Abstract
Extraintestinal pathogenic Escherichia coli (ExPEC) cause clinical infections in humans. Understanding the evolution and dissemination of ExPEC strains via potential reservoirs is important due to associated morbidity, health care costs and mortality. To further understanding this survey has examined isolates recovered from the faeces of 221 healthy dogs and 427 healthy cats. The distribution of phylogroups varied with host species, and depended on whether the animal was living in a shelter or a home. The human associated STs 69, 73, 95, 131 and 127 were prevalent, with 30.5% of cat isolates and 10.3% of dog isolates representing these ExPEC sequence types. Resistance to the antibiotics ampicillin and tetracycline was common, but resistance to other antimicrobials was negligible.
Introduction
Escherichia coli is a common member of the microbiota of the lower intestine of mammals and to a lesser extent birds [1]. The species is genetically diverse and exhibits considerable genetic substructure [2]. The species has been partitioned into eight phylogroups [3]. E. coli isolates are most likely to belong to phylogroups A, B1, B2 and D, while strains belonging to phylogroup C, E, F and Clade I are less frequently observed [1]. Strains belonging to the various phylogroups differ in their genome size, variable gene content, disease association, ecological niche, and life history characteristics [4].
Although most strains of E. coli behave as commensals of the lower intestine of vertebrates, some are able to cause intestinal and extra-intestinal disease. Strains responsible for intestinal disease have diverse phylogenetic origins [5]. However, most strains responsible for extra-intestinal diseases such as urinary tract infections (UTIs) or septicaemia are members of phylogroup B2 and to a lesser extent phylogroup D [6, 7]. The technique of multi-locus sequence typing (MLST) [8] has revealed that there are hundreds, of sequence types (STs) within phylogroups B2 and D. However despite this diversity, a very small fraction of STs, are responsible for the great majority of extra-intestinal infections. These are phylogroup B2 STs, ST73, ST95, and ST131 and the phylogroup D ST, ST69, which are largely human associated [9]. At least one (ST 95) is rarely isolated from livestock, wild birds or mammals [10]. The frequency with which these common human associated STs are observed in companion animals is unclear.
Extra-intestinal infections also occur in dogs and cats, primarily as uncomplicated UTIs. As in humans, E. coli is the most common cause of such infections in dogs [11]. Urinary tract infection is relatively uncommon in cats [12], but when it does occur, E. coli is also the most common cause. As in humans, isolates belonging to phylogroup B2 and D are more likely to be recovered from extra-intestinal infections of companion animals [13, 14]. The four STs commonly responsible for extra-intestinal infection in humans (STs 69, 73, 95 and 131) have been isolated from healthy or diseased companion animals. ST131 is commonly reported in dogs and cats [15–17] but there are few reports of the other STs: ST69 in dogs, ST73 in cats and dogs, ST127 in dogs and a single ST95 isolate in a dog [18–21]. What studies have been undertaken have largely been based on clinical isolates exhibiting resistance to particular antimicrobials, such as fluoroquinolones [15, 17, 22].
Although the human associated STs 69, 73, 95 and 131 have been observed in companion animals, there is relatively little data with which to assess the extent to which companion animals represent a zoonotic reservoir of these human associated STs. Family members, including the family pet, have been shown to be more likely to share E. coli strains than individuals not living in the same household [23]. While, in one study, an ST73 strain was found to be shared by family members, including the dog, and persisted in the family for more than three years [24].
The aim of the present study was to assess the genetic structure, diversity, as well as the frequency of the human associated STs of E. coli in the faeces of dogs and cats living in the Canberra region of Australia, and to determine the antibiotic sensitivity of the isolates recovered.
Results
E. coli diversity
E. coli was detected in 334 (78.2%) of 427 cats and in 203 (91.9%) of 221 dogs. Thus E. coli was significantly less likely to be detected in cats as compared to dogs (Contingency Table Analysis: P>X2 < 0.001).
The relative abundance of the phylogroups varied with host type (cat or dog) and differed depending on if the animal was a pet or residing in an animal shelter (Nominal Logistic Regression: Cat/Dog, P>X2 < 0.01; Pet/Shelter P>X2 < 0.001: Cat/Dog*Pet/Shelter P>X2 < 0.82) (Table 1). There was little difference in the relative abundance of phylogroup B1, D, E or F strains in cats compared to dogs, but phylogroup B2 strains were overrepresented in cats, while phylogroup A were overrepresented in dogs (Table 1). Contrasting cats and dogs living in an animal shelter with those living in a home reveals that individuals living in a shelter have lower frequency of phylogroup A strains and an increased frequency of B1 strains compared to animals not living in a shelter (Table 1). The relative abundance of phylogroups B2, D, E and F did not differ between animals living in a home versus those residing in a shelter.
