Abstract
Cardiovascular disease is a major leading cause of morbidity and mortality in the United States and elsewhere. Alterations in mitochondrial function are increasingly being recognized as a contributing factor in myocardial infarction and in patients presenting with cardiomyopathy. Recent understanding of the complex interaction of the mitochondria in regulating metabolism and cell death can provide novel insight and therapeutic targets. The purpose of this statement is to better define the potential role of mitochondria in the genesis of cardiovascular disease such as ischemia and heart failure. To accomplish this we will define the key mitochondrial processes that play a role in cardiovascular disease, which are potential targets for novel therapeutic interventions. This is an exciting time in mitochondrial research. The past decade has provided novel insight into the role of mitochondria function and their importance in complex diseases. This Statement will define the key roles that mitochondria play in cardiovascular physiology and disease, and provide insight into how mitochondrial defects can contribute to cardiovascular disease and it will also discuss potential biomarkers of mitochondrial disease and suggest potential novel therapeutic approaches.
Introduction:
The mitochondria are recognized as a key player in cardiomyocyte cell death following myocardial infarction and cardiomyopathies. Alterations in mitochondrial function are increasingly recognized in cardiovascular disease. Although it has been suggested that the failing heart is energy starved [1], the recent understanding of the complex interaction of the mitochondria in regulating metabolism and cell death provide novel insight and therapeutic targets. This bioenergetics perspective of cardiomyopathy can be understood as one manifestation of an array of different common clinical phenotypes including myopathies, neuropathies, nephropathies, endocrine disorders and metabolic diseases, aging and cancer. This is because the organs that are affected in the common “complex” diseases are the very same organs that have the highest reliance on mitochondrial function [2].
The purpose of this statement is to better define the potential role of mitochondria in the genesis of cardiovascular disease such as ischemia and heart failure (see Figure 1). To accomplish this we will define the key mitochondrial processes that play a role in cardiovascular disease, which are potential targets for novel therapeutic interventions. This is an exciting time in mitochondrial research. The past decade has provided novel insight into the role of mitochondria function and their importance in complex diseases. In section I this Statement will define the key roles that mitochondria play in cardiovascular physiology and disease. Section II will provide insight into how mitochondrial defects can contribute to cardiovascular disease and it will also discuss potential biomarkers of mitochondrial disease and suggest potential novel therapeutic approaches.
Figure 1.
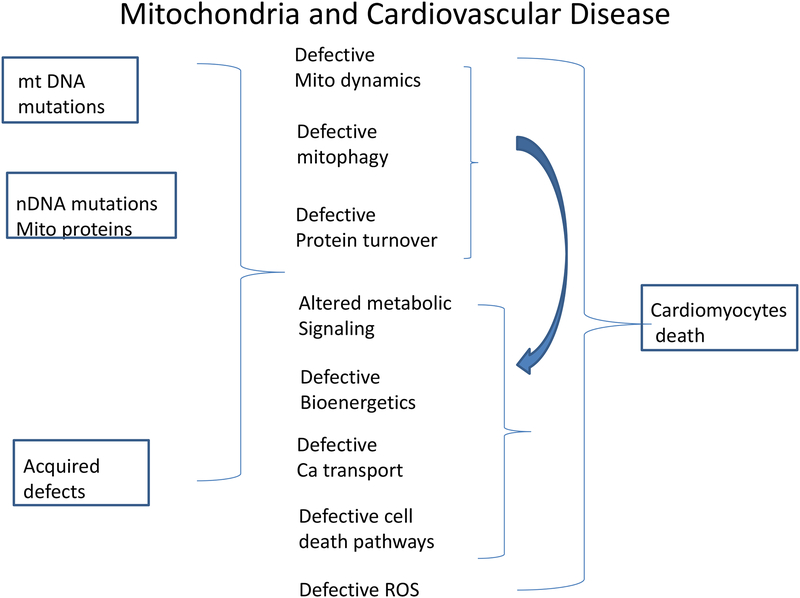
Mutations in mitochondrial proteins (either from mutation in mitochondrial DNA or nuclear DNA) or acquired defects can lead to defects in mitochondrial quality control which leads to a vicious cycle of more acquired mitochondrial defects and defects in metabolic signaling, bioenergetics, calcium transport, ROS generation and activation of cell death pathways. This leads to a vicious feed-forward cycle leading to cardiomyocyte cell death.
Mitochondria are well-known as the powerhouse of the cell and as discussed in Section I.A (Bioenergetics and Metabolism) in an active tissue such as heart they are responsible for generating most of the ATP in the cell. The role of post-translational modifications (PTMs) in the regulation of metabolism is also discussed (Section I.B). It has long been known that in addition to generating ATP, the mitochondria electron transport chain is also important in regulation of mitochondrial calcium. The recent identification of the proteins involved in regulating mitochondrial matrix calcium is providing new insights into the regulation and role of mitochondrial calcium (I.C). As discussed in Section I.D mitochondrial are also key regulators of cell death. In the process of electron transport to generate ATP, mitochondria can be a major source of reactive oxygen species (ROS) that can both contribute to cell death, and also serve as a signaling molecule (Section I.E). Because the generation of ROS can lead to damage to mitochondrial DNA and proteins it is important for the mitochondria to have mechanisms to assure quality control (Section I.F). Quality control can occur by fission/fusion to allow segregation of damaged mitochondria (Section I.F.1), mitophagy to remove damaged mitochondria (Section I.F.2) and ultimately cell death if the damage is too severe (Section I.D). Although mitophagy is important for quality control and for the removal of damaged mitochondria, based on measurement of mitochondrial protein turnover (Section I.F.4) it appears that mitochondrial proteins turnover at different rates, suggesting that under normal circumstances mitophagy is not the main driver of mitochondrial protein turnover. In fact it is suggested that the dynamics of protein turnover can provide an assessment of the physiological state. Alterations in these mitochondrial functions are important in many cardiac diseases as discussed in Section II. Section II.A discusses the mitochondrial etiology of cardiomyopathy and Section II.B discusses the role of mitochondria in cardiotoxicity. Section II.C discusses potential biomarkers for mitochondrial diseases. Taken together this AHA Statement provides a state-of-the-art assessment of the current status of basic mitochondrial biology and how alterations in mitochondria can be major contributors to complex cardiovascular diseases.
I. Mitochondrial Function
A). Generation of ATP – Bioenergetics and Metabolism
Energy demands of the heart:
The incessant energy requirements of the heart are sustained by the consumption of a mass of ATP daily that surpasses cardiac weight itself by approximately 5–10 fold [3]. This perpetual demand for energy reflects the continuous contractile functioning of the heart to sustain systemic circulation and nutrient supply. This high energy flux translates into the cardiomyocyte having a mitochondrial volume between 23% and 32% of myocellular volume [4]. Interestingly, cardiac mitochondrial density increases from man to mouse in parallel with increasing heart rate and oxygen consumption [4]. Based on the role of mitochondrial in energy transduction, it is not surprising, that any perturbations in mitochondrial energy balance, production or propagation would result in the development of cardiac pathology and/or susceptibility to injury (see Section II.A and B). However, a linear or direct correlation between mitochondrial energy metabolism and heart pathology is not clear cut. In this section, we give a brief overview of metabolism and perturbations and its consequences on myocardial function.
Electron transfer chain biology – energetics and ROS:
The final common pathway for oxidative metabolism, which generates the bulk of cardiac ATP, is the sequential passage of electrons from high (NADH or FADH2) to low (molecular oxygen) redox potentials down the electron transfer chain (ETC – complexes I through IV). This step-wise electron transfer results in the active pumping of hydrogen ions out of the mitochondrial matrix into the inter-membranous space. The ensuing electrochemical gradient generated across in the inner mitochondrial membrane facilitates the translocation of protons from the inter-membranous space through the Fo/F1 ATPase (ATP-synthase) back into the mitochondrial matrix. This proton translocation is coupled to the phosphorylation of ADP to generate ATP. Collectively these reactions constitute oxidative phosphorylation and the direct synthesis of ATP from electron-transfer encapsulates coupled respiration [5]. Being cognizant to the high-energy demand of the heart, it is not surprising that mutations in genes that encode for ETC proteins are linked to the development of cardiomyopathy (See Section II.A) [6–8]. However, it should also be recognized that the dysfunction in the electron transport chain (ETC) not only affects ATP production but can concordantly impair intracellular Ca2+ flux (see section I.C), increase the generation of reactive oxygen species (ROS) (see section I.E), and alter redox balance by altering the NAD+/NADH ratio [9, 10].
Fuel substrates and cardiac energetics in health and disease:
Mitochondrial fatty acid β-oxidation is the most efficient and predominant substrate for energy production in the normal adult human heart with glucose oxidation, glycolysis, lactate and ketones additionally contributing toward myocardial ATP production [11]. Regulation of cardiac energy metabolism is complex and determined by the summation of intracellular substrate concentrations, transcriptional rates and activity of metabolic enzymes, and metabolic demands of the myocardium [12, 13]. However, as the heart remodels in response to hypertrophy and ischemia, marked changes in cardiomyocyte substrate metabolism can occur with an ultimate effect on ATP levels in the decompensating heart (Reviewed [14–16]). The relative contribution of fatty acids diminishes with enhanced reliance on glucose utilization during the development of cardiac hypertrophy. The regulatory programs attenuating fat utilization have been extensively investigated and include regulation at the transcriptional and post-transcriptional levels [17–20]. Moreover, this reduction involves coordinate downregulation of proteins controlling fatty acid uptake by the heart and mitochondria as well as of the enzymes controlling mitochondrial fatty-acid β-oxidation (FAO); see Section I.F.3 on Protein Turnover [17, 21–24]. Enzymes involved in glycolytic pathways are up-regulated even during early stages of cardiac dysfunction in response to increased adrenergic signaling, up-regulation of fetal gene programs, and/or hypoxia [25, 26]. The shift toward glucose metabolism improves myocardial contractile efficiency by increasing the stoichiometric ratio of ATP production to oxygen consumption in addition to minimizing oxidative losses through mitochondrial respiratory chain uncoupling associated with free fatty acid metabolism [27, 28]. Abnormally high myocardial dependence on fatty acid metabolism, as seen during ischemia or high adrenergic states, increases cardiac oxygen consumption by 30–50% adjusted for equivalent stroke work indices [29, 30]. Strategies to increase glucose oxidation and decrease fatty acid metabolism may improve myocardial energy efficiency by up to 30% [31]. Although the initial shift towards glucose metabolism at progressively more advanced stages of cardiac dysfunction is physiologically adaptive, the magnitude and impact of this adaption can be significantly limited by extra-cardiac factors, specifically the development of insulin resistance. Whole body insulin resistance can affect cardiac energy metabolism, even in structurally normal hearts. Type 2 diabetic patients who have otherwise normal cardiac function regenerate phosphocreatine at a significantly lower rate after exercise compared with non-diabetic controls [32]. In patients with cardiomyopathy, the development of insulin resistance is linked to increased sympathetic signaling leading to liberation of free fatty acids from adipose tissue into the bloodstream [33]. Thus, in the failing myocardium, decreases in insulin sensitivity can lead to further reductions in glucose oxidation and deteriorations in cardiac function by depriving the heart of access to a more metabolically efficient substrate.
TCA biology and anaplerosis:
The capacity to use multiple substrates and the plasticity to switch substrate utilization enables continuous cardiac work under a wide variety of biological and pathological circumstances. Interestingly, under some pathological conditions such as severe hypertrophy, the coupling of glycolysis and pyruvate oxidation becomes disrupted with an increase in glucose oxidation that is insufficient to completely compensate for the reduced fat oxidation [24, 34]. These perturbations in substrate partitioning and selection may become associated with reduced contractile reserve and increased susceptibility to ischemia-reperfusion injury [34–36]. Partial compensation for this energy-substrate oxidation deficit has recently been identified to occur via recruitment of alternative intermediary pathways (anaplerosis) to enhance flux through the tricarboxylic acid (TCA) cycle [23]. The necessity of anaplerosis in the heart is well established, in that the mechanical performance of isolated rat hearts when exclusive precursors of acetyl-CoA are used as substrate shows progressive deterioration with rapid restoration following introduction of anaplerotic substrates [37]. Whether the disruption of anaplerosis plays a significant role in cardiac maladaptation, and or whether it may be a therapeutic target for therapy is currently unknown [27].
Metabolic modulation as a strategy for cardiac muscle pathology:
Despite the findings of altered metabolism and energetic capacity in experimental models and in patients with cardiac muscle injury and remodeling, the myriad of agents that have been directly assessed as metabolic modulators have not been found to have significant clinic benefit in the management of heart disease. The use of these agents has recently been reviewed [16], and only some of the studies are discussed here to illustrate the overall lack of efficacy, inadequate sample size and/or potential adverse effects associated with the administration of these metabolic modulators. Etomoxir is an irreversible inhibitor of mitochondrial carnitine palmitoyltransferase-1 (CPT-1) and thus results in a reduction in long chain fatty acid oxidation. An initial pilot study suggested that etomoxir may improve myocardial function in patients with heart failure [38]. However, a subsequent controlled study was stopped prematurely because the drug was associated with hepatic transaminitis [39]. Another inhibitor of CPT-1 and CPT-2 is perhexiline and one study showed improvement in MVO2 max and ejection fraction [40], however, larger studies are needed before a conclusion can be made regarding this drug in heart disease. Trimetazidine is a partial inhibitor of the β-oxidation enzyme 3-ketoacyl CoA thiolase, and is shown in small studies to improve symptoms and cardiac function in patients with heart failure [41]. Ranolazine has a similar structure to trimetazidine and is currently FDA approved for treatment of stable angina. Although it may affect FFA metabolism, its main mechanism of action may be related to inhibition of the late inward Na+ channel. In the TIMI-36 trial, ranolazine did not reduce hospitalization for HF in patients with acute coronary syndrome [42]. Finally, dichloroacetate (DCA) increases myocardial glucose utilization by inhibiting pyruvate dehydrogenase kinase, leading to increased activity of the mitochondrial pyruvate dehydrogenase. In a small study of patients with heart disease, DCA increased stroke volume and myocardial efficiency [43], however, more substantial studies are needed to characterize the role of DCA in patients with heart disease.
Creatine kinase and high energy transfer
The creatine kinase (CK) reaction is the prime energy reserve that provides a rapid source of adenosine triphosphate (ATP) and facilitates its delivery from the mitochondrial site of production to sites of use, including the myofibrils in the heart [44]. Although the heart is a high-energy consuming organ, human genetic mutations linked to creatine deficiency usually result in neurological deficits [45]. In parallel, the genetic disruption of whole body creatine synthesis in the mouse had no detrimental effects on exercise capacity, on cardiac workload or adaptation to ischemia-reperfusion injury [46], and the overexpression of the creatine transporter, or exogenous creatine supplementation in mice, again shows no cardiac benefit [47]. Nevertheless, in a large animal model, reduced CK ATP delivery was associated with impaired myocardial contractile function [48] and in human heart failure morbidity and mortality are linked to impaired CK metabolism and flux [49, 50].
B. Regulation of Function and Metabolism—The role of PTMs
It has been increasingly clear that there is cross-talk and signaling between the mitochondria, the cytosol and the nucleus. Post-translational modifications are a primary mechanism by which the mitochondrial communicate with the rest of the cell.
Acetylation:
Nutrient-overload is linked to mitochondrial dysfunction and to the cardiovascular risk factors of obesity and diabetes [27, 51]. Conceptually, perturbations in mitochondrial metabolic intermediates, such as acetyl-CoA, which itself can function as a direct post-translational substrate to modify mitochondrial proteins through acetylation, may link these pathophysiological effects [52, 53]. Additional short-chain carbon metabolic intermediates including succinyl groups and malonyl groups can also bind to, and modify protein lysine residues [45, 54]. Our knowledge of the regulation of these latter modifications is too preliminary to expand on [45, 54, 55], and this section will focus on the role acetylation, as a nutrient-dependent mechanism, in the regulation of mitochondrial function.
Enzymatic and non-enzymatic control of protein acetylation:
There are three major acetyltransferase families, and member proteins from each group have been implicated in the control of cellular homeostasis [56]. Deacetylase proteins are similarly grouped into distinct classes [57]. Class III deacetylases are NAD+-dependent, and function as sensors of the energetic status of the cell in response to the subcellular compartment levels of NAD+ and nicotinamide and/or to the ratio of NAD+:NADH [35, 58, 59]. Recent findings have highlighted novel mechanisms that regulate levels of NAD [60–62]. These enzymes are defined as sirtuins and 7 family members (Sirt1 through Sirt7) are evident in mammalia [63]. Sirt1, 2 and 3 have the most robust deacetylase activity and predominantly function in the nuclear (Sirt1), cytoplasmic (Sirt1 and 2) and mitochondrial (Sirt3) respectively. As the focus of this section is on mitochondrial PTMs, it will focus on Sirt3.
The counter-regulatory acetyltransferase enzyme system is less well characterized, although nuclear Gcn5 and p300 counter the actions of Sirt1 [64]. The process of protein acetylation in the mitochondria is even less well understood, although GCN5L1 has been identified as a critical molecular component of this program and its functional role is beginning to be explored [53, 65]. Recently, the mitochondrial protein acetyltransferase, acetyl-CoA acetyltransferase 1 (ACAT1), which functions in ketogenesis to combine two acetyl-CoA molecules [66] has been found to regulate the pyruvate dehydrogenase complex as a canonical acetyltransferase [67]. This finding may open the door to expanding our understanding of acetyltransferase functioning within mitochondria.
Concurrently, the recognition of non-enzymatic acetylation of proteins in the presence of acetyl-CoA is evident [68] and denatured mitochondrial proteins undergo acetylation in the presence of acetyl-CoA [65]. Furthermore, elevated levels of acetyl-CoA, coupled to the alkaline mitochondrial pH, have been shown to promote non-enzymatic protein acetylation [69]. This concept of non-enzymatic protein acetylation may be operational in diabetes where metabolic inflexibility, which is defined as the inability to switch from fatty acid to glucose oxidation during the transition from the fasted to fed state, results in part from the allosteric inhibition of pyruvate dehydrogenase by increased mitochondrial acetyl-CoA levels [70, 71]. The role of non-enzymatic protein acetylation has not been extensively investigated, although its’ potentially important regulatory role has been reviewed [72]. Interestingly, analysis of the mitochondrial acetylome, under various nutrient conditions and in the presence or absence of Sirt3, shows evidence compatible with non-enzymatic and enzymatic control of the mitochondrial acetylome [73].
Mitochondrial Sirt3 and the heart:
Although Sirt3 functions predominantly in mitochondria [74], data do support extra-mitochondrial deacetylase activity [75–77]. The depletion of Sirt3 has a subtle phenotype [78] which is unmasked in response to prolonged fasting,[79] following chronic perturbations in caloric intake [80–82] and in response to redox stress [83]. Numerous proteomic approaches have been employed to identify substrates of Sirt3 deacetylation and the vast majority of proteins with alternations in acetylation are found within mitochondria [73, 84, 85]. The functional characterization of these proteins show that Sirt3 mediated deacetylation regulates numerous aspects of mitochondrial function including the regulation of enzymes controlling β-oxidation, branch-chain amino acid metabolism, ketone biology, the electron transport chain, ATP production, the urea cycle [73, 79, 85, 86] and ROS catabolism [74, 87].
In light of the high energy demand of the heart and based on the Sirt3 targets characterized to date, the disruption of Sirt3 would be expected to have cardiac consequences. Despite this, young Sirt3 knockout mice do not have any obvious phenotype [88] and furthermore display normal exercise performance [89]. However, consistent with a ‘fine-tuning’ function, aging Sirt3 knockout mice develop cardiac dilatation [88], and pressure-overload results in maladaptive cardiac hypertrophy [88, 90]. The mechanisms underpinning these pathologies align with established functions attributable to Sirt3 including increased generation of ROS [88, 90]. Conversely Sirt3 overexpression promotes anti-apoptotic programs in cardiomyocytes [76] and cardiac-restricted Sirt3 transgenic mice exhibit enhanced ROS scavenging [90]. An interesting additional mechanism whereby Sirt3 deficiency could potentially contribute to the pathophysiology of cardiac hypertrophy is its regulatory role in controlling fatty acid metabolism [79]. As the loss of metabolic plasticity with the downregulation of fatty acid oxidation (FAO) is synonymous with cardiac pressure-overload mediated decompensation [12, 17], it is conceptually possible that the downregulation of FAO in Sirt3 knockout mice may play a role in the pressure-overload and aging maladaptive phenotype in the heart. However, this needs to be delineated further, as high-fat feeding has been concurrently shown to increase cardiac FAO in parallel with downregulation of Sirt3 [91].
As the regulatory control of mitochondrial protein acetylation is nutrient-level and redox-potential dependent, it is conceivable that primary perturbations within mitochondria that may modulate metabolic intermediates or redox-potential could initiate changes in the acetylome. This concept has been explored in the heart in response to genetic perturbations associated with cardiovascular pathology where disruption of frataxin, cyclophilin D and components of the electron transfer chain result in the either basal or excessive pressure-overload induced cardiac dysfunction, and are associated with reduced NAD+/NADH ratio and increased mitochondrial protein acetylation [13, 92, 93]. In primary cardiomyocytes, the frataxin and Complex I disruption of the acetylome are corrected in parallel with improvement in mitochondrial function, following Sirt3 induction [13, 92]. Although, not completely characterized, these data support the concept that the control of acetylation by intrinsic mitochondria functioning may, via a feedback loop, affect global mitochondrial functioning via mitochondrial acetylome regulation.
