Abstract
Background
Bovine tuberculosis (bTB) caused by Mycobacterium bovis is a re-emerging problem in both livestock and humans. The association of some M. bovis strains with hyper-virulence, MDR-TB and disseminated disease makes it imperative to understand the biology of the pathogen.
Methods
Mycobacterium bovis (15) among 1755 M. tuberculosis complex (MTBC) isolated between 2012 and 2014 were characterized and analyzed for associated patient demography and other risk factors. Five of the M. bovis isolates were whole-genome sequenced and comparatively analyzed against a global collection of published M. bovis genomes.
Results
Mycobacterium bovis was isolated from 3/560(0.5%) females and 12/1195(1.0%) males with pulmonary TB. The average age of M. bovis infected cases was 46.8 years (7-72years). TB patients from the Northern region of Ghana (1.9%;4/212) had a higher rate of infection with M. bovis (OR = 2.7,p = 0.0968) compared to those from the Greater Accra region (0.7%;11/1543). Among TB patients with available HIV status, the odds of isolating M. bovis from HIV patients (2/119) was 3.3 higher relative to non-HIV patients (4/774). Direct contact with livestock or their unpasteurized products was significantly associated with bTB (p<0.0001, OR = 124.4,95% CI = 30.1–508.3). Two (13.3%) of the M. bovis isolates were INH resistant due to the S315T mutation in katG whereas one (6.7%) was RIF resistant with Q432P and I1491S mutations in rpoB. M. bovis from Ghana resolved as mono-phyletic branch among mostly M. bovis from Africa irrespective of the host and were closest to the root of the global M. bovis phylogeny. M. bovis-specific amino acid mutations were detected among MTBC core genes such as mce1A, mmpL1, pks6, phoT, pstB, glgP and Rv2955c. Additional mutations P6T in chaA, G187E in mgtC, T35A in Rv1979c, S387A in narK1, L400F in fas and A563T in eccA1 were restricted to the 5 clinical M. bovis from Ghana.
Conclusion
Our data indicate potential zoonotic transmission of bTB in Ghana and hence calls for intensified public education on bTB, especially among risk groups.
Introduction
Among the Mycobacterium tuberculosis complex (MTBC), Mycobacterium bovis is the main causative agent of TB in cattle and sheep, albeit with the widest host range among other mammals including wildlife and humans [1]. M. bovis associated TB is a re-emerging global problem affecting both livestock and humans alike. The World Health Organization reported 147,000 new Bovine TB (bTB)) cases and 12,500 deaths among humans in 2016 [2]. Despite the low incidence of M. bovis associated TB (~2% globally), the mortality rate is high, especially among children and HIV co-infected patients [1,3,4]. Human-to-human transmission of M. bovis is mostly rare [5], thus human bTB is considered a zoonotic chronic disease characterized by lung infections and their draining lymph nodes as granulomatous necrotizing inflammatory disease [6,7]. Nevertheless, bTB among immunocompromised people and children are mostly extrapulmonary or disseminated affecting other organs other than the lungs and their draining lymph nodes. bTB in humans is mostly transmitted via the alimentary canal by the [4] consumption of unpasteurized dairy products from infected cattle [3,8,9] and or inhalation of aerosolised bacilli via direct contact with infected cattle and/or their carcasses [5]. However, a lack of knowledge or simply negligence of the dangers associated with being in close contact with livestock or wildlife and their unpasteurized products is apparent among some individuals who are constantly in direct contact with animals [10]. In addition, there is a growing association of M. bovis related TB cases with treatment failure due to intrinsic resistance to some commonly used anti-tuberculosis drugs [11].
Even though M. bovis, being a member of the MTBC, is genetically homogenous compared to other bacteria [12], molecular epidemiology of M. bovis infections in Great Britain has shown that they exhibit polymorphic metabolic profiles, such as differential rates of incorporation of propionate into membrane lipid components among different genotypes [13] as well as differential expression of some essential genes and accumulation of single nucleotide polymorphisms (SNPs) which could have functional implications [14].
About 85% of herds and 82% of humans in both rural and urban settings in sub Saharan Africa (SSA) live in close proximity to one another, thus driving the wide distribution of bTB compared to other global settings [15,16]. This is compounded by the inadequate sanitation practices such as the habit of sharing drinking water with beasts and consumption of non-pasteurized milk and dairy products [17–19]. Despite the economic and public health importance of bTB, little knowledge exists on the epidemiology and biology of M. bovis in relation to the human adapted MTBC (hMTBC) lineages spanning M. tuberculosis sensu stricto (Mtbss) and M. africanum (Maf) [20,21]. However, such information is critical for development of effective control tools for bTB.
We determined the prevalence of bTB among pulmonary TB patients passively reporting to selected TB diagnostic/treatment facilities in Ghana, determined potential risk factors associated with bTB in Ghana and explored genomic similarities and differences among M. bovis strains from around the globe, irrespective of the host, using whole genome sequencing.
Materials and methods
Ethical statement and participant enrolment
The Institutional Review Board (IRB) of the Noguchi Memorial Institute for Medical Research (NMIMR) with Federal Wide Assurance number FWA00001824 reviewed this study and its protocols and accordingly gave ethical clearance in support of the work.
