ABSTRACT
Hepatic fibrosis is the common end stage to a variety of chronic liver injuries and is characterized by an excessive deposition of extracellular matrix (ECM), which disrupts the liver architecture and impairs liver function. The fibrous lesions are produced by myofibroblasts, which differentiate from hepatic stellate cells (HSC). The myofibroblast’s transcriptional networks remain poorly characterized. Previous studies have shown that the Forkhead box F1 (FOXF1) transcription factor is expressed in HSCs and stimulates their activation during acute liver injury; however, the role of FOXF1 in the progression of hepatic fibrosis is unknown. In the present study, we generated αSMACreER;Foxf1fl/fl mice to conditionally inactivate Foxf1 in myofibroblasts during carbon tetrachloride-mediated liver fibrosis. Foxf1 deletion increased collagen depositions and disrupted liver architecture. Timp2 expression was significantly increased in Foxf1-deficient mice while MMP9 activity was reduced. RNA sequencing of purified liver myofibroblasts demonstrated that FOXF1 inhibits expression of pro-fibrotic genes, Col1α2, Col5α2, and Mmp2 in fibrotic livers and binds to active repressors located in promotors and introns of these genes. Overexpression of FOXF1 inhibits Col1a2, Col5a2, and MMP2 in primary murine HSCs in vitro. Altogether, FOXF1 prevents aberrant ECM depositions during hepatic fibrosis by repressing pro-fibrotic gene transcription in myofibroblasts and HSCs.
KEY WORDS: FOXF1, Hepatic fibrosis, Myofibroblast, Hepatic stellate cell, Carbon tetrachloride liver injury
Summary: The transcription factor FOXF1 is expressed in liver fibroblasts and regulates fibrosis by repressing pro-fibrotic genes. FOXF1 prevents aberrant extracellular matrix depositions during hepatic fibrosis.
INTRODUCTION
The liver is the body's filter and insults can result from a variety of infectious, toxic and metabolic agents. Hepatic fibrosis is the common end stage to a multitude of liver diseases (Civan, 2016) and is characterized by an excessive deposition of extracellular matrix (ECM) and collagen (Cheng and Mahato, 2007). Novel animal models of hepatic fibrosis are greatly needed to identify molecular mechanisms responsible for the disease pathogenesis and for the development of therapeutic agents. Hepatic stellate cells (HSC) reside in the space of Disse and are characterized by their storage of lipids when in the quiescent state (Yin et al., 2013; Croci et al., 2013). During fibrogenesis, quiescent HSCs differentiate into myofibroblasts (MF) in response to cytokine signaling from damaged hepatocytes and immune cells after liver insult. MFs secrete ECM and collagen to encapsulate the site of injury and shield the liver from plaguing insults (Cheng and Mahato, 2007). While HSCs and MFs make up only a small number of cells in liver tissue, they are the main contributors of ECM and collagen during liver repair and fibrogenesis (Brenner et al., 2012; Fausther et al., 2013). The TGF-β and PDGF signaling pathways play key roles in hepatic fibrosis and HSC activation (Makarev et al., 2016). TGF-β signaling stimulates cellular transdifferentiation of HSCs to MFs (Hellerbrand et al., 1999; Bachem et al., 1993), whereas PDGF signaling induces cellular proliferation in fibrotic foci (Wong et al., 1994; Kinnman et al., 2002).
The Forkhead Box F1 (FOXF1) transcription factor is expressed in human and murine HSCs and is important in regulating stellate cell activation after acute liver injury (Kalinichenko et al., 2003). In the advanced disease state of hepatocellular carcinoma (HCC), which is associated with significant fibrotic depositions, FOXF1 expression has been shown to be significantly decreased (Hodo et al., 2013). Foxf1−/− mice are embryonic lethal due to severe developmental abnormalities in the yolk sac and allantois (Mahlapuu et al., 2001). Murine haploinsufficiency of Foxf1 causes lung hypoplasia, loss of alveolar capillaries in the lung and gall bladder agenesis (Kalinichenko et al., 2002; Bolte et al., 2018), and was associated with delayed lung and liver repair. After acute liver injury by carbon tetrachloride (CCl4), Foxf1+/− mice exhibited diminished activation of HSCs and delayed liver repair, indicating that FOXF1 is essential for liver repair after acute liver injury (Kalinichenko et al., 2003). Foxf1 siRNA delivered to mice through nanoparticles prevented activation of HSCs and subsequent collagen deposition after cholestatic liver injury (Abshagen et al., 2015). While these studies have shown that FOXF1 is required for activation of HSCs after acute liver injury, the role of FOXF1 in MFs and in the progression of fibrotic responses remains unknown.
In the present study, we generated a novel genetic mouse model to conditionally delete Foxf1 from MFs (αSMACreER;Foxf1fl/fl). During chronic liver injury, deletion of Foxf1 in MFs exacerbated hepatic fibrosis, increased collagen deposition and stimulated expression of profibrotic genes in the liver tissue. Our studies indicate that Foxf1 expression in MFs is critical to prevent MF accumulation and collagen deposition during liver fibrosis.
RESULTS
Deletion of Foxf1 in αSMA-positive cells exacerbates CCl4-induced hepatic fibrosis
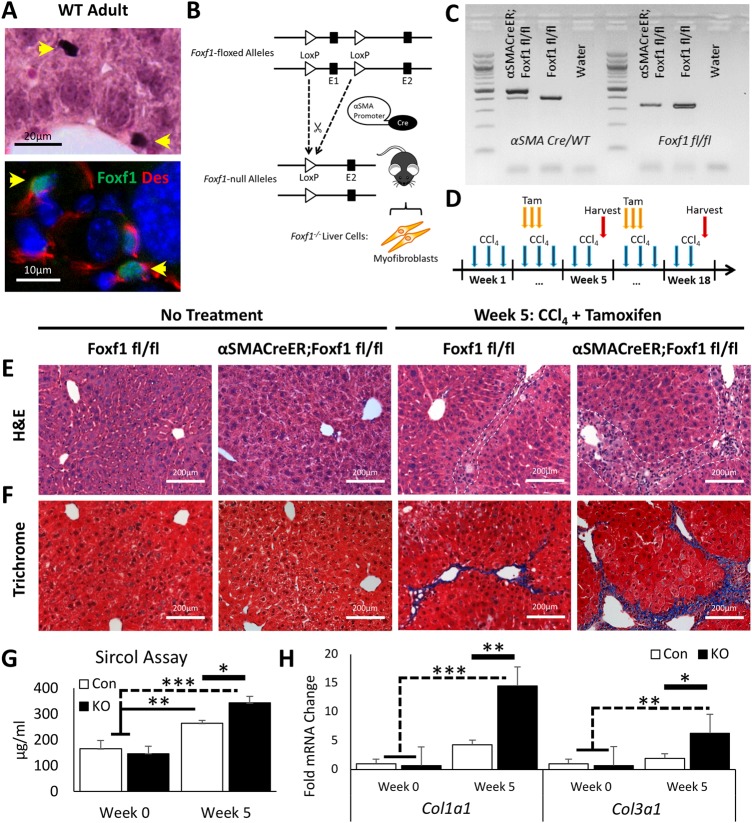
Previous studies demonstrated that FOXF1 is present in HSCs in murine developing and adult livers (Kalinichenko et al., 2003; Kim et al., 2005). Consistent with these studies, FOXF1 staining was detected in livers of e12.5-e17.5 mouse embryos as well as in mesenchyme of stomach and intestine (Fig. S1). In adult mice, FOXF1 is specifically expressed in the liver parenchyma but not in endothelial or smooth muscle cells surrounding the portal vein or hepatic artery (Kalinichenko et al., 2003) (Fig. 1A; Fig. S1), and FOXF1 staining co-localized with desmin (DES) (Fig. 1A), a known marker of HSCs (Yokoi et al., 1984). To investigate the role of Foxf1 in liver fibrosis, we utilized a conditional knockout approach. Transgenic mice containing a tamoxifen-inducible αSMA-CreER transgene and two Foxf1-floxed alleles (αSMACreER;Foxf1fl/fl) were generated by breeding αSMA-CreER and Foxf1fl/fl mice (Fig. 1B,C). Hepatic fibrosis was induced by chronic liver injury using multiple administrations of CCl4, which is known to increase fibrotic depositions and disrupt liver architecture in experimental mice (Martinez et al., 2014). Tamoxifen was given three times per week to achieve a continuous deletion of Foxf1 in αSMA-positive MFs (Fig. 1D) that derive from HSCs after liver injury (Mederacke et al., 2013). Morphological analysis of liver sections revealed increased fibrotic deposition in CCl4-treated αSMACreER;Foxf1−/− livers compared to controls as shown by H&E (Fig. 1E; Fig. S2) and Masson's Trichrome staining (Fig. 1F; Fig. S2). Increased fibrosis in αSMACreER;Foxf1−/− livers was confirmed by significant increases in collagen levels by Sircol (Fig. 1G) and hydroxyproline (Fig. S2) assays as well as by qRT-PCR for Col1α1 and Col3α1 mRNAs (Fig. 1H). Treatment with tamoxifen alone (without CCl4) did not affect liver architecture or induce liver fibrosis (Fig. S2). Thus, deletion of Foxf1 from MFs accelerates liver fibrosis after chronic liver injury.
