Abstract
The recent progress in harnessing the efficient and precise method of DNA editing provided by CRISPR/Cas9 is one of the most promising major advances in the field of gene therapy. However, the development of safe and optimally efficient delivery systems for CRISPR/Cas9 elements capable of achieving specific targeting of gene therapy to the location of interest without off-target effects is a primary challenge for clinical therapeutics. Nanoparticles (NPs) provide a promising means to meet such challenges. In this review, we present the most recent advances in developing innovative NP-based delivery systems that efficiently deliver CRISPR-Cas9 constructs and maximize their effectiveness.
Keywords: CRISPR-Cas9, gene editing, gene delivery, nanoparticle
I. Introduction
While first reported in 1987 by Ishino et al. (1), it was not until 2002 that the currently used acronym for clustered regularly-interspaced short palindromic repeats, CRISPR, started to be more widely used by scientists (2, 3). This family of DNA sequences, reported first in Escherichia coli, then later in other bacteria, including Streptococcus pyogenes, was found to be a part of the prokaryotic adaptive immune defense against bacteriophages and plasmids (4, 5), where an endogenous RNA-guided nuclease-based machinery recognizes and destroys invading foreign nucleic acid material. To explain, CRISPR loci in the DNA of bacteria and archaea comprise DNA sequence repeats separated by spacers. These spacers are derived from the fragments of foreign nucleic acid material (from phages and plasmids from a previous infection) (6) and act as the ‘memory element’ in the prokaryote’s immune defense. These sequences of repeats and spacers are usually adjacent to genes that encode for the CRISPR-associated (Cas) family of endonucleases (5). The CRISPR loci are transcribed to ultimately produce CRISPR RNA (crRNA). In the CRISPR type II system, the crRNA is base-paired with trans-activated crRNA, and this dual-RNA element, containing complimentary sequences to the exogenous (viral) DNA, ‘guides’ the Cas endonuclease protein (in this case Cas9) to the target region of the DNA and induces breaks and damage, essentially inactivating the target (7). This dual RNA guide system has now been engineered as a single guide RNA (sgRNA), sometimes referred to as gRNA, for the purposes of targeted genome editing in eukaryotic cells (7, 8). The resultant CRISPR-based genome editing systems consisting of sgRNA and Cas9 can be designed to target any specific gene by inducing double stranded DNA breaks at the targeted gene; and either knocking out mutated genes, or knocking in desired genes (7–12). Knocking in genes can be more problematic since, in addition to the delivery of the CRISPR/Cas9 components to cells, donor DNA is also required to repair mutated genes via homology directed repair. The detailed mechanisms by which these editing processes occur have been described expansively elsewhere and will not be the focus of this review (10, 12, 13).
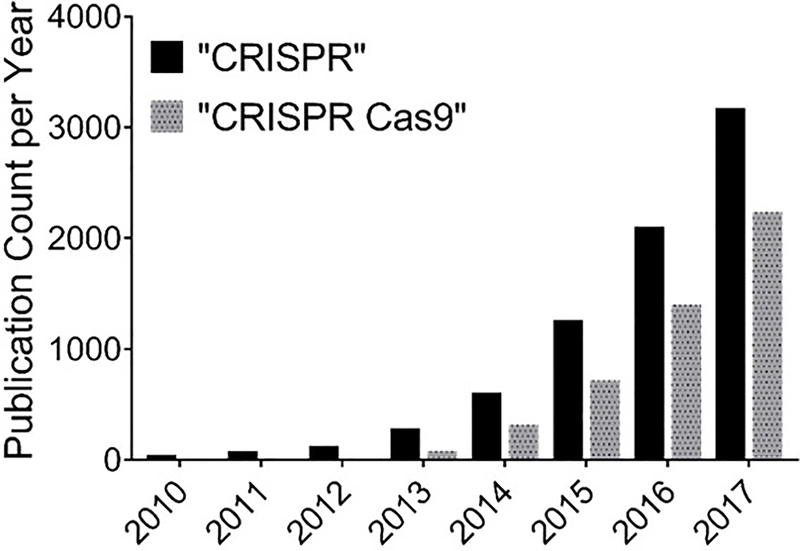
The CRISPR/Cas9 system adds a robust and versatile tool to the mammalian genome editing toolbox, which includes meganucleases, zinc finger nucleases (ZFNs), and transcription activator-like effector nucleases (TALENs). The success rates of gene editing of CRISPR/Cas9 and TALEN systems are higher than those of meganucleases and ZFNs (14). In addition, in contrast to CRISPR/Cas9 systems which requires the designing of sgRNA that compliments with the target DNA, both ZFNs and TALENs require the more demanding task of engineering of a new highly-specific protein for each target gene, since both techniques depend on protein-target DNA interaction (12). Also, unlike RNA interference (RNAi) techniques, CRISPR-Cas9 therapy is potentially a one-time treatment with lasting effects since the target is DNA as opposed to RNA (10, 15). Following the discovery of its genome editing capabilities, the number of publications that reported using CRISPR-Cas9 for gene editing has increased dramatically (Figure 1).
Figure 1.
Incidence rate of publications in the PubMed database from 2010 through December 2017 using the search terms, “CRISPR” or “CRISPR Cas9.”
In 2016, the first clinical trial using CRISPR-Cas9 therapy began in China (16). The results of this and similar studies are yet to be published in peer-reviewed articles, however, the therapy involves ex vivo genome editing of cells (knocking down PD-1 expression) that are subsequently transfused back into the patient (with metastatic lung cancer) as opposed to direct delivery of the editing apparatus to the patient. The treatment aims to improve the patient’s antitumor immune response by knocking out PD-1 gene (an immune checkpoint protein) from the patient’s own T cells. The general consensus is that direct delivery of the CRISPR/Cas9 system to patients will require a safe and efficient carrier system and, particularly for systemic delivery, this would imply the use of appropriately formulated nanocarriers or NPs. Thus, much recent research has focused on the development of novel non-viral vectors that can safely and effectively deliver the CRISPR/Cas9 elements to cells and tissues in in vitro and/or in vivo settings. NPs have emerged as an attractive option for delivering CRISPR-Cas9 based therapies for several reasons. First, they can be engineered to bind preferentially to specific types of cells or tissues, providing efficient disease-targeting capabilities (17–20). Second, they can provide protection of the loaded cargo from degradation until they reach the site of delivery (21–27). Third, NPs are also capable of delivering large-sized cargos such as plasmids and large proteins, such as CRISPR/Cas9 components. Fourth, many of the materials used for NP manufacture have very acceptable safety profiles, and they are not expected to elicit mutagenicity per se, as opposed to viral vectors (28, 29). Finally, NP manufacture generally has a very good scale-up potential, which, when added to their improved safety profiles, can facilitate their clinical translation (30).
II. CRISPR/Cas9 Delivery Systems: Strategies and Barriers
CRISPR/Cas9 components
Typically, a CRISPR/Cas9 system adopted for genome editing consists of the endonuclease, Cas9, and the specific gene-targeting sgRNA. Cas9 and sgRNA can be delivered in various forms that include: 1) the Cas9 endonuclease protein itself and the sgRNA, 2) a plasmid that encodes for both Cas9 and sgRNA, 3) two separate plasmids each encoding Cas9 or sgRNA, 4) Cas9 mRNA and sgRNA, and 5) a Cas9/sgRNA complex (usually called ribonucleoprotein (RNP)). Thus, when considering a NP-based delivery system for CRISPR/Cas9, it is important that the formulation takes into account what form the CRISPR/Cas9 components are in so as to ensure optimal compatibility. It is also important that formulation parameters designed to accommodate the particular physical properties of the cargo do not significantly compromise the NPs other desired attributes such as cargo protection, stealth, targeting or capacity to efficiently transfect cells (2). One major advantage of using a single plasmid encoding both Cas9 and sgRNA over other strategies is only requiring to load a single element (the plasmid) into the NPs, whilst other systems require either co-loading of NPs or the use of separate NP formulations (2). In addition, if required, a knock-in gene can also be integrated within the same delivered plasmid (13). However, a plasmid encoding for all of the CRISPR/Cas9 elements will have a relatively large size, and resultingly can become more difficult to deliver (29). Researchers may also wish to avoid using plasmids when translational therapies are being considered due to the plasmid, as opposed to RNA, being more persistent and “outstaying its welcome” by potentially generating off-target insertions or deletions (indels) long after the desired gene editing has been performed (31, 32). Since the ultimate goal is to deliver the Cas9/sgRNA complex to the nucleus of the host cell, RNP delivery may be regarded as the most straightforward strategy, as it requires the least amount of intracellular processing, since the functional complex has been generated prior to delivery. In addition, complexing sgRNA and the Cas9 within the RNP assembly provides significant protection for the sgRNA (33). Finally, RNPs provide transient and efficient gene editing, with relatively less off-target indels being generated (12). Each system has its advantages and disadvantages when considering them in the context of NP formulations.
Delivery approaches
One of the most commonly investigated methods of delivering nucleic acids encoding a desired protein to cells is through viral vectors (34–36). However, viral vectors, despite being very efficient at delivering the CRISPR/Cas9 cargo through cell transduction, can cause unwanted immunogenicity and mutations in the host, thus limiting their clinical translation (37, 38). Although delivery approaches such as electroporation and microinjection have been used to deliver genome editing systems such as CRISPR/Cas9 to cells, they are generally limited to being used in in vitro, or at best ex vivo, situations since in vivo delivery by these methods is, for the most part, impracticable (39). Thus, non-viral vectors, particularly NP formulations are currently being tested in preclinical situations.
NPs are usually taken up by cells through endocytosis. Most receptor-bound nanoparticles are translocated into the cells by clathrin-mediated and caveolin-mediated endocytosis, whilst non-targeting NPs may also be engulfed nonspecifically by a non-receptor-mediated form of micropinocytosis and, for certain cell types, phagocytosis (19, 40). Once NPs are taken up by endocytosis, they are located inside endosomes (pH 6.5–6.8), which develop into late endosomes (41). Late endosomes (pH 5.2–6.0) deliver their cargo to lysosomes (pH 4.5–5.2) that then destroy the engulfed material through a combination of the acidic pH and enzymatic degradation (19, 41). The escape of the NP cargo from the endosomes prior to being exposed to the harsh environment of the lysosome is crucial to maintain the integrity of the delivered cargo and to achieve the desired nuclear translocation. Many cationic nanocarriers, such as those made from polyamidoamine (PAMAM)-based dendrimers, polyethyleneimine (PEI), or chitosan, have been proposed to escape the endosomal compartment via a proton sponge mechanism (42–45). This is a process where the cationic moiety becomes protonated, thus acting as a sponge to protons, due to the acidic endosomal environment, which results in a flow of ions and water into the endosome followed by its swelling and rupture (19, 42, 46). As will be seen in this review, researchers have often applied strategies that promote endocytic escape and/or cytoplasmic entry.
Following nanocarrier translocation into the cytosol, a significant proportion of the payload has to enter the nucleus if the genetic cargo requires transcription for subsequent functional effects or if the payload, such as RNPs, needs to immediately access the target gene. Several techniques can be found in the literature that enhance nanocarrier nuclear translocation. For example, a nuclear localization signal (NLS) is a short amino acid sequence that may be used to tag macromolecule cargos to improve their nuclear targeting (47–49). Several gene delivery systems with tagged NLS have been reported, including PAMAM-, and PEI-mediated carriers (50–52). On the other hand, some cell penetrating peptides (CPPs) such as HIV-Tat (transactivator of transcription) peptide, which will be discussed more later, have the added ability to penetrate the nucleus, and have been utilized to direct macromolecules and NPs to the nucleus with considerable success (53–57). Furthermore, other researchers have adopted active targeting to increase NP nuclear localization by adding ligands that target nuclear membrane receptors to the formulation (58).
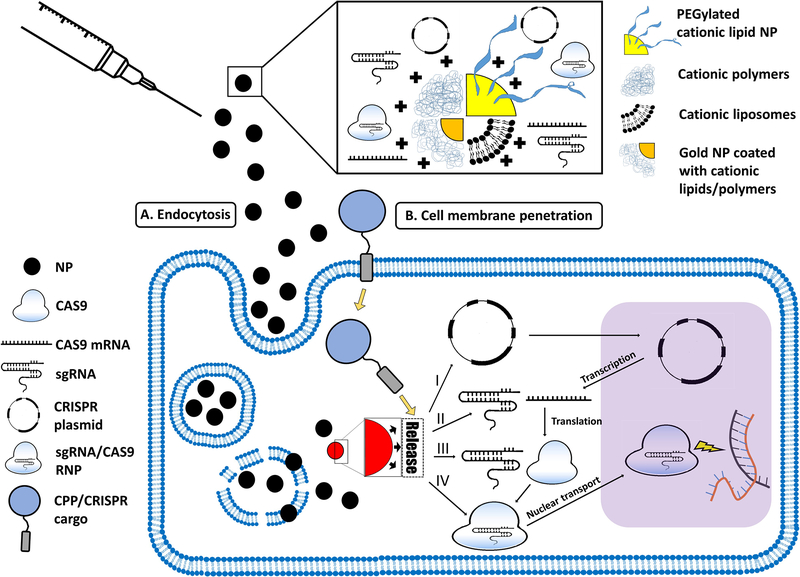
The NP-based CRISPR-Cas9 delivery strategies presented in this review include liposomal/lipid-based NPs, polymeric NPs/polyplexes, and CPPs. Figure 2 describes different forms of NPs, and the pathways of NP-mediated CRISPR/Cas9 delivery to the nucleus.
Figure 2. The pathways of NP-mediated CRISPR/Cas9 delivery to the nucleus.
Such NPs can take several forms, including PEGylated cationic lipid NPs, cationic polymers and liposomes, and cationic lipid- or polymer-coated gold NPs (top panel). The NPs may enter the cell via (A) endocytosis (in case of inorganic, or polymeric-/lipid-based NPs) or (B) direct cell membrane penetration (e.g. in the case of CPPs). While CPPs facilitate direct entry into the cytosol, other types of NPs are first engulfed into endosomes, from which the NPs have to escape into the cytosol. Once in the cytosol, NPs (regardless of their cellular uptake mechanism) need to release their CRISPR/Cas9 cargo. This cargo can be: (I) a CRISPR plasmid that has to be transported to the nucleus to be transcriptedinto sgRNA and Cas9 mRNA. Cas9 mRNA is then translated into Cas9, which is complexed with sgRNA to form the sgRNA/Cas9 RNP, which finally enters the nucleus to induce double-stranded DNA breaks at the target sites; (II) the sgRNA and Cas9 mRNA; (III) sgRNA and Cas9 protein, or (IV) sgRNA/Cas9 RNP.
