Abstract
Birds can be classified into altricial and precocial. The hatchlings of altricial birds are almost naked, whereas those of precocial birds are covered with natal down. This regulatory divergence is thought to reflect environmental adaptation, but the molecular basis of the divergence is unclear. To address this issue, we chose the altricial zebra finch and the precocial chicken as the model animals. We noted that zebra finch hatchlings show natal down growth suppressed anterior dorsal (AD) skin but partially down-covered posterior dorsal (PD) skin. Comparing the transcriptomes of AD and PD skins, we found that the feather growth promoter SHH (sonic hedgehog) was expressed higher in PD skin than in AD skin. Moreover, the data suggested that the FGF (fibroblast growth factor)/Mitogen-activated protein kinase (MAPK) signaling pathway is involved in natal down growth suppression and that FGF16 is a candidate upstream signaling suppressor. Ectopic expression of FGF16 on chicken leg skin showed downregulation of SHH, upregulation of the feather growth suppressor FGF10, and suppression of feather bud elongation, similar to the phenotype found in zebra finch embryonic AD skin. Therefore, we propose that FGF16-related signals suppress natal down elongation and cause the naked AD skin in zebra finch. Our study provides insights into the regulatory divergence in natal down formation between precocial and altricial birds.
Keywords: feather evolution, natal down, precocial bird, altricial bird
Introduction
Birds are the most diversified terrestrial vertebrates, with over 10,000 known living species (Gill and Wright 2006). Among the evolutionary novelties in birds, the feathers show the highest degree of diversity, providing an excellent model for studying how animals adapt to different environments (Prum and Brush 2002; Prum 2005; Chen et al. 2015; Strasser et al. 2015). Natal down, the downy plumage in hatchlings, is a distinct character to discriminate between altricial and precocial birds (Starck and Ricklefs 1998). Altricial hatchlings have little or no downy plumage in their skin, whereas precocial hatchlings are covered by downy feather. According to the phylogenetic distribution of altricial and precocial birds (Starck and Ricklefs 1998) and the discovery of a precocial avian embryo fossil (Zhou and Zhang 2004), the precocial phenotype is the ancestral state.
Feather distributions in avian skin have two spatial patterns: In the macropattern the feather tracts are separated by bare skin, while the micropattern shows regular spacing between individual feathers (Olivera-Martinez et al. 2004; Mou et al. 2011). The periodic skin micropatterning is achieved by the action of opposing feather growth activators and inhibitors to form a reaction diffusion mechanism (Meinhardt and Gierer 2000; Mou et al. 2011). The epithelio-mesenchymal molecular interactions between the dermis and the overlying epidermis coordinate the spatial arrangement and regular outgrowth of feathers (Hornik et al. 2005; Mou et al. 2011; Wells et al. 2012).
Many molecules that regulate feather formation have been identified. For example, Wingless (WNT)/β-catenin signaling and cDermal-1 are promoters at the early stages of skin patterning (Noramly et al. 1999; Widelitz et al. 2000; Hornik et al. 2005); FGFs (fibroblast growth factors) and SHH (sonic hedgehog) are promoters or activators, while BMPs (bone morphogenetic proteins) are inhibitors in feather placode formation (Jung et al. 1998; Mandler and Neubuser 2004; McKinnell et al. 2004; Song et al. 2004). Furthermore, the genes underlying some partial or full featherless mutants have also been characterized in chicken (Gallus gallus). The regulatory differences in BMPs cause the naked neck phenotype in chicken, and a nonsense mutation in FGF20 is associated with the featherless trait (Mou et al. 2011; Wells et al. 2012). However, the current knowledge of the molecular and cellular basis of feather morphogenesis is largely based on studies in chicken, which is a precocial bird. Little is known about the developmental biology of altricial feathers. Understanding the molecular mechanisms in natal down growth suppression in altricial birds may reveal the genetic basis underlying the altricial–precocial bird divergence.
Zebra finch (Taeniopygia guttata) and chicken are typical altricial and precocial birds, respectively. Zebra finch belongs to Passeriformes, which is the largest avian order, and all species in this order are altricial birds. Zebra finch hatchlings show both naked anterior dorsal (AD) skin and natal down covered posterior dorsal (PD) skin regions, providing a great model for studying the gene regulatory differences in natal down growth suppression and promotion. Chicken is the best studied avian model and is to date the only bird with a stable gene manipulation system (Sang 2004). Thus, zebra finch and chicken are two good models for identifying the causative elements of the natal down differences between altricial and precocial hatchlings.
In this study, we first characterized the differences in feather formation between the AD and the PD regions in zebra finch embryos. Second, we analyzed the transcriptomes of the AD and PD regions during zebra finch embryo development using RNA-seq and inferred that the SHH and FGF/MAPK signaling pathways are involved in the developmental differences between the two regions. Third, we applied the RCAS (Replication-Competent ASLV long-terminal repeat with a Splice acceptor; Hughes 2004) transformation system to chicken to functionally validate that FGF16 is a natal down growth suppressor.
Results
Two Types of Natal Down Formation in Zebra Finch Embryos
In zebra finch hatchlings, the AD, alar, caudal, and ventral regions are naked, while the PD, capital, humeral, and femoral regions are partially covered by natal down (fig. 1 and supplementary fig. S1, Supplementary Material online). In chicken hatchlings, the skin is covered by natal down. We compared the natal downs of zebra finch and chicken and found that they share similar nodes and branches (supplementary fig. S2, Supplementary Material online), suggesting that they are homologous. However, the natal down of zebra finch is softer and looser than that of chicken.
Fig. 1.

The morphologies and paraffin sections of dorsal natal down in zebra finch and chicken. (A) Dorsal view of the feather tracts in a zebra finch hatchling. Open circles and black circles denote feather buds. (B) Type I (open circles) and Type II (black circles) feather formations. (C–Q) The morphologies and the paraffin sections with H&E staining of the natal downs in AD (Type I) and PD (Type II) skins in E8 (C–G), E9 (H–L), E12 (M–Q), and D1 (R and S) zebra finch. (R) Red arrow indicates the AD region for the section in S. (T–Z) The morphologies and the paraffin sections with H&E staining of the natal downs in AD and PD skins in E12 (T–X) and D1 (Y and Z) chicken. (Y) Red arrow indicates the AD region for the section in Z. ep, epithelium; me, mesenchyme; fb, feather bud; ff, feather follicle; MDF, mature downy feather. (C, H, M, R, T) Scale bar: 2 mm, other scale bars: 100 μm.
