ABSTRACT
Cell-cell interactions that result from membrane proteins binding weakly in trans can cause accumulations in cis that suggest cooperativity and thereby an acute sensitivity to environmental factors. The ubiquitous ‘marker of self’ protein CD47 binds weakly to SIRPα on macrophages, which leads to accumulation of SIRPα (also known as SHPS-1, CD172A and SIRPA) at phagocytic synapses and ultimately to inhibition of engulfment of ‘self’ cells – including cancer cells. We reconstituted this macrophage checkpoint with GFP-tagged CD47 on giant vesicles generated from plasma membranes and then imaged vesicles adhering to SIRPα immobilized on a surface. CD47 diffusion is impeded near the surface, and the binding-unbinding events reveal cooperative interactions as a concentration-dependent two-dimensional affinity. Membrane fluctuations out-of-plane link cooperativity to membrane flexibility with suppressed fluctuations in the vicinity of bound complexes. Slight acidity (pH 6) stiffens membranes, diminishes cooperative interactions and also reduces ‘self’ signaling of cancer cells in phagocytosis. Sensitivity of cell-cell interactions to microenvironmental factors – such as the acidity of tumors and other diseased or inflamed sites – can thus arise from the collective cooperative properties of flexible membranes.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Adhesion, Vesicles, Elasticity, Cooperative binding, GPMV
Summary: Binding of membrane proteins smooths the fluctuating membrane and facilitates more binding, which can be very important for weak immunological interactions such as the interaction of macrophage SIRPα with CD47 on cancer cells.
INTRODUCTION
Cells use adhesion molecules to interact with and thereby identify other cells, but recent theoretical and simulation results indicate that the physical act of binding is highly cooperative and mediated by membrane fluctuations in a generalized law of mass action (Krobath et al., 2009). Differences in 3D versus 2D binding have been studied with cells adhering to supported lipid bilayers (Zhu et al., 2007; Tolentino et al., 2008) and with lipid vesicles bearing reconstituted binding molecules (Fenz and Sengupta, 2012; Bihr et al., 2014, Chan et al., 2007), but cell experiments are complicated by cytoskeleton-driven remodeling (Huppa et al., 2010) and liposomes lack the molecular richness of cell membranes. Here, we use cell-derived giant vesicles that lack cytoskeletal interactions (Sezgin et al., 2012), and we focus on the universally expressed protein CD47 in membranes as it mediates adhesion to SIRPα (also known as SHPS-1, CD172A and SIRPA) on a rigid surface. Wide-ranging (bio)physical experiments provide evidence of equilibration and are linked closely to multiscale simulation.
CD47 binding to SIRPα on macrophages ultimately inhibits engulfment of ‘self’ cells by macrophages (Oldenborg et al., 2000) and is now being pursued clinically in cancer therapy by antibody blockade of this ‘macrophage checkpoint’ (reviewed in Alvey and Discher, 2017). ‘Self’ cells also signal to T-cells (via interactions of PDL1 with PD1) through a phosphatase pathway also downstream of SIRPα, and antibody blockade of the PDL1-PD1 T-cell checkpoint has already proven remarkably effective in the clinic against some cancers. As with the majority of cell-cell signaling molecules, trans interactions of two integral membrane proteins in juxtaposed plasma membranes leads to a dependence of membrane adhesion on membrane conformation. An unfavorable membrane geometry such as occurs with chemically rigidified discocyte-shaped red blood cells, limits CD47 signaling and increases phagocytosis (Sosale et al., 2015a). For distances between membranes that are similar to the sizes of adhesion molecules, the membrane-membrane interaction depends on (membrane) flexibility and fluctuations as well as conformations of binding molecules (Xu et al., 2015). Studies here of a native membrane protein interacting with its immobilized receptor provides some of the first experimental evidence of cooperative binding due to suppression of membrane fluctuations. The findings should apply to immunological synapse complexes and many membrane-bound signaling complexes beyond CD47-SIRPα at the phagocytic synapse.
RESULTS AND DISCUSSION
Cell-derived vesicles retain CD47 and thermally fluctuate
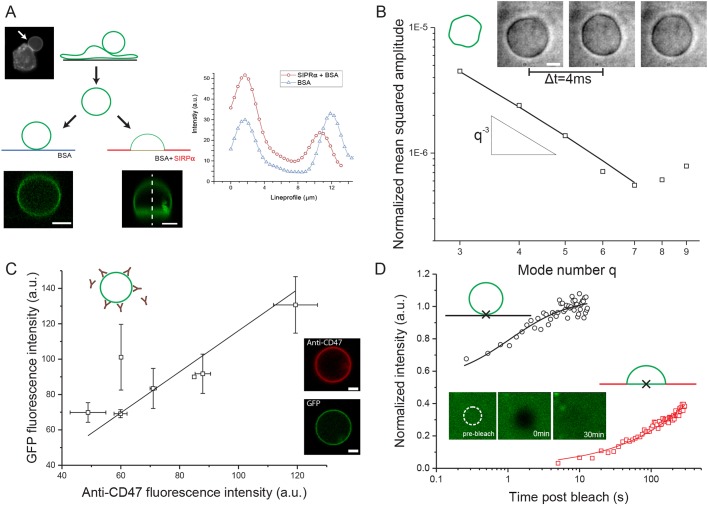
Membrane sheets of several hundred square micrometers in area were detached from the cortical cytoskeletons of CD47-GFP expressing HEK cells (Fig. 1A, left) via a method previously used to study lipid segregation (Sezgin et al., 2012). These sheets spontaneously bud and seal to form giant unilamellar vesicles that are more specifically referred to as giant plasma membrane vesicles (GPMVs). GPMVs possess a near-native composition of lipid and membrane protein (Bauer et al., 2009) and retain lipid and protein mobility and conformation (Baumgart et al., 2007; Sezgin et al., 2012; Veatch et al., 2008) as well as optically resolvable shape fluctuations (Fig. 1B, inset).
Fig. 1.
GPMV characterization and CD47-GFP diffusion. (A) GPMV formation from CD47-GFP-expressing HEK cells. Left: epi-fluorescence image of a cell with an attached GPMV (arrow). Left: epi-fluorescence image of a cell with an attached GPMV (arrow). Right: confocal microscopy z-stacks of a free GPMV on a BSA-coated coverslip (left) or an adherent GPMV on a SIRPα-coated coverslip (right). Representative fluorescent intensity traces quantified from image stacks. Lines are guides to the eye. (B) Fluctuation analyses of GPMVs are compatible with elastic sheet model of bending rigidity κ≈10±3 kBT (mean±s.d.) Inset shows phase contrast images of thermal fluctuations. Low exposure times of 200 μs and 4000 images per vesicle avoid artefacts from averaging fast fluctuations. (C) Anti-CD47 and GFP signals correlate in confocal microscopy: y=1.2x (R2=0.98). Each data point corresponds to measurement of an individual GPMV in at least two independent experiments. Error bars indicate s.d. (D) Representative recovery curve in FRAP of CD47-GFP on unbound membrane segments (black circles) with fit to free diffusion model, and after bleaching the adhering membrane segment (red squares) with fit to adhesion model. Data were normalized to the average fluorescent intensity of the unbleached area before application of the bleaching pulse. Note the logarithmic time scale. Crosses indicate bleached location on vesicles. Inset images show bleached CD47-GFP, indicating full recovery on longer timescales. White circle indicates bleached area of 1 μm in diameter. Scale bars: 5 µm.