A. Lipid-based delivery
Lipids are a diverse group of naturally occurring or synthetic organic compounds which include fatty acids and their derivatives. However, it is amphiphilic phospholipids that are the primary component of the cell membrane and liposomes. Liposomes are three-dimensional spherical structures that can encapsulate hydrophilic or hydrophobic molecules and can enter cells via endocytosis (40, 46, 59). Additionally, in the case of cationic liposomes, as opposed to classical liposomes, DNA and RNA can form strong associations via electrostatic attractions, adsorbing them onto their surfaces (60). This complexation protects against nucleic acid degradation and enhances the likelihood of DNA and RNA entering the cell for transcription and translation, respectively. Cationic liposomes and commercially available lipid-based transfection kits are commonly used for RNAi gene delivery as well as for the delivery of pDNA, thus using these liposomes/lipid-based reagents as delivery systems for CRISPR-Cas9 has potential and has been investigated as discussed below.
Commercial lipid-based reagents
Lipofectamine™, TurboFect™, and Stemfect™ are nanoparticulate-based commercial transfection kits employing cationic lipid compounds that have been used to enhance DNA and RNA delivery to cells in vitro. They have also been used to complex CRISPR-Cas9 components to increase transfection efficiency and thus DNA editing in cells. Recent research found that Lipofectamine avoids microtubule-mediated intracellular trafficking, thus circumventing lysosomal degradation that many nucleic acid cargos are exposed to when other delivery systems are used; this potentially explains the enhanced transfection efficiencies of Lipofectamine-based reagents (61). A recent product called Lipofectamine CRISPRMAX optimized for CRISPR-Cas9/sgRNA complex-based delivery showed superiority in CRISPR-mediated gene editing to the more commonly used Lipofectamine 3000 and Lipofectamine RNAiMAX, whilst also exhibiting less cytotoxicity (62). The two most common cell lines so far targeted with these agents are the human embryonic kidney cell line, HEK293 (and its variants), and the human cervical adenocarcinoma cell line, HeLa, which are both well-established cell lines for gene transfection. The aforementioned transfection kits are usually used in vitro because their safety profiles make their in vivo use impractical, although the search for safer commercial transfection reagents is still ongoing (62). In addition, once complexed with optimal amounts of pDNA for transfection, the size of the complexes can vary from 200 – 700 nm in diameter which would make them unsuitable for systemic delivery.
Stemfect™, X-tremeGENE™ and Lipofectamine 2000 were used for the transfection of HEK293 cells with pDNA encoding for sgRNA, Cas9, and GFP in two pioneering studies that exploited the genome editing capability of CRISPR/Cas9 in human cells (8, 63). In one of those studies Lipafectamine 2000 was used to demonstrate the functionality of a CRISPR/Cas9 system delivered as pDNA to HEK293FT cells (63). They also showed the potential for reducing off-target mutations by generating a form of Cas9 that acted as a nickase rather than nuclease thus prompting DNA repair through the less error prone mechanism of homology directed repair as opposed to non-homologous end joining. Although editing efficiencies were low (<20%) and there was little focus on the delivery system itself, these studies highlighted the potential of CRISPR/Cas9 for gene therapy. Another early study utilized CRISPR/Cas9 in an attempt to eradicate the latent HIV-1 genome in HIV-1-infected HeLa cells and a human CD4+ T cell line (Jurkat) (64). Although this group used Lipofectamine 2000 to transfect the separate pDNAs (one encoding sgRNA targeting the LTR of HIV; the other encoding Cas9) into HeLa cells, they used electroporation for the more clinically relevant T cell line. The reasons for using the different modes of transfection were not discussed but may have reflected the poor capacity of lipid-based reagents to transfect T lymphocytes. Nevertheless, the results in the HeLa cells demonstrated a significant, albeit marginal, effect where it was shown that reactivation of the latent virus occurred in approximately 35% of the latently HIV-infected HeLa cells that were transfected with the negative control versus 25% for those cells transfected with the CRISPR/Cas9 system targeting the LTR (64). In a separate study, a comparison of editing efficiencies of CRISPR/Cas9 RNP cargo (targeting the HPRT gene), delivered independently to HEK293FT cells in vitro by three different commercial lipid-based transfection reagents (Lipofectamine 3000, Lipofectamine 2000 and RNAiMAX), was made (65). The editing efficiencies were calculated 48 hours after transfection using a GeneArt® Genomic Cleavage Detection Kit and the percent indels were found to be higher for cells treated with either Lipofectamine 3000 or RNAiMAX (mean: 28–29%) compared to Lipofectamine 2000 (~18%).
Rolling circle transcription (RCT) is an isothermal nucleic acid amplification approach that enables the production of long repetitive RNA units that may take the form of nanocircles or NPs (66–68). The authors suggested that polymeric RNA made by RCT have demonstrated relatively stronger complexes with cationic carriers, compared to monomeric RNA, due to the high density of the negative charge of the polymerized nucleic acid material (38, 69). Ha et al. prepared polymeric sgRNA using RCT, and sgRNA units were complexed with Cas9 proteins to produce 50 nm poly-RNP NPs (38). Short interfering RNA (siRNA) sequences were incorporated within the polymeric-sgRNA sequences. SiRNA is a substrate to the endogenous ribonuclease Dicer; and once the poly-RNP nanoparticles enter the cells, they get cleaved by intracellular Dicer, yielding numerous monomeric RNP complexes. The polymeric assembly of RNP complexes provides enhanced in vivo stability, compared to monomeric RNP. The poly-RNP NP size increased to 80 nm once they were complexed with Stemfect®, which was essentially used to enhance cellular uptake as outlined by the authors (38). The zeta potential shift of the RNP nanoparticles from −12 mV to +29 mV after complexation with the cationic Stemfect reagent may in part explain the enhanced cellular uptake of the RNP/Stemfect complex compared to uncomplexed RNP NPs. In this study, all CRISPR/Cas9 constructs used were complexed with Stemfect. In GFP-expressing HeLa cells, poly sgRNA/siRNA RNPs were superior to poly sgRNA RNPs and mono sgRNA RNPs in terms of gene disruption percentage, with values of 61%, 13.5%, and 8.7%, respectively, when evaluated using a T7 Endonuclease 1 assay. Indel percentage assessment with Sanger sequencing confirmed these results, although the values were noticeably lower. The same trend was also found when the target GFP protein was assessed by Western blotting. Flow cytometry revealed that 58% of the cells became GFP-negative following transfection with poly sgRNA/siRNA RNP with Stemfect®, compared to 28% and 39% with poly sgRNA and mono sgRNA RNPs, respectively. This almost 7-fold increase in gene disruption emphasized the crucial value of polymerization of sgRNA RNP into nanoparticles using RCT, provided that a Dicer substrate (in this case, any double stranded siRNA of at least 23 base pairs regardless of gene specificity) is integrated within the polymerized RNP NPs. The crucial role of siRNA sequences integrated within the polymerized sgRNA/Cas9 system was further highlighted in the in vivo experiment using GFP-expressing HeLa tumors-bearing nude mice. Only polymeric RNPs with Dicer-sensitive siRNA significantly attenuated GFP expression in the tumors. In addition, polymerization of the RNA components was successful to preserve their integrity following injection, in contrast to the monomeric RNA RNPs, where the in vivo instability is thought to be responsible for the lack of functionality. The off-target gene disruption was found to be minimal, as no indels were found in two non-target locations that share some similar sequences with thee target genes (38).
Lipid-based and lipid-like NPs
As an alternative to commercial lipid-based reagents, lipid-based NPs, including liposomes, can be prepared in the laboratory and have frequently been used for siRNA delivery to cancers in preclinical studies, as well as having been tested in clinical trials (70). Several liposome-based formulations (e.g. Doxil®, Myocet ®, and AmBisome®) are already on the market. However, liposomes per se, aside from the commercially available kits discussed above, have been rarely used to deliver CRISPR/Cas9 components. The classical liposomal structure comprises a phospholipid bilayer forming a vesicle capable of encapsulating lipophilic drug cargos within the bilayer, or hydrophilic cargos within the aqueous core. Hydrophilic, charged molecules such as nucleic acids and RNPs can be adsorbed onto the liposomal surface if the surface is cationic. Aside from the more conventional type liposomes described above, cationic lipid-based NPs can be used to deliver CRISPR/Cas9 components. These comprise a lipidic core containing cationic constituents, including cationic lipids (e.g. 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP)) capable of complexing with negatively-charged nucleic acid molecules and RNPs (71–73). Unlike liposomes, these lipid-based NPs do not possess a phospholipid bilayer and aqueous core; alternatively they contain monolithic lipid cores and their surfaces can still be decorated with cationic moieties (71, 73). The general consensus is that lipid-based NPs, when delivered intravenously, tend to target the liver which, depending on whether one is trying to target the liver, could be considered an advantage or a drawback (74). This tropism of lipid-based NPs for the liver is because they associate with certain proteins in the serum, such as apolipoprotein, and are subsequently taken up by hepatocytes through receptor-mediated endocytosis.
A wide range of lipid-based nanocarriers have been tested independently for their ability to deliver CRISPR/Cas9 systems. In one study, Polo-like kinase-1 (Plk1), a master regulator in mitosis and overexpressed in many different types of cancer, was targeted using a lipid-gold NP formulation (75). In this study, cationic gold nanoclusters were modified with HIV-1 Tat peptide in order to improve nuclear targeting, and were complexed with Cas9 protein and pDNA encoding sgRNA (targeting Plk1) via electrostatic attraction. However, because this complex had a negatively charged surface (due to the sgRNA), a cationic lipid shell made from DOTAP, dioleoyl-phosphatidylethanolamine (DOPE), and cholesterol terminated with distearoyl-phosphatidylethanolamine (DSPE)-PEG was applied (75). The resultant NPs were spherical, 70 nm in diameter (when measured by scanning electron microscopy) and had a zeta potential of approximately +35 mV. The hydrated diameter of the particles, when measured by dynamic light scattering, was approximately 103 nm. This discrepancy is often observed, particularly for NPs with electron dense cores (76). Preliminary cell culture studies revealed that the DSPE-PEG coating improved intracellular uptake and nuclear localization compared with uncoated particles. The transfection efficiency of a human malignant melanoma cell line, A375, by the NPs (containing sgPlk1 pDNA that also encoded GFP), revealed (after 48 h): 1) approximately 55.7% of NP-treated cells were GFP-positive and 2) 26.6% cleavage of the Plk1 gene with no evidence of off-target effects at 10 predicted potential off-target locations. Corresponding Western blot analysis of PLK1 protein content revealed a 70% down-regulation compared with PBS-treated controls. BALB/c athymic nude mice challenged with A375 tumors were treated with the NPs via intratumoral injection after tumor volumes reached approximately 50 mm3. Tumor growth was significantly inhibited in mice treated with the NPs (delivered every other day for 15 days) compared with the PBS-treated control. PLK1 protein was down-regulated in tumor tissues and tumoral DNA possessed frame-shift mutations at the Plk1 gene locus (Table 1) (75). Systemic delivery was not investigated and this may have been partially due to the high positive charge of these NPs that may have limited their circulatory half-life. Issues of potential cytotoxicity need to be further investigated if this formulation is to progress toward the clinic.
