ABSTRACT
USP22, a component of the SAGA complex, is overexpressed in highly aggressive cancers, but the normal functions of this deubiquitinase are not well defined. We determined that loss of USP22 in mice results in embryonic lethality due to defects in extra-embryonic placental tissues and failure to establish proper vascular interactions with the maternal circulatory system. These phenotypes arise from abnormal gene expression patterns that reflect defective kinase signaling, including TGFβ and several receptor tyrosine kinase pathways. USP22 deletion in endothelial cells and pericytes that are induced from embryonic stem cells also hinders these signaling cascades, with detrimental effects on cell survival and differentiation as well as on the ability to form vessels. Our findings provide new insights into the functions of USP22 during development that may offer clues to its role in disease states.
KEY WORDS: USP22, SAGA, Placenta, Vascular development, TGFβ signaling, RTK receptors, Endothelial cells, Pericytes, Mouse
Highlighted Article: Global loss of the USP22 deubiquitinase, an integral subunit of the SAGA chromatin remodeler, reveals an unexpected role in the regulation of signaling cascades in endothelial and perivascular cells of the developing mouse placenta.
INTRODUCTION
Development of the mouse placenta requires specification of the trophoblast lineage at early stages of embryogenesis. The trophoblast cells that overlie the inner cell mass form the extra-embryonic ectoderm, and those away from the inner cell mass form the ectoplacental cone, immediately after implantation (Simmons et al., 2007). The extra-embryonic ectoderm gives rise to the chorionic epithelium, which will make contact with the extra-embryonic mesoderm (allantois) and eventually give rise to the labyrinth layer. Chorion-derived cells form trophoblasts and allantois-derived cells form endothelial cells. These two cell types undergo extensive intertwining and branching to create the labyrinth that facilitates nutrient exchange between mother and fetus. Defects in this process cause embryonic lethality between embryonic day (E) 9.5 and E13.5 of development (Rossant and Cross, 2001; Watson and Cross, 2005). Not surprisingly, these events require exquisite spatial and temporal control of gene expression programs, mediated by complex interactions between genetic and epigenetic factors, as mutations in these factors have detrimental effects among multiple placental layers (Hemberger, 2010). However, the molecular mechanisms that are responsible for many developmental events in the growing placenta are not well defined, as the placenta is still commonly overlooked in the phenotypic characterization of mice that bear embryonic lethal mutations, leading to an under-representation of the genes that are involved in regulating this pivotal organ (Perez-Garcia et al., 2018). Nonetheless, mouse genetic approaches have provided some important mechanistic insights to placental development, as mutations in several signaling molecules, transcription factors and epigenetic regulators cause lethality due to placental defects (Rossant and Cross, 2001; Watson and Cross, 2005).
The highly conserved SAGA complex plays important roles at multiple steps of gene transcription. SAGA houses two enzymatic activities, the Gcn5 histone acetyltransferase and the USP22 deubiquitinase (Koutelou et al., 2010), which alter histone acetylation and ubiquitination (also known as ubiquitylation) levels to regulate the expression of multiple genes (Bonnet et al., 2014; Hirsch et al., 2015). Genome-wide analyses in yeast revealed multiple gene-specific regulators that associate with SAGA, and a high dynamic range of transcripts that are regulated by SAGA, which provides evidence that SAGA is involved in cellular response events that require highly efficient transcript formation (Venters et al., 2011; Vinayachandran et al., 2018). In mice, global loss of Gcn5 causes loss of mesodermal tissues in early embryogenesis (Xu et al., 2000). Lineage-specific Gcn5 deletions have revealed its roles in neural stem cell differentiation in vivo and in vitro (Martinez-Cerdeno et al., 2012). Gcn5 also regulates retinoic acid signaling in the developing mouse diencephalon (Wilde et al., 2017) and affects multiple components of FGF signaling during embryoid body differentiation (Wang et al., 2018).
In contrast to Gcn5, the functions of USP22 during development are poorly defined. Usp22 was first described as a member of an 11-gene ‘death from cancer’ signature that was defined by gene expression microarrays (Glinsky et al., 2005). Overexpression of USP22 has since been observed by several groups in multiple cancer types (reviewed by Wang and Dent, 2014), but no clear picture has yet emerged for how this deubiquitinase might contribute to oncogenesis. The biochemical activity of USP22 against ubiquitinated histones H2B or H2A is well characterized, and USP22 also has non-histone substrates, including TRF1 (also known as TERF1) (Atanassov et al., 2009), FBP1 (Atanassov and Dent, 2011) and SIRT1 (Lin et al., 2012). When and where USP22 activity is required to regulate these or other substrates in vivo is still unclear. Usp22 deletion causes embryonic lethality in mice (Lin et al., 2012), whereas mice that harbor a hypomorphic allele of Usp22 are viable but exhibit a reduced body size (Kosinsky et al., 2015). However, the molecular basis of these phenotypes is not well defined.
To gain more insights into USP22 functions, we performed a detailed analysis of the cause of death of Usp22 null embryos. Our findings reveal that USP22 plays important roles in placenta development that are tied to multiple signaling pathways driven by TGFβ and several receptor tyrosine kinases, including VEGFR (KDR), HGFR (MET) and PDGFR (PDGFRB).
RESULTS
Usp22 loss-of-function results in vascular defects in the placental labyrinth
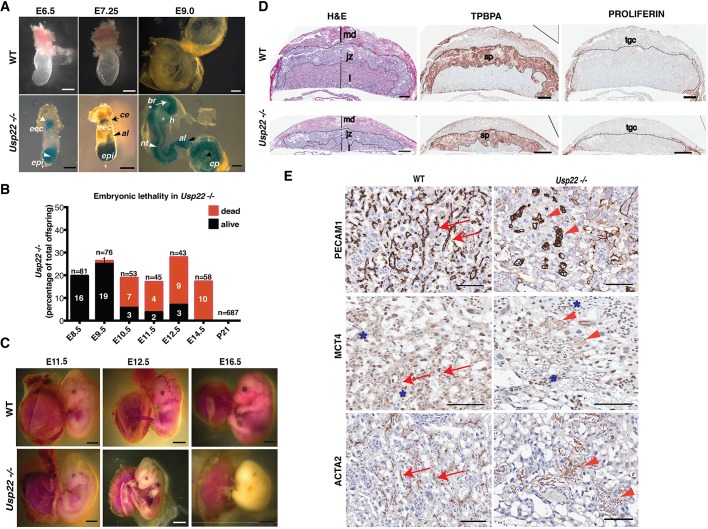
We took advantage of a β-galactosidase marker driven by the Usp22 promoter in a gene-trap allele (www.genetrap.org) (Fig. 1A and Fig. S1) to define Usp22 expression patterns within the embryo. At E6.5, Usp22 is expressed in the anterior epiblast (presumptive neuroectoderm), as well as in the posterior epiblast. A second site of expression initiates in the ectoplacental cone and is maintained in extra-embryonic ectoderm and mesoderm at E7.25. At E9, Usp22 mRNA is present throughout the brain, neural tube, heart, allantois and chorionic plate. At later stages, Usp22 expression is gradually restricted to the developing forebrain and midbrain, to cephalic and dorsal root ganglia, as well as to internal organs including the heart, testis, intestine and ribs (Fig. S2).
Fig. 1.
Loss of Usp22 results in mid-gestational embryonic lethality and vasculature defects of the developing placental labyrinth. (A) USP22 expression (indicated by β-galactosidase staining) in E6.5 embryos is detected at the ectoplacental ectoderm (eec; white arrow) and the epiblast (epi; white arrowhead); at E7.25 Usp22 expression is detected in the epiblast, chorionic ectoderm (ce; black arrow) and allantois (al; black arrowhead); and at E9 Usp22 expression is detected throughout the brain (br; white arrow), neural tube (nt; white arrowhead), heart (h; asterisk), allantois (black arrowhead) and chorionic plate (cp; black arrow). Scale bars: 100 μm. (B) Percentage of alive and dead embryos that are homozygous for Usp22 deletion, at the indicated developmental stages. (C) Extensive hemorrhages and signs of hypoxia-induced stress consistent with severe defects in extra-embryonic tissues were observed in Usp22 null embryos at E11.5, E12.5 and E16.5. Scale bars: 1 mm. (D) Histological analyses of wild-type and Usp22 mutant placentas at E14.5 showing different trophoblast cell layers, including the labyrinth (l), the spongiotrophoblasts (sp) and the trophoblast giant cells (tgc) in the junctional zone (jz), and the maternal decidua (md); defects are obvious in the developing labyrinth. Scale bars: 200 μm. (E) Histological analyses of wild-type and mutant placentas at E12.5 show that endothelial cells in the mutant (stained for PECAM1) did not form the normal tubular vessel structures seen in the wild type (red arrows), but instead clustered in unconnected, unstructured vessel-like formations (red arrowheads). MCT4, a marker for SynT-II cells that exhibits a sharp, cell-surface expression in wild-type placentas (red arrows), showed broad expression throughout the vessels in Usp22 mutants (red arrowheads), which indicates misorganization of SynT-II cells. ACTA2, which is normally expressed in pericytes that surround the fetal endothelial cells (red arrows), was also misorganized in the mutants (red arrowheads). Asterisks depict non-specific staining for MCT4 antibody in nuclei. Scale bars: 100 μm.
The Usp22 expression pattern indicates potential functions in extra-embryonic tissues early in development. Close examination of Usp22-deficient embryos revealed no abnormalities in postimplantation events, including gastrulation and neurulation. Mutant embryos survived until 11.5-14.5 days postcoitum, but died shortly thereafter, with extensive hemorrhages and signs of potential hypoxia-induced stress that are consistent with severe defects in extra-embryonic tissues (Gnarra et al., 1997) (Fig. 1B,C). Hypoxic stress response was confirmed upon RNA-seq of total RNA collected from embryos at E10.5, as several genes (Adm, Ndrg1, Car9, Egln3, P4ha1, Ier3, Pfkp) in this pathway were upregulated upon USP22 loss (Majmundar et al., 2010) (Fig. S3). These same homozygous mutant phenotypes were obtained using two independent Usp22 gene-targeting technologies (Figs S1,S4,S5) and were consistent across different genetic backgrounds (Fig. 1C and Fig. S6).
Given that Usp22 loss-of-function induces hypoxic stress in embryos and Usp22 is expressed in extra-embryonic tissues, we hypothesized that disruption of Usp22 may have deleterious consequences in the development of the placenta. The mouse placental labyrinth is the site of oxygen and nutrient exchange between the mother and the fetus, and from E11 onward it is the primary site of nutrient transport. Defects occurring in the labyrinth layer of Usp22 mutants could limit the transport of nutrients to the embryo, ultimately resulting in lethality. Therefore, we next analyzed Hematoxylin and Eosin (H&E)-stained sections that were obtained from wild-type and Usp22-deficient placentas. At embryonic day E14.5, when most Usp22 mutant embryos have died, the three layers of the placenta, including the labyrinth, the spongiotrophoblasts and the trophoblast giant cells in the junctional zone, and the maternal decidua were easily discernible (Fig. 1D) in both wild type and mutants. The trophoblast giant cell [marked with anti-proliferin (Prl2c2)] and spongiotrophoblast layers (marked with anti-TPBPA) appeared to be unaffected by Usp22 loss (Fig. 1D). However, the labyrinth layer of the Usp22 null placentas was abnormal. Examination of earlier time points (E12.5) confirmed that the cellular density of the labyrinth epithelium was decreased and the network of fetal vessels and maternal sinuses were dilated and poorly developed in the Usp22 mutants. H&E staining followed by computational morphometry also indicated that the number of fetal vessels in the labyrinth of Usp22 null embryos was significantly reduced compared with wild-type littermates (Fig. S7A,B, upper panels).