Future directions in understanding the mitochondrial acetylome:
Advances in proteomics have enhanced our understanding of both the static and dynamic alteration of the mitochondrial acetylome [73, 94]. Additionally, these studies have identified site-specific changes in lysine residue acetylation that modulate protein function, stability, localization, allosteric interactions and/or control synergistic PTM’s [79, 84, 95, 96]. Moreover, the stoichiometry of proteins and the domains surrounding substrate protein lysine residues may play important regulatory roles in the interaction of acetylase and deacetylase enzymes [73] and the further characterization of the acetylome modifying enzymes themselves may further expand our understanding of the role of acetylation in controlling mitochondrial function [53, 74, 94].
An area of some functional discrepancy has also arisen with respect to the acetylation of specific targets within a pathway compared to the global functioning of the canonical pathway in response to acetylation. This is most vividly illustrated where fatty acid oxidation is increased in the presence of excess fat and mitochondrial protein acetylation [52, 97], in contrast to studies showing direct deacetylation of lysine residues on FAO enzymes resulting in the activation of enzyme activity [79, 98]. The mechanisms underpinning these effects and whether this may be a result of tissue distinct regulatory cues needs to be explored.
Finally, although the role of acetylation in modifying individuals proteins is the main focus of this section, data is emerging to show that the overall function of mitochondrial quality control and integrity, which are also modulated by nutrient levels and redox stress, including mitochondrial turnover (mitochondrial dynamics, mitophagy and biogenesis) [53, 64, 99, 100] and redox- and proteotoxic-stress amelioration effects [101, 102] may be regulated by the mitochondrial acetylome [53]. The complexity of this regulation is further underscored where cross-talk between different PTM’s function in concert to regulate protein function, as has been shown by concomitant modifications in acetylation and phosphorylation [103].
Phosphorylation:
As recently reviewed there is extensive phosphorylation of proteins in the mitochondrial matrix as well as in the mitochondrial electron transport complexes [104, 105]. A number of recent studies have reported that there are several hundred phosphorylated proteins in cardiac mitochondria [106, 107]. There are also sex differences in phosphorylation of mitochondrial proteins [108]. Many of the phosphorylated mitochondria proteins are outer mitochondrial proteins, which are likely phosphorylated by cytosolic kinases, and have been shown to regulate mitochondrial dynamics and cell death pathways. As discussed previously [104], the occupancy or fraction of the protein that is phosphorylated may be low for many of these proteins, and it is possible that these many of these low level modifications are of little or no functional consequence. It is also unclear to what extent phosphorylation of mitochondrial matrix proteins occurs in the matrix as opposed to prior to import into the matrix. Furthermore, with the exception of the PDH and BCKDH kinase and phosphatase little is known about the kinases and phosphatases responsible for mitochondrial phosphorylation O’Rourke et al have recently reviewed the evidence for the mitochondrial localization of other kinases [105]. Furthermore, although a large number of phosphorylated mitochondrial proteins have been identified, very few phosphorylation sites have been demonstrated to alter enzyme or protein activity. It has been proposed that cAMP generated in the mitochondrial activates mitochondrial PKA to regulate ATP production [109]. However, recent studies have found that alterations in mitochondrial cAMP and PKA do not contribute significantly to acute calcium stimulation of oxidative phosphorylation [110].
Given that extensive phosphorylation has been identified in the mitochondria, it is tempting to speculate that changes in mitochondrial phosphorylation regulate mitochondrial function. However, it will be important for future studies to better define the function consequences of these sites of phosphorylation as well as defining the kinases and phosphatase that regulate their phosphorylation.
S-Nitrosylation: S-nitrosylation (SNO) is the covalent attachment of nitric oxide moiety to a protein thiol group. As recently reviewed [111–113], SNO is a redox dependent modification that suggested to alter cell function by altering protein or enzyme activity, by altering protein localization, by shielding critical cysteine residues from oxidation, by altering protein stability, by altering binding partners and by competing with other PTMs. An increase in oxidative stress leads to a decrease in protein SNO and thereby alters the SNO/ROS balance. ROS leads to the consumption of NO and thus cardiac specific overexpression of SOD leads to an increase in NO bioavailability [114]. Another mechanism by which an increase in oxidative stress reduces NO/SNO signaling is by uncoupling of NOS. Alterations in NOS signaling have been proposed to predispose one to cardiovascular disease [115]. Cardioprotection is associated with a modest increase in S-nitrosylation and the majority of the proteins that exhibit an increase in SNO are mitochondrial [116]. This may be related to the redox environment of the mitochondria. Changes in cell redox can alter the generation of NO, the lifetime or bioavailability of NO, and the reactions that lead to protein SNO and denitrosylation. A key cysteine in the mitochondrial ATP synthase was shown to undergo multiple redox modification and the extent of different modifications differed in dyssynchronous heart failure compared to cardiac resynchronization therapy [117].
C. Calcium transport
The electrochemical gradient across the inner mitochondrial membrane is the driving force for calcium transported across the mitochondria inner membrane by the recently identified [118, 119] mitochondrial calcium uniporter (MCU). Uptake into the mitochondria, of small physiological levels of calcium, is thought to regulate mitochondrial metabolism and ATP production [120–123]. In heart, an increase in contractility is mediated by an increase in the cytosolic calcium transient. The increase in cytosolic calcium is transmitted to the mitochondria via Ca uptake into mitochondria, which leads to activation of the calcium sensitive mitochondrial dehydrogenases [124] and several complexes of electron transport thereby increasing ATP production as needed for the increase in work [120]. Under pathological conditions of high cytosolic calcium (calcium overload), mitochondria are capable of taking up large amounts of calcium, which lead to the opening of the mitochondrial permeability transition pore, a large conductance channel in the inner mitochondrial membrane [125, 126] (see section I.D). The sustained opening of this transition pore is a trigger for cell death [126].
As reviewed recently the MCU exist in a multiprotein complex with several proteins that regulate its activity [127–132]. Calcium efflux from cardiac mitochondria occurs via the Na-Ca exchanger (NCXL) (see [133] for a recent review). Calcium transits the outer mitochondrial membrane via the voltage dependent anion channel (VDAC). The mitochondrial Na-Ca exchange has been shown to regulate mitochondrial calcium levels and to connect mitochondrial Ca to intracellular Na, such that the rise in Na that occurs during hypertrophy and heart failure is reported to lead to alterations in mitochondrial Ca leading to altered redox and metabolism [134, 135]. There are recent data suggesting that alterations in mitochondrial calcium can contribution to the development of arrhythmias [134, 136].
Recently several groups developed MCU knockout (MCU-KO) or mice without a functional MCU (DN-MCU) to study the role of mitochondrial Ca in modulating metabolism and cell death [137–141]. Because it is generally assumed that mPTP opening and subsequent cell death is generally initiated by calcium influx into the mitochondria via the MCU, it was hypothesized that the MCU-KO hearts would have reduced mPTP opening and reduced cell death following ischemia. There was consistency among the different groups in that mitochondria from the MCU-KO hearts did not take up Ca and did not undergo Ca activated mPTP [137, 139, 140], however there were interesting differences regarding whether these mice were protected from ischemia and reperfusion mediated death. In the mice in which MCU was knocked out or mutated before birth, the hearts did not show a decrease in infarct size following ischemia-reperfusion [137, 141]. In contrast the mice in which loss of MCU was induced in adults by tamoxifen showed smaller infarcts following ischemia and reperfusion [139, 140]. One possible explanation for these differences is that when MCU is deleted before birth that compensatory mechanisms develop which somehow modify cell death pathways such that loss of MCU is not protective. A role for compensatory mechanisms is also consistent with the observation that loss of MCU is lethal on a C57B6 background.
D). Mitochondria and cell death
Prior to the 1980s, cell death was viewed as a passive process. At odds with this concept were long-standing observations that specific cells die at specific times during development in multicellular organisms ranging from worms to mammals [142]. However, it was not until the identification of a small network of genes which modulates developmental cell death in C. elegans that the concept of “regulated cell death” came into focus [143]. By the 1990s, the descendants of these genes were recognized to also mediate apoptotic cell death in adult organisms including humans [144]. By the turn of the century, it became clear that a large proportion of necrotic cell deaths, thought to be the last bastion of passive cell death, are actually highly regulated [145–148]. In addition to apoptosis and necrosis, other regulated death programs – defined by morphology and/or the context they occur – likely exist [149], including a form of cell death associated with autophagy (“autosis”) [150]. What regulated forms of cell death share in common is that process mediated by signaling pathways whose components are constitutively present in the cell. These hardwired pathways remain inactive, however, until receipt of a “death signal” originating from outside or inside the cell.
Apoptosis and necrosis have been most intensively studied. While they share inciting death stimuli and are mediated by overlapping pathways, they differ in morphology and consequences to surrounding tissue [151]. Specifically, apoptosis is a stealth form of cell death because plasma membrane integrity is maintained until the fragmented cellular corpses are eliminated by phagocytosis. In contrast, plasma and organelle membrane breakdown is a defining feature of necrosis and may be actively mediated. The end result in necrosis is the release of inflammatory mediators that cause collateral tissue damage in a paracrine manner and through the recruitment of leukocytes. Based on traditional pathological analysis, the major form of cardiomyocyte death in the infarct zone is thought to be necrosis [152], while a delayed wave of apoptosis takes place in the peri-infarct region especially with reperfusion [153, 154]. Genetic experiments in mice have established that regulated necrosis and apoptosis both play important roles in the generation of the infarct (examples include [95, 147, 155–163]). In dilated cardiomyopathy, low - but clearly elevated - levels of cardiomyocyte apoptosis take place and are an important component in pathogenesis of this syndrome. [164]. Necrosis has also been reported to contribute to heart failure, but has been less well studied [148].
Apoptosis and necrosis can each be induced through two general pathways, one involving cell surface “death” receptors and the other the mitochondria [144, 151, 165]. Even when the signals initiate through death receptors, the mitochondria are often part of a critical amplification loop. Regardless of initiating pathway, the end-game in apoptosis is to activate caspases, a class of cystinyl proteases that cut following aspartic acid residues. Caspases then proteolyze multiple cellular substrates to bring about the demise of the cell. The molecular goal in necrosis, on the other hand, depends on the initiating pathway. Induction of necrosis through the death receptor pathway (“necroptosis”) is mediated through activation of Receptor Interacting Protein (RIP) 1 and RIP3, homologous serine/threonine kinases whose targets are an area of active investigation.
Mitochondria have been recognized as playing a central role in both apoptotic and necrotic cell death. The triggering event in mitochondrial-mediated apoptosis is permeabilization of the outer mitochondrial membrane (OMM) allowing the release of “apoptogens”, including cytochrome c, SMAC/DIABLO, Omi/HtrA2, AIF, and EndoG [165]. What these proteins share in common is that they carry out healthy functions within the mitochondria, but are toxic in the cytosolic compartment. For example, in healthy cells, cytochrome c participates in electron transport at the inner mitochondrial membrane as part of oxidative phosphorylation. In contrast, once cytosolic during apoptosis, cytochrome c binds Apaf-1 to trigger assembly of the apoptosome in which procaspase-9 is activated. OMM permeabilization during apoptosis is promoted by BAX and BAK, pro-cell death members of the BCL-2 family of proteins [166]. While it is not known precisely how these proteins bring about permeabilization (e.g. one model involves pore formation), it is clear that homo- and hetero-oligomerization is important. BAX and BAK are regulated primarily through changes in their conformations. In the case of BAX, which resides in the cytoplasm of healthy cells in an inactive conformation, conformational activation [167, 168] is brought about by direct binding of BIM or a truncated form of BID (tBid), which are members of the BH3-only arm of the BCL-2 family. The function of BH3-only proteins is to bring death signals to BAX and BAK from other pathways in the cell. Activation of BAX exposes a transmembrane domain in its 9th α-helix that has a predilection for the OMM and presumably facilitates BAX mitochondrial translocation. BAK resides constitutively in the OMM and is thought to be activated in a similar fashion, although this has been studied in less depth. Anti-apoptotic BCL-2 proteins such as BCL-2, BCL-xL, and MCL-1 inhibit BAX and BAK both by functioning as sinks for BIM and tBid and possibly also through direct interactions with BAX and BAK.
The triggering event in mitochondrial-mediated necrosis is the sustained opening of the mitochondrial permeability transition pore (mPTP) in the inner mitochondrial membrane (IMM) [151]. In healthy cells, the OMM is impermeant to apoptogens - but allows the passage of ions and small molecules. Opening of the mPTP during necrosis results in rapid dissipation of the proton gradient across the IMM that is generated by pumping of protons into the intermembrane space during oxidation of substrates in the Krebs cycle. Since this transmembrane proton gradient is needed to drive ATP synthesis, mPTP opening abruptly stops production of new ATP. To further compound this energetic deficit, ATP consumption continues largely unabated during necrosis [169]. In contrast, apoptotic cells shut down ATP-requiring functions such as DNA repair, translation, and proteasome function [170–172] and experience less reduction in ATP synthesis. A second consequence of mPTP opening during necrosis is the ingress of water down its osmotic gradient into the solute-rich mitochondrial matrix. This causes matrix swelling, resulting in expansion of the redundant IMM and sometimes rupture of the OMM, which lacks redundancy. Rupture of the OMM sets up the possibility that apoptogens may gain access to the cytoplasm in necrosis (albeit via OMM rupture rather than permeabilization) and trigger caspase activation [147]. Given the cataclysmic events resulting from cessation of ATP synthesis, the extent to which subsequent engagement of the downstream apoptosis signaling contributes to cell death in necrosis is unclear.
The composition of the mPTP has been an area of great controversy [173]. The pore has often been depicted as a complex involving the voltage dependent anion channel (VDAC) in the OMM and the adenine nucleotide translocase (ANT) in the IMM. Genetic studies, however, have demonstrated that neither VDAC [147] nor ANT [174] are required for pore opening. Similarly, the mitochondrial phosphate carrier in the IMM, more recently hypothesized to be part of the mPTP, has proven to be dispensable [175, 176]. What then is the mPTP? Recent work suggests the unanticipated result that a core component is the F1-F0 ATP synthase itself [177–179]. While these data are exciting, additional studies will be required for in vivo proof.
The best characterized stimulus for mPTP opening are increases in the concentration of Ca2+ in the mitochondrial matrix [180]. The effects of increased [Ca2+] on mPTP opening are sensitized by oxidative stress, increases in phosphate, and decreases in ATP and ADP[180–182]. These conditions operate during ischemia and/or reperfusion [183]. The binding site through which Ca2+ triggers mPTP opening is not known, however. A critical facilitator of mPTP opening is cyclophilin D, a peptidyl prolyl isomerase in the mitochondrial matrix [160, 184]. While it is known that cyclophilin D binds the F1-F0 ATP synthase [178] and it has been reported that the cyclophilin D prolyl isomerase activity is required for facilitation of mPTP opening [184], the precise mechanism is not understood. The drug cyclosporin A, which binds cyclophilin D, inhibits mPTP opening and necrosis [185]. Although not an essential component of the mPTP, ANT also functions as a positive regulator of pore opening [174]. Recently, the pro-apoptotic proteins BAX and BAK were found to be critical mediators of primary necrosis [163, 186]. Mice lacking BAX and BAK or BAX alone exhibit markedly decreased cardiac necrosis, apoptosis, and infarct size following ischemia/reperfusion in vivo [158, 163]. Analysis of BAX mutants shows that its apoptotic and necrotic functions are distinct. Current evidence supports two non-mutually exclusive models in which BAX functions as an OMM component of the mPTP and/or facilitates necrosis indirectly by promoting mitochondrial fusion.
Many questions remain concerning the mitochondrial events that mediate cell death. First, the complete composition of mPTP is not clear at this point. Second, the upstream signaling that feeds into both necrotic and apoptotic programs at the mitochondria remains incompletely understood especially in the case of ischemia/reperfusion. Third, the molecular connections linking apoptotic and necrotic programs at the mitochondria and the factors that determine how a specific cell will die are not known in any depth.
Despite these deficits in knowledge, inhibition of mitochondrial-mediated cell death has been contemplated – especially for ischemic syndromes. We will limit the discussion here to two points. First, there has been a clinical trial of cyclosporin A in 58 patients with ST-segment elevation myocardial infarction [187]. Administration of the drug at the time of percutaneous coronary intervention reduced serum creatine kinase but not troponin I. Hyperenhancement on MRI, a measure of infarct size, was reduced at day 5. At 6 months, however, there were no statistically differences in left ventricular function [188]. This study is inconclusive, but encouraging. It suggests that inhibition of mPTP opening may be an effective treatment strategy for reperfused myocardial infarction in humans, but also raises the question as to whether compounds more potent than cyclopsporin A need to be developed. Second, given that necrosis and apoptosis both contribute to the pathogenesis of myocardial infarction, selection of a therapeutic target such as BAX, which mediates both forms of cell death [163, 186], should be considered.
E. Generation of ROS
During electron transport if there is any leak of electrons it can lead to the generation of reactive oxygen species (ROS) and mitochondria are one of the major cellular sources of ROS. Mitochondria also contain antioxidant mechanisms to remove ROS. In low levels ROS can act as a signaling molecule, whereas higher levels can lead to irreversible damage to mitochondria and cells and is a major contributor to cardiovascular disease.
In mitochondria ROS formation results from sporadic, possibly undesired, reactions occurring especially at the level of the electron transport chain (ETC) [189–191]. Besides these occasional processes, mitochondria also contain enzymes that catalyze H2O2 generation as the obligatory product [192].
The ETC drives electrons from reduced coenzymes (NADH(H+) and FADH2) to oxygen that undergoes the complete reduction to water in the terminal reaction catalyzed by Complex IV (i.e., cytochrome c oxidase). A minor fraction (about 0.1%) of the electrons flowing through the ETC is suggested to cause the partial reduction of O2 into superoxide [189]. In particular, flavins or quinones of the first three complexes are able to act as single electron donors resulting in superoxide formation, especially under conditions that decrease the flow of electrons towards Complex IV where O2 is fully reduced to H2O [189, 190]. Notably, ROS formation can also result from reverse electron flow. Recently, this concept has been supported by demonstrating in vivo that succinate accumulated during ischemia is oxidized during reperfusion resulting in large ROS formation that is likely due to the reverse electron flow within complex I [193].
ROS formation is favored by high mitochondrial membrane potential (i.e., low ATP synthesis), large NADH(H+) or when electron flow is hampered by alterations in respiratory complexes. Conversely, a decrease in ROS levels should follow the acceleration in electron flow caused by mitochondrial uncoupling [194], yet conditions have been reported in which mitochondrial uncoupling and Δψm dissipation are associated with an increased ROS formation.[135, 195] According to the model of “redox-optimized ROS balance” [196] this apparent paradox might be explained by a concomitant depletion of the antioxidative capacity that would result in H2O2 accumulation despite a decreased formation of superoxide by the ETC [197].
An increased ROS formation is also associated with the uncoupling-like effect generated by opening of the mPTP. Indeed, this process has been proposed to amplify an initial oxidative stress through the so-called ROS-induced ROS release (RIRR) [191]. ROS can trigger PTP opening through oxidative modifications of mitochondrial proteins involved in PTP formation and control, such as FoF1 ATP synthase e[198] or cyclophilin D [199]. However, despite evidence that ROS formation follows PTP opening [200, 201], the underlying mechanisms have not yet been elucidated. On the other hand, a slight increase in ROS formation resulting from opening of mitochondrial K+ATP channels has been proposed to prevent mPTP opening and elicit cardioprotection [202–204]. A similar process could contribute to protection induced by preconditioning or postconditioning that is abrogated by antioxidant treatment [188, 205, 206]. Therefore, the notion that a mild ROS accumulation increases the resistance to oxidative stress [207] might be explained by opposite effects on the susceptibility to PTP opening elicited by slight and large ROS formation, respectively.
Superoxide that does not cross the inner mitochondrial membrane is rapidly dismutated into the freely permeable H2O2 by Mn-superoxide dismutase (Mn-SOD). The finding that Mn-SOD deficient mice develop ROS toxicity and dilated cardiomyopathy [208, 209], underlines the importance of ROS in this pathology and mitochondria as their source and target. This concept is further supported by the beneficial effects afforded by targeting catalase expression in mitochondria [210–212].
Besides respiratory chain complexes, several other mitochondrial enzymes have been described as potential ROS producers. These include the flavin containing glycerol-3-phosphate-, proline- and dihydroorotate-dehydrogenase at the outer leaflet of the inner mitochondrial membrane, the electron transfer flavoprotein-ubiquinone (ETF:Q) oxidoreductase system of fatty acid β-oxidation within the inner mitochondrial membrane, and pyruvate- and 2-oxoglutarate dehydrogenase within the mitochondrial matrix [213]. However, the contribution of these enzymes to the overall ROS production of mitochondria within a given cell is difficult to establish. In fact, as is also the case with respiratory complexes, loss-of-function approaches (i.e., pharmacological inhibition or genetic deletion) would inevitably hamper the physiological functions of these vital proteins jeopardizing energy metabolism, ionic homeostasis and cell viability. Convincing demonstration that mitochondria generate ROS in vivo is also provided by interventions targeting mitochondrial enzymes, such as p66Shc and monoamine oxidases (MAO) that generate H2O2 as a direct and obligatory product.
In response to various stress stimuli, the cytosolic adaptor protein p66Shc translocates to mitochondria where it catalyzes H2O2 formation by means of electron transfer from cytochrome c to oxygen [214]. Indeed, ROS generation is reduced in cells lacking p66Shc and in p66Shc−/− mice, whose lifespan is increased by 30% [215, 216]. Furthermore, genetic deletion of p66Shc protects against ischemia/reperfusion (I/R) injury and diabetes-induced cardiovascular derangements [192, 217].