Fig. 1.
Hepatic fibrosis is increased after CCl4 injury in mice with FOXF1 deficiency. (A) FOXF1 co-localizes with DES in hepatic stellate cells in adult mice. (B) Diagram demonstrates αSMA-CreER transgene with LoxP sites flanking the Foxf1 Exon 1 (encoding DNA-binding domain). (C) DNA gel shows genotypes of Foxf1fl/fl and αSMACreER;Foxf1fl/fl mice. (D) Diagram illustrates CCl4 and tamoxifen (Tam) treatment protocol. (E,F) H&E and Masson's trichrome staining show fibrotic depositions after five weeks of CCl4 treatment. Fibrosis was increased in livers from αSMACreER;Foxf1−/− mice. White dashed lines indicate fibrotic lesion boundaries. (G) Collagen deposition was quantitated using the Sircol assay. n=2 mice per group in week 0; n=4 mice per group in week 5. (H) qRT-PCR analysis demonstrates significant increases in Col1α1 and Col3α1 mRNAs in livers from αSMACreER;Foxf1−/− mice. n=3 mice per group in week 0; n=5 mice per group in week 5. Untreated livers from Foxf1fl/fl and αSMACreER;Foxf1fl/fl mice were used as normal controls. mRNAs were normalized to Actb. *P<0.05, **P<0.01, ***P<0.001.
FOXF1 expression is decreased in hepatic myofibroblasts of αSMACreER;Foxf1−/− mice
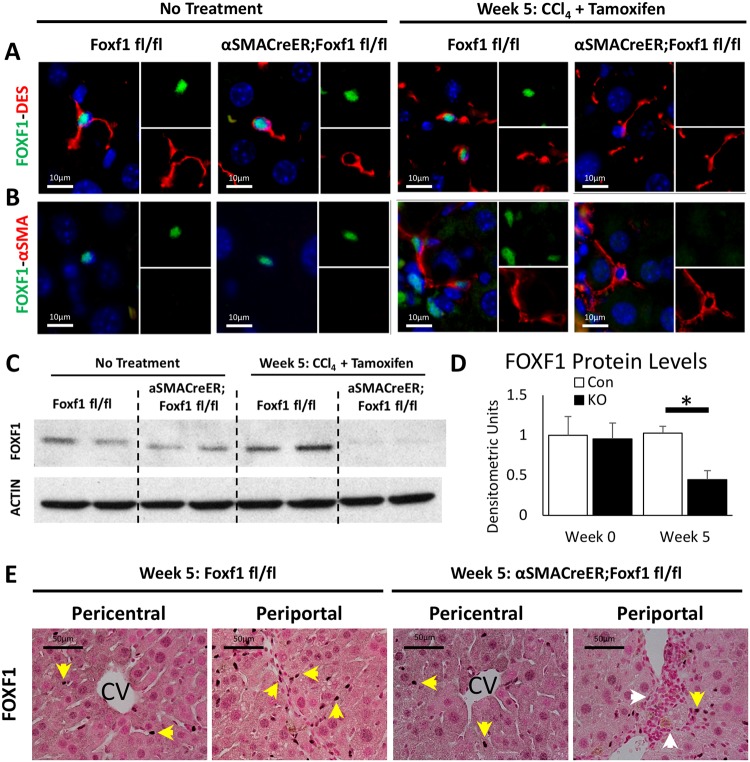
Since FOXF1 is expressed in HSCs in the liver (Kalinichenko et al., 2003), we examined the efficiency of Foxf1 deletion in our experimental model, using immunostaining for FOXF1 and DES. Without CCl4 treatment, FOXF1 was observed in cell nuclei of DES-positive stellate cells in Foxf1fl/fl and αSMACreER;Foxf1fl/fl livers (Fig. 2A). After CCl4 and Tam treatment, FOXF1 staining was reduced in DES-positive cells of αSMACreER;Foxf1−/− livers but not in Foxf1fl/fl livers (Fig. 2A). We also immunostained liver sections for FOXF1 and αSMA, a marker of MFs (Rockey et al., 2013). While αSMA was not detected in parenchyma of quiescent livers, αSMA staining was increased after CCl4 injury. FOXF1 was detected in MFs of control livers but not in αSMACreER;Foxf1−/− livers (Fig. 2B). Quantitative counts of FOXF1-expressing cells demonstrated that the number and percentage of FOXF1+ MFs (FOXF1+ αSMA+) were reduced whereas the number and percentage of FOXF1− MFs (FOXF1− αSMA+) were elevated in injured αSMACreER;Foxf1−/− livers compared to controls (Fig. S3). FOXF1 protein and mRNA were increased in CCl4-treated Foxf1fl/fl livers and purified HSCs (Fig. 2C,D; Fig. S3) but not in the αSMACreER;Foxf1−/− livers (Fig. 2C,D). The loss of FOXF1 in αSMACreER;Foxf1−/− livers occurred in periportal regions while pericentral regions were unaffected (Fig. 2E). The αSMA-CreER transgene allows for the maintained presence of FOXF1 for HSC activation (Kalinichenko et al., 2003) and only deletes FOXF1 after αSMA is expressed in MFs. Thus, αSMA-CreER transgene effectively deletes Foxf1 from hepatic MFs during CCl4-mediated chronic liver injury.
Fig. 2.
αSMA-CreER effectively deletes Foxf1 from hepatic myofibroblasts. (A,B) FOXF1 co-localizes with DES in HSCs before and after CCl4-induced injury. FOXF1 co-localizes with DES and αSMA in MFs after chronic liver injury. αSMA-CreER effectively deletes Foxf1 from MFs after Tam treatment. (C) Western blot shows total liver protein levels of FOXF1 are decreased in αSMACreER;Foxf1−/− livers after CCl4 injury. Cropped blots are presented here with full length blots presented in Fig. S12. (D) Quantification of western blot revealed a significant loss of FOXF1 in αSMACreER;Foxf1−/− livers. Quantification was averaged across four blots. FOXF1 levels were internally normalized to ACTIN for each sample. *P<0.05. (E) FOXF1 staining is detected in liver parenchyma and fibrotic regions (yellow arrows). FOXF1 staining is decreased in fibrotic regions of αSMACreER;Foxf1−/− livers (white arrows).
Deletion of Foxf1 reduces MMP9 activity in CCl4-injured livers
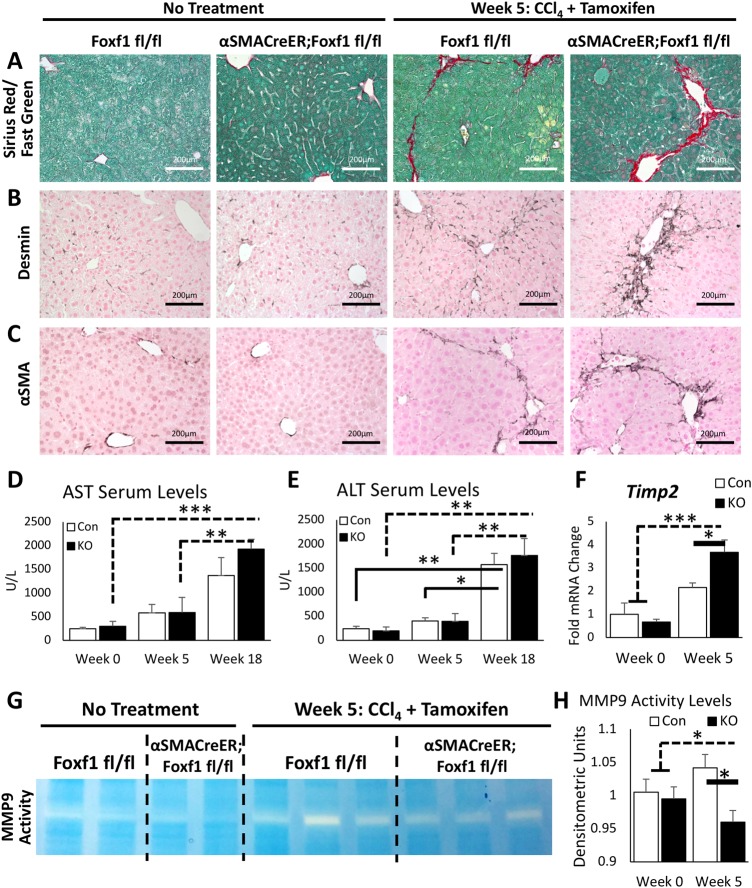
Histological staining with Sirius Red/Fast Green showed a significant increase in collagen accumulation in αSMACreER;Foxf1−/− livers after five weeks of CCl4 treatment (Fig. 3A; Fig. S4). Increased fibrosis in Foxf1-deficient livers was confirmed by immunostaining for DES and αSMA (Fig. 3B,C). To examine the consequences of extended CCl4 treatment, we treated mice with CCl4 for 18 weeks. While hepatic enzymes AST and ALT were increased in blood serum after 18 weeks of CCl4, there was no difference between CCl4 treated αSMACreER;Foxf1−/− and control mice (Fig. 3D,E). Blood serum protein (albumin, globulin) and bilirubin (direct, indirect) levels were not affected after deletion of Foxf1 (Fig. S5). Collagen accumulation was time-dependent (Fig. S6), and after 18 weeks of CCl4 treatment, resulted in widespread liver fibrosis (Fig. S7) and in rare cases, the appearance of visible tumors (Fig. S7).