To determine whether the defects in Usp22 mutant placentas were restricted to a specific cell type, we analyzed expression of cell and lineage specific markers. PECAM1 staining, which is specific for endothelial cells, again indicated a striking defect in the formation of the vasculature of the labyrinth in the mutant (Fig. 1Ε, upper panel). The endothelial cells did not form normal tubular vessel structures, but instead clustered in unconnected, unstructured vessel-like formations. MCT4 (Slc16a3), a marker for syncytiotrophoblast (SynT)-II cells that exhibits a sharp, cell-surface expression in wild-type placentas (Moreau et al., 2014; Nagai et al., 2010), showed broad expression throughout the aberrant vessels in the Usp22 mutants (Fig. 1Ε, middle panel), which indicates misorganization of SynT-II cells. Smooth-muscle actin (ACTA2), which is normally expressed in mural cells that surround the fetal endothelial cells, was also misorganized in the mutants (Fig. 1E, lower panel). Altogether these findings indicate that USP22 is required for the functional interaction of the different cell types (endothelial cells, mural cells, syncytiotrophoblasts) that form the labyrinth fetal vessels.
Usp22−/− placental failure is not due to aberrant chorioallantoic fusion or cardiovascular defects
To more precisely determine the primary cause of embryonic lethality in the Usp22 mutants, we defined the timing of death. Dead embryos were first observed at E10.5, and no live embryos were recovered past day E13.5-E14.5 (Fig. 1B). This time window, along with the phenotypes of the mutant embryos described above (Fig. 1C and Fig. S6), might be related to defects in cardiovascular development within the embryo or to defects in the maternal-embryonic circulatory system within the placenta. Therefore we examined expression of specific markers and morphological changes that are associated with such defects, focusing first on hallmark events within the placenta.
Placental development is marked by three stepwise events, beginning with the chorioallantoic fusion at E8.5. This process involves extension of the allantoic mesoderm from the embryo towards the chorionic plate to enable fusion. The interaction of adhesion molecules such as VCAM1 (Gurtner et al., 1995; Kwee et al., 1995) and its receptor α4 integrin (Yang et al., 1995) are essential for completion of fusion. Next, fetal blood vessels that are derived from the allantoic mesoderm and trophoblasts in the chorion form branches at E9, a process that depends on transcription factor Gcm1 (Anson-Cartwright et al., 2000). Finally, elaborate branching forms a functional labyrinth, in which fetal vessels are intertwined with trophoblasts in order to absorb nutrients and gases from the sinuses of maternal blood. Signaling cascades triggered by FGF family members, EGF and HGF are crucial for the regulation of these cellular interactions (Cross et al., 2003; Rossant and Cross, 2001).
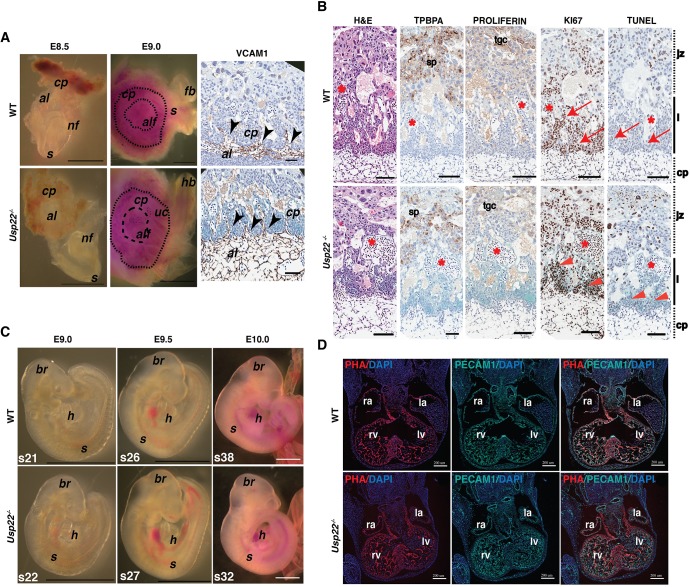
To determine whether the labyrinth defects we observe in Usp22 mutant placentas result from deregulation of the above events, we collected embryos at E8.5-E9 and carefully observed the structure of the allantois fusing into the chorionic plate. We did not detect any obvious morphological differences in this process between wild-type and Usp22 mutant embryos (Fig. 2A). Evaluation of VCAM1 expression patterns in wild-type and Usp22 mutant placentas revealed normal fusion and protrusion of allantoic mesoderm in the chorionic plate at multiple branching points in both mutant and wild-type placentas (Fig. 2A). Further analysis of placenta architecture at E9 by staining of trophoblast giant cells (marked with anti-proliferin) and spongiotrophoblast layers (marked with anti-TPBPA) again indicated both layers were well formed in wild-type and Usp22 mutant placentas. Finally, no marked changes in either proliferation (as indicated by KI67 expression) or cell death rates (as indicated by TUNEL assay) were observed in E9 placental sections (Fig. 2B).
Fig. 2.
Usp22−/− placental failure is not due to aberrant chorioallantoic fusion or cardiovascular defects. (A) Bright-field images (left) of wild-type and Usp22 mutant embryos and placentas at E8.5-E9. Histological analysis (right) of E9 wild-type and Usp22 placentas stained with VCAM1. Black arrowheads depict points of chorioallantoic fusion. Scale bars: 1 mm in bright-field images; 100 μm in histological analysis. (B) Histological analysis of E9 wild-type and Usp22 placentas stained with H&E, TPBPA, proliferin, KI67 and TUNEL assay. Similar levels of proliferation and apoptosis were observed in both wild-type (red arrows) and Usp22 mutant placentas (red arrowheads). Asterisks depict clusters of enucleated fetal red blood cells. Scale bars: 100μm. (C) Bright-field images of wild-type and Usp22 mutant embryos at E9-E10. Scale bars: 1 mm. (D) Immunofluorescence for phalloidin (PHA) and PECAM1 markers on wild-type and Usp22 mutant embryonic heart cryo sections (8 μm thick) at E11. Scale bar 200 μm. al, allantois; alf, allantoic fusion; br, brain; cp, chorionic plate; fb, forebrain; h, heart; hb, hindbrain; jz, junctional zone; l, labyrinth; la, left atrium; lv, left ventricle; nf, neural fold; ra, right atrium; rv, right ventricle; s, somites; sp, spongiotrophoblasts; tgc, trophoblast giant cells; uc, umbilical cord.
We next examined the cardiovascular system of the embryo, primarily focusing on development of the heart. Poor cardiac function or circulatory obstruction usually leads to edema, providing a good indicator of underlying heart failure (Conway et al., 2003). No edema was observed in the mutant embryos at E11.5-E12.5, which indicates normal heart function (Fig. 1C and Fig. S6). Moreover, no morphological differences were observed between wild-type and mutant embryos at E9-E10, and heartbeat and blood flow were evident in both genotypes, with no evidence of pericardial edema (Fig. 2C). As heart looping is normally completed, with formation of four heart chambers, by E10, we examined myocardium (by phalloidin staining) and endocardium (by staining for PECAM1) formation in wild-type and Usp22 mutant hearts at E11. We observed normal formation of all four heart chambers (right and left ventricle, right and left atrium) as well as the interventricular septum (Fig. 2D) in all embryos. Between E9 and E10 the myocardial tube is lined by the inner endothelium and outer myocardium, from which the trabeculation is formed. We examined ventricular myocardial trabeculation by multiphoton imaging of wild-type and Usp22 mutant hearts in whole embryos stained with phalloidin to visualize the myocardium and anti-PECAM1 to visualize the endocardium (Fig. S8) at E10. No abnormalities were observed in the organization of ventricular wall cardiomyocytes (myocardium) or endocardial cells in the Usp22 mutant hearts.
The strong expression of Usp22 in the developing allantois and the chorion in wild-type embryos, in combination with the absence of early functional heart defects (normal heart beat, blood flow, lack of edemas) in the mutants, indicates an essential role of Usp22 function in establishing embryonic-maternal circulation.
Usp22−/− placental vasculature failure is not due to intrinsic cell cycle defects or increased apoptosis
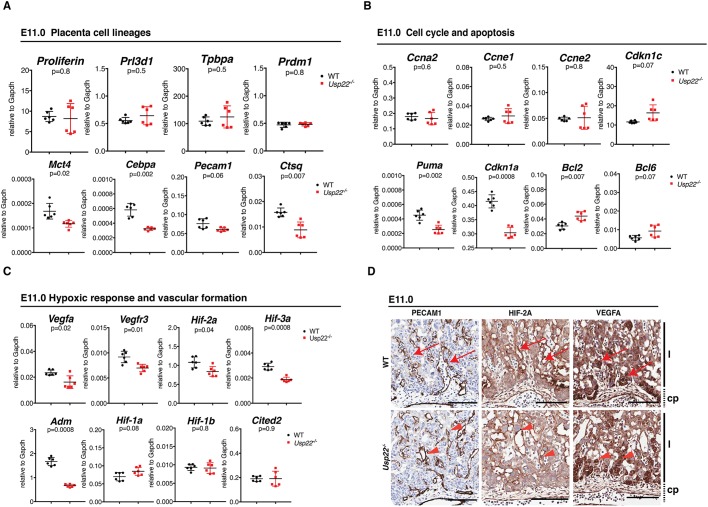
To further define the origin of the defects in the Usp22 mutants, we examined expression of trophoblast lineage differentiation markers at earlier time points. At E11, no reduction in expression of genes that are specific for trophoblast giant cells (Prl2c2, Prl3d1) or spongiotrophoblasts (Tpbpa, Prdm1) was detected. However, reduced expression of genes that are specific for labyrinth trophoblast cells, specifically syncytiotrophoblasts (Mct4, Cebpa), as well as for endothelial cells (Pecam1) and sinusoidal trophoblast giant cells (Ctsq), was observed (Fig. 3A), which indicates defective differentiation and/or specification of several cell lineages in Usp22 null placentas.
Fig. 3.
Usp22−/− placental vasculature failure is not due to intrinsic cell cycle defects or induced apoptosis. (A-C) qRT-PCR analysis of total RNA collected from wild-type and Usp22 mutant placentas at E11 showing reduced expression for labyrinth-specific cell lineages markers (Mct4, Cebpa, Pecam1 and Ctsq) (A), no obvious effect for cell cycle markers (Ccna2, Ccne1, Ccne2, Cdkn1c), a noticeable reduction for apoptotic markers (Cdkn1a, Puma) and an increase in anti-apoptotic markers (Bcl2, Bcl6) (B). Several genes that are required for vascular formation (Vegfa, Vegfr3, Hif2a, Hif3a, Adm) were downregulated in the mutant placentas, whereas hypoxic response markers (Hif1a, Hif1b, Cited2) showed no difference between wild-type and mutant placentas at E11 (C). Error bars show s.e.m. of biological replicates (n=6). P-value determined using unpaired t-test and corrected using the BH method. (D) Histological analysis of wild-type and Usp22 mutant placentas at E11 stained for PECAM1 showed that the labyrinthine layer is formed, but fetal vessels that extend from the allantoic mesenchyme appeared to be disorganized (red arrowheads) compared with the wild type (red arrows). Similar expression levels and patterns for VEGFA and HIF2A in wild-type (red arrows) and Usp22 mutant placentas (red arrowheads) indicate lack of hypoxic stress in the mutants at this stage. cp, chorionic plate; l, labyrinth. Scale bars: 100 μm.
Usp22 depletion in cultured cells has been associated with G1/M cell cycle arrest (Gennaro et al., 2018; Zhang et al., 2008b), proliferation defects (Atanassov and Dent, 2011), p53-dependent apoptosis (Lin et al., 2012) and blockage of stem cell differentiation (Sussman et al., 2013). Although no defects in proliferation or increased cell death were observed at E9.5 (Fig. 2B), we further examined cell cycle progression in the mutants by examining expression of several marker genes (Ccna2, Ccne1, Ccne2, Cdkn1c) at E11. Minimal changes in expression of these genes were detected in the mutants at this stage (Fig. 3B). Surprisingly, apoptotic markers such as Cdkn1a and Puma (Bbc3) were downregulated, rather than upregulated, in the mutant placentas, and expression of anti-apoptotic markers such as Bcl2 and Bcl6 were increased (Fig. 3B). These findings indicate that the placental abnormalities observed at E11 are not due to either decreased proliferation or increased cell death. At later times, after E12.5, a significant reduction in dividing labyrinth trophoblasts and an increase in apoptosis specifically in fetal vessels was observed (Fig. S7). These effects are likely secondary to the abnormal organization of fetal vessels that we observed at earlier stages (E11).
