Abstract
Serous ovarian cancer is the most frequent type of epithelial ovarian cancer. Despite the use of surgery and platinum‐based chemotherapy, many patients suffer from recurrence within 6 months, termed platinum resistance. Currently, the lack of relevant molecular biomarkers for the prediction of the early recurrence of serous ovarian cancers is linked to the poor prognosis. To identify an effective biomarker for early recurrence, we analyzed the genome‐wide DNA methylation status characteristic of early recurrence after treatment. The patients in The Cancer Genome Atlas (TCGA) dataset who showed a complete response after the first therapy were categorized into 2 groups: early recurrence serous ovarian cancer (ERS, recurrence ≤12 months, n = 51) and late recurrence serous ovarian cancer (LRS, recurrence >12 months, n = 158). Among the 12 differently methylated probes identified between the 2 groups, we found that ZNF671 was the most significantly methylated gene in the early recurrence group. A validation cohort of 78 serous ovarian cancers showed that patients with ZNF671 DNA methylation had a worse prognosis (P < .05). The multivariate analysis revealed that the methylation status of ZNF671 was an independent factor for predicting the recurrence of serous ovarian cancer patients both in the TCGA dataset and our cohort (P = .049 and P = .021, respectively). Functional analysis revealed that the depletion of ZNF671 expression conferred a more migratory and invasive phenotype to the ovarian cancer cells. Our data indicate that ZNF671 functions as a tumor suppressor in ovarian cancer and that the DNA methylation status of ZNF671 might be an effective biomarker for the recurrence of serous ovarian cancer after platinum‐based adjuvant chemotherapy.
Keywords: biomarker, DNA methylation, early recurrence, serous ovarian cancer, ZNF671
1. INTRODUCTION
Epithelial ovarian cancer has one of the highest tumor‐related mortality rates among women with gynecologic malignancies. Serous ovarian cancer is the most frequent (50%‐60% of epithelial ovarian cancers) and lethal type of epithelial ovarian cancer.1, 2 In addition to surgical resection, current standard treatments for serous ovarian cancer include platinum‐based and taxane‐based adjuvant chemotherapy that reduces residual tumors and prevents their recurrence.3 Although the response rate of these initial standard therapies is almost 75%, serous ovarian cancer patients frequently suffer from a recurrence of disease.4, 5
Clinically, patients without recurrence for more than 12 months after platinum chemotherapy treatment are termed “platinum sensitive.”6 The mean survival time of these patients is approximately 34 months.7, 8 By contrast, patients who suffer from recurrence within 6 months and within 6‐12 months are termed “platinum‐resistant” and “partially platinum sensitive,” respectively. The mean survival times of platinum‐resistant and partially platinum sensitive patients are approximately 12 and 18 months, respectively. Thus, the time to recurrence after chemotherapy appears to be an important factor for the prognosis of serous ovarian cancer.9 However, currently, there are no effective biomarkers available to predict the effects of chemotherapy in serous ovarian cancer patients.
Recent whole‐genome analyses revealed that epigenetic alterations including aberrant DNA methylation are common events in almost all cancers, including serous ovarian cancer.10 Aberrant DNA methylation is observed even at the early stages of tumorigenesis and this alters tumor cell lineage identity and invasion and metastasis capability throughout tumor evolution.11 Therefore, DNA methylation changes in cancer cells can be used as diagnostic markers for the existence of cancers, cancer risk, prognosis and drug sensitivity.12 Indeed, in epithelial ovarian cancer, the DNA methylation statuses of naked cuticle homolog 1 (NKD1), vascular endothelial growth factor B (VEGFB) and peroxiredoxin 2 (PRDX2) were reported to be predictors of progression free survival (PFS).13 Furthermore, DNA methylation in runt related transcription Factor 3 (RUNX3) and calcium/calmodulin dependent protein kinase II inhibitor 1 (CAMK2N1) was associated with a shorter PFS.14 However, it is still unclear whether DNA methylation status in certain genes can be used to predict patient outcome after platinum‐based adjuvant chemotherapy.
In the current study, using a public database, The Cancer Genome Atlas (TCGA), in which data of genetic mutations, mRNA expression, DNA methylation status and clinical information of serous ovarian cancer are available,10 we identified Zinc Finger Protein 671 (ZNF671) DNA methylation status as an effective biomarker, which is closely associated with serous ovarian cancer recurrence and prognosis. Furthermore, the downregulation of ZNF671 in serous ovarian cancer cells significantly increased cell proliferation, cell migration and invasion abilities. Our data shed light on the biological and clinical impact of ZNF671 on tumor behavior and its DNA methylation status as a predictive biomarker for platinum‐based adjuvant chemotherapy effectiveness in ovarian cancers.
2. MATERIALS AND METHODS
2.1. In silico DNA methylation analysis
The DNA methylation data (Infinium HumanMethylation27 BeadChip) of high‐grade serous ovarian cancer (n = 584) and normal tissues (n = 14), and corresponding clinical information, including response to first therapy and the periods from initial therapy to recurrence, were obtained from TCGA (https://genome-cancer.ucsc.edu/) (Figure 1A). The obtained DNA methylation data were further analyzed using GeneSpring GX‐13.1.1 (Agilent Technologies, Santa Clara, CA, USA) to extract significantly different probes. The DNA methylation data of serous ovarian cancers from the Infinium HumanMethylation27 BeadChip were also derived from the TCGA database. Among all the probes (n = 27 578) on the BeadChip, we excluded problematic probes containing single‐nucleotide polymorphisms (SNP) and repeat sequences;15 finally, 24 982 probes (corresponding to 14 019 genes) were further analyzed (Figure S1A). We defined DNA methylated probes as those with a β value > .2 as reported previously.15, 16 To select ovarian cancer‐specific methylated genes, we excluded probes that are methylated in the normal ovary (n = 14) and obtained ovarian cancer‐specific probes (16 482 probes corresponding to 9888 genes).
Figure 1.