Fig. 3.
Deletion of Foxf1 from myofibroblasts increases liver fibrosis and inhibits MMP9 activity. (A) Sirius Red/Fast Green staining demonstrates increased collagen deposition between portal triads in CCl4-treated αSMACreER;Foxf1−/− livers. (B,C) Immunohistochemistry shows increased staining for DES and αSMA in CCl4-treated αSMACreER;Foxf1−/− livers. (D,E) Serum enzymatic analysis demonstrates increased AST and ALT levels after chronic CCl4-induced liver injury. Foxf1 deletion does not affect AST or ALT in blood serum. For AST levels: n=3 control mice and n=4 KO mice in week 0; n=5 control mice and n=5 KO mice in week 5; n=4 control mice and n=7 KO mice in week 18. For ALT levels: n=5 control mice and n=6 KO mice in week 0; n=7 control mice and n=8 KO mice in week 5; n=4 control mice and n=7 KO mice in week 18. (F) Increased Timp2 mRNA in CCl4-treated αSMACreER;Foxf1−/− livers is found by qRT-PCR. (G) Representative zymography gel shows decreased MMP9 activity in CCl4-treated αSMACreER;Foxf1−/− livers. Cropped gel is presented here with full gel presented in Fig. S12. (H) Quantification of zymography gels reveals a significant decrease in MMP9 activity in CCl4-treated αSMACreER;Foxf1−/− livers. Quantification was averaged across three gels. *P<0.05, **P<0.01, ***P<0.001.
Since MMP9 plays an important role in collagen degradation after liver injury (Duarte et al., 2015), we evaluated mRNA expression of Mmp9 and its inhibitor, Timp2, in liver tissue. Timp2 mRNA was increased in CCl4-injured αSMACreER;Foxf1−/− livers compared to controls (Fig. 3F). Although Mmp9 mRNA was unchanged (Fig. S8), evaluation of MMP9 activity through zymography demonstrated a significant decrease in enzymatic activity of MMP9 in αSMACreER;Foxf1−/− livers after CCl4 treatment (Fig. 3G,H). Mmp8, Mmp13, Mmp16, Timp1 and Timp3 mRNA levels were not affected in Foxf1-deficient livers (Fig. S8). Thus, Foxf1 deletion from MFs increases Timp2 mRNA and reduces MMP9 activity in fibrotic livers.
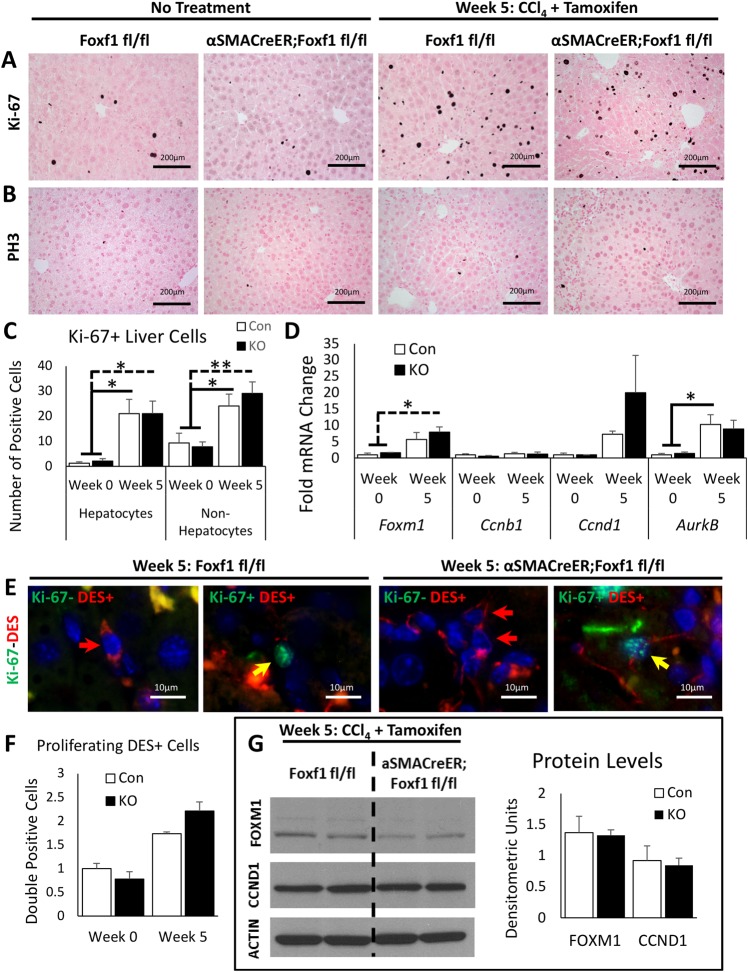
Deletion of Foxf1 does not influence cellular proliferation in fibrotic livers
We evaluated proliferation markers to investigate if the increased fibrosis in αSMACreER;Foxf1−/− livers was due to an expansion of the stromal cells. While cellular proliferation was increased after CCl4 treatment, there were no significant differences in the number of proliferating hepatocytes or non-hepatocytes between Foxf1fl/fl and αSMACreER;Foxf1−/− livers (Fig. 4A–C; Fig. S9). Hepatocytes and non-hepatocytes were identified through distinct morphological appearances (Malarkey et al., 2005) from high magnification images. mRNAs of proliferation-specific genes Foxm1, Ccnb1, Ccnd1, and AurKB (Wang et al., 2009; Kalin et al., 2011; Ren et al., 2013) were unchanged between Foxf1fl/fl and αSMACreER;Foxf1−/− livers (Fig. 4D). Proliferating HSCs and MFs were detected in CCl4-treated livers by co-localization of Ki-67 with DES (Fig. 4E) and αSMA (Fig. S9); however, there were no changes in the number of Ki-67-positive HSCs and MFs after deletion of Foxf1 (Fig. 4F). Protein levels of proliferation-specific genes FOXM1 and CCND1 were unaltered in αSMACreER;Foxf1−/− livers compared to controls (Fig. 4G). Thus, Foxf1 deletion does not affect proliferation of HSCs and MFs after chronic CCl4 liver injury.
Fig. 4.
Deletion of Foxf1 does not influence proliferation of hepatic myofibroblasts. (A) Ki-67 staining shows a significant increase in cell proliferation following CCl4-induced liver injury. No difference in Ki-67 staining is detected between Foxf1fl/fl and αSMACreER;Foxf1−/− livers. (B) PH3 staining shows no significant changes in mitotic rates between Foxf1fl/fl and αSMACreER;Foxf1−/− livers. (C) The number of Ki-67+ hepatocytes and non-hepatocytes in Foxf1fl/fl livers was similar to those in αSMACreER;Foxf1−/− livers. Numbers of Ki-67+ cells were counted in 20–25 random 200× microscope fields using n=3 mice per group in week 0 and n=7 control mice and n=6 KO mice in week 5. (D) qRT-PCR was used to measure mRNAs in whole liver RNA. mRNAs were normalized to Actb. n=3 mice per group in week 0; n=5 mice per group in week 5. (E) Co-localization of Ki-67 with DES shows the presence of Ki-67+ MFs in livers of CCl4-treated mice. (F) Quantification of co-localization of Ki-67 with DES shows no difference in the number of Ki-67+ DES+ cells in Foxf1fl/fl livers compared to αSMACreER;Foxf1−/− livers. (G) Western blot shows no significant difference in total liver protein levels of FOXM1 and CCND1 between Foxf1fl/fl and αSMACreER;Foxf1−/− livers. Cropped blots are presented here with full length blots presented in Fig. S12. *P<0.05, **P<0.01.
