Abstract
The mechanistic target of rapamycin (mTOR) is a master regulator of several crucial cellular processes, including protein synthesis, cellular growth, proliferation, autophagy, lysosomal function and cell metabolism. mTOR interacts with specific adaptor proteins to form two multiprotein complexes, called mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). In the cardiovascular system, the mTOR pathway regulates both physiological and pathological processes in the heart. It is needed for embryonic cardiovascular development and for maintaining cardiac homeostasis in post-natal life. Studies involving mTOR loss-of-function models revealed that mTORC1 activation is indispensable for the development of adaptive cardiac hypertrophy in response to mechanical overload. mTORC2 is also required for normal cardiac physiology and ensures cardiomyocyte survival in response to pressure overload. However, partial genetic or pharmacological inhibition of mTORC1 reduces cardiac remodeling and heart failure in response to pressure overload and chronic myocardial infarction. In addition, mTORC1 blockade reduces cardiac derangements induced by genetic and metabolic disorders and has been reported to extend lifespan in mice. These studies suggest that pharmacological targeting of mTOR may represent a therapeutic strategy to confer cardioprotection, although clinical evidence in support of this notion is still scarce.
This review summarizes and discusses the new evidence regarding the pathophysiological role of mTOR signaling in the cardiovascular system.
Keywords: mTOR signaling, mTORC1, mTORC2, hypertrophy, ischemia/reperfusion, heart, cardiovascular diseases
1. Introduction
The mechanistic (previously called mammalian) target of rapamycin (mTOR) is an atypical serine/threonine kinase, belonging to the phosphoinositide kinase-related kinase (PIKK) family 1-6. It is an evolutionarily conserved protein that plays a central role in the regulation of cellular physiology, metabolism and stress responses1-6.
mTOR interacts with specific adaptor proteins and forms two distinct macromolecular complexes, named mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2)6-10. The previous paradigm in mTOR biology included involvement of mTOR in the regulation of protein synthesis, cellular growth and ribosomal biogenesis, and sensing and integrating different upstream inputs, such as growth factors, nutrients, amino acids, starvation and hypoxia1-6. However, it is now known that the mTOR pathway also controls other important cellular processes, including cell survival, mitochondrial biogenesis and function, lipid synthesis and autophagy6. The signaling network of mTORC2 is less characterized than that of mTORC1. Some studies have suggested that the activities of mTORC1 and mTORC2 are strongly interconnected, with mTORC2 being less sensitive to acute rapamycin treatment than mTORC111. Previous work also indicated that mTORC2 is involved in the regulation of cell survival, growth and proliferation, and controls cell architecture and polarity 7-10, 12.
Given its numerous functions, deregulation of mTOR signaling can lead to the development of several pathologies, such as cancer, metabolic syndrome and cardiovascular diseases5, 12, 13.
Previous work has indicated that mTOR plays both adaptive and maladaptive functions in the heart. Studies conducted on mouse models with either systemic or cardiomyocyte-specific disruption of mTORC1 and mTORC2 demonstrated that mTOR signaling plays a central role in regulating the development of the cardiovascular system in embryo and in preserving cardiovascular integrity and function under unstressed conditions in the post-natal life 14-17. In addition, complete genetic disruption of mTORC1 or mTORC2 in the heart impairs the development of compensatory cardiac hypertrophy in response to stress and abrogates the ability of the heart to adapt to mechanical and ischemic injury15, 16, 18-20. In contrast, partial and selective inhibition of mTORC1 confers cardioprotection in multiple cardiac pathological conditions, indicating that pharmacological inhibition of mTORC1 may represent a potential therapeutic intervention for the treatment of cardiovascular diseases. Either genetic or pharmacological partial inhibition of mTORC1 activity reduces pathological hypertrophy in response to pressure overload and chronic myocardial infarction, thereby improving ventricular function. mTORC1 inhibition also reverses cardiac aging, as well as genetic and metabolic cardiomyopathies20-25.
This review article reports new insights into the biology of mTOR in the cardiovascular system, with a particular focus on the involvement of the mTOR complexes in different cardiac pathophysiological conditions.
2. Overview of the mTOR signaling pathway
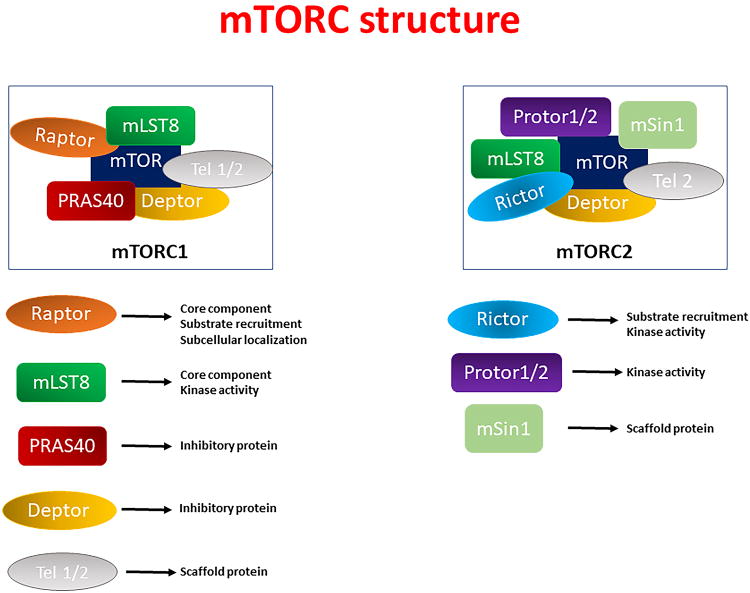
mTOR is encoded by a single gene in mammals and represents the catalytic subunit of mTORC1 and mTORC2, which are the functional homologs of the yeast TORC1 and TORC2, respectively. mTOR was discovered as a direct target of rapamycin, which binds to the FK506-binding protein of 12 kDa–rapamycin complex (FKBP12), thereby leading to mTORC1 inhibition 1-3, 6. The scaffold proteins of mTORC1 are the regulatory-associated protein of mTOR (Raptor), the mammalian lethal with SEC13 protein 8 (mLST8), the proline-rich Akt substrate of 40 kDa (PRAS40), the DEP domain containing mTOR interacting protein (DEPTOR) and Tel two interacting protein 1 (Tel2) 26-29. Raptor and mLST8, together with mTOR, represent the core components of mTORC1. Raptor is needed for substrate recruitment and for subcellular localization of mTORC1, whereas mLST8, which is associated with the mTOR catalytic domain, is required for the kinase activity 30, 31. DEPTOR and PRAS40 represent the inhibitory subunits of the complex (Figure 1)32, 33.
Figure 1. Structure of mTOR complex 1 (mTORC1) and mTORC2.
Schematic representation of the different subunits of mTORC1 and mTORC2, together with a description of their functions.
Legend: DEPTOR, DEP domain-containing mTOR-interacting protein; mLST8, mammalian lethal with sec-13 protein 8; PRAS40, proline-rich Akt substrate 40; Protor 1/2, protein observed with rictor 1/2; Raptor, regulatory-associated protein of mammalian target of rapamycin (mTOR); rictor, rapamycin insensitive companion of mTOR; Tel1/2, Tel two interacting protein 1.
mTORC2 also contains mTOR, mLST8, DEPTOR, and Tel2, but it contains the rapamycin insensitive companion of mTOR (rictor) in place of Raptor34, 35. In addition, the regulatory subunits mSin1 and protein observed with rictor (Protor1/2) have also been identified as possible mTORC2 partners (Figure 1). Moreover, several activators and inhibitors have been shown to act upstream and modulate mTOR complex activity, such as Akt (also known as protein kinase B) and adenosine monophosphate activated protein kinase (AMPK). Once modulated, mTOR transduces these signals to different downstream effectors, thereby controlling numerous cellular processes (Figures 2 and 3).
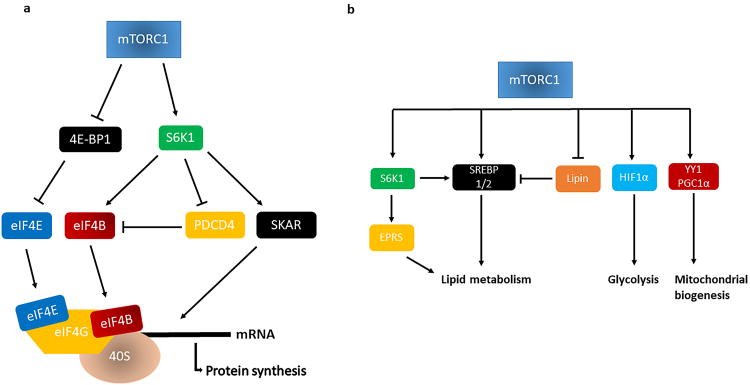
Figure 2. mTORC1 regulation of protein synthesis and cell metabolism.
Schema representing the molecular mechanisms through which mTORC1 controls protein synthesis (a) and modulates cell metabolism (b).
Legend: 40S, eukaryotic small ribosomal subunit; 4EB-P1, eukaryotic translation initiation factor 4E (eIF4E)-binding protein-1; eIF4B, eukaryotic translation initiation factor 4B; eIF4G, eukaryotic translation initiation factor 4G; EPRS, glutamylprolyl-tRNAsynthetase; HIF-1, hypoxia-inducible factor-1α; PDCD4, programmed cell death 4; S6K1, S6 kinase-1; SKAR, S6K1 Aly/REF-like substrate; SREBP1/2, sterol regulatory element-binding protein 1/2; YY1/PGC-1α, transcription factor yin-yang 1/peroxisome proliferator–activated receptor γ coactivator-1α transcriptional complex.
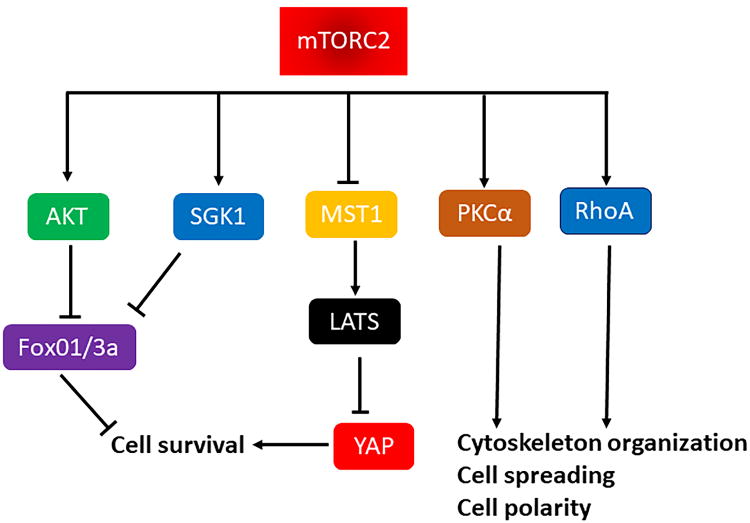
Figure 3. Main cellular functions and substrates of mTORC2.
Schema representing the main cellular functions and substrates of mTORC2.
Legend: Akt, protein kinase B; FoxO1/3, Forkhead box O1/3; PKC-α, protein kinase C-α; LATS, large tumor suppressor kinase 1; MST1, mammalian sterile 20- like kinase 1; RhoA, Ras homolog gene family, member A; SGK1, serum- and glucocorticoid-induced protein kinase-1; YAP, yes associated protein.