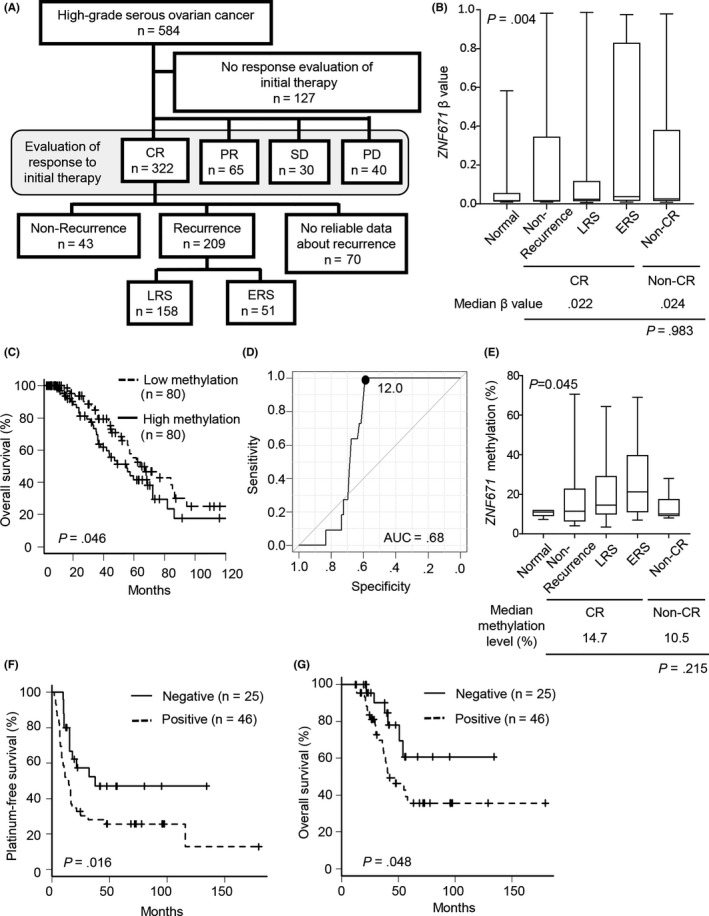
Analysis of ZNF671 in The Cancer Genome Atlas and our validation cohort. A, Schema of the grouping of high‐grade serous ovarian cancer patients by clinical information. CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease. The recurrence group was divided into early recurrent serous ovarian cancer (ERS) and late recurrent serous ovarian cancers (LRS) by platinum‐free interval (PFI). B, ZNF671 DNA methylation level of each patient group. The x‐axis indicates the group of serous ovarian cancer patients. ERS (n = 51), LRS (n = 158), non‐CR (n = 135), non‐recurrence (n = 43) and normal (n = 14), respectively. The median is indicated by a bold line inside the box whose ends denote the upper and lower quartiles. Error bars represent the 5 and 95 percentile values. C, Overall survival analysis in CR patients. Top 25% or lower 25% levels of ZNF671 DNA methylation cases were determined as high or low methylation (n = 80, each), respectively. D, A receiver operating characteristic (ROC) curve was calculated using ZNF671 DNA methylation data from normal ovaries and serous ovarian cancer in our validation cohort. The area under the curve (AUC) was .68. E, ZNF671 DNA methylation level in each patient group. The x‐axis indicates the group of serous ovarian cancer patients. ERS (n = 27), LRS (n = 20), non‐CR (n = 7), non‐recurrence (n = 24) and normal (n = 10), respectively. The median is indicated by a bold line inside the box whose ends denote the upper and lower quartiles. Error bars represent the 5 and 95 percentile values. F, Platinum‐free survival analysis in CR patients. Negative (ZNF671 methylation level <12.0, n = 25) and Positive (ZNF671 methylation level ≥ 12.0, n = 46), respectively. G, Overall survival analysis in CR patients. Negative (ZNF671 methylation level <12.0, n = 25) and positive (ZNF671 methylation level ≥ 12.0, n = 46), respectively
2.2. Clinical samples
High‐grade serous ovarian cancer samples were collected from patients who underwent surgical resection at Nagoya City University Hospital, Japan (n = 40), and at Taipei Medical University, Taiwan (n = 38). Samples were collected after appropriate institutional review board approval was received and written informed consent had been obtained. All patients received chemotherapy after surgery and the observation periods were more than 12 months. Normal ovary samples (n = 10) were collected from patients who underwent surgery for cervical cancer or cervical intraepithelial neoplasia at Nagoya City University Hospital. Grade 2 and 3 were considered high grade. The time from the last administration of chemotherapy to recurrence was defined as the platinum‐free interval (PFI).
2.3. DNA methylation analysis
DNA from frozen tissues or formalin‐fixed paraffin‐embedded (FFPE) tissues was extracted by the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) or QIAamp DNA FFPE Tissue Kit (Qiagen), respectively. DNA from cell lines was extracted using the conventional phenol‐chloroform method. To analyze DNA methylation using pyrosequencing technology (PyroMark Q24, Qiagen), the extracted DNA was treated with bisulfite using an EpiTect Plus Bisulfite Kit (Qiagen). PCR primers for pyrosequencing were designed by Pyromark Assay Design 2.0 (Qiagen) (Table S1). The mean DNA methylation level of 3 CpG of ZNF671 was calculated for further analysis.
2.4. Cell lines and 5‐aza‐2′‐deoxycytidine treatment
The serous ovarian cancer cell lines JHOS‐2, JHOS‐4 and NIH‐OVCAR3 were obtained from RIKEN BioResource Center (Tsukuba, Japan). WI‐38 and SKOV3 (epithelial ovarian cancer) were obtained from ATCC (Manassas, VA, USA). Although these cell lines were not authenticated, relatively low passage number cells were obtained. JHOS‐2 and JHOS‐4 were maintained in DMEM/Ham's F‐12 medium (Wako, Osaka, Japan), NIH‐OVCAR3 and SKOV3 were maintained in RPMI‐1640 medium (Wako), and WI‐38 was maintained in DMEM medium (Wako). All cell lines were cultured in medium containing 5% FBS (Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin‐streptomycin (Wako) at 37°C in a humidified incubator with 5% CO2. For 5‐aza‐2′‐deoxycytidine (DAC, Sigma‐Aldrich, St Louis, MO, USA) treatment, cells were treated with 500 nmol/L DAC for 3 days. Medium containing DAC was replaced every day. DNA and RNA were extracted on the 7th day following the treatment.
2.5. Quantitative RT‐PCR analysis
Total RNA from the cell lines was extracted using TRIzol (Thermo Fisher Scientific), followed by reverse‐transcription using Prime Script RT Master Mix (Takara, Kusatsu, Japan). TaqMan qPCR (Roche diagnostics, Basel, Switzerland) and SYBR Green qPCR (TOYOBO, Osaka, Japan) were performed at least in triplicate for the target genes. Expression levels of ZNF671 were normalized by GAPDH. Oligonucleotide primers used for TaqMan PCR assays and SYBR Green assays are shown in Table S1.