Interestingly, expression of the genes that encode several factors that induce vascular formation in the labyrinth, including Vegfa, Vegfr3 (Flt4), which is also expressed in lymphatic endothelial cells, Hif2a (Epas1), Hif3a and Adm were downregulated in the Usp22 mutant placentas at E11 (Fig. 3C). In contrast, histological staining for representative markers of hypoxic stress (HIF2A, VEGFA) showed no obvious difference between wild-type and mutant placentas (Fig. 3D), which suggests that the hypoxic stress response was not yet induced at this early time point, consistent with a lack of induction of Hif1a or Hif1b (Arnt) RNA expression (Fig. 3C). Expression of Cited2, a major transcriptional regulator of labyrinth vasculature that also responds to induced hypoxia, was similarly unaltered (Moreau et al., 2014; Withington et al., 2006). Altogether, these results indicate that labyrinth development initiates within the placentas of Usp22 mutants, but development is halted or delayed at a subsequent step, resulting in undervascularization.
Altered gene expression profiles in Usp22 mutant placentas reveal defects in TGFβ and RTK signal transduction
To define the molecular mechanisms that underlie the defects in labyrinth vasculogenesis in Usp22 mutants, we isolated RNA from extra-embryonic structures of wild-type and mutant placentas at E9.5 to capture early events and performed RNA sequencing (RNA-seq). We identified 1158 genes as upregulated and 1035 genes as downregulated in the Usp22-deficient placentas [P<0.01, false discovery rate (FDR)<0.05, fold change≥2].
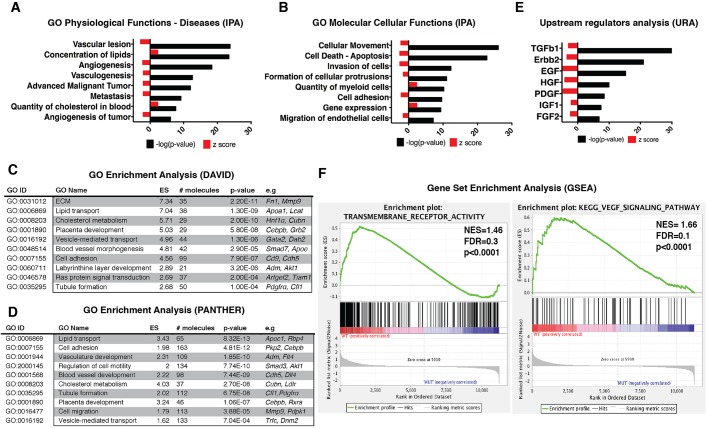
Gene ontology (GO) terms for physiological functions and diseases enriched for genes that were strongly downregulated in the Usp22 mutant placentas included processes that are related to vessel formation (vascular lesion, angiogenesis, vasculogenesis) and cancer (advanced malignant tumor, metastasis, angiogenesis of tumor) (Fig. 4A). In contrast, processes that are related to concentration and transport of lipids, as well as to the quantity of cholesterol in the blood, were consistently upregulated (Fig. 4A). The GO terms for molecular and cellular functions enriched for the downregulated genes highlighted processes including cellular movement, death, invasion, adhesion and formation of cell protrusions, which are all highly consistent with vessel formation failure (Fig. 4B). The GO terms for quantity of myeloid cells and for gene expression were upregulated. Ingenuity Pathway Analysis (IPA) software (Qiagen; www.ingenuity.com/products/ipa) was used for the in-depth processing of the RNA-seq data, and the above-mentioned GO categories were verified by two more GO analysis software packages (DAVID v6.7, PANTHER v11.0) (Fig. 4C,D).
Fig. 4.
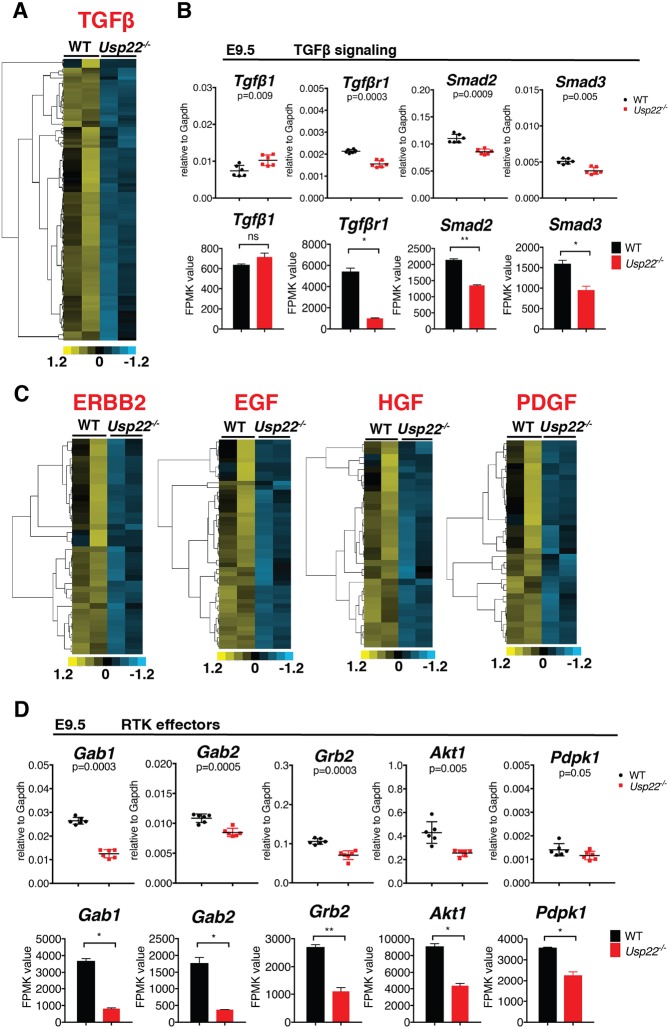
Genome-wide analysis of Usp22 mutant placentas reveals defects in vascularization, cellular movement and multiple signaling transduction pathways. (A) GO analysis of significantly affected physiological functions and diseases predicted by IPA. (B) GO analysis of significantly affected molecular and cellular functions predicted by IPA. (C,D) DAVID and PANTHER analysis software were used to find and verify the GO terms that were significantly enriched in differentially regulated gene sets produced by the RNA-seq analysis. (E) Upstream regulators analysis by IPA predicted the TGFβ1 pathway to be the most significantly inhibited (z-score: −2.900, P=4.9E-37). Several RTK pathways were predicted to be inhibited in the Usp22 mutants, including those responsive to Erbb2 (z-score: −3.369, P=1.9E-21), EGF (z-score: −4.616, P=5.2E-16), HGF (z-score: −2.051, P=1.23E-10) and PDGF (z-score: −4.560, P=3.9E-09). (F) Representative plots of gene set enrichment GSEA analysis for transmembrane receptor activity and VEGF signaling pathway. FDR, false discovery rate; NES, normalized enrichment score.
Upstream regulator analysis defined by IPA and Gene Set Enrichment Analysis (GSEA) indicated inhibition of a number of signaling pathways in the mutants (Fig. 4E,F), with the TGFβ1 pathway being the most affected (z-score: −2.900, P=4.9E-37) (Figs 4E and 5A). Tgfb1 is required for yolk sac hematopoiesis and endothelial differentiation in mice (Dickson et al., 1995). Similar to Usp22 mutants, defects in vasculogenesis in Tgfb1 mutants affect endothelial cell differentiation, rather than specification (Dickson et al., 1995). TGFβ1 receptors are also required for proper vasculogenesis (Larsson et al., 2001; Oshima et al., 1996). Expression of Tgfb1 itself is not affected in Usp22 mutants, but Tgfbr1 was significantly downregulated, as were Tgfb1 downstream transducers including Smad2 and Smad3 (Fig. 5B). Expression of Smad1 and Smad5, the two receptor-regulated Smads that do not respond to Tgfbr1, was not affected (data not shown).
Fig. 5.
TGFβ1 and RTK signal transduction is attenuated in Usp22 mutant placentas. (A,C) Heat maps of differentially expressed genes that are regulated by TGFβ1 (A) and RTK (C) signaling in wild-type and Usp22 mutant placentas at E9.5. (B,D) RNA-seq data and qRT-PCR validation of members and effectors of TGFβ1 (B) and RTK (D) signaling. Expression of Tgfb1 was not affected in Usp22 mutants, but Tgfbr1 was downregulated, as were downstream transducers (Smad2, Smad3) (B). Multiple cytoplasmic transducers of RTKs (Gab1, Gab2, Grb2, Akt1, Pdpk1) were downregulated in Usp22 mutant placentas (D). For qRT-PCR, error bars show s.e.m. of biological replicates (n=6). Bar graphs show fragments per kilobase of exon per million reads mapped (FPKM) values, represented as mean of biological replicates (n=2). Error bars show s.d. *P<0.05, calculated using unpaired t-test and corrected using the BH method.
IPA analysis of upstream regulators also indicated inhibition of several receptor tyrosine kinase (RTK) pathways in the Usp22 mutants, including those that are responsive to Erbb2 (z-score: −3.369, P=1.9E-21), EGF (z-score: −4.616, P=5.2E-16), HGF (z-score: −2.051, P=1.23E-10) and PDGF (z-score: −4.560, P=3.9E-09) (Figs 4E and 5C; Tables S1-S5). Several of these receptor tyrosine kinases, including HGFR, EGFR and PDGFR (Dackor et al., 2009; Ohlsson et al., 1999; Ueno et al., 2013), are essential for proper development of the trophoblast compartment of the labyrinth (Lindahl et al., 1997; Uehara et al., 1995). These signaling pathways mediate cellular interactions that underlie branching morphogenesis. Importantly, the block in labyrinth development that is caused by mutations in these proteins occurs after the initiation of chorioallantoic morphogenesis, consistent with the Usp22 mutant labyrinth phenotype. Moreover, downregulation of multiple cytoplasmic transducers of these RTKs, including Gab1, Gab2, Grb2, Akt1 and Pdpk1, was observed in the Usp22 mutants, which provides an explanation for the inhibition of multiple RTK pathways (Fig. 5D).
Deregulation of TGFβ and RTK signaling in Usp22 mutant placentas contributes to defective vascularization
Consistent with a role for USP22 in regulating labyrinth development, Usp22 (as indicated by β-galactosidase staining) is expressed in E9.5 placentas in the labyrinth trophoblasts of the chorion and in the mesenchymal cells of the allantois (Fig. 6A). In contrast, Usp22 expression is low in the trophoblast giant cell layer and in the spongiotrophoblast layers, which were not affected in the mutants at this stage, as confirmed by histological analysis and by analysis of expression of genes that are specific for these layers (data not shown).
Fig. 6.