To characterize the naked and downy tracts in zebra finch, we studied the feather development in zebra finch hatchlings. We focused on the dorsal tract because it showed discrete feather formation. In the AD tract and two flanks of the PD tract, the feather development does not go through the natal down stage, and the contour feathers develop directly from the feather buds around D7 (Type I; open circles in fig. 1A and B, supplementary fig. S1, Supplementary Material online). In contrast, in the middle stripe of the PD tract and other regions labeled with black circles, the feather buds formed natal down before the growth of the contour feathers, same as the natal down formation process in chicken (Type II; solid black circles in fig. 1A and B, supplementary fig. S1, Supplementary Material online).
To study the developmental differences between Type I and Type II feather buds, we compared the AD and PD regions at different stages of zebra finch and chicken embryos. In E8 zebra finch, all AD and PD tracts formed feather buds (fig. 1C–G). In E9 zebra finch, the growth of Type I feather buds was suppressed (fig. 1I and J), whereas Type II feather buds kept elongating (fig. 1K and L). In E12 zebra finch, Type I feather buds invaginated into the skin but did not elongate (fig. 1N and O), whereas the Type II feather buds invaginated into the skin and elongated (fig. 1P and Q). The phenotype of Type II feather buds in E12 zebra finch was similar to that of the AD and PD feather buds in E12 chicken, that is, the natal downs were keratinized, pigmented, and elongated (fig. 1T–X). Furthermore, in newborn zebra finches, the feather buds in the AD region developed follicle structure, but did not protrude out of the skin (fig. 1R and S). Compared with hatchling chicken embryos (fig. 1Y and Z), the down feather in the zebra finch AD region already reached the resting phase.
Anterior Dorsal Interbud Region Thickening
To dissect the phenotypic differences between zebra finch and chicken dorsal skins, paraffin sections were made to compare the histological differences. In E12 zebra finch embryos, the epithelium of interbuds in the AD skin (fig. 1N and O), where Type I feathers are formed, was significantly thicker than that of the PD skin (fig. 1P and Q), where Type II feathers are formed (AD: 21.29 ± 0.51 μm vs. PD: 12.96 ± 2.27 μm, P < 0.05, Student’s t-test). In contrast, no significant difference could be detected between AD (fig. 1S and T) and PD skins (fig. 1U and V) in E12 chicken embryos (AD: 8.75 ± 0.38 μm vs. PD: 8.71 ± 0.59 μm, P > 0.05, Student’s t-test). Moreover, the dorsal epithelia were, on average, thicker in zebra finch than in chicken (P < 0.05, Student’s t-test; supplementary fig. S3, Supplementary Material online).
To understand the temporal changes in feather development, we used the immunostaining with PCNA (proliferating cell nuclear antigen) to detect the cell proliferation regions. In AD and PD sections of zebra finch and chicken embryos at E8, E9, and E10, the PCNA signals were enriched in the epithelia of interbuds and feather buds, indicating high cell proliferation in these regions (fig. 2). In E8 and E10, the cell arrangements of interbuds were similar between AD and PD skins in both zebra finches and chickens (E8, fig. 2A–D; E10, fig. 2M–P). In E9 zebra finches, however, the epithelia of AD interbuds showed an irregular cell arrangement compared with those of PD interbuds (fig. 2E and I vs. F and J). No such divergence pattern could be detected in the same regions of E9 chicken embryos (fig. 2G and K vs. H and L). The data also showed higher PCNA signals in the epithelia of PD feather buds in E9 and E10 zebra finches (fig. 2Q) and in that of AD interbuds in E9 zebra finches (fig. 2S), while no significant difference could be detected between the two regions of chicken embryos (fig. 2R and T). To confirm the results, we used the immunostaining with the epithelia cell marker CDH1 (E-cadherin) in the zebra finch paraffin sections. Consistent with PCNA staining, the epithelia of interbud regions were thicker in AD skins than in PD skins (supplementary fig. S4, Supplementary Material online). From these histological studies, we conclude that Type I and II feather formations in zebra finch embryos undergo different growth regulation. Type I feather buds skip the elongation and downy steps, suggesting that growth suppressors exist in Type I feather buds in zebra finch embryos.
Fig. 2.

Paraffin sections with PCNA staining and the quantification of proliferating cells in chicken and zebra finch dorsal skins at E8, E9, and E10. (A–D) The anterior and posterior dorsal skin of E8 embryos. (E–H) The anterior and posterior dorsal skin of E9 embryos. (I–L) The enlargement of interbud regions of E–H. (M–P) The anterior and posterior dorsal skin of E10 embryos. (Q–T) Statistics of the proliferating cells in the epithelium of dorsal skins of zebra finch and chicken. (Q, R) The PCNA cell number per 100 μm square in feather buds of zebra finch and chicken at E8, E9, and E10. (S, T) The PCNA cell number per 100 μm square in interbuds of zebra finch and chicken at E8, E9, and E10. Scale bar: 50 μm. ep, epithelium; me, mesenchyme; fb, feather buds; **P < 0.01 (Student’s t-test).
Transcriptomes of AD and PD Regions
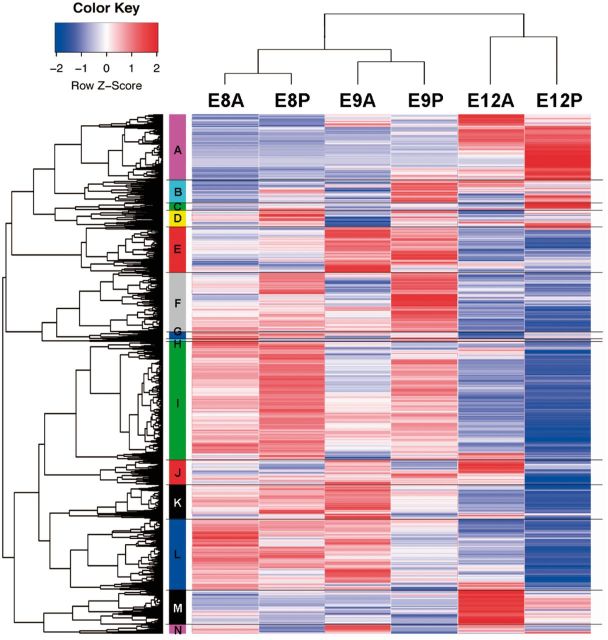
To identify the regulatory differences between Type I and II feather formations, we dissected the AD and PD skins of zebra finch embryos at E8, E9, and E12, and obtained six transcriptomes (E8A, E8P, E9A, E9P, E12A, and E12P), using RNA-seq. The sequencing reads from the six transcriptomes were mapped to the zebra finch genome (the statistics of sequencing reads are given in supplementary table S1, Supplementary Material online). Among the 18,619 annotated genes, 13,362 had an FPKM (fragments per kilobase of exon per million fragments mapped) value > 1 in at least 1 transcriptome, and they were defined as expressed genes. To evaluate the reliability of RNA-seq data, we measured the expression level of 40 randomly selected genes by the Nanostring technology. A high correlation between RNA-seq and Nanostring data suggested high reliability of the RNA-seq data (R2 = 0.83–0.89; supplementary fig. S5, Supplementary Material online).