Table 1. The relative abundance of E. coli phylogroups among 203 E. coli isolates from 221 dogs and 334 isolates from 427 cats.
| Phylogroup | Cat | Dog | ||
|---|---|---|---|---|
| Pet % (n) |
Shelter % (n) |
Pet % (n) |
Shelter % (n) |
|
| A | 15.6 (10) | 5.6 (15) | 31.8 (55) | 13.3 (4) |
| B1 | 26.6 (17) | 41.6 (112) | 33.0 (57) | 46.7 (14) |
| B2 | 40.6 (26) | 41.6 (112) | 24.3 (42) | 33.3 (10) |
| D | 6.2 (4) | 7.4 (20) | 6.4 (11) | 6.7 (2) |
| E | 0 (0) | 0.3 (1) | 1.2 (2) | 0 (0) |
| F | 10.9 (7) | 3.7 (10) | 3.5 (6) | 0 (0) |
(n), number of samples.
For cats residing in an animal shelter the relative abundance of the phylogroups did not vary with the sex of the animal (Contingency Table Analysis: P>X2 < 0.16)(data not presented). For dogs, the relative abundance of the phylogroups did not differ between the sexes (Nominal Logistic Regression: Sex, P>X2 < 0.051; Pet/Shelter P>X2 < 0.12) (data not presented).
Of the 334 isolates from cats, 30.5% represented one of the human associated STs (STs 69, 73, 95, 127, 131), a significantly greater frequency of human associated STs than was observed in dogs, as only 10.3% of the 203 isolates from dogs represented these STs (Contingency Table Analysis: P>X2 < 0.0001).
If either a cat or dog harboured a phylogroup D strain, then the likelihood that the strain was an ST69 strain did not depend on the host from which the isolate was recovered (Contingency Table Analysis; P>X2 = 0.17)(Table 2). Similarly, if either a cat or dog harboured a phylogroup B2 strain, then the likelihood that phylogroup B2 strain was an ST95, ST127 or ST131 strain did not depend on the host species from which the isolate was recovered (Contingency Table Analysis; ST95, P>X2 = 0.09; ST127, P>X2 = 0.19; ST131, P>X2 = 0.15)(Table 2). However, a phylogroup B2 strain was significantly more likely to be an ST73 strain if it was recovered from a cat rather than isolated from a dog (Contingency Table Analysis; P>X2 < 0.0001)(Table 2).
Table 2. Frequency of major human associated E. coli sequence types (STs) with respect to either the number of phylogroup D isolates (ST69), the number of B2 isolates (STs 73, 95, 127, 131) or the total number of E. coli isolates recovered from the host group.
| Phylogroup | ST | Cat (n) | % of D or B2 isolates | % of E. coli isolates | Dog (n) | % of D or B2 isolates | % of E. coli isolates |
|---|---|---|---|---|---|---|---|
| D | 69 | 8 | 33.3 | 2.4 | 2 | 15.4 | 1.0 |
| B2 | 73 | 64 | 46.4 | 19.1 | 8 | 15.4 | 3.9 |
| B2 | 95 | 24 | 17.4 | 6.9 | 4 | 7.7 | 2.0 |
| B2 | 127 | 6 | 4.3 | 1.8 | 5 | 9.6 | 2.5 |
| B2 | 131 | 1 | 0.7 | 0.3 | 2 | 3.8 | 1.0 |
ST, sequence type; n, number of samples.
Overall genotype (REP-type) diversity was high, indicating that most animals hosted a different genotype (Table 3). There were two exceptions to this pattern, phylogroup B2 and F isolates from cats. The lower levels of diversity observed among the B2 isolates was largely due to the high frequency of ST73 strains, while a single genotype represented 70% of phylogroup F isolates detected.
Table 3. E. coli genotype (REP = types) richness and diversity of E. coli isolated from cats and dogs with respect to the phylogroup membership of the isolate.
| Phylogroup | Cat | Dog | ||||
|---|---|---|---|---|---|---|
| Number of Isolates | Number of Rep Types | Simpson Diversity | Number of Isolates | Number of Rep Types | Simpson Diversity | |
| A | 25 | 22 | 0.99 | 59 | 38 | 0.98 |
| B1 | 129 | 64 | 0.98 | 71 | 67 | 0.99 |
| B2 | 138 | 71 | 0.94 | 52 | 35 | 0.97 |
| D | 24 | 21 | 0.99 | 13 | 12 | 0.99 |
| F | 17 | 6 | 0.51 | 6 | 6 | 1.00 |
Antimicrobial sensitivity
Excepting ampicillin and tetracycline, most isolates were susceptible to all of the antibiotics tested (Table 4). Among cats and dogs with a home, 62.5% of 64 isolates from cats were susceptible to both ampicillin and tetracycline, while 70.5% of 173 dog isolates were susceptible and there was no difference in the ampicillin/tetracycline sensitivity profiles of isolates from cats or dogs with a home (Contingency Table Analysis: P>X2 = 0.30), and resistance to both ampicillin/tetracycline was uncommon (3.4%). Among the dogs and cats residing in an animal shelter 65.2% of 270 isolates from cats were sensitive to both ampicillin and tetracycline, while 83.3% of 30 dog isolates were susceptible. There was a significant difference in the ampicillin/tetracycline sensitivity profiles of isolates from animals residing in a shelter (Contingency Table Analysis: P>X2 = 0.034), with dog isolates more likely to be sensitive to both antibiotics, and while 10% of isolates from cats were resistant to both ampicillin and tetracycline, none of the dog isolates were resistant to both antibiotics.