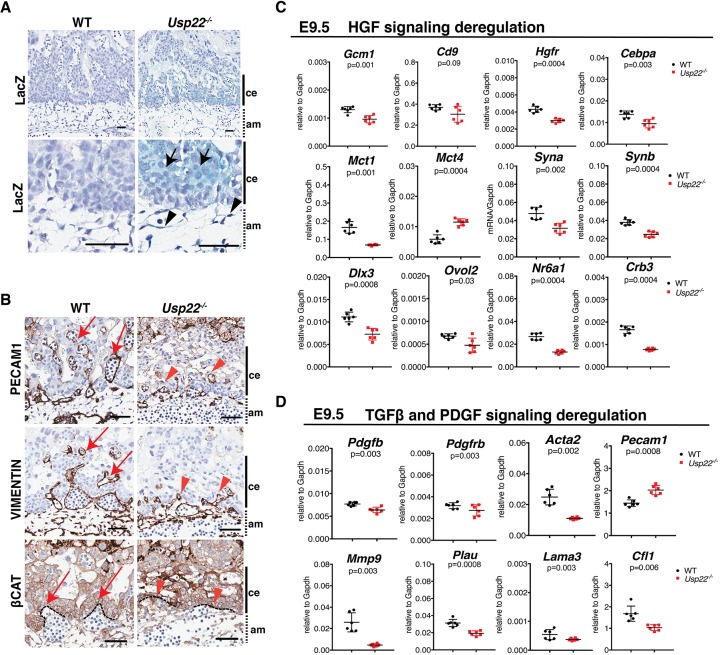
Validation of deregulation of TGFβ and RTK signaling in Usp22 mutant placentas. (A) β-galactosidase staining of placenta coronal sections at E9.5 indicates that Usp22 is expressed in trophoblasts of the chorionic plate (black arrows) and in mesenchymal cells of the allantois (black arrowheads). Scale bars: 50 μm. (B) PECAM1 and vimentin expression patterns in E9.5 wild-type and Usp22 mutant placentas indicate that fetal endothelial cells in Usp22 mutant placentas do not migrate into the chorionic epithelium (red arrowheads) as efficiently as do the wild-type cells (red arrows). β-Catenin staining indicates decreased formation of villi in the Usp22 mutant labyrinth (red arrowheads) compared with wild type (red arrows). Scale bars: 100 μm. (C,D) qRT-PCR analysis of total RNA collected from wild-type and Usp22 mutant placentas at E9.5 to detect differential expression of HGF pathway members and downstream targets (C), and TGFβ1 and PDGF pathway members and downstream targets (D). Error bars show s.e.m. of biological replicates (n=6). P-value was calculated using unpaired t-test and corrected using the BH method. am, allantoic mesoderm; ce, chorionic ectoderm.
To further connect the defects we observed in fetal vessel formation with defective TGFβ1 and RTK signaling in Usp22 mutant placentas, particularly the disorganization of endothelial cells and labyrinth trophoblast loss, we examined expression of specific markers for vascular formation that are responsive to these pathways at E9.5. Examination of sections of Usp22 mutant placentas at E9.5 indicates that embryonic vessels are unable to sprout properly into the labyrinth layer (Fig. 6B). Endothelial cells normally migrate towards angiogenic signals and then multiply and form a migration column. Migration columns lead to endothelial cell differentiation, with changes in cell shape and adhesion to form a lumen. PECAM1 and vimentin staining suggest that the endothelial cells in Usp22 mutant placentas that originate from the allantois do not migrate into the chorionic epithelium as efficiently as the wild-type cells, which results in the failure of lumen formation (Fig. 6B, upper and middle panel). The chorionic epithelium undergoes branching morphogenesis by changing the shape of cells in the basal layer that express the transcription factor Gcm1. These cells exit the cell cycle and begin to involute to create the primary villi into which fetal vessels will grow. Staining for β-catenin (Ctnnb1), which defines the chorionic trophoblasts, indicates decreased formation of villi in the Usp22 mutant labyrinths (Fig. 6B, lower panel).
Mutations in either Gcm1 or Cebpa cause delayed labyrinth development (Simmons et al., 2008). Gcm1, Cd9 and Cebpa are all downstream targets of HGF signaling in the labyrinth trophoblasts, and these genes were all downregulated in the Usp22 mutant (Fig. 6C, upper panel). Labyrinth trophoblast cells originate from the chorion, and their mitotic activity is stimulated by HGF after contact with the allantoic mesenchyme at ∼E10 (Ueno et al., 2013). Decreased mitotic activity of the trophoblasts in the labyrinth is also consistent with attenuation of HGF signaling that is observed in Usp22 mutant placentas, as verified by decreased expression of genes that are specific for both SynT-I [Syna, Mct1 (Slc16a1)] and SynT-II (Synb) (Fig. 6C, middle panel). Mct4 is the only syncytiotrophoblast marker to be upregulated in the mutant, consistent with Mct4 induction by hypoxic stress (Kay et al., 2007). The deregulation of HGF signaling was further verified by measuring expression of additional SynT markers (Dlx3, Ovol2, Nr6a1), as well as cell polarity markers such as Crb3 (Fig. 6C, lower panel).
Increased expression of endothelial markers (Pecam1) and decreased expression of markers of mural cells (Acta2) was also observed in the Usp22 mutant placentas, consistent with an imbalance of PDGF and TGFβ signaling (Fig. 6D, upper panel). PDGF receptors play a broad role in vascular development of yolk sac and placenta, as well as other tissues. Embryonic lethality in PDGF mutants is due to yolk sac and placental defects (Hellstrom et al., 1999), specifically in mural cells (French et al., 2008). Endothelial cells and mural cells of the placenta both express Tgfbr1 (Larsson et al., 2001), and embryonic lethality at ∼E9.5 of Tgfbr1 null embryos is attributed to a vascular defect both in the yolk sac and in the placenta. Nonetheless, both Usp22 and Tgfbr1-deficient placentas completed initial steps of branching morphogenesis, but vessels failed to sprout, which indicates defective angiogenesis. We also observe a migratory defect of the endothelial cells of the allantois, probably due to changes in expression of Mmp9, Plau, Lama3 and Cfl1 (Fig. 6D, lower panel). Altogether, these observations indicate that the Usp22 mutant phenotype likely reflects an accumulation of defects in multiple signaling pathways, not only that of TGFβ. The migration defects we observe in Usp22 mutant endothelial cells inside the labyrinth layer are consistent with defective TGFβ signaling, as indicated by our RNA-seq data, whereas the defects in mural cells and chorionic trophoblast cells likely result from deregulation of HGF and PDGF signaling.
Usp22 loss of function in induced endothelial cells or pericytes prevents vessel formation
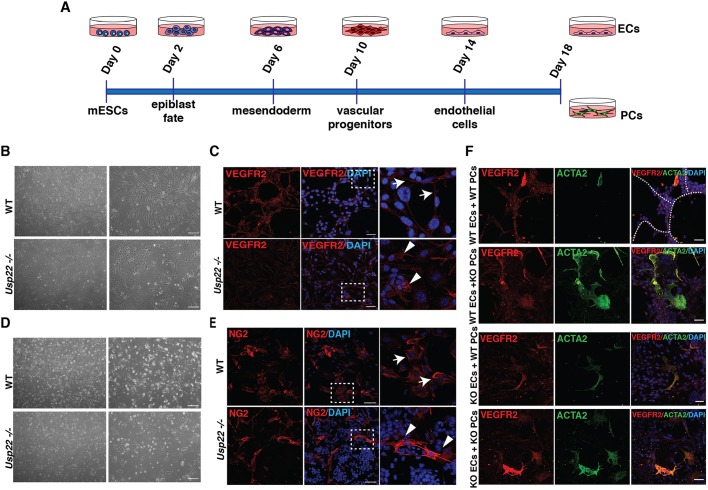
The vascular defects we observe in the Usp22 mutants establish an intriguing role for USP22 in the functional organization of endothelial cells, pericytes and labyrinth trophoblasts. Endothelial cells and pericytes are both crucial for the formation and function of the developing vasculature, but as our mutant allele affects USP22 expression in both tissues, we could not discriminate whether USP22 functions are most important in one or both of these cell types. To address this issue, we took an in vitro approach, differentiating wild-type and Usp22 null mouse embryonic stem cells (ESCs) towards a mesendodermal fate, gradually inducing differentiation towards vascular progenitors, and then finally to endothelial cells and pericytes (Fig. 7A and Fig. S9A). Generation of endothelial cells and pericytes was confirmed both by morphological observation (Fig. 7B,D and Fig. S9B) and by immunostaining with endothelial- [VEGFR2 (KDR), PECAM1] and pericyte-specific markers [NG2 (CSPG4), ACTA2, PDGFRβ] (Fig. 7C,E and Fig. S11A,B).
Fig. 7.
Usp22 loss of function in induced endothelial cells or pericytes prevents vessel formation. (A) Timeline of the differentiation course for induction of endothelial cells (ECs) and pericytes (PCs) from wild-type and Usp22 mutant ESCs. (B,D) Bright-field images of wild-type and Usp22−/− endothelial cells (B) and pericytes (D) at the final stage of the differentiation course (passage 2). Right column shows 2× magnification of left column. Scale bars: 100 μm. (C,E) Confocal imaging of VEGFR2 (C) and NG2 (E) as markers of wild-type and Usp22−/−-induced endothelial cells and pericytes, respectively (passage 4). In C, white arrows show membrane localization of VEGFR2 in wild-type induced ECs and white arrowheads show VEGFR2 mislocalization to the cytoplasm in Usp22 mutant induced ECs. E shows normal NG2 expression levels and localization both in wild-type (white arrows) and in Usp22 mutant induced pericytes (white arrowheads). Right column shows 5× magnification of the boxed area in the middle column. Scale bars: 50 μm. (F) Confocal images showing VEGFR2 (red) marking wild-type and Usp22−/−-induced endothelial cells co-cultured with indicated combinations of wild-type and Usp22−/− pericytes that are stained with ACTA2 (green) (passage 4). Sprouts are clearly visible when wild-type endothelial cells are co-cultured with wild-type pericytes (upper panel, white dotted line), but are missing in all other combinations. Scale bars: 50 μm.
Both wild-type and Usp22−/− early passage (passage 2-3) and low confluency endothelial cells and pericytes proliferated well. Closer observation of Usp22−/− endothelial cells showed that VEGFR2 and PECAM1 markers were expressed, but were mislocalized to the cytoplasm rather than the plasma membrane (Fig. 7C and Fig. S10A). At passage 5, the proliferative capacity of the mutant endothelial cells was reduced, and increased numbers of cells in G1 were observed (Fig. S10B,C). In contrast, Usp22−/− pericytes expressed NG2 similarly to wild-type cells but were more proliferative, and the cell population clearly contained a higher proportion of progenitor cells that did not express NG2. These findings suggest that loss of Usp22 limits pericyte differentiation, as fewer cells make it to mature pericytes, but those that do, express NG2 normally. Quantification of highly proliferative cells showed a significantly larger number of H3S10ph-positive cells in the Usp22−/− pericytes compared with the wild-type cells, but analysis of DNA content indicated no difference in overall cell cycle progression (Fig. S10E,F). These results indicate that USP22 loss affects endothelial cell propagation but not specification. In contrast, USP22 deletion strongly impacts pericyte differentiation. Boyden chamber assays adapted for endothelial cells revealed that USP22 mutant induced endothelial cells have a dramatic decrease in migratory capacity relative to wild-type cells (Fig. S11C,D), which is consistent with our observation of a migratory defect of endothelial cells in USP22 mutant labyrinths (Fig. 6B).
Pericytes and endothelial cells work together to form vessels, so we next performed a tube formation assay to assess functionality of these cell types. We co-cultured different combinations of wild-type or Usp22−/− endothelial cells and pericytes at early passages (passage 2-3). As expected, the combination of wild-type endothelial cells with wild-type pericytes led to formation of endothelial sprouts, which were visualized by immunostaining with VEGFR2-specific antibodies, which clearly outlined the formation of tubes (Fig. 7F). However, no combination that included either Usp22−/− endothelial cells or pericytes produced these structures, which indicates loss of USP22 in either or both cell types severely limits functional interactions between endothelial cells and pericytes, thereby crippling vessel formation.
Defective RTK and TGFβ signaling in induced endothelial cells upon USP22 loss
The phenotypes observed above may reflect deregulation of signal integration by endothelial cells and pericytes in the absence of Usp22, as was observed in the mutant placentas. Specifically, deregulation of VEGF signaling in induced endothelial cells would not inhibit their specification, but rather would cause a propagation defect (Schuh et al., 1999), as we observed in the Usp22 mutant endothelial cells. Deregulation of PDGF and TGFβ signaling in vascular progenitors is known to hinder their capacity to differentiate into pericytes (Hirschi et al., 1998), again consistent with the phenotype of Usp22 null progenitors.
