ABSTRACT
Tight junctions have been hypothesized to act as molecular fences in the plasma membrane of epithelial cells, helping to form differentiated apical and basolateral domains. While this fence function is believed to arise from the interaction of four-pass transmembrane claudins, the complexity of tight junctions has made direct evidence of their role as a putative diffusion barrier difficult to obtain. Here, we address this challenge by reconstituting claudin-4 into giant unilamellar vesicles using microfluidic jetting. We find that reconstituted claudin-4 alone can form adhesive membrane interfaces without the accessory proteins that are present in vivo. By controlling the molecular composition of the inner and outer leaflets of jetted vesicle membranes, we show that claudin-4-mediated interfaces can drive partitioning of extracellular membrane proteins with ectodomains as small as 5 nm but not of inner or outer leaflet lipids. Our findings indicate that homotypic interactions of claudins and their small size can contribute to the polarization of epithelial cells.
KEY WORDS: Cell adhesion, Epithelial cells, Polarity, Reconstitution, Tight junction, Transmembrane protein
Summary: Successful reconstitution of the tight junction protein claudin-4 drove interface formation between giant unilamellar vesicles. These interfaces were sufficient to act as molecular fences toward proteins but not lipids.
INTRODUCTION
Nowhere in biology is large-scale membrane organization more apparent than in epithelial tissue where cell surfaces are differentiated into two distinct domains, apical and basolateral (Koichi et al., 1974; Rodriguez-Boulan and Nelson, 1989). The tight junction (TJ), which controls the paracellular flux of ions, solutes and macromolecules in vertebrate epithelial cells, sits at the boundary between the apical and basolateral membrane domains (Zihni et al., 2016). Owing to its unique position, the TJ has long been implicated in the formation of a molecular ‘fence’ between the two domains (Tsukita et al., 2001), which may help to prevent apical-basolateral mixing of proteins and lipids. However, evidence for such a role is conflicting, and whether the TJ acts as a physical barrier to both protein and lipid diffusion in epithelia remains an open question.
Progress in understanding the basic properties of the TJ has been hindered by its molecular complexity. Work by Van Itallie et al. using proximity ligation and proteomics has demonstrated that >400 different proteins reside within the TJ in polarized kidney cells (Van Itallie et al., 2013). This complexity of the TJ has frustrated efforts to develop a mechanistic picture of putative fence function using traditional top-down cell biology techniques. One complementary method to address basic questions about biologically complex systems is to reconstitute key cellular components in vitro and to assay for activity. Such a bottom-up approach has not yet been applied to the TJ.
In a landmark paper, Tsukita and coworkers uncovered the membrane protein family necessary for forming intercellular pores at the TJ (Furuse et al., 1998). These proteins, the claudin family, are characterized by a four-pass architecture and are thought to interact across cells through homotypic interactions (Hou et al., 2013). Recently, the first crystal structures of transmembrane claudins have revealed one remarkable feature of claudins, namely that the height of their extracellular loops is short, <2 nm (Suzuki et al., 2014, 2017). These findings raise the question of whether close contact between claudins in trans might contribute to the putative fence function of the TJ.
Given the prevailing notion that claudins are involved in forming a diffusion barrier at the TJ (Trimble and Grinstein, 2015) and the recent finding that interfaces between membranes can exclude non-adhesive biomolecules (Schmid et al., 2016), we sought to directly test whether TJ claudins alone are capable of forming adhesive interfaces and establishing a diffusion barrier in vitro. While reconstitution of transmembrane claudins would offer a powerful way of isolating its activity and organization at interfaces, incorporation of oriented transmembrane proteins in unilamellar lipid vesicles large enough for visualizing membrane organization remains a challenge.
Here, we show that microfluidic jetting can be used to reconstitute claudins with defined orientation in a synthetic membrane. We take advantage of this approach to study both interface formation and membrane partitioning in giant unilamellar vesicles. We find that vesicles containing a classic claudin, claudin-4 (Cldn4) (Mitic et al., 2003), spontaneously form dense claudin–claudin interfaces between membranes and that these interfaces are sufficient to drive partitioning of extracellular membrane proteins, but not of lipids, demonstrating a key role for claudins, in the absence of other proteins, in TJ-mediated cell polarization.
RESULTS AND DISCUSSION
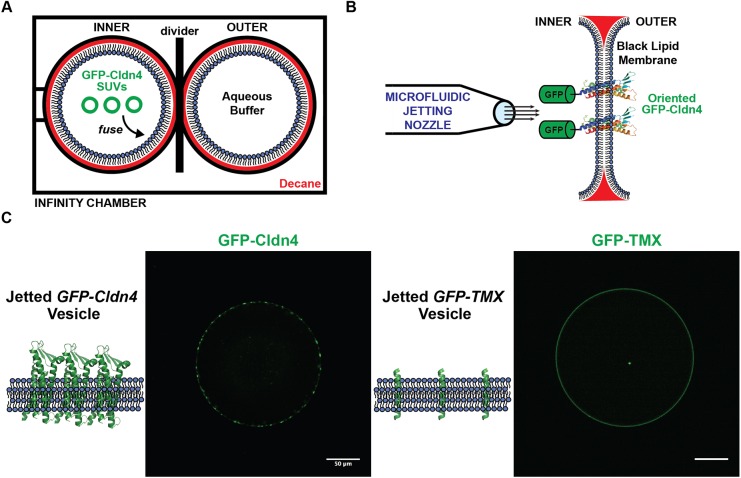
Incorporating membrane proteins into giant unilamellar vesicles (GUVs) suitable for fluorescence microscopy is a notoriously difficult problem, with limited in vitro solutions that apply to a small subset of transmembrane proteins (Cole et al., 2015; Dezi et al., 2013; Girard et al., 2004). We decided to tackle the challenge of inserting and orienting Cldn4 in GUVs by making use of microfluidic jetting (Richmond et al., 2011; Stachowiak et al., 2008) of black lipid membranes (Winterhalter, 2000). Cldn4 was selected for incorporation in unilamellar vesicles because it represents the largest claudin group, the classic claudins (Hou et al., 2013), and because it is well-characterized as a barrier claudin in vivo (Hou et al., 2010; Mitic et al., 2003; Van Itallie et al., 2001). Our strategy for asymmetric incorporation relied on (1) fusing proteoliposomes containing purified Cldn4 with lipid monolayers on one side of an infinity chamber (Fig. 1A) and (2) biasing the topology of Cldn4 during black lipid membrane formation after the removal of an acrylic divider (Fig. 1B). We hypothesized that addition of a large solubilization tag on one side of the protein would energetically bias the orientation of Cldn4 upon black lipid membrane formation. We, therefore, expressed Cldn4 with an N-terminal GFP tag in yeast cells and purified the protein in DDM detergent (Fig. S1). Since the N- and C-termini of Cldn4 are both located in the cytoplasm, we hypothesized that a large, hydrophilic domain on one side of the protein would restrict the orientation of Cldn4, ultimately orienting the GFP tag, and thus the N- and C-termini of the four-pass protein, in the lumen of the vesicle, the equivalent of the cytoplasm in cells.