2.6. Western blot analysis
The cell lysates were extracted from each epithelial ovarian cancer cell line. A total of 100 μg of protein was separated by 10% SDS/PAGE gels, transferred to nitrocellulose membranes and incubated with the following antibodies as primary antibodies: rabbit polyclonal anti‐ZNF671 (HPA046099, Atlas Antibody, Bromma, Sweden) and mouse monoclonal anti‐β‐actin (#3700, Cell Signaling Technology, Danvers, MA, USA). HRP‐linked anti‐rabbit IgG (#7074, Cell Signaling Technology) and HRP‐linked anti‐mouse IgG (#7076, Cell Signaling Technology) antibodies were used as secondary antibodies. The density of bands was quantified by ImageJ software (https://imagej.nih.gov/ij/).
2.7. RNA interference, and cell proliferation, migration and invasion assays
Cells were treated with 50 nmol/L siRNA targeting ZNF671 or negative control siRNA (Silencer Select Negative Control #1 siRNA, 4390844, Thermo Fisher Scientific) using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's protocol. ZNF671 targeting siRNA were designed by siDirect version 2.0 (http://sidirect2.rnai.jp/) (Table S1). RNA and protein were extracted at 48 hours after siRNA treatment. For cell proliferation analysis, cells were seeded in 96‐well plates at 2 × 104 cells per well and cultured for 24 hours before siRNA treatment. Cell proliferation was measured every 24 hours using a Cell Counting Kit 8 (Dojindo, Kumamoto, Japan) according to the manufacturer's protocol. For cell migration analysis, cells were seeded in 24‐well plates and cultured to create a confluent monolayer after 48 hours of siRNA treatment. A straight scratch was made on the monolayer cells using a p200 pipette tip. Cells were gently washed with PBS twice and cultured for 48 hours. Images were taken at 0 hours (immediately after the scratch) and after 48 hours incubation under a phase‐contrast microscope. The area without cells was measured using ImageJ software. The migration rate was calculated as: (the area without cells at 0 hour minus that at 48 hours) divided by (the area without cells at 0 hour). Matrigel invasion assays were performed using Corning BioCoat Matrigel Invasion Chambers (8‐μm pore, Thermo Fisher Scientific) and Corning BioCoat Control Inserts (without Matrigel, 8‐μm pore, Thermo Fisher Scientific). The lower chambers were filled with medium containing 10% FBS. After 48 hours of siRNA treatment, 2 × 105 cells were seeded into the upper chamber without FBS. Then, after 48 hours of incubation, cells attached to the bottom membrane (invasive cells) were stained with Diff‐Quick stain (Sysmex, Kobe, Japan). Stained cells were counted in at least 3 different views. The invasion rate was calculated as: the number of cells on the bottom membrane in the Matrigel chambers) divided by (the number of cells on the bottom membrane in the control chambers).
2.8. Statistical analysis
All statistical analyses were performed using EZR software (Saitama Medical Center, Saitama, Japan).17 The statistical significance of differences between 2 groups was analyzed by paired Student's t test. The statistical significance of differences among 3 groups was analyzed by one‐way ANOVA. Fisher's exact test was used to determine nonrandom associations between categorical variables. Overall survival curves were generated using the Kaplan‐Meier method with the log‐rank test. Receiver operating characteristics (ROC) analysis was performed to calculate the area under the curve (AUC) and to determine the best threshold for ZNF671 methylation positive in 78 serous ovarian cancers and 10 normal tissue specimens. All reported P‐values were 2‐sided and P < .05 was considered statistically significant.
3. RESULTS
3.1. Identification of specifically DNA methylated genes in early recurrence high‐grade serous ovarian cancers in the TCGA database
We obtained DNA methylation data for 584 patients using the TCGA database.10 All patients underwent surgical treatment followed by platinum‐based chemotherapy. After excluding patients whose clinical information of the response to the first therapy were not available, we divided the patients (n = 457) into 4 groups: complete response (CR: disappearance of all target lesions); partial response (PR: at least a 30% decrease in the sum of the longest diameter of target lesions); stable disease (SD: lack of sufficient shrinkage to qualify for PR or insufficient increase to qualify for PD); and progressive disease (PD: a 20% or greater increase in the sum of the longest diameter of target lesions) according to the response to the first therapy18 (Figure 1A). Among the 322 patients with CR, 209 patients suffered recurrence. We divided these 209 patients into 2 groups by PFI: early recurrent serous ovarian cancers (ERS, n = 51, ≤12 months) and late recurrent serous ovarian cancers (LRS, n = 158, >12 months). Forty‐three patients survived for more than 24 months without recurrence during the observation period (non‐recurrence). Information on PFI were not available for 70 patients. Patients with ERS were at a more advanced stage of disease than those with LRS and non‐recurrence (P = .023, Table S2).
Among the ovarian cancer‐specific probes (see Section 2), we identified 12 differentially methylated probes corresponding to 11 genes, whose mean β value was greater than .1 in ERS compared with LRS (Figure S1A, Table S3). Among the 12 probes, 2 probes corresponding to ZNF671 consistently showed the largest difference in β value between ERS and LRS. Interestingly, although the other candidate genes, such as CYYR1, NTM and ZFP42, also showed substantial differences between ERS and LRS (corresponding to the top 5 probes, Table S3), DNA methylation status did not affect overall survival (Figure S2).
3.2. DNA methylation of an identified methylated gene, ZNF671, as a potential biomarker for the prediction of early recurrence
ZNF671 was significantly highly methylated in ERS compared with normal tissues, non‐recurrence, LRS and non‐CR in the TCGA dataset (P = .004, Figure 1B). In addition, CR patients with high DNA methylation in ZNF671 had a worse prognosis compared with those with low DNA methylation (log‐rank, P = .046; Cox proportional hazard ratio, 1.66; 95% confidence interval, 1.01‐2.73) (Figure 1C). Interestingly, the methylation level of ZNF671 in non‐CR patients (ie, PR, SD, PD) was not significantly different from that of CR patients, suggesting that the methylation of ZNF671 is not an effective marker for the prediction of sensitivity to the first platinum treatment (P = .983 in the TCGA dataset, Figure 1B). We further examined ZNF671 DNA methylation status by pyrosequencing analysis in high‐grade serous ovarian cancers (n = 78) and normal ovarian tissues (n = 10) in our validation cohort. The 71 ovarian cancers were CR in response to the first platinum‐based adjuvant chemotherapy. Among these patients, 27, 20 and 24 patients were divided into the ERS, LRS and non‐recurrence groups, respectively (Table S4). In our validation cohort, patients with ERS were more closely associated with an advanced stage of disease and a larger residual tumor than those with LRS and non‐recurrence (P < .001 and P = .028, respectively; Table S4).