Mycobacterial isolation, drug resistance profiling and genotyping
Smear-positive sputum samples from the selected health centers in the Northern and Greater Accra regions of Ghana were decontaminated and inoculated on 2 pairs of Lowenstein Jensen (LJ) slants; one pair supplemented with 0.4% sodium pyruvate (to enhance growth of M. bovis and M. africanum (Maf)) the other with glycerol (for enhanced growth of M. tuberculosis sensu stricto (Mtbss) and incubated as previously described [22]. MTBC cells growing in confluence were harvested and heat inactivated at 95 oC for 60 min in nuclease-free water. After heat inactivation, chromosomal DNA was extracted using previously described protocol [23]. The isolates were confirmed as MTBC by PCR amplification of IS6110 and spoligotyping was carried out for lineage classification [24]. Isolates classified as M. bovis were confirmed with a large sequence polymorphism (LSP) assay using PCR detection of deleted regions of difference RD9, RD4 and RD12 [25]. Drug susceptibility testing against isoniazid (INH) and rifampicin (RIF) was carried out using the micro-plate alamar blue assay [23,26].
Whole genome sequencing and phylogenetic analysis
Whole genome sequencing of 5 candidate M. bovis isolates was carried out as previously described [27]. The 5 genomes (ERR502499; ERR502526; ERR502529; ERR502538; ERR1203064) were added to a collection of 767 previously published clinical and veterinary M. bovis genomes (S1 Table) from around the world for analysis. Sequence reads were mapped to the Mycobacterium bovis AF2122/97 reference genome (NC0002945) using BWA (minimum and maximum insert sizes of 50 and 1000 respectively) [28]. Single nucleotide polymorphisms (SNPs) were called using SAMtools mpileup and BCFtools (minimum base call quality of 50 and minimum root squared mapping quality of 30) as previously described [28,29]. Variant sites in the alignment were extracted using snp-sites [30] and a maximum likelihood phylogenetic tree was constructed using FastTree2 [31] (nucleotide general time-reversible tree). The resulting tree was annotated and rooted using iTOL [32]
Comparative mutational analysis of selected MTBC core-genes
Coordinates of 147 MTBC core genes (S2 Table) previously reported to harbour amino acid mutations with phenotypic consequence on virulence and fitness of some laboratory strains of the MTBC [33–39] were compiled from the Tuberculist database [40]. Using the fasta file of H37Rv as reference, the paired end reads of the 5 Ghanaian M. bovis genomes, 257 M. africanum [27] and global collection of 20 MTBC genomes [41] were screened for mutations within the compiled 147 core genes using ARIBA with default settings [42]. Amino acid mutations found to be present only among the 5 Ghanaian M. bovis genomes were suspected to be M. bovis specific. To confirm whether these mutations were widespread in M. bovis, the global collection of 767 clinical and veterinary M. bovis genomes (S1 Table) was screened for these specific mutations using ARIBA as described above. We further classified these amino acid mutations as M. bovis-specific if they were found in 100% of genomes in the global collection or core M. bovis mutations if found in at least 99% of genomes.
Statistical analysis
Where applicable, chi-square and Fisher’s exact tests were used to establish statistical significance. P-values less than 0.05 were considered statistically significant with 95% confidence.
Results
Demography and biological associations of TB patients infected with M. bovis
A total of 1755 MTBC isolates were obtained from 2074 smear positive TB patients (84.6% isolation rate). Among the patients from whom a MTBC was isolated, 212 (12.1%) were from the Northern region and 1543 (87.9%) from the Greater Accra region as previously described [27]. Fifteen (0.9%) of the isolates were genotyped as M. bovis whereas the remaining 1740 (99.1%) were members of the hMTBC (Mtbss and Maf). The average age of patients infected with M. bovis was 46.8 years (7 to 72 years) of which 12/1195 (1.0%) were from males compared to 3/560 (0.5%) from females (p = 0.412, OR = 1.9). Four (1.9%) of the isolates from the Northern region (n = 212) were M. bovis compared to 11/1543 (0.7%) from the Greater Accra region (p = 0.0968, OR = 2.7). Among the patients with known HIV status (893; 50.3%), 119 (13.3%) were HIV-positive compared to 774 (86.7%) HIV-negative. The incidence of bTB among HIV and non-HIV TB patients was 1.7% (2/119) and 0.5% (4/774) respectively with higher odds of isolating M. bovis from HIV patients relative to non-HIV TB patients (OR = 3.3). Six TB patients including 1 herdsman, 1 herds owner and 4 butchers representing 40% of 15 patients with history of direct contact with livestock were infected with M. bovis. This is significantly higher compared to 0.5% (9/1740) of M. bovis infected TB patients without such history (p < 0.0001, OR = 124.4, 95% CI = 30.1–508.3)
Drug resistance profile of M. bovis isolates
Most of the M. bovis isolates (13) were susceptible to all the drugs tested except two isolates resistant to INH and one isolate resistant to RIF (Table 1). The two INH resistant isolates both had the S315T mutation in katG while the RIF resistant isolate had Q432P and I1491S mutations in rpoB.
Table 1. Sensitivity of the MTBC isolates to INH and RIF.
| Drug | Total (1755) | hMTBC (1740) | M. bovis (15) | P-value | OR | 95%CI |
|---|---|---|---|---|---|---|
| INHr | 133; 7.6% | 131;7.5% | 2;13.3% | 0.3163 | 1.9 | 0.2–8.5 |
| RIFr | 16; 0.9% | 15;0.9% | 1;6.7% | 0.1288 | 8.2 | 0.2–61.0 |
| MDR | 40 (2.3%) | 40;2.3% | 0;0.0% | - | - | - |
| ANY | 189 (10.8%) | 186;10.9% | 3;20.0% | 0.2139 | 2.1 | 0.4–7.8 |
NB: ANY: Total number of isolates resistant to at least one drug.