Fig. 1.
Reconstitution of GFP–Cldn4 in jetted vesicles. (A) Infinity chamber configuration. (B) GFP–Cldn4 black lipid membrane scheme. (C) Fluorescence micrographs of single jetted vesicles containing either GFP–Cldn4 (left) or GFP–TMX (right). Scale bars: 50 μm.
To generate GUVs with oriented Cldn4, we first formed proteoliposomes with 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) and GFP–Cldn4 using established protocols (Rigaud et al., 1995). We then added GFP–Cldn4 proteoliposomes to either the inner or the outer DPhPC-stabilized droplet of an asymmetric two-droplet infinity chamber. After proteoliposomes had fused with the DPhPC monolayer, the central acrylic divider of the infinity chamber was removed and a black lipid membrane formed spontaneously. We next placed a 25-µm-diameter microfluidic nozzle close to the black lipid membranes and, upon triggering the piezoelectric actuator, formed GUVs by jetting. To test whether GFP–Cldn4 was oriented properly, we incubated both sets of GUVs with proteinase K, an enzyme capable of digesting exposed GFP (Lorenz et al., 2006). Only in the case where proteoliposomes were added to the outer chamber (corresponding to external orientation of the large, hydrophilic domain) did the GFP signal along the membrane reduce to background levels (Fig. S2). This experiment suggests that the GFP tag on the N-terminus of Cldn4 is unable to traverse the black lipid membrane and can guide Cldn4 into an oriented topology in jetted GUVs.
Next, we imaged GUVs that were generated by incubating GFP–Cldn4 proteoliposomes in the inner droplet of the infinity chamber. This geometry places the extracellular loops of Cldn4 on the outside of the vesicle and the N- and C-termini on the interior of the vesicle. First, we tested if Cldn4 would cluster in the absence of an opposing membrane. Confocal microscopy showed that GFP–Cldn4 formed large and highly variable clusters in DPhPC GUVs as evidenced by the bright puncta along the jetted membrane (Fig. 1C), consistent with the observation of cis claudin oligomers in cells (van Itallie et al., 2011; Kaufmann et al., 2012; Koval, 2013; Piontek et al., 2011; Rossa et al., 2014). To test if this clustering was specific to Cldn4, we compared the organization of Cldn4 to the membrane distribution of a synthetic transmembrane domain, TMX (Wimley and White, 2000), that should remain monomeric in lipid bilayers. After preparing TMX–GFP proteoliposomes and jetting TMX–GFP GUVs, we observed a uniform distribution of TMX along the lipid membrane (Fig. 1C). Our data lend support to the model that claudins interact in a cis configuration in lipid bilayers, even in the absence of other TJ adapter proteins.
We then investigated whether claudins in one membrane were able to form adhesive interactions with claudins in a closely apposed membrane. While claudin proteins have long been thought to interact in trans across epithelial cells, whether TJ claudins alone are sufficient for interface formation has not yet been examined experimentally. To test this, we incubated Cldn4 proteolipsomes in the inner droplet of an infinity chamber, as described above. After black lipid membranes were formed, we jetted multiple GUVs in quick succession and used confocal microscopy to image the assembly of GUVs. In contrast to the lack of interaction between jetted GUVs in the absence of adhesive proteins (Stachowiak et al., 2009), we observed adhesion between GUVs and enrichment of oriented GFP–Cldn4 at GUV–GUV interfaces (Fig. 2A). Interfaces appeared heterogeneous with respect to Cldn4 distribution; for instance, regions with both puncta and uniform GFP–Cldn4 were observed to co-exist within single interfaces. One striking feature of the GUV assemblies was the absence of close membrane contact at tri-vesicle junctions. In epithelial cells, a specific membrane protein, tricellulin, is necessary for forming tri-cellular TJ contacts (Ikenouchi et al., 2005). It appears from our experiments that Cldn4 is sufficient to form GUV–GUV interfaces but is not capable of forming tri-vesicle contacts, a finding consistent with the localization and function of TJ proteins, including the claudins, in polarized epithelial cells.
Fig. 2.