To establish the threshold of “DNA methylation positive” by pyrosequencing analysis, ROC analysis was performed between normal ovaries and serous ovarian cancer. We found that a DNA methylation level of 12.0% was the best threshold in our cohort samples, which provided high sensitivity and specificity (63.9% and 100%, respectively) to distinguish between normal ovaries and high‐grade serous ovarian cancer (Figure 1D). Consistently, ERS showed a significantly higher level of ZNF671 DNA methylation than normal ovarian tissues, no recurrence serous ovarian cancers, and LRS (P = .045) (Figure 1E). Patients with ZNF671 DNA methylation positively correlated with early recurrence (Table 1).
Table 1.
Clinical features of 78 high‐grade serous ovarian cancer patients with and without ZNF671 methylation
| Negative (%) | Positive (%) | Total (%) | P‐value | |
|---|---|---|---|---|
| Cases | 29 (37.2) | 49 (62.8) | 78 (100) | |
| Age (AV ± SD) | 56.5 ± 15.4 | 57.3 ± 11.4 | 57.1 ± 12.7 | .740 |
| Stage | ||||
| I | 5 (17.2) | 4 (8.2) | 9 (11.5) | .078 |
| II | 6 (20.7) | 4 (8.2) | 10 (12.8) | |
| III | 17 (58.6) | 32 (65.3) | 49 (62.8) | |
| IV | 1 (3.4) | 9 (18.3) | 10 (12.8) | |
| Status | ||||
| ERS | 5 (17.2) | 22 (44.9) | 27 (34.6) | .037 |
| LRS | 7 (24.1) | 13 (26.5) | 20 (25.6) | |
| Non‐recurrence | 13 (44.8) | 11 (22.4) | 24 (30.8) | |
| Non‐CR | 4 (13.8) | 3 (6.1) | 7 (9.0) | |
| Residual tumor | ||||
| No macroscopic disease | 5 (17.2) | 14 (28.6) | 19 (24.3) | .655 |
| 1‐10 mm | 0 (0) | 3 (6.1) | 3 (3.8) | |
| >10 mm | 6 (20.7) | 11 (22.4) | 17 (21.8) | |
| NA | 18 (62.1) | 21 (42.8) | 39 (50.0) | |
| ZNF671 methylation level (AV ± SD) | 8.01 ± 2.20 | 29.57 ± 17.53 | 21.56 ± 17.42 | <.001 |
AV, average; NA, not applicable; SD, standard deviation.
Early recurrence serous ovarian cancer (ERS), PFI ≤12 mo.
Late recurrence serous ovarian cancer (LRS), PFI >12 mo.
In the TCGA dataset, the incidence of recurrence was significantly associated with ZNF671 DNA methylation status (P = .013) and stage (P = .035) by univariate analysis (Table 2A). A multivariate analysis using the Cox proportional hazards model showed that ZNF671 DNA methylation status affected the incidence of recurrence of high‐grade serous ovarian cancers (hazard ratio = 1.47; 95% confidence interval = 1.00‐2.17; P = .049; Table 2A). However, stage and residual tumors were not predictive factors, although in general, advanced stage and larger residual tumors appeared to be correlated with early recurrence.
Table 2.
Univariate and multivariate analyses of recurrence of serous ovarian cancers: (A) TCGA database (n = 322, CR cases) and (B) our cohort (n = 71, CR cases)
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P‐value | HR | 95% CI | P‐value | |
| (A) | ||||||
| Age | ||||||
| <60 vs ≥60 | 1.04 | .78‐1.37 | .806 | 1.04 | .77‐1.40 | .793 |
| Stage | ||||||
| II vs III‐IV | 1.76 | 1.04‐2.98 | .035 | 1.732 | .98‐3.08 | .061 |
| Residual tumor | ||||||
| 0 mm vs 1‐10 mm | 1.39 | .97‐1.98 | .070 | 1.23 | .85‐1.78 | .270 |
| 0 mm vs >10 mm | 1.30 | .86‐1.95 | .221 | 1.11 | .72‐1.71 | .632 |
| ZNF671 methylation | ||||||
| Negative vs Positive | 1.59 | 1.10‐2.29 | .013 | 1.47 | 1.00‐2.17 | .049 |
| (B) | ||||||
| Age | ||||||
| <60 vs ≥60 | 1.47 | .81‐2.67 | .205 | 1.06 | .37‐3.06 | .915 |
| Stage | ||||||
| I‐II vs III‐IV | 10.79 | 3.31‐35.14 | <.001 | 9.64 | 2.93‐31.76 | <.001 |
| Residual tumor | ||||||
| 0 mm vs 1‐10 mm | 9.11 | 2.25‐36.8 | .002 | 3.26 | .76‐13.99 | .112 |
| 0 mm vs >10 mm | 4.20 | 1.58‐11.16 | .003 | 3.30 | 1.03‐10.52 | .044 |
| ZNF671 methylation | ||||||
| Negative vs Positive | 2.19 | 1.13‐4.23 | .019 | 4.27 | 1.24‐14.73 | .021 |
95% CI, 95% confidence interval; HR, hazard ratio; TCGA, The Cancer Genome Atlas.
In our validation cohort, patients with ZNF671 DNA methylation showed worse PFI (log‐rank, P = .016; Cox proportional hazard ratio, 2.19; 95% confidence interval, 1.13‐4.23) (Figure 1F). Univariate analysis showed that the incidence of recurrence was significantly associated with the stage, existence of residual tumor and ZNF671 DNA methylation status (Table 2B). Multivariate analysis using the Cox proportional hazards model showed that clinical stage (stage I‐II vs III‐IV; hazard ratio = 9.64; 95% confidence interval = 2.93‐31.76; P < .001), existence of larger residual tumor (>10 mm) (hazard ratio = 3.30; 95% confidence interval = 1.03‐10.52; P = .044) and ZNF671 DNA methylation status affected the incidence of recurrence of high‐grade serous ovarian cancers in our cohort (hazard ratio = 4.27; 95% confidence interval = 1.24‐14.73; P = .021; Table 2B). Taken together, ours and the TCGA data consistently showed that ZNF671 DNA methylation was an independent factor for predicting the early recurrence of ovarian cancer. Moreover, patients with ZNF671 DNA methylation had a worse overall survival rate (log‐rank, P = .048; Cox proportional hazard ratio = 2.41; 95% confidence interval = 1.03–5.93) (Figure 1G).