Table 1:
Summary of select NP formulations
| NP formulation | NP traits | CRISPR/Cas9 cargo | In vitro results (cells used) | In vivo results |
|---|---|---|---|---|
| Gold nanoclusters/Tat complexed with cargo (core): cationic lipid shell (75) | • ~70 nm d. (SEM) • ~103 nm d. (DLS) •+35 mV ZP |
Cas9 protein + pDNA encoding Plk1-targeting sgRNA – co-loaded into NP | •TE: 55.7% (A375) •EE: 26.6% (A375) Treatment: 2 μg pDNA or Cas9 protein/1×106 cells/ml |
•IT delivery of NPs significantly reduced tumor growth (>70%) in a melanoma model (A375) in athymic mice Dosing: 10 μg pDNA or Cas9 protein/mouse (7 doses over 15 days) |
| Lipid NPs: cholesterol, C14-PEG 2000, DOPE and ionizable lipid cKK-E12 formulated with cargo (77) | ~70 nm d. (DLS) (165) *1 | Pcsk9-targeting chemically modified sgRNAs + Cas9 mRNA | •IV delivery: > 80% EE (hepatocytes) •Low frequency of off-target indels •No liver toxicity or cytokine induction Dosing: 1.2 mg/kg Cas9 mRNA and 0.5 mg/kg of each sgRNA/mouse |
|
| Lipid NPs: bioreducible lipid (3-O14B), cholesterol, DOPE, and C16-PEG2000−ceramide formulated with cargo (78) | • ~292 nm d. (DLS) •−9 mV ZP |
sgRNA/Cas9 RNP targeting GFP | EE: approx. 70% (HEK293-GFP) Treatment: 25 nM RNP (6 μg/ml lipid) |
|
| Lipidoid NPs (optimal formulation: 76-O17Se) + cargo (81) | • ~314 nm d. (DLS) •−13.2 mV ZP |
sgRNA/Cas9 RNP targeting GFP | EE: approx. 50.2% (HEK293-GFP) Treatment: 25 nM RNP (3.3 mg/L lipidoids) |
|
| Cationic liposomes embedded with PSMA-specific RNA aptamer (linked to DSPE-PEG) plus cargo (86) | • ~150 nm (DLS) •+40 mV ZP |
Cas9 protein + pDNA encoding Plk1-targeting sgRNA | > 90% down-regulation of Plk1 mRNA (LNCap cells) compared to 80% down regulation by liposomes without aptamer and Lipofectamine 2000 | IV delivery (day 0) reduced LNCap tumor volumes by 1.66-fold versus 2.56-fold increase for PBS control (over days 13 – 27) in athymic mice Dosing: 40 μg liposomes-sgRNA + 40 μg Cas9 |
| PEGylated NPs comprising cationic alpha-poly-glutamate-based polypeptides (P-HNPs) plus cargo (29) |
• ~100 nm (DLS) •+20 mV ZP |
•In vitro: Cas9 pDNA + sgRNA targeting AAVS1 and/or HPRT1 • In vivo: Cas9 pDNA + sgRNA targeting Plk1 Note: pDNA and sgRNA were independently loaded into P-HNPs |
EE: 40 – 50% (HEK293T) Treatment: 1 μg/ml pDNA and 0.5 μg/ml each sgRNA |
IT delivery: 1) >71% tumor suppression 2) Improved survival (60% versus 0% (untreated/ controls)) 3) EE: 35% (mice with HeLa tumors) Dosing: P-HNPs containing 20 μg pDNA (delivered days 7,9.11,13 and 15 PTC) + P-HNPs containing 20 μg sgRNA (delivered on days 8,10,12,14, and 16 PTC) |
| Lipid NPs: biodegradable ionizable lipid (LP01), cholesterol, DSPC and PEG2k-DMG formulated with RNA cargo (N:P ratio of 4.5:1) (90) | • ~105 nm d. (DLS) | Chemically modified sgRNA (targeting TTR) and Cas9 mRNA (wt ratio of 1:1 for mRNA:sgRNA) | •IV delivery : ~ 70% EE (liver): > 97% reduction in serum TTR levels (CD-1 mice) Dosing: 3 mg/kg NPs •IV delivery: ~ 65% EE (Rats) Dosing: 5 mg/kg NPs |
|
| ZAL NPs: “ZA3-EP10” ZAL:cholesterol:PEG lipid + cargo (88) |
• ~170 nm d. (DLS) • ~−6 mV ZP |
•In vitro: sgRNA ( ~100 nt) targeting luciferase (Luc)
•In vivo: sgRNA targeting LoxP + Cas9 mRNA (wt ratio of 1:4) |
EE: 95% (HeLa-Luc-Cas9) Treatment: 7 nM sgRNA |
•EE: 1.5 −3.5% (hepatocytes) •Confocal microscopy of sectioned organs demonstrated tdTO positive cells in liver, lung and kidney (mice) •IV delivery resulted in preferential accumulation in lungs of mice Dosing: 5 mg/kg total RNA |
| Self-assembling NPs with an inorganic core (inc. protamine sulfate + cargo) and a chitosan shell with targeting moieties (58) | • ~300 nm d. •−9 mV ZP |
pDNA encoding both Cas9 and sgRNA targeting CDK11p110 gene | > 90% reduction in CDK11p110 expression by MCF-7 cells (Western blotting) Treatment: 1 μg/ml pDNA |
|
| Cationic arginine functionalized gold NPs (10 nm) + cargo (149) | 475 (± 60) nm (“nanoassemblies”) | sgRNA/Cas9glut (+NLS) RNP targeting AAVS1 gene (or PTEN gene) (1:1 molar ratio of sgRNA:Cas9) | •UE: 80–90% (HeLa) •EE: 30% (Hela/HEK-293T/RAW) Treatment: 125 nM NPs/62 nM RNP |
|
| ZIF-8 plus cargo (Loading capacity: 1.2 wt %) (153) |
• ~100 nm d. (DLS) •+5 mV ZP |
sgRNA/Cas9 RNP targeting EGFP (1:1 molar ratio of sgRNA:Cas9) | Reduced EGFP fluorescence by 37% (CHO-EGFP) Treatment: 240 nM RNP |
|
| GO-PEG-PEI NPs plus cargo (154) | • ~220 nm (DLS) •+18.5 mV |
sgRNA/Cas9 RNP targeting EGFP (1:1 molar ratio of sgRNA:Cas9) | EE: 39% (AGS-EGFP) Treatment: 30 μg/ml GO-PEG-PEI/; 100 nM RNP |
|
| PEI-coated self-assembling DNA nanoclews complexed with cargo (148) | • ~56 nm d. (DLS) •+18 mV ZP |
sgRNA/Cas9 RNP targeting EGFP | EE: 36% (U2OS-EGFP) Treatment: 8 μg/ml DNA nanoclews; 100 nM RNP |
IT delivery: ~ 25% loss of EGFP+ve cells at the site of injection (mice) Dosing: 5 μM RNP |
| PT24/Cas9 RNP NPs (32) (Ratio PT24:Cas9 RNP = 62.5:1) |
• ~270 nm (DLS) •+4 mV ZP |
sgRNA/Cas9 RNP targeting HPRT1 | EE: 30–40% (HeLa) Treatment: 160 nM RNP |
|
| Fluorinated G5DP NPs complexed with cargo (N:P ratio of 5:1) (117) | • ~150 nm (DLS) •+30 mV ZP |
pDNAs encoding sgRNA/Cas9d-VP64 targeting MASPIN | Five-fold increase in MASPIN expression over control in MCF-7 cells Treatment: 1 μg pDNAs/well |
EE = editing efficiency; TE = transfection efficiency; UE = uptake efficiency; DLS = dynamic light scattering; SEM = scanning electron microscopy; IT = intratumoral; IV = intravenous; DSPC = distearoylphosphatidyl choline; ZAL = zwitterionic amino lipid; NLS nuclear localization sequence; ZP = zeta potential; d. = diameter; nt = nucleotides; tdTO = tdTomato; wt = weight; G5DP NPs = Nanoparticles made from G5 dendronized polymers; Plk1 = polo-like kinase 1; DSPE-PEG = poly(ethylene glycol)-grafted 1,2-distearoyl-sn-glycero- 3-phosphatidylethanolamine; PTC = post tumor challenge; ZIF-8 = zeolitic imidazolate framework-8; GO = graphene oxide
size of NPs here is when no cargo was present. Authors did not provide data on size of NPs + cargo.
Recently, a valuable study was performed where intravenously injected lipid NPs were used to deliver chemically modified sgRNAs and Cas9 mRNA, resulting in > 80% editing efficiency of the targeted Pcsk9 gene (encoding proprotein convertase subtilisin/kexin type 9) in the liver in mice (74). The chemical modifications, not detailed in this review, were made to the sgRNAs targeting Pcsk9 to improve duplex stability and engender nuclease resistance. Using these RNA modifications, transfection efficiency and indel incidence were improved (77). Targeting of the Pcsk9 gene was conducted with two different chemically modified sgRNAs plus Cas9 mRNA which were formulated with cholesterol, C14-PEG 2000, DOPE and an ionizable lipid, cKK-E12, in a molecular ratio of 46.5:2.5:16:35 in order to form lipid NPs (~70 nm diameter). The aim of this study was to target a gene that had clinical relevance and to do so using NPs that could be delivered systemically and had potential to be used in the clinic. The Pcsk9 gene is a target for the treatment of familial hypocholesterolemia. Five days post intravenous injection, serum levels of Pcsk9 were undetectable, total cholesterol levels decreased by 35–40%, and 83% editing events were detected in the liver DNA with indels found primarily in hepatocytes (Table 1) (77). The editing efficiencies observed when unmodified sgRNAs were used was very poor, suggesting that the NPs themselves were not very protective against nucleases or that the improved duplex stability caused by the modifications was a very important factor in determining indel frequency. These results are highly promising from a therapeutic perspective and the authors suggest that these NP formulations may have potential for treating other liver diseases. Furthermore, it was demonstrated that these NP formulations, when delivered to mice, exhibited no liver toxicity and did not induce cytokine production, suggesting their high potential for clinical translation.
In a separate study, liposomal NPs were formulated with bioreducible lipids, cholesterol, DOPE, and C16-PEG2000-ceramide at a wt/wt ratio of 16:4:1:1 (78). The bioreducible lipids were synthesized by a Michael addition of primary or secondary amines as well as an acrylate comprising a disulfide bond and a 14-carbon hydrophobic tail. The authors asserted that the bioreducible lipids would make the NPs vulnerable to the reductive intracellular conditions experienced subsequent to endocytosis, resulting in relatively fast liposomal degradation and release of cargo. When the lipid NP formulations were tested for the ability to knock down GFP expression (with CRISPR/Cas9 RNPs as the cargo) in GFP-expressing HEK293 cells it was found that 3 formulations (varying only in head group chemical structure) were capable of knocking down GFP expression by 70% (i.e. 70% of cells were GFP negative 3 days after treatment) (Table 1). This level of knock down was comparable to that seen when the researchers used Lipofectamine 2000. What properties the head structures of the bioreducible lipids possessed that contributed to more efficient knock down when they were used in lipid NP formulations carrying RNPs was not discussed nor was it evident from comparing their chemical structures to those lipids that were formulated into lipid NPs that were less efficient at GFP knock down. Although not specifically investigated in this report, the authors suggest that the synthesis of lipids by the described combinatorial strategy can result in lower levels of toxicity and immunogenicity compared to other lipid based formulations such as Lipofectamine. This was based on their own and other researchers previous findings (79, 80). The authors nevertheless acknowledge the further need to establish the safety of these NP formulations in animal models.
Recently, the same group utilized a combinatorial library of chalcogens (O, S, Se) to prepare lipid-like NPs for CRISPR/Cas9 RNP delivery with promising results (81). Lipid-like nanoparticles are usually made from lipid-like, or lipidoid, compounds, in addition to other materials (e.g. phospholipids, cholesterol, and polyethylene glycol) (82, 83). Lipidoids are a structurally diverse library of amino-alkyl acrylate and amino-alkyl acrylamide compounds with diverse lipid tails and have shown great potential in delivering nucleic acid materials (82–85). The aforementioned chalcogen-containing lipidoids were generated by reacting lipophilic tails possessing O, S and Se ethers with various amines (R-O17X; R = amine head group; X =O, S or Se). The authors’ ultimate goal was to generate NPs capable of being used for clinical translation and thus, in these studies, were attempting to find NP formulations capable of editing efficiencies that were comparable to Lipofectamine 2000 but were less toxic. Initially, a library of lipidoid NPs (n = 51) was generated by mixing the lipidoids with a Cre recombinase protein tagged with a negatively supercharged GFP ((–30)GFP-Cre) and allowing for electrostatic interactions to yield the NPs. The reason for using (–30)GFP-Cre in the NP formulations initially was so that screening of NP uptake and functional performance of cargo could be readily assessed. Hela-DsRed cells were used which would fluoresce red upon Cre-mediated recombination (a measure of cargo functional performance), whilst the GFP tag would indicate uptake efficiency. NPs containing lipidoids with O17Se tails were most efficacious (>20% GFP+ve cells) at being taken up by the HeLa cells. Those efficacious NPs (n = 12) were then tested for Cre protein function and cytoxicity in HeLa-DsRed cells, where four formulations (containing 76-O17S, 76-O17Se, 77-O17S or 77-O17Se) exhibited high Cre-recombination levels of between 31 and 41% which were comparable or greater than that achieved with Lipofectamine 2000 (33.5%). In addition, these four formulations exhibited low cytoxicity (>80% viability) compared to Lipofectamine 2000 (~70% viability). Subsequently, Ai14 mice, containing a loxP-flanked STOP cassette that inhibits expression of tdTomato, were injected (IV) with one of three lipidoid NP formulations (including 76–017Se) containing (–30)GFP-Cre. After 20 days various organs (heart, liver, spleen, kidney and lungs) were harvested and from analyzed sections high levels for fluorescence were observed in the lungs for two of the lipidoid formulations (containing 76–017Se or 76-O17S). The reasons for the lung tropism were not known but are currently being investigated. The authors acknowledge the need for now fabricating lipidoid NPs loaded with CRISPR/Cs9 RNP that function in vivo and are capable of targeting specific organs (78). In order to confirm that the lipidoid NPs were capable of delivering functional CRISPR/Cas9 components, in vitro studies were performed using HEK293-GFP cells that demonstrated 50 – 60% GFP expression loss upon treatment with NPs carrying CRISPR/Cas9 targeting GFP (Table 1). The need to establish safety and efficiency using in vivo studies remains.
As stated earlier non-commercial-based liposomes have rarely been used to deliver CRISPR/Cas9. However, one group did manufacture cationic liposomes surface embedded with an RNA aptamer that targeted prostate cancer cells (by recognizing prostate-specific membrane antigen) for the purposes of CRISPR/Cas9 pDNA delivery (86). The ability of these liposomes (size = ~150 nm; zeta potential = +40 mV; formulated with DOTAP, cholesterol, protamine and calf thymus DNA) to specifically target prostate cancer cells was shown by competitive inhibition studies performed in vitro with LNCap cells (human prostate cancer cell line). The ability of the prostate cancer cell-targeting liposomes to knock down mRNA expression of Plk1 was demonstrated in vitro using LNCap cells and showed significantly enhanced down-regulation of mRNA compared to when Lipofectamine 2000 or non-targeting liposomes were used to deliver the CRISPR/Cas9 cargo (Table 1). Intravenous injection of the aptamer-functionalized liposomes carrying CRISPR/Cas9 pDNA targeting Plk1 in mice with LNCap tumors resulted in significant reduction in tumor volume compared to PBS treated tumors (Table 1). Although not significant, treatment with the targeting liposomes trended towards reduced tumor volumes and enhanced survival compared to the non-targeting liposomes. The degree to which the targeting liposomes accumulated in other tissues was not discussed and would have been interesting to know as well as any off-target effects that may have occurred. From a clinical translation perspective it was promising that the A10-liposome-CRISPR/Cas9 NPs did not cause a significant drop in mouse body weight, affect viability, nor display signs of immunogenicity by inducing the production of cytokines (IFN-alpha and IL-12) upon in vivo administration.
Zwitterionic NPs have gained attention due to their “stealth” properties, thereby increasing resistance to protein corona formation and circulation time in vivo (87). Zwitterionic amino lipids (ZALs) contain a zwitterionic sulfobetaine head group with an amine-rich linker to the hydrophobic tail group, and, in one particular study, were used to form zwitterionic NPs (ZAL NPs) capable of co-delivering Cas9 mRNA and sgRNA (88). One of the primary aims of the research was to develop a safe in vivo delivery option for CRISPR/Cas9. As discussed above, delivering CRISPR/Cas9 in RNA form rather than DNA form is potentially safer, and the authors here argue that zwitterionic lipids are particularly appropriate for the delivery of long nucleic acids, such as Cas9 mRNA, and the shorter sgRNA within the same NP. Preliminary in vitro screening assays (for effective delivery of sgRNA and mRNA to cells) of the many ZAL NPs generated, identified one particular formulation (ZA3-EP10; Table 1) that was subsequently tested in vivo in a range of different mouse strains. Mice that were transgenic for the Lox-Stop-Lox tdTomato cassette were treated IV (5 mg/kg RNA dose: ratio 1:4 of sgRNA:mRNA) with ZA3-EP10 carrying Cas9 mRNA and sgRNA (targeting LoxP). This NP formulation was designed to delete the stop cassette and induce tdTomato expression. Confocal microscopy revealed increased tdTomato fluorescence in the liver, kidneys and lungs after one week, and maintained expression was observed for at least two months (in the liver and kidneys), providing further confirmation that this method of delivery is reliable for Cas9-based gene editing (Table 1) (88). In vivo administration of these NPs to mice had no significant effect on body weight over the succeeding 4 days, suggesting a lack of toxicity. However, further studies into immunogenicity and toxicity are required in the future.