Phylogenetic distribution of global collection of M. bovis
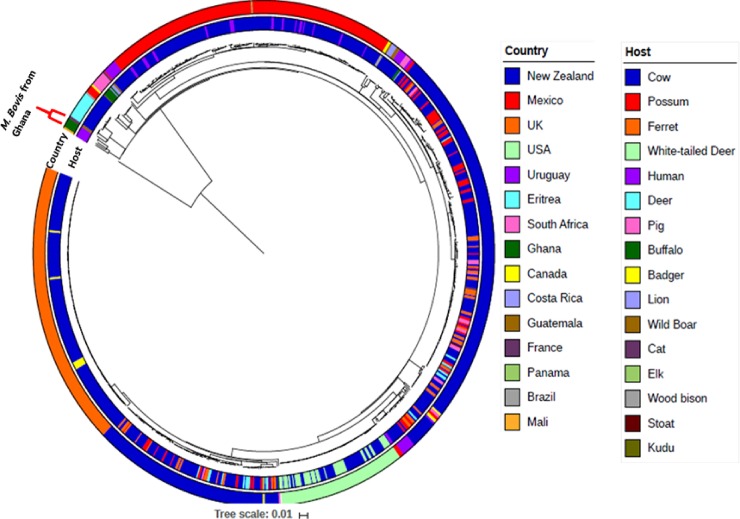
The maximum likelihood phylogenetic tree of global collection of M. bovis spanning both clinical and veterinary isolates rooted on Maf L6 as an outgroup shows random distribution of both the clinical and veterinary M. bovis (Fig 1). The majority of the global collection of M. bovis analyzed were isolated from animals (predominately cattle). The M. bovis genomes of African origin (Ghana, Eritrea and South Africa) generally clustered together closest to the root of the phylogeny irrespective of the host. Nevertheless, there were few M. bovis from South Africa which were sporadically distributed far from the root of the tree. There were 2 major clusters of M. bovis from New Zealand and one major cluster each from the United Kingdom, Mexico and the United States of America. Interestingly, the 5 Ghanaian clinical M. bovis clustered together as a monophyletic branch among the African M. bovis group (Fig 1).
Fig 1. Phylogenetic tree of the Ghanaian clinical M. bovis amidst global collection of 767 published M. bovis genomes.
The whole genome phylogeny of 767 publicly available M. bovis genomes together with 5 clinical M. bovis from Ghana rooted on M. africanum as an outgroup, shows the 5 Ghanaian clinical M. bovis genomes as a monophyletic group siting in a clade consisting mostly of other African M. bovis isolates basal to the rest of the dataset.
In silico predicted M. bovis-specific amino acid mutations
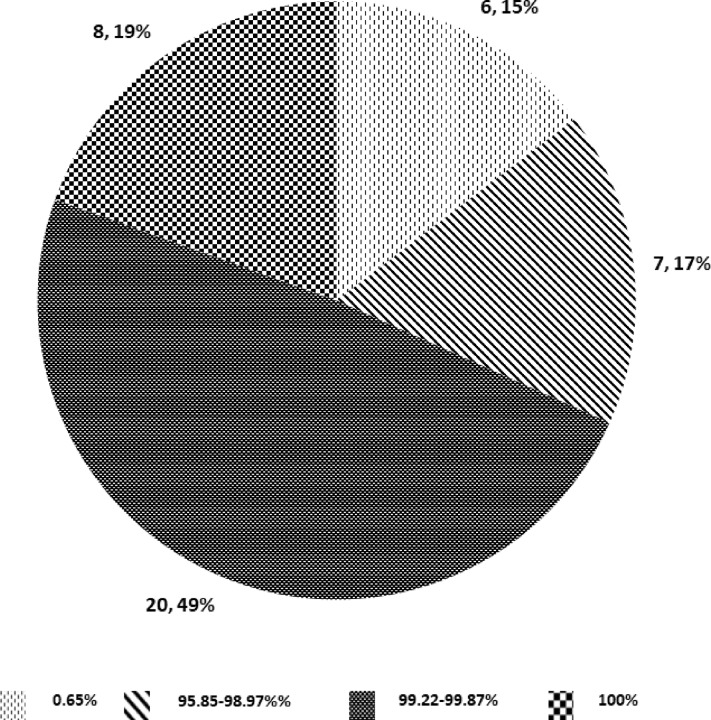
We identified 41 M. bovis restricted amino acid mutations among 32 core-genes of the 5 clinical M. bovis from Ghana when compared to 257 Maf [27] and 20 global MTBC genomes [41] (S3 Table). However, when we screened our global collection of 772 M. bovis genomes (including the 5 from Ghana), only 8 of the mutations were found in all genomes, 20 mutations in 99.22% to 99.87% of the genomes and 7 mutations in 95.85% to 98.97% of genomes. A further 6 mutations (P6T in chaA, G187E in mgtC, T35A in Rv1979c, S387A in narK1, L400F in fas and A563T in eccA1) were restricted to the 5 clinical M. bovis from Ghana (Fig 2; S4 Table; S1 Fig).
Fig 2. Distribution of selected core-gene amino acid mutations among M. bovis.