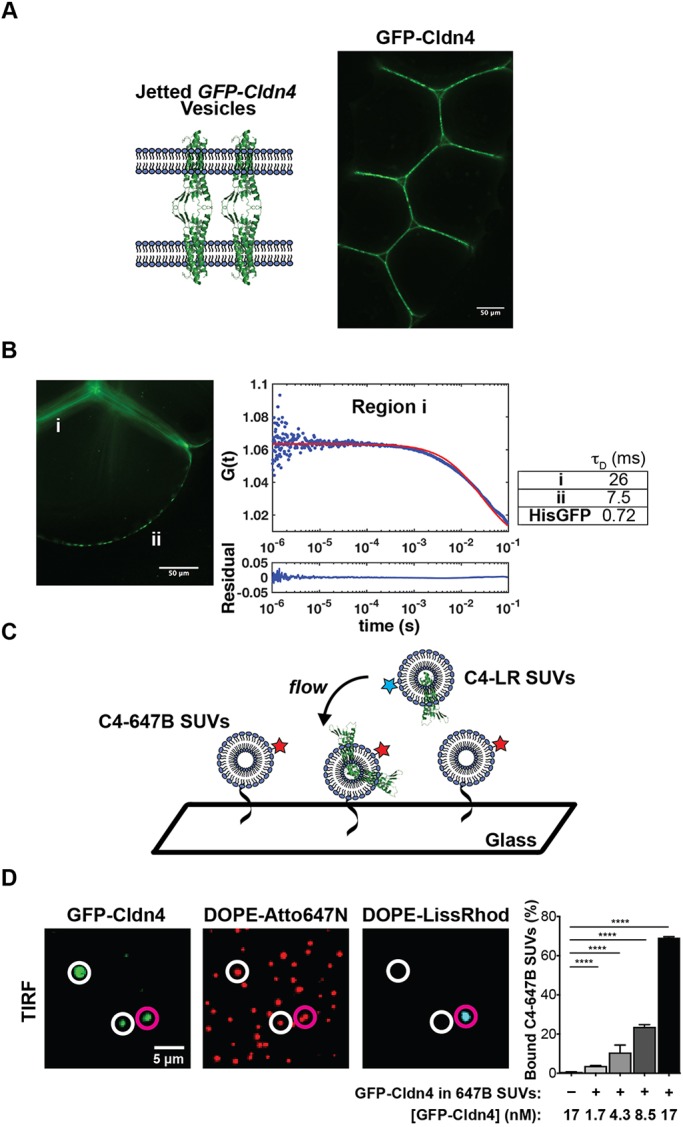
Cldn4 is sufficient to form membrane interfaces. (A) Diagram and fluorescence micrograph of jetted GUV assemblies containing GFP–Cldn4. (B) FCS autocorrelation curves were constructed for GFP–Cldn4 at interfaces, denoted region i, and at free membrane regions, denoted region ii (left). Diffusion times (right, mean, n=6 for all conditions) were then calculated for the different regions. (C) Scheme for the single SUV binding assay. (D) TIRF micrographs and quantification of the single SUV binding assay. The vesicles highlighted by magenta and white circles are bound and unbound SUVs, respectively. Data are presented as mean±s.d. (n=13, 9, 13, 12 and 10, respectively with n>500 particles counted per sample). ****P<0.0001 (two-tailed unpaired t-test). Scale bars: 50 μm (A,B), 5 μm (D).
We next turned our attention to the dynamics of GFP–Cldn4 in free membrane regions and in GUV–GUV interfaces. In the context of cell–cell contacts, previous characterization of claudin-1, a classic claudin, reported low mobility and a large immobile fraction (∼78%) for the protein, suggesting that claudins are statically captured and densely packed at the TJ in cells (Shen et al., 2008). We wanted to see whether the dynamics of Cldn4 were similar in vitro, even in the absence of other TJ proteins and an underlying cortical cytoskeleton. Fluorescence correlation spectroscopy (FCS) was employed to detect the dynamics of GFP–Cldn4 (Bacia et al., 2014) in two membrane regions of the jetted GUVs, at GUV–GUV interfaces (region i) and at free membrane regions (region ii) (Fig. 2B). We found significantly different autocorrelation curves for the two regions (Fig. S3). As our FCS measurements are based neither on a strict 2D nor a 3D configuration, and since large protein clusters are known to give rise to anomalous diffusion (Feder et al., 1996), we relied on the diffusion time parameter obtained from the autocorrelation curve to quantify and compare mobility, with higher diffusion times indicating reduced mobility. For region i, the diffusion time was 26±4.1 ms (mean±s.d.), whereas the diffusion time for region ii was 7.5±2.5 ms (Fig. 2B), reflecting the fact that the mobility of interfacial GFP–Cldn4 had been reduced by a factor of ∼3 at membrane interfaces, a result consistent with the dynamics of claudin-1 in cells. However, in contrast to in vivo studies, GFP–Cldn4 remained mobile at interfaces, which points to the role other TJ proteins play in forming static claudin structures in epithelial cells. To compare the diffusion of GFP–Cldn4 to a known protein, we jetted vesicles containing DOGS-Ni-NTA and complexed His–GFP to the outer leaflet. We found the diffusion time of His-GFP in jetted vesicles to be 0.72±0.56 ms, demonstrating that in free membrane portions of jetted vesicles, transmembrane GFP–Cldn4 formed clusters with reduced mobility compared to membrane-bound proteins.
Membrane interfaces in cells occur over a range of length scales, and, as such, we wanted to test whether the adhesive behavior of Cldn4 was dependent on the large surface area presented by GUVs. To quantify the trans binding of Cldn4 activity at smaller interfaces, we developed a single-vesicle binding assay (Fig. 2C) based on the GFP–Cldn4 proteoliposomes (100s of nm in diameter) outlined above. Two different populations of GFP–Cldn4 small unilamellar vesicles (SUVs) were prepared, one containing fluorescent DOPE-Lissamine Rhodamine B (C4-LR SUVs) and the other containing fluorescent DOPE-Atto647N and DOPE-biotin (C4-647B SUVs). Single C4-647B SUVs were tethered to glass surfaces through streptavidin and imaged using total internal reflection fluorescence (TIRF) microscopy. The GFP–Cldn4 signal from diffraction-limited Atto647N-positive particles was only observed for a sub-population of C4-647B SUVs (Fig. 2D), which is expected for the detergent-assisted insertion method (Mathiasen et al., 2014). After surface immobilization, we flowed in C4-LR SUVs and used TIRF microscopy to quantify the extent of colocalization (see Materials and Methods) of the two different fluorescent lipids as a proxy for Cldn4 binding. First, we noted that only particles positive for GFP–Cldn4 colocalized with C4-LR SUVs (Fig. 2D). Second, we found a concentration-dependent increase in bound C4-LR SUVs. No bound C4-LR SUVs were observed in the absence of C4-647B SUVs (Fig. 2D). Based on these results, purified GFP–Cldn4 appears competent for trans homotypic binding in small as well as large membrane interfaces.