Hierarchical clustering analysis clustered the 13,362 expressed genes into 14 clusters. For the six transcriptomes, three clusters were formed for the three embryonic stages, that is, transcriptomes E8A and E8P in one cluster, transcriptomes E9A and E9P in another cluster, and transcriptomes E12A and E12P in a third cluster, suggesting that regional differences in gene expression profiles were smaller than developmental stage differences (fig. 3). A description of the analysis of the 14 clusters is given in supplementary results, Supplementary Material online.
Fig. 3.
Clustering analysis of the transcriptomes and the expression heat map. Hierarchical clustering analysis clustered the 13,362 expressed coding sequences into 14 clusters (A–N; see supplementary tables for details). The expression values of each gene are shown as the scaled FPKM values across the six transcriptomes (scaled z-score: Red, upregulation; blue, downregulation).
Differential SHH Expression between Type I and II Feather Formations
According to the transcriptomic analysis above, we considered SHH a factor for growth divergence between Type I and II feather formations (supplementary results, Supplementary Material online). To study the expression profile of SHH in the zebra finch embryos, we quantified its expression levels at different embryonic stages. Quantitative Polymerase chain reaction (PCR) data showed that SHH differentially expressed between AD and PD skins at E9 and E10 (fig. 4A). The differential expression disappeared at E12, when the natal down elongation was completed (fig. 4A).
Fig. 4.
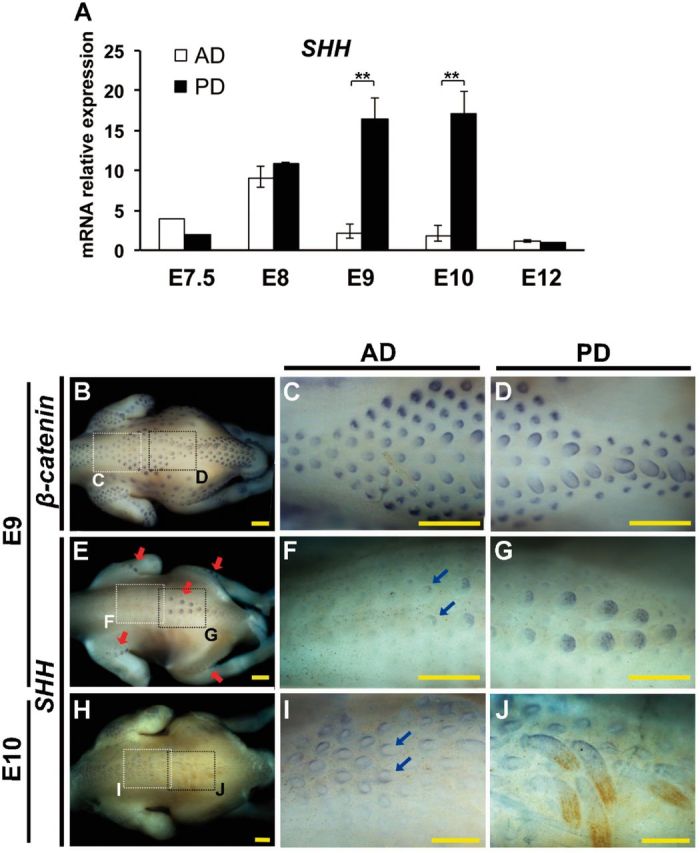
Differential expression of SHH between Type I and Type II feather formations in zebra finch. (A) Quantification of SHH mRNA expression of AD and PD skin regions at different embryonic stages. Relative expression values were given as mean ± standard deviation from at least three independent experiments. **P < 0.01 (Student’s t-test). (B–J) Whole mount in situ hybridization of β-catenin and SHH in zebra finch embryos. (B–D) β-catenin in E9 embryos. (E–G) SHH in E9 embryos. (H–J) SHH in E10 embryos. The enlargements of anterior and posterior dorsal skins were indicated by the white (C, F, and I) and black dotted-line (D, G, and J) squares in B, E and H. Red arrows indicate the expression locations of SHH. Blue arrows indicate the disruptive expression patterns of SHH. Scale bar: 0.5 mm.
To visualize the differential expression of SHH between Type I and II feather buds, whole mount in situ hybridization was conducted in the E9 zebra finch embryos, using β-catenin, a known initiation signal for feather bud formation (Noramly et al. 1999) and with little differential expression in our transcriptomes (supplementary table S3, Supplementary Material online), for the experimental control (fig. 4B–D). In E9 zebra finch embryos, the expression of SHH was restricted to the posterior end of the Type II feather buds (fig. 4E and G), the same as that in chicken feather buds (McKinnell et al. 2004), suggesting that chicken and zebra finch share homologous natal down. However, Type I feather buds showed a lower level of SHH expression than Type II feather buds (fig. 4F vs. G). Moreover, in Type I feather buds of E10 zebra finch embryos, the expression of SHH lost its regular posterior polarity (blue arrows in fig. 4F and I), implying that the function of SHH was disrupted in feather bud elongation.
FGF16 Suppresses Natal Down Growth and Thickens the Epithelium through the FGF/MAPK Pathway
From the above transcriptome analysis, we noted that FGF10, SNAI1, and TWIST2, which belong to the Gene ontology (GO) category of epithelial development, showed a higher expression level in AD skin than in PD skin in zebra finch embryos (Cluster L, fig. 3, supplementary results, Supplementary Material online), but the homologs of these genes in chicken showed little differential expression in our quantitative PCR data (supplementary fig. S6, Supplementary Material online). The same comment applies to FGF16, which is in the GO category of the MAPK signaling pathway (Cluster N, fig. 3, supplementary results, Supplementary Material online).