Phase-contrast microscopy with µs exposure times shows the amplitudes of Fourier modes q of membrane fluctuations at the equatorial plane (Fig. 1B). The q−3 dependence agrees with the Helfrich theory of elastic membranes fluctuating at finite temperature, as previously applied to pure lipid vesicles in a fluid state (Gracia et al., 2010; Lipowsky, 1991). The mechanical properties of GPMVs can thus be reduced to a few parameters, such as bending rigidity, tension and preferred curvature, that are independent of detailed molecular composition and the biological state(s) of membranes.
CD47 is properly oriented in GPMVs and functional for binding
CD47-GFP makes GPMVs uniformly fluorescent (Fig. 1A, Fig. S1A), and a red fluorescent anti-CD47, which binds to the extracellular domain of CD47, suggests proper orientation. The two signals not only correlated (Fig. 1C) but also confirm a variation of CD47 levels between GPMVs. A range of CD47 concentrations could thus be studied in adhesion experiments.
To assess CD47-SIRPα binding, we physically adsorbed the extracellular domain of SIRPα (fused to a large single domain protein, GST) or the surface-blocking agent bovine serum albumin (BSA) on coverslips (Subramanian et al., 2006). SIRPα homogenously adsorbed based on fluorescence imaging (Fig. S1B). Gravitational settling of GPMVs onto BSA-coated coverslips showed near-spherical vesicle shapes and therefore little interaction (Fig. 1A, top right). On SIRPα-coated coverslips in contrast, GPMVs deformed strongly as they adhered (Fig. 1A, bottom right): the upper unbound cap is spherical whereas the flattened lower membrane segment is where CD47 accumulated in binding the immobilized SIRPα (Fig. 1A). After initial contact between a GPMV and the SIRPα surface, CD47-GFP diffused into the adhesion zone in a ring-like fashion and became homogeneous over time (Fig. S1C). Experiments were then done on GPMVs with equilibrated and homogenous adhesion, lacking any domain formation and thus implying a single species of CD47-SIRPα adhesion complexes (Weikl et al., 2009). Diffusion dynamics in these two segments were measured by fluorescent recovery after bleaching (FRAP) of micrometer-sized spots.
2D binding affinity determined by FRAP
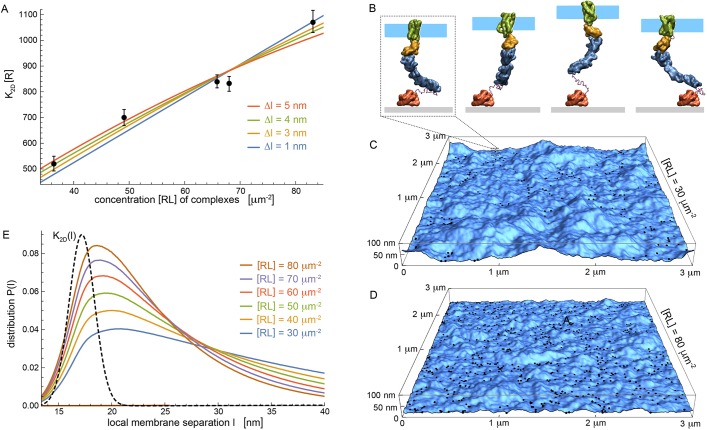
FRAP dynamics in small bleached spots differed between unbound and bound membrane segments. In unbound segments, CD47 fluorescence recovered over seconds and yielded a diffusion constant of Dcd47=0.12±0.02 µm2/s (n=3) as determined from fitting to a standard model for unhindered diffusion (Soumpasis, 1983). In stark contrast, CD47 within the adherent zone recovered ∼100-times slower (Fig. 1D). CD47-SIRPα unbinding events are likely followed by a short distance of free diffusion before re-binding (Sprague et al., 2004). Such trajectories are governed by a reaction-diffusion model (Sprague et al., 2004; see Material and Methods). By fitting the model to the measured time-intensity traces the 2D binding affinity [R]K2D=[R]kon/koff is extracted. Here, [R] is the density of (unbound) SIRPα on the glass surface. The fit to the model gives [R]K2D data for individual vesicles expressing different densities of CD47. The results are independent of the exact value of [R], but [R]∼4000 molecules/µm2 (see Fig. S1B). Strikingly, CD47-SIRPα complex concentration and binding affinity are positively correlated, which is indicative of cooperative binding (Fig. 2A). Computer simulations help to clarify the cooperative effects in CD47-SIRPα binding and unbinding.
Fig. 2.
Multiscale modeling of experiments to clarify interactions. (A) Binding constant K2D as a function of complex concentration [RL] from experiments (data points) and modeling (lines) for different values Δl of repulsive membrane-substrate interactions. Modeling is based on Eqn 1, with fit values Kmax=(956±24)/[R], (470±10)/[R], (378±8)/[R] and (326±8)/[R] for Δl=1, 3, 4, 5 nm, respectively. Binding constant K2D has units of concentration [R] of unbound SIRPα. Error bars indicate 95% confidence intervals for fitting the reaction-diffusion model±s.e.m.). Each data point corresponds to measurement of an individual GPMV in at least two independent experiments. (B) Simulation snapshots of coarse-grained CD47-SIRPα complex. Separation between fluid membrane patch (cyan) and substrate (grey) varies in simulations mainly because of conformational changes of the unstructured linker that covalently connects extracelluar SIRPα domain (blue) to substrate-bound GST (red). (C,D) Snapshots of adhering membranes with area 3×3 μm2 for respective concentrations [RL]=30, 80 μm−2 of CD47-SIRPα complexes at Δl=4 nm. Black dots indicate complexes. (E) Distributions P(l) for local separation between membrane and substrate obtained from averaging over many membrane snapshots for various [RL]. Dashed black line represents K2D(l) (arbitrary units).