2.1 mTORC1 and protein synthesis
mTORC1 is the main regulator of cellular growth in response to different environmental and intracellular conditions. Therefore, it promotes anabolic processes such as protein, nucleotide and lipid synthesis, whereas it inhibits catabolic pathways, such as autophagy 6, 7. The first step in protein synthesis in eukaryotic cells usually occurs through the cap-dependent translation process that involves interaction between a series of proteins with the 5′ end of an mRNA molecule. The first phase of translation involves the eukaryotic initiation factor (eIF) 4F, a complex formed by eIF4E, eIF4A and eIF4G. Briefly, eIF4F interacts with the 5′ end of the mRNA and the poly(A) tail, which recruits the 40S ribosomal subunit. eIF4B association with eIF4A is also required for binding of the mRNA to ribosomes36.
The main substrates of mTORC1 involved in protein synthesis include the S6 kinase-1 (S6K1) and the eukaryotic translation initiation factor 4E (eIF4E)-binding protein-1 (4E-BP1)7-10, 12. mTORC1 activates S6K1 by direct phosphorylation at Thr389. Once phosphorylated, S6K1 promotes the initiation of translation through a number of different mechanisms. It activates eIF4B, a positive regulator of the 5′ cap-binding eIF4F complex, while at the same time it inhibits programmed cell death 4 (PDCD4)37, a negative regulator of translation that prevents the incorporation of eIF4B into the translation initiation complex. Moreover, S6K1 also promotes translation through S6K1 Aly/REF-like substrate (SKAR), a component of exon-junction complexes38. In contrast, 4E-BP1 acts as a negative regulator of translation, interacting with eIF4E and inhibiting eIF4F complex assembly. Phosphorylation of 4E-BP1 at multiple residues by mTORC1 prevents 4E-BP1 interaction with eIF4E, allowing eIF4E interaction with eIF4G and, consequently, cap-dependent translation7-10, 12. In cardiomyocytes, mTORC1 is also required for protein synthesis, in which it acts through the inhibition of 4E-BP115-17, 39. mTOR gene deletion in these cells leads to a dramatic reduction of protein synthesis due to 4E-BP1 accumulation.
2.2 mTORC1 and metabolism
mTORC1 regulates the de novo synthesis of cellular membrane lipids through the activation of sterol responsive element binding protein 1/2 (SREBP 1/2) transcription factors 4, which trigger the expression of genes involved in fatty acid metabolism and cholesterol biosynthesis. SREBP 1/2 activation is also required for the oxidative pentose phosphate pathway, a process that generates metabolic intermediates needed for cellular growth 40. mTORC1 can activate SREBP1/2 directly by phosphorylation40 or indirectly, either through the S6K1 pathway or by mTORC1-mediated phosphorylation of Lipin-1, a negative regulator of SREBP 1/241. Recently, glutamylprolyl-tRNA synthetase (EPRS) has emerged as a new downstream target of the mTORC1/S6K1 pathway in the regulation of lipid metabolism. When EPRS is phosphorylated at Ser999 by S6K1, it interacts with the fatty acid transporter protein 1 (FATP1). FATP1 then translocates to lipid membranes and mediates long fatty acid uptake. However, the mechanism by which EPRS activates FATP1 requires further investigation42.
mTORC1 also enhances the expression of genes involved in mitochondrial biogenesis through activation of the transcription factor yin-yang 1 (YY1)/peroxisome proliferator–activated receptor γ coactivator-1α (PGC-1α) transcriptional complex43. In addition, mTORC1 favors cellular growth by promoting nucleotide synthesis through activation of the activating transcription factor 4 (ATF4)44. Lastly, mTORC1 promotes a metabolic shift from fatty acid oxidation toward glycolysis by enhancing the expression of hypoxia-inducible factor-1α (HIF- α), which regulates the expression of genes involved in glycolysis 40. Hearts of mice with cardiac raptor gene ablation display an increase in carbohydrate metabolism together with a decrease in the expression of fatty acid regulatory genes16.
2.3 Autophagy and lysosomal function
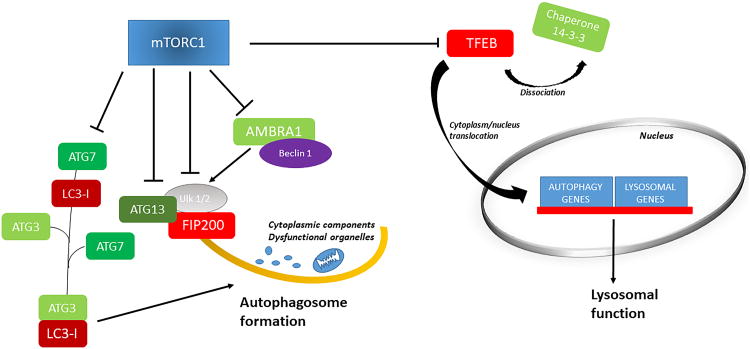
mTORC1 is an important negative regulator of autophagy (Figure 4). Autophagy is an evolutionarily conserved intracellular self-digestion mechanism that removes misfolded proteins and dysfunctional organelles45, 46. These damaged cargos are sequestered by double-membrane vesicles called autophagosomes, which ultimately fuse with lysosomes to form autophagolysosomes, in which the damaged cargoes are digested47-49. Once digested, the resulting molecular components (amino acids, lipids and carbohydrates) are recycled and reintroduced into the cellular metabolic processes. In this way, autophagy contributes to cellular homeostasis, especially during stress and nutrient deprivation. Autophagy is activated in response to cellular stress and mTORC1 inhibition contributes to autophagy activation under these conditions. mTORC1 inhibits autophagy at both the transcriptional and post-translational levels9, 12, 45, 46, impairing both autophagosome and autolysosome formation50. mTORC1 phosphorylates unc-51-like kinase (ULK) 1 and ATG13, thereby inhibiting the activity of the ULK1-ATG13-FIP200 multiprotein complex, which is crucial for autophagosome formation51. In the presence of nutrients, phosphorylation of ULK1 by mTORC1 also impairs its binding to AMPK, which phosphorylates ULK1 and activates autophagy52, 53. mTOR gene deletion in cardiomyocytes was found to be associated with autophagy activation15. The involvement of ULK1 in mediating mTOR-dependent inhibition of autophagy has been demonstrated in the hearts of cardiac raptor knockout mice and in animal models of genetic cardiomyopathy as well16, 54. In response to amino acid starvation, mTORC1 negatively regulates autophagy through inhibition of PIK3C3/VPS34 kinase, which forms a complex with ATG14 and promotes autophagosome formation. mTORC1 inhibits the ATG14-containing PIK3C3/VPS34 complex through multiple phosphorylation of ATG14 55. Previous work from Cecconi's group also showed that mTORC1 suppresses autophagy by directly phosphorylating AMBRA, a ubiquitin ligase protein of the Beclin1 complex needed for ULK1 stabilization through Lys-63-linked ubiquitination56.
Figure 4. mTORC1 modulation of autophagy and lysosomal function.
Schema representing the molecular mechanisms through which mTORC1 inhibits autophagy and lysosomal function. Legend: AMBRA1, (activating molecule in Beclin1-regulated autophagy; ATG, autophagy-related gene; FIP200, focal adhesion kinase family interacting protein of 200 kD; LC3, Light chain 3; ULK1/2, unc-51-like kinase 1/2; TFEB, transcription factor EB.
mTORC1 also transcriptionally regulates autophagy by directly regulating p73 and the transcription factor EB (TFEB). The latter regulates autophagy by enhancing the expression of autophagic proteins such as ATG7, which is fundamental for the initiation of autophagy45, 57, 58. However, mTORC1 activation in cardiomyocytes dramatically reduces ATG7 expression levels54, 59. Specifically, mTORC1 phosphorylates TFEB at Ser211, impairing its dissociation from the cytosolic chaperone 14-3-3 and consequent translocation into the nucleus, where it enhances the expression of lysosomal genes60-62. mTORC1 may also negatively regulate lysosomal biogenesis through regulation of the TFE3 and ZKSCAN3 transcription factors63, 64. In addition, mTORC1 directly impairs the activity of lysosomal proteins required for lysosomal lumen acidification, such as the proton pump v-ATPase65, thereby affecting lysosomal function and the process of autophagosome-lysosome fusion, namely autophagic flux66.
2.4 mTORC2 function: cell survival and polarity
Although mTORC2 signaling is less characterized than the mTORC1 pathway, recent evidence clearly shows its involvement in the regulation of cellular architecture and survival (Figure 2) 35, 67. The best characterized mTORC2 substrates are those belonging to the AGC protein kinase family, including Akt, serum- and glucocorticoid-induced protein kinase 1 (SGK1), and protein kinase C-α (PKC-α). Akt is phosphorylated by mTORC2 at Ser473, which enhances Thr308 phosphorylation by phosphoinositide-dependent protein kinase-1 (PDK1) and ultimately leads to Akt activation and promotion of cell survival 68, 69. mTORC2 also regulates cell survival through phosphorylation and activation of SGK1, which is known to promote cardiomyocyte survival and inhibit hypertrophy70. Cells with genetic mTORC2 disruption display an absence of SGK1 phosphorylation 71 and increased cell death69.
We recently demonstrated the existence of crosstalk between mTORC2 and mammalian sterile 20 like kinase 1 (MST1) that is involved in the regulation of cardiomyocyte survival. MST1 is a serine/threonine kinase and the main component of the Hippo pathway. The Hippo pathway is a cellular transduction cascade that negatively regulates cellular survival and growth, partially through inhibition of YAP, a transcription co-factor that promotes survival and proliferation. We showed that mTORC2 inhibits MST1 through phosphorylation at Ser438, thereby limiting its pro-death effects19. Finally, mTORC2 also controls cell polarity and cytoskeletal architecture. Cells with genetic deletion of rictor exhibit defects in cell polarity, resulting in actin-F accumulation and inhibition of cell spreading. PKC-α and Ras homolog gene family member (Rho)-A GTPase seem to be involved in cytoskeleton organization by mTORC234, 35.
3. Mechanisms of regulation of mTOR signaling and downstream targets
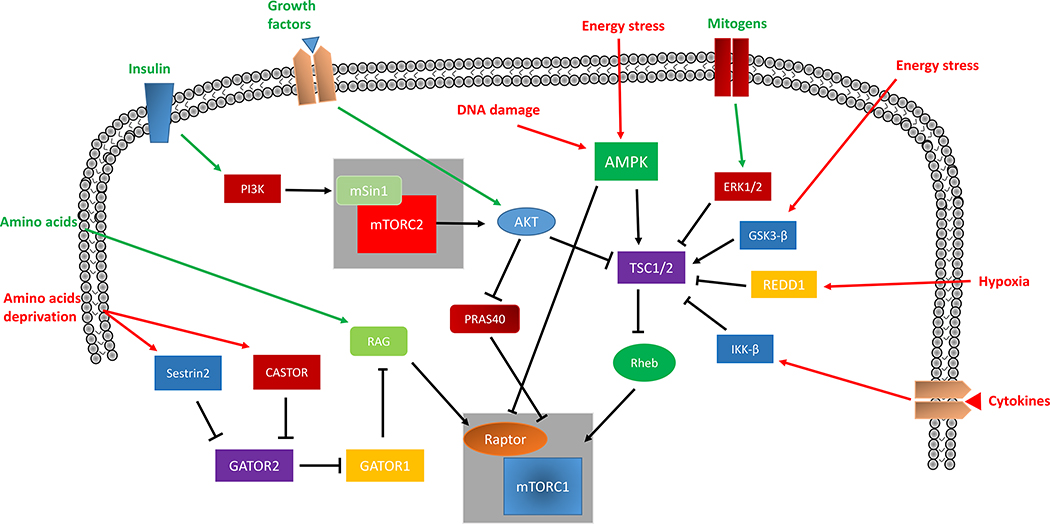
To date, several inputs are known to modulate mTOR activity. Generally, under unstressed conditions and in the presence of nutrients and growth factors, mTORC1 is activated. In contrast, mTORC1 activity is decreased in response to stress conditions such as nutrient starvation, hypoxia or DNA damage (Figure 5).