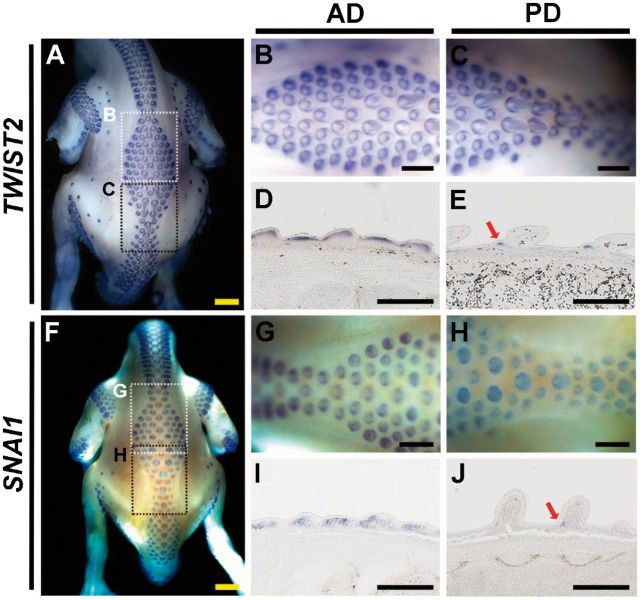
SNAI1 and TWIST2 were two highly expressed genes in the transcriptome and so were selected for whole mount in situ hybridization in zebra finch embryos. By whole mount in situ hybridization in E9 zebra finch, we found that the expression of TWIST2 was throughout the dermis of Type I feather buds (fig. 5A, B, and D), but was restricted to the anterior bottom dermis of Type II feather buds (fig. 5A, C, and E). A similar expression profile was detected in SNAI1 (Type I feather buds: fig. 5F, G, and I; Type II feather buds: fig. 5F, H, and J), which is a zinc finger transcription factor for regulating epithelial to mesenchymal transition during embryonic development (Paznekas et al. 1999). These data support the association between our predicted genes and the feather bud growth suppression.
Fig. 5.
Whole mount in situ hybridization of TWIST2 (A–E) and SNAI1 (F–J) in E9 zebra finch and the paraffin sections. (B, C) The enlargements of the dotted-line square regions in A. (G, H) The enlargements of the dotted-line square regions in F. (D, E) The paraffin sections of feather buds of B and C, respectively. (I, J) The paraffin sections of feather buds of G and H, respectively. Red arrows indicate the restrictive expression pattern of TWIST2 and SNAI1 in Type II feather buds. Scale bar: 0.5 mm.
In feather development, the MAPK signaling pathway was shown to be the major downstream pathway in response to FGFs (Lin et al. 2009). Moreover, knockout of the key component of the MAPK pathway reduced epithelium thickness in mouse (Scholl et al. 2007). These studies suggest that the FGF/MAPK pathway participates in the natal down growth suppression. FGF16 is a known upstream signal of SNAI1 in promoting ovarian cancer cell invasion through activation of the MAPK signaling pathway (Basu et al. 2014). Therefore, we hypothesized that upregulation of FGF16 in AD skin suppresses natal down growth and increases epithelium thickness.
To test this hypothesis, we utilized the RCAS retrovirus to overexpress the FGF16 gene in chicken embryos. Because injecting FGF16 cDNA into the chicken AD skin region caused high lethality (data not shown), we injected it into the legs instead. In each chicken embryo, one leg was injected with the virus carrying the FGF16 cDNA, while the other leg was used as the control. We found that FGF16 overexpressed legs exhibited a similar phenotype of the zebra finch AD skin region: Periodic feather buds were formed, but natal down elongation was suppressed (fig. 6A, C, and E). The natal down elongation in the control leg was normal (fig. 6B, D, and F). Moreover, bone formation was also influenced by FGF16 overexpression (fig. 6A), supporting a previous prediction (Laurell et al. 2014). In the paraffin sections of the skin, both the Hematoxylin and eosin stain (H&E) stain and the immunostaining with CDH1 showed thicker epithelia in the FGF16 overexpressed leg skin than in the control leg skin (fig. 6E and G vs. F and H; statistics in fig. 6J).
Fig. 6.

FGF16 overexpression suppressed the natal down growth, reduced the bone length, and increased the epithelial thickness in E12 chicken. (A, B) The chicken embryo was microinjected with RCAS-FGF16 in one leg, and the other leg was used as the control. (C, D) The enlargement images of A and B, respectively. (E, F) H&E stains of the paraffin sections of FGF16 overexpressed and control skins. (G, H) Immunochemical stain with CDH1 in the paraffin sections of the FGF16 overexpressed and the control skins. (I) AMV-3C2 staining of adjacent sections showing the RCAS virus infected regions. (J) Quantification of the epithelium thickness between the FGF16 overexpressed (white bar) and the control epithelia (black bar). ep, epithelium; me, mesenchyme; fb, feather bud. **P < 0.01 (Student’s t-test). Yellow scale bar: 1 cm, black scale bar: 100 μm.
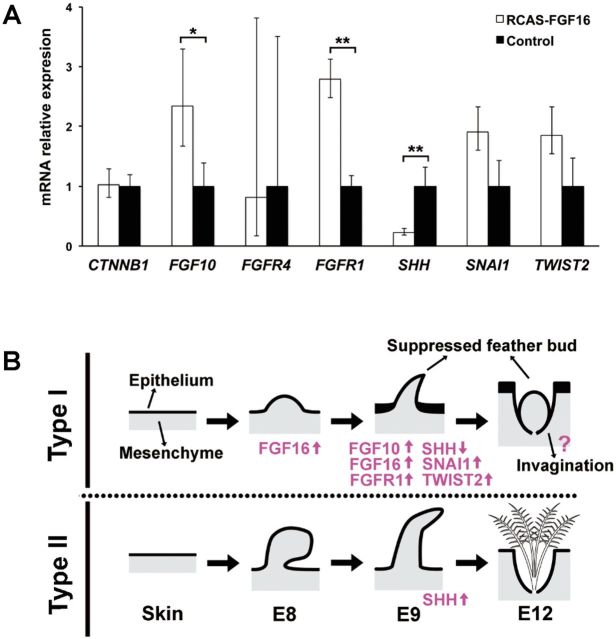
To understand how FGF16 suppresses natal down elongation, we studied the expression patterns of several genes in FGF16 overexpressed skins by quantitative PCR. The expression of FGFR1 was upregulated, whereas β-catenin (CTNNB1) and FGFR4 were not affected (fig. 7A) by FGF16 overexpression. This observation suggests interaction between FGF16 and FGFR1. Although FGFR1 was not in our list of differentially expressed genes (DEGs), the transcriptome data showed 1.6-fold higher expression of FGFR1 in the AD than in PD skin of E9 zebra finch (supplementary table S3, Supplementary Material online). Moreover, SNAI1 and TWIST2 were also upregulated in the FGF16 overexpressed skin (fig. 7A), although the differences were not significant. Interestingly, FGF10 was upregulated, while SHH was downregulated in the FGF16 overexpressed skin (fig. 7A).