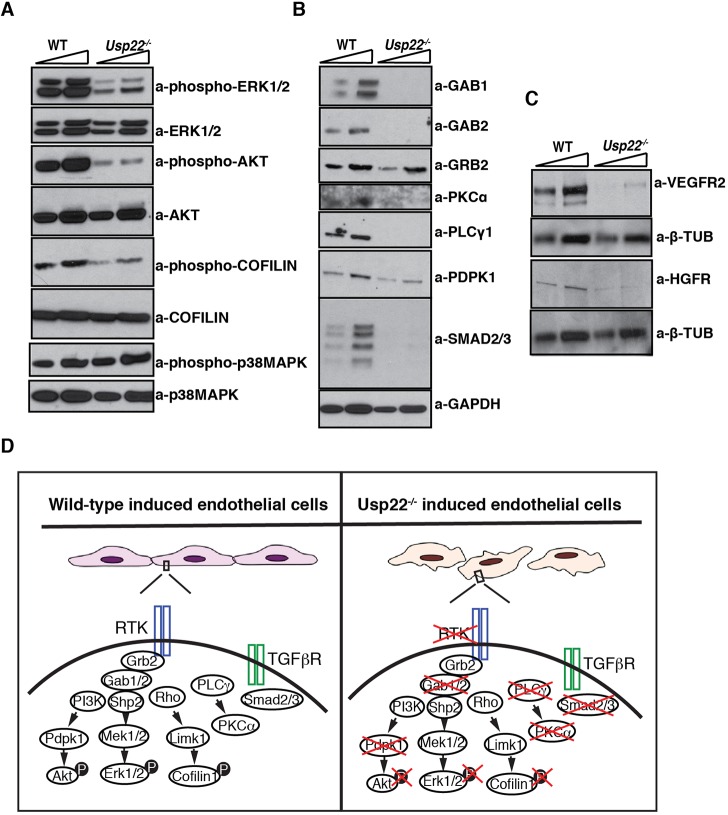
Assessment of both RTK and TGFβ signaling intermediates in Usp22 null endothelial cells revealed diminished levels of phosphorylated ERK1/2 and AKT in the Usp22−/−-induced endothelial cells, as well as phosphorylated cofilin (Fig. 8A), which is consistent with our observed effects on endothelial cell proliferation, survival and shape. Levels of signaling regulators and intermediates that were identified to be transcriptionally downregulated in Usp22−/− placentas were also strongly downregulated in the Usp22−/− endothelial cells, including Gab1, Gab2, PKCα (PRKCA), PLCγ1 and Smad2/3 (Fig. 8B). Interestingly, the levels of VEGFR2 and HGFR receptors were also affected in the Usp22−/−-induced endothelial cells (Fig. 8C). Our results identify USP22 as an indispensable regulator of signal integration in developing endothelial cells and pericytes.
Fig. 8.
Deregulation of TGFβ and RTK signaling pathways is observed in Usp22 mutant induced endothelial cells. (A) Representative immunoblots showing the decreased phosphorylated forms of ERK1/2, AKT and cofilin 1 in Usp22−/−-induced endothelial cells. (B,C) Representative immunoblots showing reduced protein levels of multiple transducers (B), and VEGFR2 and HGFR receptors (C) in Usp22−/−-induced endothelial cells. (D) Model of effects of Usp22 loss in signal transduction in induced endothelial cells. Attenuation of both RTK and TGFβ signals is due to decreased levels (or mislocalization) of the receptors, decreased levels of transducers (Gab1, Gab2, PKCα, PLCγ1, Smad2/3) and reduction in phosphorylation of the key effectors (ERK1/2, AKT and cofilin 1) that are required for the regulation of cell propagation, survival and shape.
DISCUSSION
Our findings reveal functions for USP22 in the development of the mouse placenta through effects on crucial signaling pathways. The requirement of USP22 for proper execution of multiple different signaling pathways likely explains the severity of the placental defects we observe in the Usp22 mutants and may provide new insights to the highly aggressive nature of tumors that overexpress USP22.
The effects of USP22 loss on VEGFA signaling is especially intriguing as this pathway transmits signals that are important for proliferation, migration and differentiation of vascular progenitors. VEGFA predominantly binds two receptors, VEGFR1 and VEGFR2. VEGFR2-deficient mice lack endothelial and hematopoietic cells (Shalaby et al., 1995) and die between E8.5 and E9.5 (Shalaby et al., 1997). Nevertheless, VEGFR2-deficient cells differentiate into hematopoietic and endothelial cells in culture, suggesting that VEGFR2 is required mostly for the subsequent migration and propagation of the specified cells (Schuh et al., 1999).
We used differentiated ESCs to probe the role of USP22 in the signaling cascades that are activated during lineage specification of VEGFR2-positive mesodermal progenitor cells because migration is not necessary in this context (Hirashima et al., 1999; Orlova et al., 2014; Yamashita et al., 2000). We clearly observed that Usp22 loss does not prevent vascular progenitor differentiation towards endothelial cells, as endothelial-specific markers such as PECAM1 and VE-cadherin (Cdh5) are expressed in the mutant induced endothelial cells. However, these factors exhibit abnormal localization away from the cytoplasmic membrane, and VEGFR2 itself is mislocalized and reduced in levels. The stability and internalization of VEGFR2 could be affected directly by USP22-dependent deubiquitination or indirectly by mislocalization of VE-cadherin (Lampugnani et al., 2006). Several USP deubiquitinating enzymes have been directly implicated in transmembrane receptor deubiquitination (Berthouze et al., 2009; Zhang et al., 2012). Our future work will address whether USP22 also plays such a role.
Our observation of strong decreases in phosphorylation of AKT and ERK1/2 in Usp22 null induced endothelial cells further indicates defective execution of VEGF-driven pathways. VEGFR2 induces proliferation of endothelial cells through the activation of ERK1/2 (Shu et al., 2002) and regulates cell survival through inhibition of pro-apoptotic pathways via the activation of PI3K/AKT phosphorylation (Gerber et al., 1998). A more detailed molecular analysis of the induced endothelial cells gave us further insight in the mechanistic role USP22 plays, as we found that several crucial signaling cascade effectors such as Gab1, Gab2, PKCα, PLCγ1 and PDPK1 were greatly downregulated in the absence of USP22 (model in Fig. 8D). Most of these molecules were also identified in the genome-wide transcriptional profiling of the placenta that we performed, which indicates that these factors are likely directly regulated by USP22, either transcriptionally or post-transcriptionally. USP22 has both histone and non-histone substrates, and it also functions in non-transcription related processes (Atanassov and Dent, 2011; Atanassov et al., 2009). The phenotypes we observe here, then, may reflect a combination of gene expression changes and changes in levels or activities of non-histone proteins that are regulated directly by USP22-mediated deubiquitination. Definition of such changes will require additional study.
Examination of monoubiquitination of histone H2B (H2Bub1) in Usp22 null endothelial cells revealed a decrease, rather than the expected increase, in the levels of ubiquitinated H2B relative to wild-type cells (Fig. S12). This counterintuitive result has been observed before (Atanassov et al., 2016), and it reflects redundancies and cross-regulation of two other USPs that are highly related to USP22: USP27x and USP51. USP27x and USP51 do not integrate into the SAGA complex; the phenotype of our Usp22 null mice indicate that these deubiquitinating enzymes (DUBs) cannot compensate for the loss of USP22 in the placenta. However, it is possible that USP22 loss allows greater association of Eny2 and Atxn7L3 with the other two DUBs, leading to a gain-of -function for USP27x or USP51. Additional studies will be required to define the relevant substrates of USP22 and the relative roles of these three USPs in RTK and TGFβ signaling.
A few previous studies have identified putative links between USP22 and signaling pathways in cancer cells. For example, USP22 was reported to play a role in proliferation of ovarian cancer cells that was correlated with regulation of TGFβ1 function (Ji et al., 2015). In nasopharyngeal carcinoma cells, USP22 functions in cell cycle progression were linked to modulation of the AKT/GSK3b/cyclin pathway (Zhuang et al., 2015). Interestingly, USP22 overexpression in lung adenocarcinoma invasion was correlated with upregulation of TGFβ1 and induction of epithelial-to-mesenchymal transitions (Hu et al., 2015). However, in another setting, diabetic nephropathy, USP22 overexpression in primary glomerular mesangial cells was linked to inhibition of TGFβ1 and fibronectin (Huang et al., 2015).
Unfortunately, studies to date have yielded a confusing amalgam of possible roles for USP22 in oncogenesis. Several studies have focused on potential roles for USP22 in regulating cell cycle progression through effects on oncogenes such as BMI1 (Liu et al., 2012), c-MYC (MYC) (Zhang et al., 2008a), SIRT1 (Lin et al., 2012), cyclin B1 (Lin et al., 2015), cyclin D1 (Gennaro et al., 2018) and KDM1A (Zhou et al., 2016), or on tumor suppressors such as p53 (Trp53) (Ding et al., 2015; Lin et al., 2012; Lv et al., 2011). The previously defined 11-gene ‘death from cancer’ signature that includes USP22 also includes BMI1 and downstream targets of this polycomb group protein. BMI1 overexpression was proposed to be the driving force that underlies these cancers, as BMI1 is required for self-renewal and it confers a cancer stem cell-like phenotype when overexpressed in both hematopoietic and epithelial tumors (Lessard and Sauvageau, 2003; Molofsky et al., 2003; Park et al., 2003). Our data do not exclude the possibility that USP22 might also contribute to a cancer stem cell-like phenotype when overexpressed in cancers together with BMI1. However, our results also indicate that USP22 overexpression likely has effects that are independent of BMI1.
USP22 overexpression in many different types of aggressive cancers makes it an attractive target for therapy development and, as such, it is essential to understand the full range of USP22 functions in both normal cells and in tumors. Genetic studies, as presented here, can provide unbiased definitions of the roles of USP22 in both developmental and disease contexts. Future work in mice will determine whether USP22 overexpression on its own is sufficient to drive or facilitate tumor formation or progression. Future analyses of human placentas are also needed to determine whether USP22 loss contributes to diseases such as preeclampsia or intrauterine growth restriction.
MATERIALS AND METHODS
Experimental mice
Animals were kept in regulated facilities, monitored daily and all procedures that involved animal handling were performed in accordance with the approved Institutional Animal Care and Use Committee (IACUC) protocols at the University of Texas MD Anderson Cancer Center.
We used two gene-trap ESC lines (RRS377 and XP0208) made by the Gene-Trap Consortium (www.genetrap.org). In both lines, the gene trap cassette was integrated within the first intron of Usp22, as validated by Southern blot analyses (Fig. S1A,B,D,E). We further mapped the exact integration site in RRS377 line to 428 bp downstream of exon 1 (Fig. S1C), whereas the integration site in XP0208 was 4075 bp downstream of exon 1 (Fig. S1F). We injected both lines into mouse blastocysts to generate mice bearing the gene trap alleles. Aggregation chimeras were generated with 129Ola ESCs using wild-type C57BL/6 morulae as previously described (Nagy et al., 1993). Two founder male chimeras with 80% or greater chimerism were backcrossed to wild-type C57BL6 females to produce progeny that were heterozygous for the βgeo allele. Heterozygous mice were intercrossed to produce homozygotes. Among 687 live pups born to mating between heterozygous parents from line RRS377, zero homozygous mutants (Usp22−/−) were found (Fig. S4A). Among 208 pups born to mating between heterozygous parents from line XP0208, only 15 homozygous mutants were observed, significantly less than expected from Mendelian ratios (Fig. S4D). As Usp22 transcript (Fig. S4B) and protein (Fig. S4C) were undetectable in tissues from these embryos, the Usp22 RRS377 allele is likely a null allele and will be referred to here as Usp22−/− for simplicity. On the contrary, homozygous mutant pups bearing the XP0208 allele had reduced but detectable levels of Usp22 mRNA (Fig. S4E), which indicates that this allele is hypomorphic. These mice, referred to as Usp22hyp/hyp, exhibited obvious growth retardation (Fig. S4F,G). To further confirm that loss of USP22 results in embryonic lethality, we designed and validated a pair of zinc-finger nucleases (ZFN) that targeted the start codon of Usp22. Injection of 741 zygotes (Fig. S5A,B) resulted in creation of five founder lines. Two lines were selected for additional study, and among 241 pups born to mating between heterozygous parents from these lines, no homozygous mutants were observed, which verified that the Usp22 gene is essential for mouse embryo survival (Fig. S6A,C).