Among the 41 mutations identified uniquely among 32 core-genes M. bovis, 17 were among 15 essential genes associated with important physiological processes such as lipid metabolism, cell wall and cell processes, intermediate metabolism, and cellular respiration, virulence, detoxification and virulence as well as regulatory proteins (Table 2). These include mce1A, phoT and eccA1 previously shown to be essential for the growth of Mtbss L4 strain H37Rv in primary murine macrophages [35]. In addition, mutations in other genes such as pks6, pknD, pks4 and glgP have been shown to be associated with no production of phthiocerol dimycocerosates (PDIM) among mutant strains [36], attenuation in the central nervous system of BALB/c mice [39], no production of mycolic acid derivatives (mycolipanoic, mycolipenic and mycolipodienoic acids) among mutant strains [38] and in vitro slow growth [34].
Table 2. Description of M. bovis-restricted amino acid mutations among essential genes.
| Gene | Common name | Mutation | Proportion of M. bovis | Function | Essentiality | Reference |
|---|---|---|---|---|---|---|
| Rv0169 | mce1A | P359S | 100% | virulence, detoxification, adaptation | required for survival in primary murine macrophages required for growth in C57BL/6J mouse spleen |
[35] [34] |
| Rv0405 | pks6 | A456fs | 100% | lipid metabolism | transposon mutant does not produce phthiocerol dimycocerosate (PDIM) essential gene for in Mtbss CDC1551 strain |
[36] [37] |
| Rv0820 | phoT | F35L | 100% | cell wall and cell processes | required for survival in primary murine macrophages in H37Rv | [35] |
| Rv0931c | pknD | L376fs | 99.9% | Regulatory | mutant Mtbss CDC1551 is attenuated in the central nervous system of BALB/c mice | [39] |
| Rv1181 | pks4 | D505A | 99.5% | lipid metabolism | essential gene in Mtbss CDC1551 strain mutant aggregates in liquid culture and does not produce mycolipanoic, mycolipenic, or mycolipodienoic acids |
[37] [38] |
| Rv1328 | glgP | D532G | 100% | intermediary metabolism and respiration | slow growth of Mtbss H37Rv mutant strain | [34] |
| Rv1522c | mmpL12 | S947N | 97.4% | cell wall and cell processes | essential gene for in vitro growth of Mtbss H37Rv | [43] |
| Rv1661 | pks7 | S1176P | 95.9% | lipid metabolism | essential gene for in vitro growth of Mtbss H37Rv | [43] [34] |
| Rv1662 | pks8 | A808V | 97.9% | lipid metabolism | essential gene for in vitro growth of Mtbss H37Rv | [43] [37] |
| Rv1662 | pks8 | D78Y | 97.8% | lipid metabolism | essential gene for in vitro growth of Mtbss H37Rv | [43] [34] |
| Rv1662 | pks8 | Y1469C | 99.6% | lipid metabolism | essential gene for in vitro growth of Mtbss H37Rv | [43] [34] |
| Rv2339 | mmpL9 | A44V | 99.9% | cell wall and cell processes | essential gene for in vitro growth of Mtbss H37Rv | [43] |
| Rv2524c | fas | L400F | 0.7% | lipid metabolism | essential gene in Mtbss H37Rv and CDC1551; essential gene for in vitro growth of Mtbss H37Rv |
[34] [37] [43] |
| Rv2956 | N.A | I237T | 99.6% | information pathways | essential gene for in vitro growth of Mtbss H37Rv | [43] |
| Rv3282 | N.A | A133S | 99.7% | conserved hypothetical | Mtbss H37Rv mutants are slow growing | [34] |
| Rv3666c | dppA | E451G | 97.8% | cell wall and cell processes | essential gene in Mtbss H37Rv | [34] |
| Rv3868 | eccA1 | A243V | 99.5% | cell wall and cell processes | required for survival of Mtbss H37Rv in primary murine macrophages | [35] |
Discussion
The global aim of reducing the impact of tuberculosis by the year 2030 cannot be achieved without considering the impact of zoonotic transmission and biology of M. bovis, the main causative agent of TB among cattle. The prevalence and incidence of bTB among humans is significantly lower across the globe compared to TB caused by the hMTBC [2]. Nevertheless, the association of bTB with compromised immunity and the innate resistance of M. bovis to pyrazinamide (PZA) (one of the four first line anti-TB drugs) underscore the need to adapt and implement TB control programs that encompass both bTB and TB caused by the hMTBC. Compared to other geographical regions, Africa has the highest burden of zoonotic transmission of bTB due to close contact of humans and animals (domestic and wild-life) as well as relatively poor hygienic practices [2,17,44–46]. We identified 15 M. bovis isolates among a total of 1755 MTBC isolated from pulmonary TB patients. Further molecular epidemiological analysis of these together with global collections of M. bovis and hMTBC showed (1) an association between close contact with livestock/animal carcasses and bTB infection in Ghana, (2) clustering of M. bovis of African origin close to the root of the global phylogeny and (3) the presence of M. bovis-specific amino acid mutations among both essential and non-essential core MTBC genes.
