Abstract
Autism spectrum disorder (ASD) is a complex and genetically heterogeneous neuropsychiatric disease affecting as many as 1 in 68 children. Large scale genetic sequencing of individuals along the autism spectrum has uncovered several genetic risk factors for ASD; however, understanding how, and to what extent, individual genes contribute to the overall disease phenotype remains unclear. Neuroimaging studies of ASD have revealed a wide spectrum of structural and functional perturbations that are thought to reflect, in part, the complex genetic heterogeneity underpinning ASD. These perturbations, in both preclinical models and clinical patients, were identified in preclinical genetic models and ASD patients when compared to control populations; however, few studies have directly explored intrinsic differences between the models themselves. To better understand the degree and extent to which individual genes associated with ASD differ in their contribution to global measures of white matter microstructure, diffusion tensor imaging (DTI) was acquired from three novel rat genetic models of ASD (Fmr1, Nrxn1, and Pten) and DTI parameters of fractional anisotropy, mean, axial, and radial diffusivity were measured. Subsequent whole-brain voxel-wise analysis comparing each genetic model to each other (Fmr1:Nrxn1; Fmr1:Pten; Nrxn1:Pten) identified no significant differences in any comparison for all diffusion parameters assessed (FA, AD, MD, RD).
Keywords: diffusion tensor imaging, autism spectrum disorder, Fmr1, Pten, Nrxn1, convergence
1. Introduction
Autism spectrum disorder (ASD) is a complex clinically heterogeneous neurodevelopmental disability affecting as many as 1 in 68 children (Christensen et al., 2016). While the clinical diagnosis of ASD continues to rely on behavioral classifications and patient symptomology and remains primarily understood as a behavioral disorder, emerging evidence has recast our understanding of ASD as a highly complex and heterogeneous biological disorder (Christensen et al., 2016; Edmiston et al., 2017). In particular, genome wide association (GWA) studies of ASD (Codina-solà et al., 2015) have identified numerous candidate gene-susceptibility factors as contributors to the overall disease state. These include Chd8 and Ptchd1 (Bernier et al., 2014; Tora et al., 2017) as well as epigenetic and transcriptional regulators such as Mecp2 (Witteveen et al., 2016), post-transcriptional protein modifiers and regulators such as Fmr1 (Fragile X Syndrome) (Haberl et al., 2015; Lai et al., 2016; Lee et al., 2016), Pten (PTEN hamartoma tumor syndromes and non-syndromic ASDs) (Butler, 2005; Silverman et al., 2010), and synaptic organizing and scaffolding proteins such as Shanks and neurexins (Nrxn) (Dachtler et al., 2015; Voineskos et al., 2011). While whole exome sequencing has not established any definitive causal genetic sequence variant for ASD, several highly penetrant de novo mutations – leading to partial or total loss of function – have been consistently reported in individuals with ASD that are otherwise absent in typically developing children.
To investigate the potential impact of these genetic mutations on neural microstructure, several groups have generated murine genetic knockout models to study the monogenetic influence of these candidate genes in the neuropathogenesis of ASD. Parallel neuroimaging efforts in these murine models and in humans have utilized advanced magnetic resonance imaging (MRI) techniques to quantitatively evaluate differences in neural structure between typically developing individuals and individuals with genetic mutations conferring increased susceptibility to ASD. Notable efforts toward the identification of gene-specific neuroimaging biomarkers of ASD have included volumetric analyses that have shown increased prefrontal cortex brain volume in both humans and murine models with impaired Pten function and fronto-temporal hyperconnectivity in both adolescent functional MRI studies of children with Fragile X Syndrome (FXS) and in studies involving murine hemizygous models of Fmr1.
While these and other human and murine neuroimaging studies of ASD have revealed significant differences between control and affected populations (Foss-Feig et al., 2017; Hull et al., 2017; Ismail et al., 2016; Li et al., 2017), a relative paucity has explicitly investigated the degree and extent of structural differences between cohorts of genetic models with previous work from Ellegood et. al. being the most complete neuroanatomical phenotyping study of ASD (Ellegood et al., 2015). However, no studies, to the best of our knowledge, have utilized diffusion tensor imaging (DTI) to compare differences in white matter microstructure across different genetic variants of ASD. Furthermore, no equivalent studies have been performed in a rat model of ASD. The rat model would be anticipated to provide a more translatable imaging appraisal of gene-specific changes in ASD than previously reported murine neuroimaging studies due to the closer degree of similarity in physiology and behavior between humans and rats relative to mice (Gibbs et al., 2004; Smith and Alloway, 2013). The use of rat models in our study also reflects the resurgence of rats as the model of choice in neuroscience research due to the growing use and availability of genome-editing technologies. To explore differences in brain white matter microstructure across different genetic variants of ASD, three novel monogenetic rat models of ASD were imaged with diffusion tensor brain imaging.