3.3. ZNF671 expression is regulated by DNA methylation in ovarian cancer cells
ZNF671 is a gene silenced by DNA methylation in the TCGA analysis.10 ZNF671 consists of 4 exons together with a CpG island in the promoter region (Figure 2A). We examined DNA methylation in the CpG island and the expression of ZNF671 in 4 ovarian cancer cell lines, JHOS‐2, JHOS‐4, NIH‐OVCAR3 and SKOV3, and a fibroblast cell line, WI‐38 (Figure 2B,C). ZNF671 mRNA and protein were detected in WI‐38. ZNF671 expression in normal ovarian tissues was comparable with that in WI‐38 (Figure S3). ZNF671 was highly methylated in JHOS‐4 (87%) and methylated at a low level in NIH‐OVCAR3 (33%) and SKOV3 (34%), and methylation was almost undetectable in JHOS‐2 and WI‐38. Although ZNF671 expression was almost completely silenced in JHOS‐4, it was expressed in NIH‐OVCAR3, JHOS‐2 and SKOV3 ovarian cancer cells and WI‐38, suggesting that high levels of DNA methylation in the ZNF671 promoter silenced its expression, while low‐to‐moderate levels of DNA methylation minimally affected its expression.
Figure 2.
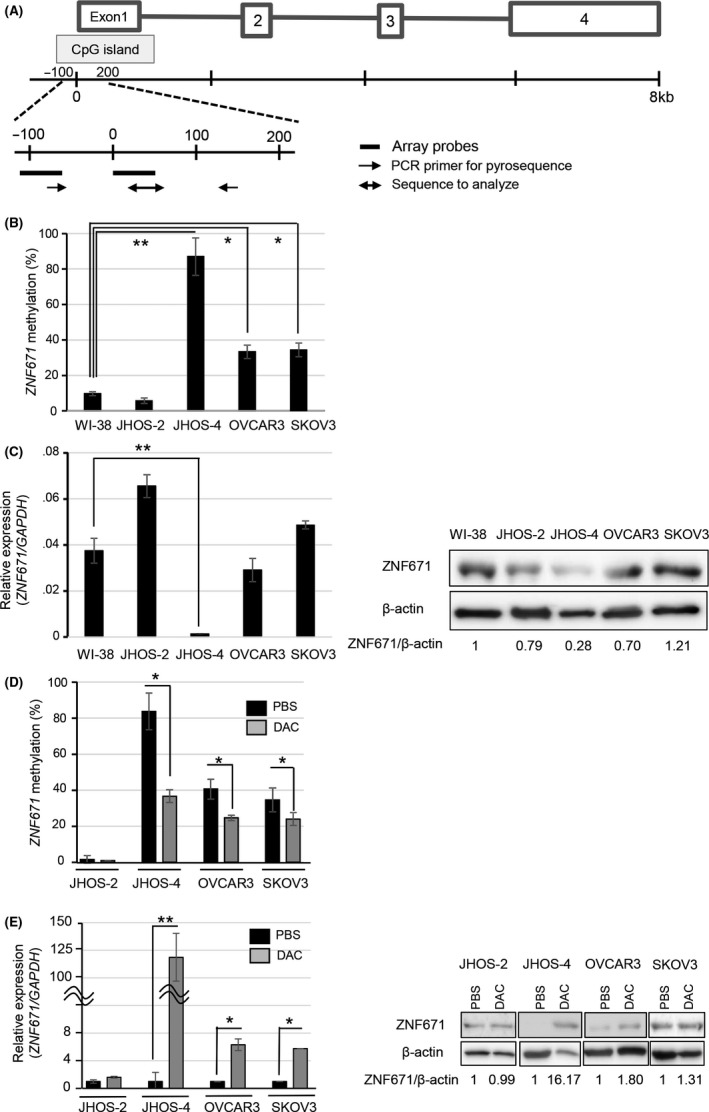
DNA methylation level and expression of ZNF671 in ovarian cancer cell lines. A, The upper schema shows the ZNF671 gene structure. Boxes and lines indicate exons and introns, respectively. ZNF671 has a CpG island in the promoter region. The lower schema shows the positions of array probes (black lines) in the The Cancer Genome Atlas database and the position of pyrosequencing (black arrows), respectively. B, DNA methylation levels of ZNF671 in ovarian cancer cell lines. DNA methylation levels were examined by pyrosequencing analysis. ZNF671 is expressed in the normal fibroblast WI‐38 cell line and we used this cell line as a control. DNA methylation levels of ZNF671 are indicated on the y‐axis. Error bars indicate the SD. *P < .05. **P < .01. C, Expression levels of ZNF671 in ovarian cancer cell lines. Relative mRNA expression levels were normalized according to GAPDH mRNA levels and indicated in the y‐axis (left panel). Error bars indicate the SD **P < .01. Protein expression levels were examined by western blotting (right panel). β‐actin protein was used as a loading control. Band densities were calculated and indicated below the western blotting images as relative values (ZNF671/β‐actin). D, DNA methylation levels of ZNF671 after PBS (black box) and 5‐aza‐2′‐deoxycytidine treatment (DAC, 500 nmol/L, gray box) in ovarian cancer cell lines. DNA methylation levels of ZNF671 are indicated on the y‐axis. Error bars indicate the SD *P < .05. E, mRNA (left) and protein (right) expression levels of ZNF671 after PBS (black box) and DAC treatment (500 nmol/L, gray box) in ovarian cancer cell lines. Relative mRNA expression levels were normalized according to GAPDH mRNA levels and are indicated in the y‐axis (left). Error bars indicate the SD **P < .01. Protein expression levels were examined by western blotting (right). β‐actin protein was used as a loading control. Band densities were calculated and are indicated below the western blotting images as relative values (ZNF671/β‐actin)
Interestingly, the inhibition of DNA methyltransferase by 5‐aza‐2‐deoxycytidine (DAC) significantly decreased DNA methylation levels and increased the expression of ZNF671 in JHOS‐4, NIH‐OVCAR3 and SKOV3, although DAC treatment dramatically increased ZNF671 expression in JHOS‐4 (118‐fold upregulated), while it moderately increased ZNF671 expression in NIH‐OVCAR3 (6‐fold upregulated) and SKOV3 (5‐fold upregulated) compared with untreated controls (Figure 2D,E). The responses of ZNF671 expression to DAC treatment were concordant with baseline DNA methylation levels in each cell line.