As a proposed therapy for transthyretin (TTR) amyloidosis, a fatal disease caused by the extrahepatic accumulation of TTR fibrils (89), Finn et al. described a lipid-based nanocarrier system that could deliver CRISPR/Cas9 to target the murine TTR gene (90). The CRISPR/Cas9 was delivered as a combination of Cas9 mRNA and chemically modified sgRNA targeting TTR. The chemical modification of the sgRNA involved end modifications with phosphorothioate. The lipid nanocarrier was formulated by microfluid mixing of the lipid solution comprising LP01 (a biodegradable synthetic proprietary lipid), cholesterol, distearoylphosphatidyl choline and PEGylated 1,2-dimyristoyl-rac-glycero-3-methylpolyoxyethylene at molar ratio of 45:44:9:2, with the CRISPR/Cas9 RNA solution at an N:P ratio of 4.5:1. The authors’ rationale for using LP01, which is readily biodegraded through its ester bonds, is that, unlike the more commonly used non-biodegradable ionizable lipids, LP01 is quickly cleared from the liver, thus avoiding potentially toxic accumulation. The resultant lipid NPs of interest were approximately 105 nm diameter and were demonstrated to be safe in both rats and mice as determined by body weight and cytokine measurements. The CRISPR/Cas9 system was delivered as sgRNA and mRNA for Cas9 translation. The lipid-based nanocarrier system achieved more than 97% reduction in the serum TTR levels in CD-1 mice when the sgRNA used was chemically modified with phosphorothioate at both ends (90). This CRISPR/Cas9 non-viral delivery system was also successful in rats, achieving editing percentages as high as ~65% when highly modified rat-specific TTR sgRNA was used (Table 1).
B. Polymeric delivery
Polymers are used extensively for drug delivery applications (25, 91–107). Positively charged polymers are advantageous in gene delivery because they easily complex with negatively charged nucleic acids via electrostatic attraction, often condensing large nucleic acids into discrete nano-sized packages (Figure 2C) (104, 106, 108, 109). They are also believed to enter cells via the same clathrin-mediated endocytosis mechanism as positively charged lipids (110).
Poly(ethylenimine) (PEI)
PEI is a cationic polymer commonly used in gene and drug delivery (108, 111, 112). Both PEI and poly(amido amine) (PAMAM; discussed below) impart cytotoxicity at high doses, however, both have also shown marked improvements in transfection efficiency over lipid-based delivery (113, 114). Much like cationic liposomes and PAMAM, the net positive charge on PEI interacts electrostatically with nucleic acids (108). The ratio of polymer to DNA is often expressed as the N/P ratio for gene transfection experiments, which is the ratio of nitrogen (amine) groups in the polymer to the phosphorus (phosphate) groups in the nucleic acid. Access of PEI nanoplexes to the cytoplasm is likely mediated by the proton sponge effect post-endocytosis (60). Progression of PEI-based formulations into the clinic has been slow, primarily due to concerns over cytotoxicity. A PEI-based nanoparticle formulation that delivers siRNA for the treatment of multiple myeloma (SNS01-T, Senesco Technologies inc.) was being evaluated in a dose-escalating open-label phase 1b/2a clinical trial (clinicaltrials.gov, identifier: NCT01435720) in 2014 (115). No further information could be found about the progress of this trial.
Reports where PEI per se was used as a delivery system for CRISPR/Cas9 are rare. One group used Polyethyleneimine Max™ (PEI Max) to transfect HEK293T cells infected with herpes simplex virus-1 (HSV-1) with CRISPR-Cas9. However, the authors did not divulge the molecular weight of the PEI used, nor was the N/P ratio mentioned (116). Also, the polyplexes generated from mixing pDNA with PEI were not characterized. Nevertheless, the study showed that transfection of HEK293T cells containing the HSV-1 genome resulted in knockdown of the target gene when using PEI Max polyplexes that comprising pDNA encoding for Cas9 and sgRNA. Two sgRNA sequences were used independently, each targeting a separate HSV-1 gene. It was found that the editing efficiency was significantly affected by the gene being targeted. Specifically, when the sgRNA targeting glycoprotein E (gE) was used an editing efficiency of 33% (of transfected cells) was observed whilst, when the thymidine kinase gene was targeted an editing efficiency of 5.8% was achieved (116). A separate study, where PEI was used as coating of a DNA nanoclew-based delivery system for CRISPR/Cas9, is discussed below.
Poly(amido amine) (PAMAM)
PAMAM has a high density of primary amine groups on the periphery of the polymer, facilitating strong complexation with DNA molecules (20, 109). Additionally, there is a high density of tertiary amines on the interior of PAMAM, which helps promote endosomal escape of DNA nanoplexes (114, 117). PAMAM is a dendrimer and the generation (G) number refers to the size of the dendrimer. When forming nucleic acid/polymer based polyplexes, as is also the case for PEI, the ratio of PAMAM to DNA or RNA is often referred to as the “N/P ratio”.
In a recent promising study, constructions of G5 PAMAM dendrons attached to a polymeric backbone composed of 2-hydroxyethyl methacrylate and glycidyl methacrylate, and referred to here as G5 dendronized polymers (G5DP), were prepared to enhance gene delivery and reduce cytotoxicity compared with the original G5 PAMAM dendrimer (117). The authors were looking for an efficient and safe delivery system for large plasmids, a situation that would apply to the delivery of a CRISPR/Cas9 system encoded by the one plasmid. The authors argued that viral vectors and liposomal complexes were less than ideal when it came to delivering large plasmids due to limited packaging capacity (for viral vectors) and rapid clearance, lability and high cytotoxicity (for lipoplexes) (118, 119). PEI, another cationic polymer, was considered excessively cytotoxic and not very efficient at delivering large plasmids (118, 120). Preliminary studies determined that the G5DPs were more effective than G1-G3 dendronized polymers at delivering pDNA to “difficult-to-transfect” cells (MCF-7: a human adenocarcinoma breast cell line) and were also more effective and less toxic than the G5 dendrimer counterpart. The authors subsequently demonstrated that fluorination of G5DPs further improved MCF-7 transfection efficiencies and that G5DP polyplexes containing large pDNA (~10 kb), expressing EGFP, were more effective at transfection than when Lipofectamine 2000 was used. Fluorination is known to improve the cellular uptake, to enhance endosomal escape, and to provide serum resistance of polyplexes formed from polymers and DNA (121). The authors then investigated the ability the G5DP constructs to deliver CRISPR tools such as sgRNA(s) and deactivated (devoid of nuclease activity) Cas9 (dCas9) bound to VP64 (a first generation transcriptional activator) (122) in order to increase the expression of the tumor suppressor mammary serine protease inhibitor (MASPIN) protein. Transfected MCF-7 cells were found to express increased MASPIN by approximately 5-fold (compared to negative control plasmid), whilst only a 2-fold increase was observed when Lipofectamine2000 was used as the delivery agent (Table 1) (117). Loss of MASPIN expression is associated with increased invasiveness and metastatic potential of breast cancer. This study demonstrated the versatility of PAMAM, the potential of G5DP constructs for delivery of CRISPR/Cas9 systems, as well as the improved transfection strength cationic polymer-based delivery systems can have over lipid-based compounds. The authors suggest this delivery platform should be suitable for translation into the clinic, however, no in vivo studies were performed.
Chitosan
Chitosan is an abundant, non-toxic, biodegradable, naturally occurring polysaccharide carrying a net positive charge, making it an attractive choice in drug and gene delivery (123). As with other cationic polymers, such as PEI, chitosan can form complexes with DNA via electrostatic interactions, however, transfection efficiencies are often comparatively lower (114). Previously, chitosan-coated poly(lactic-co-glycolic acid) NPs were used to deliver mRNA in ZFNs and TALENs to correct surfactant protein B deficiency (124), a hereditary lung disease (125), in mice. Given these findings researchers considered chitosan may be a good compositional candidate for CRISPR/Cas9 delivery systems.
Liu et al. designed self-assembling nanocarriers (size: ~300 nm; zeta potential: −9 mV) composed of an inorganic core (containing protamine sulfate, calcium carbonate and calcium phosphate), encapsulating a CRISPR/Cas9 plasmid, and a hydrophilic chitosan coating with targeting moieties. The pDNA expressed Cas9, sgRNA for CDK11 gene knockout, as well as GFP (to document transfection efficiencies) (58). Also the pDNAs were tagged with YOYO-1 so as to assess uptake efficiencies. The hydrophilic chitosan layer, consisting of biotinylated carboxymethyl chitosan and AS1411 aptamer-conjugated carboxymethyl chitosan, surface-adorned the inorganic core through electrostatic interactions. The purpose of employing protamine sulfate was not only to increase the loading efficiency of the pDNA within the nanoassemblies, due to its ability to tightly bind and condense DNA, but to facilitate nuclear delivery through a putative NLS that protamine sulfate possesses (126–128). Biotin receptors are overexpressed on many cancer cell lines (129), including the MCF-7 human breast cancer cells (130), and are not overexpressed on non-cancerous cells such as HEK293T. Also, the AS1411 aptamer targets nucleolin, which is usually overexpressed on the cell surface and in the nucleus of actively dividing cells and cancer cells, providing a means for nuclear translocation (131, 132). The target gene, CDK11p110, is a cyclin-dependent kinase gene that regulates cancer cell survival, proliferation, and growth, and is associated with poor prognosis in breast cancer patients (133). Knocking down CDK11p110 has been found to found to cause apoptosis in cancer cells (133–135). The presence of the two targeting moieties, biotin and the AS1411 aptamer, on these nanocarriers significantly enhanced uptake by MCF-7 cells, as measured by confocal microscopy and flow cytometry (YOYO-1 fluorescence) after 4 h of treatment, compared to nanocarriers containing no, or only one, of the targeting moieties. As expected, in HEK293T cells, substantial uptake was observed for all formulations, however, no significant enhancement in uptake was observed when the dual-targeting nanocarriers were used. Transfection efficiencies were not quantitatively assessed thus it was difficult to objectively arrive at conclusive comparisons based on the confocal images presented. More convincing were the cell viability and western blotting data. It was shown that the viability of the MCF-7 cells dropped substantially (< 40% viability after 48 h), when the dual targeting nanocarriers were used, compared to non-targeting nanocarriers (~60% viability). Meanwhile the cytotoxicity imposed on the HEK293T cells was negligible after 48 h of treatment (> 80% viability). Western blotting revealed that MCF-7 cells expressed > 90% less CDK11p110 than untreated control cells when the dual-targeted nanocarriers were used whilst HEK293T cells expressed approximately 35% less CDK11p110, illustrating again greater potency when targeting CDK11p110 in cancer cells versus non-cancer cells (Table 1). No in vivo studies were performed, and in order to determine the potential effectiveness of these formulations in the clinic such studies will be necessary.
C. Cell penetrating peptides and other miscellaneous delivery systems
Cell penetrating peptides (CPPs), also known as protein transduction domains, belong to a class of short peptides that can move across the cell membrane (136–138). The mechanism by which they translocate across cell membranes is not yet fully understood, however, receptor-independent direct membrane penetration, and endocytosis-based transport are among the proposed mechanisms (137). The HIV-Tat peptide, or simply Tat peptide, was the first member of this family of peptides to be discovered in 1988 (136), followed by a number of other peptides such as GALA and polyarginine. Many CPPs can be used as vectors to deliver nucleic acids and other macromolecules across the cell membrane (137, 139). Covalent attachment to protein molecules (140), electrostatic interaction (141), supramolecular assembly with the therapeutic macromolecule (32), or surface attachment onto nanocarriers (59, 139, 142) are among strategies to adopt CPP into drug and gene delivery platforms.
The first study that employed CPPs for intracellular delivery of CRISPR/Cas9 to mammalian cells was reported by Ramakrishna et al. (140). Whereas previous studies used mechanical delivery methods such as electroporation and microinjection, the authors here co-delivered Cas9 covalently conjugated to a CPP (consisting of four glycines, nine arginines, and four leucines, linked to maleimide, and abbreviated as m9R) and sgRNA (targeting C-C chemokine receptor type 5 (CCR5)) complexed with a similar CPP without the maleimide functionality (abbreviated as 9R). A cysteine residue was chemically bound to the C-terminus of Cas9 in order to promote SH-maleimide conjugation. Gene disruption efficiencies (percent indels) at the CCR5 gene locus were evaluated in a number of human cell lines, including HEK293T, HeLa, NCCIT (human pluripotent embryonal carcinoma cell line), human dermal fibroblasts, and H9 human embryonic stem cells, and the values were found to be 16%, 5.5%, 2.7%, 8.4%, and 2.3%, respectively (140). Even though the gene disruption percentages are relatively low, the importance of this work stems from it being one of the earliest studies involving non-viral vectors for CRISPR/Cas9 gene editing delivery, and it is the first report that employed CPPs for that purpose. It also showed genome editing potential in human embryonic stem cells, which if properly optimized, may have significant therapeutic applications. The potential of CRISPR/Cas9 applications in the field of human pluripotent stem cell is reviewed elsewhere (143).
Supramolecular assembly of NPs made from an amphiphilic protein and Cas9 RNPs has been described by Lostale-Seijo et al. (32). The amphiphilic protein (PTn) was composed of a cationic cell-penetrating peptide (denoted by P) covalently and independently attached to a variety of lipophilic aldehyde tails (Tn) via hydrazone bonding. The authors screened a library of NPs formulated with lipophilic aldehydes of different carbon chain lengths by assessing percent knockout of the EGFP gene in EGFP-expressing HeLa cells. Resultantly, an NP formulation using PT24, comprising an oleic aldehyde (n = 24), was found to be optimal (32). Efficient complexation between the amphiphilic protein (PT24) and Cas9 RNP was confirmed by gel electrophoresis. The particles appeared spherical when analyzed by transmission electron microscopy (TEM), possessing a dense core (with a less dense external layer), an average particle size of ~ 270 nm and a zeta potential of +4 mV. (32). The uptake of these NPs by HeLa cells was found to be primarily due to macropinocytosis and not caveolin-dependent nor clathrin-mediated endocytosis. Indels were quantified by a T7 Endonuclease 1 assay 48 h following treatment of HeLa cells with either PT24 or Lipofectamine 2000, both complexing Cas9 RNPs targeting the HPRT1 gene. It was found that editing efficiencies were similar although PT24 was marginally more efficient when lower concentrations of RNP were used, whilst Lipofectamine2000 was more efficient at higher RNP doses, however, the PT24/Cas9 RNP complexes were less cytotoxic than the Lipofectamine2000 counterpart (32). Overall, the maximum gene editing efficiency obtained with PT24/Cas9 RNP was approximately 30–40% (Table 1). Editing efficiencies of clinically relevant genes are yet to be investigated. Animal studies were not performed here and will be required in the future to assess the efficacy, toxicity and immunogenicity of PT24/Cas9 RNP complexes, thus determining the feasibility for translation into the clinic.
