PubMed] [Google Scholar]
- 28.Borregaard N, Sørensen OE, Theilgaard-Mönch K (2007) Neutrophil granules: a library of innate immunity proteins. Trends Immunol 28, 340–345. [DOI] [PubMed] [Google Scholar]
- 29.Berliner N, Hsing A, Graubert T, Sigurdsson F, Zain M, Bruno E, Hoffman R (1995) Granulocyte colony-stimulating factor induction of normal human bone marrow progenitors results in neutrophil-specific gene expression. Blood 85, 799–803. [PubMed] [Google Scholar]
- 30.Bartels M, Govers AM, Fleskens V, Lourenço AR, Pals CE, Vervoort SJ, van Gent R, Brenkman AB, Bierings MB, Ackerman SJ, van Loosdregt J, Coffer PJ (2015) Acetylation of C/EBPε is a prerequisite for terminal neutrophil differentiation. Blood 125, 1782–1792. [DOI] [PubMed] [Google Scholar]
- 31.Evrard M, Kwok IWH, Chong SZ, Teng KWW, Becht E, Chen J, Sieow JL, Penny HL, Ching GC, Devi S, Adrover JM, Li JLY, Liong KH, Tan L, Poon Z, Foo S, Chua JW, Su IH, Balabanian K, Bachelerie F, Biswas SK, Larbi A, Hwang WYK, Madan V, Koeffler HP, Wong SC, Newell EW, Hidalgo A, Ginhoux F, Ng LG (2018) Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity 48, 364–379.e8. [DOI] [PubMed] [Google Scholar]
- 32.Soler-Rodriguez AM, Zhang H, Lichenstein HS, Qureshi N, Niesel DW, Crowe SE, Peterson JW, Klimpel GR (2000) Neutrophil activation by bacterial lipoprotein versus lipopolysaccharide: differential requirements for serum and CD14. J Immunol 164, 2674–2683. [DOI] [PubMed] [Google Scholar]
- 33.Swain SD, Jutila KL, Quinn MT (2000) Cell-surface lactoferrin as a marker for degranulation of specific granules in bovine neutrophils. Am J Vet Res 61, 29–37. [DOI] [PubMed] [Google Scholar]
- 34.Deniset JF, Surewaard BG, Lee WY, Kubes P (2017) Splenic Ly6Ghigh mature and Ly6Gint immature neutrophils contribute to eradication of S. pneumonie. J Exp Med 214, 1333–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chorny A, Casas-Recasens S, Sintes J, Shan M, Polentarutti N, García-Escudero R, Walland AC, Yeiser JR, Cassis L, Carrillo J, Puga I, Cunha C, Bastos H, Rodrigues F, Lacerda JF, Morais A, Dieguez-Gonzalez R, Heeger PS, Salvatori G, Carvalho A, Garcia-Sastre A, Blander JM, Mantovani A, Garlanda C, Cerutti A (2016) The soluble pattern recognition receptor PTX3 links humoral innate and adaptive immune responses by helping marginal zone B cells. J Exp Med 213, 2167–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ball ED, McDermott J, Griffin JD, Davey FR, Davis R, Bloomfield CD (1989) Expression of the three myeloid cell-associated immunoglobulin G Fc receptors defined by murine monoclonal antibodies on normal bone marrow and acute leukemia cells. Blood 73, 1951–1956. [PubMed] [Google Scholar]
- 37.Elbim C, Monceaux V, Mueller YM, Lewis MG, François S, Diop O, Akarid K, Hurtrel B, Gougerot-Pocidalo MA, Lévy Y, Katsikis PD, Estaquier J (2008) Early divergence in neutrophil apoptosis between pathogenic and nonpathogenic simian immunodeficiency virus infections of nonhuman primates. J Immunol 181, 8613–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.