2. Methods
2.1. Animals and experimental design
Animals were housed and cared for in an AAALAC-accredited facility and all animal experiments were conducted in accordance with institutional IACUC-approved protocols and the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). Postnatal day 45 (P45) adolescent male Fmr1−/y, Nrxn1−/−, and Pten+/− rats in a Sprague Dawley background (Horizon Discovery, St. Louis, USA) and postnatal day (P45) male outbred Sprague Dawley rats (Charles River, Wilmington, MA, USA) were used in all experiments (n=4 for each genetic background; n=4 for control animals; all intra-group animals are siblings from the same litter) with the age of the animals chosen to as closely match the periadolescent period (Sengupta, 2013); male animals were chosen to avoid potentially confounding estrous effects. Each of the three genetic models were generated via zinc finger nuclease (ZFN) genome editing yielding a hemizygous, homozygous, and heterozygous genotype for the Fmr1, Nrxn1, and Pten genetic variants, respectively. Endonuclease hemizygous deletion of the Fmr1 gene mirrors the gene silencing observed with expansion of the CGG trinucleotide and monoallelic deletion of the Pten gene recapitulates the clinically encountered heterozygous genotype (the homozygous deletion is embryonic lethal). Animals were housed in a light-dark cycle, temperature- and humidity-controlled vivarium and maintained with ad libitum food and water diet supplied per institutional protocol. All experiments were performed with the experimenter blinded to genotype. At P45, animals were brought to a surgical plane of anesthesia and were transcardially perfused with ice-cold 4% paraformaldehyde (PFA). Brains were then cleanly dissected from the cranial vault, post-fixed in 4% PFA, and stored at 4C. 48 hours prior to imaging, brains were serially washed in 1X PBS to minimize the attenuating effects of fixative prior to being placed in a custom-built holder filled with Fluorinert (FC-3283, 3M, USA) to minimize magnetic susceptibility artifact. Briefly, a custom-built 10cc cylindrical container is filled with Flourinert, whereby each prepared brain is subsequently placed inside, all air evacuated, and sealed with Paraffin film (Parafilm M, Bemis, Neeah, WI, USA).. All procedures mirror previously reported methods that have been shown to have no impact on the diffusion tensor (de Guzman et al., 2016).
2.2. Ex-vivo image acquisition and image preprocessing
For ex-vivo DTI acquisition, groups of 2 brains were simultaneously imaged using a 4.7-T Agilent MRI system and 3.5-cm diameter quadrature volume RF coil (brains were placed in a side-by-side configuration within a single coil). Diffusion-weighted imaging (DWI) data were acquired employing a multi-slice spin echo sequence with the following imaging parameters: repetition time: 2000 ms; echo time: 24.17 ms; slice thickness, 0.25-mm with final isotropic voxel size of 0.25-mm. Diffusion measurements were acquired with diffusion encoded along 30 non-collinear directions at b=1200 s·mm−2 and three additional non-diffusion weighted (b=0 s·mm−2) measurements. The acquisition was averaged across two repeats resulting in a total imaging time of approximately 11 hours.
An affine registration tool (Jenkinson et al., 2002) from the FMRIB software library suite was used to co-register individual diffusion images to correct for subtle eddy current distortions. Gradient directions were then corrected for rotations (Leemans and Jones, 2009). FSL DTI output volumes were converted to NIfTI tensor and employed to first estimate a study-specific tensor template from all imaging datasets acquired from the control group. This template was then used as a target to which each subject tensor volume was spatially normalized using the DTI-TK tensor-based registration tools. DTI-TK’s registration routine, which iteratively employs rigid-body, affine, and diffeomorphic transformations, was used as it has been shown to offer improved registration results over other registration algorithms (Wang et al., 2011).
2.3. TBSS and ROI analysis
The tract-based spatial statistics (TBSS) formalism was implemented as recommended in Bach et. al. (Bach et al., 2014). The TBSS pipeline was applied utilizing the recommended parameters implemented in FSL, which includes using a 0.2 FA threshold for creating the white matter skeleton. Additionally, a permutation test with n=252 corrected for multiple comparisons and threshold-free cluster enhancement (TFCE) (Smith and Nichols, 2009) was implemented with FSL’s Randomize for inter-group comparisons with family-wise error corrected p values less than 0.05 as the threshold for significance. A standard atlas (Rumple et al., 2013) from which anatomical regions of interest (ROI) were derived and was non-linearly aligned to the group-wise template. The registration included elastic warping with a point-set based registration metric implemented in the Advanced Normalization Tools (ANTS) (Avants et al., 2011) software library. For use in the subsequent statistical analysis, mean values of the diffusion-tensor-derived indices, namely, fractional anisotropy (FA), RD (radial diffusivity), AD (axial diffusivity), and MD (mean diffusivity) were computed within each ROI for all individual samples. These values were computed in the group-wise template space. To further quantify and assess inter-subject variability, the UNC Rat Atlas was normalized to our subject common space and masked with regions-of-interest predefined by the UNC Rat Atlas (Rumple et al., 2013). Diffusion measures for all regions of interest from the atlas were extracted.
2.4. Statistical analysis
As previously detailed, TBSS data were processed per the recommended parameters implemented in FSL. Permutation tests and threshold-free cluster enhancement was implemented in FSL with p<0.05 as the threshold for significance. Statistical comparisons were performed by a two-tailed, two-sample, and unequal variance Student’s t-test (ROI analysis). A two-tailed, two-sample, and unequal variance Student’s t-test was performed comparing the mean values of FA, AD, RD, and MD in all segmented ROIs in Fmr1−/y, Nrxn1−/−, and Pten+/− rats against age-sex-matched controls and adjusted for multiple comparisons using the Benjamini-Hochberg false discovery rate (FDR) correction (FDR = 0.05). From these data, group mean and standard deviation were calculated (GraphPad, GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. Tract-based spatial statistics
In each of the three genetic models, global differences in measures of the diffusion tensor (FA, AD, MD, and RD) were assessed between experimental and control animal groups at the p<0.05 significance level (Table 1). TBSS results comparing an Fmr1 model of ASD to control animals reveal widespread changes in white matter microstructure centered principally in the frontal lobe. Specifically, we observed decreased AD, MD, and RD values bilaterally in the frontal lobe, external capsule, and corpus callosum in the Fmr1 model when compared to control animals (Fig. 1; Supplemental Fig. 1A). A bilateral decrease in both MD and RD were observed in the frontal lobe and external capsule, but additional unique decreases in RD were uncovered in the left forebrain, left midbrain, left fimbria, and right and left internal capsule (Fig. 1; Supplemental Fig. 1A; Supplemental Fig. 3). No statistically significant differences in FA were observed between the Fmr1 model and control animals.
Table 1.