The two isoforms of MAO, A and B, are flavoenzymes located in the outer mitochondrial membrane. MAOs catalyze the oxidative deamination of catecholamines, serotonin and biogenic amines generating the corresponding aldehydes, H2O2 and ammonia. H2O2 and aldehydes [206]produced by MAO have been shown to synergize in disrupting mitochondrial function associated with loss of function and viability of the heart [192]. In addition, ammonia might stimulate ROS formation by dihydrolipoyl dehydrogenase, the E3 component of pyruvate and oxoglutarate dehydrogenase [218]. Interestingly in human atrial biopsies MAO has been shown to produce 10 times more H2O2 than the respiratory chain, and its expression is correlated with an increased risk for postoperative atrial fibrillation [219]. Major advantages with investigating the role of MAO in oxidative stress are given by a defined molecular structure, specific substrates and clinically available inhibitors. However, the substrates used and the mechanisms of activation under injury conditions are still not clear. In addition, the clinical use of MAO inhibitors in cardiovascular diseases is perceived as problematic due to a hypertensive reaction occurring when selective MAO-A inhibition is combined with intake of tyramine-rich food, such as aged cheese and alcoholic beverages. Conversely, MAO-B inhibition is devoid of this potential risk [220].
The list of dedicated enzymes for ROS formation in mitochondria includes nicotinamide adenine dinucleotide phosphate oxidase 4 (NOX4) [221]. At variance from other NOX isoforms localized to the plasma membrane, NOX 4 displays a preferential localization to intracellular sites, appears to be constitutively active and generates H2O2 in preference to superoxide [222, 223]. Since the localization to cardiac mitochondria has been established based upon reactivity to antibodies that were not tested in NOX4−/− cardiomyocytes, further studies are necessary. In addition, the role in the cardiovascular system is debated, since NOX4 appears to cause both beneficial and detrimental effects in models of cardiac pressure overload [28, 29, [77, 221]
As in the rest of the cell, in mitochondria ROS generation is counterbalanced by efficacious removal systems. Besides superoxide dismutation by Mn-SOD, peroxide handling is carried out by a thiol redox system centered on glutathione (GSH and GSSG in its reduced and oxidized form, respectively) and thioredoxin (Trx) [224–226]. H2O2 is reduced into water by glutathione peroxidases (Gpx1 and 4) and peroxiredoxin 3 (Prx3) that is maintained into the active reduced form by Trx. In addition, GSH is used by the mitochondrial glutathione-S-transferase K to detoxify products of oxidative damage, such as α,β unsaturated aldehydes and alkyl hydroperoxides, and by glutaredoxin 2 that catalyzes the formation and reversal of protein-GSH mixed disulfides [225].
The oxidized forms of glutathione and Trx resulting from Gpx and Prx catalysis are reduced by the corresponding reductases at the expense of NADP(H+). The mitochondrial NADP+ pool is reduced by malic enzyme, Δψm-dependent nicotinamide nucleotide transhydrogenase, and Ca2+-modulated isocitrate deghydrogenase. Therefore, the NADP+/NADP(H+) ratio links oxidative metabolism and mitochondrial function with ROS signaling and antioxidant activities [197]. Prx3 is responsible for more than 90% of H2O2 removal in mitochondria [227]. However, since Prx3 is highly susceptible to oxidation, under conditions of severe oxidative stress such as myocardial ischemia and reperfusion [228], Gpx1 might become the major sink for H2O2.[225] Prx3 overexpression confers increased resistance to ischemia-reperfusion injury [229] lending further support to the relevance of mitochondrial-generated ROS in cardiac diseases.
The occurrence and the relevance of ROS formation in mitochondria is supported by direct methods for the in vivo detection [230], effects of targeted antioxidants and enzymes [231], and decreased ROS accumulation following inhibition or deletion of mitochondrial ROS sources [232]. The majority of studies relate mitochondrial ROS formation with cell injury suggesting that beneficial effects are afforded by preventing ROS accumulation in a wide array of cardiovascular diseases, such as ischemia/reperfusion injury, heart failure, ageing and diabetic cardiomyopathy [190, 192, 217, 233, 234].
Although it is undeniable that high levels of ROS impair function and viability of any cell type, a large body of evidence indicates that ROS generated within mitochondria are involved in signaling processes that are crucial for the optimal response to physiological and pathological stimuli [192, 235–237]. Indeed, several reports document the crucial role of mitochondrial ROS generation in a wide variety of cardiomyocyte functions. The physiological role of mitochondrial ROS is likely to be linked to post-translational modifications of proteins, especially at the level of cysteine residues [238]. For instance, S-nitrosylation has been recognized as a cardioprotective mechanism that prevents irreversible oxidation of proteins [225, 230, 239]. In addition, signaling pathways involving protein phosphorylation are modulated by oxidation of critical cysteines, especially in protein phosphatases [240]. Besides these short-term responses mitochondrial ROS are involved in long-lasting changes by acting on transcriptional factors, such as hypoxia-inducible factors (HIFs) and nuclear factor erythroid 2–related factor 2 (Nrf2) [226, 241–243].
Far from always being beneficial, a decrease in mitochondrial ROS levels could be detrimental. A large increase in glutathione content or the administration of N-acetylcysteine was shown to elicit mitochondrial oxidation and cytoxicity despite a decrease in ROS levels [244]. Suppression of mitochondrial ROS generation by mitochondrial-targeted catalase hampered autophagy worsening heart failure caused by deletion of mitofusin 2 [245]. Interventions aimed at reducing mitochondrial ROS levels, such as expression of dominant negative Nox [149], deletion of p66Shc [246], or ablation of thioredoxin-interacting protein [247], were found to exacerbate mild injury induced by ischemia-reperfusion protocols of short duration. This paradoxical notion that contrasts with protection by antioxidant treatments in prolonged episodes of ischemia-reperfusion suggests that mitochondrial ROS are involved in triggering self-defense mechanisms. Supporting this concept antioxidants abrogate the powerful protection of both ischemic pre- and post-conditioning [203, 204, 206]. In this respect, the term mitohormesis has been introduced to describe the j-shaped curve whereby low doses of mitochondrial ROS trigger beneficial adaptive responses that are replaced by detrimental processes at high doses [241, 248]. Although this concept appears to accommodate the protective effects of exercise and calorie restriction [248], especially in clinical settings methods are not available to define the threshold separating beneficial from harmful ROS levels. Other relevant issues to address are the interactions among the various ROS sources and the conditions involved in local compartmentalized ROS formation as compared to diffusion of ROS and oxidized products to the entire cell and surrounding tissues.
F). Mitochondrial Quality Control
1. Fission/Fusion/Mitochondrial Dynamics
As cells and organisms reproduce their mitochondria divide to repopulate the progeny. Mitochondria also divide and fuse back together in non-dividing, quiescent and postmitotic cells such as cardiomyocytes and neurons; however the rates in cardiomyocytes appear to be quite low. This continual fission and fusion cycle, a process also called mitochondrial dynamics, is known to be essential for the healthy maintenance of mitochondria and their host cells and organisms. Mitochondrial dynamics participate in mitophagy, apoptosis, differentiation and a variety of stress responses. The adverse consequences that interrupting in vivo cardiomyocyte mitochondrial dynamics has on mitochondrial stress, mitochondrial biogenesis, and programmed cardiomyocyte death were recently demonstrated in side-by-side comparative studies after conditional genetic deletion of either cardiac mitochondrial fusion or fission pathways [249].
Mitochondrial morphology reflects the relative rates of fission and fusion and can be visualized in fixed cells and tissues by immunostaining. Perturbing the ratio of fission and fusion rates will lead to either fragmented, punctiform mitochondria or excessively long or interconnected mitochondria. However, as mitochondrial morphology is dynamic, fission and fusion rates are best visualized in live cells. Quantification of mitochondrial fusion rates can be performed by using photoactivatable green fluorescent protein (PAGFP) that is targeted to the mitochondrial matrix. When the mito-PAGFP is activated with a laser targeted to one mitochondrion, the PAGFP fluorescence is increased about 100 times and as this mitochondrion fuses with others, the fluorescence is diluted [250]. Quantification of this dilution rate reflects the organelle fusion rate independent of the fission rate and shows remarkable difference among cell types and changes for example, early during apoptosis. An alternate approach to assess fusion utilizes two cell populations, one expressing GFP in mitochondria and another expressing red fluorescent protein (RFP) in the mitochondria. When cells from these two populations are fused with polyethylene glycol the rate that the GFP and RFP merge to form yellow fluorescence reflects the mitochondrial fusion rate [251]. These techniques have been used extensively to characterize proteins that mediate the fusion process [252] and the physiological consequences of mitochondrial dynamics [253].
The molecular machinery that mediates mitochondrial fusion utilizes large GTPases in the dynamin family [252, 254]. Mitofusins (Mfn1, Mfn2) mediate fusion of the outer mitochondrial membrane, whereas Opa1 mediates fusion of the inner mitochondrial membrane. Mfn1, Mfn2 and Opa1 all require GTPase activity for fusion activity. Mfn1 and Mfn2 span the outer mitochondrial membrane with most of the protein and the GTPase domain facing the cytosolic compartment. Opa1 is localized within the intermembrane space anchored to the inner mitochondrial membrane. Mitofusins and Opa1 usually work in concert to coordinately fuse both mitochondrial membranes. Opa1 activity is regulated by proteolytic processing but how Mfn1 and Mfn2 are regulated is not yet clear.
Mitochondrial fission utilizes a large GTPase called Drp-1, a homologue of dynamin that is well understood to mediate fission of endocytic vesicles from the plasma membrane. Like dynamin during endocytosis, Drp1 assembles into spirals that wrap around mitochondria and appear to constrict the inner and outer membranes during GTP cleavage to start the fission process. Drp1 exists free in the cytosol from where it docks to mitochondrial fission sites by interacting with outer mitochondrial membrane spanning proteins Mff, Mid49 and Mid52 [252, 254]. Fis1 is a protein required for Dnm1 (a Drp1 orthologue) mediated fission in yeast, but is not required for fission in metazoans. Instead, Fis1 in metazoans participates in mitophagy [255]. Drp1 activity is regulated by phosphorylation at several sites on the protein. Phosphorylation at some sites stimulates fission, for example during the cell cycle, and at other sites phosphorylation inhibits fission. Interestingly, endoplasmic reticulum tubules wrap around sites of mitochondria prior to their fission and may play a role in defining the site of mitochondrial fission or in assembling the fission complex at the proper location [256].
Identification of these fission and fusion proteins has allowed generation of animal models and cell culture lines for the exploration of the physiological significance of mitochondrial dynamics. Mfn1, Mfn2 and Opa1 knock out mice are all embryonic lethal suggesting that both fission and fusion are required for maintenance of mammalian embryos. However, fibroblasts generated from the embryos survive in culture although with altered mitochondrial morphology and in some cases, metabolic deficits [252]. Cardiac myocyte specific knockout of Mfn1 and 2 in adults causes cardiomyopathy. Interestingly, myocytes die after only 3–4 cycles of mitochondrial fission without opposing fusion [83]. Surprisingly, mitochondrial fusion is also linked to cardiac development. Through regulating calcium levels and calcineurin, mitochondrial dynamics control Notch signaling and stem cell differentiation into cardiomyocytes [257].
Mutations in several of the mitochondrial fusion genes have been identified to cause human disease [252, 254]. Dominant optic atrophy, the most common form of hereditary blindness, is caused by haploinsufficiency in Opa1. Thus, retinal ganglion cells are highly dependent, and more so than other human tissues, on fusion of the inner mitochondrial membrane. Another example is mutations in Mfn2 that cause Charcot-Marie-Tooth Disease Type 2A. Understanding the intriguing tissue specificity of defects from mutations in mitochondrial dynamics genes, that have what may be considered housekeeping duties, remains a major challenge in discerning the roles of mitochondria in vivo. Because of the links of mitochondrial dynamics to human and animal health, there are efforts to drug the pathway. For example, mDIVI is a small molecule that inhibits Drp1 and mitochondrial fission, which has been reported to have protective activity in numerous animal disease models including heart ischemia-reperfusion injury [258].
It is clear that mitochondria have to continually divide and fuse but what are the essential roles of mitochondrial dynamics at the molecular level? Mitochondrial fission has been linked to damage avoidance through segregation of debris within mitochondria. As discussed later in this Statement (Section I.F.2), mitophagy following fission, allows the clearance of damaged mitochondria and selective elimination of damaged proteins [253, 259]. Mitochondrial fission has been linked to apoptosis that can also function as a severe form of stress response. Mitochondrial dynamics are also required for proper transport of mitochondria to proper locations within cells. On the other hand, fusion between mitochondria is thought to allow compensation to help rescue organelles from damage by the exchange of proteins and RNAs from one mitochondrion to another [260]. Mitochondria accumulate mtDNA deletions and mutations over time and these mutations can generate mitochondrial proteins that are dysfunctional or misfolded. If mitochondria did not fuse, dysfunctional proteins could lead to serious loss of function consequences. However, fusion with another mitochondrion, that may have mutations in other genes, would allow compensation between organelles and avoid serious consequences of mutation accumulation. Mitochondrial hyperfusion is a broadly active stress response that may facilitate such compensation [261].
2. Mitophagy and mitochondrial autophagy: mechanisms, overlap, and distinctions
Mitophagy, literally meaning “eating mitochondria”, is the term applied to the cellular mechanism for identifying and selectively eliminating dysfunctional mitochondria as part of the overall mitochondrial quality control process [262]. Mitophagy is essential to sequester and remove senescent or damaged mitochondria that could otherwise accumulate and become sources of cytotoxic ROS (see Section I.E). Whereas the distal components of the mitophagy pathway, ie autophagosomal engulfment of mitochondria and their transfer to lysosomes for degradation and component recycling, are shared with macroautophagy, the proximal events that detect and select dysfunctional organelles for targeted elimination are highly specific for mitophagy. Two central proteins driving this detection/selection process are the cytosolic E3 ubiquitin ligase Parkin [263] and the mitochondrial kinase PINK1[56], encoded by genes (PARK2 and PARK7, respectively) for which loss of function mutations have been linked to autosomal recessive forms of Parkinson’s disease [264].
The discovery that PINK1 and Parkin interact to promote mitochondrial fitness [265, 266], and elucidation of mitochondrial stabilization of PINK1 as the initiating event in mitophagy [267, 268], were central to our current understanding of the mechanisms underlying mitochondrial quality control. Briefly, healthy mitochondria maintain an electrochemical inner membrane gradient, Δψm that drives ATP production by the electron transport complex (see Section I.A). Senescent mitochondria are unable to sustain a normal Δψm, and damaged mitochondria may completely dissipate Δψm, resulting in depolarization. Mitochondrial Δψm status is a key determinant of PINK1-Parkin pathway activity: Healthy fully polarized mitochondria import and rapidly degrade PINK1, maintaining low kinase activity. Upon depolarization however, PINK1 degradation is suppressed [268], thereby increasing its abundance and promoting multiple PINK1 kinase-mediated events including Parkin translocation to [269], and activation at, [270, 271] mitochondrial outer membranes. At the mitochondrion Parkin ubiquitylates dozens of mitochondrial proteins [272], thereby promoting autophagosomal engulfment of the damaged organelle. The overall result for the cell is selective mitophagic destruction of depolarized mitochondria.
Mitophagy is inextricably linked to mitochondrial dynamism, i.e. mitochondrial fission and fusion. In many cells mitochondria form highly interconnected reticular networks that are constantly remodeling through periodic fission and fusion. However, in adult cardiac myocytes mitochondria fission/fusion are rare [273]. For this reason, in hearts it is likely that mitochondrial dynamism is dispensable for morphometric remodeling, but nevertheless plays an important role in cardiac mitophagic quality control through the process of dynamin-related protein (Drp)1-mediated asymmetric fission [274]. Accordingly, a mitochondrion in the early stages of senescence or one that has sustained moderate damage will segregate its dysfunctional components into one of the two daughter organelles generated by a fission event. The damaged (and therefore depolarized) daughter mitochondrion will be promptly identified as such and removed via PINK1-Parkin mediated mitophagy, whereas the healthy daughter will rejoin the cellular mitochondria pool, likely by fusing with other similarly fit mitochondria. The particular role for Parkin-dependent versus Parkin-independent or “alternate” mitophagy mechanisms in healthy and diseased hearts is only beginning to be investigated [275, 276].
Mitofusins (Mfn) 1 and 2, so designated because they promote physical tethering between mitochondria and subsequent GTPase-dependent mitochondrial fusion, also have a role in mitophagy. In addition to promoting fusion of the healthy daughters after asymmetric fission (see above), PINK1 kinase stabilization in damaged mitochondria results in phosphorylation of Mfn2 on two domains, thus conferring Parkin binding activity to this mitochondrial outer membrane protein and facilitating both Parkin translocation and its subsequent ubiquitination of mitochondrial proteins [55]. For this reason, hearts deficient in Mfn2 that do not exhibit defects in mitochondrial fusion (because Mfn1 is still present) instead develop a defect in mitochondrial quality control [55, 277].
There are surprising consequences of the mechanistic involvement of mitochondrial dynamism in mitochondrial quality control. For example, if mitophagy is malfunctioning but dynamism is intact, then the process of asymmetric fission will generate a highly dysfunctional daughter organelle that cannot be removed through the usual quality control process. Instead, the improperly retained damaged mitochondrion can fuse with, and by exchanging cellular components thereby damage, normal mitochondria within the same cell. An example of fusion-mediated mitochondrial contagion was recently uncovered in Parkin-deficient Drosophila fruit fly heart tubes [245]. In this model, because fusion contributed to the spread of mitochondrial damage, heart failure was attenuated by cardiomyocyte-specific suppression of Drosophila mitofusin (MARF).
An important role for PINK1-Parkin mediated mitophagy in normal functioning of the nervous system is clear from Parkinson’s disease [278]. Surprisingly, while genetic suppression of PINK1 or Parkin in fruit flies is detrimental to mitochondrial fitness and normal functioning of neural tissue, skeletal muscle, and myocardium, [279] germ-line gene ablation of the orthologous mouse genes evokes only modest phenotypes [280]. In mouse hearts, germ line ablation of PINK1 and Parkin appears to produce only mild and slowly progressive basal cardiac dysfunction, but increased sensitivity to ischemic injury [84, 93, 281]. Likewise, cardiomycyte-specific Parkin ablation in adult mice provoked no detectable phenotype, and conditional cardiac Parkin overexpression had no detectable adverse consequences (Song M et al Circ Res 2015, epub ahead of print). Thus, it is possible that PINK1-Parkin mediated mitophagy is relatively unimportant to mitochondrial homeostasis in normal mammalian hearts. On the other hand, the absence of notable nervous system dysfunction (i.e. Parkinson’s disease phenotypes) in these same mice [280], evidence of compensatory upregulation of alternate E3 ligases in hearts of germ-line Parkin knockout mice [245], and the cardiomyopathy that is evoked by cardiomyocyte-specific interruption of PINK1-Parkin signaling (through Mfn2 ablation) [55, 277] suggest that this pathway of mitochondrial quality control may indeed be important under specific and as yet incompletely understood circumstances. The true role of PINK1 and Parkin in mammalian hearts may only be uncovered by creating new experimental models and/or by additional human genetic testing for rare PINK1 and Parkin mutations in clinical cardiac disease. It is also worth mentioning that Parkin can also regulate fat uptake [282].
If it is correct that maintaining mitochondrial quality is essential to cell health, then the absence of damaging mouse phenotypes in PINK1 and Parkin knockout mice, and focal degeneration of dopaminergic neurons (rather than larger system-wide effects) in Parkinson’s disease linked to human PINK1 or Parkin mutations, suggest the presence of one or more alternate pathways of mitochondrial quality control [283]. Indeed, mitochondria can be eliminated independent of PINK1 and Parkin by an autophagic mechanism that employs pro-apoptotic Bcl2 family proteins Nix and Bnip3 to target dysfunctional mitochondria and connect them to autophagosomes [284]. Conceptually, this mechanism resembles so-called “mitoptosis”, in which opening of the mitochondrial permeability transition pore activates mitochondrial autophagy [285]. In this context, Nix and Bnip3 accumulate on damaged mitochondria, facilitate the permeability transition, and promote mitochondrial autophagy by serving as mitochondrial adaptor proteins that bind to autophagosomal LC3 or GABARAP [286–289]. Although Nix and Bnip3 are widely recognized for their pro-apoptotic effects in cardiac failure following pressure overload hypertrophy and myocardial infarction, respectively [290–293], the possibility that they also promote homeostatic mitochondrial autophagy in hearts merits further investigation.
Dissipation of Δψm, aka mitochondrial depolarization, contributes to the signal for PINK1 stabilization and initiation of Parkin-mediated mitophagy. Evidence is accumulating that ROS, which are also markers of mitochondrial dysfunction, can play a similar role in Parkin-independent mitochondrial autophagy [277, 294]. In vivo disruption of cardiomyocyte Parkin signaling by ablating its Mfn2 mitochondrial receptor evokes a cardiomyopathy. As expected, normalization of ROS with mitochondrial-directed catalase improves this mitophagic cardiomyopathy. In contrast, super-suppression of ROS (with mitoCAT expressed at higher levels) is detrimental, both accelerating and exacerbating the cardiomyopathy [277]. These findings reveal an essential signaling function for mitochondria-derived ROS in compensatory mitochondrial autophagy pathways induced when the Parkin pathway is interrupted.