Could the ability of claudin to establish interfaces between membranes contribute to the fence function of TJs in the absence of other TJ proteins? Recently, our laboratory has shown that the molecular dimensions of synthetic adhesive proteins can lead to exclusion of non-binding proteins from membrane interfaces (Schmid et al., 2016). We wondered whether this type of exclusion is also relevant to the TJ, since the extracellular portion of claudin was shown to be remarkably short in height (<2 nm) by crystallography (Suzuki et al., 2014, 2017) in stark contrast to the size of E-cadherin (>19 nm) of the adherens junctions (Harrison et al., 2011). Early work on the TJ pointed to a barrier able to prevent the diffusion of apical components, both proteins and lipids, to the basolateral surface of polarized epithelial cells (Diamond, 1977), but debate has continued with respect to the precise role TJs play in segregating apical and basolateral surfaces. Refinements to the early barrier model were made in light of additional findings: only outer leaflet lipids appeared to experience a barrier (Dragsten et al., 1981; van Meer and Simons, 1986) and the bulky glycoproteins and glycolipids were especially affected by a fence (Spiegel et al., 1985). After their discovery as the functional transmembrane unit of the TJ (Furuse et al., 1998), claudins were accordingly assigned a role as proteins mediating the diffusion barrier between the apical and basolateral domains (Trimble and Grinstein, 2015). However, one paper questioned this model and provided evidence of cell polarity in isolated single cells, suggesting that TJs are not needed to partition membrane components (Baas et al., 2004). Even more recently, Tsukita and coworkers found that mammary epithelial cells lacking claudin strands appeared to display signs of polarity (Umeda et al., 2006). To shed light on these varied findings and isolate the influence of claudins on establishing a diffusion barrier, we examined Cldn4-mediated interfaces formed between jetted vesicles for evidence of protein and lipid partitioning.
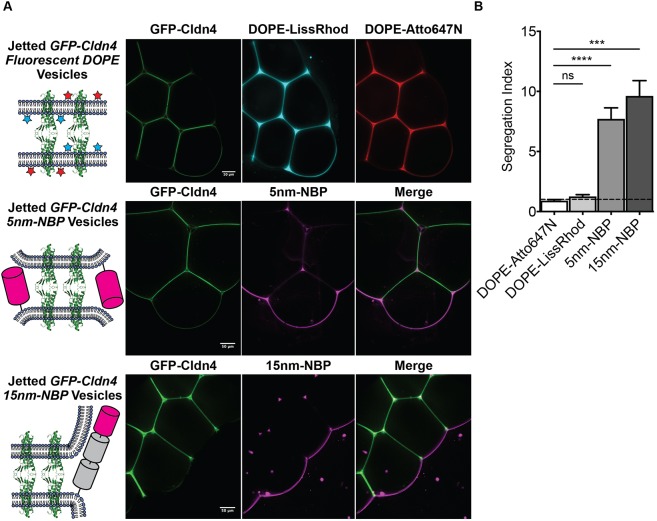
We jetted GFP–Cldn4 GUVs from two different black lipid membrane configurations. In the first configuration, the inner droplet was incubated with GFP–Cldn4 proteoliposomes containing the far-red fluorescent lipid DOPE-Atto647N, while the outer droplet was incubated with liposomes containing the red fluorescent lipid DOPE-Lissamine Rhodamine B (LissRhod). In the second configuration, the inner droplet was incubated with GFP–Cldn4 proteoliposomes as before, and the outer droplet was incubated with liposomes containing DOGS-Ni-NTA lipid. For the first configuration, black lipid membranes were formed and again GUVs were jetted in rapid succession, generating vesicles with different fluorophores in the inner and outer leaflets of the GUVs. Following Cldn4-mediated assembly, we did not observe segregation of the two different lipids from interfaces, indicating that Cldn4 alone does not exclude lipids, on either leaflet, from intermembrane junctions (Fig. 3A,B). With the second configuration, we also formed black lipid membranes, but before jetting, we incubated the outer leaflet with either of two different sized non-binding proteins (NBPs), one 5 nm in height (5nm-NBP) and the other 15 nm (15nm-NBP) (Schmid et al., 2016). We took advantage of His-tagged versions of NBPs to isolate the effect of ectodomain size and avoid the potentially convoluting effects of hydrophobic mismatch (Milovanovic et al., 2015) on partitioning. Next, we jetted multiple vesicles and imaged the resulting assemblies. For vesicles containing either the 5 or the 15 nm NBPs, we observed dramatic segregation of the non-binding protein at Cldn4-mediated interfaces, with complete exclusion for the 15 nm protein (Fig. 3A,B). Notably, this is the first in vitro example of an adhesion protein, namely claudin, forming a membrane interface capable of extensive exclusion of a 5 nm protein. These results, therefore, suggest that claudins alone may be able to partition most apical- and basolateral-domain proteins, especially bulky apical glycoproteins. Our data may also help explain the spatial separation between the TJ and the other epithelial junctions (i.e. adherens junction and desmosomes), which are formed by adhesive proteins with large molecular dimensions (e.g. E-cadherin and desmogleins, respectively).
Fig. 3.
Cldn4–Cldn4 interfaces drive the segregation of membrane-bound proteins. (A) Diagrams and fluorescent micrographs of jetted GFP–Cldn4 GUVs containing fluorescent lipids (upper), fluorescent 5 nm NBPs (middle) or fluorescent 15 nm NBPs (bottom). (B) Quantification of extent of segregation for inner leaflet lipids, DOPE-Atto647N, outer leaflet lipids, DOPE-LissRhod, and 5 nm NBP and 15 nm NBPs. Segregation Index=[Fluorescence Intensityfree membrane/(Fluorescence Intensityinterface/2)]. Data are presented as mean±s.d. (n=5 for all conditions). ***P<0.001; ****P<0.0001; ns, not significant (two-tailed unpaired t-test). Scale bars: 50 μm.