Genomic PCR
Genomic DNA was prepared from embryonic yolk sac or mouse tail. Tissues were lysed through incubation with Proteinase K (x Manufacturer x) overnight at 55°C, followed by phenol extraction. DNA was precipitated by adding isopropanol and was washed in 70% ethanol. All embryos and mice were genotyped with specific primers that distinguished the mutant allele from the wild-type allele (Table S7). Mutation detection analysis in the ZFN alleles was performed using the Cel-I assay (Surveyor Mutation Detection Kit, Transgenomic). Briefly, 10 µl of PCR products, produced with primers that flanked the deletion generated by the ZFN pairs, were incubated using the following program: 95°C for 10min, a gradient from 95°C to 85°C with a decrease of 2°C every sec and a second gradient from 85°C to 25°C with a decrease of 0.1°C per sec. We then added 1 μl each of nuclease S and enhancer (Transgenomic) to digest the above reaction at 42°C for 30 min. The mixture was resolved on a 10% polyacrylamide TBE gel (Bio-Rad).
Southern blot hybridization
Genomic DNA (6 μg) was digested overnight in 30 units of restriction enzyme [SacI (New England Biolabs) for USP22 RRS377 allele and NcoI (New England Biolabs) for USP22 XP0208 allele] made up to 35 μl using 1× NEBuffer (1.1 for SacI and 3.1 for NcoI), and an additional 10 units of enzyme were added for 2-3 h on the second day. Samples were loaded on 0.8% agarose gels in Tris-acetate-EDTA (TAE) buffer and run at 26 V overnight. The gels were then denatured for 1 h in 1.5 M NaCl/0.5 M NaOH, neutralized for 40 min in 0.1 M Tris-HCl (pH 7.5)/0.5 M NaCl, washed with 20× SSC, and transferred for 24 h with 20× SSC on positively charged membranes (Roche). Probes were labeled with DIG-dUTP by PCR using the PCR DIG Probe Synthesis Kit (Roche). The membranes were UV-crosslinked, rinsed with water, air dried and prehybridized and hybridized with a 5′ probe or 3′ probe according to the manufacturer's instructions, using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche). The probes were heat denatured, added into the hybridization buffer and membranes were rotated overnight at temperatures calculated according to the manufacturer's instructions. The washing buffer (0.2× SSC, 0.5% SDS) was pre-warmed to 68°C, and the membranes were washed twice for 15 min at 68°C under shaking. Next, the probe-bound DNA fragments were detected using the DIG High Prime DNA Labeling and Detection Starter Kit II and X-ray films were exposed to the probed membrane to obtain the desired signal, following the manufacturer's instructions.
Conceptus dissections and X-gal staining
Conceptuses were dissected at various gestational ages: noon of the day that a vaginal plug was detected was defined as E0.5. For X-gal staining, specimens were fixed for 15 to 30 min in 1% formaldehyde, 0.2% glutaraldehyde, 0.02% NP-40, 5 mM EGTA, 2 mM MgCl2, 0.1 M sodium phosphate (pH 7.3). Specimens were stained whole for 4-24 h at 37°C in 0.1% X-gal, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 in buffer [0.02% NP-40, 0.01% deoxycholate, 2 mM MgCl2, 0.1 M sodium phosphate (pH 7.3)]. Following X-gal staining, conceptuses were post-fixed with 4% paraformaldehyde (PFA) for 12 h at 4°C.
Histological analysis
Conceptuses were fixed in 4% PFA for 30-120 min at 4°C, equilibrated in 15% sucrose in PBS for 4 h followed by 30% sucrose in PBS for 12 h at 4°C. Samples were post-fixed in 10% neutral buffered formalin for 24-48 h then transferred to 70% ethanol, dehydrated in an ethanol series to 100% ethanol, cleared in xylene substitute and infiltrated with paraffin. Samples were then embedded in paraffin, sectioned at 4 μm and put onto slides for staining with H&E or for immunoperoxidase staining with different antibodies (Table S6). For every marker analyzed at least three biological replicates were used, unless stated otherwise. Slides stained with H&E were further analyzed using surface morphometry, as described in Philipp et al. (2002).
RNA isolation and qRT-PCR
Embryos and placentas of different developmental stages were carefully dissected and snap frozen. Total RNA from embryos or tissues was extracted using RNeasy Plus Mini Kit (Qiagen). qRT-PCRs were measured using the 7500 Fast Real-Time PCR System (Applied Biosystems) using the SYBR Green method. The sequences for the gene-specific primers used are listed in Table S8. The relative expression of RNAs was calculated using the comparative Ct method with Gapdh as an internal control. Data are presented as normalized individual data points. Significant P values were corrected using the Benjamini–Hochberg (BH) method.
Western blotting
Embryos were carefully dissected, snap frozen, pulverized with a mortar and pestle in liquid nitrogen and resuspended in RIPA buffer. The extracts were dissolved in Laemmli buffer and boiled at 95°C for 5 min. The lysates (10-20 μg of total protein) were analyzed using western blotting with the anti-USP22 antibody described in Atanassov et al. (2016). Protein extraction from cultured cells was carried out using RIPA buffer. Western blots were carried out using 20-30 µg of protein. The antibodies used are listed in Table S6.
Whole transcriptome sequencing (RNA-seq)
For the experiment, total RNA from E9.5 wild-type and Usp22 mutant placentas was extracted using the RNeasy Plus Mini Kit. The purified RNA was treated with TURBO DNase (Thermo Fisher Scientific) and used for library preparation. Briefly, 1 µg of total RNA was used to prepare RNA-seq libraries with the Illumina TruSeq Stranded Total RNA Kit according to the manufacturer's protocol. The libraries were then sequenced using 2×75 base paired-end protocol on the Illumina HiSeq 2500; 35-77 million pairs of reads were generated per sample. Each pair of reads represents a cDNA fragment from the library. For mapping, the reads were mapped to the mouse genome (mm10) using TopHat (version 2.0.10) (Kim et al., 2013). By reads, the overall mapping rate is 85-94%; 73-92% fragments have both ends mapped to the mouse genome. For differential expression, the number of fragments in each known gene from the RefSeq database (Pruitt et al., 2014) (downloaded from UCSC Genome Browser on May 23, 2014) was enumerated using htseq-count from HTSeq package (version 0.6.0) (Anders et al., 2015). Genes with fewer than ten fragments in all the samples were removed before differential expression analysis. The differential expression between conditions was statistically assessed using the R/Bioconductor package DESeq (version 1.18.0) (Anders and Huber, 2010). Genes with FDR ≤0.05 and length >200 bp were described as differentially expressed. For the gene function and pathway analysis, data were analyzed using IPA software. Heat maps were generated using Cluster v3.0 and Java TreeView v1.1.6r4.
Functional enrichment analysis
For RNA-seq functional enrichment analysis, the GSEA pre-rank test was used to test the enrichment of gene sets for differentially expressed genes in mutant versus wild-type placentas (Subramanian et al., 2005). DAVID v6.7 (Huang et al., 2007) and PANTHER v11.0 (Mi et al., 2013) were also used to perform functional enrichment analysis of differentially expressed genes for target pathway analysis. Significant P values were adjusted using the Bonferroni method.
ESC culture, differentiation, tube formation assay and transwell migration assay
ESCs were routinely grown on gelatin-coated plates in Dulbecco's Modified Eagle's medium (DMEM)/high glucose medium supplemented with 15% (v/v) ESC-screened fetal bovine serum, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 1% (v/v) penicillin/streptomycin, 0.1 mM β-mercaptoethanol, 1000 U/ml of LIF/ESGRO, 1 μM PD0325901 and 3 μM 1-Azakenpaullone, and passaged with 0.25% trypsin every 2-3 days. Induced endothelial cells and pericytes were generated using the protocol of Orlova et al. (2014) with some modifications for mouse ESCs. Briefly, mouse ESCs were cultured on ColIV-coated plates for 2 days in DFNK medium supplemented with B27, N2 and ROCK inhibitor (Tocris Bioscience) (2.5 µg/ml). On day 2 the medium was replaced with MEDF (DMEM-based low serum medium) for 24 h, then supplemented with activin A (25 ng/ml) for two additional days to induce differentiation towards mesendoderm. On day 6, medium was replaced with MEDF supplemented with VEGF 164 (50 ng/ml) and SB431542 (10 µM) (Stemcell Technologies) to induce differentiation towards vascular progenitors. The medium was replaced every day after removing the dead cells carefully using a PBS wash. On day 10 medium was replaced using an endothelial differentiation medium that was supplemented with VEGF 164 (30 ng/ml) and basic FGF (20 ng/ml) (Sigma-Aldrich) for 3-4 days. Finally, on day 14 the medium was replaced with pericyte differentiation medium that was supplemented with PDGF-BB (4 ng/ml) and TGFβ3 (2 ng/ml) for 3-4 more days to induce pericyte fate. Induced endothelial cells and pericytes were maintained in endothelial and pericyte maintenance medium respectively. The cells were used from passages 2-5. Both the differentiated endothelial cells and the pericytes were seeded and cultured at low confluency and experiments were performed within 48 h of the passage. For the tube formation assay, endothelial cells and pericytes were trypsinized, counted and resuspended in human endothelial/serum-free growth medium (EC-SFM). The cells were diluted to a concentration of 1×105 cells/ml and were combined in a ratio of 1:4 endothelial cells to pericytes. They were placed in an angiogenesis μ-slide (ibidi) that was coated with ColIV. The cells were placed in the incubator at 37°C and 5% CO2. They were fed at day 3 with EC-SFM and they were fixed and stained for imaging at day 8. The transwell migration assay of induced endothelial cells was performed as previously described (Chen, 2005). Briefly, the migration assay was performed in Transwell inserts in 24-well plates (Corning). We placed 1×105 cells in 500 μl EC-SFM in the upper chamber, and 750 μl of the same medium containing 20 ng/ml of VEGF was placed in the lower chamber. The plates were incubated for 4 h at 37°C in 5% CO2. Cells were washed with 600 μl PBS twice and fixed in 100% methanol for 5 min. Non-migrating cells in the upper chambers were removed with a cotton swab; migrated cells were stained with 0.5% crystal violet for 20 min and photographed under an optical microscope (Stemi 2000c, Zeiss). Five random fields were chosen in each well and the cell number was quantified manually.
Cell cycle analysis
Approximately 4×105 cells for each sample were harvested by trypsinization, washed in PBS, centrifuged at 500 g for 5 min and pellets were resuspended in PBS. To each sample 100% ice cold ethanol was added dropwise while vortexing, and samples were incubated overnight at −20°C. Cells were harvested by centrifugation at 400 g for 2 min, supernatant was discarded and pellets resuspended in PBS. Cells were treated with 20 µg/ml RNase A (Roche) for 30 min at 37°C and the cell suspension was moved to flow cytometry tubes, passing the suspension through cell strainer caps. Propidium iodide was then added to each sample (20 µg/ml final concentration) and samples were analyzed using a BD LSRFortessa cell analyzer (BD Biosciences).