Table 4. Antibiotic disc diffusion test results for 334 E. coli isolates from cats and 203 isolates from dogs.
| Antibiotic | Cat % resistant (n) |
Dog % resistant (n) |
|---|---|---|
| Ampicillin | 31.1 (104) | 20.2(41) |
| Tetracycline | 13.2 (44) | 9.9 (20) |
| Chloramphenicol | 0.3 (1) | 0 |
| Ceftazidine | 0.3 (1) | 0.5 (1) |
| Nalidixic acid | 0.3 (1) | 0.5 (1) |
| Gentamicin | 0 | 0.5 (1) |
| Ciprofloxacin | 0 | 0.5 (1) |
| Ertapenem | 0 | 0 |
(n), number of samples
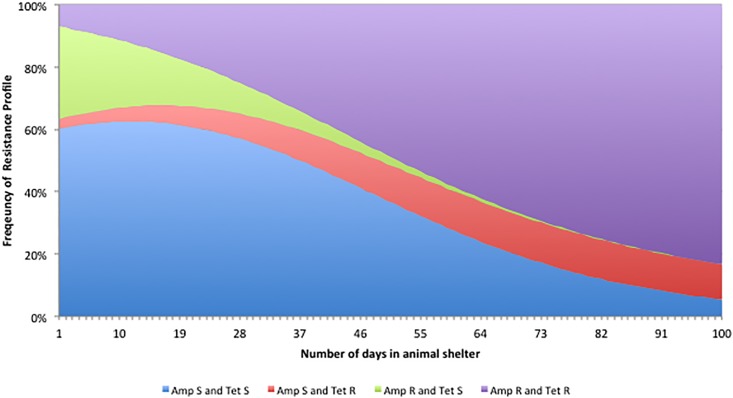
The frequency of isolates susceptible to both ampicillin and tetracycline declined significantly with the number of days the cat spent in the animal shelter, while the frequency of isolates resistant to both antibiotics increased (Nominal Logistic Regression: P>X2 = 0.004)(Fig 1). There was a modest increase in the frequency of isolates resistant to just tetracycline (Fig 1).
Fig 1. Predicted change in the frequency of E. coli isolates from cats resistant to ampicillin, tetracycline, or both antimicrobials as a function of the numbers of days the cat had resided in the animal shelter.
Discussion
Few other studies have examined the relative abundance of the E. coli phylogroups in the faeces of healthy dogs and cats, and comparison of these studies reveals little concordance among studies (Table 5). The relative abundance of phylogroups in human faecal samples has been shown to vary with geographic locality [25]: in samples taken from humans living in temperate climates/developed countries, isolates belonging to phylogroup B2 are most common, while phylogroup A or B1 strains dominate in samples from people living in tropical climates/developing countries [25, 26]. All of the studies investigating the genetic structure of E. coli in healthy dogs were in temperate climate/developed country locations. While, typical dog food diets may vary between Japan and the other countries, it seems unlikely that there are large differences among the diets fed to dogs in Australia, Canada and the United Kingdom. The present study has shown differences in phylogroup structure depending on if the dogs were household pets or residing in an animal shelter. The dogs sampled in the Canadian and UK studies were household pets. In healthy cats living in Toronto, Canada, faecal E. coli phylogroup representation was 76.2%, 14.3%, 9.5% and 21% respectively for A, B1, B2 and D phylogroups [27]. Phylogroup representation of E. coli isolated from rectal swabs of healthy cats in Iran (n = 90) of groups A, B1, B2 and D was 66.7%, 1.2%, 13.4% and 18.9% [28]. By contrast, the frequency of phylogroup B1 and B2 strains in Australian cats was much higher (Table 1). Further studies are required if we are to understand the differences in phylogroup structure of E. coli populations isolated from a single host species sampled from different localities.
Table 5. Relative abundance of E. coli phylogroups isolated from healthy dogs living in different countries.
REP typing revealed a high diversity amongst dominant isolates. Overall there were 185 unique DNA fingerprints among 334 cat isolates, and 158 unique types among the 201 dog isolates. This result is comparable with that reported by [32] with 48 unique types among 108 isolates from 37 cats, and 106 types among 196 isolates from 71 dogs.
The human associated ExPEC STs represent about a third of all E. coli isolates recovered from human faecal samples in the Canberra region [[10]; unpublished data] (Table 6). Dogs are as likely as people to carry ST127 strains, but substantially less likely to carry strains belonging to the other human associated STs (Table 6). By contrast, while cats are less likely to harbour ST95, ST131 or ST69 strains, they are as likely to harbour ST127 strains and are almost twice as likely to carry ST73 strains compared to carriage of these STs in humans (Table 6).
Table 6. Frequency with which STs 69, 73, 95, 127, and 131 are detected in cats, dogs and people living the in Canberra, ACT region of Australia.
| Phylogroup | ST | Human % of E. coli isolates |
Cat % of E. coli isolates |
Dog % of E. coli isolates |
|---|---|---|---|---|
| D | 69 | 4.6 | 2.4 | 1.0 |
| B2 | 73 | 10.0 | 19.1 | 3.9 |
| B2 | 95 | 9.8 | 7.2 | 2.0 |
| B2 | 127 | 2.0 | 1.8 | 2.5 |
| B2 | 131 | 7.8 | 0.3 | 1.0 |
All of the human associated ExPEC STs have been isolated from healthy or diseased dogs, but the great majority of the previous studies focused on clinical isolates, used antibiotic selection (especially fluoroquinolones), or both. ST131 is the most common ExPEC ST reported in dogs and cats [15, 16, 33] but there are a few reports of the other STs: ST69 in dogs, ST73 in cats and dogs, ST127 in dogs and a single ST95 isolate in a dog [18–20].