Whole-brain voxel-wise TBSS results displaying FWE-corrected p values
| DTI measure | Control>Fmr1−/y | Fmr1−/ >control | Control>Nrxn1−/- | Nrxn1−/−>control | Control>Pten+/− | Pten+/−>control |
|---|---|---|---|---|---|---|
| FA | 0.0571 | 0.1 | 0.2143 | 0.0429* | 0.7 | 0.0143* |
| MD | 0.025* | 0.3286 | 0.0143* | 0.5286 | 0.0143* | 0.3857 |
| AD | 0.0286* | 0.1571 | 0.0857 | 0.4714 | 0.0286* | 0.5143 |
| RD | 0.0143* | 0.6429 | 0.0143* | 0.214 | 0.0143* | 0.6429 |
Statistically significant FWE-corrected permutation tests comparing whole-brain voxel-wise analysis, p < 0.05.
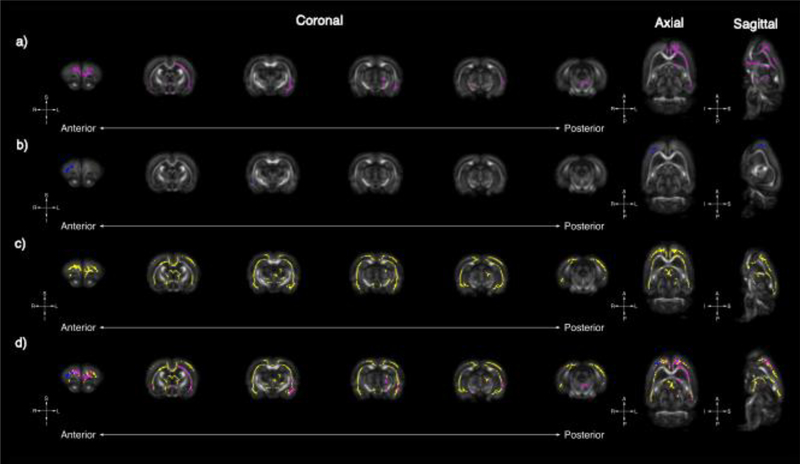
Figure 1.
Tract-based spatial statistics uncovers global changes in RD in Fmr1, Nrxn1, and Pten models of ASD. Whole-brain voxel-wise tract-based spatial statistics reveal significant areas of decreased RD in Fmr1 (A, pink), Nrxn1 (B, blue), and Pten (C, yellow) models of ASD. Six representative coronal sections (left [anterior] to right [posterior]) reveal significant overlapping regions of decreased in RD for all three models in the frontal lobe (D); however, unique gene-specific areas of RD change are noted caudally. Uniquely observed in the Fmr1 model are decreases in RD in the left external capsule whereas unique changes in the Pten model are observed in the frontal lobe, genu of the corpus callosum, and fimbria.
TBSS analyses from Nrxn1 animals yielded significantly greater FA values than controls in small clusters of voxels in the left hemisphere in the frontal lobe, brainstem, and external capsule (Fig. 2; Supplemental Figs. 1B, 2). Furthermore, MD values were significantly decreased in the left external capsule (Supplemental Fig. 3) and RD values were significantly decreased in the right frontal lobe and right thalamus and left external capsule (Fig. 1). Decreases in AD for Nrxn1 trended towards but did not reach statistical significance after family-wise error correction for multiple comparisons (p=0.08). Substantial differences in white matter structural properties were also detected when comparing our Pten model to control animals. These include significantly decreased AD in the right external capsule and decreased MD in nearly the entire right external capsule as well as portions of the frontal lobe (Supplemental Fig. 1C, Supplemental Fig. 3). Additionally, regions encompassing the right external capsule and right amygdala showed significantly greater FA when compared to controls (Fig. 2, Supplemental Fig. 2). The most robust differences in the Pten model were reflected by significantly reduced RD values in the external capsule, frontal lobe, and corpus callosum (Fig. 1).
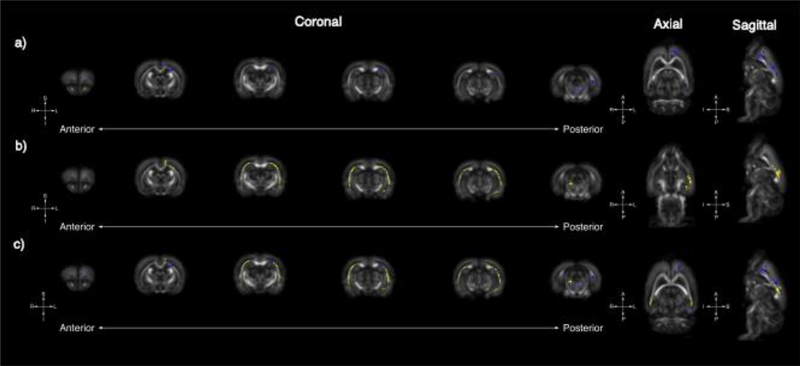
Figure 2.
Tract-based spatial statistics reveals global changes in FA in Nrxn1 and Pten models of ASD: Whole-brain voxel-wise tract-based spatial statistics reveal significant areas of increased FA in Nrxn1 (A, blue) and Pten (B, yellow) models of ASD. Six representative coronal sections (left [anterior] to right [posterior]) reveals unique FA changes in the Nrxn1 model, which are observable medially in anterior regions encompassing the frontal lobe as well as the medial structures of the brain stem. In contrast, increases in FA in the Pten model are most concentrated bilaterally along medial margins of the external capsule. There are also overlapping small areas of increased FA medially in the external capsule and frontal lobe in both the Nrxn1 and Pten models (C).