RNA-seq analysis identified direct FOXF1 target genes critical for ECM deposition and hepatic fibrosis
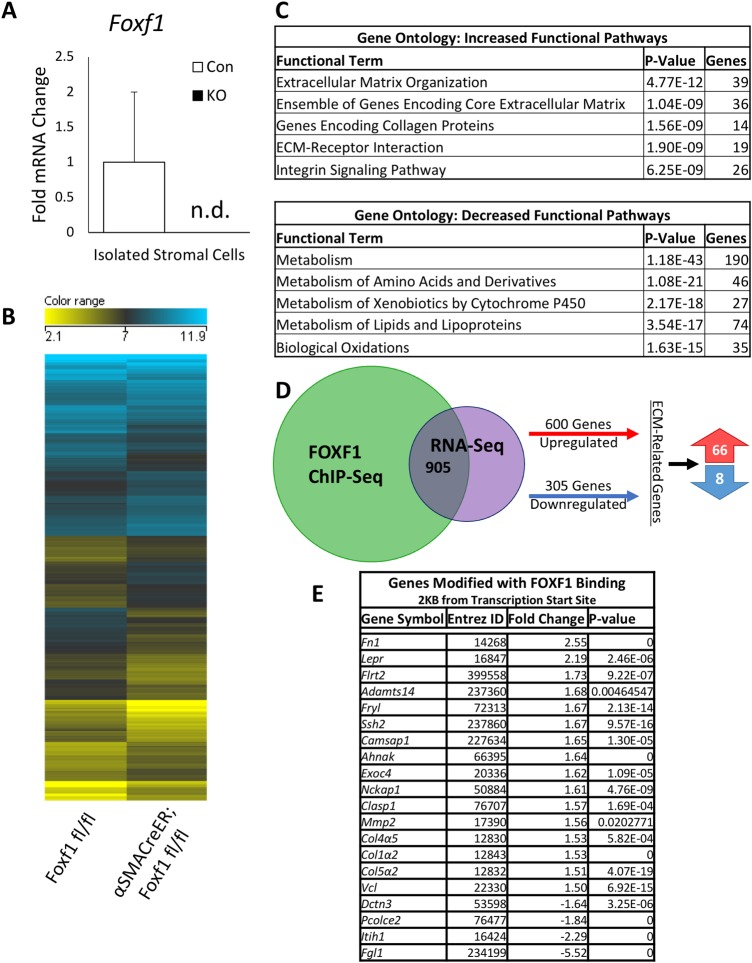
In order to identify FOXF1 target genes, RNA-seq (GEO accession GSE123726) was performed on primary hepatic stromal cells (containing MFs and HSCs) isolated from CCl4-treated Foxf1fl/fl and αSMACreER;Foxf1−/− livers. Purified cells expressed Des and Acta2, but lacked hepatocyte (Nikoozad et al., 2014) and Kupffer cell (Yang et al., 2013) markers (Fig. S10). Foxf1 mRNA was lost in isolated αSMACreER;Foxf1−/− stromal cells (Fig. 5A), a finding consistent with efficient deletion of Foxf1 from CCl4-treated livers. The RNA-seq was used to compare differential gene expression patterns between the Foxf1fl/fl and αSMACreER;Foxf1−/− stromal cells. The differential gene expression in the two groups are represented in a heat map (Fig. 5B). Gene ontology demonstrated that increased functional pathways for the αSMACreER;Foxf1−/− mice were related to ECM regulation, while decreased functional pathways included normal liver functions and metabolism (Fig. 5C). RNA-seq analysis was cross referenced with FOXF1 ChIP-seq analysis (GEO accession GSE100149). 905 genes were common between RNA-seq and ChIP-seq (Fig. 5D), which include 74 genes related to ECM deposition and fibrosis. ChIP-seq proximity analysis revealed that 20 of these ECM genes had FOXF1 binding sites within 2KB of the transcription start site (Fig. 5E).
Fig. 5.
FOXF1 deletion alters expression of pro-fibrotic genes in hepatic myofibroblasts. (A) qRT-PCR analysis of primary hepatic stromal cells submitted for RNA sequencing shows that Foxf1 mRNA is not detected (n.d.) in αSMACreER;Foxf1−/− livers. Samples were pooled for further analysis. (B) Heat map shows differentially expressed genes in stromal cells from Foxf1fl/fl and αSMACreER;Foxf1−/− livers after chronic CCl4-induced hepatic injury as identified by RNA-seq analysis. (C) Biological processes influenced by the deletion of Foxf1 were identified using ToppFunn. P-values and number of genes are listed for each classification. (D) 905 overlapping genes were identified between ChIP-seq (GEO accession GSE100149) and RNA-seq (GEO accession GSE123726) data, of which 74 were ECM-related genes. (E) Table shows 20 ECM-related genes identified by ChIP-seq and RNA-seq (FOXF1 binding was analyzed within 2 KB from transcriptional start site).
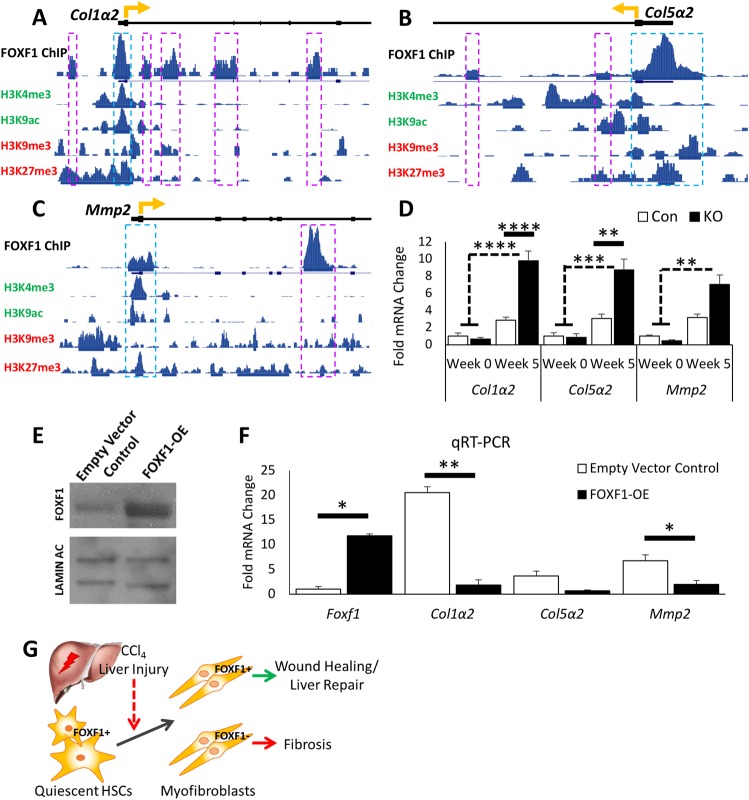
Interestingly, Col1α2, Col5α2 and Mmp2 were among the 20 ECM-related genes that had FOXF1 binding sites within the gene loci (Fig. S11, Table S1). COL1α2 and COL5α2 are common ECM components in fibrotic livers (Mak et al., 2016), whereas MMP2 is a collagenase that is increased during liver fibrosis and associated with disease progression (Benyon et al., 1996). Expression of Col1α2, Col5α2 and Mmp2 mRNAs were increased in CCl4-treated αSMACreER;Foxf1−/− livers as shown by RNA-seq and qRT-PCR (Fig. 5E, Fig. 6D), suggesting a negative regulation by FOXF1. The presence of gene silencing histone methylation marks H3K9me3 and H3K27me3 (Dong and Weng, 2013; Bernstein et al., 2006) in FOXF1-binding promoter regions (Fig. 6A–C) is consistent with negative regulation of these genes by FOXF1. In order to confirm the regulation of Col1α2, Col5α2 and Mmp2 by FOXF1, we overexpressed FOXF1 in isolated murine HSCs (Fig. 6E). Lentiviral-mediated overexpression of FOXF1 decreased Col1α2 and Mmp2 in vitro (Fig. 6F). Thus, FOXF1 negatively regulates expression of pro-fibrotic genes in MFs. Altogether, FOXF1 expression in myofibroblasts is essential to inhibit liver fibrosis after chronic liver injury (Fig. 6G).
Fig. 6.