Immunofluorescence, confocal imaging and multiphoton image acquisition of embryonic hearts
For confocal microscopy imaging of cultured cells and heart cryosections, cells or embryos were washed in PBS and fixed in 2% PFA for 5-15 min at 4°C, then washed briefly with methanol followed by incubation in pre-chilled (at −20°C) methanol for 5 min at −20°C. Alternatively, phalloidin-stained cells or embryos were fixed in 4% PFA for 15 min at room temperature followed by permeabilization with 1% Triton X-100. Cells were washed with PBS 3× for 5 min each. Nonspecific binding was blocked by adding blocking solution (0.5% gelatin in PBS) for 30 min at room temperature. The blocked cells were incubated with primary antibodies (Table S6) diluted in washing media (0.1% gelatin, 1% goat or donkey serum in PBS) overnight at 4°C. After five washes in washing buffer, cells were incubated for 40 min with secondary antibodies. DAPI staining was performed after washing off the secondary antibodies. Cells on coverslips were mounted using Prolong Gold (Life Technologies) and cells in wells were mounted with Slowfade Gold (Life Technologies). Laser scanning confocal microscopy was performed using a Zeiss LSM880 and 20× (0.8 NA) or 40× oil (1.4 NA) Plan/Apo objectives and with a pinhole aperture of 1-1.5 AU. For multiphoton imaging, whole embryos were permeabilized with 0.5% Triton and stained using the same antibodies and conditions except primary antibody staining was performed over 1-2 days in PBS with 0.5% Triton X-100, and secondary antibody and DAPI staining were performed overnight. Embryos were mounted in a viewing dish in a minimal volume of melted and cooled (45°C) 0.5% Ultrapure agarose (Invitrogen) prepared in sterile ddH2O. Two-photon images of embryonic hearts were acquired using an upright Leica TCS SP8 DIVE multiphoton microscope with Insight X3 (Spectra-Physics) infrared laser and a 25×/0.95 NA FLUOTAR water immersion objective. For acquisition of images to encompass the lateral and axial dimensions of the E10 heart (∼1200×800 μm), mosaic tiling with a 10% overlap was employed over the designated region of interest with a z-stack depth range from 414-280 μm and a z-interval of 1.7 μm. Sequential scanning was performed using the 1040 nm fixed line to excite AF568 (phalloidin) and the tunable laser line for AF488 (PECAM1) and DAPI using 950 nm and 750 nm, respectively. The emission was captured separately on non-descanned hybrid (HyD) detectors. Stitching to create a seamless mosaic was performed using Leica LASX software, and Imaris 9.1 (Bitplane) was used to render 3D volumes and visualize vasculature and heart tissue using the green (PECAM1) and red (phalloidin) channels, respectively. Wild-type and Usp22 mutants were processed in parallel using the same parameters and the Imaris Blend and Orthoslicer features.
Histone purification from wild-type and Usp22 mutant induced endothelial cells
Total histones from the induced endothelial cells were prepared using a Histone Purification Mini Kit (Active Motif). Laemmli buffer was added and the lysates were analyzed using western blotting.
Supplementary Material
Acknowledgements
We thank Dr Jan Parker-Thornburg and Debra Hollowel for assistance with mouse line generation. We thank Amanda Martin for animal husbandry, and Tim Macatee and Pam Whitney for excellent technical support. We also thank Dr Ondine Cleaver for critical reading of the manuscript. Sequencing was done by the University of Texas MD Anderson Cancer Center (MDACC) Science Park NGS Core, supported by Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Support Grants RP120348 and RP170002. Bioinformatics support was made possible by CPRIT Core Facility Support Grants RP120348 and RP170002 and by support from the MDACC Center for Cancer Epigenetics. Animal services were supported by the MDACC Support Grant CA16672 from the National Cancer Institute. Histological analysis was performed at the Science Park histology core, partially supported by the MDACC Center for Environmental and Molecular Carcinogenesis. Confocal and multiphoton imaging and flow cytometry were performed at the Science Park Flow Cytometry and Cell Imaging Core and supported by CPRIT Core Facility Grant RP170628.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.K., D.G.T., S.Y.R.D.; Methodology: E.K., L.W., A.C.S., H.-P.C., X.K., K.L., Y.L., J.S., C.R.J., A.S., M.W., Y.C.C., B.S.A., S.Y.R.D.; Software: E.K., H.-P.C., Y.L., J.S.; Validation: E.K., L.W., A.C.S., H.-P.C., X.K., K.L., Y.L., J.S., C.R.J., A.S., M.W., Y.C.C., B.S.A.; Formal analysis: L.W., A.C.S., X.K., Y.L., C.R.J.; Investigation: E.K., L.W., A.C.S., C.R.J., M.W., S.Y.R.D.; Resources: B.S.A.; Data curation: E.K., L.W., D.G.T., S.Y.R.D.; Writing - original draft: E.K., S.Y.R.D.; Writing - review & editing: E.K., L.W., A.C.S., C.R.J., D.G.T.; Visualization: E.K.; Supervision: S.Y.R.D.; Project administration: E.K., S.Y.R.D.; Funding acquisition: S.Y.R.D.
Funding
This work was largely supported by the National Institutes of Health grants R01GM096472 and R01HD094400 to S.Y.R.D. A.C.S. was funded through a fellowship from the Cancer Prevention Research Institute of Texas Training Program (RP170067). Deposited in PMC for release after 12 months.
Data availability
RNA-seq data have been deposited in GEO under accession number GSE126069.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.174037.supplemental
References
- Anders S. and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson-Cartwright L., Dawson K., Holmyard D., Fisher S. J., Lazzarini R. A. and Cross J. C. (2000). The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25, 311-314. 10.1038/77076 [DOI] [PubMed] [Google Scholar]
- Atanassov B. S. and Dent S. Y. (2011). USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 12, 924-930. 10.1038/embor.2011.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov B. S., Evrard Y. A., Multani A. S., Zhang Z., Tora L., Devys D., Chang S. and Dent S. Y. (2009). Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell 35, 352-364. 10.1016/j.molcel.2009.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov B. S., Mohan R. D., Lan X., Kuang X., Lu Y., Lin K., McIvor E., Li W., Zhang Y., Florens L. et al. (2016). ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol. Cell 62, 558-571. 10.1016/j.molcel.2016.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthouze M., Venkataramanan V., Li Y. and Shenoy S. K. (2009). The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 28, 1684-1696. 10.1038/emboj.2009.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J., Wang C.-Y., Baptista T., Vincent S. D., Hsiao W.-C., Stierle M., Kao C.-F., Tora L. and Devys D. (2014). The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 28, 1999-2012. 10.1101/gad.250225.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. C. (2005). Boyden chamber assay. Methods Mol. Biol. 294, 15-22. [DOI] [PubMed] [Google Scholar]
- Conway S. J., Kruzynska-Frejtag A., Kneer P. L., Machnicki M. and Koushik S. V. (2003). What cardiovascular defect does my prenatal mouse mutant have, and why? Genesis 35, 1-21. 10.1002/gene.10152 [DOI] [PubMed] [Google Scholar]
- Cross J. C., Simmons D. G. and Watson E. D. (2003). Chorioallantoic morphogenesis and formation of the placental villous tree. Ann. N. Y. Acad. Sci. 995, 84-93. 10.1111/j.1749-6632.2003.tb03212.x [DOI] [PubMed] [Google Scholar]
- Dackor J., Li M. and Threadgill D. W. (2009). Placental overgrowth and fertility defects in mice with a hypermorphic allele of epidermal growth factor receptor. Mamm. Genome 20, 339-349. 10.1007/s00335-009-9189-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson M. C., Martin J. S., Cousins F. M., Kulkarni A. B., Karlsson S. and Akhurst R. J. (1995). Defective haematopoiesis and vasculogenesis in transforming growth factor-beta 1 knock out mice. Development 121, 1845-1854. [DOI] [PubMed] [Google Scholar]
- Ding F., Bao C., Tian Y., Xiao H., Wang M., Xie X., Hu F. and Mei J. (2015). USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. Int. J. Mol. Sci. 16, 307-320. 10.3390/ijms16010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French W. J., Creemers E. E. and Tallquist M. D. (2008). Platelet-derived growth factor receptors direct vascular development independent of vascular smooth muscle cell function. Mol. Cell. Biol. 28, 5646-5657. 10.1128/MCB.00441-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro V. J., Stanek T. J., Peck A. R., Sun Y., Wang F., Qie S., Knudsen K. E., Rui H., Butt T., Diehl J. A. et al. (2018). Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc. Natl. Acad. Sci. USA 115, E9298-E9307. 10.1073/pnas.1807704115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber H. P., McMurtrey A., Kowalski J., Yan M., Keyt B. A., Dixit V. and Ferrara N. (1998). Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3'-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J. Biol. Chem. 273, 30336-30343. 10.1074/jbc.273.46.30336 [DOI] [PubMed] [Google Scholar]
- Glinsky G. V., Berezovska O. and Glinskii A. B. (2005). Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115, 1503-1521. 10.1172/JCI23412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra J. R., Ward J. M., Porter F. D., Wagner J. R., Devor D. E., Grinberg A., Emmert-Buck M. R., Westphal H., Klausner R. D. and Linehan W. M. (1997). Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 94, 9102-9107. 10.1073/pnas.94.17.9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner G. C., Davis V., Li H., McCoy M. J., Sharpe A. and Cybulsky M. I. (1995). Targeted disruption of the murine VCAM1 gene: essential role of VCAM-1 in chorioallantoic fusion and placentation. Genes Dev. 9, 1-14. 10.1101/gad.9.1.1 [DOI] [PubMed] [Google Scholar]
- Hellstrom M., Kalen M., Lindahl P., Abramsson A. and Betsholtz C. (1999). Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047-3055. [DOI] [PubMed] [Google Scholar]
- Hemberger M. (2010). Genetic-epigenetic intersection in trophoblast differentiation: implications for extraembryonic tissue function. Epigenetics 5, 24-29. 10.4161/epi.5.1.10589 [DOI] [PubMed] [Google Scholar]
- Hirashima M., Kataoka H., Nishikawa S., Matsuyoshi N. and Nishikawa S. (1999). Maturation of embryonic stem cells into endothelial cells in an in vitro model of vasculogenesis. Blood 93, 1253-1263. [PubMed] [Google Scholar]
- Hirsch C. L., Coban Akdemir Z., Wang L., Jayakumaran G., Trcka D., Weiss A., Hernandez J. J., Pan Q., Han H., Xu X. et al. (2015). Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 29, 803-816. 10.1101/gad.255109.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi K. K., Rohovsky S. A. and D'Amore P. A. (1998). PDGF, TGF-beta, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J. Cell Biol. 141, 805-814. 10.1083/jcb.141.3.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Yang D., Zhang H., Liu W., Zhao Y., Lu H., Meng Q., Pang H., Chen X., Liu Y. et al. (2015). USP22 promotes tumor progression and induces epithelial-mesenchymal transition in lung adenocarcinoma. Lung Cancer 88, 239-245. 10.1016/j.lungcan.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Tan Q., Collins J. R., Alvord W. G., Roayaei J., Stephens R., Baseler M. W., Lane H. C. and Lempicki R. A. (2007). The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, R183 10.1186/gb-2007-8-9-r183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.-P., Chen C., Hao J., Huang J.-Y., Liu P.-Q. and Huang H.-Q. (2015). AGEs-RAGE system down-regulates Sirt1 through the ubiquitin-proteasome pathway to promote FN and TGF-beta1 expression in male rat glomerular mesangial cells. Endocrinology 156, 268-279. 10.1210/en.2014-1381 [DOI] [PubMed] [Google Scholar]
- Ji M., Shi H., Xie Y., Zhao Z., Li S., Chang C., Cheng X. and Li Y. (2015). Ubiquitin specific protease 22 promotes cell proliferation and tumor growth of epithelial ovarian cancer through synergy with transforming growth factor beta1. Oncol. Rep. 33, 133-140. 10.3892/or.2014.3580 [DOI] [PubMed] [Google Scholar]