3. Protein turnover independent of mitophagy
As discussed in Section I.F.2, damaged mitochondria can be removed by mitophagy. However individual mitochondrial proteins can also be damaged and may underlie several pathologic phenotypes [295]. It is therefore paramount that the renewal, or turnover, of proteins within mitochondria is sustained in times of enhanced cellular stress, as failure to maintain normal protein turnover may lead to accumulation of damaged/misfolded proteins and may underlie various disease etiologies. Protein turnover has been deemed “a missing dimension” in proteomics [296], as quantitative proteomic measurements typically involve the profiling of static protein abundance between different disease states or conditions. Recent advances in protein dynamics methodologies have enabled the simultaneous measurement of individual proteins comprising entire mitochondrial [244, 297–299] proteomes. A recent study provided the first assessment of global mitochondrial proteome kinetic signatures in a disease model of cardiac remodeling, which demonstrated that protein turnover rates are under independent control indicative of diverse regulatory processes driving remodeling of the mitochondria in disease [297].
Protein turnover measurements rely on the ability to track the rate at which individual proteins are being replaced by de novo synthesized proteins. Several methodologies have been utilized for these measurements, and all involve the introduction of an isotope precursor into a living system in order to mark individual proteins and determine their longevity in cells. For an excellent, comprehensive review on strategies used to measure protein turnover, including the experimental model, stable isotope label, labeling protocol, relative isotope abundance transition, and calculation of turnover rate, see [300].
Protein turnover rate is evaluated by tracking the integration or loss of a label into a protein pool. Proteins exhibit a diverse range of half-lives, with housekeeping proteins tending towards longer half-lives and regulatory proteins towards shorter half-lives. Thus, sampling times following initiation of labeling must cover an adequate range of time in order to accurately model proteins exhibiting both fast and slow rates of turnover. Moreover, the number of sampling timepoints is directly correlated to the accuracy of the labeling trajectory. An additional consideration for complex organisms is the slow equilibration of the stable isotope label (e.g., 2H) in a precursor pool (e.g., body water), which is incompletely labeled in in vivo labeling strategies and requires complex analysis to determine precursor pool enrichment. While each stable isotope methodology has its strengths and weaknesses, heavy water labeling has distinct advantages for translational research in that at low enrichment levels it is safe for humans over years [301], it is easy to maintain constant enrichment levels of 2H in body water following 2H2O intake[302, 303], and it is the most cost effective stable isotope. Detailed methods and equations underlying computational analysis are outlined in [244], and the automated software, ProTurn[297] is available at http://www.heartproteome.org/proturn/.
Four recent studies have interrogated protein dynamics in mitochondria in whole animals or humans, using oral consumption of either deuterated leucine-labeled protein [298] or drinking water [244, 297, 299]. Two recent studies investigated mitochondrial protein turnover changes is cardiac pathologies. A translational study by Lam et al. [297], in which 2H2O labeling was employed to investigate changes in protein dynamics of cardiac proteins in mouse and blood proteins in both mouse and human. Turnover rates for 496 human plasma proteins, spanning a 50-fold range of turnover rates from albumin (half-life 18.3 days) to IGF2 (half-life 8 hours) were determined. Mice undergoing cardiac remodeling via chronic isoproterenol infusion were compared to controls, and turnover rates for 2,964 mouse proteins were assessed in mouse cardiac mitochondrial and cytosolic fractions and in blood. Rates were highly diverse and ranged from <1 day to >3 weeks (100-fold range). Cytosolic proteins turned over approximately 10% per day (average half-life of 6.5 days) and mitochondrial proteins approximately 5% per day (average half-life 15 days). Consistent with enhanced protein synthesis occurring in cardiac hypertrophy during remodeling, turnover rates in isoproterenol-treated mice averaged 1.2 times faster than in wild-type hearts with turnover rate increases detected in 972 proteins (>1.3 fold) and decreases in 216 proteins. In contrast, isoproterenol withdrawal, or reverse remodeling, led to an average of 20% decrease in protein turnover rate. Importantly, this study identified proteins exhibiting supernormal elevations or attenuations in protein turnover during cardiac remodeling, suggesting that these proteins may be pathogenic in the cardiac remodeling process. Proteins involved in mitochondrial dynamics showed heterogeneous results, in that some mitochondrial dynamics proteins exhibited elevated turnover (MIRO1/2, LONP and PHB), while others were unchanged (MFN1/2 and FIS1). Subunits residing in the same respiratory complex (ETC I-V) displayed widespread changes, thereby giving insight into proteins that may be rate-limiting factors in complex formation and highlighting the important finding that synthesis-degradation cycles of proteins within the same functional group (i.e. complexes, organelles) are under independent control. Interestingly, turnover measurements also unveiled markedly enhanced turnover in virtually all glycolytic enzymes (HKI half-life decreased from 16.7 to 9.8 days; GAPDH 10.8 to 6.7 days; phosphoglycerate mutase-1 12.2 to 6.9 days) in the absence of any changes in static protein abundance, thus providing mechanistic insights for supporting the alteration in substrate utilization, from fatty acids to glucose, known to occur in cardiac disease. Importantly, these novel mechanistic insights into cardiac remodeling are masked in measurements of static protein abundance, highlighting the unique biological dimension unveiled by protein turnover measurements. Lastly, a study by Shekar et al. [299], used 2H2O-labeling in rats to measure turnover in heart failure from two cardiac mitochondrial subpopulations, subsarcolemmal (SSM) and interfibrillar (IFM). This group investigated the hypothesis that mitochondrial protein synthesis (and thus, oxidative capacity) is decreased in transverse aorta constriction (TAC)-induced heart failure, and IFM exhibit more pronounced detriments than SSM. Results from this study showed an overall decrease in mitochondrial content in IFM, but not SSM mitochondrial populations, coordinate with a more pronounced detriment in basal and stimulated respiratory rate in IFM.
The physiological and pathophysiological implications gleaned from turnover measurements of individual proteins within functional mitochondrial subproteomes are many. Dynamic equilibria of proteins fluctuate in times of cellular stimulation or altered cellular stress, and protein turnover measurements provide a missing dimension of protein behavior that will enable mechanistic studies on the governance of protein synthesis and degradation. Very little is known regarding the interplay of these processes in tuning the abundance of individual proteins or protein complexes in the cell. Global turnover measurements with individual protein resolution, now possible for all detectable proteins within the mitochondrial proteome, have indicated that diverse regulatory mechanisms exist in metabolic pathways. Furthermore, it is now understood that protein half-life is an exquisitely regulated cellular parameter that is correlated with phenotype, however is mostly disassociated with static protein abundance. Protein dynamics provide a unique understanding of biological regulation of mitochondrial proteins, that when related to static protein abundance and mRNA expression, can inform on the poorly understood process of protein degradation in basal and diseased conditions.
Mitochondrial protein dynamics measurements have high translational significance regarding mitochondrial biomarker discovery and treatment of mitochondrial diseases. Mitochondrial diseases are a heterogeneous class of conditions that need sensitive and specific biomarkers for their accurate diagnosis and prognostic assessment. The advantages of using protein turnover measurements, rather than the commonly-employed protein abundance, for early detection of pathophysiological states has been discussed [304] and clearly demonstrated [305, 306]. The sensitivity of protein dynamics is in many ways superior to static protein abundance, in that alterations in turnover measurements will likely be more pronounced and may precede alterations in static protein abundance in pathologic progression of diseases. Protein turnover rate is also a critical factor in biomarker discovery (see Section II.C), as markers with lower clearance rates within the systemic circulation would be more robust indicators of disease states. Protein dynamics measurements will also unveil novel disease mechanisms (i.e., protein degradation insufficiencies), that will spawn the development of novel therapeutic classes.
Moreover, questions underlying the relationship between protein turnover and abundance remain. For example, certain mitochondrial proteins exhibit rapid turnover rates (and thus, consume a sizeable amount of ATP for their renewal) with little to no change in static abundance. Observations such as these may begin to challenge the conventional school of thought that the abundance of a protein present in a cell directly correlates with its magnitude of impact in the cell. While several educated guesses have been put forth regarding questions like these, scientific evidence is lacking.
G). Signaling by release of mitochondrial DNA and proteins
As discussed in Section I.D, mitochondria are involved in the intrinsic pathway of apoptosis where they release soluble proteins, including cytochrome c from the intermembrane space into the cytosol to initiate caspase activation [307]. The release of these proteins is a consequence of mitochondrial outer membrane permeabilization. Other examples for functional molecules released from mitochondria include 1) ROS to activate hypoxic gene expression, ROS-dependent MAP kinase and damaged macromolecules including DNA and proteins, 2) calcium which participates calcium crosstalk between mitochondria and the plasma membrane and between mitochondria and the endoplasmic/sarcoplasmic reticulum and ATP from apoptotic and necrotic cells as a danger signal [308]. Recently, it has been reported that mitochondria DNA and proteins are involved in innate immunity. In this section, we will focus on the role of mitochondria in inflammation [309].
The immune system is activated not only by microorganisms infection, but also by endogenous molecules [310]. The endogenous molecules are separated from immune system sensors by plasma membrane and compartmentalization within the cell. However, endogenous molecules are released into circulation during necrosis or into the cytoplasm during degradation of organelles. These endogenous molecules that can induce inflammatory responses are referred to as damaged-associated molecular patterns (DAMPs). Pattern recognition receptors (PPRs) are responsible for sensing the presence of microorganisms by recognizing structures that are conserved among microbial species, which are called pathogen-associated molecular patterns (PAMP). Moreover, PRRs are attributable to recognizing DAMP. To date, four different classes of PRR families have been identified including transmembrane proteins such as Toll-like receptors (TLR) and C-type lectin receptors, as well as cytoplasmic sensors such as Retinoic acid-inducible gene (RIG)-I-like receptors and NOD-like receptors (NLRs). In addition to PRRs, inflammasomes are multiprotein complexes which contribute to the intracellular identification of potentially harmful substances, bacteria, or viruses. Inflammasomes are comprised of three major components, a characteristic scaffolding protein, the small adapter molecule ASC, and procaspase-1 which is responsible for the activation of pro-inflammatory cytokines. Those scaffolding proteins include NOD-like receptor family, pyrin domain containing 1 (NLRP1), Absent in melanoma 2 (AIM2), NLR family CARD domain containing protein 4 (NLRC4), RIG-I and NLRP3, and each protein forms the corresponding inflammasome in concert with ASC and procaspase 1.
Since mitochondria are evolutionary endosymbionts derived from bacteria, mitochondria still have many morphological and biochemical features of their bacterial ancestors, including a double-membrane, membrane lipid (cardiolipin), unmethylated CpG motifs in mitochondrial DNA, absence of histones and the ability to form N-formyl peptides, which are synthesized by the use of separate sets of rRNAs and tRNAs encoded by the mitochondrial genome [311]. Unmethylated CpG motifs and N-formyl peptides (NFPs) are inflammatogenic and mitochondrial DAMPs.
In response to pressure overload, mitochondria are damaged and damaged mitochondria are degraded by autophagy or mitophagy (see section I.F.2). Among PRRs, TLR9 senses unmethylated CpG motifs in bacteria and virus. Mitochondrial DNA is degraded by DNase II in the autolysosome, which is an acidic DNase and localized in the lysosome. When the induction of autophagy is insufficient in pressure-overloaded hearts, mitochondrial DNA escapes from autophagy-mediated degradation and binds to TLR9 to induce inflammatory responses cell-autonomously in cardiomyocytes, myocarditis, and dilated cardiomyopathy. However, it is reported that depletion of autophagic proteins promotes cytosolic translocation of mitochondrial DNA and caspase-1-dependent cytokines mediated by the NALP3 inflammasome in response to lipopolysaccharide in macrophages [312]. In addition to TLR9, the NALP3 inflammasome might be involved in inflammation in the failing heart. The role of autophagy in innate immunity may be cell type-dependent.
In the case with myocardial infarction, where necrosis is a main feature of cell death, mtDNA is released into circulation. The serum from patients after coronary intervention contains mitochondrial DNA [313]. Cellular disruption by trauma releases mitochondrial molecules including DNA into circulation to activate neutrophils and cause systemic inflammation [314]. Thus, it is possible that mitochondrial DNA in circulation after myocardial infarction may contribute to the inflammatory responses in infarct hearts.
Mitochondrial NFPs are released from degenerating mitochondria at tissue damage [314, 315] and recognized by formyl peptide receptors (FPRs), which have evolved to mediate phagocyte migration to sites of tissue injury. Although the role of NFPs-FRPs signaling pathway in heart diseases, specially myocardial infarction remains to be elucidated, it is possibility that the pathway may play a role in inflammation in infarct hearts.
The newly discovered role of mitochondria as DAMP-containing organelles places mitochondria in a central position as initiators and modulators of sterile inflammation in failing or infarct hearts. In case of pressure-overloaded hearts, autophagy regulates degradation of mitochondrial DNA and resultant inflammation in cardiomyocytes. In infarct hearts accompanied by necrosis, mitochondrial DNA released into circulation could activate and recruit various inflammatory cells in the lesion.
II). Mitochondria and Cardiovascular Disease
A). Mitochondrial myopathies-Mitochondrial etiology of cardiomyopathy
Cardiomyocytes have among the highest concentrations on mitochondria of any human cell. Because of the high mitochondrial ATP demands of the heart, relatively subtle defects in the mitochondrial ATP generating apparatus, oxidative phosphorylation (OXPHOS), can preferentially affect cardiac function.
If we rearrange the classification of the common diseases based on bioenergetics rather than anatomy, it becomes clear that all of the complex diseases can be envisioned as having the same underlying pathophysiological basis, partial bioenergetics dysfunction. Since the mitochondria are assembled from between 1000 and 2000 nuclear DNA (nDNA) genes plus thousands of copies of the maternally-inherited mitochondrial DNA (mtDNA) genes and the mitochondria both process the calories in our diet into usable energy and are acutely sensitive to a wide range of toxins, it follows that perturbation of mitochondrial bioenergetics can readily explain the etiology of the full range of common clinical disease symptoms (Figure 2)[2].
Figure 2:
Bioenergetic paradigm for degenerative and metabolic diseases, cancer, and aging. Mitochondrial OXPHOS can be perturbed by nDNA genetic alterations and/or epigenomic regulation, by mtDNA ancient adaptive of recent deleterious mutations, or by variation in the availability of calories and in caloric demands. Alterations in mitochondrial structure and function can impair OXPHOS, which in turn can reduce energy production, alter cellular redox state, increase ROS production, deregulate Ca2+ homeostasis, and ultimately activate the mtPTP, leading to apoptosis. These and other consequences of OXPHOS perturbation can destabilize mtDNA. This results in progressive accumulation of somatic mtDNA mutations and decline of mitochondrial function, which accounts for aging and the delayed-onset and progressive course of degenerative diseases. As energy output declines, the most energetic tissues are preferentially affected, resulting in degenerative diseases of the heart, muscle, nervous system, and kidney. Aberrant mitochondrial caloric metabolism also leads to metabolic deregulation, endocrine dysfunction, and symptoms such as diabetes, obesity, and cardiovascular disease. Energetic failure of apoptosis can result in the release into the blood stream of mitochondrial antigenic cardiolipin, N-formylmethionine polypeptides, and mtDNA (mitochondrial damage associated molecular patterns (DAMPs) [311, 314, 361, 362]) can initiate the inflammatory response, contributing to autoimmune diseases (e.g., multiple sclerosis and type I diabetes) and possibly also to the inflammatory component of late-onset degenerative diseases. Finally, cancer cells must manage energy resources to permit rapid replication. Figure from [2].
The nDNA coded genes relevant to mitochondrial function include the approximately 1000 proteins located within the mitochondrion [316] plus all of the genes involved in regulating cellular bioenergetics including the signal transduction enzymes (AMPK, SIRTUINs, etc.); nuclear receptor transcription factors (peroxisome proliferator-activated receptor (PPAR) family, the heteromeric partners of the PPARPs (RXR), the PPAR gamma coactivator-1-alpha (PGC-1alpha) family of co-activators, etc.); environmentally regulated transcription factors (HIF1α, NFκB, etc.), nutrient sensing systems (PTEN, TSC, mTOR, etc.); regulators of mtDNA biogenesis (replication [POLG & Trinkle], transcription [POLRMT] and translation [mitochondrial ribosomal proteins and elongation factors]); and those chromatin remodeling systems that permit nDNA-mitochondrial gene expression. Because of their high energetic demand the tissues most commonly affected by partial mitochondrial dysfunction are the heart, brain, muscle, renal, and endocrine systems [2, 317–320].
While the nDNA codes for the great majority of mitochondrial proteins, the mtDNA codes for the thirteen most critical OXPHOS polypeptides plus the 22 tRNAs and two rRNAs for their expression. OXPHOS generates energy by coupling electron transport (complexes I, II, and IV) with ATP synthesis (complex V) through the electrochemical gradient.
Severe mitochondrial diseases can result from homozygous mutations in nDNA-coded mitochondrial genes [318, 321, 322] or severe mtDNA mutations. Milder mitochondrial disease can result from heterozygous nDNA mutations, from mild mtDNA mutations, or from severe mtDNA mtations that are heteroplasmic, the mixture within the cytoplasmic of mutant and normal mtDNA. Milder mtDNA mutations result in symptoms when approximating homoplasmic (pure mutant) while more severe mutations an lead to disease when heteroplasmic.
There are three classes of clinically relevant mtDNA variants: ancient adaptive variants, recent deleterious mutations, and somatic mutations that accumulate in tissues during development and with age. Ancient adaptive mutations have accumulated along radiating maternal lineages as women migrated out-of-Africa to populate Eurasian and the Americas. A subset of these mtDNA mutations changed OXPHOS function and human physiology permitting descendent populations to adaptation of the new environments. mtDNAs harboring these locally beneficial variants became regionally enriched by adaptive selection, and as their descendants acquired additional mutations a group of related haplotypes developed known collectively as a haplogroup. While beneficial in one environment, these variants can be maladaptive in anther environment. Various mtDNA haplogroups have now been correlated with a broad spectrum of diseases including Alzheimer and Parkinson disease, macular degeneration, psychiatric disorders, stoke, diabetes, cardiovascular disease, sepsis, asthma, AIDs progression, various forms of cancer, types of athletic performance, and longevity [2, 320, 322–324].
Recent deleterious mutations continually arise within modern female lineages. Hundreds of such pathogenic mutations have been identified and are catalogued in our mtDNA database, MITOMAP[325]. An example of a frequently homoplasmic “mild” pathogenic mutation is the common Leber Hereditary Optic Neuropathy (LHON) complex I gene missense mutation, ND4 nt 11778 G>A (R340H) [322, 326, 327]. Examples of more severe heteroplasmic mtDNA mutations are the tRNALys nt 8344 A>G mutations which can manifest as hearing loss at low heteropmasmy but cardiomyopathy and Myoclonic Epilepsy and Ragged Red Fiber (MERRF) disease at higher heteroplasmy [328, 329] and the tRNALeu(UUR) nt 3243A>G mutation which at 50–90% mutant heteroplasmy can manifest as myopathy and cardiomyopathy [330] or stereotypically as Mitochondrial Encephalomyopathy, Lactic Acidosis and Stroke-Like Episodes (MELAS) [331] but at 10–30% mutant can present as Type I or Type II diabetes [332, 333] or autism [334]. Somatic mtDNA mutations accumulate throughout life, progressively eroding mitochondrial function. The rate of accumulation of these mutation can be modulated by nDNA gene variants [40, 335–337] resulting in degenerative diseases such as Alzheimer and Parkinson Disease [338–340], and like can be induced by environmental challenges.
A major reason for the complexity of mitochondrial diseases is the reliance of every aspect of cellular function on energy flux. Thus, the energy status of the mitochondrion must be monitored by every functional change in the cell. This is accomplished by all cellular functions being either driven or regulated my mitochondrial high energy or metabolic intermediated. The importance of this mitochondrial signaling to the nuclear-cytosolic epigenome and signal transduction systems has been demonstrated by examining the cellular and transcriptional changes that occur in cells with the same nucleus by different percentages of the tRNALeu(UUR) nt3243G mutation. This revealed cellular structure and gene transcription changed in discrete phases in response to the progressive increase in percentage of mutant mtDNA, relative to homoplasmic normal mtDNAs, with 20–30% 3243G mutant having one transcriptional profile, 50–90% mutant a second, and 100% a third. Thee phase shifts in the transcriptome exactly correspond to the changes in patient phenotypes that are associated with this mutation, 10–30% diabetes and autism, 50–90% cardiomyopathy and MELAS, 100% perinatal lethality [341].
mtDNA Variation and Cardiomyopathy:
Cardiomyopathy has been associated with all three classes of mtDNA variants, ancient adaptive mutations, recent maternal mutations, and somatic mutations. Ancient adaptive mutations have been linked to increased risk of metabolic and cardiovascular disease [342, 343] and mice heterozygous for the mitochondrial antioxidant enzyme, MnSOD (Sod2) are prone to hypertension [344].
Recent deleterious mtDNA mutations have repeatedly been linked to cardiomyopathy [322]. While heteroplasmic mtDNA mutations are commonly accepted as causal for cardiomyopathy, homoplasmic mutations are more difficult to distinguish from ancient adaptive variants which may or may not be contributory. One approach to identify potentially pathological homoplasmic mutations is to first determine the mtDNA haplogroup using MITOMAP and MITOMASTER [325] in association with PHYLOTREE [345]. Then determine if the putatively deleterious homoplasmic mutation has been previously observed with that haplogroup in the normal population. If then it is increasingly likely that the homoplasmic variant may be contributory to the disease [346]. The pathogenicity of the mutation must still be confined by functional tests in cybrids [347]. However, it is also possible that a variant has been observed before but in a different haplogroup context. Such variants may be deleterious on the wrong mtDNA background and thus might contribute to the diseases [348]. Proof that a homoplasmic mtDNA mutation can cause cardiomyopathy comes from the generation of a mouse that harbors an mtDNA missense mutation in the COI nt 6598T>C V421A and develops hypertrophic cardiomyopathy [349].