Figure 5. Schematic overview of the upstream signaling modulators of mTORC1 and mTORC2.
Signaling network regulating mTORC1 and mTORC2 activity. Arrows indicate the effects of different conditions or cellular events on mTOR activity. Red arrows represent maladaptive input signals, whereas green arrows represent adaptive input signals.
Legend: AMPK, adenosine monophosphate activated protein kinase; Akt, protein kinase B; CASTOR 1/2, cellular arginine sensor for mTORC1 1/2; ERK1/2, extracellular signal regulated kinase 1/2; GATOR 1/2, GAP activity towards Rags 1/2; GSK-3β, glycogen synthase kinase-3β; IKKβ, inhibitor of NF-κB kinase-β; PI3K, phosphoinositide 3 kinase; PRAS40, proline-rich Akt substrate 40; Rag, Ras-related GTPase; Raptor, regulatory-associated protein of mTOR; REDD1, regulated in development and DNA damage response 1; Rheb, Ras homolog enriched in brain; TSC1/2, tuberous sclerosis protein 1/2.
3.1 Growth factors
The Akt and AMPK pathways are the major upstream regulators of mTORC1. Akt is activated by nutrient-rich conditions and growth factors, and once activated Akt phosphorylates and inhibits PRAS40 and the tuberous sclerosis protein (TSC) 1/2, which are endogenous mTORC1 inhibitors7-10, 72, 73. TSC1/2 inhibit the small GTP-binding protein Ras homolog enriched in brain (Rheb), a positive regulator of mTORC174-77. TSC1/2 and Rheb regulate mTORC1 on the lysosomal membrane, where mTORC1 seems to be primarily localized74-77. Excessive lysosomal accumulation of TSC2 is also responsible for mTOR inhibition, as shown in the heart in the presence of Pompe disease78. On the other hand, upon phosphorylation and inhibition by Akt, TSC1/2 dissociate from the lysosomal membrane, preventing it from interacting with and hydrolyzing Rheb- GTP74. Extracellular signal regulated kinase 1/2 (ERK 1/2) and inhibitor of nuclear factor κB (NF-kB) kinase β (IKKβ) were also reported to mediate TSC1/2 inhibition in response to growth factors79, 80.
3.2 Energy, hypoxia and amino acids
During energy stress, nutrient deprivation and hypoxia, mTORC1 activity is reduced, leading to a shutdown of anabolic processes. AMPK was shown to inhibit mTORC1 in response to stress either directly, through the phosphorylation of Raptor with a subsequent impairment of complex assembly, or indirectly, through the activation of TSC1/277, 81-83. During energy stress, mTORC1 is inactivated by glycogen synthase kinase (GSK) 3β, which also activates TSC1/281. Activation of AMPK and GSK-3β and inhibition of Rheb also lead to mTORC1 inhibition in cardiomyocytes in response to energy deprivation47, 59, 84. p38-regulated/activated kinase also contributes to Rheb inhibition during energy deprivation, independently of TSC2. Similarly, a stress response protein named regulated in DNA damage and development 1 (REDD1) activates TSC1/2 during hypoxia independently of AMPK85. In addition, Miyamoto's group recently demonstrated that hexokinase-II (HK-II), a protein involved in the first step of glycolysis, increases autophagy and diminishes cell death in cardiomyocytes and non-cardiomyocyte cells during glucose deprivation through direct interaction with and inhibition of mTORC186. We recently demonstrated that mTOR activity is also regulated through a redox mechanism. mTOR is oxidized at Cys1483 in response to oxidative stress in cardiomyocytes, and this modification reduces its activity. Thioredoxin-1 binds to mTOR and reduces its oxidation, thereby preserving its activity, which is crucial for promoting cardiomyocyte survival under pro-oxidative conditions87.
Interestingly, accumulating lines of evidence have recently demonstrated that amino acids are crucial regulators of mTORC1 activity through the control of Rag GTPases. Specific cytoplasmic and lysosomal amino acids maintain Rag in an active conformation, allowing it to positively interact with Raptor, leading to translocation of mTORC1 to lysosomal membranes and its subsequent activation by Rheb32, 88. This evidence indicates that simultaneous Rag and Rheb activation is required for mTORC1 activation. It was also demonstrated that the lysosomal amino acid transporter SLC38A9 interacts with Rag and acts as a sensor of arginine levels, together with the lysosomal v-ATPase, to control mTORC1 activity89-91. In contrast, Sestrin2, a leucine sensor, has emerged as an mTORC1 inhibitor during amino acid deprivation. In fact, in the absence of leucine, Sestrin2 deactivates GAP activity towards Rags (GATOR) 2, a positive regulator of mTORC192, 93. Once inhibited, GATOR2 fails to bind GATOR1, which in turn deactivates mTORC1 through Rag inhibition. In fact, GATOR1 acts as the GAP protein of Rag GTPases. Similarly, the cellular arginine sensor for mTORC1 (CASTOR1) inhibits mTORC1 through the GATOR1/2 pathway in response to low arginine levels94, 95. A recent work also demonstrated that Rag A/B play an important role in the regulation of lysosomal function in cardiomyocytes96.
3.3 Regulators of mTORC2
mTORC2 activity is also modulated by multiple upstream stimuli, including insulin/phosphoinositide 3 kinase (PI3K) signaling and growth factors97. In contrast, it appears to be insensitive to nutrient deprivation8, 12. However, the regulatory subunit mSin1 inhibits mTORC2 catalytic activity in the absence of insulin98 and TSC1/2 complex may also inhibit mTORC299. Interestingly, mTORC1 acts as a negative regulator of mTORC2, through inhibition of the insulin/phosphoinositide 3 kinase (PI3K) pathway68, 69, 100. This action is mediated by the activation of growth factor receptor-bound protein 10 (Grb10), a negative regulator of the insulin/IGF-1 pathway101, 102, and by S6K1, which phosphorylates and promotes the degradation of insulin receptor substrate 1 (IRS1)103, 104. Recently, an interaction between mTORC2 and an atypical variant of the calcineurin Aβ gene was shown in mouse embryonic stem cells, which led to activation of the AKT/GSK3β/β-catenin pathway and cellular differentiation towards the mesodermal lineage105.
4. mTOR in the heart
In recent years, the mTOR complexes have emerged as crucial regulators of cardiovascular embryonic development, homeostasis and adaptation to stress. In general, mTORC1 and mTORC2 are essential for the preservation of cardiac structure, growth and vascular integrity in both prenatal and postnatal stages. They are also required for cardiac adaptation to mechanical stress, contributing to the development of compensatory hypertrophy and limiting cardiomyocyte death (Table 1). However, mTORC1 activation in the heart during chronic stress has also been shown to have multiple maladaptive effects, such as the promotion of pathological hypertrophy, misfolded protein accumulation and energy stress. In fact, partial inhibition of mTORC1 activity was shown to reduce cardiovascular damage in response to pressure overload, acute and chronic ischemic injury and aging, and in both genetic and metabolic cardiomyopathies (Table 1).
Table 1.
Cardiac effects of genetic and pharmacological mTOR modulation during development, physiology and stress.
| Cardiac condition | mTOR modulation/ animal model | Cellular modifications and cardiac outcomes | Reference |
|---|---|---|---|
| Development/physiological conditions | Constitutive cardiac mTOR gene deletion | ↑ Early mortality, heart failure, cardiac dilatation, cardiac dysfunction | 14 |
| Constitutive cardiac rictor gene deletion | ↓ Cardiac function over time | 19 | |
| Inducible cardiac mTOR gene deletion during adulthood | ↑ Apoptosis, mitochondrial dysfunction, autophagy, sarcomere disarray ↓ Protein synthesis ↑ Early mortality, heart failure, cardiac dilatation, cardiac dysfunction |
15,16 | |
| Constitutive cardiac mTOR gene deletion at early post-natal stage | ↑ Apoptosis ↓ Protein synthesis ↑ Early mortality, hypoxia, cardiac dilatation |
39 | |
| Constitutive cardiac Rheb gene deletion | ↓ Protein synthesis ↑ Early mortality, heart failure, cardiac growth, cardiac dysfunction |
17 | |
| Aging | mTOR inhibition using rapamycin | ↑ Energy metabolism ↓ Heart inflammation, cardiac fibrosis |
22 |
| Systemic GSK-3α gene deletion | ↓ Autophagy ↑ Sarcomere disarray ↑ Early mortality, cardiac hypertrophy, cardiac dysfunction during aging |
107 | |
| Cardiac hypertrophy | Inducible cardiac mTOR gene deletion during adulthood and pressure overload | ↓ Protein synthesis ↓ Compensatory hypertrophy ↑ Cardiac dysfunction, heart failure |
15,16 |
| mTOR inhibition using rapamycin and pressure overload | ↑ Cardiac function ↓ Cardiac hypertrophy |
110 | |
| mTOR inhibition using rapamycin and TAC | ↓ Cardiac hypertrophy, cardiac dysfunction | 113 | |
| mTOR inhibition using Akt inhibitor and volume overload | ↓ Eccentric hypertrophy | 114 | |
| Constitutive heterozygous cardiac Rheb gene deletion and pressure overload | ↓ Cardiac hypertrophy, cardiac fibrosis | 23 | |
| Cardiac PRAS40 overexpression and pressure overload | ↓ Cardiac hypertrophy, cardiac fibrosis ↑ Cardiac function |
115 | |
| Systemic TSC1 overexpression and isoproterenol administration | ↓ Cardiac hypertrophy ↑ Cardiac function |
116 | |
| Epigenetic mTOR inhibition and pressure overload | ↓ Cardiac hypertrophy ↑ Cardiac function |
119,120 | |
| Constitutive cardiac rictor gene deletion and pressure overload | ↑ Apoptosis ↑ Cardiac dilatation ↓ Compensatory hypertrophy |
19 | |
| Inducible cardiac rictor gene deletion during adulthood and pressure overload | ↑ Cardiac dysfunction, cardiac dilatation | 18 | |
| Ischemic injury | Cardiac Rheb overexpression during prolonged ischemia | ↓ Autophagy ↑ Apoptosis ↑ Infarct size |
59 |
| Cardiac dominant negative GSK-3β overexpression and systemic heterozygous GSK-3β deletion during prolonged ischemia | ↓ Autophagy ↑ Ischemic injury |
84 | |
| Cardiac PRAS40 overexpression during chronic myocardial infarction | ↓ Apoptosis ↓ Ischemic injury, cardiac remodeling ↑ Cardiac function |
20 | |
| mTOR inhibition using everolimus and chronic myocardial infarction | ↑ Autophagy ↓ Cardiac remodeling, infarct size, cardiac dilatation ↑ Cardiac function |
25 | |
| mTOR inhibition using rapamycin and S6K inhibitor and myocardial infarction | ↓ Apoptosis ↓ Ischemic injury |
128 | |
| Cardiac rictor gene deletion and myocardial infarction | ↑ Cardiac remodeling, cardiac dysfunction | 20 | |
| Cardiac dominant negative GSK-3β overexpression and systemic heterozygous GSK-3β deletion and I/R | ↓ Reperfusion injury | 84 | |
| Genetic disorders | mTOR inhibition using rapamycin in a model of cardiomyopathy caused by mutation in Laminin A/C gene | ↑ Autophagy ↓ Cardiac remodeling ↑ Cardiac function |
54,138 |
| mTOR inhibition using rapamycin in a mouse model of LEOPARD syndrome | ↓ Cardiac hypertrophy, myocardial disarray | 136 | |
| Metabolic disorders | mTOR inhibition using rapamycin in high-fat diet-induced obesity | ↑ Autophagy ↑ Cardiac function |
141 |
| Systemic Akt2 knockout in high-fatdiet-induced obesity | ↑ Autophagy ↑ Cardiac function |
142 | |
| mTOR inhibition using rapamycin in type II diabetic mice | ↓ Plasma glucose, triglyceride levels, oxidative stress ↑ Cardiac function |
145 | |
| Cardiac PRAS40 overexpression in diabetic mice | ↑ Metabolic function, cardiac function ↓ Cardiac hypertrophy |
146 |
Legend: Akt, protein kinase B; GSK, glycogen synthase kinase; I/R, ischemia/reperfusion; mTOR, mechanistic target of rapamycin; PRAS40, proline-rich Akt substrate 40; Rheb, Ras homolog enriched in brain; rictor, rapamycin insensitive companion of mTOR; S6K, S6 kinase; TAC, transverse aortic constriction; TSC1, tuberous sclerosis protein 1.