The finding of a significant association between bTB and close contact with animals (p < 0.0001) suggests zoonotic transmission and this calls for the implementation of preventive policies and strategies to reduce zoonotic transmission of TB among these high-risk groups [44]. This observation also calls for intensive education to create awareness of the disease about the risks of infection, the detection of infected animals/carcasses and prevention among farmers, butchers and the general population. Further emphasis should be placed on training butchers and animal handlers on the importance of adequate infection control measures, including the use of personal protective equipment (PPE) and the disposal of infected organs to avoid transmission of bTB among such personnel. An experienced butcher suffering from bTB in Australia gave an account of slaughtering many animals suspected of bTB and further cutting out the lungs for over 35 years without any proper precaution [47]. Also, some butchers in Nigeria, suffering from bTB, admitted eating visibly infected parts of the lung of cattle out of ignorance in order to convince customers to buy meat [48]. These instances highlight the importance of public education in the fight against bTB. This education should include veterinarians because there are instances of these professionals getting infected with bTB due to a lack of precautionary measures during execution of their work as was the case of a veterinary surgeon who suffered cutaneous bTB after performing several examinations without proper PPE [49].
Our observation also confirms the importance of the test and slaughter (TS) control strategy for bTB. In addition to pasteurisation of dairy products, bTB has been controlled in developed countries due to the successful implementation of the TS policy of all infected cattle and compensation of affected farmers by governments [50]. However, this has not been implemented in SSA partly due to the costs involved. Nevertheless, our findings call for a reconsideration of the TS strategy and mass vaccination for bTB control in SSA and Governments must respond to this call.
We found the proportion of M bovis infected patients among participants from the Northern region (1.9%) of Ghana to be relatively higher (OR = 2.7) compared to those from the Greater Accra region of Ghana (0.6%). The Northern region is home to over 70% of the national cattle population [51], confirming the observation that there is a relationship between close animal contact and bTB. Even though we found no clear association between the M. bovis isolates and drug resistance and HIV infection, the proportions were relatively higher than among the hMTBC. However, the lack of association may be because of the relatively limited number of M. bovis isolates thus further investigation using a larger number of isolates is required.
The global phylogeny of M. bovis clusters most of the M. bovis of African origin at the root of the tree (Fig 1) which might be an indication that they are closest to the progenitor of this successful member of the MTBC with the widest host range. However, the limited number of genomes from Africa does not allow inference of ancestry. With the exception of the five clinical M. bovis from Ghana which clustered as a monophyletic branch at the base of the tree, the random distribution of M. bovis irrespective of the speciation of the host underscores the wide host range of M. bovis and indicates that there is no specific host adaptation. However, the geographical distribution may suggest transmission of specific clones within certain geographical locations which agrees with earlier reports [52–54].
The identification and implications of M. bovis-specific amino acid mutations among genes such as mce1A, phoT and eccA1 [35], pks6 [36,38] as well as glgP [34] highlights the potential attenuated virulence of M. bovis relative to the hMTBC [55]. It would be interesting to test the effects of these mutations on fitness of mutants using ex vivo human cell lines or in vivo bovine models. In addition to the potential phenotypic implications of the identified mutations among essential genes, the 8 M. bovis-specific mutations could be utilized in developing either a rapid lateral flow diagnostic tool or a PCR-based tool specific for differential diagnosis of bTB among TB patients to advice an appropriate treatment regimen since M. bovis is innately resistant to pyrazinamide, a component of the DOTS regimen.
The scarcity of M. bovis genomes from Africa limited our ability to infer ancestry of the Ghanaian clinical isolates. Nevertheless, our data indicates a potential zoonotic transmission of bTB hence highlights the need for public education among people at risk. Moreover, the identified M. bovis-specific mutations could be utilized in the development of rapid diagnostic assays for differential diagnosis of bTB.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Distribution of the M. bovis restricted amino acid mutations on the midpoint-rooted maximum-likelihood phylogeny of 772 global collection of M. bovis genomes. Mutation present and absent are represented by the red and white blocks respectively.
(TIF)
Acknowledgments
We would like to thank the Ghana Health Service, the national tuberculosis control program and especially the clinicians and lab technicians of the Korle-Bu teaching hospital (Accra), La general hospital (Accra), Tamale teaching hospital (Tamale) and Baptist medical center (Nalerigu) for their relentless support. Most importantly, we thank all the TB patients who consented to be part of this study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Wellcome Trust Intermediate Fellowship awarded to DYM (Grant Number 097134/Z/11/Z). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Müller B, Dürr S, Alonso S, Hattendorf J, Laisse CJM, Parsons SDC, et al. (2013) Zoonotic Mycobacterium bovis—induced tuberculosis in humans. Emerg Infect Dis 19: 899–908. Available: http://wwwnc.cdc.gov/eid/article/19/6/pdfs/12-0543.pdf. 10.3201/eid1906.120543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (2017) Global Tuberculosis Report 2017. Geniva.