De novo and somatic mtDNA mutations also cause cardiac disease. Spontaneous single event heteroplasmic deletions in the mtDNA can cause the Kearns-Sayre Syndrome which frequently presents with cardiac conduction defects and heart block in association with chronic external ophthalmoplegia (CPEO) [322]. The accumulation of a heterogenous array of somatic mtDNA mutations has also been shown to be associated with cardiomyopathy in the hearts of heart transplantation patients, as monitored using the common mtDNA 5 kb deletion [6, 350].
nDNA Mitochondrial Gene Mutations and Cardiomyopathy:
Cardiomyopathy is one of the primary presentations of boys with Barth Syndrome which results from mutations in the nDNA-coded X-linked Tafazzin gene required for cardiolipin metabolism [351]. Cardiomyopathy is also the primary symptom of homozygous mutations that inactivate the chromosome 4 heart-muscle-brain isoform of the adenine nucleotide translocator (ANT1)[352, 353]. The ANTs, of which there are four in humans, exchange mitochondrial ATP for cytoplasmic ADP across the mitochondrial inner membrane. ANT1 inactivating mutations result in mitochondrial cardiomyopathy and myopathy [354–356]. This has been confirmed by the generation of a mouse lacking the Ant1 gene which also developes mitochondrial myopathy and hypertrophic cardiomyopathy. The Ant1-deficient mouse has a partial defect in cardiac mitochondrial ATP production, since the heart also expresses a second ANT isoform, Ant2. The Ant1-deficient mice develop lactic acidosis, mitochondrial ROS production, and a striking increase in the accumulation of cardiac mtDNA somatic mutations [209, 357]. The hypertrophic cardiomyopathy of these mice progresses to dilated cardiomyopathy as the mice age [358]. Presumably this is because of the age-related accumulation of somatic mutations which exacerbate the inherited nuclear Ant1 mutation.
Patients that develop dilated cardiomyopathy associated with myocarditis develop antibodies to ANT [359, 360]. These antibodies may be either the initiation and/or promoting factor in developing the cardiomyopathy. In either case, a mitochondrial bioenergetic defect cause by viral infection or preexisting mitochondrial genetic defect which impairs mitochondrial energetics could inhibit the energy demanding apoptotic process permitting the release of mitochondrial antigens (DAMPs)[cardiolipin, mtDNA, N-formyl methionine initiated polypeptides, ANT1, etc.] [311, 314, 361, 362] into the blood stream where the can initiate a cardiac inflammatory response.
Mutations in many other mitochondrial encoded genes have been reported to cardiovascular disease. For example, defects in the mitochondrial matrix protein frataxin are involed in Friedreichs ataxia [363]. Defects in the mitochondrial phosphate transporter have also been associated with cardiac defects [364].
nDNA-mtDNA Interactions and Cardiomyopathy:
The genetic complexity of cardiac diseases is further enhanced by the potential of the deleterious interaction of nDNA and mtDNA genetic variants. This pathogenic nuclear-mitochondrial interaction was first demonstrated by the discovery that mutations in the nDNA-coded mtDNA polymerase γ gene can resulted in autosomal dominant multiple mtDNA deletion syndrome, the mtDNA damage causing CPEO and potentially cardiac symptoms [322, 337]. Mitochondrial disease has also been shown to result from the interaction nDNA-coded partial mitochondrial genetic defects and homoplasmic mtDNA mutations. This was exemplified by in a family segregating a missense mutation (G32R) in the X-linked complex I NDUFA1 gene resulting in an approximately 30%reduction in complex I plus a pair of non-haplogroup-associated homoplasmic missense mutations in the mtDNA complex I genes ND1 3308T>C (M21T) and ND5 12599T>C (M88T). Both the X-chromosomel and the mtDNA in boys are inherited form the mother resulting in affected boys along the maternal lineage [365].
The severity of cardiac disease resulting from a nDNA mitochondrial gene defect can also be modulated by the inheritance of otherwise normal mtDNA haplogroup. In a 13 generation pedigree segregating a frameshift mutation in the ANT1 gene, multiple homozygous patients were identified with mitochondrial myopathy and cardiomyopathy. However, the severity of the cardiomyopathy varied strikingly with some individuals progressing to dilated cardiomyopathy necessitating heart transplantation while others maintained a relatively stable hypertrophic cardiomyopathy. Sequencing of the mtDNAs from the homozygous Ant1 frameshift patients revealed that those that progressed to heart transplant had mtDNA haplogroup U2 while those that manifest stable hypertrophic cardiomyopathy were haplogroup H [356].
This augmentation of deleterious nDNSA mutations by mtDNA variants may be a more general phenomenon. Analysis of the mtDNAs of cardiomyopathy patients with mutations in the nDNA-coded cardiac contractile apparatus proteins have been found to contain potentially contributory mtDNA mutations [366, 367].
Conclusion:
The high energetic demands of the heart are primarily met by mitochondrial OXPHOS. Hence, it follows that defects in mitochondrial bioenergetics should preferentially affect the heart. This is proven by the fact that cardiomyopathy is the most obvious phenotypic manifestation in both humans and mice harboring null mutations in the heart-muscle-brain isoform of the ANT. If mutations in nDNA-coded mitochondrial genes can generate cardiomyopathy, it follows that mtDNA mutations causing partial OXPHOS defects should also contribute to cardiomyopathy. This is supported by the observations that mtDNA haplogroups and de novo mtDNA mutations can augment the deleterious consequences of nDNA mutations. If inherited homoplasmic or heteroplasmic mtDNA mutations can cause cardiomyopathy, the mitochondrial dysfunction resulting from the accumulation of somatic mtDNA mutations could also cause cardiomyopathy. This leads to the conclusion that the age-related accumulation of mtDNA mutations which augment inherited or acquired mitochondrial defects, in addition to the effects of secondary inflammation from the release of mitochondrial DAMPs, may provide a coherent conceptual framework for understanding the etiology of many forms of mtDNA mutations which augment inherited or acquired mitochondrial defects, in addition to the effects of secondary inflammation from the release of mitochondrial DAMPs (see Section I.G), may provide a coherent conceptual framework for understanding the etiology of many forms of cardiomyopathy.
B). Cardiotoxicity
Anticancer treatments have improved significantly over the past few years. However, despite the improvement in their target effects on cancer cells, anticancer treatments have also been associated with an increase in the incidence of side effects. One of the major side effects of anti-cancer drugs is their toxicity towards cardiac muscle cells. This cardiotoxicity can manifest at an early stage of therapy (within days) or many years after treatment. Thus, patients undergoing these treatments should be monitored closely. More importantly, patients at high risk should be identified before treatment is started in order to reduce morbidity from cardiotoxicity. This would require close collaboration between cardiologists and oncologists.
Two forms of cardiac damage have been characterized. One form, typically seen with anthracyclines, is associated with irreversible damage and death of cardiac cells, while the second form generally occurs with protein kinase inhibitors, and is associated with reversible myocardial dysfunction. These two forms are discussed below.
Anthracycline-related Cardiotoxicity (Type 1):
Anthracyclines have been used for the treatment of various forms of cancer for several decades. They constitute one of the major successes in the field of cancer. For example, in pediatric oncology, the 5-year survival rate has increased from ~30% in the 1960s to 70–80% today [368, 369], and >50% of childhood cancer patients have received anthracyclines [370]. Different statistics have been published on the incidence of anthracycline -mediated cardiotoxicity, ranging from ~1% to up to 48% [371]. However, a strong correlation exists between the incidence of cardiotoxicity and the dosage of the drug [372].
Although the effects of anthracyclines on cancer prevention is thought to be mainly through inhibition of DNA replication, RNA replication, DNA cross linking and topoisomearses [373], their cardiotoxic effects appear to be through distinct mechanisms. The pathophysiology of anthracycline-mediated cardiotoxicity is likely multi-factorial and multiple mechanisms have been proposed. It has been suggested that anthracyclines induces an increase in reactive oxygen species (ROS) production [374, 375]. According to this model, oxidation of the aglycone portion of doxorubicin (DOX) results in the formation of a semiquinone radical, which can rapidly revert to its parent compound by using O2 as an electron acceptor [376]. This futile redox cycle leads to the formation of superoxide, which is converted to H2O2 spontaneously or by superoxide dismutase. Subsequently, H2O2 may be converted to highly toxic hydroxyl radicals in the presence of heavy metals, such as iron, through the Fenton reaction. In addition, DOX can interact with iron directly to form a DOX-Fe complex [376, 377]. In addition to production of ROS, other mechanisms including mitochondrial dysfunction and depletion of energy, induction of apoptosis and changes in topoisomerase IIb activity have been proposed [378, 379].
Although different mechanisms have been proposed for the cardiotoxicity of anthracycline, the mitochondrial effects are likely the major contributor to this disorder. Mitochondrial dysfunction, including mitochondrial swelling, mitochondrial cristae disruption and accumulation of myelin figures have been observed after treatment with anthracyclines. There is also evidence that anthracyclines penetrates into the mitochondria (although the mechanism for this transport is not clear). After translocation of anthracyclines into the mitochondria, it can then interact with a number of molecules, including: 1) mitochondrial DNA (mtDNA). Anthracyclines can cause damage by inducing large scale deletions within mtDNA, form covalent bonds with mtDNA after transformation of the quinone ring to quinone methide, and cause damage to mtDNA indirectly by producing ROS [380]. 2) Electron transport chain (ETC)/oxidative phosphorylation (OXPHOS). Anthracyclines can cause damage to ETC and OXPHOS proteins through ROS production or by forming a complex with cardiolipins present in the mitochondrial membrane [381, 382]. The latter can also cause an increase in ROS or may lead to cardiolipin dysfunction, which is needed for normal activity of several enzymes in the ETC/OXPHOS pathway. 3) Mitochondrial permeability transition pore (mPTP). mPTP plays a major role in the regulation of cell death [383]. Anthracyclines have been shown to induce mPTP opening and cyclosporine A (which inhibits mPTP by binding to cyclophilin D) prevents mitochondrial failure and cell killing [384]. 4) Mitochondrial iron. Iron has been known to mediate some of the cardiotoxic effects of anthracyclines, however, iron chelators were shown not to be effective against anthracycline-mediated cardiotoxicity. It is now demonstrated that anthracyclines cause mitochondrial iron accumulation and that a reduction in mitochondrial iron is protective against the cardiotoxic effects of anthracyclines [385].
Drugs for other disorders:
Several drugs that are currently routinely used for common diseases also have cardiac side effects. For example, it is now known that glitazones can worsen heart failure, and their mechanism of action may be through modulation of the activity of peroxisome proliferator-activated receptor (PPAR) proteins in the heart [386]. Although PPARs regulate metabolic processes, they may also have an effect on the mitochondria through indirect mechanisms.
C). Biomarkers
Primary mitochondrial diseases constitute a broad spectrum of disorders affecting multiple tissues and organ systems. It is estimated that mitochondrial diseases affect 1 in 5000 individuals [387], however it is suspected that 1 in 250 may be more accurate [388], as many cases likely go undiagnosed. Clinical diagnosis of mitochondrial diseases is complicated by diverse phenotypical manifestations, in part due to variably affected genomes (i.e., mitochondrial or nuclear), organs (e.g., muscle, liver) and age of onset. Thus, clinical protocols required for definitive diagnosis are labor-intensive and routinely involve invasive procedures with variable success. It would therefore be of great clinical use to discover biomarkers in accessible biofluids (e.g., blood, urine) that specifically inform on primary mitochondrial diseases. Mitochondrial-derived biomarkers, including mitochondrial DNA (mtDNA) (see Section I.G), proteins and metabolites, have been explored, however these have exhibited limited specificity, sensitivity and diagnostic/prognostic accuracy. Current markers can only add weight to the likelihood that a patient may have a primary mitochondrial disorder, and at best may only substantiate a more invasive diagnostic testing regimen.
MtDNA sequencing from intact cells acquired through tissue biopsy (most commonly skeletal muscle) in symptomatic patients constitutes the most definitive current measure for diagnosing mitochondrial disease. However, this method is invasive and is not amenable to routine screening or suitable for high-risk populations. The non-invasive quantification of cell-free mtDNA in plasma has gained substantial interest for diagnosing certain cancers [389, 390] and for other clinical scenarios such as predicting ICU mortality [391], but has not yet been successfully interrogated for the diagnosis of primary mitochondrial disorders. Mitochondrial proteins and metabolites quantified in accessible biofluids have received limited interest; however show great promise as biomarkers of mitochondrial diseases in that they are highly specific and sensitive indicators of underlying metabolic changes.
Improved strategies are warranted for the specific diagnosis of mitochondrial disorders, and biomarkers are most promising to this effort, as disease-specific mitochondrial-derived proteins, metabolites, or mtDNA would likely offer superior sensitivity and specificity in diagnosis/risk assessment and could be accurately and consistently measured across all clinical settings. Below is an overview of mitochondrial-derived markers (mtDNA, proteins and metabolites) that have been the subject of intense investigation.
Blood and urine are readily accessible biofluids acquired with minimal invasiveness. Blood can be centrifuged to separate aqueous plasma or serum from blood cells. Mitochondrial-derived biomarkers found in plasma/serum are cell-free circulating (cfc)-mtDNA, metabolites and proteins. Thrombocytes and leukocytes, but not erythrocytes, contain mitochondria, thus mtDNA and proteins can be obtained from intact blood cells in the more dense fraction. However, since mitochondrial genomes are heteroplasmic and primary mitochondrial diseases can affect various organs, disease markers will likely be tissue-derived, rather than blood cell-derived; thus, cell-free markers in plasma likely represent a more relevant and unbiased assessment of mitochondrial disorders. Urine would also provide an unbiased source of metabolites. Cfc-mtDNA in plasma and serum has been a subject of intense interest as a biomarker of disease since the first published study showing a mutation in cfc-mtDNA in patients with type-2 diabetes mellitus [392]. Mt-DNA is a 16.5 kbp circular strand of DNA, and hundreds to thousands of copies of mtDNA can be found in a single cell. Though the precise physiology is unclear, it is thought that cfc-mtDNA enters the bloodstream through cell necrosis, apoptosis or active secretion[393]. Cfc-mtDNA in plasma exists in both particle-associated and free forms; and methodology to measure amounts in whole blood are exquisitely dependent upon preparatory protocols used (e.g., size exclusion filtration), indicating that cfc-mtDNA exists in various conformations [394]. Mt-DNA can be reliably assayed using a quantitative real-time PCR approach, which exhibits a dynamic range of 5 orders of magnitude and a sensitivity of detection down to one copy of mtDNA [394]. Thus, mtDNA possess favorable physical characteristics for a good candidate biomarker of various physiological states. However, while cfc-mtDNA has shown promise for early detection and tumor classification in oncology patients [389, 390], its role in diagnosis and prognosis of mitochondrial diseases remains to be determined.
Despite the sophisticated and sensitive technologies we have available to measure mitochondrial-derived biomarkers, no non-invasive biomarkers for mitochondrial disorders have materialized from the discovery phase to the clinic. One of the most prominent reasons for this is that studies aimed at biomarker discovery have been poorly designed, having little clinical translational potential [395]. Plasma biomarker studies should ideally proceed through an evolution of systematic tests that appropriately credential markers for clinical use, [see [396]]. A gap exists between biomarker discovery studies, where thousands of biomarkers exhibit differential profiles in case versus controls, and clinical tests, where assays are optimized and standardized for rapid and efficient measurement in the clinic. Within this gap is the narrowing and prioritizing of likely biomarker candidates, followed by rigorous steps of validation. Despite the overall lack of clinically available biomarkers, there are promising candidates that have surfaced in recent studies that, with appropriate validation strategies in large clinical cohorts, may add value or replace current clinical markers and diagnostic/prognostic regimens. A study by Suomalainen et al.[397] found that serum fibroblast growth factor 21 (FGF-21) was a sensitive and specific marker for primary muscle-manifesting respiratory chain deficiencies in adults and children. This study employed 67 patients with mitochondrial diseases diagnosed by muscular biopsy and DNA analysis, 34 non-mitochondrial neurological disease controls and 74 healthy controls. FGF-21 had superior diagnostic accuracy to conventionally-used, more non-specific mitochondrial disease indicators, including lactate, pyruvate, lactate-to-pyruvate ratio and creatine kinase. A later prospective study supported these results and indicated that FGF-21 is a superiorly sensitive biomarker for diagnosing both mtDNA and nuclear DNA encoded mitochondrial diseases [398]. Taken together, these results showed that FGF-21 may provide a first-tier, non-invasive biomarker for diagnosing mitochondrial diseases that may diminish the need for invasive muscular biopsies. Rigorous validation using large, well-characterized patient cohorts is warranted to verify the utility of FGF-21 in the clinic. Another recent study by Enns et al.[399] examined GSH and GSSH protein levels in whole blood from 58 control and 59 patients with primary mitochondrial disease of varying etiologies, including Leigh syndrome, electron transport chain abnormalities, mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes, mtDNA deletion syndrome, and mtDNA depletion syndrome. Quantitative mass spectrometric analysis determined that in patients with mitochondrial disease, redox status was significantly more oxidized (lower GSH and higher GSSH), showing the greatest change in patients that were hospitalized with metabolic crisis. Hence, GSH and GSSH may be clinically useful biomarkers for all primary mitochondrial disease subtypes, and should be rigorously evaluated in clinical trials for demarcating clinical prognosis and patient response to redox-modulating treatments.
In conclusion, the low diagnostic/prognostic accuracy of primary mitochondrial diseases warrants intense research on mitochondrial biomarkers. The proper experimental design utilizing cutting edge omics technologies will undoubtedly propel this area of research in fields of study is the best response to the challenge of mitochondrial biomarker realization.
III. Summary and Recommendations:
The data in the literature show that mitochondria play a key role in cardiovascular disease, specifically in the response to myocardial ischemia and the transition to heart failure. The accumulation of defects in mitochondrial electron transport, ion transport, metabolism, redox regulation, and mitochondrial quality control lead to a feed forward cycle of further acquired defects. Ultimately the mitochondria can no longer meet the high energetic demands of the cardiac cell and that coupled with an increase activation of cell death pathways leads to the death of the myocytes. The role of mitochondrial defects in heart failure and other cardiovascular diseases needs to be considered and evaluated.
There are a number of gaps in our understanding that need to be addressed in future studies. We need a better understanding of the effects of post-translational modifications on protein function. In recent years we have made great strides in defining mitochondrial post-translational modification, but the consequences of the modification is in many cases poorly understood. We need additional information on the mechanisms by which mitochondria regulate apoptosis and necroptosis. We need to define the mitochondrial permeability transition pore. The F1F0-ATPase has been proposed as the pore; this need to be verified by other groups and the mechanism of its regulation needs to be defined. We need to define the redox sensitive component of this pore. We need to more completely define the mechanisms that regulate mitochondrial dynamics and elucidate how this regulates cell function and metabolism. We also need additional information on the mechanisms that regulate mitochondrial turnover at the level of mitochondrial proteins and the organelle. What are the signals for turnover of mitochondrial proteins and how is this accomplished?
Additionally, the interrelationship between mitochondria and other intracellular compartments (endoplasmic reticulum, lysosomes etc.) and intracellular structures (mitochondria associated membranes) to regulate mitochondrial function and overall cellular homeostasis, are increasingly being recognized [400–402]. Our understanding of how mitochondrial function and pathophysiology integrate within this more complex intracellular environment will also be necessary to enable the modulation of quality-control programs to sustain cardiomyocyte homeostasis and stress resistance.