In a recent study, Wang et al. prepared PEGylated NPs (size: ~100 nm; zeta potential: ~+20 mV), referred to as P-HNPs, using a synthetic cationic α-helical poly-glutamate-based polypeptide, poly(γ−4-((2-(piperidin-1-yl) ethyl) aminomethyl) benzyl-L-glutamate) (PPABLG), for the delivery of two plasmids independently encoding Cas9 and sgRNA (29). The Cas9 encoding plasmid either encoded a GFP fusion protein or was tagged with the fluorescent YO-YO-1 tag for transfection and uptake studies, respectively. This class of α-helical synthetic poly-glutamates, previously utilized for various nucleic acid delivery applications (144–147), exhibit several intriguing features, including high cell membrane penetration capability that surpasses that of the HIV-Tat peptide, and enhanced endosomal escape; with both properties attributed to the stable α-helical structure and the strong cationic charge of these peptides (29, 145). Importantly, unlike other CPPs, these polypeptides have been found to be resistant to most proteases (29). The NPs were PEGylated by mixing polyethylene glycol 2000-polythymine 40 conjugate (PEG2k-T40) with the plasmid prior to the addition of PPABLG. It is worth noting that PEGylation decreased NP-induced cytotoxicity in HEK293T cells in vitro, and significantly improved the nanoplex stability in serum-containing media, the latter trait suggesting the potential for these P-HNPs to possess stealth properties in vivo, however, this was not tested in these studies. Surface-coating of the NPs with PEG, instead of adding it to the plasmid, was found to significantly decrease the transfection efficiency of the Cas9-GFP plasmid-loaded NPs, most likely because surface PEG masked the cellular membrane penetrating capability of PPABLG. Cellular uptake studies revealed that, as well as relying partially on endocytosis, a large proportion (36%) of the P-HNPs entered the cells by an energy-independent, endocytic-independent pathway. Indirect evidence suggested that the P-HNPs entering cells in an endocytic-independent manner directly accessed the cytoplasm by punching holes in the outer membrane. The genome editing efficiencies of these P-HNPs independently loaded with sgRNA or Cas9 pDNA were investigated. First, the authors obtained comparable editing efficiencies when they either targeted AAVS1 gene (42.5%) or the HPRT1 gene (46%) in HEK293T cells using the relevant sgRNA for each gene, or when they co-delivered both sgRNAs in the same NP (41.9% for the AAVS1 gene and 47.3% for the HPRT1 gene) (Table 1). Finally, a P-HNP system independently loaded with Cas9 pDNA and sgRNA that targeted the survival gene Plk1 significantly suppressed the growth of HeLa tumors in nude mice, and significantly extended the median survival of such mice, following 10 intratumoral injections over the range of 10 days (Table 1) (29). This P-HNP-based therapy was shown to not cause any reduction in mouse body weights compared to control mice, indicating a lack of toxicity. Nevertheless, further in vivo studies evaluating toxicity and immunogenicity are required if progression into clinical studies is to occur.
Self-assembling DNA nanoclews are yarn-like structures that are formed by rolling circle amplification. Nanoclews were investigated as a novel delivery system for sgRNA/Cas9 RNPs. sgRNA/Cas9 RNPs were complexed with DNA nanoclews containing sequences complementary to the 5’ end of the sgRNA (targeting EGFP) in order to promote reversible complexation. Subsequent to complexation, PEI (linear PEI Max; MW: 40,000) was then added in order to facilitate endosomal escape presumably via the proton sponge effect. After coating with PEI, the nanoplexes had a positive charge (zeta potential: +18 mV; size: ~56 nm). The editing efficiency of these nanoplexes was assessed in vitro by transfecting U2OS-EGFP cells (a human bone osteosarcoma cell line expressing EGFP) and was demonstrated to be 36% (Table 1). When PEI alone was used to complex the sgRNA/Cas9 RNPs the editing efficiency was only 5% (148). It was determined that uptake of the complexes was primarily due to lipid raft-dependent (i.e. caveolae-mediated) endocytosis and micropinocytosis and that these complexes exhibited no cytotoxicity. Intratumoral (U2OS-EGFP cells) injection of the nanoplexes targeting EGFP demonstrated reduction in EGFP expressing cells proximal to the injection site (10 days post-injection) as determined by analysis of frozen tumor sections using fluorescence microscopy (Table 1). Future in vivo studies examining the toxicity and immunogenicity of nanoclews complexed with sgRNA/Cas9 RNPs are required in order to assess their potential for clinical translation.
Inorganic nanomaterials can also be chemically modified in order to utilize them for nucleic acid delivery. In one study, cationic arginine-functionalized gold nanoparticles (ArgNPs) were used to deliver Cas9 RNPs, comprising chemically modified Cas9 protein and sgRNA (149). Gold nanoparticles were functionalized with arginine. Cas9 and sgRNA (1:1 molar ratio) were mixed with the ArgNPs, resulting in “self-assembled nanoassemblies”. It should be noted that although the authors refer to them as nanoassemblies they were in actuality approximately 475 nm in diameter, which technically implicates them as microparticles rather than NPs as defined by the International Union of Applied Chemistry (150). However, they were self-assembled particles made from nanosized components (ArgNP: 10 nm; Cas9: 7.5 nm; sgRNA: 5.5 nm). The Cas9 used was either modified with a glutamate peptide tag at the N-terminus (Cas9glut) or remained unmodified. The main purpose of the glutamate peptide tag was to impart a negatively charged region to the highly positive Cas9, thus enabling stronger binding to the positively-charged arginine residues on the ArgNPs (149). Also, the C-terminus of the Cas9glut molecule was modified by the addition of a nuclear localization signal (NLS) to improve nuclear delivery. Surprisingly, in vitro cellular uptake studies using HeLa cells revealed that these nanoassemblies entered into the cytoplasm directly through cholesterol-mediated membrane fusion, rather than by endocytosis, as determined through the use of inhibitors of both processes of cell entry (149). The authors do not expand on the specificities as to what properties these nanoassemblies possessed that would have contributed to their cytosolic delivery by membrane fusion. Particle-based drug delivery via non-endocytic pathways has proven to very difficult and usually requires the presence of virally derived fusogenic peptides/proteins (151). Nevertheless, efficient uptake and nuclear delivery were reported to have been achieved. Genome editing efficiency values in the range of 23–30% were obtained following the incubation of the nanoassemblies using a variety cell lines (Table 1)(149). Although these nanoassemblies are not appropriate for IV delivery (primarily due to their size), the authors argue that they could still be of use in many in vitro situations. There were no animal studies performed to determine the effectiveness of these nanoassemblies, which will ultimately be necessary if they are to move forward into the clinic. More promisingly, in terms of potential applications for in vivo therapy, one group developed a gold NP-based system capable of simultaneously delivering Cas9 RNP, sgRNA and donor DNA and efficiently correcting the mutated gene responsible for Duchenne muscular dystrophy DMDD) in mice (152). The delivery system, CRISPR-Gold, also contained an endosomal disruptive cationic polymer, PAsp(DET). Manufacture of the CRISPR-Gold involved coating gold NPs with thiol-terminated DNA which was then hybridized to the donor DNA and then Cas9 RNP was allowed to absorb to the NPs. Then the NPs were coated with silica and subsequently PAsp(DET) was added. One drawback of CRISPR-Gold was the tendency to aggregate once placed in solution for times extending beyond 5 minutes, making them inappropriate for IV administration. In vitro experiments using HEK cells suggested the mechanism of uptake of CRISPR-Gold was via caveolae/raft-mediated endocytosis and that the uptake depended on the presence of PAsp(DET). DMD is a lethal disease resulting from mutations in the dystrophin gene and for which there is not current effective remedy. Using C57/BL/10ScSn-Dmdmdx/J (mdx) mice, which possess a nonsense mutation in the dystrophin gene, it was shown that one intramuscular injection of CRISPR-Gold (formulated to target the dystrophin gene) at 6 mg/kg was capable of a 5.4% correction rate of the mutated dystrophin gene to the wild type (152). In addition, enhanced dystrophin protein expression occurred at the site of the injected muscle and the mice exhibited enhanced strength using clinically relevant assays (152).
Metal-organic frameworks (MOFs) have been investigated as delivery systems for CRISPR/Cas9. MOFs, which are a subclass of coordination polymers, generally comprise metal ions that are complexed with organic ligands, potentially forming 3-dimensional structures containing empty pockets of tunable volumes that are capable of hosting molecular cargo. Recently, zeolitic imidazolate framework-8, a coordination of Zn++ and 2-methylimidazole, and a subclass of MOFs were used to deliver Cas9 protein and sgRNA targeting EGFP to Chinese hamster ovary cells expressing EGFP (CHO-EGFP) (153). It was shown that this nanoformulation ( ~100 nm diameter and +5 mV) could reduce EGFP fluorescence levels by 37% within 4 days of incubation with CHO-EGFP in vitro (Table 1). In addition, aside from being biocompatible, it was shown that the nanostructures were stable at neutral pH but sensitive to degradation at the acidic/endosomal pH of 5.5. In order to demonstrate the potential for translation of this nanoformulation into the clinic, it will need to be tested in animal models for efficacy, toxicity and immunogenicity.
Another potential candidate for CRISPR/Cas9 delivery was investigated in vitro that involved NPs made from dual functionalized graphene oxide (GO), PEG, and PEI (154). The resultant NPs were formed by modifying planar GO with PEG, then covalently linking to PEI and complexing with sgRNA (targeting EGFP) and Cas9 protein via self-assembly. Using a human gastric adenocarcinoma cell line expressing EGFP, AGS-EGFP, it was shown the manufactured NPs ( ~220 nm and +18.5 mV) had an editing efficiency of 39% (Table 1). Promisingly, it was also shown that the carrier component of the NPs (GO-PEG-PEI) could protect the cargo (sgRNA/Cas9) from enzymatic hydrolysis. The authors suggest the potential for these types of NPs to be used in vivo, however, as yet they have not presented any animal-based efficacy or toxicity studies, which will be required if these formulation are to progress into the clinic.
D. The potential for successful clinical translation of CRISPR/Cas9 delivery systems
While many of the CRISPR/Cas9 delivery systems described above are novel formulations, many other similar NP formulations have been previously employed to deliver siRNA, plasmids, and other therapeutic molecules. Table 2 enlists some examples of such formulations that are currently being evaluated in clinical trials, and that contain components that are also used to deliver CRISPR/Cas9 molecules. As shown, treatment- or vehicle-related toxicity issues are not severe, with the most adverse events reported being grade 1 or 2. As detailed above, some of the novel formulations delivering CRISPR/Cas9 have undergone preliminary tests for toxicity in in vivo murine models where promising results were obtained. Nevertheless, each of these formulations will likely require further preclinical assessments of in vivo toxicity, immunogenicity and efficacy if they are to progress to clinical trials. Many of these novel systems are comprised of familiar components that have independently exhibited strong biocompatibility and have been either FDA-approved. For example, FDA-approved PEG has been used in many clinical formulations (155, 156). Many lipid-based NP formulations delivering siRNA, chemotherapy or pDNA have recently entered into clinical trials and the results from these trials could well inform their potential for use as vectors for CRISPR/Cas9 components (157, 158) (159) (160). Formulations comprising gold NPs, such as Auroshell, have been recognized by the FDA as medical devices and have been assessed for safety in clinical settings with promising results (161). Nevertheless, further biodistribution, clearance and cytotoxicity studies in a clinical context are required if the full therapeutic potential of gold NPs are to be evaluated. Finally, CPPs have been extensively tested for safety in a range of clinical trials over the past decade with favorable outcomes (162).
Table 2:
Examples of clinical trials in which several nucleic acid molecules or other active moieties are being delivered by formulations that are also applied to deliver CRISPR/Cas 9 systems. No significant or severe treatment- or formulation-related toxicities were reported with most formulations.