- 3.Thoen C, LoBue P, de Kantor I (2006) The importance of Mycobacterium bovis as a zoonosis. Vet Microbiol 112: 339–345. Available: https://www.sciencedirect.com/science/article/pii/S0378113505004086?via%3Dihub. Accessed 23 November 2018. 10.1016/j.vetmic.2005.11.047 [DOI] [PubMed] [Google Scholar]
- 4.Grange JM (2001) Mycobacterium bovis infection in human beings. Tuberculosis 81: 71–77. Available: http://www.ncbi.nlm.nih.gov/pubmed/11463226. Accessed 23 November 2018. 10.1054/tube.2000.0263 [DOI] [PubMed] [Google Scholar]
- 5.Evans JT, Smith EG, Banerjee A, Smith RM, Dale J, Innes JA, et al. (2007) Cluster of human tuberculosis caused by Mycobacterium bovis: evidence for person-to-person transmission in the UK. Lancet 369: 1270–1276. Available: http://linkinghub.elsevier.com/retrieve/pii/S0140673607605984. Accessed 8 November 2016. 10.1016/S0140-6736(07)60598-4 [DOI] [PubMed] [Google Scholar]
- 6.Pesciaroli M, Alvarez J, Boniotti MB, Cagiola M, Di Marco V, Marianelli C, et al. (2014) Tuberculosis in domestic animal species. Res Vet Sci 97: S78–S85. Available: https://www.sciencedirect.com/science/article/pii/S0034528814001623?via%3Dihub. Accessed 23 November 2018. 10.1016/j.rvsc.2014.05.015 [DOI] [PubMed] [Google Scholar]
- 7.Domingo M, Vidal E, Marco A (2014) Pathology of bovine tuberculosis. Res Vet Sci 97: S20–S29. Available: https://www.sciencedirect.com/science/article/pii/S0034528814000927?via%3Dihub. Accessed 23 November 2018. 10.1016/j.rvsc.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 8.Rodwell TC, Moore M, Moser KS, Brodine SK, Strathdee SA (2008) Tuberculosis from Mycobacterium bovis in binational communities, United States. Emerg Infect Dis 14: 909–916. Available: http://www.ncbi.nlm.nih.gov/pubmed/18507901. Accessed 23 November 2018. 10.3201/eid1406.071485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritacco V, Sequeira MD, De Kantor IN (2008) Human Tuberculosis Caused by Mycobacterium Bovis in Latin America and the Caribbean. Mycobacterium Bovis Infect Anim Humans Second Ed 14: 13–17. 10.1002/9780470344538.ch3 [DOI] [Google Scholar]
- 10.Nuru A, Mamo G, Zewude A, Mulat Y, Yitayew G, Admasu A, et al. (2017) Preliminary investigation of the transmission of tuberculosis between farmers and their cattle in smallholder farms in northwestern Ethiopia: a cross-sectional study. BMC Res Notes 10: 31 Available: http://www.ncbi.nlm.nih.gov/pubmed/28061860. Accessed 23 November 2018. 10.1186/s13104-016-2349-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vazquez-Chacon CA, Martínez-Guarneros A, Couvin D, González-Y-Merchand JA, Rivera-Gutierrez S, Escobar-Gutierrez A, et al. (2015) Human multidrug-resistant Mycobacterium bovis infection in Mexico. Tuberculosis (Edinb) 95: 802–809. Available: http://www.sciencedirect.com/science/article/pii/S1472979215207482. [DOI] [PubMed] [Google Scholar]
- 12.Smith NH, Gordon S V., de la Rua-Domenech R, Clifton-Hadley RS, Hewinson RG (2006) Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat Rev Microbiol 4: 670–681. Available: http://www.nature.com/doifinder/10.1038/nrmicro1472. Accessed 26 July 2017. 10.1038/nrmicro1472 [DOI] [PubMed] [Google Scholar]
- 13.Winder CL, Gordon S V., Dale J, Hewinson RG, Goodacre R (2006) Metabolic fingerprints of Mycobacterium bovis cluster with molecular type: implications for genotype-phenotype links. Microbiology 152: 2757–2765. Available: http://mic.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.28986-0. Accessed 23 November 2018. 10.1099/mic.0.28986-0 [DOI] [PubMed] [Google Scholar]
- 14.Golby P, Nunez J, Witney A, Hinds J, Quail M A, Bentley S, et al. (2013) Genome-level analyses of Mycobacterium bovis lineages reveal the role of SNPs and antisense transcription in differential gene expression. BMC Genomics 14: 710 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3856593&tool=pmcentrez&rendertype=abstract. Accessed 31 October 2016. 10.1186/1471-2164-14-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosivi O, Grange JM, Daborn CJ, Raviglione MC, Fujikura T, Cousins D, et al. (1998) Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg Infect Dis 4: 59–70. Available: http://www.ncbi.nlm.nih.gov/pubmed/9452399. Accessed 27 March 2018. 10.3201/eid0401.980108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humblet M-F, Boschiroli ML, Saegerman C (2009) Classification of worldwide bovine tuberculosis risk factors in cattle: a stratified approach. Vet Res 40: 50 Available: http://www.ncbi.nlm.nih.gov/pubmed/19497258. Accessed 23 November 2018. 10.1051/vetres/2009033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boukary AR, Thys E, Rigouts L, Matthys F, Berkvens D, Mahamadou I, et al. (2012) Risk Factors Associated with Bovine Tuberculosis and Molecular Characterization of Mycobacterium bovis Strains in Urban Settings in Niger. Transbound Emerg Dis 59: 490–502. 10.1111/j.1865-1682.2011.01302.x [DOI] [PubMed] [Google Scholar]
- 18.Cleaveland S, Shaw DJ, Mfinanga SG, Shirima G, Kazwala RR, Eblate E, et al. (2007) Mycobacterium bovis in rural Tanzania: Risk factors for infection in human and cattle populations. Tuberculosis 87: 30–43. Available: https://www.sciencedirect.com/science/article/pii/S1472979206000497?via%3Dihub. Accessed 23 November 2018. 10.1016/j.tube.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 19.Kang’ethe EK, Ekuttan CE, Kimani VN (2007) Investigation of the prevalence of bovine tuberculosis and risk factors for human infection with bovine tuberculosis among dairy and non-dairy farming neighbour households in Dagoretti Division, Nairobi, Kenya. East Afr Med J 84: S92–5. Available: http://www.ncbi.nlm.nih.gov/pubmed/18338728. Accessed 23 November 2018. [DOI] [PubMed] [Google Scholar]