FOXF1 binds to DNA regulatory regions of Col1α2, Col5α2 and Mmp2. (A–C) ChIP-seq shows FOXF1 binding near the transcriptional start sites in Col1α2, Col5α2 and Mmp2 gene loci. Histone modification marks of enhancers (H3K4me3, H3K9ac) and repressors (H3K9me3, H3K27me3) are aligned with FOXF1-binding regions. Significant areas of FOXF1 binding are marked with boxes, with blue boxes indicating the binding site is within gene promoter region. Gene transcriptional start sites are marked with a directional yellow arrow. (D) qRT-PCR analysis shows the significant increase of Col1α2, Col5α2 and Mmp2 mRNAs in the isolated stromal cells of CCl4-treated αSMACreER;Foxf1−/− livers. For Col1α2 and Col5α2: n=3 mice per group in week 0; n=5 mice per group in week 5. For Mmp2: n=3 mice per group in week 0; n=6 control mice and n=4 KO mice in week 5. (E) Western blot shows increase in FOXF1 expression in isolated HSCs after FOXF1-overexpression. Cropped blots are presented here with full length blots presented in Fig. S12. (F) qRT-PCR shows an increase of Foxf1 mRNA and a decrease of Col1α2, Col5α2 and Mmp2 mRNAs in isolated HSCs after FOXF1-overexpression. (G) Diagram of hepatic fibrosis in Foxf1-deficient mice shows that the loss of FOXF1 promotes ECM deposition and exacerbated fibrosis after CCl4-treatment. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
DISCUSSION
Myofibroblast activation is a key mechanism in the development of hepatic fibrosis. However, transcriptional regulation of myofibroblasts during liver fibrogenesis remains poorly characterized. In the present study, we found that the deletion of Foxf1 in MFs during chronic CCl4-mediated injury exacerbated liver fibrosis, increased collagen deposition and stimulated expression of pro-fibrotic genes. ECM-related proteins were identified as novel FOXF1 transcriptional targets, suggesting that FOXF1 plays an important role in the regulation of ECM and collagen deposition during the progression of hepatic fibrosis.
Previous studies have focused on the role of FOXF1 in acute liver injury using a single CCl4 administration to Foxf1+/− mice. These studies demonstrated that FOXF1 is necessary for HSC activation to promote liver repair (Kalinichenko et al., 2003). CCl4-treated Foxf1+/− mice exhibited diminished collagen depositions and increased mortality after the liver injury (Kalinichenko et al., 2003). A recently published model of Foxf1-silencing using a lipid-based nanoparticle system to deliver Foxf1 siRNA to the liver demonstrated attenuated collagen deposition when Foxf1 siRNA was delivered 48 h prior to bile duct ligation (Abshagen et al., 2015). It is likely that Foxf1 siRNA inhibited FOXF1 signaling in hepatic stellate cells, decreasing their activation and subsequent collagen depositions into the liver tissue, a finding consistent with previous studies using Foxf1+/− mice (Kalinichenko et al., 2003). Recently, a model of chronic hepatic injury using CCl4-injections, similar to the present study, was unsuccessful in silencing Foxf1 expression using the same lipid based system to deliver Foxf1 siRNA (Abshagen et al., 2015, 2017). This method involved four weeks of IP CCl4-injections before two weeks of treatment with Foxf1 siRNA (Abshagen et al., 2017). It is possible that the lack of Foxf1 silencing was due to inability of nanoparticles to target hepatic MFs. In the current study, we utilized a conditional genetic mouse model to delete Foxf1 in MFs during CCl4-induced hepatic fibrosis which shares multiple histological similarities with human disease (Masugi et al., 2018; Bataller and Brenner, 2005). Interestingly, the loss of Foxf1 in MFs resulted in increased collagen deposition, causing severe fibrotic lesions between hepatic portal triads in αSMACreER;Foxf1−/− livers. Our studies suggest that FOXF1 inhibits production of collagen and ECM during the progression of liver fibrosis. Increased fibrosis in Foxf1-deficient mice was associated with the appearance of liver tumors, a finding consistent with increased tumor formation in patients with liver cirrhosis (EASL-EORTC et al., 2018). Our studies suggest that maintaining Foxf1 expression can be beneficial in patients with advanced liver fibrosis to inhibit fibrotic responses and decrease the risk of liver tumorigenesis.
In the present study, collagens were significantly increased in αSMACreER;Foxf1−/− livers after chronic CCl4-treatment. Desmin and αSMA were both increased in αSMACreER;Foxf1−/− livers; however, there were no differences in the number of proliferating cells between Foxf1fl/fl and αSMACreER;Foxf1−/− livers. Previously, FOXF1 has been shown to stimulate cell proliferation in lung endothelial cells (Ren et al., 2014; Bolte et al., 2017) and in rhabdomyosarcoma tumor cells (Milewski et al., 2017). Surprisingly, we found that deletion of Foxf1 from MFs does not affect their proliferation during liver fibrogenesis. It is possible that FOXF1 requires additional co-activator or co-repressor proteins (that are not present in MFs) to regulate cellular proliferation. Additionally, we found an increase in Timp2 expression with a decrease in MMP9 activity in αSMACreER;Foxf1−/− livers. Since it is well-known that TIMPs and MMPs regulate ECM depositions to balance the scaring and healing processes during fibrosis (Duarte et al., 2015), it is possible that the loss of Foxf1 alters the TIMP/MMP balance to allow accumulation of collagens without the degradation mechanisms necessary for proper wound healing. Interestingly, MMP9 has been implicated in HSC to MF transdifferentiation (Han et al., 2007) in addition to its roles in ECM degradation (Duarte et al., 2015; Kurzepa et al., 2014). Therefore, decreased MMP9 activity can contribute to increased liver fibrosis in αSMACreER;Foxf1−/− mice. Surprisingly, FOXF1 was increased in activated HSCs compared to quiescent HSCs. It is possible that FOXF1 is differentially regulated in HSCs compared to hepatic MFs, and that after liver injury, FOXF1 protects HSCs from differentiating into MFs through transcriptional repression of profibrotic genes.
Consistent with increased fibrosis in Foxf1-deficient livers, RNA-seq analysis revealed increased ECM-related functional pathways in a purified stromal cell population. Comparison with FOXF1 ChIP-seq data revealed 20 novel transcriptional targets of FOXF1, which include Col1α2, Col5α2 and Mmp2, expression of which was increased in Foxf1-deficient cells.
COL1α2 is one of the most abundant ECM components in the liver along with COL1α1 and COL3α1 (Lai et al., 2011). COL5α2 is highly expressed with Collagens 1 and 3 and is important in regulating the assembly and structure of these collagens in the fibrotic matrix (Moriya et al., 2011). MMP2 acts as a collagenase, known to be activated during hepatic fibrosis (Benyon et al., 1996). In addition to increased mRNA levels of the genes in FOXF1-deficient cells, we found multiple FOXF1 binding sites within their gene promoter region and introns, suggesting direct transcriptional repression. This hypothesis is supported by the presence of H3K4me3 and H3K9ac, histone modifications associated with transcriptional repression (Dong and Weng, 2013; Rea et al., 2000), at FOXF1 binding sites. In summary, we have developed a novel genetic mouse model to study the role of FOXF1 in MFs during chronic liver injury. Using this model, we demonstrated that Foxf1 expression in MFs is necessary to inhibit hepatic fibrosis and maintain the balance of collagen depositions, through transcriptional repression of pro-fibrotic genes.
MATERIALS AND METHODS
Mice
The Foxf1fl/fl mouse line was previously generated and bred into the C57Bl/6 mouse background (Ren et al., 2014; Cai et al., 2016). Foxf1fl/fl mice were bred with αSMA-CreER mice (Jackson Laboratory, 029925; Wendling et al., 2009) to generate αSMACreER;Foxf1fl/fl mice (Black et al., 2018). αSMACreER;Foxf1fl/fl mice were bred with Foxf1fl/fl mice and male pups were genotyped and used for all experiments at the age of 6–8 weeks. The following primers were used for genotyping: αSMA-CreER sense: 5′ TGCAACGAGTGATGAGGTTCGC 3′ and anti-sense: 5′ GATCCTGGCAATTTCGGCTATACG 3′; αSMA-WT sense 5′ GGTTTCTATTGCTACCAAGAGACAT 3′ and anti-sense: 5′ TGCACCAAACCCTGGACTAAGCAT 3′; Foxf1fl/fl sense: 5′ GCTTTGTCTCCAAGCGCTGC 3′ and anti-sense: 5′ TTCAGATCTGAGAGTGGCAGCTTC 3′. Foxf1fl/fl littermates were used as controls. To activate the conditional Foxf1 knockout, tamoxifen (Tam) was given via intraperitoneal injection (40 mg/kg of body weight; Sigma-Aldrich) three days in a row at the beginning of each week starting at week 2 over the course of the chronic liver injury period. Hepatic injury was induced by intraperitoneal injections of carbon tetrachloride (CCl4; 1 μl/g of body weight 20% v/v; Sigma-Aldrich; diluted in sunflower seed oil) three times a week every other day over the course of the chronic liver injury period. The levels of aminotransferases AST and ALT, proteins albumin and globulin, and direct and indirect bilirubin were determined by serological analysis of blood serum as previously described (Sun et al., 2017; Ren et al., 2010). All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Cincinnati Children's Research Foundation and the NIH IACUC Guidebook. All experiments were covered under our animal protocol (IACUC2016-0038). The Cincinnati Children's Research Foundation Institutional Animal Care and Use Committee is an AAALAC and NIH accredited institution (NIH Insurance #8310801).