| Name/Clinical trial identifier (NCT) # | Therapeutic molecule | Target/purpose | Vehicle/vector | Route of administration, dose, and dosage a | Reported adverse events a |
|---|---|---|---|---|---|
| DOTAP:chol-TUSC2/ NCT00059605 (166, 167) | Plasmid DNA | Restoration of the activity of the tumor suppressor gene TUSC2/FUS1 | DOTAP (cationic phospholipid) and cholesterol-based nanoparticles | Six dose levels ranging from 0.01 to 0.09 mg/kg given as a single intravenous infusion every 3 weeks. |
Phase I study (in 31 patients with recurrent, and/or metastatic lung cancer) • IV injection of DOTAP:Chol-TUSC2 at escalating doses was found to be safe and well-tolerated. • The Maximum tolerated dose (MTD) was found to be 0.06 mg/kg. • Premedication with dexamethasone and diphenhydramine was mandated. • Dose-limiting toxicity (DLT) was detected in 2 patients, and manifested as grade 3 hypophosphatemia at 0.06 and 0.09 mg/kg. |
|
PNT2258/ NCT01191775 NCT01733238 (168–171) |
24-base single-stranded DNA oligodeoxy-nucleotide | BCL-2 oncogene | Smarticles® liposomes (approximately 130 nm, containing pH sensitive lipids that are cationic during manufacture and anionic during circulation). |
Phase I study Intravenous infusion of 10 dose levels (ranging from 1 to 150 mg/m2) on an escalating dose design. Single dose was administered daily for 5 days on 21-day cycles. |
Phase I study (in 22 patients with advanced metastatic solid tumors) • PNT2258 is well-tolerated with doses ranging from 1 up to 150 mg/m2. • No unmanageable stage 3 or 4 adverse events. • No MTD was reached. • Decrease in lymphocytes and platelet count was manageable. • Dose limiting toxicity (DLT), manifested as a transient increase in aspartate aminotransferase, was reported with the highest dose (150 mg/m2) Phase II study (in patients with relapsed or refractory non-Hodgkin’s lymphoma) • PNT2258 was found to be well-tolerated. • No grade 3 or 4 adverse events reported. |
| SNS01-T/ NCT01435720 (115, 172, 173) | siRNA | Eukaryotic Translation Initiation Factor 5A (eIF5A) | Polyethyleneimine (PEI) nanoparticles | Intravenous infusion of 0.0125, 0.05, 0.2, and 0.375 mg/kg twice a week for six weeks. |
Phase I/II study dose escalating study
• Treatment-related adverse events were reported in all patients. • Grade 3 adverse events were reported in 50% of patients with no dose relationship found, with thrombocytopenia as the only grade 3 adverse event reported in one or more patient. • There was no reported treatment-related deaths, while one incident of DLT (grade 4 infusion reaction) was reported in one patient. • Treatment was discontinued and the treatment protocol was amended to add pretreatment with corticosteroids, antihistamine, and acetaminophen (obligatory), and opioid analgesic if clinically needed. |
|
BC-819/ NCT01878188 NCT00826150 (174–177) |
Plasmid DNA | H19 oncofetal gene | Polyethyleneimine (PEI) |
Phase I/IIa study One Intraperitoneal injection/week for 6–9 weeks at maximum at doses up to 240 mg. |
Phase I/IIa study (in 14 patients with recurrent ovarian/peritoneal cancer) • BC-819 was found to be safe and well-tolerated when doses up to 240 mg were administered intraperitoneally to cancer patients. • No dose limiting toxicity (DLT) was observed. • Grade 1 and 2 adverse events were observed in 4 patients. No grade 3 adverse events were recorded. Phase I/II study • Escalating-dose Intravesical administration was found to be safe and well-tolerated in patients with superficial bladder cancer. |
|
Atu027/ NCT00938574 NCT01808638 (178–183) |
siRNA | Protein kinase N3 (PKN3) gene in the vascular endothelial cells. | Cationic lipoplexes, containing fusigenic and PEGylated lipid components (AtuPLEX) |
Phase I study One single dose received, followed by a twice weekly infusions over a 28-days cycle, according to a dose-escalation design with doses ranging from 0.001 to 0.336 mg/kg. |
Phase I study (34 patients with advanced refractory solid tumors) • Atu027 was safe and well-tolerated in patients with advanced solid tumors. • No MTD was reached. • One patient showed DLT related to Atu027 administration (increased lipase, grade 3 adverse event). • Three deaths were reported, and none of them was attributed to Atu027. • Most adverse events reported were grade 1 and 2 (low). Ten percent of patients showed grade 3 adverse events. • No corticosteroid or antihistamine premedication was required. • Phase Ib/IIa study in combination with gemcitabine (23 patients with advanced pancreatic carcinoma) • The combination between Atu027 and gemcitabine was well-tolerated in patients with advanced pancreatic cancer. • Most adverse events were consistent with those reported with gemcitabine. |
| DCR-MYC/ NCT02110563 (184, 185) | siRNA | MYC oncogene | Encore® Lipid Nanoparticles | IV infusion on day 1 and day 8 over a 3-weeks cycle (week 3 is a rest period). Cycles continue until unacceptable toxicity emerges. The starting dose is 0.1 mg/kg with a dose escalation of 100, 50, or 25% increase depending on toxicity. |
Phase I study • DCR-MYC doses up to 1 mg/kg were found to be safe and well-tolerated in advanced cancer patients. • Adverse events were all of low grade (grade 1 and 2). One patient developed a grade 3 infusion-related adverse event, and one patient experienced a grade 3 DLT (transient rise in AST). Efficacy results did not encourage further clinical investigation and thus the clinical development of DCR-MYC project was discontinued. |
|
p28/ NCT00914914 NCT01975116 (186–189) |
p28 | Wild type and mutant P53 | Cancer-cell killing cell penetrating peptide (p28), a synthetic 28-peptide fragment of azurin. |
Phase I study in adult patients Intravenous infusion, three times/week for three weeks, followed by a rest period of 2 weeks. Dose escalation design was followed at doses of 0.83, 1.66, 2.5, 3.33, and 4.16 mg/kg. |
Phase I study in adult patients with stage IV advanced solid tumors • Cumulative doses up to 140 mg/kg were well-tolerated and showed no immunogenicity (no antibody response to the p28 protein) or significant toxicity. • No MTD or No-Observed-Adverse-Effect-Level (NOAEL) was reached. • Only 15% of adverse events reported for all patients during the whole period of treatment (multiple visits) were of grade 3. None of them were considered directly-related to p28. Phase I study in pediatric patients with recurrent and progressive central nervous system tumors • In general, the treatment was well-tolerated in pediatric patients with solid CNS tumors. • The most-common treatment-related adverse event was grade 1 infusion reaction (transient). • One patient experienced 2 events of DLT (grade 4 neutropenia and thrombocytopenia). |
The columns entitled ‘Route of administration, dose, and dosage’ and ‘Reported adverse events’ both report only selected examples of the clinical trials that employ the associated treatment, regardless of their status. They do not report information about all the clinical trials found in Clinicaltrials.gov, or any other source, about that particular treatment.
Aside from issues of toxicity and antigen independent immunogenicity, another potential barrier to CRISPR/Cas9 is that a large percent of the human population possess pre-existing immunity to Cas9 (163) (primarily derived from the common human commensals, Staphylococcus aureus or Streptococcus pyogenes). This can potentially lead to antibody-mediated neutralization of the CRISPR/Cas9 system when Cas9 is delivered in protein form. Thus, NP formulations that protect the cargo from being recognized by the host’s Cas9-specific antibodies would be ultimately desired. An additional concern related to pre-existing immunity is the potential for cell-mediated based immune eradication of cells expressing Cas9. To explain, cells targeted with CRISPR/Cas9, where the Cas9 is delivered in nucleic acid forms, can be potentially recognized and eliminated by pre-existing Cas9-specific cytotoxic T cells, since most cells express cell surface MHC class I and will therefore present Cas9 epitopes to the host’s immune system. One way of decreasing the chances of this occurring would be to deliver the Cas9 to the target cells in protein form rather than in nucleic acid form as this would result in significantly reduced presentation of Cas9 epitopes in association with MHC class I and therefore the target cells would be less readily detectable by the host’s pre-existing Cas9-specific cytotoxic T cells.
Finally, a major concern for using CRISPR/Cas9 is the pathogenic potential caused by off-target or on-target mutations (164). Whilst NP based delivery may enhance targeting to certain tissues and promote delivery to the nucleus, at this stage the delivery vehicle itself will unlikely be capable of resolving this important issue which needs serious attention prior to using CRISPR/Cas9 as a therapeutic tool.
III. Conclusion and Future Perspectives
This review article has summarized many of the novel NP-based approaches currently being investigated as potential modes for delivering CRISPR/Cas9 gene editing systems to cells. Many of the early studies used commercial lipid-based delivery systems primarily to establish the feasibility of delivering CRISPR/Cas9 to cells. Subsequently, however, many researchers have developed novel NP carriers with the ultimate goal of these formulations being capable of safely and efficiently delivering CRISPR/Cas9 based therapeutics directly to patients in order to ablate a specific disease. Some of the studies compared editing efficiencies of the novel NP delivery systems with commercial lipid-based delivery systems such as Lipofectamine 2000, often revealing comparable results, however, the novel NP-based strategies were usually shown to be less cytotoxic. Also, whilst only a few of the NP formulations were tested by intravenous administration, many of those yet to be tested are being designed with this potential application in mind. Thus, while commercial lipid-based delivery systems may still have some clinical applications where ex vivo delivery of CRISPR/Cas9 will suffice, in order to progress to direct treatment of patients such delivery systems are not practical, and the need for appropriate NP-based formulations are required.
Many researchers have addressed the potential of delivering NP-based CRISPR-Cas9 as a therapeutic by assessing editing and phenotypic efficacies in vivo, with very promising results (Table 1). The finding that lipid-based NPs could essentially ablate expression of a target gene (expressed by hepatocytes) when administered intravenously in a murine model is very promising, particularly for liver diseases, given the tropism of lipid-based NPs for the liver. Some of the formulations were tested for anticancer activity in vivo by intratumoral injection of NP formulations using murine tumor models, achieving promising results. While intratumoral injection is a useful tool to confirm the proof of concept of many experimental anti-cancer therapeutics, its potential for clinical translation is limited to certain situations or tumor types. Due to the metastatic nature of cancer, other administration routes, particularly IV, are more clinically relevant and delivery of NP formulations need to be tested preclinically using these routes in order to assess safety and efficiency.
Although CRISPR/Cas9 is still in its infancy as a potential therapeutic, the future is looking promising with clinical trials, albeit ex vivo based therapy, having already begun. The finding that different formulations can have a predilection for a particular organ such as the lung or the liver (Table 1) should prove to be very useful when treating organ-specific pathologies and can only be further improved upon by the generation of actively targeting NP formulations. Of course, assiduous monitoring of off-target effects in vivo will be an imperative as preclinical investigations move forward into the clinic.
Acknowledgements
B. E. G. acknowledges fellowship support from the Alfred P. Sloan Foundation, the University of Iowa Graduate College, and the National GEM Consortium. E. J. Devor acknowledges support from the University of Iowa Department of Obstetrics and Gynecology Research Development Fund. A.K.S. acknowledges support from the National Cancer Institute at the National Institutes of Health (5P30CA086862) and the Lyle and Sharon Bighley Chair of Pharmaceutical Sciences.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest in the preparation and submission of this manuscript.
V. References
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–75. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–12. [DOI] [PubMed] [Google Scholar]
- 5.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–70. [DOI] [PubMed] [Google Scholar]
- 6.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–82. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens CJ, Kashentseva E, Everett W, Kaliberova L, Curiel DT. Targeted in vivo knock-in of human alpha-1-antitrypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Ther. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- 11.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–64. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Hu S, Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials. 2018;171:207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16(6):387–99. [DOI] [PubMed] [Google Scholar]
- 15.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cyranoski D First trial of CRISPR in people. Nature. 2016;535(7613):476–7. [DOI] [PubMed] [Google Scholar]
- 17.Xu Z, Chen L, Gu W, Gao Y, Lin L, Zhang Z, et al. The performance of docetaxel-loaded solid lipid nanoparticles targeted to hepatocellular carcinoma. Biomaterials. 2009;30(2):226–32. [DOI] [PubMed] [Google Scholar]
- 18.Aldayel AM, Naguib YW, O’Mary HL, Li X, Niu M, Ruwona TB, et al. Acid-Sensitive Sheddable PEGylated PLGA Nanoparticles Increase the Delivery of TNF-alpha siRNA in Chronic Inflammation Sites. Mol Ther Nucleic Acids. 2016;5(7):e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naguib YW, Cui Z. Nanomedicine: the promise and challenges in cancer chemotherapy. Adv Exp Med Biol. 2014;811:207–33. [DOI] [PubMed] [Google Scholar]
- 20.Rajasekaran D, Srivastava J, Ebeid K, Gredler R, Akiel M, Jariwala N, et al. Combination of Nanoparticle-Delivered siRNA for Astrocyte Elevated Gene-1 (AEG-1) and All-trans Retinoic Acid (ATRA): An Effective Therapeutic Strategy for Hepatocellular Carcinoma (HCC). Bioconjug Chem. 2015;26(8):1651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Givens BE, Geary SM, Salem AK. Nanoparticle-based CpG-oligonucleotide therapy for treating allergic asthma. Immunotherapy. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebeid K, Meng X, Thiel KW, Do AV, Geary SM, Morris AS, et al. Synthetically lethal nanoparticles for treatment of endometrial cancer. Nat Nanotechnol. 2018;13(1):72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wongrakpanich A, Morris AS, Geary SM, Joiner MA, Salem AK. Surface-modified particles loaded with CaMKII inhibitor protect cardiac cells against mitochondrial injury. Int J Pharm. 2017;520(1–2):275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris AS, Sebag SC, Paschke JD, Wongrakpanich A, Ebeid K, Anderson ME, et al. Cationic CaMKII Inhibiting Nanoparticles Prevent Allergic Asthma. Mol Pharm. 2017;14(6):2166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi VB, Adamcakova-Dodd A, Jing X, Wongrakpanich A, Gibson-Corley KN, Thorne PS, et al. Development of a poly (lactic-co-glycolic acid) particle vaccine to protect against house dust mite induced allergy. AAPS J. 2014;16(5):975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi VB, Geary SM, Salem AK. Biodegradable particles as vaccine delivery systems: size matters. AAPS J. 2013;15(1):85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salem AK, Searson PC, Leong KW. Multifunctional nanorods for gene delivery. Nat Mater. 2003;2(10):668–71. [DOI] [PubMed] [Google Scholar]
