Abstract
Model-based phylogenetic reconstructions increasingly consider spatial or phenotypic traits in conjunction with sequence data to study evolutionary processes. Alongside parameter estimation, visualization of ancestral reconstructions represents an integral part of these analyses. Here, we present a complete overhaul of the spatial phylogenetic reconstruction of evolutionary dynamics software, now called SpreaD3 to emphasize the use of data-driven documents, as an analysis and visualization package that primarily complements Bayesian inference in BEAST ( http://beast.bio.ed.ac.uk , last accessed 9 May 2016). The integration of JavaScript D3 libraries ( www.d3.org , last accessed 9 May 2016) offers novel interactive web-based visualization capacities that are not restricted to spatial traits and extend to any discrete or continuously valued trait for any organism of interest.
Keywords: SpreaD3, SPREAD, BEAST, phylogenetics, phylogeography, Bayesian inference, visualization, Java, JavaScript, D3, Google Earth, KML, software, JSON, GeoJSON.
Evolutionary reconstructions that integrate various sources of information alongside sequence data are becoming increasingly widespread in statistical phylogenetics. Phylogenetic diffusion processes have been proposed to model the evolution of both discrete and continuous traits without the need to assume that the tree is known without error ( Ronquist 2004 ; Sanmartín et al. 2008 ; Lemey et al. 2009 , 2010 ). For instance, both MrBayes ( Ronquist et al. 2012 ) and BEAST ( Drummond et al. 2012 ) offer the capacity to simultaneously infer sequence and trait evolutionary processes, with the latter focusing entirely on time-calibrated phylogenies. These approaches have traditionally focused on spatial traits (e.g., in the context of phylogeography or biogeography), and the ability to infer spatiotemporal history has become particularly popular in infectious disease research. Although trait evolution can be readily visualized by mapping characters to phylogenetic nodes, spatial phylogenetic processes are visualized most explicitly by projecting trees in geographic space. This motivated the development of the SPREAD software ( Bielejec et al. 2011 ), which was mostly used as a tool to convert a summary tree or posterior set of trees with BEAST annotations to Keyhole Markup Language (KML) for visualization and animation of spatial diffusion through time in virtual globe software such as Google Earth. Here we present SpreaD3, an entirely new implementation of this software that mainly serves two purposes: To enable modern and interactive web-based visualization and to generalize this to any phylogenetic trait history of interest.
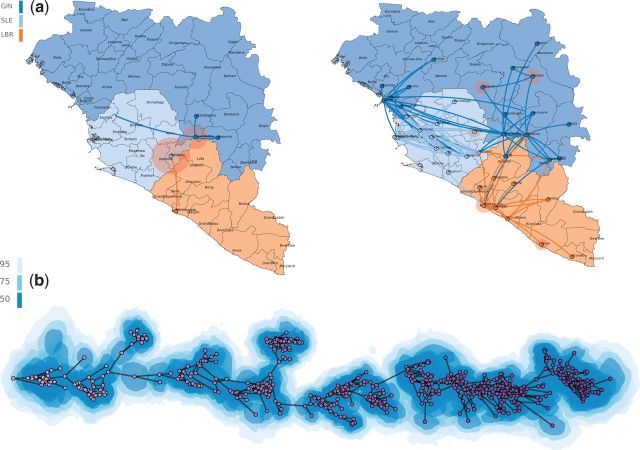
Apart from a graphical user interface to set up the web-based visualization, we also provide a command line interface for easy scripting purposes. A typical SpreaD3 analysis proceeds in two steps: The first step consists in parsing and analyzing the BEAST output in order to create a data file conforming to Java Script Object Notation (JSON) format. As with the previous version of the software, SpreaD3 can process a summary tree, typically a “maximum clade credibility tree,” with either discrete ( fig. 1 a ) or continuous trait annotations, a posterior distribution of trees with continuous trait annotations, or a log file containing a posterior distribution of rate indicators from a Bayesian stochastic search variable selection procedure. All the styling and display choices are delegated to the second rendering step, which avoids the need to reprocess the estimates when only a single element of the visualization needs to be modified.
Fig. 1.
Examples of D3 visualizations in SpreaD3. ( a ) Different stages of phylogeographic history of Ebola virus spread in western Africa under a discrete diffusion model. Tree branches are colored according to their parental locations, and circular polygon area is proportional to the number of tree lineages maintaining that location. ( b ) Antigenic drift dynamics of the influenza H3N2 inferred using a continuous diffusion model. Polygons surrounding branches denote 50%, 75%, and 95% highest posterior density intervals, giving a glimpse into the uncertainty of the inference.