Cooperative binding by suppression of membrane fluctuations
Our multiscale modeling approach combines: (i) simulations of a coarse-grained CD47-SIRPα complex (Fig. 2B) to determine the effective spring constant of the complex, and (ii) simulations of large membrane segments adhering via CD47-SIRPα complexes, modeled as elastic springs (Fig. 2C,D). In simulations of a CD47-SIRPα complex, the transmembrane domain of CD47 is embedded in a small membrane patch parallel to the substrate and connected by a short flexible linker to its extracellular domain. This domain is bound to SIRPα in the correct orientation based on the crystal structure of the complex (Protein Databank, PDB-2JJS). SIRPα is connected via a linker to substrate-bound GST in the experiments. Flexible linkers and protein interactions have been successfully applied to several multi-protein complexes (Kim and Hummer, 2008), including ERK2-HePTP (Francis et al., 2011) and ESCRT complexes (Boura et al., 2011, Boura et al., 2012). Variation of the separations between the membrane patch and substrate observed in simulations (Fig. S2) result from the molecular architecture and flexibility of the CD47-SIRPα complex. The mean length (l0=17.2 nm) of these separations is the preferred membrane-substrate separation of the complex, the standard deviation σ=1.2 nm reflects flexibility and an effective spring constant kS=kBT/σ2 of the complex, where kB is Boltzmann's constant and T is temperature (∼300 K).
In our simulations of large adhering membrane segments (Fig. 2C,D), membranes are elastic surfaces with bending rigidity κ=10kBT as measured experimentally, and discretized into quadratic patches of size 15×15 nm2 (Weikl and Lipowsky, 2006). Membrane patches that contain CD47-SIRPα complexes are bound to the substrate with separation l0 and spring constant kS. Repulsive interactions between the protein layer on the substrate and the membrane are taken into account by allowing only local separations l>(l0–Δl) between membrane patches and substrate, with Δl>0. From simulations, we obtain distributions P(l) of local separations that depend on concentration [RL] of CD47-SIRPα complexes (Fig. 2E) because bound complexes constrain membrane shape fluctuations reflected by P(l).
From the distributions P(l), and from the preferred length l0 and spring constant σ of the complexes, we obtain the binding constant K2D from Xu et al. (2015):
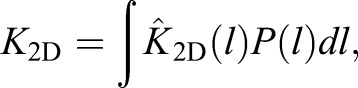 |
(1) |
with 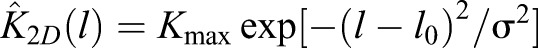 up to a pre-factor Kmax, which we treat as a fit parameter. K2D increases with bond concentration [RL] (Fig. 2A), because distributions P(l) become narrow and shift towards l0 with increasing [RL] (Fig. 2E). Modeling results agree with experimental data points for different values of Δl for the repulsion between membrane and protein-coated substrate (Fig. 2A). We vary Δl because neither the thickness of protein layers nor the role(s) of GPMV proteins in steric repulsion are known. Strong increase of K2D with [RL] reflects cooperative binding of CD47-SIRPα, which results from smoothing of membrane fluctuations by bound complexes (Krobath et al., 2009; Hu et al., 2013). Smoothing facilitates formation of additional CD47-SIRPα complexes.
up to a pre-factor Kmax, which we treat as a fit parameter. K2D increases with bond concentration [RL] (Fig. 2A), because distributions P(l) become narrow and shift towards l0 with increasing [RL] (Fig. 2E). Modeling results agree with experimental data points for different values of Δl for the repulsion between membrane and protein-coated substrate (Fig. 2A). We vary Δl because neither the thickness of protein layers nor the role(s) of GPMV proteins in steric repulsion are known. Strong increase of K2D with [RL] reflects cooperative binding of CD47-SIRPα, which results from smoothing of membrane fluctuations by bound complexes (Krobath et al., 2009; Hu et al., 2013). Smoothing facilitates formation of additional CD47-SIRPα complexes.
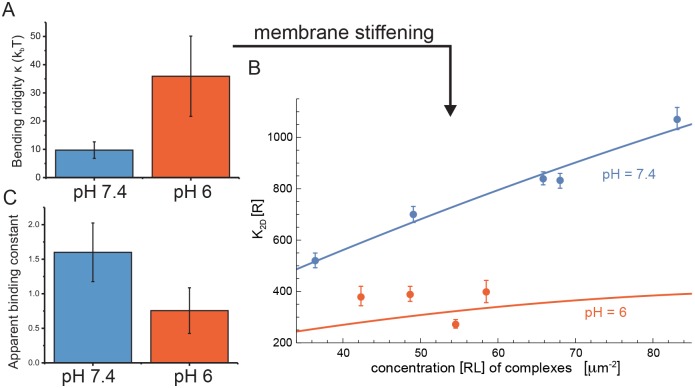
Slight acidity inhibits CD47-SIRPα cooperativity and favors phagocytosis
To further validate our model, we explored the effects of membrane fluctuations (and hence cooperativity) in relation to the bending rigidity of the membrane. We discovered that subjecting cells to a slight acidity of pH 6 induces stiffening of the GPMV (Fig. 3A). In both simulations and experiments, such rigidification led to a substantial decrease in cooperativity (Fig. 3B). Simulations and experiments are linked via the measured bending rigidity, which lumps the complex cell (plasma membrane) response to a pH change into a single parameter. Absolute values of [R]K2D data points are also significantly reduced, as confirmed by an independent but more approximate determination of K2D using fluorescent intensities of adherent and free membrane segments (Fig. 3C). Some alternative explanations for the decreased 2D binding affinity can be ruled out by further experiments: first, binding of soluble SIRPα to red blood cells did not exhibit a strong pH dependence in the 3D binding affinity (Fig. S3A). Second, density changes in absorbed protein [R] do not change with pH, perhaps because of a change in conformational and rotational free-energy loss of CD47 and/or SIRPα during binding (Xu et al., 2015). Regardless of the exact cause of the 2- to 4-fold decrease in 2D binding affinity with decreased pH, the loss of cooperativity is likely to be more important for signaling, because CD47 densities vary over one to two orders of magnitude across otherwise healthy cells and yet CD47 signaling remains effective (Subramanian et al., 2007). Changes in the collective binding affinity of CD47 for SIRPα could ultimately inhibit a cell's ability to efficiently signal through this pathway.
Fig. 3.
Acidosis conditions (pH 6) affect membrane flexibility and interactions. (A) Membrane fluctuations reveal 3.6-fold stiffening of GPMV membranes upon pH shift (n=9 GPMVs per condition). (B) Binding constant K2D as a function of the complex concentration [RL] from experiments (data points) and simulation (lines) at pH 7.4 and 6, respectively, for the parameter value Δl=4 nm of the repulsive membrane-substrate interactions. Model uses measured bending rigidities κ=10 kBT at pH 7.4 and κ=40 kBT at pH 6 and varies the maximal binding constant Kmax in Eqn 1. At pH 7.4, the best-fit Kmax=(378±8)/[R] (R2=0.997) for Δl=4 nm (see Fig. 2D). At pH 6, Kmax=(74±8)/[R] (R2=0.997) for Δl=4 nm, and the best-fit value depends weakly on Δl. Error bars are 95% confidence intervals of the fit to the reaction-diffusion model (±s.e.m.). Each data point corresponds to measurement of an individual GPMV in at least two independent experiments. (C) Apparent binding constant at low CD47 (∼40±10/µm2) independently obtained from fluorescence analysis (Fig. S1) is consistent with FRAP in B (n=5 GPMVs per condition). Error bars are s.d. unless indicated and differences between distributions are significant (P<0.05, t-test).