There is concern that antimicrobial resistance in E. coli harboured by companion animal strains may transfer to humans further limiting treatment options for ExPEC infections in humans. While admitting that there is no direct evidence of transmission of antimicrobial resistance from E. coli in pets to humans, several authors [34–36] point out that the close relationship of humans and their companion animals provides opportunities for sharing strains; that resistant bacteria and genes have been recovered from healthy pets [35]; that the same classes of antimicrobials are used in human and veterinary medicine [37]; and cases of possible ExPEC transmission from pet animals to humans have been reported [38, 39].
Screening of dominant E. coli isolates in this study found phenotypic resistance to ampicillin in 31.1% cat isolates and 20.2% of dog isolates and to tetracycline in 13.2% cat isolates and 9.9% dog isolates. Resistance to ampicillin and tetracycline is common and this finding is consistent with most previous studies. A recent survey of E. coli faecal isolates from dogs visiting veterinarians in the UK revealed that 37.2% of isolates were resistant to ampicillin and 30% were resistant to tetracycline [40]. A survey of dogs and cats in Portugal found a much lower prevalence of ampicillin resistance in E. coli faecal isolates with 16.7% of cat isolates resistant and 7.7% of dog isolates resistant: levels of resistance to tetracycline were 20.5% for faecal isolates from dog isolates and 18.2% for cat faecal isolates [35]. A survey of healthy kennel dogs in Belgium found 12% resistance to ampicillin and 17% resistance to tetracycline and very low or not detectable levels of resistance to chloramphenicol, enrofloxacin and gentamicin [41]. A Canadian study of clinical E. coli isolates from canine UTI infections found low levels of resistance to all antibiotics, with ampicillin at 8.8% and tetracycline at 7.1% and no extended-spectrum beta-lactamases were detected [42]. Other recent studies of clinical E. coli isolates from UTI infections in dogs and cats reported resistance to a wider range of antibiotics, including quinolones and extended-spectrum beta-lactamases (ESBL) [43, 44].
Direct contact with companion animals living in close contact with their owners, has been implicated as a source of human infection with extended spectrum beta-lactamase (ESBLs) producing E. coli [23, 45]. However in this study very few E. coli isolates from dogs and cats were resistant to third generation cephalosporins, based on screening using ceftazidime. This antibiotic is considered one of the third generation cephalosporins with the highest sensitivity for ESBL detection [46]. The ESBL susceptibility found in this study contrasts with reports elsewhere in Australia and overseas of high levels of ESBL resistance in E. coli isolated from both dogs and cats [17, 40, 47–54]. However these studies used selective media or clinical isolates from diverse sites.
In common with another report which didn’t use antibiotic selective media [55], there was no evidence of resistance to carbapenems among isolates from Canberra dogs and cats.
The frequency of E. coli isolates from cats resistant to both ampicillin and tetracycline, increased with the time animals had resided in the animal shelter prior to being sampled. Tetracyclines (doxycycline) were commonly used to empirically treat cats residing in the shelter showing symptoms of feline upper respiratory infection (causal agents include feline herpesvirus, calicivirus, Mycoplasma felis, Chlamydophila felis and Bordatella bonchiseptica) and secondary bacterial infection as recommended by the Australian Infectious Diseases Advisory Panel—Antibiotics (http://www.ava.com.au/sites/default/files/AVA_website/pdfs/AIDAP%20prescribing%20guidelines.pdf). Isolates resistant to ampicillin, tetracycline or both were observed among strains belonging to phylogroups A, B1, B2, D and F. Such outcomes suggest that the evolution of isolates resistant to both antibiotics may have been due to the transfer of an R plasmid among isolates belonging to different lineages, rather than independent evolution events occurring in isolates belonging to each of the phylogroups. Sequential sampling of the same individuals through their stay at the animal shelter coupled with whole genome sequencing of the isolates would be required to determine how the frequency of isolates resistant to both antibiotics arises.
This study has shown that companion animals do carry E. coli isolates belonging to one of the human associated lineages. However considerable substructure is known to occur within clonal complexes 73, 95, and 131 [10, 56, 57]. Clonal complex 95 consists of at least five well-defined subgroups and while CC95 strains can be isolated from poultry and humans [10], the great majority of isolates from poultry belong to just one of the CC95 subgroups. Such an outcome suggests that there may be some degree of ‘host preference’ exhibited by isolates belonging to the same clonal complex. Substructure is also present in ST131 [57] and ST73 [56] but clear host specificities have not been described to date [58, 59]. Whole genome sequencing studies are required in order to determine the degree of difference exhibited by isolates belonging to the same clonal complex but isolated from different host species.