The most recent evidence regarding the role of mTOR complexes in the cardiovascular system will be discussed in the following paragraphs.
4.1 mTORC1 and the heart under unstressed conditions
mTOR regulates cardiac development and function both during embryogenesis and post-natal life14, 15, 69. Mice with systemic deletion of mTORC1 components die in utero and display multiple cardiac and vascular derangements 5. Similarly, live born mice with constitutive cardiac-specific mTOR gene disruption suffer from cardiac dilatation, heart failure and metabolic derangements14. Similar results were observed in adult mice with inducible cardiomyocyte-restricted deletion of mTOR and Raptor genes, in which cardiac dilatation, dysfunction, heart failure and early mortality were observed soon after induction, together with sarcomere disarray, apoptosis, autophagy and mitochondrial dysfunction15. Cardiac-specific deletion of the Rheb gene was also associated with mTORC1 inhibition and rapid development of dilated cardiomyopathy, resulting in early mortality within 10 days after birth, although cardiomyocyte apoptosis was not observed in this model. Mechanistically, the detrimental effects of cardiac mTORC1 disruption in the postnatal stage appear to be partially mediated by 4E-BP1 accumulation15, with consequent impairment of protein translation and a lack of fundamental protein production within cardiomyocytes. In support of this notion, simultaneous genetic deletion of 4E-BP1 partially reversed the dramatic phenotype observed in mTOR and Rheb knockout mice15-17. In a recent study, mice with early postnatal cardiac deletion of the mTOR gene displayed heart dilatation, cardiac fibrosis, apoptosis and heart failure, and died after three weeks. Apoptosis was associated with the accumulation of p53 and ankyrin repeat domain 1. These mice also displayed JNK activation, HIF-1α downregulation and a reduction of cardiac myoglobin content. These data suggest that mTOR in the early postnatal myocardium may be required for cardiomyocyte growth and oxygen supply maintenance39. On the other hand, no differences were observed in the phosphorylation status of the co-transcription factor YAP1; the activities of other components of the Hippo pathway, such as MST1/2 and LATS1/2, were not evaluated.
While these data indicate that complete disruption of the mTOR pathway is catastrophic for the heart, accumulating lines of evidence have demonstrated that partial inhibition of mTORC1 activity is beneficial for the heart during the aging process, likely due to reductions in energy expenditure and misfolded protein accumulation and to the activation of autophagy. Both partial genetic and pharmacological mTORC1 inhibition were shown to extend lifespan in lower organisms and in mammals21, 22, 24. Pharmacological inhibition of mTOR with rapamycin reduced age-related heart inflammation and fibrosis, and improved energy metabolism22. Similarly, caloric restriction was shown to improve diastolic function, reduce mTORC1 activity and activate autophagy in the aged rat106. Moreover, GSK-3α knockout mice displayed premature death and age-related cardiac abnormalities, such as hypertrophy and sarcomere disruption, as a result of mTOR activation and autophagy inhibition107. Interestingly, short-term caloric restriction and rapamycin reversed cardiac hypertrophy and diastolic dysfunction in aged hearts. A global proteomic analysis after these treatments revealed a marked attenuation in age-dependent protein oxidation and ubiquitination, accompanied by an upregulation of mitochondrial proteins, such as those involved in the electron transport chain, citric acid cycle and fatty acid metabolism108.
4.2 mTORC1 and cardiac hypertrophy
mTORC1 is activated in response to hypertrophic signals, such as pressure overload, β-adrenergic stimulation, angiotensin II and IGF-1, and plays both adaptive and maladaptive roles under these conditions. On the one hand, mTORC1 is required for the development of compensatory cardiac hypertrophy and maintenance of cardiac function in response to pressure overload. Mice with inducible cardiac-specific mTOR and raptor gene deletion develop cardiac dysfunction and heart failure without compensatory hypertrophy in response to transverse aortic constriction15, 16. However, it should be pointed out that whether a compensatory form of hypertrophy that favors cardiac adaptation to mechanical stress actually exists is currently under question 109.
On the other hand, deregulated mTORC1 activation in the heart leads to the development of pathological hypertrophy. In fact, it has been repeatedly demonstrated that partial genetic or pharmacological inhibition of mTORC1 is beneficial in various pro-hypertrophic conditions 110. However, it should be noted that mTORC1 activation alone does not appear to be sufficient to induce cardiac hypertrophy. In fact, previous work showed that mTOR overexpression does not induce significant changes in ventricular mass 111, 112. This evidence suggests that mTOR activation contributes to the development of cardiac hypertrophy together with other signaling pathways only in the presence of hypertrophic stimuli.
In a murine model of transverse aortic constriction (TAC)-induced cardiac hypertrophy, rapamycin reduced hypertrophy and improved cardiac function113. Similarly, pharmacological mTORC1 inhibition was found to reduce eccentric hypertrophy during volume overload induced by arteriovenous fistula in the abdominal aorta114. Mice with heterozygous cardiac deletion of Rheb showed reduced cardiac hypertrophy and fibrosis23. Cardiac overexpression of PRAS40 was also reported to reduce hypertrophy and fibrosis, improving cardiac function in response to pressure overload115. Moreover, mice with systemic TSC1 overexpression driven by the ubiquitin C promoter displayed decreased mTORC1 activity in the heart, preserved cardiac function and reduced hypertrophy in response to isoproterenol in vivo116.
Multiple mechanisms contribute to the regulation of mTORC1 activation in the heart in response to hypertrophic stimuli and control the development of pathological hypertrophy.
Cardiac-specific deletion of Folliculin (FLCN), a tumor suppressor and negative regulator of mTORC1 activity, caused cardiac hypertrophy and dysfunction, which were reversed by rapamycin treatment117. Cardiac deletion of TSC2 also resulted in cardiac dysfunction and cardiomyocyte hypertrophy, associated with an impairment of autophagic flux, which were reversed by pharmacological autophagy reactivation118.
Epigenetics also play a role in the regulation of mTOR during hypertrophy. Both genetic and pharmacological inhibition of class I histone deacetylases (HDACs) suppress pathological cardiac hypertrophy in mice subjected to pressure overload through TSC2 activation119. Interestingly, the heart-enriched long non-coding (lnc)RNA named ‘Cardiac hypertrophy-associated epigenetic regulator’ (Chaer) has recently emerged as a positive epigenetic regulator during cardiac hypertrophy development under the control of mTORC1. Cardiac disruption of Chaer reduced pathological cardiac hypertrophy and improved heart function in response to pressure overload. Chaer interacts with and inhibits the histone methyltransferase Polycomb Repressor Complex 2 (PRC2) in an mTORC1-dependent manner, reducing the level of histone methylation of pro-hypertrophic genes, thereby inducing their transcription120.
MicroRNAs also appear to regulate cardiac hypertrophy by modulating mTOR activity. Mice with cardiac-specific overexpression of miR-211 showed cardiac dysfunction and heart failure, associated with reduced autophagy and increased mTOR activity. miR-211 downregulates the cyclin-dependent kinase (CDK) inhibitor p27, which in turn activates mTOR through CDK2-dependent mechanisms, thereby leading to heart failure121. Cardiac-specific miR-199a transgenic mice also developed spontaneous cardiac hypertrophy associated with a reduction of autophagy, through downregulation of GSK-3β and subsequent mTORC1 activation122. In contrast, mTOR is a direct target of miR-99a, which was shown to attenuate cardiac hypertrophy and reduce the mTOR/P70/S6K pathway in response to pressure overload123. Of note, in most of these studies the direct contribution of mTOR modulation to the effects of different epigenetic modifications was not clarified.
The valosin-containing protein (VCP), belonging to the type II AAA (ATPases associated with various cellular activities) ATPase family, modulates mTORC1 during pressure overload. VCP-overexpressing mice show a reduction of left ventricular hypertrophy and mTORC1 inhibition124. The DNA-damage-inducible transcript 4-like (DDiT4L) regulates mTOR activity in cardiomyocytes and is highly expressed in the heart of a murine model of pathological hypertrophy compared to a model of physiological hypertrophy. Overexpression of DDiT4L in the heart inhibits mTORC1, activates mTORC2, and stimulates autophagy, and these effects are associated with a subtle reduction of systolic function125. The specific contribution of mTOR complexes to the effects induced by DDiT4L was not investigated, but the authors found that heterozygous Beclin-1 gene deletion reversed the phenotype of the transgenic mice, suggesting that autophagy activation may contribute to the detrimental cardiac effects induced by DDiT4L overexpression. Recently, two novel obscurins (Obsc), Obsc40 and Obsc80, which are localized at the intercalated disc, have emerged as negative modulators of cardiomyocyte cell size and adhesion through inhibition of the mTOR pathway126. Additionally, p38γ and p38δ, two MAPKs involved in the regulation of stress response, modulate both physiological and pathological hypertrophy by enhancing mTOR activation through phosphorylation and consequent proteasome-mediated degradation of DEPTOR127.
4.3 mTORC1 and ischemic damage
An increasing body of evidence suggests that mTORC1 also regulates cardiac adaptation to energy deprivation and ischemia. We previously demonstrated that mTORC1 activity is reduced in cardiomyocytes under these conditions through inhibition of Rheb59. Reactivation of Rheb/mTORC1 inhibits autophagy through ATG7 downregulation and promotes cardiomyocyte death59. In line with this evidence, we also found that inhibition of cardiac GSK-3β during prolonged myocardial ischemia attenuates ischemia-induced mTORC1 inhibition, inhibits autophagy and increases ischemic injury84. This detrimental effect of GSK-3β inhibition was reversed by rapamycin treatment. Overall, these results suggest that inhibition of the mTORC1 pathway is an adaptive response in cardiomyocytes, where it promotes cell survival-promoting mechanisms, such as autophagy, energy preservation, and reduction of misfolded protein accumulation 59.
mTORC1 activation also contributes to pathological cardiac remodeling in response to chronic ischemic injury 20, 25. Attenuation of mTORC1 activity with RAD01 treatment activates autophagy and reduce chronic post-infarction cardiac remodeling25. Administration of rapamycin and S6K inhibitors during myocardial infarction also reduces cardiac ischemic damage and apoptosis through a PDK1-Akt-dependent pathway 128. Interestingly, mTORC1 inhibition through overexpression of PRAS40 reduces post-infarction cardiac remodeling and improves cardiac function through activation of the mTORC2-Akt pathway20. Accordingly, cardiac-specific knockdown of rictor exacerbated cardiac remodeling and dysfunction after myocardial infarction20. These data demonstrate that pharmacological inhibition of mTORC1 attenuates chronic ischemic remodeling and highlight the importance of a proper balance between mTORC1 and mTORC2 activities in the regulation of the cardiac adaptation to stress.