In summary, we have reconstituted claudin-mediated membrane interfaces in GUVs by means of microfluidic jetting of black lipid membranes. With this system, we addressed one aspect of a long-standing question in epithelial biology, namely whether claudins alone form a physical barrier that impedes protein and lipid diffusion in polarized epithelial tissue. By chemically engineering the molecular environment at membrane interfaces in vitro, we have found that Cldn4 can join GUVs together to build assemblies with non-uniform distribution of components. Our data point to a model where the molecular dimensions of claudin proteins limit the ability of proteins of ∼5 nm and larger to translocate past the interface, but do not limit lipids embedded in either leaflet. In vivo, this physical segregation model of protein partitioning at TJs may be further augmented to restrict lipid mixing through the formation of claudin strands, which could act as a barrier to lipid diffusion due to claudin membrane density, and thus excluded volume effects. The ability to control the orientation of claudin proteins in GUVs reported here opens up possibilities for more complex reconstitutions and questions, such as examining how TJ plaque proteins, for example, the zonula occludens (ZO) proteins, further tune the partitioning of membrane proteins and lipids at interfaces.
MATERIALS AND METHODS
General methods
All of the chemical reagents were of analytical grade, obtained from commercial suppliers and used without further purification, unless otherwise noted. Proteinase K was purchased from New England Biolabs. 1,2-diphytanoyl-sn-glycero-3-phophoscholine (DPhPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl], nickel salt (DOGS-Ni-NTA), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl), sodium salt (DOPE-biotin), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)-2000], ammonium salt (DSPE-PEG-biotin), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000], ammonium salt (DOPE-PEG) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(Lissamine Rhodamine B sulfonyl), ammonium salt (DOPE-Lissamine Rhodamine B) were obtained from Avanti Polar Lipids. 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine labeled with Atto 647N (DOPE-Atto647N) was purchased from Sigma-Aldrich.
Fluorescence imaging was carried out on a Ti Eclipse microscope (Nikon) equipped with a CSU-X spinning disk confocal module (Yokogawa) and a Zyla sCMOS camera (Andor). Fluorescence micrographs of GUVs were acquired with either a 10× (Nikon, NA 0.3) or a 20× objective (Nikon, NA 0.45). TIRF imaging was performed on the Ti Eclipse microscope (Nikon) using a 60× objective (Nikon, NA 1.49 TIRF) and an iXon Ultra EM-CCD camera (Andor).
Fluorescence correlation spectroscopy (FCS) data sets were acquired on a custom-built set-up with an inverted Nikon Eclipse TE2000 microscope (Nikon) equipped with a 100× oil immersion objective (Nikon, NA 1.49 TIRF) and a 488 nm CW laser as an excitation source (Sapphire, Coherent). Several turning mirrors were used to center and shape the laser as it was focused into a solution of Atto488. Fluorescence emission was collected on an avalanche photodiode (Excelitas, SPCM-AQR-14), and photon arrival times were measured with the counter module of an NI DAQ board (National Instruments, PCI-6321). Six 20 s photon streams were collected for each position on GUVs. Photon data were analyzed with custom MATLAB (MathWorks) scripts (available upon request).
Protein expression and purification
The cDNA encoding full-length human Cldn4 was cloned into the pBV-3 vector (a kind gift from Dr. Peng Yuan, Washington University) in frame with the gene encoding the GFP protein to create an N-terminal fusion protein. The construct was expressed in Pichia pastoris. Cells were disrupted by milling (Retsch MM400) and resuspended in lysis buffer containing 50 mM Tris-HCl pH 8.0 and 500 mM NaCl. Lysate was extracted with 2% (w/v) n-dodecyl β-d-maltopyranoside (DDM, Anatrace) for 2 h with stirring at 4°C and then centrifuged for 1 h at 30,000 g. Supernatant was added to cobalt-charged resin (G-Biosciences), and the suspension was mixed by inversion for 3 h. Resin was then washed with ten column volumes of buffer containing 20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM imidazole and 4 mM DDM, and eluted with buffer containing 250 mM imidazole. The eluted proteins were concentrated and further purified by gel filtration using a Superose 6 column (GE Healthcare) pre-equilibrated with buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM dithiothreitol (DTT) and 1 mM DDM. Peak fractions corresponding to the monomeric Cldn4 proteins were collected and concentrated to ∼8.5 µM.
TMX, a synthetic transmembrane domain bearing the sequence WNALAAVAAALAAVAAALAAVAA, was selected for its reported ability to insert into lipid membranes (Wimley and White, 2000). We cloned TMX into the bacterial expression vector pRSETa (Invitrogen) with a C-terminal eGFP fusion and an N-terminal fusion consisting of a 6× His tag, a maltose-binding protein (MBP) and a TEV cleavage site. Both N-terminal MBP and C-terminal eGFP fusions enhance the solubility of the intervening hydrophobic stretch, allowing us to express it in the cytoplasm of Escherichia coli. DNA fragments for each of these modules were amplified using PCR (MBP, eGFP) or synthesized as a gBlock fragment (TMX) and inserted into pRSETa between NheI and EcoRI sites. For protein production, 1–2 l of cells were grown to an optical density at 600 nm (OD600) of 0.7, induced with 250 µM IPTG and cultured overnight at 16°C. After 16 h, cells were harvested by centrifugation (5,000 g for 10 min) and resuspended in lysis buffer consisting of 25 mM HEPES, pH 7.5, 50 mM NaCl, 0.5 mM TCEP, 1% Triton X-100, and supplemented with PMSF and DNase I. After briefly pulse-sonicating and incubating for 30 min at 4°C, lysed cells were pelleted to remove cell debris and the lysate was circulated over a 5 ml MBPTrap HP column for 2 h. Following binding, the detergent concentration was gradually reduced to 0.1% and bound protein was eluted using 10 mM maltose. Finally, cleavage with TEV protease was performed overnight at 4°C using a 1:20 molar ratio of TEV to TMX. Following cleavage, residual MBP, TEV and uncleaved protein was removed by passing the protease-treated mixture over a HisTrap column.
Infinity chamber fabrication
Infinity chambers were fabricated by first cutting 4.5-mm-thick acrylic sheets (McMaster-Carr) using a laser cutter (Versa Laser). The inner and outer chambers were fabricated to be 4 mm in diameter and separated by a 0.15-mm-wide slot. 1.7 mm holes were then drilled into the side of the inner chamber. Subsequently, a thin 0.2 mm acrylic coverslip was cemented (TAP Plastics, Acrylic Cement) to the inner chamber, and a no. 1.5 glass coverslip coated in poly-L-lysine was glued (Norland Optical Adhesive 60) to the bottom of the outer chamber for imaging. After placing a thin 0.2 mm acrylic divider between the two chambers, latex (McMaster-Carr) was glued (Gorilla Super Glue) over the inner chamber hole and pierced with a 23G needle.