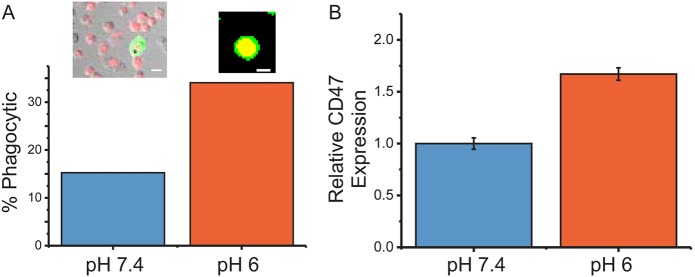
Tumor tissues commonly experience acidosis (Tannock and Rotin, 1989). Opsonization of lung cancer-derived cells (A549) with an IgG to present an ‘eat me’ signal to macrophages normally does not efficiently engage phagocytosis because of expression of CD47 on the cancer cell surface (Sosale et al., 2015b). However, opsonization under acidic conditions caused an increase in phagocytosis (Fig. 4A), consistent with the biophysical data, showing diminished cooperativity and binding capacity of CD47 at slightly acidic conditions of pH 6 (Fig. 3B,C). When A549 cancer cells are cultured long term at pH 6, CD47 levels were elevated (Fig. 4B), indicating a cellular signaling cascade that works against reduction in ‘self’ signaling by upregulation of CD47. Together, these data suggest that the measured 2D binding affinities reveal physiologically relevant parameters that would have been missed by using a classical binding affinity assay.
Fig. 4.
Phagocytosis and CD47 levels change with acidosis. (A) Phagocytic uptake assay of opsonized A549 (red in insets) cells by THP-1 macrophages (green). Yellow indicates overlapping cells, and full overlap indicates completed phagocytosis. Shift to pH 6 increases such engulfment. Left inset shows phase-contrast images overlaid with fluorescence. Scale bars: 10 μm. Data from two independent experiments (n=90 cells for pH 7, n=47 for pH 6). (B) CD47 levels increase on viable A549 cells under acidosis conditions. Experiment was performed with three technical replicates. Error bars are ±s.e.m. Differences between distributions are significant (P<0.05, t-test).
Conclusion
Here, we studied protein complex formation on reconstituted cytoskeleton-free plasma membrane and demonstrated that membrane shape fluctuations on nanoscales lead to cooperative binding. In contrast to our plasma membrane vesicles, cellular plasma membranes are bound to a cytoskeleton that confines shape fluctuations on length scales larger than the cytoskeletal mesh size (∼50-100 nm) (Morone et al., 2006), but hardly affects fluctuations on lateral scales of tens of nanometers (the typical distance between neighboring bound complexes) that lead to cooperative binding (Weikl et al., 2009; Hu et al., 2013). Therefore, these nanoscale thermal fluctuations, and thus the binding cooperativity, should persist in living cells. Cell membrane fluctuations appear to be driven by active processes and by thermal motion (Biswas et al., 2017; Turlier et al., 2016; Monzel et al., 2015), and both can enhance binding cooperativity. In general, fluctuations and displacements of single, unbound membranes should be distinguished from distance fluctuations between two membranes adhering via protein complexes (Lipowsky, 1995). The distance fluctuations of two adhering membranes or of one membrane adhering to a surface are constrained by the protein complexes on length scales larger than the typical distance between neighboring complexes, which leads to binding cooperativity (Fig. 2). GPMVs are known to exhibit many aspects of native cell membranes, and yet the membrane mechanical properties can be described using a just few parameters that connect experiment to theory and simulation. Membrane fluctuations are reduced when the plasma membrane is stiffened by a pH shift to pH 6, which can be induced by tumor tissue acidosis (Fig. 3) (Tannock and Rotin, 1989). In experiments on plasma membrane vesicles, multiscale simulation and phagocytosis, we consistently find reduced CD47 signaling (Fig. 4A). Clearly, enhanced phagocytosis at pH 6 is not caused exclusively by the diminished CD47 (K2D) binding affinity. However, the model system presented here can be used to address the contribution to total cell signaling from binding affinities of individual signaling components in absence of the cytoskeleton, while presenting a physiological membrane-bound environment. Finally, we speculate that that reduction in CD47 signaling by membrane rigidification selects for the elevated levels of CD47 in many tumors. Reduction of CD47 signaling would otherwise tend to enhance phagocytosis.
Reductionist approaches to cell adhesion with giant vesicles derived from plasma membranes provide, more generally, attractive models of cytoskeleton-independent adhesion that can be thermodynamically equilibrated. CD47 remains integrated, properly oriented, and functional in plasma membrane-derived vesicles. A theoretical prediction of cooperative binding and hence a non-linear law of mass action is confirmed through studies of the interaction of CD47 with its cognate ligand SIRPα. We also varied the membrane bending rigidity of extracted plasma vesicles. Our multiscale simulation and experimental approaches identify a few intuitive parameters, which describe binding, unbinding and cooperative effects for a wide range of membrane-bound complexes. Binding and signaling in cell-cell interactions are not only determined by the relative affinity of the complexes but also indirectly by changes such as the membrane fluctuation spectrum. Our method of reconstituted adhesion should have many applications where affinities of cell signaling complexes are investigated. Ultimately, convolution of membrane fluctuations with binding of adhesion molecules provides an elegant way to control activation and clustering for cell signaling.
MATERIALS AND METHODS
Generation of CD47-GFP HEK cell line and cell culture
HEK 293T cells were transduced with a lentivector encoding CD47-GFP and puromycin resistance transgenes. Lentiviral transduction was carried out over 72 h with a MOI of 5 and followed by puromycin selection (3 μg/μl from Gibco by Life Technologies) for 3 weeks. Cells were then cultured under standard condition in DMEM (without Phenol Red; Millipore-Sigma), 10% FBS and 1% penicillin-streptomycin and split every 2-3 days. CD47-GFP HEK 293 and A549 cell lines were authenticated and screened for contamination using American Type Culture Collection (ATCC) standards (Tech Bulletin number 8: TB-0111-00-02, 2010). HEK 293 and A549 cell lines were acquired from ATCC.
GPMV Isolation
Before experiments, CD47-GFP HEK cells were plated under identical conditions in T-25 culture flasks and cultured for 3 days until cells were 90% confluent. GPMVs were isolated according to a published protocol (Sezgin et al., 2012). HEK cells were incubated with 2 mM DTT, 25 mM paraformaldehyde at 37°C for 1 h. For experiments in which the pH was varied, the GPMV buffer was adjusted to the desired pH on the day of the experiment.