In contrast, the role of mTORC1 in response to reperfusion injury is still debated. Although previous work demonstrated that pharmacological inhibition of mTORC1 before the ischemic phase reduces final ischemia/reperfusion damage129, other studies showed that mTORC1 inhibition is ineffective when administered at the time of reperfusion injury84. Prolonged rapamycin treatment also decreases cardiac function and increases myocardial necrosis in healthy pigs, although these effects may be mediated by mTORC2 downregulation since long-term rapamycin treatment also impairs mTORC2 activity130. Rapamycin abrogates the beneficial effects of ischemic preconditioning131, despite the fact that mTORC1 is already inhibited in response to ischemic preconditioning through G9a-mediated hypermethylation of the H3K9 site132. Overall, these data may suggest that mTORC1 activation is adaptive in response to reperfusion injury. In support of this notion, we previously found that heterozygous deletion of the GSK-3β gene reduces reperfusion injury through mTORC1 activation84. Cardiac-specific overexpression of Creb binding protein/p300–interacting transactivator with ED-rich carboxy-terminal domain (CITED)-4 also reduces cardiac remodeling after ischemia/reperfusion injury through activation of the mTORC1 pathway133. In addition, mice with mTOR overexpression in cardiomyocytes displayed reduced necrosis, inflammation and cardiac dysfunction after ischemia/reperfusion134. These results are in accordance with previous compelling data from Kirshembaum's group demonstrating that mTOR overexpression reduces cardiomyocyte death in hypoxic conditions through the activation of NF-kB and downregulation of BCL2/adenovirus E1B 19 kDa protein–interacting protein 3 (Bnip3), a mitochondrial death gene135. However, the specific contributions of mTORC1 and mTORC2 in the beneficial effects exerted by mTOR overexpression were not evaluated in these studies. mTORC1 activation during reperfusion injury may be beneficial through several mechanisms. It may reduce maladaptive autophagy47, it may limitmPTP opening 84 and it may also promote mitochondrial biogenesis and function43, which is important for mitochondrial turnover, new energy production and functional cardiac recovery.
4.4 mTORC1 in genetic cardiomyopathies
Dysregulation of the mTOR pathway was also found to be associated with several genetic forms of cardiomyopathy. mTORC1 is hyperactivated in the heart of a mouse model of LEOPARD disease, which is characterized by hypertrophic cardiomyopathy, fibrosis and cardiac dysfunction. Rapamycin administration reduced hypertrophic cardiomyopathy and myocardial disarray136. Consistent with this result, mTOR activity was found to be increased in a variety of models of hypertrophic cardiomyopathies, including those caused by mutations in the TRIM63 gene, encoding for Muscle RING finger protein 1 (MuRF1), and in the Laminin A/C gene54, 137, 138. In the latter model, pharmacological inhibition of mTORC1 activated autophagy, reduced cardiac remodeling and improved cardiac function54, 138. In addition, de novo S75Y mutation of RagC GTP-binding protein is associated with the development of syndromic fetal dilated cardiomyopathy. This mutation leads to a reduction of RagC activity and its overexpression increases mTORC1 activity, making cells insensitive to amino acid deprivation in vitro139. This evidence corroborates previous evidence demonstrating that cardiac disruption of RagA/B proteins leads to a massive hypertrophic cardiomyopathy mimicking lysosomal storage disease. Finally, high levels of mTORC1 activation in human myocardial biopsies are associated with increased myocardial fibrosis and a worse prognosis in patients with non-ischemic cardiomyopathy, indicating that the impact of mTORC1 activation on the development and progression of genetic cardiomyopathies is clinically relevant140.
4.5 mTORC1 in metabolic cardiomyopathy
Cardiac activation of mTORC1 also contributes to the development and progression of cardiac abnormalities induced by metabolic derangements, such as diabetes, obesity and metabolic syndrome. In obese mice, mTOR activation impairs autophagosome formation, causing a reduction of cardiac functionality, which is rescued by rapamycin administration 141. Similarly, deregulated activation of the Akt2/mTORC1 pathway contributes to the development of cardiac dysfunction and autophagic flux abnormalities in response to high fat diet consumption 142. Cardiac activation of the mTOR pathway is also responsible for the suppression of autophagosome formation in the hearts of ob/ob mice, independently of IGF1/Akt signaling143. mTOR activation is observed in the heart of a swine model of metabolic syndrome and is correlated with a reduction of autophagy, with increased apoptosis and with the development of cardiac dysfunction 144. In type II diabetic mice, mTOR inhibition by chronic administration of rapamycin improves cardiac function, with a significant reduction of plasma glucose, insulin and triglyceride levels, and an attenuation of oxidative stress 145. In addition, mTORC1 inhibition by PRAS40 treatment prevents the development of high fat diet-induced diabetic cardiomyopathy, by improving metabolic function and insulin receptor signaling 146. Metabolic syndrome and diabetes are often associated with the development of renal insufficiency, and mTOR inhibition attenuates cardiac fibrosis in uremic cardiomyopathy147.
mTORC1 activation is also responsible for the reduction of myocardial tolerance to stress induced by metabolic defects. We previously found that ischemia-induced activation of autophagy is inhibited in the hearts of mice with high fat diet-induced metabolic syndrome through a dysregulation of Rheb/mTORC1 signaling. Either pharmacological or genetic mTORC1 inhibition rescues autophagy and decreases infarct size after ischemia in these animals59. Rapamycin also protects against ischemia/reperfusion injury through activation of the STAT3 signaling pathway in diabetic hearts148.
Interestingly, increased circulating levels of branched-chain amino acids (BCAA) can usually be observed in patients with metabolic abnormalities149. Increased BCAA levels may contribute to the activation of myocardial mTORC1 and to the increased myocardial susceptibility to ischemic injury. In this regard, oral BCAA administration exacerbates cardiac hypertrophy and remodeling in response to chronic myocardial infarction, and these detrimental effects are alleviated by rapamycin administration150.
4.6 The direct role of mTOR in cardiac fibroblasts
As described in the previous paragraphs, deregulated mTOR activation in cardiomyocytes often leads to the development of myocardial fibrosis. However, the direct role of mTOR in cardiac fibroblasts has been poorly investigated. A number of different profibrotic stimuli lead to mTOR activation in cardiac fibroblasts, and mTOR upregulation in this context is associated with an increase in collagen synthesis, fibroblast proliferation and transformation of fibroblasts into myofibroblasts151. mTOR was also found to mediate the cardiac profibrotic effects induced by the epidermal growth factor-like growth factor (HB-EGF). In the hearts of transgenic mice overexpressing HB-EGF, the Akt and mTOR signaling pathways were upregulated in cardiac fibroblasts, together with S6K. Furthermore, in cultured cardiac fibroblasts treated with HB-EGF, Akt-mTOR pathway activation correlated with collagen synthesis and fibroblast proliferation152. Similarly, in rat cardiac fibroblasts, angiotensin II induced the transformation of cardiac fibroblasts into myofibroblasts, which was associated with mTOR activation and AMPK downregulation153. In addition, a previous study demonstrated that knockdown of Sestrin 1 in cardiac fibroblasts leads to mTOR activation, promotes fibroblast proliferation, enhances collagen synthesis and stimulates connective tissue growth factor (CTGF) production, both at baseline and in response to angiotensin II treatment154. It should be pointed out that the direct contribution of mTOR signaling to fibroblast activation and collagen synthesis was not completely demonstrated in these studies.
4.7 The role of mTORC2 in cardiac homeostasis and stress
mTORC2 appears to be essential for normal cardiac development and for the maintenance of postnatal cardiac structure and function. Global deletion of rictor is embryonically lethal, although rictor-/- embryos die significantly later than raptor-/- ones69, suggesting that mTORC2 is important in a later stage of embryonic development with respect to mTORC1. Nonetheless, rictor-/- embryos show marked cardiovascular abnormalities34. Mice with constitutive cardiac-specific rictor gene deletion are born normally but develop signs of cardiac dysfunction at 6 months of age19, in keeping with previous evidence that heterozygous rictor gene deletion reduces life span155. On the other hand, cardiac deletion of the rictor gene in adulthood does not significantly affect baseline cardiac function18.
mTORC2 disruption also impairs the ability of the heart to adapt to stress. We previously found that mice with α-MHC-Cre-mediated cardiac deletion of the rictor gene develop cardiac dilation, fibrosis and apoptosis in response to 4 weeks of pressure overload. These mice also display an attenuated left ventricular hypertrophic response. We found that mTORC2 interacts with and negatively regulates MST1 by phosphorylating it at Ser438 in the SARAH domain, which is important for MST1 dimerization and activation. As a result, the activity of MST1 was significantly increased in the hearts of rictor knockout mice compared to in controls, and inhibition of this molecule in vivo, by the overexpression of a dominant negative form of MST1, rescued their phenotype19. Mice with tamoxifen-induced cardiomyocyte-specific rictor gene deletion develop cardiac dysfunction and dilation in response to 1 week of pressure overload, confirming that mTORC2 plays a pivotal role in cardiac adaptation to mechanical stress. Interestingly, these mice do not show significant differences in total ventricular weight with respect to controls, suggesting a similar hypertrophic response18. However, this result might also be interpreted as a defect in the development of compensatory hypertrophy, since ventricular weight should be higher in rictor knockout mice as a consequence of the presence of cardiac dysfunction and dilation18. In support of this latter hypothesis, a recent work demonstrated that stromal interaction molecule 1 (STIM1) is required for the development of compensatory hypertrophy and preservation of cardiac function in response to pressure overload through direct activation of the mTORC2/Akt pathway and inhibition of GSK-3β, a regulator of cardiomyocyte growth156.
mTORC2 also appears to limit ischemic injury. Volkers et al. previously demonstrated that cardiac rictor gene knockdown accelerates cardiac dysfunction and remodeling in response to chronic myocardial infarction, suggesting that mTORC2 exerts important anti-remodeling effects in response to myocardial infarction20. The cardioprotective effects of ischemic preconditioning appear to be mediated by activation of the mTORC2/Akt pathway. In fact, dual mTORC inhibitors abrogated the beneficial effects of ischemic preconditioning but rapamycin did not 157. Likewise, mTORC2 has been shown to mediate the cardioprotective effects of hydrogen sulphide in response to ischemia/reperfusion in rats158.
5. Perspectives
Several aspects of the pathophysiological functions of mTOR in the cardiovascular system have been characterized, but still many others need to be understood.
mTOR complexes exert both adaptive and maladaptive functions and future studies are encouraged to understand the mechanisms underlying these dual functions, which may depend upon the condition and the cell type in which the activity of mTOR complexes is modulated. The level of activation and inhibition may also determine whether the function of mTOR is physiological or pathological. However, it is also likely that the cellular effects of mTOR signaling may depend upon its regulators and upstream signals or may be determined by the specific downstream targets that are modulated given the specific condition and timepoint. For this reason, it is important to identify other regulators and effectors of mTOR, as well as adaptor proteins. In addition, the subcellular localization of mTOR under different conditions needs to be determined. This information may help identify new therapeutic targets that would selectively stimulate the beneficial effects of the mTOR pathway while inhibiting the detrimental ones.
In particular, the molecular mechanisms through which mTORC1 inhibition exerts beneficial effects during cardiac stress need to be clearly dissected. Autophagy activation, a reduction of misfolded protein accumulation, energy-saving mechanisms or a combination of all these mechanisms may be involved in these beneficial effects. It is also possible that the mechanisms underlying the beneficial effects of mTORC1 inhibition during cardiac stress are condition-dependent. Future studies are warranted to address these issues.