Fig. 7.
The quantification of the candidate genes for natal down growth suppression, and the summary diagram of Type I and Type II feather formations. (A) The gene expressions in the FGF16 overexpressed (white bar) and control (black bar) skins in chicken were compared by quantitative PCR. Relative expression values were given as mean ± standard deviation from at least three independent experiments. *P < 0.05; **P < 0.01 (Student’s t-test). (B) The summary diagram of Type I and Type II feather formations, and the involved transcription factor genes.
To test the relationship between FGF10 and FGF16, FGF10 was overexpressed in the dorsal skins of chicken embryos, resulting in the suppression of the natal down formation (supplementary fig. S7A–D, Supplementary Material online), but the expression of FGF16 was not affected (supplementary fig. S7E, Supplementary Material online). These observations suggest that FGF10 is not a regulator but a target of FGF16. Thus, we conclude that FGF16 suppresses the natal down elongation through the FGF/MAPK pathway (FGF10, FGFR1, SNAI1, and TWIST2) and the downregulation of SHH (fig. 7B).
Discussion
In this study, we first defined two types of natal down formation in the dorsal skin of zebra finch, in contrast to only one type in chicken. The absence of natal down in Type I feather formation signifies the altricial phenotype in zebra finch. Previous studies found that the naked skins in the sc/sc and naked neck chicken were due to the abolishment of feather bud formation (Mou et al. 2011; Wells et al. 2012). However, in zebra finch hatchlings we found typical feather buds in some regions of the skin (fig. 1), suggesting that a different regulatory mechanism suppresses feather growth. Moreover, according to the expression patterns of SHH (fig. 4), the difference between AD and PD skin regions at the developmental stages studied is not due to heterochrony because AD skin never grows natal down. We utilized the comparative transcriptome approach to infer that molecules in the FGF/MAPK pathway are involved in the natal down growth suppression and epithelial thickening, leading to naked AD skin regions in zebra finch hatchlings.
FGFs are key players in the processes of proliferation and differentiation of a wide variety of animal cells and tissues (Ornitz and Itoh 2001). In feather elongation, FGFs may play two opposite functions. Some, such as FGF2 and FGF4 (Widelitz et al. 1996; Song et al. 2004), may induce or promote feather growth, while others, such as FGF10 and FGF16, may play a suppressor role (Tao et al. 2002; Yue et al. 2012). Overexpression of FGF10 thickens the epithelium, upregulates NCAM, and downregulates SHH. FGF10 suppresses the chicken natal down growth through the epithelium/mesenchyme signaling interaction (Tao et al. 2002), leading to a phenotype similar to that in zebra finch AD skin in which periodic feather germs are formed, but feather elongation is suppressed.
The natal down growth suppressors showed functional conservation between different skin regions and between avian species. In chicken, FGF10 was shown to suppress the natal down growth in the leg skin previously (Tao et al. 2002) and in dorsal skin in this study (supplementary fig. S7, Supplementary Material online). In our transcriptome analysis, FGF10 expressed higher in AD skin than in PD skin of zebra finch embryos (supplementary table S3, Supplementary Material online), suggesting a role in natal down growth suppression. On the other hand, FGF16 expressed higher in AD skin than in PD skin of zebra finch embryos (supplementary table S3, Supplementary Material online), and suppressed the natal down elongation in the leg skin of chicken embryo, suggesting a role in natal down growth suppression in altricial hatchlings. However, due to experimental limitations in zebra finch, we are unable to overexpress or knock down FGF16 in zebra finch. Furthermore, due to the low expression of FGF16 in zebra finch (FPKM value = 1–8), we were unable to distinguish noise from the true signal in situ hybridization. Therefore, we cannot rule out the possibility that the natal down growth was indirectly suppressed due to the wide range of FGF16 overexpression by the RCAS system. More experimental innovations are needed to address these issues.
TWIST2 is known to be a feather growth initiator, but overexpression of TWIST2 induced thickened dermis with normally shaped ectopic feather buds (Hornik et al. 2005). There are two possible explanations for its role in natal down suppression. First, other molecules such as SNAI1 that showed coexpression with TWIST2 may work in a combined action manner. As suggested by a previous study (Oh et al. 2004), the combined action of modest inhibitors can abolish the function of MAPKs. Moreover, the differential expression of TWIST2 might be the consequence, but not the cause of feather bud growth suppression. The continued expression of TWIST2 in Type I feather buds in E9 zebra finch might be due to a pleiotropic effect of FGF16 overexpression.
We should also point out that the developmental process of natal down is diverse among altricial birds. For example, in most finches, the natal down development is finished at hatchlings, but in the parrots, the natal down growth continues after hatching (our observation, data not shown). Furthermore, when we mapped the altricial and precocial phenotypes onto the recently published avian phylogeny (Zhang et al. 2014), we found that the altricial–precocial transition occurred multiple times in the past 70 million years, as previously proposed (Starck and Ricklefs 1998). Although the precocial phenotype is considered ancestral to the altricial phenotype, some precocial orders, such as ciconiiformes and gruiformes, are clustered with altricial lineages, while some altricial orders, such as cuculiformes and apodiformes, are clustered with precocial lineages (Starck and Ricklefs 1998; Zhang et al. 2014). Thus, different mechanisms may act in the natal down growth regulation in birds. Whether the FGF/MAPK signaling pathway is utilized as the natal down growth suppressor in all altricial birds needs to be investigated.
The feather bud elongation in AD skin of zebra finch embryos stopped at around E9, and the phenotype of the suppressed feather bud is similar to that in FGF16 overexpressed chicken skins (figs. 1N and 6C). However, the epithelium invagination and feather follicle formation still proceed in the AD skin of E12 zebra finch embryos (fig. 1O), but not in FGF16 overexpressed chicken skins (fig. 6E). This difference suggests that overexpression FGF16 may suppress invagination and follicle formation or the FGF/MAPK pathway is not the only factor for natal down growth suppression. More works remain to be investigated to identify the whole regulatory network of altricial feather suppression.
The natal down divergence between altricial and precocial hatchlings is thought to be associated with heat transfer and conservation (Starck and Ricklefs 1998; Bicudo 2010). In altricial hatchlings, most of their body heat is conferred by the parents, and the naked dorsal skin is thought to be associated with heat transduction (Starck and Ricklefs 1998). The cornified epidermal keratinocytes, such as the feathers of birds and the hairs of mammals, are essential for the adaptation of the terrestrial animals (Strasser et al. 2015). We found that epithelial thickening is a phenotype in featherless AD skin of zebra finch hatchlings. In the naked mole-rat, lack of fur is compensated by a thicker epidermal layer and a marked reduction in sweat glands (Daly and Buffenstein 1998). Similar mechanisms might be shared between these naked organisms for environmental adaptation.