Materials and methods
Source of samples
Faecal samples from 221 dogs and 427 cats in the city of Canberra and its surrounds (Australian Capital Territory, Australia) were collected from 2015 to 2017. Cat samples were collected from an animal shelter and short-term boarding facilities. Dog samples were either collected by the owner, from domestic dogs at a dog park, at short-term boarding kennels or at longer-term kennels; including dogs housed at an animal shelter. Host age and body mass was not available for most animals, but the isolates were generally taken from adults.
E. coli isolation
Fresh faecal material was dilution streaked onto MacConkey agar plates [60] and incubated overnight at 35°C. One well isolated lactose-positive colony from each sample was transferred to Simmons citrate and Luria Bertani agar plates [60] and incubated overnight at 35°C. Putative E. coli isolates (lactose positive and citrate negative) were confirmed to be E. coli by genetic analysis (see below). Isolates were inoculated into 5 ml of lysogeny broth (LB) and incubated at 36°C with shaking for 18 h. A 1 ml aliquot of this suspension was combined with 0.5 ml sterile glycerol and stored at -80°C.
E. coli characterisation
The isolation procedure yielded 203 isolates from 221 dogs and 334 isolates from 427 cats. DNA was extracted from an overnight LB culture of each isolate using DNAzol genomic DNA isolation reagent (Invitrogen) according to the manufacturer’s instructions. Ethanol precipitated DNA was resuspended in Tris-EDTA buffer and this template DNA was stored at -20C.
DNA fingerprints of different strains were obtained using polymerase chain reaction (PCR) to amplify Repetitive Extragenic Palindromic (REP) elements and Enterobacterial Repetitive Intergenic Consensus (ERIC) sequences [61, 62]. PCR amplification was performed in an Applied Biosystems 2720 Thermal Cycler. REP PCR reaction mix (20 μl) contained about 10 ng of DNA template, 1 x PCR polymerisation buffer (Fisher Biotech), MgCl2 at 3.5 mM, N6(CGG)4 primer [62] at 0.4 mM and 1.0 U MyTaq Red DNA polymerase (Bioline). Cycling conditions were as follows: denaturation at 95°C for 3 min, followed by amplification for 35 cycles of 95°C for 1 min and 72°C for 3 min, followed by a final extension step at 72°C for 8 min. The ERIC PCR reaction mix (20 μl) contained approximately 10 ng of DNA template, 1 x PCR polymerisation buffer (Bioline), ERIC1 and ERIC2 [61] each at 0.4 mM and 1.0 U MyTaq Red DNA polymerase (Bioline). ERIC cycling conditions were as follows: 2 min initial denaturation at 95°C, followed by 30 cycles of 3 s at 94°C, 30 s at 92°C, annealing at 50°C for 1 min and extension at 65°C for 8 min. PCR products were separated and visualised on 1.2% agarose gels in 1-Trisborate-EDTA (TBE) buffer using ethidium bromide staining.
Isolates were assigned to a phylogroup using the method of Clermont [3]. The reaction mix (20 μl) contained about 10 ng of DNA template, 1 x MyTaq Red PCR buffer (Bioline), Primers (arpA.f, arpA.r, Chua.1b, Chua.2, Yja.1b, Yja.2b, Tsp.1b and Tsp.2 each at 0.4 mM and 1.0 U MyTaq Red DNA polymerase (Bioline). Cycling conditions were as follows: 5 min initial denaturation at 94°C, followed by 30 cycles of 5 s at 94°C, 25 s at 59°C; with a final extension of 5 min at 72°C. PCR products were separated and visualised on 1.2% agarose gels in 1-Trisborate-EDTA (TBE) buffer using ethidium bromide staining.
Phylogroup B2 and D isolates were screened for the presence of 4 human associated sequence types (STs) 69, 73, 95 and 131 using the method of Doumith [9]. The method of Clermont [63] was used to detect whether phylogroup B2 isolates could be assigned to any of the nine main E. coli phylogroup B2 lineages involved in human extra-intestinal infection strains.
Antimicrobial susceptibility
All isolates were screened using the BD BBL Sensi-Disc procedure (http://legacy.bd.com/ds/technicalCenter/inserts/8840621(201107).pdf) on Mueller-Hinton agar plates (Merck) and using BBL Sensi-Disc antimicrobial susceptibility test discs (Becton, Dickinson and Company). The antimicrobials representing eight key antimicrobial classes were ampicillin (10 μg), ciprofloxacin (5 μg), tetracycline (30 μg), chloramphenicol (30 μg), gentamicin (120 μg), ertapenem (10 μg), nalidixic acid (30 μg) and ceftazidine (30 μg). The zone of inhibition diameters were measured using the instrument ProtoCol 3 (Synbiosis). The E. coli strains were classified as susceptible, intermediate or resistant to an antimicrobial, based on their zone diameters after 18 to 24 hours incubation at 35°C using BBL Sensi-Disc breakpoints.
Host metadata and REP type designation, phylogroup, human-associated ST, and antimicrobial susceptibility profile for all Escherichia coli isolates are presented in S1 Table.