3.4. ZNF671 suppresses tumor cell growth, migration and invasion in ovarian cancer
To clarify the function of ZNF671 in ovarian cancers, we investigated the effects of inhibiting ZNF671 by siRNA targeting 2 different siRNA sequences in SKOV3, JHOS‐2 and NIH‐OVCAR3 (Figure 3 and Figure S4A), which moderately express ZNF671. Cell growth was significantly upregulated by the depletion of ZNF671 expression in JHOS‐2 and NIH‐OVCAR3 (P < .05), while a minimum effect on cell growth was observed in SKOV3, although the significant depletion of ZNF671 expression by siRNA was verified in all cell lines, suggesting that ZNF671 may have a dominant role in cell growth suppression in JHOS‐2 and NIH‐OVCAR3 ovarian cancer cells (Figure 3 and Figure S4B).
Figure 3.
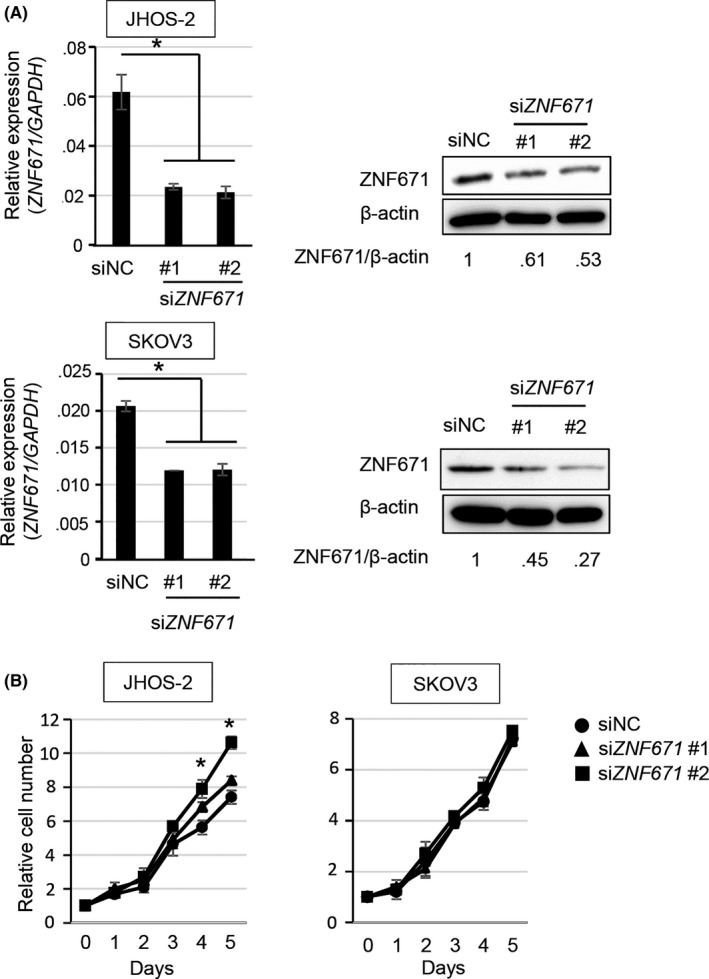
Effect of ZNF671 on cell proliferation in ovarian cancer cell lines. A, Expression levels of ZNF671 treated with si‐negative control (siNC) and siZNF671 #1 and #2 in JHOS‐2 and SKOV3 cells. Relative mRNA expression levels were normalized according to GAPDH mRNA levels and indicated in the y‐axis (left). Error bars indicate the SD *P < .05. Protein expression levels were examined by western blotting (right). β‐actin protein was used as a loading control. Band densities were calculated and indicated below the western blotting images as relative values (ZNF671/β‐actin). B, Effects of siZNF671 on cell proliferation in JHOS‐2 and SKOV3. The y‐axis indicates the relative number of cells to that of day 0. Error bars indicate the SD *P < .05
Cell migration and invasion are important pathological processes for the infiltrative and destructive growth of malignant tumors. A scratch assay was performed to address the effects of ZNF671 on cell migration in ovarian cancer cells. The depletion of ZNF671 expression significantly increased the cell migration of JHOS‐2 and SKOV3 (P < .05) (Figure 4). The cell migration capacity of NIH‐OVCAR3 could not be addressed because most cells were easily detached from the dishes after scratching. Next, we performed a Matrigel invasion assay. JHOS‐2 was less invasive compared with SKOV3 and NIH‐OVCAR3, which showed intrinsic invasive ability.19, 20, 21 Cell invasion was significantly increased by ZNF671 depletion in the SKOV3 and NIH‐OVCAR3 cell line, whereas ZNF671 depletion had a minimal effect on cell invasion by JHOS‐2 (Figure 4 and Figure S4C). Taken together, these data indicate that ZNF671 consistently showed tumor suppressor activity in ovarian cancer cells via multiple processes.
Figure 4.