Additional studies are encouraged to better characterize the cardiac phenotypes of mice with loss of function of different components of the mTOR complexes in response to specific stress conditions. These results are fundamental to understanding the specific effects of mTOR complexes in various contexts. In fact, numerous studies have used pharmacological compounds to modify the activity of mTORC1 and mTORC2 or studied the effects of modulation of their upstream regulators. However, these approaches may not be specific, generating some confusion in the interpretation of the results. In particular, characterization of the phenotypes of mTORC1 and mTORC2 knockout models in response to myocardial ischemia/reperfusion may help to clarify the role of mTOR complexes in this condition.
In addition, several pharmacological modulators of mTOR have been developed in the last years, particularly mTORC1 inhibitors. However, most of them also have multiple non-specific targets and display side effects. Therefore, it is important to find highly selective and safe modulators that may be suitable for the treatment of cardiovascular diseases. To date, mTOR inhibitors have already been tested in several clinical settings. Rapalogs, such as everolimus and temsirolimus, have shown efficacy against certain types of cancer, such as renal cell carcinoma and mantle cell lymphoma159, 160. A new class of dual PI3K/mTOR inhibitors, called ATP competitive inhibitors, have also been tested in phase I and phase II clinical trials in cancer patients, but their toxicity has emerged as a major problem related to their use161. On the other hand, more selective mTOR inhibitors, such as Torin1, did not show significant toxicity in phase I clinical trials161.
mTOR inhibitors have also been largely tested in patients with coronary artery disease, as pharmacological components of drug-eluting stents, which are used for the treatment of coronary artery narrowing due to atherosclerotic plaques. The release of mTOR inhibitors by these devices inhibits vascular smooth muscle cell proliferation and reduces the rate of stent restenosis after implantation. Of note, a clinical trial is currently recruiting participants with the objective of evaluating the effect of oral administration of everolimus on the infarct size after an acute myocardial infarction (NCT01529554). In addition, several trials tested the effects of mTOR inhibitors after cardiac transplantation162, 163. Additional trials investigating the effects of mTOR inhibitors for the treatment of heart failure, cardiac remodeling and genetic or metabolic cardiomyopathy may also be organized in the future. However, dual mTOR inhibitors may not be completely beneficial for the treatment of cardiac diseases. In fact, preclinical studies demonstrated that mTORC2 exerts beneficial effects in the heart during stress and that abrogation of these functions by dual mTOR inhibitors may result in a lack of efficacy or even harmful effects. Very few selective modulators of mTORC2 are available. Pharmacological mTORC2 activation may also represent a potential therapeutic approach for the treatment of cardiovascular diseases although this hypothesis needs to be fully tested in preclinical studies.
The direct role of the mTOR pathway in cardiac fibroblasts and vascular cells also requires further investigation.
Finally, the activity of mTOR complexes should be evaluated in human specimens from patients with cardiovascular diseases in order to evaluate its impact on cardiovascular outcomes.
Acknowledgments
The authors thank Daniela Zablocki for critical reading of the manuscript. This work was supported in part by U.S. Public Health Service Grants HL67724, HL91469, HL102738, HL112330 and AG23039 (J.S.) and by the Leducq Foundation Transatlantic Network of Excellence (J.S.). This work is also partially supported by a grant from the Italian Ministry of Health to SS (GR-2013-02355401).
Abbreviation List
- 4EB-P1
eukaryotic translation initiation factor 4E (eIF4E)-binding protein-1
- Akt
protein kinase B
- AMBRA1
activating molecule in Beclin1-regulated autophagy
- AMPK
adenosine monophosphate activated protein kinase
- ATG
autophagy-related gene
- CASTOR 1/2
cellular arginine sensor for mTORC1 1/2
- DEPTOR
DEP domain-containing mTOR-interacting protein
- eIF4
eukaryotic translation initiation factor 4
- FIP200
focal adhesion kinase family interacting protein of 200 kD
- FKBP12
FK506-binding protein of 12 kDa–rapamycin complex
- FoxO1/3
Forkhead box O1/3
- GATOR 1/2
GAP activity towards Rags 1/2
- GSK-3β
glycogen synthase kinase-3β
- LC3
Light chain 3
- mLST8
mammalian lethal with sec-13 protein 8
- mTOR
mammalian or mechanistic target of rapamycin
- mTORC1
mTOR complex 1
- mTORC2
mTOR complex 2
- PGC-1α
peroxisome proliferator–activated receptor γ coactivator-1α
- PI3K
phosphoinositide 3 kinase
- PKC-α
protein kinase C-α
- PRAS40
proline-rich Akt substrate 40
- Protor 1/2
protein observed with rictor 1/2
- Raptor
regulatory-associated protein of mammalian target of rapamycin (mTOR)
- Rheb
Ras homolog enriched in brain
- Rictor
rapamycin insensitive companion of mTOR
- S6K1
S6 kinase-1
- SGK1
serum- and glucocorticoid-induced protein kinase-1
- SREBP 1/2
sterol regulatory element-binding protein 1/2
- Tel1/2
Tel two interacting protein 1
- TFEB
transcription factor EB
- TSC1/2
tuberous sclerosis protein 1/2
- ULK1/2
unc-51-like kinase 1/2
Footnotes
Conflict of interests: The authors declare that they have no competing interests.
References
- 1.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–8. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 2.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 3.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–22. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 4.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–52. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–64. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 7.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–65. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–9. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–45. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 12.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Pires KM, Whitehead KJ, Olsen CD, Wayment B, Zhang YC, Bugger H, Ilkun O, Litwin SE, Thomas G, Kozma SC, Abel ED. Mechanistic target of rapamycin (Mtor) is essential for murine embryonic heart development and growth. PLoS One. 2013;8:e54221. doi: 10.1371/journal.pone.0054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Contu R, Latronico MV, Zhang J, Zhang JL, Rizzi R, Catalucci D, Miyamoto S, Huang K, Ceci M, Gu Y, Dalton ND, Peterson KL, Guan KL, Brown JH, Chen J, Sonenberg N, Condorelli G. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest. 2010;120:2805–16. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shende P, Plaisance I, Morandi C, Pellieux C, Berthonneche C, Zorzato F, Krishnan J, Lerch R, Hall MN, Rüegg MA, Pedrazzini T, Brink M. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123:1073–82. doi: 10.1161/CIRCULATIONAHA.110.977066. [DOI] [PubMed] [Google Scholar]
- 17.Tamai T, Yamaguchi O, Hikoso S, Takeda T, Taneike M, Oka T, Oyabu J, Murakawa T, Nakayama H, Uno Y, Horie K, Nishida K, Sonenberg N, Shah AM, Takeda J, Komuro I, Otsu K. Rheb (Ras homologue enriched in brain)-dependent mammalian target of rapamycin complex 1 (mTORC1) activation becomes indispensable for cardiac hypertrophic growth after early postnatal period. J Biol Chem. 2013;288:10176–87. doi: 10.1074/jbc.M112.423640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shende P, Xu L, Morandi C, Pentassuglia L, Heim P, Lebboukh S, Berthonneche C, Pedrazzini T, Kaufmann BA, Hall MN, Rüegg MA, Brink M. Cardiac mTOR complex 2 preserves ventricular function in pressure-overload hypertrophy. Cardiovasc Res. 2016;109:103–14. doi: 10.1093/cvr/cvv252. [DOI] [PubMed] [Google Scholar]
- 19.Sciarretta S, Zhai P, Maejima Y, Del Re DP, Nagarajan N, Yee D, Liu T, Magnuson MA, Volpe M, Frati G, Li H, Sadoshima J. mTORC2 regulates cardiac response to stress by inhibiting MST1. Cell Rep. 2015;11:125–36. doi: 10.1016/j.celrep.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Völkers M, Konstandin MH, Doroudgar S, Toko H, Quijada P, Din S, Joyo A, Ornelas L, Samse K, Thuerauf DJ, Gude N, Glembotski CC, Sussman MA. Mechanistic target of rapamycin complex 2 protects the heart from ischemic damage. Circulation. 2013;128:2132–44. doi: 10.1161/CIRCULATIONAHA.113.003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flynn JM, O'Leary MN, Zambataro CA, Academia EC, Presley MP, Garrett BJ, Zykovich A, Mooney SD, Strong R, Rosen CJ, Kapahi P, Nelson MD, Kennedy BK, Melov S. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12:851–62. doi: 10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Cao Y, Nie J, Liu H, Lu S, Hu X, Zhu J, Zhao X, Chen J, Chen X, Yang Z, Li X. Genetic and pharmacological inhibition of Rheb1-mTORC1 signaling exerts cardioprotection against adverse cardiac remodeling in mice. Am J Pathol. 2013;182:2005–14. doi: 10.1016/j.ajpath.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, Lago CU, Zhang S, DuBois W, Ward T, deCabo R, Gavrilova O, Mock B, Finkel T. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4:913–20. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buss SJ, Muenz S, Riffel JH, Malekar P, Hagenmueller M, Weiss CS, Bea F, Bekeredjian R, Schinke-Braun M, Izumo S, Katus HA, Hardt SE. Beneficial effects of Mammalian target of rapamycin inhibition on left ventricular remodeling after myocardial infarction. J Am Coll Cardiol. 2009;54:2435–46. doi: 10.1016/j.jacc.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 26.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 28.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 29.Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura S, Natsume T, Mizushima N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285:20109–16. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–4. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 31.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 32.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–86. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 35.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–8. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 36.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–71. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 38.Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–13. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 39.Mazelin L, Panthu B, Nicot AS, Belotti E, Tintignac L, Teixeira G, Zhang Q, Risson V, Baas D, Delaune E, Derumeaux G, Taillandier D, Ohlmann T, Ovize M, Gangloff YG, Schaeffer L. mTOR inactivation in myocardium from infant mice rapidly leads to dilated cardiomyopathy due to translation defects and p53/JNK-mediated apoptosis. J Mol Cell Cardiol. 2016;97:213–25. doi: 10.1016/j.yjmcc.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–20. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arif A, Terenzi F, Potdar AA, Jia J, Sacks J, China A, Halawani D, Vasu K, Li X, Brown JM, Chen J, Kozma SC, Thomas G, Fox PL. EPRS is a critical mTORC1-S6K1 effector that influences adiposity in mice. Nature. 2017;542:357–361. doi: 10.1038/nature21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–40. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 44.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sciarretta S, Zhai P, Volpe M, Sadoshima J. Pharmacological modulation of autophagy during cardiac stress. J Cardiovasc Pharmacol. 2012;60:235–41. doi: 10.1097/FJC.0b013e3182575f61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 48.