Supporting information
(XLSX)
Acknowledgments
This study could not have been undertaken without the assistance of the many people who allowed me to sample their pets and shelter animals. Their help was essential to the success of this study and is very much appreciated.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Gordon DM, Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003;149(12):3575–86. [DOI] [PubMed] [Google Scholar]
- 2.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nature Reviews Microbiology. 2010;8(3):207–17. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 3.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environmental microbiology reports. 2013;5(1):58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- 4.Gordon D. The Influence of Ecological Factors on the Distribution and the Genetic Structure of Escherichia coli. EcoSal Plus. 2004;1(1). [DOI] [PubMed] [Google Scholar]
- 5.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clinical microbiology reviews. 2013;26(4):822–80. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, et al. The link between phylogeny and virulence inEscherichia coli extraintestinal infection. Infection and immunity. 1999;67(2):546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JR, Delavari P, Kuskowski M, Stell AL. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. The Journal of infectious diseases. 2001;183(1):78–88. 10.1086/317656 [DOI] [PubMed] [Google Scholar]
- 8.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Sciences. 1998;95(6):3140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doumith M, Day M, Ciesielczuk H, Hope R, Underwood A, Reynolds R, et al. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. Journal of clinical microbiology. 2015;53(1):160–6. 10.1128/JCM.02562-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon DM, Geyik S, Clermont O, O’Brien CL, Huang S, Abayasekara C, et al. Fine-Scale Structure Analysis Shows Epidemic Patterns of Clonal Complex 95, a Cosmopolitan Escherichia coli Lineage Responsible for Extraintestinal Infection. Msphere. 2017;2(3):e00168–17. 10.1128/mSphere.00168-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson MF, Litster AL, Platell JL, Trott DJ. Canine bacterial urinary tract infections: New developments in old pathogens. The veterinary journal. 2011;190(1):22–7. 10.1016/j.tvjl.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 12.Litster A, Thompson M, Moss S, Trott D. Feline bacterial urinary tract infections: An update on an evolving clinical problem. The Veterinary Journal. 2011;187(1):18–22. 10.1016/j.tvjl.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 13.Hutton TA, Innes GK, Harel J, Garneau P, Cucchiara A, Schifferli DM, et al. Phylogroup and virulence gene association with clinical characteristics of Escherichia coli urinary tract infections from dogs and cats. Journal of Veterinary Diagnostic Investigation. 2018;30(1):64–70. 10.1177/1040638717729395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infection, Genetics and Evolution. 2011;11(3):654–62. 10.1016/j.meegid.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 15.Guo S, Brouwers HJ, Cobbold RN, Platell JL, Chapman TA, Barrs VR, et al. Fluoroquinolone-resistant extraintestinal pathogenic Escherichia coli, including O25b-ST131, isolated from faeces of hospitalized dogs in an Australian veterinary referral centre. The Journal of antimicrobial chemotherapy. 2013;68(5):1025–31. 10.1093/jac/dks515 . [DOI] [PubMed] [Google Scholar]
- 16.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, et al. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrobial agents and chemotherapy. 2011;55(8):3782–7. 10.1128/AAC.00306-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewers C, Grobbel M, Stamm I, Kopp PA, Diehl I, Semmler T, et al. Emergence of human pandemic O25: H4-ST131 CTX-M-15 extended-spectrum-β-lactamase-producing Escherichia coli among companion animals. Journal of antimicrobial chemotherapy. 2010;65(4):651–60. 10.1093/jac/dkq004 [DOI] [PubMed] [Google Scholar]
- 18.Ewers C, Bethe A, Semmler T, Guenther S, Wieler L. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clinical Microbiology and Infection. 2012;18(7):646–55. 10.1111/j.1469-0691.2012.03850.x [DOI] [PubMed] [Google Scholar]
- 19.Johnson JR, Johnston B, Clabots CR, Kuskowski MA, Roberts E, DebRoy C. Virulence genotypes and phylogenetic background of Escherichia coli serogroup O6 isolates from humans, dogs, and cats. Journal of clinical microbiology. 2008;46(2):417–22. 10.1128/JCM.00674-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamang MD, Nam H-M, Jang G-C, Kim S-R, Chae MH, Jung S-C, et al. Molecular characterization of extended-spectrum β-lactamase and plasmid mediated AmpC β-lactamase-producing Escherichia coli isolated from stray dogs from Korea. Antimicrobial agents and chemotherapy. 2012:AAC. 05598–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Thungrat K, Boothe DM. Multilocus sequence typing and virulence profiles in uropathogenic Escherichia coli isolated from cats in the United States. PloS one. 2015;10(11):e0143335 10.1371/journal.pone.0143335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Platell JL, Cobbold RN, Johnson JR, Trott DJ. Clonal group distribution of fluoroquinolone-resistant Escherichia coli among humans and companion animals in Australia. Journal of antimicrobial chemotherapy. 2010;65(9):1936–8. 10.1093/jac/dkq236 [DOI] [PubMed] [Google Scholar]
- 23.Johnson JR, Owens K, Gajewski A, Clabots C. Escherichia coli colonization patterns among human household members and pets, with attention to acute urinary tract infection. The Journal of infectious diseases. 2008;197(2):218–24. 10.1086/524844 [DOI] [PubMed] [Google Scholar]