Purification and absorption of SIRPα on glass slides
The soluble extracellular domain of SIRPα was purified as previously described (Seiffert et al., 1999). The resulting protein was tagged with GST and we refer to the resulting SIRPαEx-GST as to ‘SIRPα’. SIRPα at a concentration of 34 nM was then incubated on clean glass slides (2× rinsing with ethanol and ultra-pure water or a 5 min exposure to oxygen plasma yielded the same results) for 1 h in phosphate-buffered saline solution at room temperature. Unbound SIRPα was washed away and uncovered glass surface was blocked by incubation with 1% BSA, 0.05% Tween-20 for 1 h at room temperature. The coated glass slides were then washed in GPMV buffer at the desired pH and immediately covered with GPMV suspension.
Confocal imaging, FRAP experiments and image analysis
Confocal images were obtained on a Leica SP8 system, dyes were excited using the 488 and 638 laser lines and fluorescence was collected (495-600 nm and 650-750 nm). To avoid cross-talk between the channels, two-color images were always obtained sequentially. FRAP experiments were performed on the same system. A circle of 1 µm in diameter was bleached using the 488 laser for two consecutive frames and recovery was observed under normal imaging conditions for 60 frames and 0.2 frames per second. The data were corrected for bleaching by dividing the intensity obtained in the bleached spot by the intensity of an unbleached area about 10-15 µm away from the bleaching spot. The background level was subtracted as estimated from the first frame after bleaching. To estimate the diffusion of free CD47, GPMVs were bleached on the upper pole and recovery curves were fitted using the standard model for free diffusion (Soumpasis, 1983). For the bound membrane segment fluorescence recovery, a reaction diffusion model was used (Sprague et al., 2004):
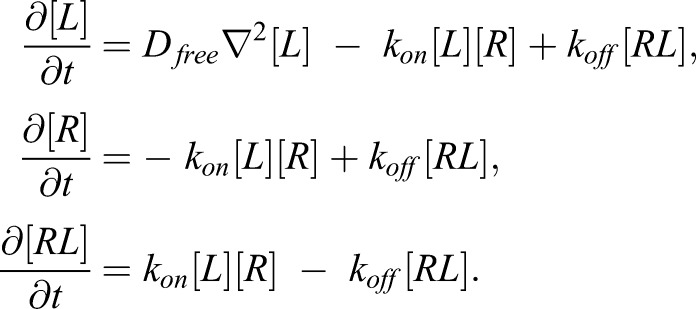 |
(2) |
Here [L], [R] and [RL] are the concentrations of free CD47-GFP, surface-immobilized SIPRA molecules and bound complexes respectively. 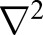 has the usual meaning of the Laplace operator and Dfree, kon and koff are parameters of the model. The data was subsequently fitted to the model using the routine MATLAB invlap (Karl J. Hollenbeck, 1998) and nlinfit (MATLAB, 2014a). Note that within the bleached spot there are only about 30-80 CD47 molecules; hence in the first seconds of recovery only a few GFP molecules contribute to the signal. The data is inherently noisy and we found that the recovered absolute values for [R]kon and koff depend significantly on the starting values of the fitting routine. However, the ratio of these rates is robust; in other words [R]K2D is obtained with higher degree of confidence. The error in fitting the individual fluorescent recovery trajectories was estimated following the method Müller et al. (2009). The value of the best fit of [R]kon and koff is varied around the best estimate (in the least-square sense) until the deviation is found to be outside the bounds set by the χ2 distribution, effectively giving the 95% confidence intervals.
has the usual meaning of the Laplace operator and Dfree, kon and koff are parameters of the model. The data was subsequently fitted to the model using the routine MATLAB invlap (Karl J. Hollenbeck, 1998) and nlinfit (MATLAB, 2014a). Note that within the bleached spot there are only about 30-80 CD47 molecules; hence in the first seconds of recovery only a few GFP molecules contribute to the signal. The data is inherently noisy and we found that the recovered absolute values for [R]kon and koff depend significantly on the starting values of the fitting routine. However, the ratio of these rates is robust; in other words [R]K2D is obtained with higher degree of confidence. The error in fitting the individual fluorescent recovery trajectories was estimated following the method Müller et al. (2009). The value of the best fit of [R]kon and koff is varied around the best estimate (in the least-square sense) until the deviation is found to be outside the bounds set by the χ2 distribution, effectively giving the 95% confidence intervals.
Fluorescence intensities were quantified either by a line profile on the vesicle equator (to obtain the correlation between GFP and anti-CD47), a line profile on the x,z contour in the case of adhering vesicles, both lines were 1 μm wide, as shown in Fig. S1. We used the peak of the resulting intensity distribution as an indicator of dye concentration in the membrane. Using this method, we could quantify both the fluorescence intensity in the bound and unbound membrane segments, Ibound and Ifree respectively. We found that the value [RL]=G(Ibound−Ifree) is the most robust way to quantify CD47-SIRPα complex densities using fluorescence confocal microscopy, presumably because small variation or defects in GFP tagging are eliminated this way. Here, G is the proportionally factor to convert fluorescent intensities to number density per membrane area and is determined as described below. Plots of [RL] vs [L]∼Ifree were in general too scattered for detailed analysis and we used these values only as a consistency check as shown in Fig. 3C. To assess the CD47 density between RBC and GPMVs we compared the maximum intensity projection at the upper surface of labelled RBC to GPMVs as obtained by high-resolution confocal microscopy (63×1.2 water immersion lens). To convert fluorescence intensities to CD47 concentration, we incubated red blood cells (Fig. S3B), which naturally express CD47 (about 250 molecules/µm2; Tsai et al., 2010), together with the GPMVs and fluorescent CD47 antibodies. The average anti-CD47 fluorescence of RBC cells and co-incubated GPMVs was determined from x,y confocal sections of circular shape (∼1 µm2) at poles and the average number density per area of CD47 molecules on the GPMV population was determined to be about 100 CD47/µm2. The factor G was subsequently assessed from the average CD47 coverage and the average GFP fluorescence intensity in the GPMV population, as determined from line profiles in x,z confocal scans on free GPMVs obtained as described above.
Immunocytochemistry
CD47 on GPMVs was labelled by incubation of B6H12 Alexa Fluor 647 mouse anti-human antibody (BD Biosciences, 561249; 1:100) for 1 h at room temperature (RT) according to the manufacturer's protocol. RBCs were isolated from a healthy male individual, washed in PBS and incubated in the same conditions as the GPMVs with the antibody. In some experiments, SIRPα was labelled on the glass surface by incubating with rabbit anti-GST Alexa Fluor 488 (Invitrogen) at 20 μg/ml for 1 h at RT and subsequent washing.