3.2. Biological overlap in genetic models of autism spectrum disorder
An initial analysis of our TBSS results comparing each genetic model of ASD to control animals uncovers significant transformations of the diffusion tensor that are attributable to each genetic variant. When these changes are superimposed on each other, the TBSS maps from the three genetic variants demonstrate a similar pattern of change to the diffusion tensor both in the direction and spatial distribution of changes, particularly in the corpus callosum and external capsule differing only in the overall magnitude of these changes in the diffusion tensor in each genetic variant. For example, while Nrxn1 and Pten both show significantly increased FA and decreased RD in both the corpus callosum and external capsule relative to controls, the extent of observed change is greater in the Pten model. In addition to regions of common and converging change, there are equally interesting divergent areas of change in the diffusion tensor, which may reflect region-specific genetic influences in white matter structural integrity. These include confluent bilateral increases in FA in the external capsule in the Pten model, which are less apparent in the Nrxn1 model and are entirely absent in the Fmr1 model (Fig. 2). FA comparisons between groups also reveal unique confluent areas of change in FA in left frontal lobe of the Nrxn1 model that are absent in the other experimental models (Fig. 2). And while all genetic variants demonstrate areas of significantly decreased MD, the spatial distribution of these changes differs; right temporal lobe changes in MD cover a greater volume in the Pten group compared to the other experimental models and focal changes in MD appear in the Nrxn1 model in the mesial left frontal lobe that are not present in the other experimental groups (Supplemental Fig. 3).
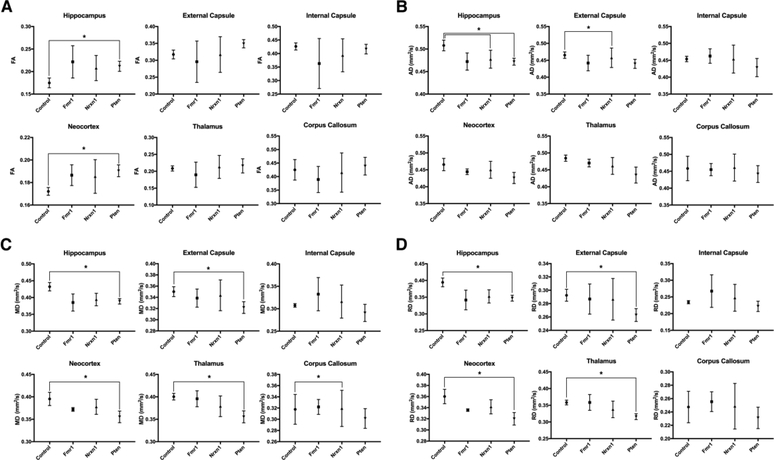
Region of interest (ROI) analysis between control and experimental groups (control:Fmr1; control:Nrxn1; control:Pten) was additionally performed to determine whether monogenetic perturbations lead to differences in diffusion signal in the hippocampus, thalamus, frontal lobe, corpus callosum, and internal and external capsule (Fig. 3). There were no statistical differences after Bonferroni correction in diffusion scalar measures in controls relative to Fmr1. Comparing diffusion measures in the assessed regions reveals significant decreases in AD in the hippocampus (p=0.0014) and external capsule (p=0.0003) in the Nrxn1 group compared to control. AD in the corpus callosum of Nrxn1 was also significantly decreased compared to control (p=0.0079). When comparing control to Pten, there were significant differences after correcting for multiple comparisons for all diffusion scalar measures in several regions. Relative to control, Pten showed increased FA in the hippocampus (p=0.0032) and frontal lobe (p=0.0019) and decreased AD in the hippocampus (p=0.0035). Significant decreases in MD in Pten relative to control were observed in the hippocampus (p=0.0018), external capsule (p=0.0065), thalamus (p=0.0024), and frontal lobe (p=0.0067). Similarly, there were significant decreases in both RD in Pten relative to control in the hippocampus (p=0.0016), external capsule (p=0.0041), frontal lobe (p=0.0003), and thalamus (p=0.0036).
Figure 3.
Representative plots of FA (A), AD (B), MD (C), and RD (D) displaying the mean ± S.D. values for 6 regions of interest for control, Fmr1, Nrxn1, and Pten. Statistical differences, after Bonferroni correction, were identified between control and both Nrxn1 and Pten genetic variants in several regions of interest across all diffusion measures (indicated by starred (*) brackets). After correcting for multiple comparisons, intragroup comparisons between each genetic model within each region of interest showed no statistically significant difference in any of the four diffusion tensor metrics calculated. N.B. Units of measure for MD, AD, and RD are [10−3 mm2/s].
3.3. Convergent white matter microstructural change in genetic models of autism spectrum disorder
In an extension of our analysis, we sought to uncover regions of shared comorbidity in ASD by directly comparing the brains of each of our three genetic variants of ASD to each other. To these ends, whole-brain voxel-wise TBSS was performed comparing each genetic knockout model cohort to each other. Surprisingly, in each of the comparisons (Fmr1:Nrxn1; Fmr1:Pten; Nrxn1:Pten), no voxels of significant change were identified across all of the four major diffusion tensor values. In an extension (and to secondarily confirm) these surprising TBSS results, all subject scans were then masked and segmented into the 28 major regions of interest (ROI) present in the UNC Rat Brain Atlas (Rumple et al., 2013) and the four major diffusion tensor values were subsequently calculated for each ROI. After correcting for multiple comparisons, no significant intra-ROI differences were identified between the genetic variants for FA, MD, RD, or AD (Fig. 3). To further verify that the results of our tract-based spatial statistics and ROI analysis were not driven by single sample variations skewing intragroup tensor values, an intragroup sample quality assurance analysis was performed. As before, after scans were masked and segmented, six major ROIs present in the UNC Rat Brain Atlas (Rumple et al., 2013) (hippocampal formation, external capsule, internal capsule, frontal lobe, thalamus, corpus callosum) were selected and the mean and standard deviation of all tensor values in each ROI were then calculated (Table 2). Overall, our analysis reveals low intragroup variability across each ROI. Exemplars of the low variance seen in our imaging samples include measures of RD in the frontal lobe across all genetic variants in the present study: Fmr1 frontal lobe mean RD = 0.34 ± 0.002 10−3 mm2/s; Nrxn1 frontal lobe mean RD = 0.34 ± 0.01 10−3 mm2/s; Pten frontal lobe mean RD = 0.32 ± 0.01 10−3 mm2/s.