- 24.Reeves PR, Liu B, Zhou Z, Li D, Guo D, Ren Y, et al. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PloS one. 2011;6(10):e26907 10.1371/journal.pone.0026907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escobar-Páramo P, Grenet K, Le Menac’h A, Rode L, Salgado E, Amorin C, et al. Large-scale population structure of human commensal Escherichia coli isolates. Applied and environmental microbiology. 2004;70(9):5698–700. 10.1128/AEM.70.9.5698-5700.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blyton MD, Cornall SJ, Kennedy K, Colligon P, Gordon DM. Sex-dependent competitive dominance of phylogenetic group B2 Escherichia coli strains within human hosts. Environmental microbiology reports. 2014;6(6):605–10. [DOI] [PubMed] [Google Scholar]
- 27.White A, Sibley K, Sibley C, Wasmuth J, Schaefer R, Surette M, et al. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Applied and environmental microbiology. 2011;77(21):7620–32. 10.1128/AEM.05909-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akhtardanesh B, Ghanbarpour R, Ganjalikhani S, Gazanfari P, editors. Determination of antibiotic resistance genes in relation to phylogenetic background in Escherichia coli isolates from fecal samples of healthy pet cats in Kerman city. Veterinary Research Forum; 2016: Faculty of Veterinary Medicine, Urmia University, Urmia, Iran. [PMC free article] [PubMed] [Google Scholar]
- 29.Harada K, Okada E, Shimizu T, Kataoka Y, Sawada T, Takahashi T. Antimicrobial resistance, virulence profiles, and phylogenetic groups of fecal Escherichia coli isolates: A comparative analysis between dogs and their owners in Japan. Comparative immunology, microbiology and infectious diseases. 2012;35(2):139–44. 10.1016/j.cimid.2011.12.005 [DOI] [PubMed] [Google Scholar]
- 30.Schmidt VM, Pinchbeck GL, Nuttall T, McEwan N, Dawson S, Williams NJ. Antimicrobial resistance risk factors and characterisation of faecal E. coli isolated from healthy Labrador retrievers in the United Kingdom. Preventive veterinary medicine. 2015;119(1):31–40. [DOI] [PubMed] [Google Scholar]
- 31.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of theEscherichia coli phylogenetic group. Applied and environmental microbiology. 2000;66(10):4555–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson LK, Brown MB, Carruthers EA, Ferguson JA, Dombek PE, Sadowsky MJ. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Applied and environmental microbiology. 2004;70(8):4478–85. 10.1128/AEM.70.8.4478-4485.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson JR, Miller S, Johnston B, Clabots C, DebRoy C. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. Journal of clinical microbiology. 2009;47(11):3721–5. 10.1128/JCM.01581-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guardabassi L, Schwarz S, Lloyd DH. Pet animals as reservoirs of antimicrobial-resistant bacteria. Journal of Antimicrobial Chemotherapy. 2004;54(2):321–32. 10.1093/jac/dkh332 [DOI] [PubMed] [Google Scholar]
- 35.Costa D, Poeta P, Sáenz Y, Coelho AC, Matos M, Vinué L, et al. Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Veterinary microbiology. 2008;127(1):97–105. [DOI] [PubMed] [Google Scholar]
- 36.Lloyd DH. Reservoirs of antimicrobial resistance in pet animals. Clinical Infectious Diseases. 2007;45(Supplement_2):S148–S52. [DOI] [PubMed] [Google Scholar]
- 37.Shaban R, Simon G, Trott D, Turnidge J, Jordan D. Surveillance and reporting of antimicrobial resistance and antibiotic usage in animals and agriculture in Australia. Canberra, Australia: Department of Agriculture, The Australian Government; 2014. [Google Scholar]
- 38.Johnson JR, Clabots C, Kuskowski MA. Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. Journal of clinical microbiology. 2008;46(12):4078–82. 10.1128/JCM.00980-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Damborg P, Nielsen SS, Guardabassi L. Escherichia coli shedding patterns in humans and dogs: insights into within-household transmission of phylotypes associated with urinary tract infections. Epidemiology and infection. 2009;137(10):1457–64. 10.1017/S095026880900226X [DOI] [PubMed] [Google Scholar]
- 40.Wedley AL, Dawson S, Maddox TW, Coyne KP, Pinchbeck GL, Clegg P, et al. Carriage of antimicrobial resistant Escherichia coli in dogs: Prevalence, associated risk factors and molecular characteristics. Veterinary microbiology. 2017;199:23–30. 10.1016/j.vetmic.2016.11.017 [DOI] [PubMed] [Google Scholar]
- 41.De Graef E, Decostere A, Devriese L, Haesebrouck F. Antibiotic resistance among fecal indicator bacteria from healthy individually owned and kennel dogs. Microbial drug resistance (Larchmont, NY). 2004;10(1):65–9. [DOI] [PubMed] [Google Scholar]
- 42.Courtice R, Sniatynski M, Rubin J. Antimicrobial resistance and beta-lactamase production of Escherichia coli causing canine urinary tract infections: Passive surveillance of laboratory isolates in Saskatoon, Canada, 2014. The Canadian veterinary journal = La revue veterinaire canadienne. 2016;57(11):1166 [PMC free article] [PubMed] [Google Scholar]
- 43.Zogg AL, Zurfluh K, Schmitt S, Nüesch-Inderbinen M, Stephan R. Antimicrobial resistance, multilocus sequence types and virulence profiles of ESBL producing and non-ESBL producing uropathogenic Escherichia coli isolated from cats and dogs in Switzerland. Veterinary microbiology. 2018;216:79–84. 10.1016/j.vetmic.2018.02.011 [DOI] [PubMed] [Google Scholar]
- 44.Saputra S, Jordan D, Mitchell T, San Wong H, Abraham RJ, Kidsley A, et al. Antimicrobial resistance in clinical Escherichia coli isolated from companion animals in Australia. Veterinary microbiology. 2017;211:43–50. 10.1016/j.vetmic.2017.09.014 [DOI] [PubMed] [Google Scholar]
- 45.Marques C, Belas A, Franco A, Aboim C, Gama LT, Pomba C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. Journal of Antimicrobial Chemotherapy. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.CDC. Laboratory Detection of Extended-Spectrum β-Lactamases (ESBLs) 2010. https://www.cdc.gov/hai/settings/lab/lab_esbl.html.