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, Finkbeiner S, Fueyo-Margareto J, Gewirtz D, Jäättelä M, Kroemer G, Levine B, Melia TJ, Mizushima N, Rubinsztein DC, Simonsen A, Thorburn A, Thumm M, Tooze SA. A comprehensive glossary of autophagy-related molecules and processes (2nd edition) Autophagy. 2011;7:1273–94. doi: 10.4161/auto.7.11.17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–93. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, MacKay VL, An EH, Strong R, Ladiges WC, Rabinovitch PS, Kaeberlein M, Kennedy BK. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. 2012;4:144ra103. doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9:1983–95. doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–16. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 57.Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol. 2008;28:5951–64. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, Condorelli G, Sadoshima J. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–46. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–14. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chauhan S, Goodwin JG, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–66. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 66.Choi YJ, Park YJ, Park JY, Jeong HO, Kim DH, Ha YM, Kim JM, Song YM, Heo HS, Yu BP, Chun P, Moon HR, Chung HY. Inhibitory effect of mTOR activator MHY1485 on autophagy: suppression of lysosomal fusion. PLoS One. 2012;7:e43418. doi: 10.1371/journal.pone.0043418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Braz JC, Gregory K, Pathak A, Zhao W, Sahin B, Klevitsky R, Kimball TF, Lorenz JN, Nairn AC, Liggett SB, Bodi I, Wang S, Schwartz A, Lakatta EG, DePaoli-Roach AA, Robbins J, Hewett TE, Bibb JA, Westfall MV, Kranias EG, Molkentin JD. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat Med. 2004;10:248–54. doi: 10.1038/nm1000. [DOI] [PubMed] [Google Scholar]
- 68.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 69.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–71. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 70.Aoyama T, Matsui T, Novikov M, Park J, Hemmings B, Rosenzweig A. Serum and glucocorticoid-responsive kinase-1 regulates cardiomyocyte survival and hypertrophic response. Circulation. 2005;111:1652–9. doi: 10.1161/01.CIR.0000160352.58142.06. [DOI] [PubMed] [Google Scholar]
- 71.García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–85. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 72.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–15. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Sekulić A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM, Abraham RT. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 2000;60:3504–13. [PubMed] [Google Scholar]
- 74.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–85. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–34. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–68. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 77.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 78.Lim JA, Li L, Shirihai OS, Trudeau KM, Puertollano R, Raben N. Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol Med. 2017;9:353–370. doi: 10.15252/emmm.201606547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–93. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 80.Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, He X, Hung JY, Lai CC, Ding Q, Su JL, Yang JY, Sahin AA, Hortobagyi GN, Tsai FJ, Tsai CH, Hung MC. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–55. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 81.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–68. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 82.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–9. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 84.Zhai P, Sciarretta S, Galeotti J, Volpe M, Sadoshima J. Differential roles of GSK-3β during myocardial ischemia and ischemia/reperfusion. Circ Res. 2011;109:502–11. doi: 10.1161/CIRCRESAHA.111.249532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roberts DJ, Tan-Sah VP, Ding EY, Smith JM, Miyamoto S. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol Cell. 2014;53:521–33. doi: 10.1016/j.molcel.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oka SI, Hirata T, Suzuki W, Naito D, Chen Y, Chin A, Yaginuma H, Saito T, Nagarajan N, Zhai P, Bhat S, Schesing K, Shao D, Hirabayashi Y, Yodoi J, Sciarretta S, Sadoshima J. Thioredoxin-1 maintains mTOR function during oxidative stress in cardiomyocytes. J Biol Chem. 2017 doi: 10.1074/jbc.M117.807735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jung J, Genau HM, Behrends C. Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol Cell Biol. 2015;35:2479–94. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rebsamen M, Pochini L, Stasyk T, de Araújo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, Filipek PA, Huber KV, Bigenzahn JW, Heinz LX, Kraft C, Bennett KL, Indiveri C, Huber LA, Superti-Furga G. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–81. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, Wang T, Bar-Peled L, Zoncu R, Straub C, Kim C, Park J, Sabatini BL, Sabatini DM. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–94. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, Karin M, Budanov AV. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014;9:1281–91. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165:153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016;536:229–33. doi: 10.1038/nature19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim YC, Park HW, Sciarretta S, Mo JS, Jewell JL, Russell RC, Wu X, Sadoshima J, Guan KL. Rag GTPases are cardioprotective by regulating lysosomal function. Nat Commun. 2014;5:4241. doi: 10.1038/ncomms5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–68. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 98.Liu P, Gan W, Chin YR, Ogura K, Guo J, Zhang J, Wang B, Blenis J, Cantley LC, Toker A, Su B, Wei W. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov. 2015;5:1194–209. doi: 10.1158/2159-8290.CD-15-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–15. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009;29:5657–70. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–6. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–23. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–6. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 105.Gómez-Salinero JM, López-Olañeta MM, Ortiz-Sánchez P, Larrasa-Alonso J, Gatto A, Felkin LE, Barton PJ, Navarro-Lérida I, Ángel Del Pozo M, García-Pavía P, Sundararaman B, Giovinazo G, Yeo GW, Lara-Pezzi E. The Calcineurin Variant CnAβ1 Controls Mouse Embryonic Stem Cell Differentiation by Directing mTORC2 Membrane Localization and Activation. Cell Chem Biol. 2016;23:1372–1382. doi: 10.1016/j.chembiol.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 106.Shinmura K, Tamaki K, Sano M, Murata M, Yamakawa H, Ishida H, Fukuda K. Impact of long-term caloric restriction on cardiac senescence: caloric restriction ameliorates cardiac diastolic dysfunction associated with aging. J Mol Cell Cardiol. 2011;50:117–27. doi: 10.1016/j.yjmcc.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 107.Zhou J, Freeman TA, Ahmad F, Shang X, Mangano E, Gao E, Farber J, Wang Y, Ma XL, Woodgett J, Vagnozzi RJ, Lal H, Force T. GSK-3α is a central regulator of age-related pathologies in mice. J Clin Invest. 2013;123:1821–32. doi: 10.1172/JCI64398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, Beyer RP, MacCoss MJ, Rabinovitch PS. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13:529–39. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schiattarella GG, Hill TM, Hill JA. Is Load-Induced Ventricular Hypertrophy Ever Compensatory? Circulation. 2017;136:1273–1275. doi: 10.1161/CIRCULATIONAHA.117.030730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shioi T, McMullen JR, Tarnavski O, Converso K, Sherwood MC, Manning WJ, Izumo S. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation. 2003;107:1664–70. doi: 10.1161/01.CIR.0000057979.36322.88. [DOI] [PubMed] [Google Scholar]
- 111.Song X, Kusakari Y, Xiao CY, Kinsella SD, Rosenberg MA, Scherrer-Crosbie M, Hara K, Rosenzweig A, Matsui T. mTOR attenuates the inflammatory response in cardiomyocytes and prevents cardiac dysfunction in pathological hypertrophy. Am J Physiol Cell Physiol. 2010;299:C1256–66. doi: 10.1152/ajpcell.00338.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shen WH, Chen Z, Shi S, Chen H, Zhu W, Penner A, Bu G, Li W, Boyle DW, Rubart M, Field LJ, Abraham R, Liechty EA, Shou W. Cardiac restricted overexpression of kinase-dead mammalian target of rapamycin (mTOR) mutant impairs the mTOR-mediated signaling and cardiac function. J Biol Chem. 2008;283:13842–9. doi: 10.1074/jbc.M801510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T, Izumo S. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation. 2004;109:3050–5. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- 114.Ikeda M, Ide T, Fujino T, Matsuo Y, Arai S, Saku K, Kakino T, Oga Y, Nishizaki A, Sunagawa K. The Akt-mTOR axis is a pivotal regulator of eccentric hypertrophy during volume overload. Sci Rep. 2015;5:15881. doi: 10.1038/srep15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Völkers M, Toko H, Doroudgar S, Din S, Quijada P, Joyo AY, Ornelas L, Joyo E, Thuerauf DJ, Konstandin MH, Gude N, Glembotski CC, Sussman MA. Pathological hypertrophy amelioration by PRAS40-mediated inhibition of mTORC1. Proc Natl Acad Sci U S A. 2013;110:12661–6. doi: 10.1073/pnas.1301455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang HM, Diaz V, Walsh ME, Zhang Y. Moderate lifelong overexpression of tuberous sclerosis complex 1 (TSC1) improves health and survival in mice. Sci Rep. 2017;7:834. doi: 10.1038/s41598-017-00970-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hasumi Y, Baba M, Hasumi H, Huang Y, Lang M, Reindorf R, Oh HB, Sciarretta S, Nagashima K, Haines DC, Schneider MD, Adelstein RS, Schmidt LS, Sadoshima J, Marston Linehan W. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum Mol Genet. 2014;23:5706–19. doi: 10.1093/hmg/ddu286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taneike M, Nishida K, Omiya S, Zarrinpashneh E, Misaka T, Kitazume-Taneike R, Austin R, Takaoka M, Yamaguchi O, Gambello MJ, Shah AM, Otsu K. mTOR Hyperactivation by Ablation of Tuberous Sclerosis Complex 2 in the Mouse Heart Induces Cardiac Dysfunction with the Increased Number of Small Mitochondria Mediated through the Down-Regulation of Autophagy. PLoS One. 2016;11:e0152628. doi: 10.1371/journal.pone.0152628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morales CR, Li DL, Pedrozo Z, May HI, Jiang N, Kyrychenko V, Cho GW, Kim SY, Wang ZV, Rotter D, Rothermel BA, Schneider JW, Lavandero S, Gillette TG, Hill JA. Inhibition of class I histone deacetylases blunts cardiac hypertrophy through TSC2-dependent mTOR repression. Sci Signal. 2016;9:ra34. doi: 10.1126/scisignal.aad5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, Gao C, Yokota T, Ang YS, Li S, Cass A, Vondriska TM, Li G, Deb A, Srivastava D, Yang HT, Xiao X, Li H, Wang Y. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Su M, Wang J, Wang C, Wang X, Dong W, Qiu W, Wang Y, Zhao X, Zou Y, Song L, Zhang L, Hui R. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 2015;22:986–99. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Z, Song Y, Liu L, Hou N, An X, Zhan D, Li Y, Zhou L, Li P, Yu L, Xia J, Zhang Y, Wang J, Yang X. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ. 2017;24:1205–1213. doi: 10.1038/cdd.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Q, Xie J, Wang B, Li R, Bai J, Ding L, Gu R, Wang L, Xu B. Overexpression of microRNA-99a Attenuates Cardiac Hypertrophy. PLoS One. 2016;11:e0148480. doi: 10.1371/journal.pone.0148480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou N, Ma B, Stoll S, Hays TT, Qiu H. The valosin-containing protein is a novel repressor of cardiomyocyte hypertrophy induced by pressure overload. Aging Cell. 2017;16:1168–1179. doi: 10.1111/acel.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Simonson B, Subramanya V, Chan MC, Zhang A, Franchino H, Ottaviano F, Mishra MK, Knight AC, Hunt D, Ghiran I, Khurana TS, Kontaridis MI, Rosenzweig A, Das S. DDiT4L promotes autophagy and inhibits pathological cardiac hypertrophy in response to stress. Sci Signal. 2017;10 doi: 10.1126/scisignal.aaf5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ackermann MA, King B, Lieberman NAP, Bobbili PJ, Rudloff M, Berndsen CE, Wright NT, Hecker PA, Kontrogianni-Konstantopoulos A. Novel obscurins mediate cardiomyocyte adhesion and size via the PI3K/AKT/mTOR signaling pathway. J Mol Cell Cardiol. 2017;111:27–39. doi: 10.1016/j.yjmcc.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.González-Terán B, López JA, Rodríguez E, Leiva L, Martínez-Martínez S, Bernal JA, Jiménez-Borreguero LJ, Redondo JM, Vazquez J, Sabio G. p38γ and δ promote heart hypertrophy by targeting the mTOR-inhibitory protein DEPTOR for degradation. Nat Commun. 2016;7:10477. doi: 10.1038/ncomms10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Di R, Wu X, Chang Z, Zhao X, Feng Q, Lu S, Luan Q, Hemmings BA, Li X, Yang Z. S6K inhibition renders cardiac protection against myocardial infarction through PDK1 phosphorylation of Akt. Biochem J. 2012;441:199–207. doi: 10.1042/BJ20110033. [DOI] [PubMed] [Google Scholar]