- 28.Wang HX, Li M, Lee CM, Chakraborty S, Kim HW, Bao G, et al. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem Rev. 2017;117(15):9874–906. [DOI] [PubMed] [Google Scholar]
- 29.Wang HX, Song Z, Lao YH, Xu X, Gong J, Cheng D, et al. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc Natl Acad Sci U S A. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paliwal R, Babu RJ, Palakurthi S. Nanomedicine scale-up technologies: feasibilities and challenges. AAPS PharmSciTech. 2014;15(6):1527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glass Z, Lee M, Li Y, Xu Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018;36(2):173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lostale-Seijo I, Louzao I, Juanes M, Montenegro J. Peptide/Cas9 nanostructures for ribonucleoprotein cell membrane transport and gene edition. Chem Sci. 2017;8(12):7923–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mekler V, Minakhin L, Semenova E, Kuznedelov K, Severinov K. Kinetics of the CRISPR-Cas9 effector complex assembly and the role of 3’-terminal segment of guide RNA. Nucleic Acids Res. 2016;44(6):2837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiCarlo JE, Deeconda A, Tsang SH. Viral Vectors, Engineered Cells and the CRISPR Revolution. Adv Exp Med Biol. 2017;1016:3–27. [DOI] [PubMed] [Google Scholar]
- 35.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess GT, Tycko J, Yao D, Bassik MC. Methods and Applications of CRISPR-Mediated Base Editing in Eukaryotic Genomes. Mol Cell. 2017;68(1):26–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M, Glass ZA, Xu Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017;24(3):144–50. [DOI] [PubMed] [Google Scholar]
- 38.Ha JS, Lee JS, Jeong J, Kim H, Byun J, Kim SA, et al. Poly-sgRNA/siRNA ribonucleoprotein nanoparticles for targeted gene disruption. J Control Release. 2017;250:27–35. [DOI] [PubMed] [Google Scholar]
- 39.Mout R, Ray M, Lee YW, Scaletti F, Rotello VM. In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges. Bioconjug Chem. 2017;28(4):880–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M, et al. Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery. Gene Ther. 2007;14(8):682–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF. Targeting receptor-mediated endocytotic pathways with nanoparticles: rationale and advances. Adv Drug Deliv Rev. 2013;65(1):121–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–63. [DOI] [PubMed] [Google Scholar]
- 43.Vercauteren D, Rejman J, Martens TF, Demeester J, De Smedt SC, Braeckmans K. On the cellular processing of non-viral nanomedicines for nucleic acid delivery: mechanisms and methods. J Control Release. 2012;161(2):566–81. [DOI] [PubMed] [Google Scholar]
- 44.Sonawane ND, Szoka FC Jr., Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–31. [DOI] [PubMed] [Google Scholar]
- 45.Richard I, Thibault M, De Crescenzo G, Buschmann MD, Lavertu M. Ionization behavior of chitosan and chitosan-DNA polyplexes indicate that chitosan has a similar capability to induce a proton-sponge effect as PEI. Biomacromolecules. 2013;14(6):1732–40. [DOI] [PubMed] [Google Scholar]
- 46.Cardarelli F, Pozzi D, Bifone A, Marchini C, Caracciolo G. Cholesterol-dependent macropinocytosis and endosomal escape control the transfection efficiency of lipoplexes in CHO living cells. Mol Pharm. 2012;9(2):334–40. [DOI] [PubMed] [Google Scholar]
- 47.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39(3 Pt 2):499–509. [DOI] [PubMed] [Google Scholar]
- 48.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Xian L, Xing H, Yu J, Yang Z, Yang T, et al. Factors influencing the nuclear targeting ability of nuclear localization signals. J Drug Target. 2016;24(10):927–33. [DOI] [PubMed] [Google Scholar]
- 50.Hu Q, Wang J, Shen J, Liu M, Jin X, Tang G, et al. Intracellular pathways and nuclear localization signal peptide-mediated gene transfection by cationic polymeric nanovectors. Biomaterials. 2012;33(4):1135–45. [DOI] [PubMed] [Google Scholar]
- 51.Lee J, Jung J, Kim YJ, Lee E, Choi JS. Gene delivery of PAMAM dendrimer conjugated with the nuclear localization signal peptide originated from fibroblast growth factor 3. Int J Pharm. 2014;459(1–2):10–8. [DOI] [PubMed] [Google Scholar]
- 52.Park E, Cho HB, Takimoto K. Effective gene delivery into adipose-derived stem cells: transfection of cells in suspension with the use of a nuclear localization signal peptide-conjugated polyethylenimine. Cytotherapy. 2015;17(5):536–42. [DOI] [PubMed] [Google Scholar]
- 53.Pan L, He Q, Liu J, Chen Y, Ma M, Zhang L, et al. Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J Am Chem Soc. 2012;134(13):5722–5. [DOI] [PubMed] [Google Scholar]
- 54.Malhotra M, Tomaro-Duchesneau C, Prakash S. Synthesis of TAT peptide-tagged PEGylated chitosan nanoparticles for siRNA delivery targeting neurodegenerative diseases. Biomaterials. 2013;34(4):1270–80. [DOI] [PubMed] [Google Scholar]
- 55.Nitin N, LaConte L, Rhee WJ, Bao G. Tat peptide is capable of importing large nanoparticles across nuclear membrane in digitonin permeabilized cells. Ann Biomed Eng. 2009;37(10):2018–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ammosova T, Jerebtsova M, Beullens M, Lesage B, Jackson A, Kashanchi F, et al. Nuclear targeting of protein phosphatase-1 by HIV-1 Tat protein. J Biol Chem. 2005;280(43):36364–71. [DOI] [PubMed] [Google Scholar]
- 57.Sandgren S, Cheng F, Belting M. Nuclear targeting of macromolecular polyanions by an HIV-Tat derived peptide. Role for cell-surface proteoglycans. J Biol Chem. 2002;277(41):38877–83. [DOI] [PubMed] [Google Scholar]
- 58.Liu BY, He XY, Xu C, Xu L, Ai SL, Cheng SX, et al. A Dual-Targeting Delivery System for Effective Genome Editing and In Situ Detecting Related Protein Expression in Edited Cells. Biomacromolecules. 2018. [DOI] [PubMed] [Google Scholar]
- 59.Asai T, Tsuzuku T, Takahashi S, Okamoto A, Dewa T, Nango M, et al. Cell-penetrating peptide-conjugated lipid nanoparticles for siRNA delivery. Biochem Biophys Res Commun. 2014;444(4):599–604. [DOI] [PubMed] [Google Scholar]
- 60.Nelson CE, Gersbach CA. Engineering Delivery Vehicles for Genome Editing. Annu Rev Chem Biomol Eng. 2016;7:637–62. [DOI] [PubMed] [Google Scholar]
- 61.Cardarelli F, Digiacomo L, Marchini C, Amici A, Salomone F, Fiume G, et al. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci Rep. 2016;6:25879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu X, Liang X, Xie H, Kumar S, Ravinder N, Potter J, et al. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol Lett. 2016;38(6):919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. [DOI] [PubMed] [Google Scholar]
- 66.Daubendiek SL, Kool ET. Generation of catalytic RNAs by rolling transcription of synthetic DNA nanocircles. Nat Biotechnol. 1997;15(3):273–7. [DOI] [PubMed] [Google Scholar]
- 67.Daubendiek SL, Ryan K, Kool ET. Rolling-Circle RNA Synthesis: Circular Oligonucleotides as Efficient Substrates for T7 RNA Polymerase. J Am Chem Soc. 1995;117(29):7818–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim H, Park Y, Lee JB. Self-assembled Messenger RNA Nanoparticles (mRNA-NPs) for Efficient Gene Expression. Sci Rep. 2015;5:12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 2012;11(4):316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharma B, Crist RM, Adiseshaiah PP. Nanotechnology as a Delivery Tool for Precision Cancer Therapies. AAPS J. 2017. [DOI] [PubMed] [Google Scholar]
- 71.del Pozo-Rodriguez A, Delgado D, Solinis MA, Pedraz JL, Echevarria E, Rodriguez JM, et al. Solid lipid nanoparticles as potential tools for gene therapy: in vivo protein expression after intravenous administration. Int J Pharm. 2010;385(1–2):157–62. [DOI] [PubMed] [Google Scholar]
- 72.Radaic A, de Paula E, de Jesus MB. Factorial Design and Development of Solid Lipid Nanoparticles (SLN) for Gene Delivery. J Nanosci Nanotechnol. 2015;15(2):1793–800. [DOI] [PubMed] [Google Scholar]
- 73.Sune-Pou M, Prieto-Sanchez S, El Yousfi Y, Boyero-Corral S, Nardi-Ricart A, Nofrerias-Roig I, et al. Cholesteryl oleate-loaded cationic solid lipid nanoparticles as carriers for efficient gene-silencing therapy. Int J Nanomedicine. 2018;13:3223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18(7):1357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang P, Zhang L, Xie Y, Wang N, Tang R, Zheng W, et al. Genome Editing for Cancer Therapy: Delivery of Cas9 Protein/sgRNA Plasmid via a Gold Nanocluster/Lipid Core-Shell Nanocarrier. Advanced Science. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng T, Bott S, Huo Q. Techniques for Accurate Sizing of Gold Nanoparticles Using Dynamic Light Scattering with Particular Application to Chemical and Biological Sensing Based on Aggregate Formation. ACS Appl Mater Interfaces. 2016;8(33):21585–94. [DOI] [PubMed] [Google Scholar]
- 77.Yin H, Song CQ, Suresh S, Wu Q, Walsh S, Rhym LH, et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci U S A. 2016;113(11):2868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang M, Alberti K, Sun S, Arellano CL, Xu Q. Combinatorially designed lipid-like nanoparticles for intracellular delivery of cytotoxic protein for cancer therapy. Angew Chem Int Ed Engl. 2014;53(11):2893–8. [DOI] [PubMed] [Google Scholar]
- 81.Li Y, Yang T, Yu Y, Shi N, Yang L, Glass Z, et al. Combinatorial library of chalcogen-containing lipidoids for intracellular delivery of genome-editing proteins. Biomaterials. 2018. [DOI] [PubMed] [Google Scholar]
- 82.Li B, Dong Y. Preparation and Optimization of Lipid-Like Nanoparticles for mRNA Delivery. Methods Mol Biol. 2017;1632:207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li B, Luo X, Deng B, Giancola JB, McComb DW, Schmittgen TD, et al. Effects of local structural transformation of lipid-like compounds on delivery of messenger RNA. Sci Rep. 2016;6:22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107(5):1864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhen S, Takahashi Y, Narita S, Yang YC, Li X. Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome. Oncotarget. 2017;8(6):9375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pombo Garcia K, Zarschler K, Barbaro L, Barreto JA, O’Malley W, Spiccia L, et al. Zwitterionic-coated “stealth” nanoparticles for biomedical applications: recent advances in countering biomolecular corona formation and uptake by the mononuclear phagocyte system. Small. 2014;10(13):2516–29. [DOI] [PubMed] [Google Scholar]
- 88.Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, et al. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl. 2017;56(4):1059–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Panayiotou E, Papacharalambous R, Antoniou A, Christophides G, Papageorgiou L, Fella E, et al. Genetic background modifies amyloidosis in a mouse model of ATTR neuropathy. Biochem Biophys Rep. 2016;8:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22(9):2227–35. [DOI] [PubMed] [Google Scholar]
- 91.Langer R, Folkman J. Polymers for the sustained release of proteins and other macromolecules. Nature. 1976;263(5580):797–800. [DOI] [PubMed] [Google Scholar]
- 92.Leelakanok N, Geary SM, Salem AK. Antitumor Efficacy and Toxicity of 5-Fluorouracil-Loaded Poly(Lactide Co-glycolide) Pellets. J Pharm Sci. 2018;107(2):690–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Coleman MC, Goetz JE, Brouillette MJ, Seol D, Willey MC, Petersen EB, et al. Targeting mitochondrial responses to intra-articular fracture to prevent posttraumatic osteoarthritis. Sci Transl Med. 2018;10(427). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wafa EI, Geary SM, Goodman JT, Narasimhan B, Salem AK. The effect of polyanhydride chemistry in particle-based cancer vaccines on the magnitude of the anti-tumor immune response. Acta Biomater. 2017;50:417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khorsand B, Elangovan S, Hong L, Dewerth A, Kormann MS, Salem AK. A Comparative Study of the Bone Regenerative Effect of Chemically Modified RNA Encoding BMP-2 or BMP-9. AAPS J. 2017;19(2):438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salem AK. Nanoparticles in vaccine delivery. AAPS J. 2015;17(2):289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Makkouk A, Joshi VB, Wongrakpanich A, Lemke CD, Gross BP, Salem AK, et al. Biodegradable microparticles loaded with doxorubicin and CpG ODN for in situ immunization against cancer. AAPS J. 2015;17(1):184–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geary SM, Hu Q, Joshi VB, Bowden NB, Salem AK. Diaminosulfide based polymer microparticles as cancer vaccine delivery systems. J Control Release. 2015;220(Pt B):682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wongrakpanich A, Adamcakova-Dodd A, Xie W, Joshi VB, Mapuskar KA, Geary SM, et al. The absence of CpG in plasmid DNA-chitosan polyplexes enhances transfection efficiencies and reduces inflammatory responses in murine lungs. Mol Pharm. 2014;11(3):1022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gross BP, Wongrakpanich A, Francis MB, Salem AK, Norian LA. A therapeutic microparticle-based tumor lysate vaccine reduces spontaneous metastases in murine breast cancer. AAPS J. 2014;16(6):1194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joshi VB, Geary SM, Carrillo-Conde BR, Narasimhan B, Salem AK. Characterizing the antitumor response in mice treated with antigen-loaded polyanhydride microparticles. Acta Biomater. 2013;9(3):5583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hong L, Wei N, Joshi V, Yu Y, Kim N, Krishnamachari Y, et al. Effects of glucocorticoid receptor small interfering RNA delivered using poly lactic-co-glycolic acid microparticles on proliferation and differentiation capabilities of human mesenchymal stromal cells. Tissue Eng Part A. 2012;18(7–8):775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krishnamachari Y, Geary SM, Lemke CD, Salem AK. Nanoparticle delivery systems in cancer vaccines. Pharm Res. 2011;28(2):215–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Intra J, Salem AK. Rational design, fabrication, characterization and in vitro testing of biodegradable microparticles that generate targeted and sustained transgene expression in HepG2 liver cells. J Drug Target. 2011;19(6):393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Intra J, Salem AK. Fabrication, characterization and in vitro evaluation of poly(D,L-lactide-co-glycolide) microparticles loaded with polyamidoamine-plasmid DNA dendriplexes for applications in nonviral gene delivery. J Pharm Sci. 2010;99(1):368–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang XQ, Intra J, Salem AK. Comparative study of poly (lactic-co-glycolic acid)-poly ethyleneimine-plasmid DNA microparticles prepared using double emulsion methods. J Microencapsul. 2008;25(1):1–12. [DOI] [PubMed] [Google Scholar]
- 107.Abbas AO, Donovan MD, Salem AK. Formulating poly(lactide-co-glycolide) particles for plasmid DNA delivery. J Pharm Sci. 2008;97(7):2448–61. [DOI] [PubMed] [Google Scholar]
- 108.Intra J, Salem AK. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo. J Control Release. 2008;130(2):129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang XQ, Intra J, Salem AK. Conjugation of polyamidoamine dendrimers on biodegradable microparticles for nonviral gene delivery. Bioconjug Chem. 2007;18(6):2068–76. [DOI] [PubMed] [Google Scholar]