References:
- [1].Ingwall JS, Weiss RG. Is the failing heart energy starved? On using chemical energy to support cardiac function. Circulation research. 2004;95:135–45. [DOI] [PubMed] [Google Scholar]
- [2].Wallace DC. Mitochondrial bioenergetic etiology of disease. J Clin Invest. 2013;123:1405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Opie L Fuels: aerobic and anaerobic metabolism In: Opie LH, editor. The Heart, Physiology, from Cell to Circulation, 4th edition Philadelphia: Lippincott-Raven; 2004. p. 306–54. [Google Scholar]
- [4].Schaper J, Meiser E, Stammler G. Ultrastructural morphometric analysis of myocardium from dogs, rats, hamsters, mice, and from human hearts. CircRes. 1985;56:377–91. [DOI] [PubMed] [Google Scholar]
- [5].Skulachev VP. Uncoupling: new approaches to an old problem of bioenergetics. Biochimica et biophysica acta. 1998;1363:100–24. [DOI] [PubMed] [Google Scholar]
- [6].Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutation Research. 1992;275:169–80. [DOI] [PubMed] [Google Scholar]
- [7].Santorelli FM, Mak SC, El-Schahawi M, Casali C, Shanske S, Baram TZ, et al. Maternally inherited cardiomyopathy and hearing loss associated with a novel mutation in the mitochondrial tRNA(Lys) gene (G8363A). Am J Hum Genet. 1996;58:933–9. [PMC free article] [PubMed] [Google Scholar]
- [8].Arbustini E, Diegoli M, Fasani R, Grasso M, Morbini P, Banchieri N, et al. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. The American journal of pathology. 1998;153:1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Grad LI, Sayles LC, Lemire BD. Introduction of an additional pathway for lactate oxidation in the treatment of lactic acidosis and mitochondrial dysfunction in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Neubauer S The failing heart--an engine out of fuel. The New England journal of medicine. 2007;356:1140–51. [DOI] [PubMed] [Google Scholar]
- [11].Taegtmeyer H Energy metabolism of the heart: from basic concepts to clinical applications. Curr Probl Cardiol. 1994;19:59–113. [DOI] [PubMed] [Google Scholar]
- [12].Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiological reviews. 2010;90:207–58. [DOI] [PubMed] [Google Scholar]
- [13].Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC Jr., Suthammarak W, Gong G, et al. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell metabolism. 2013;18:239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sack MN, Kelly DP. The energy substrate switch during development of heart failure: gene regulatory mechanisms (Review). IntJMolMed. 1998;1:17–24. [DOI] [PubMed] [Google Scholar]
- [15].Opie LH, Sack MN. Metabolic plasticity and the promotion of cardiac protection in ischemia and ischemic preconditioning. JMolCell Cardiol. 2002;34:1077–89. [DOI] [PubMed] [Google Scholar]
- [16].Ardehali H, Sabbah HN, Burke MA, Sarma S, Liu PP, Cleland JG, et al. Targeting myocardial substrate metabolism in heart failure: potential for new therapies. European journal of heart failure. 2012;14:120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sack MN, Rader TA, Park S, Bastin J, McCune SA, Kelly DP. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation. 1996;94:2837–42. [DOI] [PubMed] [Google Scholar]
- [18].Sack MN, Disch DL, Rockman HA, Kelly DP. A role for Sp and nuclear receptor transcription factors in a cardiac hypertrophic growth program. Proc Natl Acad Sci. 1997;94:6438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-alpha during cardiac hypertrophic growth. The Journal of clinical investigation. 2000;105:1723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circulation research. 2004;95:568–78. [DOI] [PubMed] [Google Scholar]
- [21].Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. [DOI] [PubMed] [Google Scholar]
- [22].van der Vusse GJ, van Bilsen M, Glatz JF. Cardiac fatty acid uptake and transport in health and disease. Cardiovascular research. 2000;45:279–93. [DOI] [PubMed] [Google Scholar]
- [23].Sorokina N, O’Donnell JM, McKinney RD, Pound KM, Woldegiorgis G, Lanoue KF, et al. Recruitment of compensatory pathways to sustain oxidative flux with reduced carnitine palmitoyltransferase I activity characterizes inefficiency in energy metabolism in hypertrophied hearts. Circulation. 2007;115:2033–41. [DOI] [PubMed] [Google Scholar]
- [24].Allard MF, Schonekess B, Henning SL, English DR, Lopaschuck GD. Contribution of oxidative metabolism and glycolysis to ATP production in hypertrophied hearts. Am J Physiol. 1994;267:H742–H50. [DOI] [PubMed] [Google Scholar]
- [25].Taegtmeyer H, Overturf ML. Effects of moderate hypertension on cardiac function and metabolism in the rabbit. Hypertension. 1988;11:416–26. [DOI] [PubMed] [Google Scholar]
- [26].Kantor PF, Robertson MA, Coe JY, Lopaschuk GD. Volume overload hypertrophy of the newborn heart slows the maturation of enzymes involved in the regulation of fatty acid metabolism. Journal of the American College of Cardiology. 1999;33:1724–34. [DOI] [PubMed] [Google Scholar]
- [27].Lu Z, Scott I, Webster BR, Sack MN. The emerging characterization of lysine residue deacetylation on the modulation of mitochondrial function and cardiovascular biology. CircRes. 2009;105:830–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sack MN. Mitochondrial depolarization and the role of uncoupling proteins in ischemia tolerance. CardiovascRes. 2006;72:210–9. [DOI] [PubMed] [Google Scholar]
- [29].Boehm EA, Jones BE, Radda GK, Veech RL, Clarke K. Increased uncoupling proteins and decreased efficiency in palmitate-perfused hyperthyroid rat heart. American journal of physiology Heart and circulatory physiology. 2001;280:H977–83. [DOI] [PubMed] [Google Scholar]
- [30].Korvald C, Elvenes OP, Myrmel T. Myocardial substrate metabolism influences left ventricular energetics in vivo. American journal of physiology Heart and circulatory physiology. 2000;278:H1345–51. [DOI] [PubMed] [Google Scholar]
- [31].Chavez PN, Stanley WC, McElfresh TA, Huang H, Sterk JP, Chandler MP. Effect of hyperglycemia and fatty acid oxidation inhibition during aerobic conditions and demand-induced ischemia. American journal of physiology Heart and circulatory physiology. 2003;284:H1521–7. [DOI] [PubMed] [Google Scholar]
- [32].Scheuermann-Freestone M, Madsen PL, Manners D, Blamire AM, Buckingham RE, Styles P, et al. Abnormal cardiac and skeletal muscle energy metabolism in patients with type 2 diabetes. Circulation. 2003;107:3040–6. [DOI] [PubMed] [Google Scholar]
- [33].Nikolaidis LA, Sturzu A, Stolarski C, Elahi D, Shen YT, Shannon RP. The development of myocardial insulin resistance in conscious dogs with advanced dilated cardiomyopathy. Cardiovascular research. 2004;61:297–306. [DOI] [PubMed] [Google Scholar]
- [34].Anderson PG, Allard MF, Thomas GD, Bishop SP, Digerness SB. Increased ischemic injury but decreased hypoxic injury in hypertrophied rat hearts. Circulation research. 1990;67:948–59. [DOI] [PubMed] [Google Scholar]
- [35].Allard MF, Wambolt RB, Longnus SL, Grist M, Lydell CP, Parsons HL, et al. Hypertrophied rat hearts are less responsive to the metabolic and functional effects of insulin. Am J Physiol Endocrinol Metab. 2000;279:E487–93. [DOI] [PubMed] [Google Scholar]
- [36].Lydell CP, Chan A, Wambolt RB, Sambandam N, Parsons H, Bondy GP, et al. Pyruvate dehydrogenase and the regulation of glucose oxidation in hypertrophied rat hearts. Cardiovascular research. 2002;53:841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Russell RR 3rd, Taegtmeyer H. Pyruvate carboxylation prevents the decline in contractile function of rat hearts oxidizing acetoacetate. The American journal of physiology. 1991;261:H1756–62. [DOI] [PubMed] [Google Scholar]
- [38].Schmidt-Schweda S, Holubarsch C. First clinical trial with etomoxir in patients with chronic congestive heart failure. Clinical science. 2000;99:27–35. [PubMed] [Google Scholar]
- [39].Holubarsch CJ, Rohrbach M, Karrasch M, Boehm E, Polonski L, Ponikowski P, et al. A double-blind randomized multicentre clinical trial to evaluate the efficacy and safety of two doses of etomoxir in comparison with placebo in patients with moderate congestive heart failure: the ERGO (etomoxir for the recovery of glucose oxidation) study. Clinical science. 2007;113:205–12. [DOI] [PubMed] [Google Scholar]
- [40].Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–4. [DOI] [PubMed] [Google Scholar]
- [41].Fragasso G, Palloshi A, Puccetti P, Silipigni C, Rossodivita A, Pala M, et al. A randomized clinical trial of trimetazidine, a partial free fatty acid oxidation inhibitor, in patients with heart failure. Journal of the American College of Cardiology. 2006;48:992–8. [DOI] [PubMed] [Google Scholar]
- [42].Morrow DA, Scirica BM, Karwatowska-Prokopczuk E, Murphy SA, Budaj A, Varshavsky S, et al. Effects of ranolazine on recurrent cardiovascular events in patients with non-ST-elevation acute coronary syndromes: the MERLIN-TIMI 36 randomized trial. JAMA. 2007;297:1775–83. [DOI] [PubMed] [Google Scholar]
- [43].Wargovich TJ, MacDonald RG, Hill JA, Feldman RL, Stacpoole PW, Pepine CJ. Myocardial metabolic and hemodynamic effects of dichloroacetate in coronary artery disease. The American journal of cardiology. 1988;61:65–70. [DOI] [PubMed] [Google Scholar]
- [44].Ingwall JS, Kramer MF, Fifer MA, Lorell BH, Shemin R, Grossman W, et al. The creatine kinase system in normal and diseased human myocardium. The New England journal of medicine. 1985;313:1050–4. [DOI] [PubMed] [Google Scholar]
- [45].Alcaide P, Merinero B, Ruiz-Sala P, Richard E, Navarrete R, Arias A, et al. Defining the pathogenicity of creatine deficiency syndrome. Hum Mutat. 2011;32:282–91. [DOI] [PubMed] [Google Scholar]
- [46].Lygate CA, Aksentijevic D, Dawson D, ten Hove M, Phillips D, de Bono JP, et al. Living without creatine: unchanged exercise capacity and response to chronic myocardial infarction in creatine-deficient mice. Circulation research. 2013;112:945–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Aksentijevic D, Zervou S, Faller KM, McAndrew DJ, Schneider JE, Neubauer S, et al. Myocardial creatine levels do not influence response to acute oxidative stress in isolated perfused heart. PloS one. 2014;9:e109021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Weiss RG, Gerstenblith G, Bottomley PA. ATP flux through creatine kinase in the normal, stressed, and failing human heart. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bottomley PA, Panjrath GS, Lai S, Hirsch GA, Wu K, Najjar SS, et al. Metabolic rates of ATP transfer through creatine kinase (CK Flux) predict clinical heart failure events and death. Sci Transl Med. 2013;5:215re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Boudina S, Abel ED. Diabetic cardiomyopathy, causes and effects. Rev Endocr Metab Disord. 2010;11:31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like Protein 1 (GCN5L1) Controls Mitochondrial Content through Coordinated Regulation of Mitochondrial Biogenesis and Mitophagy. The Journal of biological chemistry. 2014;289:2864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chen Y, Dorn GW 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Deas E, Plun-Favreau H, Wood NW. PINK1 function in health and disease. EMBO molecular medicine. 2009;1:152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Riccio A New endogenous regulators of class I histone deacetylases. Science signaling. 2010;3:pe1. [DOI] [PubMed] [Google Scholar]
- [58].Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. JBiolChem. 2002;277:45099–107. [DOI] [PubMed] [Google Scholar]
- [59].Cargile BJ, Bundy JL, Grunden AM, Stephenson JL Jr. Synthesis/degradation ratio mass spectrometry for measuring relative dynamic protein turnover. Anal Chem. 2004;76:86–97. [DOI] [PubMed] [Google Scholar]
- [60].Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell metabolism. 2015;22:31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Stein LR, Imai SI. The dynamic regulation of NAD metabolism in mitochondria. Trends EndocrinolMetab. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. BiochemBiophysResCommun. 2000;273:793–8. [DOI] [PubMed] [Google Scholar]
- [64].Sack MN, Finkel T. Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Boisvert FM, Ahmad Y, Gierlinski M, Charriere F, Lamont D, Scott M, et al. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics. 2012;11:M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Balasse EO, Fery F. Ketone body production and disposal: effects of fasting, diabetes, and exercise. Diabetes Metab Rev. 1989;5:247–70. [DOI] [PubMed] [Google Scholar]
- [67].Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, et al. Tyr Phosphorylation of PDP1 Toggles Recruitment between ACAT1 and SIRT3 to Regulate the Pyruvate Dehydrogenase Complex. Molecular cell. 2014;53:534–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Paik WK, Pearson D, Lee HW, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochimica et biophysica acta. 1970;213:513–22. [DOI] [PubMed] [Google Scholar]
- [69].Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Muoio DM, Noland RC, Kovalik JP, Seiler SE, Davies MN, DeBalsi KL, et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell metabolism. 2012;15:764–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schrenk DF, Bisswanger H. Measurements of electron spin resonance with the pyruvate dehydrogenase complex from Escherichia coli. Studies on the allosteric binding site of acetyl-coenzyme A. European journal of biochemistry / FEBS. 1984;143:561–6. [DOI] [PubMed] [Google Scholar]
- [72].Ghanta S, Grossman R, Brenner C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: chemical and metabolic logic of acetyl-lysine modifications. Critical Reviews in Biochemistry & Molecular Biology. 2013;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, et al. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. JCell Biochem. 2010;110:238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. MolCell Biol. 2008;28:6384–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, et al. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PloS one. 2010;5:e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. MolCell Biol. 2007;27:8807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. The Biochemical journal. 2011;433:505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, et al. SIRT3 Deficiency and Mitochondrial Protein Hyperacetylation Accelerate the Development of the Metabolic Syndrome. Molecular cell. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Chen Y, Liu Y, Dorn GW 2nd. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circulation research. 2011;109:1327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A. 2011;108:9572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hallows WC, Yu W, Smith BC, Devires MK, Ellinger JJ, Someya S, et al. Sirt3 Promotes the Urea Cycle and Fatty Acid Oxidation during Dietary Restriction. Molecular cell. 2011;41:139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. ProcNatlAcadSciUSA. 2008;105:14447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular cell. 2010;40:893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY). 2010;2:914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL 3rd, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY). 2009;1:771–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. JClinInvest. 2009;119:2758–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Alrob OA, Sankaralingam S, Ma C, Wagg CS, Fillmore N, Jaswal JS, et al. Obesity-induced lysine acetylation increases cardiac fatty acid oxidation and impairs insulin signalling. Cardiovascular research. 2014;103:485–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Wagner GR, Pride PM, Babbey CM, Payne RM. Friedreich’s ataxia reveals a mechanism for coordinate regulation of oxidative metabolism via feedback inhibition of the SIRT3 deacetylase. Hum Mol Genet. 2012;21:2688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].East DA, Campanella M. Ca2+ in quality control: an unresolved riddle critical to autophagy and mitophagy. Autophagy. 2013;9:1710–9. [DOI] [PubMed] [Google Scholar]
- [95].Pyo JO, Nah J, Kim HJ, Chang JW, Song YW, Yang DK, et al. Protection of cardiomyocytes from ischemic/hypoxic cell death via Drbp1 and pMe2GlyDH in cardio-specific ARC transgenic mice. The Journal of biological chemistry. 2008;283:30707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Xiong Y, Guan KL. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol. 2012;198:155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Jing E, O’Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, et al. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62:3404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, et al. Sirtuin 3 (SIRT3) Protein Regulates Long-chain Acyl-CoA Dehydrogenase by Deacetylating Conserved Lysines Near the Active Site. The Journal of biological chemistry. 2013;288:33837–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Samant SA, Zhang HJ, Hong Z, Pillai VB, Sundaresan NR, Wolfgeher D, et al. SIRT3 Deacetylates and Activates OPA1 To Regulate Mitochondrial Dynamics during Stress. Molecular and cellular biology. 2014;34:807–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Tseng AH, Shieh SS, Ling Wang D. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free radical biology & medicine. 2013. [DOI] [PubMed] [Google Scholar]
- [101].Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyl transferase program; a novel role for GCN5L1. The Biochemical journal. 2012;443:627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Papa L, Germain D. SirT3 regulates a novel arm of the mitochondrial unfolded protein response. Molecular and cellular biology. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Parker BL, Shepherd NE, Trefely S, Hoffman NJ, White MY, Engholm-Keller K, et al. Structural Basis for Phosphorylation and Lysine Acetylation Cross-talk in a Kinase Motif Associated with Myocardial Ischemia and Cardioprotection. The Journal of biological chemistry. 2014;289:25890–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Covian R, Balaban RS. Cardiac mitochondrial matrix and respiratory complex protein phosphorylation. American journal of physiology Heart and circulatory physiology. 2012;303:H940–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].O’Rourke B, Van Eyk JE, Foster DB. Mitochondrial protein phosphorylation as a regulatory modality: implications for mitochondrial dysfunction in heart failure. Congestive heart failure. 2011;17:269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Deng N, Zhang J, Zong C, Wang Y, Lu H, Yang P, et al. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Molecular & cellular proteomics: MCP. 2011;10:M110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Boja ES, Phillips D, French SA, Harris RA, Balaban RS. Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. Journal of proteome research. 2009;8:4665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lagranha CJ, Deschamps A, Aponte A, Steenbergen C, Murphy E. Sex differences in the phosphorylation of mitochondrial proteins result in reduced production of reactive oxygen species and cardioprotection in females. CircRes. 2010;106:1681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell metabolism. 2009;9:265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Covian R, French S, Kusnetz H, Balaban RS. Stimulation of oxidative phosphorylation by calcium in cardiac mitochondria is not influenced by cAMP and PKA activity. Biochimica et biophysica acta. 2014;1837:1913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Murphy E, Kohr M, Menazza S, Nguyen T, Evangelista A, Sun J, et al. Signaling by S-nitrosylation in the heart. Journal of molecular and cellular cardiology. 2014;73:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Murphy E, Kohr M, Sun J, Nguyen T, Steenbergen C. S-nitrosylation: a radical way to protect the heart. Journal of molecular and cellular cardiology. 2012;52:568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Chung HS, Wang SB, Venkatraman V, Murray CI, Van Eyk JE. Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circulation research. 2013;112:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Obal D, Dai S, Keith R, Dimova N, Kingery J, Zheng YT, et al. Cardiomyocyte-restricted overexpression of extracellular superoxide dismutase increases nitric oxide bioavailability and reduces infarct size after ischemia/reperfusion. Basic Res Cardiol. 2012;107:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hare JM, Stamler JS. NO/redox disequilibrium in the failing heart and cardiovascular system. J Clin Invest. 2005;115:509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. CircRes. 2007;101:1155–63. [DOI] [PubMed] [Google Scholar]
- [117].Wang SB, Foster DB, Rucker J, O’Rourke B, Kass DA, Van Eyk JE. Redox regulation of mitochondrial ATP synthase: implications for cardiac resynchronization therapy. Circulation research. 2011;109:750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Glancy B, Willis WT, Chess DJ, Balaban RS. Effect of calcium on the oxidative phosphorylation cascade in skeletal muscle mitochondria. Biochemistry. 2013;52:2793–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Williams GS, Boyman L, Chikando AC, Khairallah RJ, Lederer WJ. Mitochondrial calcium uptake. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Williams GS, Boyman L, Lederer WJ. Mitochondrial Calcium and the Regulation of Metabolism in Heart. Journal of molecular and cellular cardiology. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Liu T, O’Rourke B. Regulation of mitochondrial Ca2+ and its effects on energetics and redox balance in normal and failing heart. Journal of bioenergetics and biomembranes. 2009;41:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Denton RM, McCormack JG, Edgell NJ. Role of calcium ions in the regulation of intramitochondrial metabolism. Effects of Na+, Mg2+ and ruthenium red on the Ca2+-stimulated oxidation of oxoglutarate and on pyruvate dehydrogenase activity in intact rat heart mitochondria. The Biochemical journal. 1980;190:107–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Hunter DR, Haworth RA. The Ca2+-induced membrane transition in mitochondria. I. The protective mechanisms. ArchBiochemBiophys. 1979;195:453–9. [DOI] [PubMed] [Google Scholar]
- [126].Di Lisa F, Carpi A, Giorgio V, Bernardi P. The mitochondrial permeability transition pore and cyclophilin D in cardioprotection. BiochimBiophysActa. 2011;1813:1316–22. [DOI] [PubMed] [Google Scholar]