Table 2.
Group mean diffusion measures in six regions of interest demonstrate low intragroup sample variability
| FA | AD | RD | MD | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hip. | EC | Neo. | CC | Hip. | EC | Neo. | CC | Hip. | EC | Neo. | CC | Hip. | EC | Neo. | CC | |
| Fmrl | 0.22 ± 0.03 | 0.30 ± 0.06 | 0.19 ± 0.01 | 0.39 ± 0.05 | 0.47 ± 0.02 | 0.44 ± 0.02 | 0.44 ± 0.01 | 0.45 ± 0.02 | 0.34 ± 0.03 | 0.29 ± 0.02 | 0.34 ± 0.002 | 0.26 ± 0.01 | 0.39 ± 0.03 | 0.34 ± 0.02 | 0.37 ± 0.004 | 0.32 ± 0.01 |
| Nrxnl | 0.29 ± 0.04 | 0.28 ± 0.05 | 0.20 ± 0.02 | 0.36 ± 0.05 | 0.51 ± 0.02 | 0.49 ± 0.04 | 0.46 ± 0.02 | 0.55 ± 0.05 | 0.33 ± 0.02 | 0.33 ± 0.02 | 0.34 ± 0.01 | 0.31 ± 0.03 | 0.39 ± 0.01 | 0.35 ± 0.02 | 0.38 ± 0.02 | 0.33 ± 0.04 |
| Pten | 0.25 ± 0.03 | 0.27 ± 0.03 | 0.20 ± 0.004 | 0.38 ± 0.03 | 0.46 ± 0.01 | 0.44 ± 0.01 | 0.43 ± 0.02 | 0.47 ± 0.04 | 0.32 ± 0.01 | 0.30 ± 0.02 | 0.32 ± 0.01 | 0.26 ± 0.02 | 0.36 ± 0.01 | 0.34 ± 0.02 | 0.35 ± 0.01 | 0.33 ± 0.02 |
Values are mean ± SD. Each column corresponds to each diffusion measure in a region of interest derived from the P72 UNC Atlas.
FA=fractional anisotropy; AD=axial diffusivity; RD=radial diffusivity; MD=mean diffusivity; Hip=hippocampus; EC=external capsule; Neo=neocortex; CC=corpus callosum.
4. Discussion
ASD is a genetically and phenotypically heterogeneous neurodevelopmental disorder characterized by communication deficits, restricted interests, and repetitive behaviors. While there exists ample evidence linking many genetic variants to the clinical phenotypes seen in the ASD patient population, the precise manner through which individual genetic variants contribute to the overall disease phenotype remains largely opaque (Christensen et al., 2016). Recent advances in neuroimaging techniques such as DTI have begun to unpack the complex relationship between these susceptibility genes and their related neuroimaging phenotypes within the clinical ASD population. Progress toward identifying salient biomarkers for ASD, however, has stalled, which has largely been attributed to the inherent heterogeneity of the ASD patient population and is further underscored by the conspicuous absence of a true neuroimaging biomarker in ASD. As an illustrative example of the challenges encountered in clinical populations, several studies have investigated the impact of Fmr1 on global measures of white matter structural integrity. While some studies have reported an increase in FA in tracts such as the uncinate fasciculus and inferior longitudinal fasciculus (Green et al., 2015; Hall et al., 2016), others have reported alterations in the white matter of the genu and splenium of the corpus callosum and the middle cerebellar peduncle (Filley et al., 2015) without recapitulating the findings in the uncinate fasciculus and inferior longitudinal fasciculus. The heterogeneity of studies such as these highlights the challenge of interpreting human imaging studies, even those with pre-identified genetically homogenous patient populations. Furthermore, it emphasizes the importance of considering the potential influence of epistatic and environmental factors in determining the overall clinical and neuroimaging disease phenotype.
Recent advances in genome editing technology have allowed for the novel generation of monogenetic rat models of ASD. The models of ASD used herein (Pten−/+, Fmr1−/y, and Nrxn1−/−) were generated with ZFNs (zinc finger nucleases), which are dual-function artificial restriction enzymes containing both a zinc-finger binding domain as well as a DNA-cleavage domain that enable precise manipulation of complex genomes previously inaccessible to genetic manipulation. The advantages of these monogenetic rat models of ASD over analogous murine models lies in the closer physiological similarity to and more analogous behavioral characteristics found in rats (Gibbs et al., 2004; Smith and Alloway, 2013) relative to humans. Therefore, the rat would be anticipated to provide a more translatable imaging appraisal of gene-specific changes in ASD when compared to previously reported murine neuroimaging studies. Our study represents one of the first reports of rat diffusion tensor imaging analysis of three salient genetic rat models of ASD and contributes to our understanding of single-gene contributions to white matter structural integrity free of potentially confounding environmental and epistatic effects encountered in clinical studies and populations. The three models described herein (Fmr1−/y [decreased production of FMRP, a critical protein in synaptic development], Nxrn1−/− [cell surface receptor binding neuroligins to form Ca2+-dependent neurexin/neuroligin complexes at synapses], and Pten−/+ (PI3K-Akt-mTOR pathway disruption)) were selected as each model demonstrates disparate molecular mechanisms that contribute to the overall ASD phenotype. That we find a similar degree of microstructural change in all of our models further extends prior work examining volumetric and morphometric differences between different murine models of ASD (Ellegood et al., 2015) to quantitative diffusion MRI and further buttresses our emerging understanding of the neurostructural similarities between genetic models of ASD.