Phagocytosis assay
A549 Cells were opsonized with a rabbit anti-human RBC antibody (Rockland, 109-4139 polyclonal; Alvey et al., 2017) at final concentration of 250 µg/5 ml for 1 h at RT. Cells were washed and labelled with the dye PKH26 (Sigma-Aldrich) according to the manufacturer's protocol. THP1 cells were left to adhere in standard cell 6-well culture plates in RPMI/PMA medium (Millipore-Sigma; initial concentration 4×105 cells per well) for 48 h. A549 cells were then added to the wells and incubated at either pH 7.4 or pH 6. After 75 min, wells were digested with trypsin so that ‘uneaten’ A549 cells were removed by trypsinization. THP-1 were labelled by incubation with CD11b APC antibody (Biolegend, 101211, clone M1/70; Alvey et al., 2017) at 2 ng/ml for 1 h at RT. Finally, cells were fixed by incubation with 4% paraformaldehyde. Imaging was performed using a 10× objective on Leica SP8 system as described below. Images were processed by a Gaussian filter, edges of cells were detected and thresholded to obtain binary images of cells (ImageJ v.1.50b). Overlap of the different channels was then calculated per cell.
3D binding affinity assay using RBCs
Fresh RBCs were washed and incubated with SIRPα (final concentration 34 nM) for 1 h at RT in PBS adjusted to pH 7.4 or pH 6. GST was labelled as described before. RBCs were pelleted and unbound protein was washed away. RBCs were then imaged and the mean fluorescence in the center of a RBC was measured in a circle of 6 µm diameter.
Quantification of CD47 expression during long-term acidosis
A549 cells were seeded and grown until confluent for 3 days in full F12 medium (supplemented with 25 mM HEPES) in the absence of carbon dioxide. Cells were cultured either at pH 7.4 or pH 6. Cells were then detached non-enzymatically, washed and resuspended in PBS supplemented with 5% FBS. CD47 was labeled by incubation for 1 h at RT using 3 μl Alexa Fluor 647 human CD47 antibody (BD Biosciences, 561249, clone B6H12; Alvey et al., 2017), 0.5 µl Hoechst 33342 (Thermo Fisher, H3570) and 1 µl of 7-amino-actinomycin D (Sigma, A9400-1MG) per 300 μl cell suspension. Cells were then washed, resuspended in Flow buffer (5% FBS in PBS), and transferred to flow cytometry tubes. Cell suspension was analyzed at a flow rate of <1000 cells per second for 1 min. For analysis, aggregates were removed from data using Hoechst intensity and forward scatter signal (FSC). Dead cells, identified as 7ADD+ and low FSC, were also removed from analysis. Quantification of CD47 intensity was recorded as the geometric mean of a given population of cells.
Fluctuation spectroscopy
We measured the bending rigidity of the GPMVs by fluctuation analysis of the thermally induced motion of the membrane. Details of the method are published elsewhere (Gracia et al., 2010). Experiments were performed on an Axiovert 135 microscope (Zeiss, Germany) using a 40× objective in phase contrast mode. Imaging was done with a fast digital camera HG-100K (Redlake, San Diego, CA) using a mercury lamp HBO W/2 as a light source. We acquired a total of 4000 snapshots per vesicle with exposure time of 200 µs. Only vesicles with clearly visible fluctuations and no visible defects were considered for analysis.
Molecular modeling and coarse-grained simulations of the CD47-SIRPα complex
A structural model of the full-length protein CD47 in complex with the SIRPα-GST construct was built using PDB structures 2JJS, 2WNG and 1GTA for the CD47 ectodomain, the SIRPα ectodomain and the GST domain, respectively. The transmembrane domain of CD47 (comprising residues 117 to 275) was modeled using EVfold (Hopf et al., 2012) and positioned in the membrane environment using the PPM server (Lomize et al., 2006). The ectodomains of CD47 and SIRPα as well as the GST domain were next positioned beneath the membrane (see Fig. 2B) using VDM (Humphrey et al., 1996). The interface between CD47 and SIRPα was taken to be as in the PDB structure 2JJS. The 33-residue peptide segment connecting SIRPα with GST, the 5-residue linker between the two domains of CD47, and the 7-residue loop in the C-terminal Ig-like domain of SIRPα (comprising residues 288 to 294) were built into the structural model using MODELLER (Fiser et al., 2000). The resulting structural model was taken as the initial conformation for molecular simulations. Equilibrium conformations of a single CD47-SIRPα complex were simulated using the coarse-grained model of Kim and Hummer (2008). The model was adapted to the CD47-SIRPα complex in the following way. Four protein domains were represented as rigid bodies: the trans-membrane domain of CD47, the ectodomain of CD47, the ectodomain of SIRPα and the GST domain. The interactions between the individual domains were treated at the residue level and described as in the Kim–Hummer model. A bias potential was applied to 45 inter-domain contacts of the ectodomains of CD47 and SIRPα to ensure the correct binding interface as observed in the crystal structure of the CD47-SIRPα ectodomain complex (PDB code 2JJS). The inter-domain contacts were determined using an overlap criterion (Różycki et al., 2014). The bias potential was taken to have a form of the Lennard–Jones potential. The depth of the Lennard–Jones potential was set to 0.67 kT to prevent dissociation of the CD47-SIRPα complex. The GST domain was positioned on a flat substrate with the linker connecting GST to SIRPα oriented away from the substrate and the transmembrane domain of CD47 was embedded in a membrane (see Fig. 2B). The membrane was assumed to be flat and parallel to the substrate surface. Both the 33-residue peptide segment connecting SIRPα with GST and the 5-residue linker joining the two domains of CD47 were represented as chains of amino acid beads with appropriate bending, stretching and torsion potentials (Kim and Hummer, 2008). The interactions between the flexible linkers and the rigid domains as well as the interactions between the membrane and the proteins were simulated as in the original model of Kim–Hummer. A harmonic potential was applied between Cys15 (located in the extracellular domain of CD47) and Cys245 (positioned in the transmembrane domain of CD47) to mimic the disulfide bond.
Monte Carlo (MC) simulations
The basic MC moves of the CD47 and SIRPα ectodomains were random rotations and translations in three dimensions. For the transmembrane domain of CD47, the basic MC moves were translations parallel to the membrane surface and rotations around the axis perpendicular to the membrane surface. For the linker peptides, crank-shaft moves are employed to enhance sampling in addition to local MC moves on each amino acid bead. The separation between the substrate surface and the membrane was varied in additional MC moves. In the MC simulations, 105 configurations taken every 103 MC sweeps were generated for analysis. The simulations were performed at room temperature (T=300 K). The ectodomains of CD47 and SIRPα were observed to remain in the bound state throughout the simulation runs. The basic MC moves in our simulations of large adhering membrane segments were variations of the local separations one of all membrane patches, and hopping of the harmonic bonds between neighboring patches (Weikl and Lipowsky, 2006). The bonds are taken to be mobile because positions of CD47-SIRPα complexes in the experiments vary due to binding and unbinding events. Simulations were performed with up to 8×107 MC steps per membrane patch.