- 129.Das A, Salloum FN, Durrant D, Ockaili R, Kukreja RC. Rapamycin protects against myocardial ischemia-reperfusion injury through JAK2-STAT3 signaling pathway. J Mol Cell Cardiol. 2012;53:858–69. doi: 10.1016/j.yjmcc.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lassaletta AD, Elmadhun NY, Zanetti AV, Feng J, Anduaga J, Gohh RY, Sellke FW, Bianchi C. Rapamycin treatment of healthy pigs subjected to acute myocardial ischemia-reperfusion injury attenuates cardiac functions and increases myocardial necrosis. Ann Thorac Surg. 2014;97:901–7. doi: 10.1016/j.athoracsur.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hausenloy DJ, Mocanu MM, Yellon DM. Cross-talk between the survival kinases during early reperfusion: its contribution to ischemic preconditioning. Cardiovasc Res. 2004;63:305–12. doi: 10.1016/j.cardiores.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 132.Gidlöf O, Johnstone AL, Bader K, Khomtchouk BB, O'Reilly JJ, Celik S, Van Booven DJ, Wahlestedt C, Metzler B, Erlinge D. Ischemic Preconditioning Confers Epigenetic Repression of Mtor and Induction of Autophagy Through G9a-Dependent H3K9 Dimethylation. J Am Heart Assoc. 2016;5 doi: 10.1161/JAHA.116.004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bezzerides VJ, Platt C, Lerchenmüller C, Paruchuri K, Oh NL, Xiao C, Cao Y, Mann N, Spiegelman BM, Rosenzweig A. CITED4 induces physiologic hypertrophy and promotes functional recovery after ischemic injury. JCI Insight. 2016;1 doi: 10.1172/jci.insight.85904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu Z, Xu X, Hu X, Fassett J, Zhu G, Tao Y, Li J, Huang Y, Zhang P, Zhao B, Chen Y. PGC-1 alpha regulates expression of myocardial mitochondrial antioxidants and myocardial oxidative stress after chronic systolic overload. Antioxid Redox Signal. 2010;13:1011–22. doi: 10.1089/ars.2009.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dhingra R, Gang H, Wang Y, Biala AK, Aviv Y, Margulets V, Tee A, Kirshenbaum LA. Bidirectional regulation of nuclear factor-κB and mammalian target of rapamycin signaling functionally links Bnip3 gene repression and cell survival of ventricular myocytes. Circ Heart Fail. 2013;6:335–43. doi: 10.1161/CIRCHEARTFAILURE.112.000061. [DOI] [PubMed] [Google Scholar]
- 136.Marin TM, Keith K, Davies B, Conner DA, Guha P, Kalaitzidis D, Wu X, Lauriol J, Wang B, Bauer M, Bronson R, Franchini KG, Neel BG, Kontaridis MI. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121:1026–43. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen SN, Czernuszewicz G, Tan Y, Lombardi R, Jin J, Willerson JT, Marian AJ. Human molecular genetic and functional studies identify TRIM63, encoding Muscle RING Finger Protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2012;111:907–19. doi: 10.1161/CIRCRESAHA.112.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Choi JC, Wu W, Muchir A, Iwata S, Homma S, Worman HJ. Dual specificity phosphatase 4 mediates cardiomyopathy caused by lamin A/C (LMNA) gene mutation. J Biol Chem. 2012;287:40513–24. doi: 10.1074/jbc.M112.404541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Long PA, Zimmermann MT, Kim M, Evans JM, Xu X, Olson TM. De novo RRAGC mutation activates mTORC1 signaling in syndromic fetal dilated cardiomyopathy. Hum Genet. 2016;135:909–917. doi: 10.1007/s00439-016-1685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yano T, Shimoshige S, Miki T, Tanno M, Mochizuki A, Fujito T, Yuda S, Muranaka A, Ogasawara M, Hashimoto A, Tsuchihashi K, Miura T. Clinical impact of myocardial mTORC1 activation in nonischemic dilated cardiomyopathy. J Mol Cell Cardiol. 2016;91:6–9. doi: 10.1016/j.yjmcc.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 141.Guo R, Zhang Y, Turdi S, Ren J. Adiponectin knockout accentuates high fat diet-induced obesity and cardiac dysfunction: role of autophagy. Biochim Biophys Acta. 2013;1832:1136–48. doi: 10.1016/j.bbadis.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu X, Hua Y, Nair S, Sreejayan N, Zhang Y, Ren J. Akt2 knockout preserves cardiac function in high-fat diet-induced obesity by rescuing cardiac autophagosome maturation. J Mol Cell Biol. 2013;5:61–3. doi: 10.1093/jmcb/mjs055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pires KM, Buffolo M, Schaaf C, David Symons J, Cox J, Abel ED, Selzman CH, Boudina S. Activation of IGF-1 receptors and Akt signaling by systemic hyperinsulinemia contributes to cardiac hypertrophy but does not regulate cardiac autophagy in obese diabetic mice. J Mol Cell Cardiol. 2017;113:39–50. doi: 10.1016/j.yjmcc.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li ZL, Woollard JR, Ebrahimi B, Crane JA, Jordan KL, Lerman A, Wang SM, Lerman LO. Transition from obesity to metabolic syndrome is associated with altered myocardial autophagy and apoptosis. Arterioscler Thromb Vasc Biol. 2012;32:1132–41. doi: 10.1161/ATVBAHA.111.244061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Das A, Durrant D, Koka S, Salloum FN, Xi L, Kukreja RC. Mammalian target of rapamycin (mTOR) inhibition with rapamycin improves cardiac function in type 2 diabetic mice: potential role of attenuated oxidative stress and altered contractile protein expression. J Biol Chem. 2014;289:4145–60. doi: 10.1074/jbc.M113.521062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Völkers M, Doroudgar S, Nguyen N, Konstandin MH, Quijada P, Din S, Ornelas L, Thuerauf DJ, Gude N, Friedrich K, Herzig S, Glembotski CC, Sussman MA. PRAS40 prevents development of diabetic cardiomyopathy and improves hepatic insulin sensitivity in obesity. EMBO Mol Med. 2014;6:57–65. doi: 10.1002/emmm.201303183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Haller ST, Yan Y, Drummond CA, Xie J, Tian J, Kennedy DJ, Shilova VY, Xie Z, Liu J, Cooper CJ, Malhotra D, Shapiro JI, Fedorova OV, Bagrov AY. Rapamycin Attenuates Cardiac Fibrosis in Experimental Uremic Cardiomyopathy by Reducing Marinobufagenin Levels and Inhibiting Downstream Pro-Fibrotic Signaling. J Am Heart Assoc. 2016;5 doi: 10.1161/JAHA.116.004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Das A, Salloum FN, Filippone SM, Durrant DE, Rokosh G, Bolli R, Kukreja RC. Inhibition of mammalian target of rapamycin protects against reperfusion injury in diabetic heart through STAT3 signaling. Basic Res Cardiol. 2015;110:31. doi: 10.1007/s00395-015-0486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sciarretta S, Boppana VS, Umapathi M, Frati G, Sadoshima J. Boosting autophagy in the diabetic heart: a translational perspective. Cardiovasc Diagn Ther. 2015;5:394–402. doi: 10.3978/j.issn.2223-3652.2015.07.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang W, Zhang F, Xia Y, Zhao S, Yan W, Wang H, Lee Y, Li C, Zhang L, Lian K, Gao E, Cheng H, Tao L. Defective branched chain amino acid catabolism contributes to cardiac dysfunction and remodeling following myocardial infarction. Am J Physiol Heart Circ Physiol. 2016;311:H1160–H1169. doi: 10.1152/ajpheart.00114.2016. [DOI] [PubMed] [Google Scholar]
- 151.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac Fibrosis: The Fibroblast Awakens. Circ Res. 2016;118:1021–40. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lian H, Ma Y, Feng J, Dong W, Yang Q, Lu D, Zhang L. Heparin-binding EGF-like growth factor induces heart interstitial fibrosis via an Akt/mTor/p70s6k pathway. PLoS One. 2012;7:e44946. doi: 10.1371/journal.pone.0044946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ai F, Chen M, Yu B, Yang Y, Xu G, Gui F, Liu Z, Bai X, Chen Z. Berberine regulates proliferation, collagen synthesis and cytokine secretion of cardiac fibroblasts via AMPK-mTOR-p70S6K signaling pathway. Int J Clin Exp Pathol. 2015;8:12509–16. [PMC free article] [PubMed] [Google Scholar]
- 154.Sun G, Xue R, Yao F, Liu D, Huang H, Chen C, Li Y, Zeng J, Zhang G, Dong Y, Liu C. The critical role of Sestrin 1 in regulating the proliferation of cardiac fibroblasts. Arch Biochem Biophys. 2014;542:1–6. doi: 10.1016/j.abb.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 155.Lamming DW, Mihaylova MM, Katajisto P, Baar EL, Yilmaz OH, Hutchins A, Gultekin Y, Gaither R, Sabatini DM. Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell. 2014;13:911–7. doi: 10.1111/acel.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bénard L, Oh JG, Cacheux M, Lee A, Nonnenmacher M, Matasic DS, Kohlbrenner E, Kho C, Pavoine C, Hajjar RJ, Hulot JS. Cardiac Stim1 Silencing Impairs Adaptive Hypertrophy and Promotes Heart Failure Through Inactivation of mTORC2/Akt Signaling. Circulation. 2016;133:1458–71. doi: 10.1161/CIRCULATIONAHA.115.020678. discussion 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yano T, Ferlito M, Aponte A, Kuno A, Miura T, Murphy E, Steenbergen C. Pivotal role of mTORC2 and involvement of ribosomal protein S6 in cardioprotective signaling. Circ Res. 2014;114:1268–80. doi: 10.1161/CIRCRESAHA.114.303562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou Y, Wang D, Gao X, Lew K, Richards AM, Wang P. mTORC2 phosphorylation of Akt1: a possible mechanism for hydrogen sulfide-induced cardioprotection. PLoS One. 2014;9:e99665. doi: 10.1371/journal.pone.0099665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Carew JS, Kelly KR, Nawrocki ST. Mechanisms of mTOR inhibitor resistance in cancer therapy. Target Oncol. 2011;6:17–27. doi: 10.1007/s11523-011-0167-8. [DOI] [PubMed] [Google Scholar]
- 160.Huang S, Bjornsti MA, Houghton PJ. Rapamycins: mechanism of action and cellular resistance. Cancer Biol Ther. 2003;2:222–32. doi: 10.4161/cbt.2.3.360. [DOI] [PubMed] [Google Scholar]
- 161.Pallet N, Legendre C. Adverse events associated with mTOR inhibitors. Expert Opin Drug Saf. 2013;12:177–86. doi: 10.1517/14740338.2013.752814. [DOI] [PubMed] [Google Scholar]
- 162.Eisen HJ, Tuzcu EM, Dorent R, Kobashigawa J, Mancini D, Valantine-von Kaeppler HA, Starling RC, Sørensen K, Hummel M, Lind JM, Abeywickrama KH, Bernhardt P, Group RBS. Everolimus for the prevention of allograft rejection and vasculopathy in cardiac-transplant recipients. N Engl J Med. 2003;349:847–58. doi: 10.1056/NEJMoa022171. [DOI] [PubMed] [Google Scholar]
- 163.Mancini D, Pinney S, Burkhoff D, LaManca J, Itescu S, Burke E, Edwards N, Oz M, Marks AR. Use of rapamycin slows progression of cardiac transplantation vasculopathy. Circulation. 2003;108:48–53. doi: 10.1161/01.CIR.0000070421.38604.2B. [DOI] [PubMed] [Google Scholar]