Individuals with mutations in Pten, a well-established ASD neural susceptibility gene in the mTOR signaling pathway (Eng, 2003), have been associated with nonspecific multifocal white matter hyperintensities, enlarged perivascular spaces, and macrocephaly (Vanderver et al., 2014). However, to the best of our knowledge, no studies have yet examined the impact of Pten on measures of white matter microstructure with diffusion tensor imaging. To examine the influence of Pten mutations on white matter microstructure in the rat, we conducted whole-brain voxel-wise TBSS analysis, which reveals confluent areas of increased FA and decreased MD, AD, and RD values in the Pten model compared to controls. These include areas of decreased RD and concomitantly increased FA throughout most of the frontal lobe and in major white matter tracts such as the corpus callosum and the external capsule as well as the cerebellum (Fig. 1, 2) that are consistent with recent studies showing MR findings in these same regions (Balci et al., 2018; Frazier et al., 2015).
In addition to presenting the DTI findings of a Pten+/− model of ASD, we also explored changes in white matter in the Nrxn1−/− model of ASD. Closely related human functional MRI analyses with genetic variants in contactin-associated protein-like 2 (CNTNAP2, a neurexin superfamily member) provide the closest analogous imaging study, which demonstrate altered frontal connectivity in individuals with neurodevelopmental disorders such as ASD (Scott-Van Zeeland et al., 2010). In line with these results, our TBSS analyses also reveal marked increases in FA predominantly in the frontal lobe of the Nrxn1 knockout model. Additionally, we observed significant reductions in global MD and RD throughout the frontal lobe as well as in the corpus callosum as well as in the external capsule. The widespread, confluent changes in measures of the diffusion tensor throughout the brain likely reflect the cytoarchitectural consequences of neurexin-1 deletion given the critical role neurexins play in establishing and mediating GABAergic and glutamatergic synaptic function through postsynaptic neuroligins (Südhof, 2008). It is interesting to note that the observed increase of FA in both our Nrxn1 and Pten models is a somewhat surprising finding and stands in contrast to findings typically observed in idiopathic ASD in clinical populations. However, numerous studies have also highlighted how these changes are reversed in young children (Bashat et al., 2007; Bode et al., 2011; Weinstein et al., 2011). While the underlying mechanism for these changes are not well understood, this observation is critical given the age of the animals used in our study (P45) frames the same developmental time period as the young children in the aforementioned studies, and further highlights an important translational neuroimaging parallel between our preclinical results and currently available clinical studies. That each of these three genetic models yields similar results similarly suggests that while the genetic susceptibilities are somewhat disparate, each gene may be contributing and impacting neural microstructure via common neurodevelopmental pathways.
Prior preclinical murine DTI studies involving a Fmr1−/y mouse model of autism report reduced structural integrity in the splenium of the corpus callosum and forceps minor of the corpus callosum (Haberl et al., 2015). Whole-brain voxel-wise TBSS and tractography analysis performed in our study revealed no significant differences in FA between Fmr1−/y and control animals; however, we did see significant changes in other DTI metrics (AD, RD, MD) in the TBSS analysis. These findings suggest important biological transformations to the diffusion tensor exist that ultimately are balanced to a degree such that no changes in FA were observed. These differences in AD, RD, and MD were observed in the frontal lobe, forebrain, external capsule, and fimbria, which correlate with prior areas of FA change in the mouse and human ASD population (Ingalhalikar et al., 2011; Kumar et al., 2012; Lai et al., 2016). While unexpected, the absence of significant FA change in the Fmr1 analysis highlights the limitations of modeling these complex neuropsychiatric disorders in a rodent model and likely reflects important biological differences in physiology, environmental conditions, and development between small animal models (mouse to rat) and from rodents to humans. The lack of significant changes in FA in the whole-brain TBSS analysis also highlights the importance of drawing conclusions from multiple measures of diffusion results rather than relying FA as a faithful representative summary measure of the diffusion tensor (Alexander et al., 2000).
In an extension of previous morphometric studies comparing murine genetic variants of ASD (most notably by Ellegood et. al.), our study also found no significant differences in white matter microstructure between our three genetic models of ASD following both TBSS and ROI analysis. With a growing appreciation for the biological overlap among numerous neuropsychiatric diseases including ASD, schizophrenia, and bipolar disorder (Carroll and Owen, 2009; Forstner et al., 2017), our study’s findings reinforce a growing understanding that a comprehensive understanding of disease comorbidity and shared biology may be helpful, especially within newly developed frameworks such as the National Institute of Mental Health (NIMH) Research Domain Criteria (RDoC) (Gold et al., 2016). Especially in the context of ASD, which involves a wide and profoundly diverse spectrum of genetic contributions to the disease phenotype, new evidence has demonstrated how diverse genetic perturbations can lead to phenotypic convergence at multiple biological levels in a complex neuropsychiatric disorder (Parikshak et al., 2016). Mirroring these molecular findings, our TBSS analysis, in addition to our ROI analysis, confirms and recapitulates these molecular insights along global measures of white matter structural integrity. While each genetic variant independently demonstrates areas of significant change when compared to control animals, when compared to each other, these three common genetic variants of ASD show remarkable similarity to each other with no areas of statistically significant difference. These data highlight that while there exists the potential for gene-specific neuroimaging biomarkers in ASD, the relative degree of change between each variant is small and mirrors other examples of phenotypic convergence that have also been seen in studies comparing brain morphometry across multiple childhood psychiatric disorders (Gold et al., 2016) and in other morphological studies of ASD (Ellegood et al., 2015).
There are some features of our study that limit the interpretation of our data. As this is a pilot study, one concern would be the relatively small number of replicates used in each of our imaging group analyses. Although we do present secondary analyses to demonstrate that there exists a low degree of variance in our data (Table 2) with no single sample skewing our results, the variance is not zero. Thus, the small sample size emphasizes the pilot nature of our work but provides a springboard for future experiments with larger sample sizes to validate our findings as well as other experimental work with similar hypotheses. The absence of a female animals in our pilot work also precludes an examination of whether our results would be generalizable to females and future work with female animals will help determine if structural convergence is a phenomenon that is seen in both sexes. Lastly, recent work describing in situ gene expression levels mapped to neural microstructural changes in the 16p11.2 hemideletion seen in ASD and ADHD have shown that sex-specific differential gene expression that demonstrates high correlation to underlying deficits in white matter microstructure (Kumar et al., 2018). Such a granular detailed gene-expression map is currently not available for the rat (or for these models) but future work investigating concomitant changes in gene expression with voxel-wise changes in neural microstructure would be informative and provide a greater understanding of the association between gene expression and imaging findings not only in these models of ASD but in other genetic models of neuropsychiatric illness.