- Kay H. H., Zhu S. and Tsoi S. (2007). Hypoxia and lactate production in trophoblast cells. Placenta 28, 854-860. 10.1016/j.placenta.2006.11.011 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. and Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinsky R. L., Wegwitz F., Hellbach N., Dobbelstein M., Mansouri A., Vogel T., Begus-Nahrmann Y. and Johnsen S. A. (2015). Usp22 deficiency impairs intestinal epithelial lineage specification in vivo. Oncotarget 6, 37906-37918. 10.18632/oncotarget.5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E., Hirsch C. L. and Dent S. Y. R. (2010). Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22, 374-382. 10.1016/j.ceb.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee L., Baldwin H. S., Shen H. M., Stewart C. L., Buck C., Buck C. A. and Labow M. A. (1995). Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development 121, 489-503. [DOI] [PubMed] [Google Scholar]
- Lampugnani M. G., Orsenigo F., Gagliani M. C., Tacchetti C. and Dejana E. (2006). Vascular endothelial cadherin controls VEGFR-2 internalization and signaling from intracellular compartments. J. Cell Biol. 174, 593-604. 10.1083/jcb.200602080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Goumans M. J., Sjostrand L. J., van Rooijen M. A., Ward D., Leveen P., Xu X., ten Dijke P., Mummery C. L. and Karlsson S. (2001). Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J. 20, 1663-1673. 10.1093/emboj/20.7.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J. and Sauvageau G. (2003). Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255-260. 10.1038/nature01572 [DOI] [PubMed] [Google Scholar]
- Lin Z., Yang H., Kong Q., Li J., Lee S.-M., Gao B., Dong H., Wei J., Song J., Zhang D. D. et al. (2012). USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46, 484-494. 10.1016/j.molcel.2012.03.024 [DOI] [PubMed] [Google Scholar]
- Lin Z., Tan C., Qiu Q., Kong S., Yang H., Zhao F., Liu Z., Li J., Kong Q., Gao B. et al. (2015). Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov. 1, 15028 10.1038/celldisc.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl P., Johansson B. R., Leveen P. and Betsholtz C. (1997). Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242-245. 10.1126/science.277.5323.242 [DOI] [PubMed] [Google Scholar]
- Liu Y.-L., Jiang S.-X., Yang Y.-M., Xu H., Liu J.-L. and Wang X.-S. (2012). USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem. Biophys. 62, 229-235. 10.1007/s12013-011-9287-0 [DOI] [PubMed] [Google Scholar]
- Lv L., Xiao X.-Y., Gu Z.-H., Zeng F.-Q., Huang L.-Q. and Jiang G.-S. (2011). Silencing USP22 by asymmetric structure of interfering RNA inhibits proliferation and induces cell cycle arrest in bladder cancer cells. Mol. Cell. Biochem. 346, 11-21. 10.1007/s11010-010-0585-4 [DOI] [PubMed] [Google Scholar]
- Majmundar A. J., Wong W. J. and Simon M. C. (2010). Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294-309. 10.1016/j.molcel.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cerdeno V., Lemen J. M., Chan V., Wey A., Lin W., Dent S. R. and Knoepfler P. S. (2012). N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS ONE 7, e39456 10.1371/journal.pone.0039456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A. and Thomas P. D. (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377-D386. 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A. V., Pardal R., Iwashita T., Park I.-K., Clarke M. F. and Morrison S. J. (2003). Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425, 962-967. 10.1038/nature02060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau J. L., Artap S. T., Shi H., Chapman G., Leone G., Sparrow D. B. and Dunwoodie S. L. (2014). Cited2 is required in trophoblasts for correct placental capillary patterning. Dev. Biol. 392, 62-79. 10.1016/j.ydbio.2014.04.023 [DOI] [PubMed] [Google Scholar]
- Nagai A., Takebe K., Nio-Kobayashi J., Takahashi-Iwanaga H. and Iwanaga T. (2010). Cellular expression of the monocarboxylate transporter (MCT) family in the placenta of mice. Placenta 31, 126-133. 10.1016/j.placenta.2009.11.013 [DOI] [PubMed] [Google Scholar]
- Nagy A., Rossant J., Nagy R., Abramow-Newerly W. and Roder J. C. (1993). Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90, 8424-8428. 10.1073/pnas.90.18.8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R., Falck P., Hellstrom M., Lindahl P., Bostrom H., Franklin G., Ahrlund-Richter L., Pollard J., Soriano P. and Betsholtz C. (1999). PDGFB regulates the development of the labyrinthine layer of the mouse fetal placenta. Dev. Biol. 212, 124-136. 10.1006/dbio.1999.9306 [DOI] [PubMed] [Google Scholar]
- Orlova V. V., van den Hil F. E., Petrus-Reurer S., Drabsch Y., Ten Dijke P. and Mummery C. L. (2014). Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat. Protoc. 9, 1514-1531. 10.1038/nprot.2014.102 [DOI] [PubMed] [Google Scholar]
- Oshima M., Oshima H. and Taketo M. M. (1996). TGF-beta receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev. Biol. 179, 297-302. 10.1006/dbio.1996.0259 [DOI] [PubMed] [Google Scholar]
- Park I.-K., Qian D., Kiel M., Becker M. W., Pihalja M., Weissman I. L., Morrison S. J. and Clarke M. F. (2003). Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302-305. 10.1038/nature01587 [DOI] [PubMed] [Google Scholar]
- Perez-Garcia V., Fineberg E., Wilson R., Murray A., Mazzeo C. I., Tudor C., Sienerth A., White J. K., Tuck E., Ryder E. J. et al. (2018). Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 555, 463-468. 10.1038/nature26002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp M., Brede M. E., Hadamek K., Gessler M., Lohse M. J. and Hein L. (2002). Placental alpha(2)-adrenoceptors control vascular development at the interface between mother and embryo. Nat. Genet. 31, 311-315. 10.1038/ng919 [DOI] [PubMed] [Google Scholar]
- Pruitt K. D., Brown G. R., Hiatt S. M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C. M., Hart J., Landrum M. J., McGarvey K. M. et al. (2014). RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42, D756-D763. 10.1093/nar/gkt1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. and Cross J. C. (2001). Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2, 538-548. 10.1038/35080570 [DOI] [PubMed] [Google Scholar]
- Schuh A. C., Faloon P., Hu Q. L., Bhimani M. and Choi K. (1999). In vitro hematopoietic and endothelial potential of flk-1(−/−) embryonic stem cells and embryos. Proc. Natl. Acad. Sci. USA 96, 2159-2164. 10.1073/pnas.96.5.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F., Rossant J., Yamaguchi T. P., Gertsenstein M., Wu X.-F., Breitman M. L. and Schuh A. C. (1995). Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62-66. 10.1038/376062a0 [DOI] [PubMed] [Google Scholar]
- Shalaby F., Ho J., Stanford W. L., Fischer K.-D., Schuh A. C., Schwartz L., Bernstein A. and Rossant J. (1997). A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell 89, 981-990. 10.1016/S0092-8674(00)80283-4 [DOI] [PubMed] [Google Scholar]
- Shu X., Wu W., Mosteller R. D. and Broek D. (2002). Sphingosine kinase mediates vascular endothelial growth factor-induced activation of ras and mitogen-activated protein kinases. Mol. Cell. Biol. 22, 7758-7768. 10.1128/MCB.22.22.7758-7768.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. G., Fortier A. L. and Cross J. C. (2007). Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 304, 567-578. 10.1016/j.ydbio.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Simmons D. G., Natale D. R. C., Begay V., Hughes M., Leutz A. and Cross J. C. (2008). Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 135, 2083-2091. 10.1242/dev.020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman R. T., Stanek T. J., Esteso P., Gearhart J. D., Knudsen K. E. and McMahon S. B. (2013). The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2). J. Biol. Chem. 288, 24234-24246. 10.1074/jbc.M113.469783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara Y., Minowa O., Mori C., Shiota K., Kuno J., Noda T. and Kitamura N. (1995). Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 373, 702-705. 10.1038/373702a0 [DOI] [PubMed] [Google Scholar]
- Ueno M., Lee L. K., Chhabra A., Kim Y. J., Sasidharan R., Van Handel B., Wang Y., Kamata M., Kamran P., Sereti K. I. et al. (2013). c-Met-dependent multipotent labyrinth trophoblast progenitors establish placental exchange interface. Dev. Cell 27, 373-386. 10.1016/j.devcel.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters B. J., Wachi S., Mavrich T. N., Andersen B. E., Jena P., Sinnamon A. J., Jain P., Rolleri N. S., Jiang C., Hemeryck-Walsh C. et al. (2011). A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 41, 480-492. 10.1016/j.molcel.2011.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayachandran V., Reja R., Rossi M. J., Park B., Rieber L., Mittal C., Mahony S. and Pugh B. F. (2018). Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res. 28, 357-366. 10.1101/gr.226761.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. and Dent S. Y. (2014). Functions of SAGA in development and disease. Epigenomics 6, 329-339. 10.2217/epi.14.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Koutelou E., Hirsch C., McCarthy R., Schibler A., Lin K., Lu Y., Jeter C., Shen J., Barton M. C. et al. (2018). GCN5 regulates FGF signaling and activates selective MYC target genes during early embryoid body differentiation. Stem Cell Reports 10, 287-299. 10.1016/j.stemcr.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E. D. and Cross J. C. (2005). Development of structures and transport functions in the mouse placenta. Physiology 20, 180-193. 10.1152/physiol.00001.2005 [DOI] [PubMed] [Google Scholar]
- Wilde J. J., Siegenthaler J. A., Dent S. Y. R. and Niswander L. A. (2017). Diencephalic size is restricted by a novel interplay between GCN5 acetyltransferase activity and retinoic acid signaling. J. Neurosci. 37, 2565-2579. 10.1523/JNEUROSCI.2121-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withington S. L., Scott A. N., Saunders D. N., Lopes Floro K., Preis J. I., Michalicek J., Maclean K., Sparrow D. B., Barbera J. P. M. and Dunwoodie S. L. (2006). Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev. Biol. 294, 67-82. 10.1016/j.ydbio.2006.02.025 [DOI] [PubMed] [Google Scholar]
- Xu W., Edmondson D. G., Evrard Y. A., Wakamiya M., Behringer R. R. and Roth S. Y. (2000). Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26, 229-232. 10.1038/79973 [DOI] [PubMed] [Google Scholar]
- Yamashita J., Itoh H., Hirashima M., Ogawa M., Nishikawa S., Yurugi T., Naito M., Nakao K. and Nishikawa S.-I. (2000). Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature 408, 92-96. 10.1038/35040568 [DOI] [PubMed] [Google Scholar]
- Yang J. T., Rayburn H. and Hynes R. O. (1995). Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development 121, 549-560. [DOI] [PubMed] [Google Scholar]
- Zhang X.-Y., Pfeiffer H. K., Thorne A. W. and McMahon S. B. (2008a). USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 7, 1522-1524. 10.4161/cc.7.11.5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Varthi M., Sykes S. M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A. W., Berger S. L. and McMahon S. B. (2008b). The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29, 102-111. 10.1016/j.molcel.2007.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhou F., Drabsch Y., Gao R., Snaar-Jagalska B. E., Mickanin C., Huang H., Sheppard K. A., Porter J. A., Lu C. X. et al. (2012). USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-beta type I receptor. Nat. Cell Biol. 14, 717-726. 10.1038/ncb2522 [DOI] [PubMed] [Google Scholar]
- Zhou A., Lin K., Zhang S., Chen Y., Zhang N., Xue J., Wang Z., Aldape K. D., Xie K., Woodgett J. R. et al. (2016). Nuclear GSK3beta promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat. Cell Biol. 18, 954-966. 10.1038/ncb3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y.-J., Liao Z.-W., Yu H.-W., Song X.-L., Liu Y., Shi X.-Y., Lin X.-D. and Zhou T.-C. (2015). ShRNA-mediated silencing of the ubiquitin-specific protease 22 gene restrained cell progression and affected the Akt pathway in nasopharyngeal carcinoma. Cancer Biol. Ther. 16, 88-96. 10.4161/15384047.2014.987029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.