This rendering step maintains support for KML output, but SpreaD3 is now also equipped with the capacity to create in-browser visualizations facilitated by the data-driven documents (D3; Bostock et al. 2011 ) JavaScript (JS) libraries. During the rendering step, a background geographic map in GeoJSON format ( www.geojson.org ) can be loaded, on which phylogeographic estimates can be overlaid ( fig. 1 a ). However, SpreaD3 can project trees in any arbitrary space, which allows for example producing two-dimensional plots of influenza antigenic evolution based on Bayesian multidimensional scaling estimates ( Bedford et al. 2014 ) ( fig. 1 b ). Such plots accommodate both mean estimates in continuous space as well as their uncertainty. Using the same generic visualization approach, any one-dimensional phenotypic trait evolving on a tree can also be plotted as a function of time (for an example, we refer to our online tutorial: http://rega.kuleuven.be/cev/ecv/software/SpreaD3 , last accessed 9 May 2016). JSON files created by different analyses can also be merged into a single visualization, allowing to combine any estimates of interest.
SpreaD3 employs a set of JS scripts for D3 rendering and creates a webpage in a user-specified location; the software seamlessly integrates with the default browser by automatically loading the visualization upon creation. In the browser, the user has interactive control over different visualization components based on the grammar of graphics ( Wilkinson 2005 ; Wickham 2010 ). Visualization settings can be based on the attributes associated with each component and color choices are provided by ColorBrewer palettes ( Harrower and Brewer 2003 ). The temporal dimension can be controlled by a time slider, and tree projections over time can be animated, paused, fast-forwarded, or re-winded. The in-browser visualization encoded in a stand-alone HTML document can be readily embedded in webpages, blogs, or social media and can be viewed on mobile devices. We also envisage these becoming interactive figures in online journals. Expert users can further fine-tune visualizations using the built-in JS console in modern browsers.
Compiled, runnable packages targeting all major platforms along with a tutorial and supplementary data are hosted at http://rega.kuleuven.be/cev/ecv/software/SpreaD3 , last accessed 9 May 2016. SpreaD3 is licensed under the GNU Lesser GPL and its source code is freely available from its repository: https://github.com/phylogeography/SpreaD3 , last accessed 9 May 2016.
Acknowledgments
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 278433-PREDEMICS and ERC grant agreement no. 260864, from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement nos. 634650-VIROGENESIS and 643476-COMPARE, from the National Institutes of Health (R01 AI107034, R01 HG006139, and LM011827), and the National Science Foundation (IIS 1251151 and DMS 1264153). F.B. is supported by a postdoctoral mandate from the Research Fund KU Leuven. G.B. acknowledges support from a Research Grant of the Research Foundation - Flanders (FWO; Fonds Wetenschappelijk Onderzoek - Vlaanderen).
References
- Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, McCauley JW, Russell CA, Smith DJ, Rambaut A. 2014. . Integrating influenza antigenic dynamics with molecular evolution . Elife 3 : e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F, Rambaut A, Suchard MA, Lemey P. 2011. . Spread: spatial phylogenetic reconstruction of evolutionary dynamics . Bioinformatics 27 ( 20 ): 2910 – 2912 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock M, Ogievetsky V, Heer J. 2011. . D3: data-driven documents . IEEE Trans Vis Comput Graph . 17 : 2301 – 2039 . [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. . Bayesian phylogenetics with BEAUti and the BEAST 1.7 . Mol Biol Evol . 29 ( 8 ): 1969 – 1973 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrower M, Brewer CA. 2003. . Colorbrewer.org: an online tool for selecting color schemes for maps . Cartogr J . 40 : 27 – 37 . [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. . Bayesian phylogeography finds its roots . PLoS Comput Biol . 5 ( 9 ): e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Welch JJ, Suchard MA. 2010. . Phylogeography takes a relaxed random walk in continuous space and time . Mol Biol Evol . 27 ( 8 ): 1877 – 1885 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. 2004. . Bayesian inference of character evolution . Trends Ecol Evol . 19 : 475 – 481 . [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. . MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space . Syst Biol . 61 ( 3 ): 539 – 542 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín I, van der Mark P, Ronquist F. 2008. . Inferring dispersal: a Bayesian approach to phylogeny-based island biogeography, with special reference to the Canary Islands . J Biogeogr . 35 : 428 – 449 . [Google Scholar]
- Wickham H. 2010. . A layered grammar of graphics . J Comput Graph Stat . 19 ( 1 ): 3–28. [Google Scholar]
- Wilkinson L. 2005. . The grammar of graphics (statistics and computing). Secaucus (NJ: ): Springer-Verlag New York, Inc; . [Google Scholar]