The evolution of feathers was so successful as to enable the birds to become the most diverse amniotes. However, like the recurrent losses of limb or eye in animal evolution (Lande 1978; Protas et al. 2011), feather evolution is not unidirectional. Fossil records showed that most ancestral birds had flight feathers on their legs, but this phenotype is rare in modern birds (Dhouailly 2009; Zheng et al. 2013). The loss of the leg flight feather might have enhanced flight ability (Dial et al. 2008). Our study provided another case of feather growth suppression. Our view is that the feather growth suppression during Type I feather formation is due to the overexpression of specific suppressors, but not due to the functional loss of the feather growth promotors. The evolution of feather growth suppressors implies that feather growth may sometimes lower the species fitness. Furthermore, the saved energy of feather growth can be allocated to the development of other organs, such as the post hatch fast brain growth (Starck and Ricklefs 1998). Together, these evolutionary novelties may have made the passerine birds the most diverse avian species.
Materials and Methods
Ethics Statement
All the animal experiments in this study were conducted according to the protocol approved by the Institutional Animal Care and Use Committees of National Chung Hsing University (Taichung, Taiwan).
Eggs and Animals
Pairs of adult zebra finches were purchased from a breeder in Tainan, Taiwan, and their fertilized eggs were collected for the study. The white leghorn chicken was used as the precocial bird model to avoid blocking the signal from in situ hybridization by feather pigmentation. The pathogen-free fertilized chicken eggs were obtained from the farm of National Chung Hsing University. All the eggs used were incubated at 38 °C and in 65% relative humidity until the specific stages. The stages and corresponding incubation days of zebra finch embryos followed the description of Murray et al. (2013), and the stages and corresponding incubation days of chicken embryos followed the description of Hamburger and Hamilton (1992). The corresponding stages between chicken and zebra finch embryos followed the supplementary description of Abzhanov et al. (2004). The chicken and zebra finch showed similar development within E12.
Paraffin Section and Immunohistochemistry
The chicken and zebra finch embryos were fixed in 4% paraformaldehyde at 4 °C overnight. Paraffin sections of 5 μm were conducted following the procedure of Chuong (1998). For immunohistochemical stain, PCNA antibody was purchased from Chemicon (CBL407), AMV-3C2 antibody was from Developmental Studies Hybridoma Bank, and CDH1 antibody was from BD Biosciences (610182). 4',6-diamidino-2-phenylindole (DAPI) was used to visualize the nuclei.
Tissue Total RNA Isolation
The dissected skin was immersed at 4 °C overnight for penetration by RNALater solution (Ambion) and then stored at −20 °C before isolation of total RNA. After thawing, the samples were homogenized by MagNA Lyzer (Roche). Total RNA was extracted using the MasterPure Complete DNA and RNA Purification kit (Epicentre). The 30 min DNase1 treatment was carried out at room temperature as described in the manual to remove the DNA thoroughly.
Quantitative PCR
To quantify the candidate gene expressions, the cDNAs were synthesized from the total RNA by QuaniTect Reverse Transcription kit (Qiagen). Each cDNA sample containing SYBR green (KAPA SYBR FAST qPCR kit) was run on LightCycler 480 (Roche) under the appropriate conditions. Quantification of the TATA box binding protein RNA was used to normalize target gene expression levels. All the PCR primers are listed in supplementary table S11, Supplementary Material online.
mRNA Whole Mount In Situ Hybridization
Gene-specific fragments were amplified from RNA extracted from dorsal skins of chicken and zebra finch embryos and subsequently cloned into pGEM-T Easy vector system (Promega, A1360). Both antisense and sense RNA probes were made by in vitro transcription according to manufacturer’s instructions (Roche, Cat #11277073910). Whole mount in situ hybridization was performed using nonradioactive in situ hybridization according to the procedure described in Chuong et al. (1996). PCR primers for the cDNA amplifications are listed in supplementary table S12, Supplementary Material online.
Stranded RNA Sequencing
Total RNA concentrations from six samples (E8A, E8P, E9A, E9P, E12A, E12P; five zebra finch individuals for each sample) were measured by Qubit fluorometer (Invitrogen USA), and quality was assessed by BioAnalyzer 2100 RNA Nano kit (Agilent, USA). The samples had RNA integrity number (RIN) values ranging from 8.7 to 9.3. Three microgram of total RNA from each sample was used for library construction using the TruSeq Stranded mRNA Sample Prep Kit (Illumina, USA), following the manufacturer’s instructions with some modifications described below. Briefly, poly-A mRNAs were purified and fragmented at 94 °C for 7.5 min. The first-strand cDNAs were synthesized by random priming, and the second strand was synthesized using Deoxynucleotide (dNTP) mixture containing Deoxyuridine Triphosphate (dUTP) in place of Deoxythymidine triphosphate (dTTP). The double-stranded cDNA fragments were then processed by end repair, 3′-adenylation, and ligation to barcoded adaptors. The dUTP-containing second-strand cDNAs were degraded by uracil-specific excision reagent enzyme (NEB, USA), and the remaining first-strand cDNA fragments were amplified by 12 cycles of PCR using KAPA HiFi HotStart ReadyMix PCR Kit (Kapa Biosystems, USA). The final libraries were cleaned up by Agencourt AMPure XP beads (Beckman Coulter, USA), and the concentrations were determined by Qubit RNA assay (Invitrogen). The library profiles showed a distribution at 250–700 bp as visualized by BioAnalyzer High Sensitivity DNA kit (Agilent, USA). The molar concentrations were normalized by Quantitative real time polymerase chain reaction (qPCR) using the Kapa SYBR Fast qPCR Kit Master mix against the Library Quantification DNA Standards (Kapa Biosystems, USA) on LightCycler 480 (Roche, USA). Deep sequencing of paired-end 2 × 101 nt was carried out on HiSeq2500 (Illumina) by six-plex pooling in a total of three lanes. The Illumina library construction and sequencing was conducted by High Throughput Genomics Core of the Biodiversity Research Center, Academia Sinica, Taiwan.
Data Processing and Reads Mapping
Low-quality bases and reads were removed by using Trimomatic version 0.30 (Bolger et al. 2014) according to the following procedure: 1) Remove adaptors; 2) remove leading low quality bases below quality 3; 3) remove trailing low quality bases below quality 3; 4) scan the read with a 4-base wide sliding window, cutting when the average quality per base drops below 20; and 5) drop trimmed reads below 36 bases long. Besides, we trimmed all the paired-end sequencing reads from both ends of each cDNA fragment to 99 bp to reduce sequencing errors.