In sum, high-resolution diffusion tensor imaging of transgenic models of ASD can provide insight into the relationship between genetic variants and aberrant white matter microstructure in ASD and ultimately highlights the structural convergence in genetic models of a complex neuropsychiatric disorder. Future work with other genetic variants of ASD will continue to shed light on the impact of genotype on global measures of neural microstructure and will help refine our understanding of the shared neuroimaging features in ASD.
Supplementary Material
Highlights:
Diffusion tensor imaging study comparing white matter microstructure between multiple genetic models of autism.
Parallel microstructural changes exist in multiple genetic models of autism buttressing an emerging understanding of neurostructural phenotypic convergence in psychiatric disease.
Acknowledgements:
The authors would like to thank Beth Rauch for outstanding imaging support through the Small Animal Imaging Facility at the University of Wisconsin Carbone Cancer Center and University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520. Funding: J-P.J.Y was supported by UW-Madison School of Medicine and Public Health and Department of Radiology, the Brain and Behavior Research Foundation (NARSAD) Young Investigator Grant and the University of Wisconsin Institute for Clinical and Translational Research KL2 Scholars Program (NCATS UL1TR000427); DTI analysis tools and pipelines were developed with partial support from NIH U54 HD090256 IDDRC Core Grant to the Waisman Center; J.G.G is supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1256259.
Footnotes
Conflicts of interest: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Alexander AL, Hasan K, Kindlmann G, Parker DL, Tsuruda JS, 2000. A geometric analysis of diffusion tensor measurements of the human brain. Magn. Reson. Med 44, 283–291. [DOI] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC, 2011. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044. 10.1016/j.neuroimage.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M, Laun FB, Leemans A, Tax CMW, Biessels GJ, Stieltjes B, Maier-Hein KH, 2014. Methodological considerations on tract-based spatial statistics (TBSS). Neuroimage 100, 358–369. 10.1016/j.neuroimage.2014.06.021 [DOI] [PubMed] [Google Scholar]
- Balci TB, Davila J, Lewis D, Boafo A, Sell E, Richer J, Nikkel SM, Armour CM, Tomiak E, Lines MA, Sawyer SL, 2018. Broad spectrum of neuropsychiatric phenotypes associated with white matter disease in PTEN hamartoma tumor syndrome. Am. J. Med. Genet. B. Neuropsychiatr. Genet 177, 101–109. 10.1002/ajmg.b.32610 [DOI] [PubMed] [Google Scholar]
- Bashat D. Ben, Kronfeld-duenias V, Zachor DA, Ekstein PM, Hendler T, Tarrasch R, Even A, Levy Y, Ben L, 2007. Accelerated maturation of white matter in young children with autism : A high b value DWI study. Hum. Brain Mapp. J 37, 40–47. 10.1016/j.neuroimage.2007.04.060 [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, Schuurs-Hoeijmakers JH, Fichera M, Bosco P, Buono S, Alberti A, Failla P, Peeters H, Steyaert J, Vissers LELM, Francescatto L, Mefford HC, Rosenfeld JA, Bakken T, O’Roak BJ, Pawlus M, Moon R, Shendure J, Amaral DG, Lein E, Rankin J, Romano C, de Vries BBA, Katsanis N, Eichler EE, 2014. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode MK, Mattila M, Kiviniemi V, Rahko J, Moilanen I, Ebeling H, Tervonen O, Nikkinen J, 2011. White matter in autism spectrum disorders – evidence of impaired fiber formation 1169–1174. [DOI] [PubMed]
- Butler MG, 2005. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet 42, 318–321. 10.1136/jmg.2004.024646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll LS, Owen MJ, 2009. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 1, 102 10.1186/gm102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DL, Baio J, Braun KVN, Bilder D, Charles J, Constantino JN, Daniels J, Durkin MS, Fitzgerald RT, Kurzius-Spencer M, Lee L-C, Pettygrove S, Robinson C, Schulz E, Wells C, Wingate MS, Zahorodny W, Yeargin-Allsopp M, 2016. Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR. Surveill. Summ 65, 1–23. 10.15585/mmwr.ss6503a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codina-solà M, Rodríguez-santiago B, Homs A, Santoyo J, Rigau M, Aznar-laín G, Campo M, Gener B, Gabau E, Botella MP, Gutiérrez-arumí A, Antiñolo G, Pérez-jurado LA, Cuscó I, 2015. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders.??? 1–16. 10.1186/s13229-015-0017-0 [DOI] [PMC free article] [PubMed]
- Dachtler J, Ivorra JL, Rowland TE, Lever C, Rodgers RJ, Clapcote SJ, 2015. Heterozygous deletion of α-neurexin I or α-neurexin II results in behaviors relevant to autism and schizophrenia. Behav. Neurosci 129, 765–76. 10.1037/bne0000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guzman AE, Wong MD, Gleave JA, Nieman BJ, 2016. Variations in post-perfusion immersion fixation and storage alter MRI measurements of mouse brain morphometry. Neuroimage 142, 687–695. 10.1016/j.neuroimage.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Edmiston E, Ashwood P, Water J. Van De, 2017. Review Autoimmunity, Autoantibodies, and Autism Spectrum Disorder. Biol. Psychiatry 81, 383–390. 10.1016/j.biopsych.2016.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J, Anagnostou E, Babineau BA, Crawley JN, Lin L, Genestine M, DiCicco-Bloom E, Lai JKY, Foster JA, Penagarikano O, Geschwind DH, Pacey LK, Hampson DR, Laliberte CL, Mills AA, Tam E, Osborne LR, Kouser M, Espinosa-Becerra F, Xuan Z, Powell CM, Raznahan A, Robins DM, Nakai N, Nakatani J, Takumi T, van Eede MC, Kerr TM, Muller C, Blakely RD, Veenstra-VanderWeele J, Henkelman RM, Lerch JP, 2015. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol. Psychiatry 20, 118–125. 10.1038/mp.2014.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C, 2003. PTEN: One gene, Many syndromes. Hum. Mutat 22, 183–198. 10.1002/humu.10257 [DOI] [PubMed] [Google Scholar]