- 110.Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC, et al. Cellular uptake of nanoparticles: journey inside the cell. Chem Soc Rev. 2017;46(14):4218–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wongrakpanich A, Wu M, Salem AK. Correlating intracellular nonviral polyplex localization with transfection efficiency using high-content screening. Biotechnol Prog. 2015;31(6):1685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Atluri K, Seabold D, Hong L, Elangovan S, Salem AK. Nanoplex-Mediated Codelivery of Fibroblast Growth Factor and Bone Morphogenetic Protein Genes Promotes Osteogenesis in Human Adipocyte-Derived Mesenchymal Stem Cells. Mol Pharm. 2015;12(8):3032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ganas C, Weiss A, Nazarenus M, Rosler S, Kissel T, Rivera Gil P, et al. Biodegradable capsules as non-viral vectors for in vitro delivery of PEI/siRNA polyplexes for efficient gene silencing. J Control Release. 2014;196:132–8. [DOI] [PubMed] [Google Scholar]
- 114.Li L, He ZY, Wei XW, Gao GP, Wei YQ. Challenges in CRISPR/CAS9 Delivery: Potential Roles of Nonviral Vectors. Hum Gene Ther. 2015;26(7):452–62. [DOI] [PubMed] [Google Scholar]
- 115.Francis SM, Taylor CA, Tang T, Liu Z, Zheng Q, Dondero R, et al. SNS01-T modulation of eIF5A inhibits B-cell cancer progression and synergizes with bortezomib and lenalidomide. Mol Ther. 2014;22(9):1643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suenaga T, Kohyama M, Hirayasu K, Arase H. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol Immunol. 2014;58(9):513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kretzmann JA, Ho D, Evans CW, Plani-Lam JHC, Garcia-Bloj B, Mohamed AE, et al. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem Sci. 2017;8(4):2923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–55. [DOI] [PubMed] [Google Scholar]
- 119.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–93. [DOI] [PubMed] [Google Scholar]
- 120.Campeau P, Chapdelaine P, Seigneurin-Venin S, Massie B, Tremblay JP. Transfection of large plasmids in primary human myoblasts. Gene Ther. 2001;8(18):1387–94. [DOI] [PubMed] [Google Scholar]
- 121.Wang M, Liu H, Li L, Cheng Y. A fluorinated dendrimer achieves excellent gene transfection efficacy at extremely low nitrogen to phosphorus ratios. Nat Commun. 2014;5:3053. [DOI] [PubMed] [Google Scholar]
- 122.Beerli RR, Segal DJ, Dreier B, Barbas CF 3rd. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95(25):14628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Worthington KS, Green BJ, Rethwisch M, Wiley LA, Tucker BA, Guymon CA, et al. Neuronal Differentiation of Induced Pluripotent Stem Cells on Surfactant Templated Chitosan Hydrogels. Biomacromolecules. 2016;17(5):1684–95. [DOI] [PubMed] [Google Scholar]
- 124.Mahiny AJ, Dewerth A, Mays LE, Alkhaled M, Mothes B, Malaeksefat E, et al. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat Biotechnol. 2015;33(6):584–6. [DOI] [PubMed] [Google Scholar]
- 125.Wilder MA. Surfactant Protein B Deficiency in Infants With Respiratory Failure. The Journal of Perinatal & Neonatal Nursing. 2004;18(1):61–7. [DOI] [PubMed] [Google Scholar]
- 126.Liu Y, Wang T, He F, Liu Q, Zhang D, Xiang S, et al. An efficient calcium phosphate nanoparticle-based nonviral vector for gene delivery. Int J Nanomedicine. 2011;6:721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ge X, Zhang Q, Cai Y, Duan S, Chen S, Lv N, et al. PEG-PCL-DEX polymersome-protamine vector as an efficient gene delivery system via PEG-guided self-assembly. Nanomedicine (Lond). 2014;9(8):1193–207. [DOI] [PubMed] [Google Scholar]
- 128.Puras G, Martinez-Navarrete G, Mashal M, Zarate J, Agirre M, Ojeda E, et al. Protamine/DNA/Niosome Ternary Nonviral Vectors for Gene Delivery to the Retina: The Role of Protamine. Mol Pharm. 2015;12(10):3658–71. [DOI] [PubMed] [Google Scholar]
- 129.Yang W, Cheng Y, Xu T, Wang X, Wen LP. Targeting cancer cells with biotin-dendrimer conjugates. Eur J Med Chem. 2009;44(2):862–8. [DOI] [PubMed] [Google Scholar]
- 130.Vineberg JG, Zuniga ES, Kamath A, Chen YJ, Seitz JD, Ojima I. Design, synthesis, and biological evaluations of tumor-targeting dual-warhead conjugates for a taxoid-camptothecin combination chemotherapy. J Med Chem. 2014;57(13):5777–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koutsioumpa M, Papadimitriou E. Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat Anticancer Drug Discov. 2014;9(2):137–52. [DOI] [PubMed] [Google Scholar]
- 132.Sinclair JF, O’Brien AD. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J Biol Chem. 2002;277(4):2876–85. [DOI] [PubMed] [Google Scholar]
- 133.Zhou Y, Han C, Li D, Yu Z, Li F, Li F, et al. Cyclin-dependent kinase 11(p110) (CDK11(p110)) is crucial for human breast cancer cell proliferation and growth. Sci Rep. 2015;5:10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu X, Gao Y, Shen J, Yang W, Choy E, Mankin H, et al. Cyclin-Dependent Kinase 11 (CDK11) Is Required for Ovarian Cancer Cell Growth In Vitro and In Vivo, and Its Inhibition Causes Apoptosis and Sensitizes Cells to Paclitaxel. Mol Cancer Ther. 2016;15(7):1691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Feng Y, Sassi S, Shen JK, Yang X, Gao Y, Osaka E, et al. Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system. J Orthop Res. 2015;33(2):199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoyer J, Neundorf I. Peptide vectors for the nonviral delivery of nucleic acids. Acc Chem Res. 2012;45(7):1048–56. [DOI] [PubMed] [Google Scholar]
- 137.Guo Z, Peng H, Kang J, Sun D. Cell-penetrating peptides: Possible transduction mechanisms and therapeutic applications. Biomed Rep. 2016;4(5):528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Radis-Baptista G, Campelo IS, Morlighem JRL, Melo LM, Freitas VJF. Cell-penetrating peptides (CPPs): From delivery of nucleic acids and antigens to transduction of engineered nucleases for application in transgenesis. J Biotechnol. 2017;252:15–26. [DOI] [PubMed] [Google Scholar]
- 139.Li H, Tsui TY, Ma W. Intracellular Delivery of Molecular Cargo Using Cell-Penetrating Peptides and the Combination Strategies. Int J Mol Sci. 2015;16(8):19518–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24(6):1020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yamashita H, Kato T, Oba M, Misawa T, Hattori T, Ohoka N, et al. Development of a Cell-penetrating Peptide that Exhibits Responsive Changes in its Secondary Structure in the Cellular Environment. Sci Rep. 2016;6:33003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jeong EJ, Choi M, Lee J, Rhim T, Lee KY. The spacer arm length in cell-penetrating peptides influences chitosan/siRNA nanoparticle delivery for pulmonary inflammation treatment. Nanoscale. 2015;7(47):20095–104. [DOI] [PubMed] [Google Scholar]
- 143.Chaterji S, Ahn EH, Kim DH. CRISPR Genome Engineering for Human Pluripotent Stem Cell Research. Theranostics. 2017;7(18):4445–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gabrielson NP, Lu H, Yin L, Li D, Wang F, Cheng J. Reactive and bioactive cationic alpha-helical polypeptide template for nonviral gene delivery. Angew Chem Int Ed Engl. 2012;51(5):1143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.He H, Zheng N, Song Z, Kim KH, Yao C, Zhang R, et al. Suppression of Hepatic Inflammation via Systemic siRNA Delivery by Membrane-Disruptive and Endosomolytic Helical Polypeptide Hybrid Nanoparticles. ACS Nano. 2016;10(2):1859–70. [DOI] [PubMed] [Google Scholar]
- 146.Lu H, Wang J, Bai Y, Lang JW, Liu S, Lin Y, et al. Ionic polypeptides with unusual helical stability. Nat Commun. 2011;2:206. [DOI] [PubMed] [Google Scholar]
- 147.Zheng N, Song Z, Yang J, Liu Y, Li F, Cheng J, et al. Manipulating the membrane penetration mechanism of helical polypeptides via aromatic modification for efficient gene delivery. Acta Biomater. 2017;58:146–57. [DOI] [PubMed] [Google Scholar]
- 148.Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54(41):12029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, et al. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11(3):2452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vert M, Doi Y, Hellwich KH, Hess M, Hodge P, Kubisa P, et al. Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure Appl Chem. 2012;84(2):377–408. [Google Scholar]
- 151.Yang J, Bahreman A, Daudey G, Bussmann J, Olsthoorn RC, Kros A. Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes. ACS Cent Sci. 2016;2(9):621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1:889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Alsaiari SK, Patil S, Alyami M, Alamoudi KO, Aleisa FA, Merzaban JS, et al. Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. J Am Chem Soc. 2018;140(1):143–6. [DOI] [PubMed] [Google Scholar]
- 154.Yue H, Zhou X, Cheng M, Xing D. Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing. Nanoscale. 2018;10(3):1063–71. [DOI] [PubMed] [Google Scholar]
- 155.Senevirathne SA, Washington KE, Biewer MC, Stefan MC. PEG based anti-cancer drug conjugated prodrug micelles for the delivery of anti-cancer agents. Journal of Materials Chemistry B. 2016;4(3):360–70. [DOI] [PubMed] [Google Scholar]
- 156.Li W, Zhan P, De Clercq E, Lou H, Liu X. Current drug research on PEGylation with small molecular agents. Progress in Polymer Science. 2013;38(3):421–44. [Google Scholar]
- 157.Xu C-f, Wang J. Delivery systems for siRNA drug development in cancer therapy. Asian Journal of Pharmaceutical Sciences. 2015;10(1):1–12. [Google Scholar]
- 158.Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioeng Transl Med. 2016;1(1):10–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zuckerman JE, Davis ME. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat Rev Drug Discov. 2015;14(12):843–56. [DOI] [PubMed] [Google Scholar]
- 160.Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials (Basel). 2017;7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stern JM, Kibanov Solomonov VV, Sazykina E, Schwartz JA, Gad SC, Goodrich GP. Initial Evaluation of the Safety of Nanoshell-Directed Photothermal Therapy in the Treatment of Prostate Disease. Int J Toxicol. 2016;35(1):38–46. [DOI] [PubMed] [Google Scholar]
- 162.Guidotti G, Brambilla L, Rossi D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol Sci. 2017;38(4):406–24. [DOI] [PubMed] [Google Scholar]
- 163.Charlesworth CT, Deshpande PS, Dever DP, Dejene B, Gomez-Ospina N, Mantri S, et al. Identification of Pre-Existing Adaptive Immunity to Cas9 Proteins in Humans. bioRxiv. 2018.
- 164.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A. 2014;111(11):3955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lu C, Stewart DJ, Lee JJ, Ji L, Ramesh R, Jayachandran G, et al. Phase I clinical trial of systemically administered TUSC2(FUS1)-nanoparticles mediating functional gene transfer in humans. PLoS One. 2012;7(4):e34833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Phase I Study of IV DOTAP: Cholesterol-Fus1 in Non-Small-Cell Lung Cancer [Available from: https://ClinicalTrials.gov/show/NCT00059605.
- 168.Study of PNT2258 for Treatment of Relapsed or Refractory Non-Hodgkin’s Lymphoma [Available from: https://ClinicalTrials.gov/show/NCT01733238.
- 169.A Study of PNT2258 in Patients With Advanced Solid Tumors [Available from: https://ClinicalTrials.gov/show/NCT01191775.
- 170.Harb WA, Lakhani N, Logsdon A, Steigelman M, Smith-Green H, Gaylor S, et al. The BCL2 Targeted Deoxyribonucleic Acid Inhibitor (DNAi) PNT2258 Is Active in Patients with Relapsed or Refractory Non-Hodgkin’s Lymphoma. Blood. 2014;124(21):1716. [Google Scholar]
- 171.Tolcher AW, Rodrigueza WV, Rasco DW, Patnaik A, Papadopoulos KP, Amaya A, et al. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73(2):363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Safety and Tolerability Study of SNS01-T in Relapsed or Refractory B Cell Malignancies (Multiple Myeloma, B Cell Lymphoma, or Plasma Cell Leukemia (PCL) [Available from: https://ClinicalTrials.gov/show/NCT01435720.
- 173.Siegel DS, McDonald A, Novitzky N, Bensinger W, Craig M, van Rhee F, et al. Mature Results of a Phase 1–2 Open-Label, Dose-Escalation Study of Intravenous SNS01-T in Patients (pts) with Relapsed or Refractory B-Cell Malignancies. Blood. 2014;124(21):4464. [Google Scholar]
- 174.Pilot Study of BC-819/PEI and BCG in Patients With Superficial Transitional Cell Bladder Carcinoma [Available from: https://ClinicalTrials.gov/show/NCT01878188.
- 175.Phase 1/2a Study of DTA-H19 in Advanced Stage Ovarian Cancer [Available from: https://ClinicalTrials.gov/show/NCT00826150.
- 176.Lavie O, Edelman D, Levy T, Fishman A, Hubert A, Segev Y, et al. A phase 1/2a, dose-escalation, safety, pharmacokinetic, and preliminary efficacy study of intraperitoneal administration of BC-819 (H19-DTA) in subjects with recurrent ovarian/peritoneal cancer. Arch Gynecol Obstet. 2017;295(3):751–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sidi AA, Ohana P, Benjamin S, Shalev M, Ransom JH, Lamm D, et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J Urol. 2008;180(6):2379–83. [DOI] [PubMed] [Google Scholar]
- 178.Study With Atu027 in Patients With Advanced Solid Cancer [Available from: https://ClinicalTrials.gov/show/NCT00938574.
- 179.Atu027 Plus Gemcitabine in Advanced or Metastatic Pancreatic Cancer (Atu027-I-02) [Available from: https://ClinicalTrials.gov/show/NCT01808638.
- 180.Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68(23):9788–98. [DOI] [PubMed] [Google Scholar]
- 181.Schultheis B, Strumberg D, Kuhlmann J, Wolf M, Link K, Seufferlein T, et al. A phase Ib/IIa study of combination therapy with gemcitabine and Atu027 in patients with locally advanced or metastatic pancreatic adenocarcinoma. Journal of Clinical Oncology. 2016;34(4_suppl):385-. [Google Scholar]
- 182.Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O, et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol. 2014;32(36):4141–8. [DOI] [PubMed] [Google Scholar]
- 183.Silence T. Pancreatic cancer study Atu027-I-02 Interim analysis 2015. [Available from: https://www.silence-therapeutics.com/media/1263/atu027-phase-2a-pancreatic-cancer-interim-analysis.pdf.
- 184.Phase I, Multicenter, Dose Escalation Study of DCR-MYC in Patients With Solid Tumors, Multiple Myeloma, or Lymphoma [Available from: https://ClinicalTrials.gov/show/NCT02110563.
- 185.Tolcher AW, Papadopoulos KP, Patnaik A, Rasco DW, Martinez D, Wood DL, et al. Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. Journal of Clinical Oncology. 2015;33(15_suppl):11006-. [Google Scholar]
- 186.Safety Study of a Cell Penetrating Peptide (p28) to Treat Solid Tumors That Resist Standard Methods of Treatment [Available from: https://ClinicalTrials.gov/show/NCT00914914.
- 187.p28 in Treating Younger Patients With Recurrent or Progressive Central Nervous System Tumors [Available from: https://ClinicalTrials.gov/show/NCT01975116.
- 188.Lulla RR, Goldman S, Beattie C, Yamada T, Pollack I, Fisher PG, et al. Phase 1 trial of p28 (NSC745104), a non-HDM2 mediated peptide inhibitor of p53 ubiquitination in children with recurrent or progressive CNS tumors: A final report from the Pediatric Brain Tumor Consortium. Journal of Clinical Oncology. 2015;33(15_suppl):10059-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Warso MA, Richards JM, Mehta D, Christov K, Schaeffer C, Rae Bressler L, et al. A first-in-class, first-in-human, phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours. Br J Cancer. 2013;108(5):1061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]