The zebra finch genome (version Taeniopygia_guttata.taeGut3.2.4) and its gene annotations were downloaded from Ensembl FTP. The processed sequencing reads were mapped to the genome using Tophat version 2.0.8 (Trapnell et al. 2009), and its embedded aligner Bowtie version 2.1.0 (Langmead et al. 2009) with the following parameters: -N 3 –read-edit-dist 3 –no-novel-juncs –library-type fr-firststrand. The normalized expression levels of genes, represented by FPKMs (Mortazavi et al. 2008), were generated by Cufflinks version 2.1.1 (Trapnell et al. 2013) with the following parameters: –max-bundle-frags 1012 –multi-read-correct –library-type fr-firststrand.
Clustering Analysis and Identification of DEGs
In the clustering analysis, a gene is said to be expressed if its FPKM value is higher than 1 in at least one transcriptome. All expressed genes were hierarchically clustered by weighted pair-group method with arithmetic mean method in the R package. The clustering is shown in figure 3, with each clustered cluster in different color. The cut-off for the cluster analysis is given in supplementary figure S8, Supplementary Material online. We identified the DEGs through three sets of comparisons. Gene expressions between AD and PD skin samples in E8 and E9 libraries were compared. To increase the power of detecting the DEGs with low expression, the transcriptomes of E8 and E9 AD skins were used as the AD replicate, while the transcriptomes of E8 and E9 of PD skins were used as the PD replicate. These two replicates were compared with each other (E8A + E9A vs. E8P + E9P). Here we skipped the samples in E12 because the natal down growth was stopped.
The DEGs from the comparisons were computed by NOISeq (Tarazona et al. 2011). Only the genes with q > 0.7 were defined as DEGs (Liu et al. 2013).
Gene Set Enrichment and Pathway Analysis
To search the possible pathways involved in natal down growth regulation, the Ensemble gene ID of the expressed genes were converted to the ID of their chicken homologs and input into g:Profiler (Reimand et al. 2007; Reimand et al. 2011), a web-based toolset for functional profiling of gene lists from large-scale experiments. Biological process, cellular component, molecular function, reactome, and human phenotype were used as the data set. The P-value of the gene enrichment was corrected by Benjamini–Hochberg false discovery rate. Only the gene ontology with the corrected P < 0.05 was used in further analyses.
Functional Studies
For the generation of proviral constructs, full-length FGF10 and FGF16 cDNA PCR products were cloned into the pCR8/GW/TOPO Gateway entry vector (Invitrogen, Carlsbad, CA) and sequenced. The cDNAs were transferred into a Gateway compatible RCASBP-Y DV vector through an recombination between attL and attR sites (see Gateway® Cloning system) (LR) recombination reaction (Loftus et al. 2001). Virus was made according to Chuong (1998) concentrated by ultracentrifugation. For an in vivo assay, RCAS virus directing the expression of the candidate genes was injected into the leg or AD skins in E3 chicken embryos. Samples were harvested at E12. Five independent experiments were conducted for each candidate gene. The primer pairs for the full-length coding sequence amplification were listed in supplementary table S13, Supplementary Material online.
Supplementary Material
Acknowledgments
We thank Drs Wen-Lang Fan, Chih-Feng Chen, and John Wang for their advice. We also thank the NGS Core Facility, Biodiversity Research Center, Academia Sinica, for transcriptome sequencing and National Center for High-performance Computing of National Applied Research Laboratories of Taiwan and National Research Program for Biopharmaceuticals (MOST 104-2325-B-492-001) for providing a computational biology platform. This study was supported by MOST 104-2621-B-001-003-MY3.
Additional Information
Accession codes: The RNA-Seq data have been deposited in the National Center for Biotechnology Information under the accession code PRJNA296752.
References
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. 2004. Bmp4 and morphological variation of beaks in Darwin's finches. Science 305:1462–1465. [DOI] [PubMed] [Google Scholar]
- Basu M, Mukhopadhyay S, Chatterjee U, Roy SS. 2014. FGF16 promotes invasive behavior of SKOV-3 ovarian cancer cells through activation of mitogen-activated protein kinase (MAPK) signaling pathway. J Biol Chem. 289:1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bicudo JEPW. 2010. Ecological and environmental physiology of birds. Oxford: Oxford University Press. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Foley J, Tang PC, Li A, Jiang TX, Wu P, Widelitz RB, Chuong CM. 2015. Development, regeneration, and evolution of feathers. Annu Rev Anim Biosci. 3:169–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong CM. 1998. Molecular basis of epithelial appendage morphogenesis. Austin (TX: ): R.G. Landes. [Google Scholar]
- Chuong CM, Widelitz RB, Ting-Berreth S, Jiang TX. 1996. Early events during avian skin appendage regeneration: dependence on epithelial-mesenchymal interaction and order of molecular reappearance. J Invest Dermatol. 107:639–646. [DOI] [PubMed] [Google Scholar]
- Daly TJ, Buffenstein R. 1998. Skin morphology and its role in thermoregulation in mole-rats, Heterocephalus glaber and Cryptomys hottentotus. J Anat. 193(Pt 4):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouailly D. 2009. A new scenario for the evolutionary origin of hair, feather, and avian scales. J Anat. 214:587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dial KP, Jackson BE, Segre P. 2008. A fundamental avian wing-stroke provides a new perspective on the evolution of flight. Nature 451:985–989. [DOI] [PubMed] [Google Scholar]
- Gill FB, Wright MT. 2006. Birds of the world: recommended English names. Princeton (PA: ): Princeton University Press. [Google Scholar]
- Hamburger V, Hamilton HL. 1992. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 195:231–272. [DOI] [PubMed] [Google Scholar]
- Hornik C, Krishan K, Yusuf F, Scaal M, Brand-Saberi B. 2005. cDermo-1 misexpression induces dense dermis, feathers, and scales. Dev Biol. 277:42–50. [DOI] [PubMed] [Google Scholar]
- Hughes SH. 2004. The RCAS vector system. Folia Biol (Praha). 50:107–119. [PubMed] [Google Scholar]
- Jung HS, Francis-West PH, Widelitz RB, Jiang TX, Ting-Berreth S, Tickle C, Wolpert L, Chuong CM. 1998. Local inhibitory action of BMPs and their relationships with activators in feather formation: implications for periodic patterning. Dev Biol. 196:11–23. [DOI] [PubMed] [Google Scholar]