Histology and immunohistochemistry
Paraffin-embedded liver sections were used for H&E staining, immunohistochemistry (IHC), or immunofluorescence (IF) as previously described (Ren et al., 2010; Kalinichenko et al., 2003; Wang et al., 2003). The following antibodies were used for immunostaining: FOXF1 (1:1000 IHC, 1:200 IF, R&D Systems), DES (1:500 IHC, 1:100 IF; Santa Cruz Technologies), αSMA (1:10,000 IHC, 1:5000 IF; Sigma-Aldrich), Ki-67 (1:1000 IHC, 1:200 IF; Thermo Fisher Scientific), Ki-67 (1:200 IF; BD Biosciences), and PH3 (1:10,000 IHC; Santa Cruz Technologies). Antibody-antigen complexes were detected using biotinylated secondary antibodies followed by avidin-biotin-horseradish peroxidase complex and 3,3′diaminobenzidine substrate (Vector Labs) as previously described (Kalinichenko et al., 2003; Ren et al., 2010; Wang et al., 2012). Sections were counterstained with Nuclear Fast Red (Vector Labs). For immunofluorescence imaging, secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen/Molecular Probes) were used as described (Ustiyan et al., 2012; Wang et al., 2010). Cell nuclei were counterstained with DAPI (Vector Labs). Masson's Trichrome (BD Biosciences) and Sirius Red/Fast Green (Chondrex, Inc.) specialty stains were performed according to the manufacturer’s protocols. Brightfield images were obtained using a Zeiss AxioImage.A2 microscope. Fluorescent images were obtained using a Zeiss AxioPlan 2 microscope.
qRT-PCR, western blot, and zymography
The caudate lobe of the liver was halved and used for RNA and protein studies. RNA was isolated using RNA Stat-60 (Tel-Test, Inc.) according to the manufacturer’s protocol and was reverse transcribed using the High Capacity Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. mRNAs of specific genes were measured by qRT-PCR using TaqMan probes (Applied Biosystems; Table S2) and the StepOnePlus Real-Time PCR system (Applied Biosystems) as described (Bolte et al., 2011, 2015, 2012; Wang et al., 2010). mRNAs were normalized to Actb. Protein extracts were isolated using cell lysis buffer as previously described (Pradhan et al., 2016) and used for either western blot analysis with Pierce ECL western blotting substrate (Thermo Fisher Scientific) or gel zymography (NOVEX) according to the manufacturer’s protocols. The following antibodies were used for protein blots: FOXF1 (1:1000, R&D Systems) (Bolte et al., 2017; Black et al., 2018; Ren et al., 2014), ACTIN (1:2000; Santa Cruz Biotechnology) (Pradhan et al., 2016), FOXM1 (1:3000; Santa Cruz Biotechnology) (Pradhan et al., 2016; Bolte et al., 2011), CCND1 (1:1000; Cell Signaling Technology) (Milewski et al., 2017). Protein band intensities were determined by ImageJ software and were normalized to ACTIN.
Hepatic stellate cell isolation, transfection
Hepatic stellate cells were isolated from male C57Bl/6-WT mice (40–50 g), purified using Nycodenz gradient, and cultured as previously described (Pradhan et al., 2016; Dangi et al., 2012; Sumpter et al., 2012). Quiescent HSCs were harvested at day two after cell culture (Reinehr et al., 1998). After ten days in culture, activated MFs were harvested (Reinehr et al., 1998). The pMIEG3 bicistronic retroviral vector was used for FOXF1 protein overexpression as previously described (Pradhan et al., 2016). The cells were transfected as previously described (Singh et al., 2008). mRNAs in isolated MFs were normalized to 18 s (Eukaryotic 18S rRNA Endogenous Control; Applied Biosciences). Protein in isolated MFs were normalized to LAMIN AC (1:10,000; Santa Cruz Biotechnology) (Pradhan et al., 2016).
RNA sequencing
RNA was isolated from HSC/MF population purified from CCl4-treated Foxf1fl/fl and αSMACreER;Foxf1−/− livers using a differential plating method (Giassetti et al., 2016) that we modified. Briefly, liver cell suspension was plated on tissue culture dishes (Corning) and incubated for 2 h at 37°C. Supernatant and non-adherent cells were washed off and the adherent cell population was collected for experiments. Samples were pooled to generate the libraries using the TruSeq RNA library preparation kit and were sequenced on an Illumina HiSeq 2000 sequencer, generating approximately 10 M high quality single end reads (75 base-long reads). Alignment was performed using the Tophat/Cufflink pipeline (Trapnell et al., 2009, 2010). Finally, cuffmerge tool was used to generate Binary Alignment/Map files (BAM files) (Roberts et al., 2011). BAM files of RNA-seq data were analyzed using Avadis® NGS Version 1.3.0 software. Reads were filtered to remove: (1) duplicate reads, (2) non-primary-matched reads, and (3) reads with alignment scores <95. Quantification was performed on the filtered reads against the RefSeq annotation. Data normalization was performed with the DESeq package. The sequencing depth was estimated by the read count of the gene with the median read count ratio across all genes. The method was based on the negative binomial distribution, which allows for less restrictive variance parameter assumptions than does the Poisson distribution. The false discovery rate was calculated according to the Benjamini and Hochberg algorithm (Benjamini and Hochberg, 1995). Genes with expression altered by a factor of 1.5 and a false discovery rate of 0.05 in Foxf1fl/fl cells compared with αSMACreER;Foxf1−/− cells were selected for gene set enrichment analysis using ToppGene Suite. Hierarchical clustering was performed by Ward's method using Euclidean distance metric. RNA-seq data are available at GEO accession GSE123726. RNA-seq data were compared to previously published ChIP-seq data (GEO accession GSE100149) using a two-way Venn diagram.
Statistical analysis
Statistical significance differences in measured variables between control and experimental groups were assessed with a Student's t-test (two-tailed) or one-way analysis of variance (ANOVA) with Bonferroni post hoc test as appropriate. P<0.05 was considered to be significant, with P<0.05 indicated with *, P<0.01 indicated with **, P<0.001 indicated with ***, and P<0.0001 indicated with ****. Values for all measurements were expressed as mean±s.e. of mean.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: H.M.F., V.V.K.; Methodology: H.M.F.; Software: H.M.F.; Validation: H.M.F., N.D., V.V.K.; Formal analysis: H.M.F., C.B., N.D., A.S., Y.Z., C.R.G., T.V.K., V.V.K.; Investigation: H.M.F., C.B., A.S., Y.Z., C.R.G.; Resources: H.M.F.; Data curation: H.M.F.; Writing - original draft: H.M.F., V.V.K.; Writing - review & editing: H.M.F., C.B., N.D., A.S., Y.Z., C.R.G., T.V.K., V.V.K.; Visualization: H.M.F., V.V.K.; Supervision: H.M.F., V.V.K.; Project administration: H.M.F., V.V.K.; Funding acquisition: T.V.K., V.V.K.
Funding
We would like to acknowledge the following funding sources: National Institutes of Health Grants [HL84151 (V.V.K.), HL123490 (V.V.K.), HL141174 (V.V.K.), and HL132849 (T.V.K.)].