- [127].Sancak Y, Markhard AL, Kitami T, Kovacs-Bogdan E, Kamer KJ, Udeshi ND, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kamer KJ, Mootha VK. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO reports. 2014;15:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Harrington J, Murphy E. The mitochondrial calcium uniporter: Mice cn live and die without it. Journal of molecular and cellular cardiology. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell metabolism. 2013;17:976–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Patron M, Checchetto V, Raffaello A, Teardo E, Vecellio Reane D, Mantoan M, et al. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Molecular cell. 2014;53:726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Finkel T, Menazza S, Holmstrom KM, Parks RJ, Liu J, Sun J, et al. The Ins and Outs of Mitochondrial Calcium. Circulation research. 2015;116:1810–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Boyman L, Williams GS, Khananshvili D, Sekler I, Lederer WJ. NCLX: the mitochondrial sodium calcium exchanger. Journal of molecular and cellular cardiology. 2013;59:205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Liu T, Takimoto E, Dimaano VL, DeMazumder D, Kettlewell S, Smith G, et al. Inhibiting mitochondrial Na+/Ca2+ exchange prevents sudden death in a Guinea pig model of heart failure. Circulation research. 2014;115:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Kohlhaas M, Liu T, Knopp A, Zeller T, Ong MF, Bohm M, et al. Elevated cytosolic Na+ increases mitochondrial formation of reactive oxygen species in failing cardiac myocytes. Circulation. 2010;121:1606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Shimizu H, Schredelseker J, Huang J, Lu K, Naghdi S, Lu F, et al. Mitochondrial Ca(2+) uptake by the voltage-dependent anion channel 2 regulates cardiac rhythmicity. eLife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Pan X, Liu J, Nguyen T, Liu C, Sun J, Teng Y, et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nature cell biology. 2013;15:1464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wu Y, Rasmussen TP, Koval OM, Joiner ML, Hall DD, Chen B, et al. The mitochondrial uniporter controls fight or flight heart rate increases. Nature communications. 2015;6:6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Luongo TS, Lambert JP, Yuan A, Zhang X, Gross P, Song J, et al. The Mitochondrial Calcium Uniporter Matches Energetic Supply with Cardiac Workload during Stress and Modulates Permeability Transition. Cell reports. 2015;12:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Kwong JQ, Lu X, Correll RN, Schwanekamp JA, Vagnozzi RJ, Sargent MA, et al. The Mitochondrial Calcium Uniporter Selectively Matches Metabolic Output to Acute Contractile Stress in the Heart. Cell reports. 2015;12:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Rasmussen TP, Wu Y, Joiner MA, Koval OM, Wilson NR, Luczak ED, et al. Inhibition of MCU forces extramitochondrial adaptations governing physiological and pathological stress responses in heart. Proceedings of the National Academy of Sciences of the United States of America. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anatomy and embryology. 1990;181:195–213. [DOI] [PubMed] [Google Scholar]
- [143].Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–29. [DOI] [PubMed] [Google Scholar]
- [144].Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. [DOI] [PubMed] [Google Scholar]
- [145].Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nature immunology. 2000;1:489–95. [DOI] [PubMed] [Google Scholar]
- [146].Xu K, Tavernarakis N, Driscoll M. Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca(2+) release from the endoplasmic reticulum. Neuron. 2001;31:957–71. [DOI] [PubMed] [Google Scholar]
- [147].Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nature cell biology. 2007;9:550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Nakayama H, Chen X, Baines CP, Klevitsky R, Zhang X, Zhang H, et al. Ca2+− and mitochondrial-dependent cardiomyocyte necrosis as a primary mediator of heart failure. The Journal of clinical investigation. 2007;117:2431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell death and differentiation. 2015;22:58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Liu Y, Shoji-Kawata S, Sumpter RM Jr., Wei Y, Ginet V, Zhang L, et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Kung G, Konstantinidis K, Kitsis RN. Programmed necrosis, not apoptosis, in the heart. Circulation research. 2011;108:1017–36. [DOI] [PubMed] [Google Scholar]
- [152].Jennings RB, Sommers HM, Smyth GA, Flack HA, Linn H. Myocardial necrosis induced by temporary occlusion of a coronary artery in the dog. Archives of pathology. 1960;70:68–78. [PubMed] [Google Scholar]
- [153].Gottlieb RA, Burleson KO, Kloner RA, Babior BM, Engler RL. Reperfusion injury induces apoptosis in rabbit cardiomyocytes. The Journal of clinical investigation. 1994;94:1621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Fliss H, Gattinger D. Apoptosis in ischemic and reperfused rat myocardium. Circulation research. 1996;79:949–56. [DOI] [PubMed] [Google Scholar]
- [155].Brocheriou V, Hagege AA, Oubenaissa A, Lambert M, Mallet VO, Duriez M, et al. Cardiac functional improvement by a human Bcl-2 transgene in a mouse model of ischemia/reperfusion injury. The journal of gene medicine. 2000;2:326–33. [DOI] [PubMed] [Google Scholar]
- [156].Jeremias I, Kupatt C, Martin-Villalba A, Habazettl H, Schenkel J, Boekstegers P, et al. Involvement of CD95/Apo1/Fas in cell death after myocardial ischemia. Circulation. 2000;102:915–20. [DOI] [PubMed] [Google Scholar]
- [157].Chen Z, Chua CC, Ho YS, Hamdy RC, Chua BH. Overexpression of Bcl-2 attenuates apoptosis and protects against myocardial I/R injury in transgenic mice. American journal of physiology Heart and circulatory physiology. 2001;280:H2313–20. [DOI] [PubMed] [Google Scholar]
- [158].Hochhauser E, Kivity S, Offen D, Maulik N, Otani H, Barhum Y, et al. Bax ablation protects against myocardial ischemia-reperfusion injury in transgenic mice. American journal of physiology Heart and circulatory physiology. 2003;284:H2351–9. [DOI] [PubMed] [Google Scholar]
- [159].Lee P, Sata M, Lefer DJ, Factor SM, Walsh K, Kitsis RN. Fas pathway is a critical mediator of cardiac myocyte death and MI during ischemia-reperfusion in vivo. American journal of physiology Heart and circulatory physiology. 2003;284:H456–63. [DOI] [PubMed] [Google Scholar]
- [160].Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, et al. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–8. [DOI] [PubMed] [Google Scholar]
- [161].Toth A, Jeffers JR, Nickson P, Min JY, Morgan JP, Zambetti GP, et al. Targeted deletion of Puma attenuates cardiomyocyte death and improves cardiac function during ischemia-reperfusion. American journal of physiology Heart and circulatory physiology. 2006;291:H52–60. [DOI] [PubMed] [Google Scholar]
- [162].Chua CC, Gao J, Ho YS, Xiong Y, Xu X, Chen Z, et al. Overexpression of IAP-2 attenuates apoptosis and protects against myocardial ischemia/reperfusion injury in transgenic mice. Biochimica et biophysica acta. 2007;1773:577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Whelan RS, Konstantinidis K, Wei AC, Chen Y, Reyna DE, Jha S, et al. Bax regulates primary necrosis through mitochondrial dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Wencker D, Chandra M, Nguyen K, Miao W, Garantziotis S, Factor SM, et al. A mechanistic role for cardiac myocyte apoptosis in heart failure. The Journal of clinical investigation. 2003;111:1497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annual review of physiology. 2010;72:19–44. [DOI] [PubMed] [Google Scholar]
- [166].Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Molecular cell. 2010;37:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Molecular cell. 2010;40:481–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. The Journal of experimental medicine. 1997;185:1481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Soldani C, Scovassi AI. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: an update. Apoptosis: an international journal on programmed cell death. 2002;7:321–8. [DOI] [PubMed] [Google Scholar]
- [171].Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Caspase activation inhibits proteasome function during apoptosis. Molecular cell. 2004;14:81–93. [DOI] [PubMed] [Google Scholar]
- [172].Saelens X, Festjens N, Parthoens E, Vanoverberghe I, Kalai M, van Kuppeveld F, et al. Protein synthesis persists during necrotic cell death. The Journal of cell biology. 2005;168:545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Karch J, Molkentin JD. Identifying the components of the elusive mitochondrial permeability transition pore. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10396–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Kokoszka JE, Waymire KG, Levy SE, Sligh JE, Cai J, Jones DP, et al. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Gutierrez-Aguilar M, Douglas DL, Gibson AK, Domeier TL, Molkentin JD, Baines CP. Genetic manipulation of the cardiac mitochondrial phosphate carrier does not affect permeability transition. Journal of molecular and cellular cardiology. 2014;72:316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kwong JQ, Davis J, Baines CP, Sargent MA, Karch J, Wang X, et al. Genetic deletion of the mitochondrial phosphate carrier desensitizes the mitochondrial permeability transition pore and causes cardiomyopathy. Cell death and differentiation. 2014;21:1209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Bonora M, Bononi A, De Marchi E, Giorgi C, Lebiedzinska M, Marchi S, et al. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell cycle. 2013;12:674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Giorgio V, von Stockum S, Antoniel M, Fabbro A, Fogolari F, Forte M, et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Alavian KN, Beutner G, Lazrove E, Sacchetti S, Park HA, Licznerski P, et al. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Crompton M, Costi A, Hayat L. Evidence for the presence of a reversible Ca2+-dependent pore activated by oxidative stress in heart mitochondria. The Biochemical journal. 1987;245:915–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Crompton M, Ellinger H, Costi A. Inhibition by cyclosporin A of a Ca2+-dependent pore in heart mitochondria activated by inorganic phosphate and oxidative stress. The Biochemical journal. 1988;255:357–60. [PMC free article] [PubMed] [Google Scholar]
- [182].Halestrap AP. Calcium-dependent opening of a non-specific pore in the mitochondrial inner membrane is inhibited at pH values below 7. Implications for the protective effect of low pH against chemical and hypoxic cell damage. The Biochemical journal. 1991;278 (Pt 3):715–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Griffiths EJ, Halestrap AP. Mitochondrial non-specific pores remain closed during cardiac ischaemia, but open upon reperfusion. The Biochemical journal. 1995;307 (Pt 1):93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–62. [DOI] [PubMed] [Google Scholar]
- [185].Griffiths EJ, Halestrap AP. Further evidence that cyclosporin A protects mitochondria from calcium overload by inhibiting a matrix peptidyl-prolyl cis-trans isomerase. Implications for the immunosuppressive and toxic effects of cyclosporin. The Biochemical journal. 1991;274 (Pt 2):611–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Karch J, Kwong JQ, Burr AR, Sargent MA, Elrod JW, Peixoto PM, et al. Bax and Bak function as the outer membrane component of the mitochondrial permeability pore in regulating necrotic cell death in mice. eLife. 2013;2:e00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Piot C, Croisille P, Staat P, Thibault H, Rioufol G, Mewton N, et al. Effect of cyclosporine on reperfusion injury in acute myocardial infarction. The New England journal of medicine. 2008;359:473–81. [DOI] [PubMed] [Google Scholar]
- [188].Mewton N, Croisille P, Gahide G, Rioufol G, Bonnefoy E, Sanchez I, et al. Effect of cyclosporine on left ventricular remodeling after reperfused myocardial infarction. Journal of the American College of Cardiology. 2010;55:1200–5. [DOI] [PubMed] [Google Scholar]
- [189].Murphy MP. How mitochondria produce reactive oxygen species. BiochemJ. 2009;417:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Chen YR, Zweier JL. Cardiac mitochondria and reactive oxygen species generation. Circulation research. 2014;114:524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiological reviews. 2014;94:909–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Carpi A, Menabò R, Kaludercic N, Pelicci P, Di Lisa F, Giorgio M. The cardioprotective effects elicited by p66(Shc) ablation demonstrate the crucial role of mitochondrial ROS formation in ischemia/reperfusion injury. BiochimBiophysActa. 2009;1787:774–80. [DOI] [PubMed] [Google Scholar]
- [193].Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Korshunov SS, Skulachev VP, Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS letters. 1997;416:15–8. [DOI] [PubMed] [Google Scholar]
- [195].Aon MA, Cortassa S, Marban E, O’Rourke B. Synchronized whole cell oscillations in mitochondrial metabolism triggered by a local release of reactive oxygen species in cardiac myocytes. JBiolChem. 2003;278:44735–44. [DOI] [PubMed] [Google Scholar]
- [196].Aon MA, Cortassa S, O’Rourke B. Redox-optimized ROS balance: a unifying hypothesis. Biochimica et biophysica acta. 2010;1797:865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Nickel A, Kohlhaas M, Maack C. Mitochondrial reactive oxygen species production and elimination. Journal of molecular and cellular cardiology. 2014;73:26–33. [DOI] [PubMed] [Google Scholar]
- [198].Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? NatRevMolCell Biol. 2007;8:722–8. [DOI] [PubMed] [Google Scholar]
- [199].Cambridge SB, Gnad F, Nguyen C, Bermejo JL, Kruger M, Mann M. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J Proteome Res. 2011;10:5275–84. [DOI] [PubMed] [Google Scholar]
- [200].Zorov DB, Filburn CR, Klotz LO, Zweier JL, Sollott SJ. Reactive oxygen species (ROS)-induced ROS release: a new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. JExpMed. 2000;192:1001–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Juhaszova M, Wang S, Zorov DB, Nuss HB, Gleichmann M, Mattson MP, et al. The identity and regulation of the mitochondrial permeability transition pore: where the known meets the unknown. Annals of the New York Academy of Sciences. 2008;1123:197–212. [DOI] [PubMed] [Google Scholar]
- [202].Andrukhiv A, Costa AD, West IC, Garlid KD. Opening mitoKATP increases superoxide generation from complex I of the electron transport chain. AmJPhysiol Heart CircPhysiol. 2006;291:H2067–H74. [DOI] [PubMed] [Google Scholar]
- [203].Pain T, Yang XM, Critz SD, Yue Y, Nakano A, Liu GS, et al. Opening of mitochondrial K(ATP) channels triggers the preconditioned state by generating free radicals. CircRes. 2000;87:460–6. [DOI] [PubMed] [Google Scholar]
- [204].Forbes RA, Steenbergen C, Murphy E. Diazoxide-induced cardioprotection requires signaling through a redox-sensitive mechanism. Circulation research. 2001;88:802–9. [DOI] [PubMed] [Google Scholar]
- [205].Barja G Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. JBioenergBiomembr. 1999;31:347–66. [DOI] [PubMed] [Google Scholar]
- [206].Penna C, Rastaldo R, Mancardi D, Raimondo S, Cappello S, Gattullo D, et al. Post-conditioning induced cardioprotection requires signaling through a redox-sensitive mechanism, mitochondrial ATP-sensitive K+ channel and protein kinase C activation. Basic ResCardiol. 2006;101:180–9. [DOI] [PubMed] [Google Scholar]
- [207].Vanden Hoek TL, Becker LB, Shao Z, Li C, Schumacker PT. Reactive oxygen species released from mitochondria during brief hypoxia induce preconditioning in cardiomyocytes. JBiolChem. 1998;273:18092–8. [DOI] [PubMed] [Google Scholar]
- [208].Ahmad S, Singh N, Glazer RI. Role of AKT1 in 17beta-estradiol- and insulin-like growth factor I (IGF-I)-dependent proliferation and prevention of apoptosis in MCF-7 breast carcinoma cells. BiochemPharmacol. 1999;58:425–30. [DOI] [PubMed] [Google Scholar]
- [209].Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci USA. 1999;96:4820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–11. [DOI] [PubMed] [Google Scholar]
- [211].Anderson EJ, Lustig ME, Boyle KE, Woodlief TL, Kane DA, Lin CT, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. The Journal of clinical investigation. 2009;119:573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Dai DF, Chen T, Szeto H, Nieves-Cintron M, Kutyavin V, Santana LF, et al. Mitochondrial targeted antioxidant Peptide ameliorates hypertensive cardiomyopathy. Journal of the American College of Cardiology. 2011;58:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Brand MD. The sites and topology of mitochondrial superoxide production. Experimental gerontology. 2010;45:466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–33. [DOI] [PubMed] [Google Scholar]
- [215].Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–13. [DOI] [PubMed] [Google Scholar]
- [216].Affaitati A, Cardone L, De Cristofaro T, Carlucci A, Ginsberg MD, Varrone S, et al. Essential role of A-kinase anchor protein 121 for cAMP signaling to mitochondria. JBiolChem. 2003;278:4286–94. [DOI] [PubMed] [Google Scholar]
- [217].Camici GG, Schiavoni M, Francia P, Bachschmid M, Martin-Padura I, Hersberger M, et al. Genetic deletion of p66(Shc) adaptor protein prevents hyperglycemia-induced endothelial dysfunction and oxidative stress. ProcNatlAcadSciUSA. 2007;104:5217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Kareyeva AV, Grivennikova VG, Cecchini G, Vinogradov AD. Molecular identification of the enzyme responsible for the mitochondrial NADH-supported ammonium-dependent hydrogen peroxide production. FEBS letters. 2011;585:385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Anderson EJ, Efird JT, Davies SW, O’Neal WT, Darden TM, Thayne KA, et al. Monoamine oxidase is a major determinant of redox balance in human atrial myocardium and is associated with postoperative atrial fibrillation. Journal of the American Heart Association. 2014;3:e000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Weinreb O, Bar-Am O, Amit T, Chillag-Talmor O, Youdim MB. Neuroprotection via pro-survival protein kinase C isoforms associated with Bcl-2 family members. FASEB J. 2004;18:1471–3. [DOI] [PubMed] [Google Scholar]
- [221].Ago T, Kuroda J, Pain J, Fu C, Li H, Sadoshima J. Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. CircRes. 2010;106:1253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Martyn KD, Frederick LM, von Loehneysen K, Dinauer MC, Knaus UG. Functional analysis of Nox4 reveals unique characteristics compared to other NADPH oxidases. Cellular signalling. 2006;18:69–82. [DOI] [PubMed] [Google Scholar]
- [223].Montezano AC, Touyz RM. Oxidative stress, Noxs, and hypertension: experimental evidence and clinical controversies. Annals of medicine. 2012;44 Suppl 1:S2–16. [DOI] [PubMed] [Google Scholar]
- [224].Berndt C, Lillig CH, Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. AmJPhysiol Heart CircPhysiol. 2007;292:H1227–H36. [DOI] [PubMed] [Google Scholar]
- [225].Chalmers S, Caldwell ST, Quin C, Prime TA, James AM, Cairns AG, et al. Selective uncoupling of individual mitochondria within a cell using a mitochondria-targeted photoactivated protonophore. JAmChemSoc. 2012;134:758–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Cortassa S, O’Rourke B, Aon MA. Redox-Optimized ROS Balance and the relationship between mitochondrial respiration and ROS. Biochimica et biophysica acta. 2014;1837:287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Cox AG, Winterbourn CC, Hampton MB. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. The Biochemical journal. 2010;425:313–25. [DOI] [PubMed] [Google Scholar]
- [228].Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell DeathDiffer. 2009;16:1093–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Matsushima S, Ide T, Yamato M, Matsusaka H, Hattori F, Ikeuchi M, et al. Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation. 2006;113:1779–86. [DOI] [PubMed] [Google Scholar]
- [230].Chouchani ET, Methner C, Nadtochiy SM, Logan A, Pell VR, Ding S, et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nature medicine. 2013;19:753–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Smith RA, Hartley RC, Cocheme HM, Murphy MP. Mitochondrial pharmacology. Trends PharmacolSci. 2012;33:341–52. [DOI] [PubMed] [Google Scholar]
- [232].Di Lisa F, Kaludercic N, Carpi A, Menabò R, Giorgio M. Mitochondrial pathways for ROS formation and myocardial injury: the relevance of p66(Shc) and monoamine oxidase. Basic ResCardiol. 2009;104:131–9. [DOI] [PubMed] [Google Scholar]
- [233].Bugger H, Abel ED. Mitochondria in the diabetic heart. CardiovascRes. 2010;88:229–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Lesnefsky EJ, Gudz TI, Moghaddas S, Migita CT, Ikeda-Saito M, Turkaly PJ, et al. Aging decreases electron transport complex III activity in heart interfibrillar mitochondria by alteration of the cytochrome c binding site. JMolCellCardiol. 2001;33:37–47. [DOI] [PubMed] [Google Scholar]
- [235].Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95. [DOI] [PubMed] [Google Scholar]
- [236].Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, et al. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free RadicBiolMed. 2008;45:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Molecular cell. 2012;48:158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Finkel T From sulfenylation to sulfhydration: what a thiolate needs to tolerate. Science signaling. 2012;5:pe10. [DOI] [PubMed] [Google Scholar]
- [239].Prime TA, Blaikie FH, Evans C, Nadtochiy SM, James AM, Dahm CC, et al. A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. ProcNatlAcadSciUSA. 2009;106:10764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–70. [DOI] [PubMed] [Google Scholar]
- [241].Jakob R, Beutner G, Sharma VK, Duan Y, Gross RA, Hurst S, et al. Molecular and functional identification of a mitochondrial ryanodine receptor in neurons. Neuroscience letters. 2014;575:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annual review of physiology. 2014;76:39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in biochemical sciences. 2014;39:199–218. [DOI] [PubMed] [Google Scholar]
- [244].Kim TY, Wang D, Kim AK, Lau E, Lin AJ, Liem DA, et al. Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics. 2012;11:1586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Bhandari P, Song M, Chen Y, Burelle Y, Dorn GW 2nd. Mitochondrial contagion induced by Parkin deficiency in Drosophila hearts and its containment by suppressing mitofusin. Circulation research. 2014;114:257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Akhmedov A, Montecucco F, Braunersreuther V, Camici GG, Jakob P, Reiner MF, et al. Genetic deletion of the adaptor protein p66Shc increases susceptibility to short-term ischaemic myocardial injury via intracellular salvage pathways. European heart journal. 2014. [DOI] [PubMed] [Google Scholar]
- [247].Spindel ON, Berk BC. Redox redux: protecting the ischemic myocardium. The Journal of clinical investigation. 2012;122:30–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Ristow M Unraveling the truth about antioxidants: mitohormesis explains ROS-induced health benefits. Nature medicine. 2014;20:709–11. [DOI] [PubMed] [Google Scholar]
- [249].Song M, Mihara K, Chen Y, Scorrano L, Dorn GW 2nd. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell metabolism. 2015;21:273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. The Journal of cell biology. 2004;164:493–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Legros F, Lombes A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Molecular biology of the cell. 2002;13:4343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nature reviews Molecular cell biology. 2014;15:634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, et al. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Molecular biology of the cell. 2014;25:145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Kasahara A, Cipolat S, Chen Y, Dorn GW 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science.342:734–7. [DOI] [PubMed] [Google Scholar]
- [258].Sharp WW, Fang YH, Han M, Zhang HJ, Hong Z, Banathy A, et al. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2014;28:316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6400–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. The EMBO journal. 2009;28:1589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature reviews Molecular cell biology. 2011;12:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Healy DG, Abou-Sleiman PM, Wood NW. PINK, PANK, or PARK? A clinicians’ guide to familial parkinsonism. The Lancet Neurology. 2004;3:652–62. [DOI] [PubMed] [Google Scholar]
- [265].Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. [DOI] [PubMed] [Google Scholar]
- [266].Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. [DOI] [PubMed] [Google Scholar]
- [267].Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. The Journal of cell biology. 2010;191:933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–6. [DOI] [PubMed] [Google Scholar]