Distance of membrane to substrate in molecular simulations
The separation between the membrane and substrate was identified by measuring the distance in the z-direction between the C-terminal residue of the short inter-domain linker in CD47 and the plane at which the GST domain was placed. The histogram of the separations obtained from the MC simulations is shown in Fig. S2. The key quantities obtained from this histogram are the mean value l0=17.2 nm, which is the preferred membrane-substrate separation of the complex, and the standard deviation σ=1.2 nm, which reflects the flexibility of the complex. Visual inspection of the simulation structures reveals that variations in the separation between membrane and substrate are related mainly to the flexibility of the long linker between SIRPα and GST. Other joints between the rigid domains appear to be relatively stiff. First, in CD47, the transmembrane domain is connected to the ectodomain by a short linker and by the disulfide bond between Cys15 and Cys245. These two connections, together with steric effects between the protein and the membrane, effectively restrain the orientation of the ectodomain relative to the transmembrane domain. Second, the relative orientation of the ectodomains of CD47 and SIRPα is restrained by the geometry of their interface. In fact, only small fluctuations around the orientation given by the crystal structure were observed in the simulations.
Supplementary Material
Acknowledgements
Some data and Materials and Methods in this paper form part of the PhD thesis of Jan Steinkühler, Technischen Universität Berlin, 2016.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.S., B.R., R.L., T.R.W., D.E.D.; Methodology: J.S., B.R., C.A., T.R.W., R.D., D.E.D.; Software: J.S., B.R., R.L., T.R.W.; Validation: J.S., B.R., T.R.W.; Formal analysis: J.S., B.R., C.A., R.L., T.R.W.; Investigation: J.S., B.R., C.A., T.R.W., R.D., D.E.D.; Resources: B.R., R.L., T.R.W., R.D., D.E.D.; Data curation: B.R., T.R.W.; Writing - original draft: J.S., T.R.W., D.E.D.; Writing - review & editing: J.S., B.R., C.A., R.L., T.R.W., R.D., D.E.D.; Visualization: B.R., R.L., T.R.W.; Supervision: R.L., T.R.W., R.D., D.E.D.; Project administration: R.L., T.R.W., R.D., D.E.D.; Funding acquisition: B.R., R.L., T.R.W., R.D., D.E.D.
Funding
Financial support for J.S. from the Deutsche Forschungsgemeinschaft (DFG) (IRTG-1524) is gratefully acknowledged. B.R. was supported by the National Science Centre, Poland (2016/21/B/NZ1/00006). This work is part of the MaxSynBio consortium which was jointly funded by the Federal Ministry of Education and Research of Germany and the Max Planck Society. Financial support for C.A. and D.E.D. is from the National Institutes of Health (National Cancer Institute U54-CA193417, National Heart Lung and Blood Institute R01-HL124106, R21-HL128187), and a National Science Foundation Materials Science and Engineering Center grant to the University of Pennsylvania. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.216770.supplemental
References
- Alvey C. and Discher D. E. (2017). Engineering macrophages to eat cancer: from “marker of self” CD47 and phagocytosis to differentiation. J. Leukoc. Biol. 102, 31-40. 10.1189/jlb.4RI1216-516R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvey C. M., Spinler K. R., Irianto J., Pfeifer C. R., Hayes B., Xia Y., Cho S., Dingal P. C. P. D., Hsu J., Smith L. et al. (2017). SIRPα-inhibited, marrow-derived macrophages engorge, accumulated, and differentiate in antibody targeted regression of solid tumors. Curr. Biol. 27, 2065-2077. 10.1016/j.cub.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer B., Davidson M. and Orwar O. (2009). Proteomic analysis of plasma membrane vesicles. Angew. Chem. Int. Ed. 48, 1656-1659. 10.1002/anie.200803898 [DOI] [PubMed] [Google Scholar]
- Baumgart T., Hammond A. T., Sengupta P., Hess S. T., Holowka D. A., Baird B. A. and Webb W. W. (2007). Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl Acad. Sci. USA 104, 3165-3170. 10.1073/pnas.0611357104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihr T., Fenz S., Sackmann E., Merkel R., Seifert U., Sengupta K. and Smith A.-S. (2014). Association rates of membrane-coupled cell adhesion molecules. Biophys. J. 107, L33-L36. 10.1016/j.bpj.2014.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A., Alex A. and Sinha B. (2017). Mapping cell membrane fluctuations reveals their active regulation and transient heterogeneities. Biophys. J. 113, 1768-1781. 10.1016/j.bpj.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E., Różycki B., Herrick D. Z., Chung H. S., Vecer J., Eaton W. A., Cafiso D. S., Hummer G. and Hurley J. H. (2011). Solution structure of the ESCRT-I complex by small-angle X-ray scattering, EPR, and FRET spectroscopy. Proc. Natl Acad. Sci. USA 108, 9437-9442. 10.1073/pnas.1101763108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E., Różycki B., Chung H. S., Herrick D. Z., Canagarajah B., Cafiso D. S., Eaton W. A., Hummer G. and Hurley J. H. (2012). Solution structure of the ESCRT-I and -II supercomplex: implications for membrane budding and scission. Structure 20, 874-886. 10.1016/j.str.2012.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.-H. M., Lenz P. and Boxer S. G. (2007). Kinetics of DNA-mediated docking reactions between vesicles tethered to supported lipid bilayers. Proc. Natl Acad. Sci. USA 104, 18913-18918. 10.1073/pnas.0706114104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenz S. F. and Sengupta K. (2012). Giant vesicles as cell models. Integr. Biol. 4, 982-995. 10.1039/c2ib00188h [DOI] [PubMed] [Google Scholar]
- Fiser A., Do R. K. G. and Šali A. (2000). Modeling of loops in protein structures. Protein Sci. 9, 1753-1773. 10.1110/ps.9.9.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D. M., Różycki B., Tortajada A., Hummer G., Peti W. and Page R. (2011). Resting and active states of the ERK2:HePTP complex. J. Am. Chem. Soc. 133, 17138-17141. 10.1021/ja2075136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia R. S., Bezlyepkina N., Knorr R. L., Lipowsky R. and Dimova R. (2010). Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Mat. 6, 1472-1482. 10.1039/b920629a [DOI] [Google Scholar]
- Hopf T. A., Colwell L. J., Sheridan R., Rost B., Sander C. and Marks D. S. (2012). Three-dimensional structures of membrane proteins from genomic sequencing. Cell 149, 1607-1621. 10.1016/j.cell.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Lipowsky R. and Weikl T. R. (2013). Binding constants of membrane-anchored receptors and ligands depend strongly on the nanoscale roughness of membranes. Proc. Natl Acad. Sci. USA 110, 15283-15288. 10.1073/pnas.1305766110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A. and Schulten K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33-38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Huppa J. B., Axmann M., Mortelmaier M. A., Lillemeier B. F., Newell E. W., Brameshuber M., Klein L. O., Schutz G. J. and Davis M. M. (2010). TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963-967. 10.1038/nature08746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C. and Hummer G. (2008). Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J. Mol. Biol. 375, 1416-1433. 10.1016/j.jmb.2007.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krobath H., Różycki B., Lipowsky R. and Weikl T. R. (2009). Binding cooperativity of membrane adhesion receptors. Soft Mat. 5, 3354-3361. 10.1039/b902036e [DOI] [Google Scholar]