Data availability
The RNA sequencing microarray data presented in this article has been uploaded to Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (NCBI) and the accession number is GSE123726.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.039800.supplemental
References
- Abshagen K., Brensel M., Genz B., Roth K., Thomas M., Fehring V., Schaeper U. and Vollmar B. (2015). Foxf1 siRNA delivery to hepatic stellate cells by DBTC lipoplex formulations ameliorates fibrosis in livers of bile duct ligated mice. Curr. Gene Ther. 15, 215-227. 10.2174/1566523215666150126114634 [DOI] [PubMed] [Google Scholar]
- Abshagen K., Rotberg T., Genz B. and Vollmar B. (2017). No significant impact of Foxf1 siRNA treatment in acute and chronic CCl4 liver injury. Exp. Biol. Med. (Maywood) 242, 1389-1397. 10.1177/1535370217716425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem M., Sell K.-M., Melchior R., Kropf J., Eller T. & Gressner A. (1993). Tumor necrosis factor alpha (TNFα) and transforming growth factor β1 (TGFβ1) stimulate fibronectin synthesis and the transdifferentiation of fat-storing cells in the rat liver into myofibroblasts. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 63, 123-130. [DOI] [PubMed] [Google Scholar]
- Bataller R. and Brenner D. A. (2005). Liver fibrosis. J. Clin. Invest. 115, 209-218. 10.1172/JCI24282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. and Hochberg Y. (1995). Controlling the false discovery rate-a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol. 57, 289-300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Benyon R. C., Iredale J. P., Goddard S., Winwood P. J. and Arthur M. J. (1996). Expression of tissue inhibitor of metalloproteinases 1 and 2 is increased in fibrotic human liver. Gastroenterology 110, 821-831. 10.1053/gast.1996.v110.pm8608892 [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315-326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Black M., Milewski D., Le T., Ren X., Xu Y., Kalinichenko V. V. and Kalin T. V. (2018). FOXF1 inhibits pulmonary fibrosis by preventing CDH2-CDH11 cadherin switch in myofibroblasts. Cell Rep 23, 442-458. 10.1016/j.celrep.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C., Zhang Y., Wang I.-C., Kalin T. V., Molkentin J. D. and Kalinichenko V. V. (2011). Expression of foxm1 transcription factor in cardiomyocytes is required for myocardial development. PLoS ONE 6, e22217 10.1371/journal.pone.0022217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C., Zhang Y., York A., Kalin T. V., Schultz J. E. J., Molkentin J. D. and Kalinichenko V. V. (2012). Postnatal ablation of foxm1 from cardiomyocytes causes late onset cardiac hypertrophy and fibrosis without exacerbating pressure overload-induced cardiac remodeling. PLoS ONE 7, e48713 10.1371/journal.pone.0048713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C., Ren X., Tomley T., Ustiyan V., Pradhan A., Hoggatt A., Kalin T. V., Herring B. P. and Kalinichenko V. V. (2015). Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J. Biol. Chem. 290, 7563-7575. 10.1074/jbc.M114.609487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C., Flood H. M., Ren X., Jagannathan S., Barski A., Kalin T. V. and Kalinichenko V. V. (2017). FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci. Rep. 7, 10690 10.1038/s41598-017-11175-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C., Whitsett J. A., Kalin T. V. and Kalinichenko V. V. (2018). Transcription factors regulating embryonic development of pulmonary vasculature. Adv. Anat. Embryol. Cell Biol. 228, 1-20. 10.1007/978-3-319-68483-3_1 [DOI] [PubMed] [Google Scholar]
- Brenner D. A., Kisseleva T., Scholten D., Paik Y. H., Iwaisako K., Inokuchi S., Schnabl B., Seki E., De Minicis S., Oesterreicher C. et al. (2012). Origin of myofibroblasts in liver fibrosis. Fibrogenesis Tissue Repair 5, S17-S17 10.1186/1755-1536-5-S1-S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Bolte C., Le T., Goda C., Xu Y., Kalin T. V. and Kalinichenko V. V. (2016). FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci. Signal. 9, ra40-ra40 10.1126/scisignal.aad1899 [DOI] [PubMed] [Google Scholar]
- Cheng K. and Mahato R. I. (2007). Gene modulation for treating liver fibrosis. Crit. Rev. Ther. Drug Carrier Syst. 24, 93-146. 10.1615/CritRevTherDrugCarrierSyst.v24.i2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civan J. (2016). Hepatic and Biliary Diseases: Hepatic Fibrosis. Merck Manual: Professional Version. Kenilworth, NJ, USA: Merck & Co., Inc. [Google Scholar]
- Croci I., Byrne N. M., Choquette S., Hills A. P., Chachay V. S., Clouston A. D., O'moore-Sullivan T. M., Macdonald G. A., Prins J. B. and Hickman I. J. (2013). Whole-body substrate metabolism is associated with disease severity in patients with non-alcoholic fatty liver disease. Gut 62, 1625-1633. 10.1136/gutjnl-2012-302789 [DOI] [PubMed] [Google Scholar]
- Dangi A., Sumpter T. L., Kimura S., Stolz D. B., Murase N., Raimondi G., Vodovotz Y., Huang C., Thomson A. W. and Gandhi C. R. (2012). Selective expansion of allogeneic regulatory T cells by hepatic stellate cells: role of endotoxin and implications for allograft tolerance. J. Immunol. 188, 3667-3677. 10.4049/jimmunol.1102460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. and Weng Z. (2013). The correlation between histone modifications and gene expression. Epigenomics 5, 113-116. 10.2217/epi.13.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte S., Baber J., Fujii T. and Coito A. J. (2015). Matrix metalloproteinases in liver injury, repair and fibrosis. Matrix Biol. 44, 147-156. 10.1016/j.matbio.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EASL-EORTC, LIVER, E. A. F. S. O. T. & CANCER, E. O. F. R. A. T. O. (2018). EASL clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 69, 182-236. 10.1016/j.jhep.2018.03.019 [DOI] [PubMed] [Google Scholar]
- Fausther M., Lavoie E. G. and Dranoff J. A. (2013). Contribution of myofibroblasts of different origins to liver fibrosis. Curr. Pathobiol. Rep. 1, 225-230. 10.1007/s40139-013-0020-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giassetti M. I., Goissis M. D., De Barros F. R. O., Bruno A. H., Assumpção M. E. O. and Visintin J. A. (2016). Comparison of diverse differential plating methods to enrich bovine spermatogonial cells. Reprod. Domest. Anim. 51, 26-32. 10.1111/rda.12641 [DOI] [PubMed] [Google Scholar]
- Han Y.-P., Yan C., Zhou L., Qin L. and Tsukamoto H. (2007). A matrix metalloproteinase-9 activation cascade by hepatic stellate cells in trans-differentiation in the three-dimensional extracellular matrix. J. Biol. Chem. 282, 12928-12939. 10.1074/jbc.M700554200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerbrand C., Stefanovic B., Giordano F., Burchardt E. R. and Brenner D. A. (1999). The role of TGFβ1 in initiating hepatic stellate cell activation in vivo. J. Hepatol. 30, 77-87. 10.1016/S0168-8278(99)80010-5 [DOI] [PubMed] [Google Scholar]
- Hodo Y., Honda M., Tanaka A., Nomura Y., Arai K., Yamashita T., Sakai Y., Yamashita T., Mizukoshi E., Sakai A. et al. (2013). Association of interleukin-28B genotype and hepatocellular carcinoma recurrence in patients with chronic hepatitis C. Clin. Cancer Res. 19, 1827-1837. 10.1158/1078-0432.CCR-12-1641 [DOI] [PubMed] [Google Scholar]
- Kalin T. V., Ustiyan V. and Kalinichenko V. V. (2011). Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle 10, 396-405. 10.4161/cc.10.3.14709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko V. V., Zhou Y., Shin B., Stolz D. B., Watkins S. C., Whitsett J. A. and Costa R. H. (2002). Wild-type levels of the mouse Forkhead Box f1 gene are essential for lung repair. Am. J. Physiol. Lung Cell. Mol. Physiol. 282, L1253-L1265. 10.1152/ajplung.00463.2001 [DOI] [PubMed] [Google Scholar]
- Kalinichenko V. V., Bhattacharyya D., Zhou Y., Gusarova G. A., Kim W., Shin B. and Costa R. H. (2003). Foxf1 +/− mice exhibit defective stellate cell activation and abnormal liver regeneration following CCl4 injury. Hepatology 37, 107-117. 10.1053/jhep.2003.50005 [DOI] [PubMed] [Google Scholar]
- Kim I.-M., Zhou Y., Ramakrishna S., Hughes D. E., Solway J., Costa R. H. and Kalinichenko V. V. (2005). Functional characterization of evolutionarily conserved DNA regions in forkhead box f1 gene locus. J. Biol. Chem. 280, 37908-37916. 10.1074/jbc.M506531200 [DOI] [PubMed] [Google Scholar]
- Kinnman N., Goria O., Wendum D., Gendron M. C., Rey C., Poupon R. and Housset C. (2001). Hepatic stellate cell proliferation is an early platelet-derived growth factor-mediated cellular event in rat cholestatic liver injury. Lab Invest. 81, 1709-1716. [DOI] [PubMed] [Google Scholar]
- Kurzepa J., Madro A., Czechowska G., Kurzepa J., Celiński K., Kazmierak W. and Slomka M. (2014). Role of MMP-2 and MMP-9 and their natural inhibitors in liver fibrosis, chronic pancreatitis and non-specific inflammatory bowel diseases. Hepatobiliary Pancreat. Dis. Int. 13, 570-579. 10.1016/S1499-3872(14)60261-7 [DOI] [PubMed] [Google Scholar]