- [272].Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Dorn GW II KR. The mitochondrial dynamism-mitophagy-cell death interactome: Multiple roles performed by members of a mitochondrial molecular ensemble. Circul Res. 2014. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Song M, Gong G, Burelle Y, Gustafsson AB, Kitsis RN, Matkovich SJ, et al. Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. Circulation research. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. The EMBO journal. 2014;33:2798–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Song M, Chen Y, Gong G, Murphy E, Rabinovitch PS, Dorn GW 2nd. Super-suppression of mitochondrial reactive oxygen species signaling impairs compensatory autophagy in primary mitophagic cardiomyopathy. Circulation research. 2014;115:348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiological reviews. 2011;91:1161–218. [DOI] [PubMed] [Google Scholar]
- [279].Guo M Drosophila as a model to study mitochondrial dysfunction in Parkinson’s disease. Cold Spring Harbor perspectives in medicine. 2012;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010;66:646–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Siddall HK, Yellon DM, Ong SB, Mukherjee UA, Burke N, Hall AR, et al. Loss of PINK1 increases the heart’s vulnerability to ischemia-reperfusion injury. PloS one. 2013;8:e62400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. The Journal of clinical investigation. 2011;121:3701–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: a question of fate? Cell death and differentiation. 2005;12 Suppl 2:1484–9. [DOI] [PubMed] [Google Scholar]
- [284].Dorn GW 2nd. Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. Journal of cardiovascular translational research. 2010;3:374–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Skulachev VP. Programmed death phenomena: from organelle to organism. Annals of the New York Academy of Sciences. 2002;959:214–37. [DOI] [PubMed] [Google Scholar]
- [286].Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–8. [DOI] [PubMed] [Google Scholar]
- [287].Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO reports. 2010;11:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. The Journal of biological chemistry. 2010;285:27879–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. The Journal of biological chemistry. 2012;287:19094–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, et al. Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nature medicine. 2002;8:725–30. [DOI] [PubMed] [Google Scholar]
- [291].Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. The Journal of clinical investigation. 2007;117:2825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW 2nd. Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Chaanine AH, Gordon RE, Kohlbrenner E, Benard L, Jeong D, Hajjar RJ. Potential role of BNIP3 in cardiac remodeling, myocardial stiffness, and endoplasmic reticulum: mitochondrial calcium homeostasis in diastolic and systolic heart failure. Circulation Heart failure. 2013;6:572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, et al. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. American journal of physiology Heart and circulatory physiology. 2009;296:H470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. Embo J. 2008;27:306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Pratt JM, Petty J, Riba-Garcia I, Robertson DH, Gaskell SJ, Oliver SG, et al. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1:579–91. [DOI] [PubMed] [Google Scholar]
- [297].Lam MP, Wang D, Lau E, Liem DA, Kim AK, Ng DC, et al. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J Clin Invest. 2014;124:1734–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Hsieh EJ, Shulman NJ, Dai DF, Vincow ES, Karunadharma PP, Pallanck L, et al. Topograph, a software platform for precursor enrichment corrected global protein turnover measurements. Mol Cell Proteomics. 2012;11:1468–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Shekar KC, Li L, Dabkowski ER, Xu W, Ribeiro RF Jr., Hecker PA, et al. Cardiac mitochondrial proteome dynamics with heavy water reveals stable rate of mitochondrial protein synthesis in heart failure despite decline in mitochondrial oxidative capacity. Journal of molecular and cellular cardiology. 2014;75:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Claydon AJ, Beynon R. Proteome dynamics: revisiting turnover with a global perspective. Mol Cell Proteomics. 2012;11:1551–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Busch R, Neese RA, Awada M, Hayes GM, Hellerstein MK. Measurement of cell proliferation by heavy water labeling. Nat Protoc. 2007;2:3045–57. [DOI] [PubMed] [Google Scholar]
- [302].Raman A, Schoeller DA, Subar AF, Troiano RP, Schatzkin A, Harris T, et al. Water turnover in 458 American adults 40–79 yr of age. Am J Physiol Renal Physiol. 2004;286:F394–401. [DOI] [PubMed] [Google Scholar]
- [303].Messmer BT, Messmer D, Allen SL, Kolitz JE, Kudalkar P, Cesar D, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115:755–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Zhang Y, Reckow S, Webhofer C, Boehme M, Gormanns P, Egge-Jacobsen WM, et al. Proteome scale turnover analysis in live animals using stable isotope metabolic labeling. Anal Chem. 2011;83:1665–72. [DOI] [PubMed] [Google Scholar]
- [305].Rao PK, Rodriguez GM, Smith I, Li Q. Protein dynamics in iron-starved Mycobacterium tuberculosis revealed by turnover and abundance measurement using hybrid-linear ion trap-Fourier transform mass spectrometry. Anal Chem. 2008;80:6860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Pupim LB, Flakoll PJ, Ikizler TA. Nutritional supplementation acutely increases albumin fractional synthetic rate in chronic hemodialysis patients. J Am Soc Nephrol. 2004;15:1920–6. [DOI] [PubMed] [Google Scholar]
- [307].Vaux DL. Apoptogenic factors released from mitochondria. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2011;1813:546–50. [DOI] [PubMed] [Google Scholar]
- [308].Chandel N Mitochondria as signaling organelles. BMC Biology. 2014;12:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Nakayama H, Otsu K. Translation of hemodynamic stress to sterile inflammation in the heart. Trends in Endocrinology & Metabolism. 2013;24:546–53. [DOI] [PubMed] [Google Scholar]
- [310].Arnoult D, Soares F, Tattoli I, Girardin SE. Mitochondria in innate immunity 2011. [DOI] [PMC free article] [PubMed]
- [311].Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends in Immunology. 2011;32:157–64. [DOI] [PubMed] [Google Scholar]
- [312].Nakahira K, Haspel JA, Rathinam VAK, Lee S-J, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Bliksøen M, Mariero LH, Ohm IK, Haugen F, Yndestad A, Solheim S, et al. Increased circulating mitochondrial DNA after myocardial infarction. International Journal of Cardiology. 2012;158:132–4. [DOI] [PubMed] [Google Scholar]
- [314].Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Carp H Mitochondrial N-formylmethionyl proteins as chemoattractants for neutrophils. The Journal of Experimental Medicine. 1982;155:264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. http://www.broadinstitute.org/pubs/MitoCarta/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Path. 2010;5:297–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Wallace DC. Mitochondria and cancer. Nature Reviews Cancer. 2012;12:685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Wallace DC. Bioenergetics in human evolution and disease:Implications for the origins of biological complexity and the missing genetic variation of common diseases. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med. 2012;366:1132–41. [DOI] [PubMed] [Google Scholar]
- [322].Wallace DC, Lott MT, Procaccio V. Mitochondrial Medicine: The Mitochondrial Biology and Genetics of Metabolic and Degenerative Diseases, Cancer, and Aging In: Rimoin DL, Pyeritz RE, Korf BR, editors. Emery and Rimoin’s Principles and Practice of Medical Genetics. 6th ed. Philadelphia: Churchill Livingstone Elsevier; 2013. [Google Scholar]
- [323].Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E, Diez-Sanchez C, Montoya J, Lopez-Perez MJ, et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19:3343–53. [DOI] [PubMed] [Google Scholar]
- [324].Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MR, Mirra SS, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–84. [DOI] [PubMed] [Google Scholar]
- [325].MITOMAP. A Human Mitochondrial Genome Database. http://www.mitomap.org. 2013. [DOI] [PMC free article] [PubMed]
- [326].Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–30. [DOI] [PubMed] [Google Scholar]
- [327].Sadun AA, La Morgia C, Carelli V. Leber’s Hereditary Optic Neuropathy. Curr Treat Options Neurol. 2011;13:109–17. [DOI] [PubMed] [Google Scholar]
- [328].Wallace DC, Zheng X, Lott MT, Shoffner JM, Hodge JA, Kelley RI, et al. Familial mitochondrial encephalomyopathy (MERRF): Genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55:601–10. [DOI] [PubMed] [Google Scholar]
- [329].Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 1990;61:931–7. [DOI] [PubMed] [Google Scholar]
- [330].Heddi A, Stepien G, Benke PJ, Wallace DC. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J Biol Chem. 1999;274:22968–76. [DOI] [PubMed] [Google Scholar]
- [331].Goto Y, Nonaka I, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–3. [DOI] [PubMed] [Google Scholar]
- [332].van den Ouweland JM, Lemkes HHP, Ruitenbeek W, Sandkjujl LA, deVijlder MF, Struyvenberg PAA, et al. Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet. 1992;1:368–71. [DOI] [PubMed] [Google Scholar]
- [333].Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Pons R, Andreu AL, Checcarelli N, Vila MR, Engelstad K, Sue CM, et al. Mitochondrial DNA abnormalities and autistic spectrum disorders. Journal of Pediatrics. 2004;144:81–5. [DOI] [PubMed] [Google Scholar]
- [335].Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–23. [DOI] [PubMed] [Google Scholar]
- [336].Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–11. [DOI] [PubMed] [Google Scholar]
- [337].Zeviani M, Bresolin N, Gellera C, Bordoni A, Pannacci M, Amati P, et al. Nucleus-driven multiple large-scale deletions of the human mitochondrial genome: a new autosomal dominant disease. Am J Hum Genet. 1990;47:904–14. [PMC free article] [PubMed] [Google Scholar]
- [338].Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci USA. 2004;101:10726–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Coskun PE, Wyrembak J, Derbereva O, Melkonian G, Doran E, Lott IT, et al. Systemic mitochondrial dysfunction and the etiology of Alzheimer’s disease and down syndrome dementia. J Alzheimers Dis. 2010;20 Suppl 2:S293–S310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Coskun P, Wyrembak J, Schriner SE, Chen HW, Marciniack C, LaFerla F, et al. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2012;1820:553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Picard M, Zhang J, Hancock S, Derbeneva O, Golhar R, Golik P, et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci USA. 2014;111:E4033–E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, Matsuo H, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. 2007;80:407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Nishigaki Y, Yamada Y, Fuku N, Matsuo H, Segawa T, Watanabe S, et al. Mitochondrial haplogroup N9b is protective against myocardial infarction in Japanese males. Hum Genet. 2007;120:827–36. [DOI] [PubMed] [Google Scholar]
- [344].Rodriguez-Iturbe B, Sepassi L, Quiroz Y, Ni Z, Wallace DC, Vaziri ND. Association of mitochondrial SOD deficiency with salt-sensitive hypertension and accelerated renal senescence. J Appl Physiol. 2007;102:255–60. [DOI] [PubMed] [Google Scholar]
- [345].van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat (Online). 2009;30:E386–E94. [DOI] [PubMed] [Google Scholar]
- [346].Zaragoza MV, Brandon MC, Diegoli M, Arbustini E, Wallace DC. Mitochondrial cardiomyopathies: how to identify candidate pathogenic mutations by mitochondrial DNA sequencing, MITOMASTER and phylogeny. Eur J Hum Genet. 2011;19:200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Brown MD, Trounce IA, Jun AS, Allen JC, Wallace DC. Functional analysis of lymphoblast and cybrid mitochondria containing the 3460, 11778, or 14484 Leber’s Hereditary Optic Neuropathy mtDNA mutation. J Biol Chem. 2000;275:39831–6. [DOI] [PubMed] [Google Scholar]
- [348].Ji F, Sharpley MS, Derbeneva O, Alves LS, Qian P, Wang Y, et al. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci USA. 2012;109:7391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Fan W, Waymire K, Narula N, Li P, Rocher C, Coskun PE, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Corral-Debrinski M, Stepien G, Shoffner JM, Lott MT, Kanter K, Wallace DC. Hypoxemia is associated with mitochondrial DNA damage and gene induction. Implications for cardiac disease. JAMA. 1991;266:1812–6. [PubMed] [Google Scholar]
- [351].Phoon CK, Acehan D, Schlame M, Stokes DL, Edelman-Novemsky I, Yu D, et al. Tafazzin knockdown in mice leads to a developmental cardiomyopathy with early diastolic dysfunction preceding myocardial noncompaction. J Am Heart Assoc. 2012;1:jah3–e000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Li K, Warner CK, Hodge JA, Minoshima S, Kudoh J, Fukuyama R, et al. A human muscle adenine nucleotide translocator gene has four exons, is located on chromosome 4, and is differentially expressed. J Biol Chem. 1989;264:13998–4004. [PubMed] [Google Scholar]
- [353].Li K, Hodge JA, Wallace DC. OXBOX, a positive transcriptional element of the heart-skeletal muscle ADP/ATP translocator gene. J Biol Chem. 1990;265:20585–8. [PubMed] [Google Scholar]
- [354].Palmieri L, Alberio S, Pisano I, Lodi T, Meznaric-Petrusa M, Zidar J, et al. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum Mol Genet. 2005;14:3079–88. [DOI] [PubMed] [Google Scholar]
- [355].Echaniz-Laguna A, Chassagne M, Ceresuela J, Rouvet I, Padet S, Acquaviva C, et al. Complete loss of expression of the ANT1 gene causing cardiomyopathy and myopathy. J Med Genet. 2012;49:146–50. [DOI] [PubMed] [Google Scholar]
- [356].Strauss KA, Dubiner L, Simon M, Zaragoza M, Sengupta PP, Li P, et al. Severity of cardiomyopathy associated with adenine nucleotide translocator-1 deficiency correlates with mtDNA haplogroup. Proc Natl Acad Sci USA. 2013;110:3253–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [357].Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/skeletal muscle isoform of the adenine nucleotide translocator. Nat Genet. 1997;16:226–34. [DOI] [PubMed] [Google Scholar]
- [358].Narula N, Zaragoza MV, Sengupta PP, Li P, Haider N, Verjans J, et al. Adenine nucleotide translocase 1 deficiency results in dilated cardiomyopathy with defects in myocardial mechanics, histopathological alterations, and activation of apoptosis. JACC Cardiovascular Imaging. 2011;4:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [359].Dorner A, Schulze K, Rauch U, Schultheiss HP. Adenine nucleotide translocator in dilated cardiomyopathy: pathophysiological alterations in expression and function. Mol Cell Biochem. 1997;174:261–9. [PubMed] [Google Scholar]
- [360].Dorner A, Schultheiss HP. Adenine nucleotide translocase in the focus of cardiovascular diseases. Trends Cardiovasc Med. 2007;17:284–90. [DOI] [PubMed] [Google Scholar]
- [361].Zhang Q, Itagaki K, Hauser CJ. Mitochondrial DNA is released by shock and activates neutrophils via p38 map kinase. Shock. 2010;34:55–559. [DOI] [PubMed] [Google Scholar]
- [362].Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [363].Anzovino A, Lane DJ, Huang ML, Richardson DR. Fixing frataxin: ‘ironing out’ the metabolic defect in Friedreich’s ataxia. British journal of pharmacology. 2014;171:2174–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [364].Seifert EL, Ligeti E, Mayr JA, Sondheimer N, Hajnoczky G. The mitochondrial phosphate carrier: Role in oxidative metabolism, calcium handling and mitochondrial disease. Biochemical and biophysical research communications. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [365].Potluri P, Davila A, Ruiz-Pesini E, Mishmar D, O’Hearn S, Hancock S, et al. A novel NDUFA1 mutation leads to a progressive mitochondrial complex I-specific neurodegenerative disease. Mol Genet Metab. 2009;96:189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Arbustini E, Diegoli M, Fasani R, Grasso M, Morbini P, Banchieri N, et al. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am J Pathol. 1998;153:1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Arbustini E, Fasani R, Morbini P, Diegoli M, Grasso M, Dal Bello B, et al. Coexistence of mitochondrial DNA and beta myosin heavy chain mutations in hypertrophic cardiomyopathy with late congestive heart failure [published erratum appears in Heart 1999 Mar;81(3):330]. Heart. 1998;80:548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Gatta G, Capocaccia R, Coleman MP, Ries LA, Berrino F. Childhood cancer survival in Europe and the United States. Cancer. 2002;95:1767–72. [DOI] [PubMed] [Google Scholar]
- [369].Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA: a cancer journal for clinicians. 2006;56:106–30. [DOI] [PubMed] [Google Scholar]
- [370].Krischer JP, Epstein S, Cuthbertson DD, Goorin AM, Epstein ML, Lipshultz SE. Clinical cardiotoxicity following anthracycline treatment for childhood cancer: the Pediatric Oncology Group experience. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1997;15:1544–52. [DOI] [PubMed] [Google Scholar]
- [371].Todaro MC, Oreto L, Qamar R, Paterick TE, Carerj S, Khandheria BK. Cardioncology: state of the heart. International journal of cardiology. 2013;168:680–7. [DOI] [PubMed] [Google Scholar]
- [372].Yeh ET, Bickford CL. Cardiovascular complications of cancer therapy: incidence, pathogenesis, diagnosis, and management. Journal of the American College of Cardiology. 2009;53:2231–47. [DOI] [PubMed] [Google Scholar]
- [373].Ewer MS, Ewer SM. Cardiotoxicity of anticancer treatments: what the cardiologist needs to know. Nature reviews Cardiology. 2010;7:564–75. [DOI] [PubMed] [Google Scholar]
- [374].Davies KJ, Doroshow JH. Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J Biol Chem. 1986;261:3060–7. [PubMed] [Google Scholar]
- [375].Doroshow JH, Davies KJ. Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J Biol Chem. 1986;261:3068–74. [PubMed] [Google Scholar]
- [376].Keizer HG, Pinedo HM, Schuurhuis GJ, Joenje H. Doxorubicin (adriamycin): a critical review of free radical-dependent mechanisms of cytotoxicity. Pharmacology & therapeutics. 1990;47:219–31. [DOI] [PubMed] [Google Scholar]
- [377].Myers CE, Gianni L, Simone CB, Klecker R, Greene R. Oxidative destruction of erythrocyte ghost membranes catalyzed by the doxorubicin-iron complex. Biochemistry. 1982;21:1707–12. [DOI] [PubMed] [Google Scholar]
- [378].Mordente A, Meucci E, Silvestrini A, Martorana GE, Giardina B. Anthracyclines and mitochondria. Advances in experimental medicine and biology. 2012;942:385–419. [DOI] [PubMed] [Google Scholar]
- [379].Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, et al. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat Med. 2012;18:1639–42. [DOI] [PubMed] [Google Scholar]
- [380].Lebrecht D, Walker UA. Role of mtDNA lesions in anthracycline cardiotoxicity. Cardiovascular toxicology. 2007;7:108–13. [DOI] [PubMed] [Google Scholar]
- [381].Goormaghtigh E, Huart P, Brasseur R, Ruysschaert JM. Mechanism of inhibition of mitochondrial enzymatic complex I-III by adriamycin derivatives. Biochim Biophys Acta. 1986;861:83–94. [DOI] [PubMed] [Google Scholar]
- [382].Pointon AV, Walker TM, Phillips KM, Luo J, Riley J, Zhang SD, et al. Doxorubicin in vivo rapidly alters expression and translation of myocardial electron transport chain genes, leads to ATP loss and caspase 3 activation. PloS one. 2010;5:e12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [383].Montaigne D, Marechal X, Preau S, Baccouch R, Modine T, Fayad G, et al. Doxorubicin induces mitochondrial permeability transition and contractile dysfunction in the human myocardium. Mitochondrion. 2011;11:22–6. [DOI] [PubMed] [Google Scholar]
- [384].Zhou S, Starkov A, Froberg MK, Leino RL, Wallace KB. Cumulative and irreversible cardiac mitochondrial dysfunction induced by doxorubicin. Cancer research. 2001;61:771–7. [PubMed] [Google Scholar]
- [385].Ichikawa Y, Ghanefar M, Bayeva M, Wu R, Khechaduri A, Naga Prasad SV, et al. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J Clin Invest. 2014;124:617–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [386].Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. The New England journal of medicine. 2007;356:2457–71. [DOI] [PubMed] [Google Scholar]
- [387].Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders--past, present and future. Biochim Biophys Acta. 2004;1659:115–20. [DOI] [PubMed] [Google Scholar]
- [388].Liang C, Ahmad K, Sue CM. The broadening spectrum of mitochondrial disease: shifts in the diagnostic paradigm. Biochim Biophys Acta. 2014;1840:1360–7. [DOI] [PubMed] [Google Scholar]
- [389].Ellinger J, Albers P, Muller SC, von Ruecker A, Bastian PJ. Circulating mitochondrial DNA in the serum of patients with testicular germ cell cancer as a novel noninvasive diagnostic biomarker. BJU Int. 2009;104:48–52. [DOI] [PubMed] [Google Scholar]
- [390].Fernandes J, Michel V, Camorlinga-Ponce M, Gomez-Delgado A, Maldonado C, De Reuse H, et al. Circulating mitochondrial DNA level, a non-invasive biomarker for the early detection of gastric cancer. Cancer Epidemiol Biomarkers Prev. 2014. [DOI] [PubMed] [Google Scholar]
- [391].Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577; discussion e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [392].Zhong S, Ng MC, Lo YM, Chan JC, Johnson PJ. Presence of mitochondrial tRNA(Leu(UUR)) A to G 3243 mutation in DNA extracted from serum and plasma of patients with type 2 diabetes mellitus. J Clin Pathol. 2000;53:466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [393].Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. [DOI] [PubMed] [Google Scholar]
- [394].Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, Rainer TH, et al. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem. 2003;49:719–26. [DOI] [PubMed] [Google Scholar]
- [395].Pfeffer G, Horvath R, Klopstock T, Mootha VK, Suomalainen A, Koene S, et al. New treatments for mitochondrial disease-no time to drop our standards. Nat Rev Neurol. 2013;9:474–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [396].Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol. 2011;29:635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Suomalainen A, Elo JM, Pietilainen KH, Hakonen AH, Sevastianova K, Korpela M, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011;10:806–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [398].Davis RL, Liang C, Edema-Hildebrand F, Riley C, Needham M, Sue CM. Fibroblast growth factor 21 is a sensitive biomarker of mitochondrial disease. Neurology. 2013;81:1819–26. [DOI] [PubMed] [Google Scholar]
- [399].Enns GM, Moore T, Le A, Atkuri K, Shah MK, Cusmano-Ozog K, et al. Degree of glutathione deficiency and redox imbalance depend on subtype of mitochondrial disease and clinical status. PLoS One. 2014;9:e100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [400].Hoppins S, Nunnari J. Cell Biology. Mitochondrial dynamics and apoptosis--the ER connection. Science. 2012;337:1052–4. [DOI] [PubMed] [Google Scholar]
- [401].Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature reviews Molecular cell biology. 2013;14:283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [402].Ngoh GA, Papanicolaou KN, Walsh K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. The Journal of biological chemistry. 2012;287:20321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]