Proteoliposome preparation
Proteolipsomes were generated according to established procedures (Martens et al., 2007). Briefly, GFP–Cldn4 in 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM DTT and 1 mM DDM was diluted 1:1 with buffer containing 2% octyl β-D-glucopyranoside (OGP), 25 mM HEPES pH 7.5 and 150 mM NaCl to a final volume of 80 µl. 20 µl of 10 mM DPhPC SUVs in 25 mM HEPES, pH 7.5, 150 mM NaCl was added to the protein-detergent solution. The mixture was incubated for 15 min at room temperature. OGP-solubilized DPhPC/GFP–Cldn4 was then diluted 4-fold with 25 mM HEPES pH 7.5, 150 mM NaCl and 0.5 mM TCEP and dialyzed overnight against buffer containing 25 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP and 10 g of BioBeads (Thermo Scientific). A similar procedure was used to prepare TMX–GFP proteoliposomes and DOPC-based proteoliposomes for the single SUV binding assay.
Black lipid membrane formation
A planar bilayer between the inner and outer compartments of the infinity chamber was formed by following the protocol developed by Richmond et al. (Richmond et al., 2011). In brief, a 15 µl volume of a freshly prepared 25 mg/ml solution of DPhPC in n-decane was first applied to the central acrylic divider in the infinity chamber. Subsequently, 40 µl of a 5-fold dilution of the GFP–Cldn4 proteoliposome solution was added to the inner chamber, and a 40 µl volume of buffer containing 25 mM HEPES pH 7.5, 150 mM NaCl and 0.5 mM of TCEP was added to the outer chamber. Solutions in the infinity chamber were incubated for 15 min at room temperature, 45 min at 4°C and finally for 5 min at room temperature. The acrylic divider was subsequently removed and, after 15 min, oil evacuation and black lipid membrane formation was monitored using brightfield microscopy.
Microfluidic jetting of black lipid membranes
GUVs were formed by placing a microfluidic jetting nozzle (Microfab Technologies, single-jet microdispensing device with 25 µm orifice) filled with a 10% OptiPrep (Sigma-Aldrich), 25 mM HEPES pH 7.5, 150 mM NaCl solution in close proximity (<200 µm) to GFP–Cldn4 planar bilayers. This solution ultimately constitutes the lumen of the jetted GUVs and has a matched osmolarity, but different density, compared to the outer buffer. The piezoelectric actuator was controlled by a waveform generator (Agilent) and an amplifier (Krohn-Hite) with pulse train envelopes designed by a custom MATLAB script (available upon request). For jetting of GFP–Cldn4 GUVs, the actuator was triggered by an increasing parabolic envelope defined by 40 trapezoidal bursts at 15 kHz, 3 µs rise and fall times, a 30 µs hold time, and a maximum voltage of 15–30 V. Planar bilayer deformation and GUV formation were monitored via brightfield microscopy with a high-speed camera (Photron, 1024PCI). GUVs formed by microfluidic jetting sunk to the poly-L-lysine-coated coverglass due to the density mismatch between the interior of the GUVs and the surrounding buffer. To determine the orientation of GFP–Cldn4 in GUVs, a volume of 1 µl of 800 units/ml solution of proteinase K was added to the outer buffer after GUV formation and incubated with jetted GUVs for 30 min.
Single SUV binding assay
Proteoliposomes containing GFP–Cldn4 were prepared as described above. Two separate populations of GFP–Cldn4 proteoliposomes were generated for the single SUV assay: one with a lipid composition of DOPC (99.8%), DOPE-Atto647N (0.1%) and DOPE-biotin (0.1%), and the other with a lipid composition of DOPC (99.9%) and DOPE-Lissamine Rhodamine B (0.1%). Both sets of proteoliposomes were purified and diluted by gel filtration with a S400 HR gel filtration column (GE Healthcare) to remove lipid aggregates before use. To construct a sandwich channel, glass slides were piranha cleaned and treated with poly-L-lysine-g-PEG to passivate the surface. Glass coverslips, on the other hand, were RCA cleaned. The two glass surfaces were adhered to one another with double-sided tape, and an immobile supported lipid bilayer (SLB) was formed on the glass coverslip by incubating the chamber with a 1 mg/ml solution of SUVs containing DOPC (97%), DOPE-PEG (2.94%), and DOPE-PEG-biotin (0.06%) for 10 min. DOPE-PEG acts to passivate the coverslip surface, while DOPE-PEG-biotin acts as tethering points for single SUVs. The channel was washed with a volume of 200 µl of buffer containing 25 mM HEPES pH 7.5, 150 mM NaCl, and then incubated with a 50 µg/ml solution of streptavidin (Life Technologies) for 5 min. The channel was washed with buffer and then incubated with purified proteoliposomes containing GFP–Cldn4, DOPC, DOPE-Atto647N and DOPE-biotin for 10 min. The channel was subsequently washed with buffer and then either imaged by TIRF microscopy or incubated with the second set of proteoliposomes containing GFP–Cldn4, DOPC, and DOPE-Lissamine Rhodamine B at various dilutions and followed by washing. To determine the percentage of bound DOPE-Atto647N-containing proteoliposomes, a custom MATLAB script was used (available upon request). Thresholding was first applied to each fluorescence channel. Fluorescent particles were then located and their centroids were determined using a peak-finding algorithm. Positions of particles positive for both DOPE-Atto647N and GFP-Cldn4 were first saved and then compared to positions of DOPE-Lissamine Rhodamine B-positive particles to calculate the percentage of bound DOPE-Atto647N-containing proteoliposomes. At least ∼500 particles/sample were analyzed for each data point.