- 47.Rubin JE, Pitout JD. Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Veterinary microbiology. 2014;170(1):10–8. [DOI] [PubMed] [Google Scholar]
- 48.Huber H, Zweifel C, Wittenbrink M, Stephan R. ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Veterinary microbiology. 2013;162(2–4):992–6. 10.1016/j.vetmic.2012.10.029 [DOI] [PubMed] [Google Scholar]
- 49.Ljungquist O, Ljungquist D, Myrenås M, Rydén C, Finn M, Bengtsson B. Evidence of household transfer of ESBL-/pAmpC-producing Enterobacteriaceae between humans and dogs–a pilot study. Infection ecology & epidemiology. 2016;6(1):31514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carvalho A, Barbosa A, Arais L, Ribeiro P, Carneiro V, Cerqueira A. Resistance patterns, ESBL genes, and genetic relatedness of Escherichia coli from dogs and owners. brazilian journal of microbiology. 2016;47:150–8. 10.1016/j.bjm.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pomba C, Rantala M, Greko C, Baptiste KE, Catry B, Van Duijkeren E, et al. Public health risk of antimicrobial resistance transfer from companion animals. Journal of Antimicrobial Chemotherapy. 2017;72(4):957–68. 10.1093/jac/dkw481 [DOI] [PubMed] [Google Scholar]
- 52.Qekwana DN, Phophi L, Naidoo V, Oguttu JW. Antimicrobial resistance among Escherichia coli isolates from dogs presented with urinary tract infections at a veterinary teaching hospital in South Africa. BMC veterinary research. 2018;14(1):228 10.1186/s12917-018-1552-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zogg AL, Simmen S, Zurfluh K, Stephan R, Schmitt SN, Nüesch-Inderbinen M. High prevalence of extended-spectrum β-lactamase producing Enterobacteriaceae among clinical isolates from cats and dogs admitted to a veterinary hospital in Switzerland. Frontiers in veterinary science. 2018;5:62 10.3389/fvets.2018.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rusdi B, Laird T, Abraham R, Ash A, Robertson ID, Mukerji S, et al. Carriage of critically important antimicrobial resistant bacteria and zoonotic parasites amongst camp dogs in remote Western Australian indigenous communities. Scientific reports. 2018;8(1):8725 10.1038/s41598-018-26920-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nam H-M, Lee H-S, Byun J-W, Yoon S-S, Jung S-C, Joo Y-S, et al. Prevalence of antimicrobial resistance in fecal Escherichia coli isolates from stray pet dogs and hospitalized pet dogs in Korea. Microbial Drug Resistance. 2010;16(1):75–9. 10.1089/mdr.2009.0125 [DOI] [PubMed] [Google Scholar]
- 56.Kallonen T, Brodrick HJ, Harris SR, Corander J, Brown NM, Martin V, et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome research. 2017;27(8):1437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zakour NLB, Alsheikh-Hussain AS, Ashcroft MM, Nhu NTK, Roberts LW, Stanton-Cook M, et al. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. MBio. 2016;7(2):e00347–16. 10.1128/mBio.00347-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, et al. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS genetics. 2014;10(12):e1004776 10.1371/journal.pgen.1004776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McNally A, Oren Y, Kelly D, Pascoe B, Dunn S, Sreecharan T, et al. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS genetics. 2016;12(9):e1006280 10.1371/journal.pgen.1006280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Power DA, McCuen PJ. Manual of BBL products and laboratory procedures: Becton Dickinson Microbiology Systems; 1988.
- 61.Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic acids research. 1991;19(24):6823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adamus-Bialek W, Wojtasik A, Majchrzak M, Sosnowski M, Parniewski P. (CGG) 4-based PCR as a novel tool for discrimination of uropathogenic Escherichia coli strains: comparison with enterobacterial repetitive intergenic consensus-PCR. Journal of clinical microbiology. 2009;47(12):3937–44. 10.1128/JCM.01036-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clermont O, Christenson JK, Daubié A-S, Gordon DM, Denamur E. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. Journal of microbiological methods. 2014;101:24–7. 10.1016/j.mimet.2014.03.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.