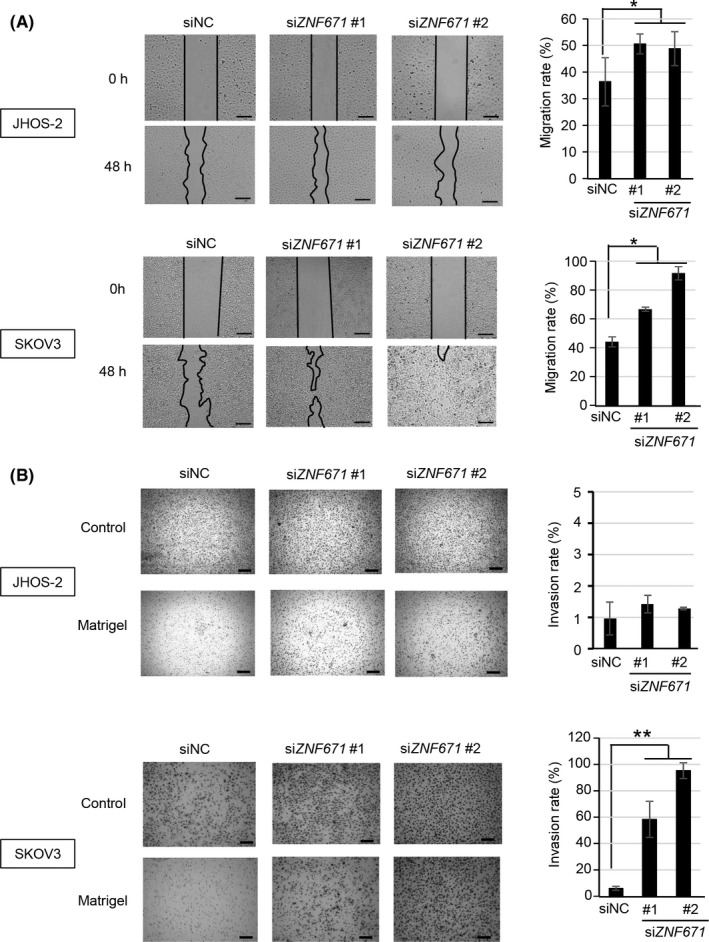
Effect of ZNF671 on cell migration and invasion in ovarian cancer cell lines. A, Effect of siZNF671 on cell migration in JHOS‐2 (left upper) and SKOV3 (left lower). The images were taken at 0 hours (immediately after scratch) and 48 hours. The black lines show the areas without cells. Scale bar indicates 200 μm. Cell migration rates of JHOS‐2 (right upper) and SKOV3 (right lower) were calculated and are shown. Error bars indicate the SD *P < .05. B, Effect of siZNF671 on cell invasion in JHOS‐2 (left upper) and SKOV3 (left lower). Controls indicate wells with no Matrigel basement membrane, while Matrigel indicates wells with Matrigel basement membranes. Scale bar indicates 200 μm. The cell invasion rate of JHOS‐2 (right upper) and SKOV3 (right lower) was calculated and is shown. Error bars indicate the SD **P < .01
4. DISCUSSION
The current study used publicly available datasets to identify silencing of ZNF671 in our cohort and ovarian cancer cell lines by DNA methylation and to show this was biologically associated with the malignant phenotype of high‐grade serous ovarian cancers, indicating that ZNF671 methylation might be an effective predictor for early recurrence after platinum‐based chemotherapy. High‐grade serous ovarian cancers tend to have a poor survival rate due to residual tumors and recurrence of disease after surgery combined with platinum‐based chemotherapy. Currently, there are no effective markers to predict the early recurrence of serous ovarian cancer. Serum level of CA125, a classical marker, is widely used for diagnosis and monitoring the response to treatment and recurrence of epithelial ovarian cancer. However, due to its low sensitivity for recurrence, the diagnosis of recurrence by elevated levels of serum CA125 and subsequent comprehensive treatment did not show significant evidence of survival benefit.22, 23
Therefore, extensive studies to discover biomarkers of platinum resistance have been initiated to develop new therapeutic strategies.24 A recent integrated analysis of miRNA profiling, real‐time PCR and immunohistochemical staining revealed that Smad2 phosphorylation and high expression levels of miR‐181a‐5p might identify ovarian cancers with a poor outcome and low response to platinum‐based neoadjuvant chemotherapy.25 DNA methylation was proven to be a prognostic biomarker of serous ovarian cancers. A study of DNA methylation microarray analysis identified 112 methylated loci that predicted progression‐free survival after platinum‐based chemotherapy with high accuracy.26 These markers appeared to be strong biomarkers to predict progression‐free survival after platinum‐based chemotherapy; however, given the requirement of complex processes to acquire accurate results, simple methods with a minimum number of markers are still required. Multivariate analysis both in the TCGA dataset and our cohort demonstrated that the DNA methylation status of ZNF671 was an independent factor to predict the recurrence of high‐grade serous ovarian cancers in patients who developed CR after platinum‐based chemotherapy. This indicates the potential usefulness of this biomarker in clinical serous ovarian cancer settings. In the previous study, the size of residual tumors was also associated with the incidence of poor prognosis of ovarian cancers.27, 28 However, in the current study, we found that the existence of a larger residual tumor (>10 mm) affected the incidence of recurrence of high‐grade serous ovarian cancers in our cohort but not in the TCGA dataset.
ZNF671 contains Krüppel‐associated box (KRAB) and C2H2‐type zinc finger domains and functions as a transcription factor, although the precise roles of this protein have not been fully elucidated. In previous reports, ZNF671 functioned as a tumor suppressor in urothelial cancer and nasopharyngeal cancer.29, 30 In addition, the hypermethylation of ZNF671 was observed in several cancers, such as clear cell renal carcinomas and urothelial cancers.31, 32 In particular, the DNA methylation of ZNF671 was reported to be a potential biomarker for the relapse of disease in urothelial cancer.29 In ovarian cancer, ZNF671 is 1 of 168 genes whose expressions are silenced by DNA methylation in clinical cases.10 In the current study, we found that ZNF671 was significantly highly methylated in early recurrence ovarian cancer after platinum‐based chemotherapy. The depletion of ZNF671 expression induced a more aggressive phenotype with strong migration and infiltration abilities, together with increased proliferative abilities in ovarian cancer cells. Although further studies are required, our study and previous studies indicated that ZNF671 has tumor suppressive functions and the silencing of ZNF671 by DNA methylation is associated with cancer progression, which may be linked to the early recurrence of serous ovarian cancer. Of note, our study indicated that the methylation of ZNF671 was not an effective marker for the prediction of sensitivity to the first platinum treatment. Indeed, ZNF671 methylation status did not significantly affect patient prognosis when all patients (ie, non‐CR and CR) were examined (Figure S1B,C).
In conclusion, we showed that ZNF671 DNA methylation status may be a useful biomarker for prediction of the early recurrence of serous ovarian cancer after treatment with surgery and platinum‐based combination chemotherapy, which will hopefully improve the treatment of this devastating disease.
CONFLICT OF INTEREST
The authors disclose no potential conflicts of interest.