- 20.Cadmus S, Palmer S, Okker M, Dale J, Gover K, Smith N, et al. (2006) Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J Clin Microbiol 44: 29–34. Available: http://www.ncbi.nlm.nih.gov/pubmed/16390943. Accessed 6 October 2016. 10.1128/JCM.44.1.29-34.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller B, Steiner B, Bonfoh B, Fané A, Smith NH, Zinsstag J (2008) Molecular characterisation of Mycobacterium bovis isolated from cattle slaughtered at the Bamako abattoir in Mali. BMC Vet Res 4: 26 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2483712&tool=pmcentrez&rendertype=abstract. Accessed 1 April 2014. 10.1186/1746-6148-4-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asante-Poku A, Yeboah-Manu D, Otchere ID, Aboagye SY, Stucki D, Hattendorf J, et al. (2015) Mycobacterium africanum Is Associated with Patient Ethnicity in Ghana. PLoS Negl Trop Dis 9: e3370 Available: http://www.ncbi.nlm.nih.gov/pubmed/25569290. Accessed 9 January 2015. 10.1371/journal.pntd.0003370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otchere ID, Asante-poku A, Osei-Wusu S, Baddoo A, Sarpong E, Ganiyu AH, et al. (2016) Detection and characterization of drug-resistant conferring genes in Mycobacterium tuberculosis complex strains: A prospective study in two distant regions of. Tuberculosis (Edinb) 99: 147–154. Available: http://www.sciencedirect.com/science/article/pii/S1472979216301329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamerbeek J, Schouls L, Kolk a, van Agterveld M, van Soolingen D, Kuijper S et al. (1997) Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35: 907–914. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=229700&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren RM, Richardson M, Sampson SL, van der Spuy GD, Bourn W, Hauman JH, et al. (2001) Molecular evolution of Mycobacterium tuberculosis: phylogenetic reconstruction of clonal expansion. Tuberculosis 81: 291–302. Available: http://www.ncbi.nlm.nih.gov/pubmed/11584597. Accessed 12 October 2017. 10.1054/tube.2001.0300 [DOI] [PubMed] [Google Scholar]
- 26.Franzblau SG, Witzig RS, Mclaughlin JC, Torres P, Madico G, Hernandez A, et al. (1998) Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol 36: 362–366. Available: https://www.ncbi.nlm.nih.gov/pubmed/9466742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otchere ID, Coscollá M, Sánchez-Busó L, Asante-Poku A, Brites D, Loiseau C, et al. (2018) Comparative genomics of Mycobacterium africanum Lineage 5 and Lineage 6 from Ghana suggests distinct ecological niches. Sci Rep 8: 11269 Available: http://www.nature.com/articles/s41598-018-29620-2. Accessed 30 July 2018. 10.1038/s41598-018-29620-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, et al. (2010) Evolution of MRSA during hospital transmission and intercontinental spread. Science 327: 469–474. Available: http://www.ncbi.nlm.nih.gov/pubmed/20093474. Accessed 1 November 2018. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. (2016) SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb genomics 2: e000056 Available: http://www.ncbi.nlm.nih.gov/pubmed/28348851. Accessed 23 November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490 Available: http://www.ncbi.nlm.nih.gov/pubmed/20224823. Accessed 23 November 2018. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. Available: https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkw290. Accessed 1 November 2018. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sassetti CM, Rubin EJ (2003) Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100: 12989–12994. Available: http://www.ncbi.nlm.nih.gov/pubmed/14569030. Accessed 23 November 2018. 10.1073/pnas.2134250100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sassetti CM, Boyd DH, Rubin EJ (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48: 77–84. Available: http://doi.wiley.com/10.1046/j.1365-2958.2003.03425.x. Accessed 23 November 2018. [DOI] [PubMed] [Google Scholar]
- 35.Rengarajan J, Bloom BR, Rubin EJ (2005) Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A 102: 8327–8332. Available: http://www.pnas.org/content/102/23/8327.long. Accessed 26 November 2015. 10.1073/pnas.0503272102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waddell SJ, Chung GA, Gibson KJC, Everett MJ, Minnikin DE, Besra GS, et al. (2005) Inactivation of polyketide synthase and related genes results in the loss of complex lipids in Mycobacterium tuberculosis H37Rv. Lett Appl Microbiol 40: 201–206. Available: http://doi.wiley.com/10.1111/j.1472-765X.2005.01659.x. Accessed 23 November 2018. 10.1111/j.1472-765X.2005.01659.x [DOI] [PubMed] [Google Scholar]
- 37.Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, et al. (2003) A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 100: 7213–7218. Available: http://www.ncbi.nlm.nih.gov/pubmed/12775759. Accessed 23 November 2018. 10.1073/pnas.1231432100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubey VS, Sirakova TD, Kolattukudy PE (2002) Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol Microbiol 45: 1451–1459. Available: http://doi.wiley.com/10.1046/j.1365-2958.2002.03119.x. Accessed 23 November 2018. [DOI] [PubMed] [Google Scholar]