- Lai K. K. Y., Shang S., Lohia N., Booth G. C., Masse D. J., Fausto N., Campbell J. S. and Beretta L. (2011). Extracellular matrix dynamics in hepatocarcinogenesis: a comparative proteomics study of PDGFC transgenic and pten null mouse models. PLoS Genet. 7, e1002147 10.1371/journal.pgen.1002147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M., Ormestad M., Enerback S. and Carlsson P. (2001). The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development 128, 155-166. [DOI] [PubMed] [Google Scholar]
- Mak K. M., Png C. Y. M. and Lee D. J. (2016). Type V collagen in health, disease, and fibrosis. Anat. Rec. 299, 613-629. 10.1002/ar.23330 [DOI] [PubMed] [Google Scholar]
- Makarev E., Izumchenko E., Aihara F., Wysocki P. T., Zhu Q., Buzdin A., Sidransky D., Zhavoronkov A. and Atala A. (2016). Common pathway signature in lung and liver fibrosis. Cell Cycle 15, 1667-1673. 10.1080/15384101.2016.1152435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkey D. E., Johnson K., Ryan L., Boorman G. and Maronpot R. R. (2005). New insights into functional aspects of liver morphology. Toxicol. Pathol. 33, 27-34. 10.1080/01926230590881826 [DOI] [PubMed] [Google Scholar]
- Martinez A. K., Maroni L., Marzioni M., Ahmed S. T., Milad M., Ray D., Alpini G. and Glaser S. S. (2014). Mouse models of liver fibrosis mimic human liver fibrosis of different etiologies. Curr. Pathobiol. Rep. 2, 143-153. 10.1007/s40139-014-0050-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masugi Y., Abe T., Tsujikawa H., Effendi K., Hashiguchi A., Abe M., Imai Y., Hino K., Hige S., Kawanaka M. et al. (2018). Quantitative assessment of liver fibrosis reveals a nonlinear association with fibrosis stage in nonalcoholic fatty liver disease. Hepatol. Commun. 2, 58-68. 10.1002/hep4.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke I., Hsu C. C., Troeger J. S., Huebener P., Mu X., Dapito D. H., Pradere J.-P. and Schwabe R. F. (2013). Fate-tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its etiology. Nat. Commun. 4, 2823-2823 10.1038/ncomms3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski D., Pradhan A., Wang X., Cai Y., Le T., Turpin B., Kalinichenko V. V. and Kalin T. V. (2017). FoxF1 and FoxF2 transcription factors synergistically promote Rhabdomyosarcoma carcinogenesis by repressing transcription of p21(Cip1) CDK inhibitor. Oncogene 36, 850-862. 10.1038/onc.2016.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya K., Bae E., Honda K., Sakai K., Sakaguchi T., Tsujimoto I., Kamisoyama H., Keene D. R., Sasaki T. and Sakai T. (2011). A fibronectin-independent mechanism of collagen fibrillogenesis in adult liver remodeling. Gastroenterology 140, 1653-1663. 10.1053/j.gastro.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoozad Z., Ghorbanian M. T. and Rezaei A. (2014). Comparison of the liver function and hepatic specific genes expression in cultured mesenchymal stem cells and hepatocytes. Iran. J. Basic Med. Sci. 17, 27-33. [PMC free article] [PubMed] [Google Scholar]
- Pradhan A., Ustiyan V., Zhang Y., Kalin T. V. and Kalinichenko V. V. (2016). Forkhead transcription factor FoxF1 interacts with Fanconi anemia protein complexes to promote DNA damage response. Oncotarget 7, 1912-1926. 10.18632/oncotarget.6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O'carroll D., Strahl B. D., Sun Z.-W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D. et al. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593-599. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Reinehr R. M., Kubitz R., Peters-Regehr T., Bode J. G. and Häussinger D. (1998). Activation of rat hepatic stellate cells in culture is associated with increased sensitivity to endothelin 1. Hepatology 28, 1566-1577. 10.1002/hep.510280617 [DOI] [PubMed] [Google Scholar]
- Ren X., Zhang Y., Snyder J., Cross E. R., Shah T. A., Kalin T. V. and Kalinichenko V. V. (2010). Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol. Cell. Biol. 30, 5381-5393. 10.1128/MCB.00876-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Shah T. A., Ustiyan V., Zhang Y., Shinn J., Chen G., Whitsett J. A., Kalin T. V. and Kalinichenko V. V. (2013). FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Mol. Cell. Biol. 33, 371-386. 10.1128/MCB.00934-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Ustiyan V., Pradhan A., Cai Y., Havrilak J. A., Bolte C. S., Shannon J. M., Kalin T. V. and Kalinichenko V. V. (2014). FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ. Res. 115, 709-720. 10.1161/CIRCRESAHA.115.304382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pimentel H., Trapnell C. and Pachter L. (2011). Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325-2329. 10.1093/bioinformatics/btr355 [DOI] [PubMed] [Google Scholar]
- Rockey D. C., Weymouth N. and Shi Z. (2013). Smooth muscle α Actin (Acta2) and myofibroblast function during hepatic wound healing. PLoS ONE 8, e77166 10.1371/journal.pone.0077166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T. R., Ali A. M., Busygina V., Raynard S., Fan Q., Du C.-H., Andreassen P. R., Sung P. and Meetei A. R. (2008). BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 22, 2856-2868. 10.1101/gad.1725108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter T. L., Dangi A., Matta B. M., Huang C., Stolz D. B., Vodovotz Y., Thomson A. W. and Gandhi C. R. (2012). Hepatic stellate cells undermine the allostimulatory function of liver myeloid dendritic cells via STAT3-dependent induction of IDO. J. Immunol. 189, 3848-3858. 10.4049/jimmunol.1200819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Ren X., Wang I.-C., Pradhan A., Zhang Y., Flood H. M., Han B., Whitsett J. A., Kalin T. V. and Kalinichenko V. V. (2017). The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci. Signal. 10, eaai8583 10.1126/scisignal.aai8583 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., Van Baren M. J., Salzberg S. L., Wold B. J. and Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511-515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V., Wert S. E., Ikegami M., Wang I.-C., Kalin T. V., Whitsett J. A. and Kalinichenko V. V. (2012). Foxm1 transcription factor is critical for proliferation and differentiation of Clara cells during development of conducting airways. Dev. Biol. 370, 198-212. 10.1016/j.ydbio.2012.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bhattacharyya D., Dennewitz M. B., Kalinichenko V. V., Zhou Y., Lepe R. and Costa R. H. (2003). Rapid hepatocyte nuclear translocation of the Forkhead Box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr. 11, 149-162. 10.3727/000000003108749044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.-C., Meliton L., Ren X., Zhang Y., Balli D., Snyder J., Whitsett J. A., Kalinichenko V. V. and Kalin T. V. (2009). Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS ONE 4, e6609 10.1371/journal.pone.0006609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.-C., Zhang Y., Snyder J., Sutherland M. J., Burhans M. S., Shannon J. M., Park H. J., Whitsett J. A. and Kalinichenko V. V. (2010). Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes clara cell hyperplasia. Dev. Biol. 347, 301-314. 10.1016/j.ydbio.2010.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.-C., Snyder J., Zhang Y., Lander J., Nakafuku Y., Lin J., Chen G., Kalin T. V., Whitsett J. A. and Kalinichenko V. V. (2012). Foxm1 mediates cross talk between kras/mitogen-activated protein kinase and canonical Wnt pathways during development of respiratory epithelium. Mol. Cell. Biol. 32, 3838-3850. 10.1128/MCB.00355-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling O., Bornert J.-M., Chambon P. and Metzger D. (2009). Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 47, 14-18. 10.1002/dvg.20448 [DOI] [PubMed] [Google Scholar]
- Wong L., Yamasaki G., Johnson R. J. and Friedman S. L. (1994). Induction of beta-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. J. Clin. Investig. 94, 1563-1569. 10.1172/JCI117497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-Y., Chen J.-B., Tsai T.-F., Tsai Y.-C., Tsai C.-Y., Liang P.-H., Hsu T.-L., Wu C.-Y., Netea M.-G., Wong C.-H. and et al. (2013). CLEC4F is an inducible C-type lectin in F4/80-positive cells and is involved in alpha-galactosylceramide presentation in liver. PLoS ONE 8, e65070 10.1371/journal.pone.0065070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Evason K. J., Asahina K. and Stainier D. Y. R. (2013). Hepatic stellate cells in liver development, regeneration, and cancer. J. Clin. Invest. 123, 1902-1910. 10.1172/JCI66369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi Y., Namihisa T., Kuroda H., Komatsu I., Miyazaki A., Watanabe S. and Usui K. (1984). Immunocytochemical detection of desmin in fat-storing cells (Ito Cells). Hepatology 4, 709-714. 10.1002/hep.1840040425 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.