Supporting information
Mase S, Shinjo K, Totani H, et al. ZNF671 DNA methylation as a molecular predictor for the early recurrence of serous ovarian cancer. Cancer Sci. 2019;110:1105–1116. 10.1111/cas.13936
REFERENCES
- 1. Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamagami W, Aoki D. Annual report of the committee on gynecologic oncology, the Japan Society of Obstetrics and Gynecology. J Obstet Gynaecol Res. 2015;41:1861‐1869. [DOI] [PubMed] [Google Scholar]
- 3. Bookman MA, Brady MF, McGuire WP, et al. Evaluation of new platinum‐based treatment regimens in advanced‐stage ovarian cancer: a Phase III Trial of the Gynecologic Cancer Intergroup. J Clin Oncol. 2009;27:1419‐1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burger RA, Brady MF, Bookman MA, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473‐2483. [DOI] [PubMed] [Google Scholar]
- 5. Katsumata N, Yasuda M, Isonishi S, et al. Long‐term results of dose‐dense paclitaxel and carboplatin versus conventional paclitaxel and carboplatin for treatment of advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer (JGOG 3016): a randomised, controlled, open‐label trial. Lancet Oncol. 2013;14:1020‐1026. [DOI] [PubMed] [Google Scholar]
- 6. Harries M, Gore M. Part II: Chemotherapy for epithelial ovarian cancer‐treatment of recurrent disease. Lancet Oncol. 2002;3:537‐545. [DOI] [PubMed] [Google Scholar]
- 7. Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced‐stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aghajanian C, Goff B, Nycum LR, Wang YV, Husain A, Blank SV. Final overall survival and safety analysis of OCEANS, a phase 3 trial of chemotherapy with or without bevacizumab in patients with platinum‐sensitive recurrent ovarian cancer. Gynecol Oncol. 2015;139:10‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eisenhauer EA, Vermorken JB, van Glabbeke M. Predictors of response to subsequent chemotherapy in platinum pretreated ovarian cancer: a multivariate analysis of 704 patients [see comments]. Ann Oncol. 1997;8:963‐968. [DOI] [PubMed] [Google Scholar]
- 10. Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat Rev Cancer. 2011;11:726‐734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:459‐466. [DOI] [PubMed] [Google Scholar]
- 13. Dai W, Zeller C, Masrour N, Siddiqui N, Paul J, Brown R. Promoter CpG island methylation of genes in key cancer pathways associates with clinical outcome in high‐grade serous ovarian cancer. Clin Cancer Res. 2013;19:5788‐5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Häfner N, Steinbach D, Jansen L, Diebolder H, Dürst M, Runnebaum IB. RUNX3 and CAMK2N1 hypermethylation as prognostic marker for epithelial ovarian cancer. Int J Cancer. 2016;138:217‐228. [DOI] [PubMed] [Google Scholar]
- 15. Campan M, Moffitt M, Houshdaran S, et al. Genome‐scale screen for DNA methylation‐based detection markers for ovarian cancer. PLoS ONE. 2011;6:e28141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanda Y. Investigation of the freely available easy‐to‐use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228‐247. [DOI] [PubMed] [Google Scholar]
- 19. Wang Y, Yan S, Liu X, et al. miR‐1236‐3p represses the cell migration and invasion abilities by targeting ZEB1 in high‐grade serous ovarian carcinoma. Oncol Rep. 2014;31:1905‐1910. [DOI] [PubMed] [Google Scholar]
- 20. Huo W, Zhao G, Yin J, et al. Lentiviral CRISPR/Cas9 vector mediated miR‐21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer. 2017;8:57‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park YA, Lee JW, Kim HS, et al. Tumor suppressive effects of bromodomain‐containing protein 7 (BRD7) in epithelial ovarian carcinoma. Clin Cancer Res. 2014;20:565‐575. [DOI] [PubMed] [Google Scholar]
- 22. Rustin GJ, Nelstrop AE, Tuxen MK, Lambert HE. Defining progression of ovarian carcinoma during follow‐up according to CA 125: a North Thames Ovary Group Study. Ann Oncol. 1996;7:361‐364. [DOI] [PubMed] [Google Scholar]
- 23. Rustin GJ, van der Burg ME, Griffin CL, et al. Early versus delayed treatment of relapsed ovarian cancer (MRC OV05/EORTC 55955): a randomised trial. Lancet. 2010;376:1155‐1163. [DOI] [PubMed] [Google Scholar]
- 24. van Zyl B, Tang D, Bowden NA. Biomarkers of platinum resistance in ovarian cancer: what can we use to improve treatment. Endocr Relat Cancer. 2018;25:R303‐R318. [DOI] [PubMed] [Google Scholar]
- 25. Petrillo M, Zannoni GF, Beltrame L, et al. Identification of high‐grade serous ovarian cancer miRNA species associated with survival and drug response in patients receiving neoadjuvant chemotherapy: a retrospective longitudinal analysis using matched tumor biopsies. Ann Oncol. 2016;27:625‐634. [DOI] [PubMed] [Google Scholar]
- 26. Wei SH, Balch C, Paik HH, et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006;12:2788‐2794. [DOI] [PubMed] [Google Scholar]
- 27. Wimberger P, Lehmann N, Kimmig R, Burges A, Meier W, Du Bois A. Prognostic factors for complete debulking in advanced ovarian cancer and its impact on survival. An exploratory analysis of a prospectively randomized phase III study of the Arbeitsgemeinschaft Gynaekologische Onkologie Ovarian Cancer Study Group (AGO‐OVAR). Gynecol Oncol. 2007;106:69‐74. [DOI] [PubMed] [Google Scholar]
- 28. Nick AM, Coleman RL, Ramirez PT, Sood AK. A framework for a personalized surgical approach to ovarian cancer. Nat Rev Clin Oncol. 2015;12:239‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeh CM, Chen PC, Hsieh HY, et al. Methylomics analysis identifies ZNF671 as an epigenetically repressed novel tumor suppressor and a potential non‐invasive biomarker for the detection of urothelial carcinoma. Oncotarget. 2015;6:29555‐29572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J, Wen X, Liu N, et al. Epigenetic mediated zinc finger protein 671 downregulation promotes cell proliferation and tumorigenicity in nasopharyngeal carcinoma by inhibiting cell cycle arrest. J Exp Clin Cancer Res. 2017;36:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arai E, Chiku S, Mori T, et al. Single‐CpG‐resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis. 2012;33:1487‐1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian Y, Arai E, Gotoh M, Komiyama M, Fujimoto H, Kanai Y. Prognostication of patients with clear cell renal cell carcinomas based on quantification of DNA methylation levels of CpG island methylator phenotype marker genes. BMC Cancer. 2014;14:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