- Lande R. 1978. Evolutionary mechanisms of limb loss in tetrapods. Evolution 32:73–92. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell T, Nilsson D, Hofmeister W, Lindstrand A, Ahituv N, Vandermeer J, Amilon A, Anneren G, Arner M, Pettersson M, et al. 2014. Identification of three novel FGF16 mutations in X-linked recessive fusion of the fourth and fifth metacarpals and possible correlation with heart disease. Mol Genet Genomic Med. 2:402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CM, Jiang TX, Baker RE, Maini PK, Widelitz RB, Chuong CM. 2009. Spots and stripes: pleomorphic patterning of stem cells via p-ERK-dependent cell chemotaxis shown by feather morphogenesis and mathematical simulation. Dev Biol. 334:369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WY, Chang YM, Chen SC, Lu CH, Wu YH, Lu MY, Chen DR, Shih AC, Sheue CR, Huang HC, et al. 2013. Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc Natl Acad Sci U S A. 110:3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Larson DM, Watkins-Chow D, Church DM, Pavan WJ. 2001. Generation of RCAS vectors useful for functional genomic analyses. DNA Res. 8:221–226. [DOI] [PubMed] [Google Scholar]
- Mandler M, Neubuser A. 2004. FGF signaling is required for initiation of feather placode development. Development 131:3333–3343. [DOI] [PubMed] [Google Scholar]
- McKinnell IW, Turmaine M, Patel K. 2004. Sonic Hedgehog functions by localizing the region of proliferation in early developing feather buds. Dev Biol. 272:76–88. [DOI] [PubMed] [Google Scholar]
- Meinhardt H, Gierer A. 2000. Pattern formation by local self-activation and lateral inhibition. Bioessays 22:753–760. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 5:621–628. [DOI] [PubMed] [Google Scholar]
- Mou C, Pitel F, Gourichon D, Vignoles F, Tzika A, Tato P, Yu L, Burt DW, Bed'hom B, Tixier-Boichard M, et al. 2011. Cryptic patterning of avian skin confers a developmental facility for loss of neck feathering. PLoS Biol. 9:e1001028.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JR, Varian-Ramos CW, Welch ZS, Saha MS. 2013. Embryological staging of the Zebra Finch, Taeniopygia guttata. J Morphol. 274:1090–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noramly S, Freeman A, Morgan BA. 1999. beta-catenin signaling can initiate feather bud development. Development 126:3509–3521. [DOI] [PubMed] [Google Scholar]
- Oh HM, Choi SC, Lee HS, Chun CH, Seo GS, Choi EY, Lee HJ, Lee MS, Yeom JJ, Choi SJ, et al. 2004. Combined action of extracellular signal-regulated kinase and p38 kinase rescues Molt4 T cells from nitric oxide-induced apoptotic and necrotic cell death. Free Radic Biol Med. 37:463–479. [DOI] [PubMed] [Google Scholar]
- Olivera-Martinez I, Viallet JP, Michon F, Pearton DJ, Dhouailly D. 2004. The different steps of skin formation in vertebrates. Int J Dev Biol. 48:107–115. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. 2001. Fibroblast growth factors. Genome Biol. 2:REVIEWS3005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paznekas WA, Okajima K, Schertzer M, Wood S, Jabs EW. 1999. Genomic organization, expression, and chromosome location of the human SNAIL gene (SNAI1) and a related processed pseudogene (SNAI1P). Genomics 62:42–49. [DOI] [PubMed] [Google Scholar]
- Protas ME, Trontelj P, Patel NH. 2011. Genetic basis of eye and pigment loss in the cave crustacean, Asellus aquaticus. Proc Natl Acad Sci U S A. 108:5702–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prum RO. 2005. Evolution of the morphological innovations of feathers. J Exp Zool B Mol Dev Evol. 304:570–579. [DOI] [PubMed] [Google Scholar]
- Prum RO, Brush AH. 2002. The evolutionary origin and diversification of feathers. Q Rev Biol. 77:261–295. [DOI] [PubMed] [Google Scholar]
- Reimand J, Arak T, Vilo J. 2011. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 39:W307–W315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. 2007. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35:W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang H. 2004. Prospects for transgenesis in the chick. Mech Dev. 121:1179–1186. [DOI] [PubMed] [Google Scholar]
- Scholl FA, Dumesic PA, Barragan DI, Harada K, Bissonauth V, Charron J, Khavari PA. 2007. Mek1/2 MAPK kinases are essential for mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell. 12:615–629. [DOI] [PubMed] [Google Scholar]
- Song HK, Lee SH, Goetinck PF. 2004. FGF-2 signaling is sufficient to induce dermal condensations during feather development. Dev Dyn. 231:741–749. [DOI] [PubMed] [Google Scholar]
- Starck JM, Ricklefs RE. 1998. Avian growth and development: evolution within the altricial-precocial spectrum. New York: Oxford University Press. [Google Scholar]
- Strasser B, Mlitz V, Hermann M, Tschachler E, Eckhart L. 2015. Convergent evolution of cysteine-rich proteins in feathers and hair. BMC Evol Biol. 15:82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Yoshimoto Y, Yoshioka H, Nohno T, Noji S, Ohuchi H. 2002. FGF10 is a mesenchymally derived stimulator for epidermal development in the chick embryonic skin. Mech Dev. 116:39–49. [DOI] [PubMed] [Google Scholar]
- Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. 2011. Differential expression in RNA-seq: a matter of depth. Genome Res. 21:2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells KL, Hadad Y, Ben-Avraham D, Hillel J, Cahaner A, Headon DJ. 2012. Genome-wide SNP scan of pooled DNA reveals nonsense mutation in FGF20 in the scaleless line of featherless chickens. BMC Genomics 13:257.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widelitz RB, Jiang TX, Lu J, Chuong CM. 2000. beta-catenin in epithelial morphogenesis: conversion of part of avian foot scales into feather buds with a mutated beta-catenin. Dev Biol. 219:98–114. [DOI] [PubMed] [Google Scholar]
- Widelitz RB, Jiang TX, Noveen A, Chen CW, Chuong CM. 1996. FGF induces new feather buds from developing avian skin. J Invest Dermatol. 107:797–803. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jiang TX, Wu P, Widelitz RB, Chuong CM. 2012. Sprouty/FGF signaling regulates the proximal-distal feather morphology and the size of dermal papillae. Dev Biol. 372:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GJ, Li C, Li QY, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhou Z, Wang X, Zhang F, Zhang X, Wang Y, Wei G, Wang S, Xu X. 2013. Hind wings in Basal birds and the evolution of leg feathers. Science 339:1309–1312. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhang F. 2004. A precocial avian embryo from the Lower Cretaceous of China. Science 306:653.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.