- Lipowsky R. (1991). The conformation of membranes. Nature 349, 475-481. 10.1038/349475a0 [DOI] [PubMed] [Google Scholar]
- Lipowsky R. (1995). Generic interactions of flexible membranes. Handbook Biol. Phy. 1, 521-602. 10.1016/S1383-8121(06)80004-7 [DOI] [Google Scholar]
- Lomize M. A., Lomize A. L., Pogozheva I. D. and Mosberg H. I. (2006). OPM: orientations of proteins in membranes database. Bioinformatics 22, 623-625. 10.1093/bioinformatics/btk023 [DOI] [PubMed] [Google Scholar]
- Monzel C., Schmidt D., Kleusch C., Kirchenbüchler D., Seifert U., Smith A. S., Sengupta K. and Merkel R. (2015). Measuring fast stochastic displacements of bio-membranes with dynamic optical displacement spectroscopy. Nat. Commun. 6, 8162 10.1038/ncomms9162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morone N., Fujiwara T., Murase K., Kasai R. S., Ike H., Yuasa S., Usukura J. and Kusumi A. (2006). Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J. Cell Biol. 174, 851-862. 10.1083/jcb.200606007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K. P., Erdel F., Caudron-Herger M., Marth C., Fodor B. D., Richter M., Scaranaro M., Beaudouin J., Wachsmuth M. and Rippe K. (2009). Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys. J. 97, 2876-2885. 10.1016/j.bpj.2009.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg P.-A., Zheleznyak A., Fang Y.-F., Lagenaur C. F., Gresham H. D. and Lindberg F. P. (2000). Role of CD47 as a marker of self on red blood cells. Science 288, 2051-2054. 10.1126/science.288.5473.2051 [DOI] [PubMed] [Google Scholar]
- Różycki B., Mioduszewski Ł. and Cieplak M. (2014). Unbinding and unfolding of adhesion protein complexes through stretching: Interplay between shear and tensile mechanical clamps. Proteins 82, 3144-3153. 10.1002/prot.24674 [DOI] [PubMed] [Google Scholar]
- Seiffert M., Cant C., Chen Z., Rappold I., Brugger W., Kanz L., Brown E. J., Ullrich A. and Bühring H.-J. (1999). Human signal-regulatory protein is expressed on normal, but not on subsets of leukemic myeloid cells and mediates cellular adhesion involving its counterreceptor CD47. Blood 94, 3633-3643. [PubMed] [Google Scholar]
- Sezgin E., Kaiser H.-J., Baumgart T., Schwille P., Simons K. and Levental I. (2012). Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protocols 7, 1042-1051. 10.1038/nprot.2012.059 [DOI] [PubMed] [Google Scholar]
- Sosale N. G., Rouhiparkouhi T., Bradshaw A. M., Dimova R., Lipowsky R. and Discher D. E. (2015a). Cell rigidity and shape override CD47's “self”-signaling in phagocytosis by hyperactivating myosin-II. Blood 125, 542-552. 10.1182/blood-2014-06-585299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosale N. G., Spinler K. R., Alvey C. and Discher D. E. (2015b). Macrophage engulfment of a cell or nanoparticle is regulated by unavoidable opsonization, a species-specific ‘Marker of Self’ CD47, and target physical properties. Curr. Opin. Immunol. 35, 107-112. 10.1016/j.coi.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumpasis D. M. (1983). Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41, 95-97. 10.1016/S0006-3495(83)84410-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B. L., Pego R. L., Stavreva D. A. and Mcnally J. G. (2004). Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86, 3473-3495. 10.1529/biophysj.103.026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Parthasarathy R., Sen S., Boder E. T. and Discher D. E. (2006). Species- and cell type-specific interactions between CD47 and human SIRPα. Blood 107, 2548-2556. 10.1182/blood-2005-04-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Boder E. T. and Discher D. E. (2007). Phylogenetic divergence of CD47 interactions with human signal regulatory protein α reveals locus of species specificity: implications for the binding site. J. Biol. Chem. 282, 1805-1818. 10.1074/jbc.M603923200 [DOI] [PubMed] [Google Scholar]
- Tannock I. F. and Rotin D. (1989). Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 49, 4373-4384. [PubMed] [Google Scholar]
- Tolentino T. P., Wu J., Zarnitsyna V. I., Fang Y., Dustin M. L. and Zhu C. (2008). Measuring diffusion and binding kinetics by contact area FRAP. Biophys. J. 95, 920-930. 10.1529/biophysj.107.114447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R. K., Rodriguez P. L. and Discher D. E. (2010). Self inhibition of phagocytosis: the affinity of ‘Marker of Self’ CD47 for SIRPα dictates potency of inhibition but only at low expression levels. Blood Cells Mol. Dis. 45, 67-74. 10.1016/j.bcmd.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlier H., Fedosov D. A., Audoly B., Auth T., Gov N. S., Sykes C., Joanny J. F., Gompper G. and Betz T. (2016). Equilibrium physics breakdown reveals the active nature of red blood cell flickering. Nat. Phys. 12, 513-519. 10.1038/nphys3621 [DOI] [Google Scholar]
- Veatch S. L., Cicuta P., Sengupta P., Honerkamp-Smith A., Holowka D. and Baird B. (2008). Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 3, 287-293. 10.1021/cb800012x [DOI] [PubMed] [Google Scholar]
- Vernon-Wilson E. F., Kee W.-J., Willis A. C., Barclay A. N., Simmons D. L. and Brown M. H. (2000). CD47 is a ligand for rat macrophage membrane signal regulatory protein SIRP (OX41) and human SIRPα1. Eur. J. Immunol. 30, 2130-2137. [DOI] [PubMed] [Google Scholar]
- Weikl T. R. and Lipowsky R. (2006). Membrane adhesion and domain formation. Adv. Planar Lipid Bilayers Liposomes 5, 63-127. 10.1016/S1554-4516(06)05004-6 [DOI] [Google Scholar]
- Weikl T. R., Asfaw M., Krobath H., Różycki B. and Lipowsky R. (2009). Adhesion of membranes via receptor-ligand complexes: Domain formation, binding cooperativity, and active processes. Soft Mat. 5, 3213-3224. 10.1039/b902017a [DOI] [Google Scholar]
- Xu G.-K., Hu J., Lipowsky R. and Weikl T. R. (2015). Binding constants of membrane-anchored receptors and ligands: a general theory corroborated by Monte Carlo simulations. J. Chem. Phys. 143, 243136 10.1063/1.4936134 [DOI] [PubMed] [Google Scholar]
- Zhu D.-M., Dustin M. L., Cairo C. W. and Golan D. E. (2007). Analysis of two-dimensional dissociation constant of laterally mobile cell adhesion molecules. Biophys. J. 92, 1022-1034. 10.1529/biophysj.106.089649 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.