- 39.Be NA, Lamichhane G, Grosset J, Tyagi S, Cheng Q, Kim KS, et al. (2018) Murine Model to Study the Invasion and Survival of Mycobacterium tuberculosis in the Central Nervous System. 198 10.1086/592447 [DOI] [PubMed] [Google Scholar]
- 40.Lew JM, Kapopoulou A, Jones LM, Cole ST (2011) TubercuList—10 years after. Tuberculosis (Edinb) 91: 1–7. 10.1016/j.tube.2010.09.008 [DOI] [PubMed] [Google Scholar]
- 41.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. (2010) Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42: 498–503. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2883744&tool=pmcentrez&rendertype=abstract. Accessed 11 July 2014. 10.1038/ng.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, et al. (2017) ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genomics. Available: http://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000131.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffin JE, Gawronski JD, DeJesus MA., Ioerger TR, Akerley BJ, Sassetti CM (2011) High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7: 1–9. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3182942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vayr F, Martin-Blondel G, Savall F, Soulat J-M, Deffontaines G, Herin F (2018) Occupational exposure to human Mycobacterium bovis infection: A systematic review. PLoS Negl Trop Dis 12: e0006208 Available: https://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.0006208. Accessed 23 November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayele WY, Neill SD, Zinsstag J, Weiss MG, Pavlik I (2004) Bovine tuberculosis: an old disease but a new threat to Africa. Int J Tuberc Lung Dis. 8(8):924–937. Available: https://www.ingentaconnect.com/content/iuatld/ijtld/2004/00000008/00000008/art00002. Accessed 23 November 2018. [PubMed] [Google Scholar]
- 46.Boukari AR, Chaibou M, Marichatou H, Vias GF (2007) Caractérisation des systèmes de production laitière et analyse des stratégies de valorisation du lait en milieu rural et périurbain au Niger: cas de la communauté urbaine de Niamey et de la commune rurale de Filingué. Rev d’élevage médecine vétérinaire des pays Trop 60: 113 Available: http://revues.cirad.fr/index.php/REMVT/article/view/9963. Accessed 23 November 2018. [Google Scholar]
- 47.Ingram PR, Bremner P, Inglis TJ, Murray RJ, Cousins D V (2010) Zoonotic tuberculosis: on the decline. Commun Dis Intell Q Rep 34: 339–341. Available: http://www.ncbi.nlm.nih.gov/pubmed/21090190. Accessed 23 November 2018. [PubMed] [Google Scholar]
- 48.Hambolu D, Freeman J, Taddese HB (2013) Predictors of bovine TB risk behaviour amongst meat handlers in Nigeria: a cross-sectional study guided by the health belief model. PLoS One 8: e56091 Available: http://www.ncbi.nlm.nih.gov/pubmed/23409127. Accessed 23 November 2018. 10.1371/journal.pone.0056091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Twomey DF, Collins R, Cranwell MP, Crawshaw TR, Higgins RJ, Dean GS et al. (2012) Controlling tuberculosis in a llama (Lama glama) herd using clinical signs, tuberculin skin testing and serology. Vet J 192: 246–248. Available: https://www.sciencedirect.com/science/article/pii/S1090023311001882?via%3Dihub. Accessed 23 November 2018. 10.1016/j.tvjl.2011.05.014 [DOI] [PubMed] [Google Scholar]
- 50.Zamri-Saad M, Kamarudin MI (2016) Control of animal brucellosis: The Malaysian experience. Asian Pac J Trop Med 9: 1136–1140. Available: https://www.sciencedirect.com/science/article/pii/S1995764516304679. Accessed 21 March 2018. 10.1016/j.apjtm.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 51.Ministry of Food and Agriculture-Ghana (2004) Livestock development in Ghana policies and Strategies: 1–122. [Google Scholar]
- 52.Rodriguez-Campos S, Schürch AC, Dale J, Lohan AJ, Cunha MV, Botelho A, et al. (2012) European 2—A clonal complex of Mycobacterium bovis dominant in the Iberian Peninsula. Infect Genet Evol 12: 866–872. Available: 10.1016/j.meegid.2011.09.004. Accessed 31 October 2016. 10.1016/j.meegid.2011.09.004 [DOI] [PubMed] [Google Scholar]
- 53.Smith NH (2012) The global distribution and phylogeography of Mycobacterium bovis clonal complexes. Infect Genet Evol 12: 857–865. Available: https://www.sciencedirect.com/science/article/pii/S1567134811003170?via%3Dihub. Accessed 11 October 2018. 10.1016/j.meegid.2011.09.007 [DOI] [PubMed] [Google Scholar]
- 54.Smith NH, Berg S, Dale J, Allen A, Rodriguez S, Romero B, et al. (2011) European 1: A globally important clonal complex of Mycobacterium bovis. Infect Genet Evol 11: 1340–1351. Available: https://www.sciencedirect.com/science/article/pii/S156713481100133X?via%3Dihub. Accessed 11 October 2018. 10.1016/j.meegid.2011.04.027 [DOI] [PubMed] [Google Scholar]
- 55.Villarreal-Ramos B, Berg S, Whelan A, Holbert S, Carreras F, Salguero FJ, et al. (2018) Experimental infection of cattle with Mycobacterium tuberculosis isolates shows the attenuation of the human tubercle bacillus for cattle. Sci Rep 8: 894 Available: https://www.nature.com/articles/s41598-017-18575-5.pdf. Accessed 27 March 2018. 10.1038/s41598-017-18575-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Distribution of the M. bovis restricted amino acid mutations on the midpoint-rooted maximum-likelihood phylogeny of 772 global collection of M. bovis genomes. Mutation present and absent are represented by the red and white blocks respectively.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.