Jetted GUV segregation assay
For the jetted GUV segregation assay, four separate proteoliposome/liposomes were formulated as described above: proteoliposomes containing GFP-Cldn4/DPhPC, proteoliposomes containing GFP-Cldn4/DPhPC/DOPE-Atto647N (0.1%), liposomes containing DPhPC/DOGS-Ni-NTA (2.5%), and liposomes containing DPhPC/DOPE-Lissamine Rhodamine B (0.1%). Using the fabricated infinity chamber, a 15 µl volume of a freshly prepared 25 mg/ml DPhPC solution in n-decane was applied to the central acrylic divider. A 40 µl volume of a 5-fold dilution of proteoliposomes containing either GFP–Cldn4/DPhPC/DOPE-Atto647N or GFP–Cldn4/DPhPC was incubated in the inner chamber, while a 40 µl volume of a 5-fold dilution of liposomes containing either DPhPC/DOPE-Lissamine Rhodamine B or DPhPC/DOGS-Ni-NTA was incubated in the outer chamber simultaneously. After the acrylic divider was removed, a black lipid membrane was formed over the course of 15 min. For the fluorescent lipid experiment, GFP–Cldn4 GUVs were jetted in rapid succession with DOPE-Atto647N embedded in the inner leaflet and DOPE-Lissamine Rhodamine B embedded in the outer leaflet. For the fluorescent protein experiment, after planar bilayer formation, a volume of 1 µl of a 20 µM solution of either the 5 nm or the 15 nm His-tagged mCherry NBP was added to the outer chamber and incubated for 30 min. GUVs were then generated in rapid succession to produce assemblies of GFP–Cldn4 interfaces.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: B.B., D.A.F.; Methodology: B.B., S.S., M.D.V.; Formal analysis: B.B., S.S.; Investigation: B.B.; Resources: S.S., J.W., J.H.; Data curation: B.B.; Writing - original draft: B.B., D.A.F.; Writing - review & editing: B.B., S.S., M.D.V., J.W., J.H., D.A.F.; Supervision: J.H., D.A.F.; Project administration: D.A.F.; Funding acquisition: B.B., J.H., D.A.F.
Funding
This work was supported by grants from the National Institutes of Health (R01GM114344 and R01DK084059). B.B. was supported by a NIH Ruth L. Kirschstein NRSA fellowship from the NIH (1F32GM115091). S.S. was supported by a fellowship from the Life Sciences Research Foundation. M.D.V. was supported by a CASI fellowship from the Burroughs Wellcome Fund. D.A.F. is a Chan Zuckerberg Biohub Investigator. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.221556.supplemental
References
- Baas A. F., Kuipers J., van der Wel N. N., Batlle E., Koerten H. K., Peters P. J. and Clevers H. C. (2004). Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457-466. 10.1016/S0092-8674(04)00114-X [DOI] [PubMed] [Google Scholar]
- Bacia K., Haustein E. and Schwille P. (2014). Fluorescence correlation spectroscopy: principles and applications. Cold Spring Harb. Protoc. 2014, 709-725. 10.1101/pdb.top081802 [DOI] [PubMed] [Google Scholar]
- Cole C. M., Brea R. J., Kim Y. H., Hardy M. D., Yang J. and Devaraj N. K. (2015). Spontaneous reconstitution of functional transmembrane proteins during bioorthogonal phospholipid membrane synthesis. Angew. Chem. Int. Ed. 54, 12738-12742. 10.1002/anie.201504339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezi M., Cicco A. D., Bassereau P. and Lévy D. (2013). Detergent-mediated incorporation of transmembrane proteins in giant unilamellar vesicles with controlled physiological contents. Proc. Natl. Acad. Sci. 110, 7276-7281. 10.1073/pnas.1303857110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond J. M. (1977). Twenty-first Bowditch lecture. The epithelial junction: bridge, gate, and fence. Physiologist 20, 10-18. [PubMed] [Google Scholar]
- Dragsten P. R., Blumenthal R. and Handler J. S. (1981). Membrane asymmetry in epithelia: is the tight junction a barrier to diffusion in the plasma membrane? Nature 294, 718-722. 10.1038/294718a0 [DOI] [PubMed] [Google Scholar]
- Feder T. J., Brust-Mascher I., Slattery J. P., Baird B. and Webb W. W. (1996). Constrained diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys. J. 70, 2767-2773. 10.1016/S0006-3495(96)79846-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Fujita K., Hiiragi T., Fujimoto K. and Tsukita S. (1998). Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141, 1539-1550. 10.1083/jcb.141.7.1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard P., Pécréaux J., Lenoir G., Falson P., Rigaud J.-L. and Bassereau P. (2004). A new method for the reconstitution of membrane proteins into giant unilamellar vesicles. Biophys. J. 87, 419-429. 10.1529/biophysj.104.040360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison O. J., Jin X., Hong S., Bahna F., Ahlsen G., Brasch J., Wu Y., Vendome J., Felsovalyi K., Hampton C. M. et al. (2011). The extracellular architecture of adherens junctions revealed by crystal structures of Type I cadherins. Structure 19, 244-256. 10.1016/j.str.2010.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Renigunta A., Yang J. and Waldegger S. (2010). Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc. Natl. Acad. Sci. USA 107, 18010-18015. 10.1073/pnas.1009399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Rajagopal M. and Yu A. S. L. (2013). Claudins and the kidney. Annu. Rev. Physiol. 75, 479-501. 10.1146/annurev-physiol-030212-183705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J., Furuse M., Furuse K., Sasaki H., Tsukita S. and Tsukita S. (2005). Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 171, 939-945. 10.1083/jcb.200510043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann R., Piontek J., Grüll F., Kirchgessner M., Rossa J., Wolburg H., Blasig I. E. and Cremer C. (2012). Visualization and quantitative analysis of reconstituted tight junctions using localization microscopy. PLoS ONE 7, e31128 10.1371/journal.pone.0031128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koichi K., Michiya F. and Makoto N. (1974). Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim. Biophys. Acta BBA - Lipids Lipid Metab. 369, 222-233. 10.1016/0005-2760(74)90253-7 [DOI] [PubMed] [Google Scholar]
- Koval M. (2013). Differential pathways of claudin oligomerization and integration into tight junctions. Tissue Barriers 1, e24518 10.4161/tisb.24518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz H., Hailey D. W., Wunder C. and Lippincott-Schwartz J. (2006). The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nat. Protoc. 1, 276-279. 10.1038/nprot.2006.42 [DOI] [PubMed] [Google Scholar]
- Martens S., Kozlov M. M. and McMahon H. T. (2007). How synaptotagmin promotes membrane fusion. Science 316, 1205-1208. 10.1126/science.1142614 [DOI] [PubMed] [Google Scholar]
- Mathiasen S., Christensen S. M., Fung J. J., Rasmussen S. G. F., Fay J. F., Jorgensen S. K., Veshaguri S., Farrens D. L., Kiskowski M., Kobilka B. et al. (2014). Nanoscale high-content analysis using compositional heterogeneities of single proteoliposomes. Nat. Methods 11, 931-934. 10.1038/nmeth.3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milovanovic D., Honigmann A., Koike S., Göttfert F., Pähler G., Junius M., Müllar S., Diederichsen U., Janshoff A., Grubmüller H. et al. (2015). Hydrophobic mismatch sorts SNARE proteins into distinct membrane domains. Nat. Commun. 6, 5984 10.1038/ncomms6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitic L. L., Unger V. M. and Anderson J. M. (2003). Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci.. 12, 218-227. 10.1110/ps.0233903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek J., Fritzsche S., Cording J., Richter S., Hartwig J., Walter M., Yu D., Turner J. R., Gehring C., Rahn H.-P. et al. (2011). Elucidating the principles of the molecular organization of heteropolymeric tight junction strands. Cell. Mol. Life Sci. 68, 3903-3918. 10.1007/s00018-011-0680-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond D. L., Schmid E. M., Martens S., Stachowiak J. C., Liska N. and Fletcher D. A. (2011). Forming giant vesicles with controlled membrane composition, asymmetry, and contents. Proc. Natl. Acad. Sci. USA 108, 9431-9436. 10.1073/pnas.1016410108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud J.-L., Pitard B. and Levy D. (1995). Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim. Biophys. Acta 1231, 223-246. 10.1016/0005-2728(95)00091-V [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E. and Nelson W. J. (1989). Morphogenesis of the polarized epithelial cell phenotype. Science 245, 718-725. 10.1126/science.2672330 [DOI] [PubMed] [Google Scholar]
- Rossa J., Ploeger C., Vorreiter F., Saleh T., Protze J., Günzel D., Wolburg H., Krause G. and Piontek J. (2014). Claudin-3 and claudin-5 protein folding and assembly into the tight junction are controlled by non-conserved residues in the transmembrane 3 (TM3) and extracellular loop 2 (ECL2) segments. J. Biol. Chem. 289, 7641-7653. 10.1074/jbc.M113.531012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid E. M., Bakalar M. H., Choudhuri K., Weichsel J., Ann H., Geissler P. L., Dustin M. L. and Fletcher D. A. (2016). Size-dependent protein segregation at membrane interfaces. Nat. Phys. 12, 704-711. 10.1038/nphys3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Weber C. R. and Turner J. R. (2008). The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J. Cell Biol. 181, 683-695. 10.1083/jcb.200711165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel S., Blumenthal R., Fishman P. H. and Handler J. S. (1985). Gangliosides do not move from apical to basolateral plasma membrane in cultured epithelial cells. Biochim. Biophys. Acta BBA - Biomembr. 821, 310-318. 10.1016/0005-2736(85)90101-4 [DOI] [PubMed] [Google Scholar]
- Stachowiak J. C., Richmond D. L., Li T. H., Liu A. P., Parekh S. H. and Fletcher D. A. (2008). Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc. Natl. Acad. Sci. USA 105, 4697-4702. 10.1073/pnas.0710875105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak J. C., Richmond D. L., Li T. H., Brochard-Wyart F. and Fletcher D. A. (2009). Inkjet formation of unilamellar lipid vesicles for cell-like encapsulation. Lab. Chip. 9, 2003-2009. 10.1039/b904984c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Nishizawa T., Tani K., Yamazaki Y., Tamura A., Ishitani R., Dohmae N., Tsukita S., Nureki O. and Fujiyoshi Y. (2014). Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344, 304-307. 10.1126/science.1248571 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Tani K. and Fujiyoshi Y. (2017). Crystal structures of claudins: insights into their intermolecular interactions. Ann. N. Y. Acad. Sci. 1397, 25-34. 10.1111/nyas.13371 [DOI] [PubMed] [Google Scholar]
- Trimble W. S. and Grinstein S. (2015). Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol. 208, 259-271. 10.1083/jcb.201410071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S., Furuse M. and Itoh M. (2001). Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2, 285-293. 10.1038/35067088 [DOI] [PubMed] [Google Scholar]
- Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M. and Tsukita S. (2006). ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126, 741-754. 10.1016/j.cell.2006.06.043 [DOI] [PubMed] [Google Scholar]
- Van Itallie C., Rahner C. and Anderson J. M. (2001). Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J. Clin. Invest. 107, 1319-1327. 10.1172/JCI12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie C. M., Mitic L. L. and Anderson J. M. (2011). Claudin-2 Forms homodimers and is a component of a high molecular weight protein complex. J. Biol. Chem. 286, 3442-3450. 10.1074/jbc.M110.195578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie C. M., Aponte A., Tietgens A. J., Gucek M., Fredriksson K. and Anderson J. M. (2013). The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks. J. Biol. Chem. 288, 13775-13788. 10.1074/jbc.M113.466193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G. and Simons K. (1986). The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J. 5, 1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W. C. and White S. H. (2000). Designing transmembrane alpha-helices that insert spontaneously. Biochemistry (Mosc) 39, 4432-4442. 10.1021/bi992746j [DOI] [PubMed] [Google Scholar]
- Winterhalter M. (2000). Black lipid membranes. Curr. Opin. Colloid Interface Sci. 5, 250-255. 10.1016/S1359-0294(00)00063-7 [DOI] [Google Scholar]
- Zihni C., Mills C., Matter K. and Balda M. S. (2016). Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 17, 564-580. 10.1038/nrm.2016.80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.