- Filley CM, Brown MS, Onderko K, Ray M, Bennett RE, Berry-Kravis E, Grigsby J, 2015. White matter disease and cognitive impairment in FMR1 premutation carriers. Neurology 84, 2146–2152. 10.1212/WNL.0000000000001612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstner AJ, Hecker J, Hofmann A, Maaser A, Reinbold CS, Mühleisen TW, Leber M, Strohmaier J, Degenhardt F, Treutlein J, Mattheisen M, Schumacher J, Streit F, Meier S, Herms S, Hoffmann P, Lacour A, Witt SH, Reif A, Müller-Myhsok B, Lucae S, Maier W, Schwarz M, Vedder H, Kammerer-Ciernioch J, Pfennig A, Bauer M, Hautzinger M, Moebus S, Schenk LM, Fischer SB, Sivalingam S, Czerski PM, Hauser J, Lissowska J, Szeszenia-Dabrowska N, Brennan P, McKay JD, Wright A, Mitchell PB, Fullerton JM, Schofield PR, Montgomery GW, Medland SE, Gordon SD, Martin NG, Krasnov V, Chuchalin A, Babadjanova G, Pantelejeva G, Abramova LI, Tiganov AS, Polonikov A, Khusnutdinova E, Alda M, Cruceanu C, Rouleau GA, Turecki G, Laprise C, Rivas F, Mayoral F, Kogevinas M, Grigoroiu-Serbanescu M, Becker T, Schulze TG, Rietschel M, Cichon S, Fier H, Nöthen MM, 2017. Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS One 12, e0171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss-Feig JH, Adkinson BD, Lisa Ji J, Yang G, Srihari VH, McPartland JC, Krystal JH, Murray JD, Anticevic A, 2017. Searching for Cross-diagnostic Convergence: Neural Mechanisms Governing Excitation and Inhibition Balance in Schizophrenia and Autism Spectrum Disorders. Biol. Psychiatry 81, 848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, Eng C, 2015. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol. Psychiatry 20, 1132–1138. 10.1038/mp.2014.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R. a, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera, Holt R. a, Adams MD, Amanatides PG, BadenTillson H, Barnstead M, Chin S, Evans C. a, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee H-M, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song X-Z, Li B, Liu Y, Qin X, Cawley S, Cooney a J., D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler D. a, Zhang Z, Bailey J. a, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee Y-A, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson S. a, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone E. a, Payseur B. a, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner P. a, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F, 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521. 10.1038/nature02426 [DOI] [PubMed] [Google Scholar]
- Gold AL, Brotman MA, Adleman NE, Lever SN, Steuber ER, Fromm SJ, Mueller SC, Pine DS, Leibenluft E, 2016. Comparing Brain Morphometry Across Multiple Childhood Psychiatric Disorders. J. Am. Acad. Child Adolesc. Psychiatry 55, 1027–1037.e3. 10.1016/j.jaac.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T, Barnea-Goraly N, Raman M, Hall SS, Lightbody AA, Bruno JL, Quintin EM, Reiss AL, 2015. Specific effect of the fragile-X mental retardation-1 gene (FMR1) on white matter microstructure. Br J Psychiatry 207, 143–148. 10.1192/bjp.bp.114.151654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberl MG, Zerbi V, Veltien A, Ginger M, Heerschap A, Frick A, 2015. Structural-functional connectivity deficits of neocortical circuits in the Fmr1−/y mouse model of autism. Sci. Adv 1, e1500775–e1500775. 10.1126/sciadv.1500775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SS, Dougherty RF, Reiss AL, 2016. NeuroImage : Clinical Pro fi les of aberrant white matter microstructure in fragile X syndrome. YNICL 11, 133–138. 10.1016/j.nicl.2016.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull JV, Jacokes ZJ, Torgerson CM, Irimia A, Van Horn JD, 2017. Resting-State Functional Connectivity in Autism Spectrum Disorders: A Review. Front. Psychiatry 7, 205 10.3389/FPSYT.2016.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M, Parker D, Bloy L, Roberts TPL, Verma R, 2011. Diffusion based Abnormality Markers of Pathology: Towards Learned Diagnostic Prediction of ASD. Neuroimage 57, 918–927. 10.1038/jid.2014.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail MMT, Keynton RS, Mostapha MMMO, ElTanboly AH, Casanova MF, Gimel’farb GL, El-Baz A, 2016. Studying Autism Spectrum Disorder with Structural and Diffusion Magnetic Resonance Imaging: A Survey. Front. Hum. Neurosci 10, 211 10.3389/fnhum.2016.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S, 2002. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841. 10.1016/S1053-8119(02)91132-8 [DOI] [PubMed] [Google Scholar]
- Kumar M, Kim S, Pickup S, Chen R, Fairless AH, Ittyerah R, Abel T, Brodkin ES, Poptani H, 2012. Longitudinal in-vivo diffusion tensor imaging for assessing brain developmental changes in BALB/cJ mice, a model of reduced sociability relevant to autism. Brain Res. 1455, 56–67. 10.1016/j.brainres.2012.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar VJ, Grissom NM, McKee SE, Schoch H, Bowman N, Havekes R, Kumar M, Pickup S, Poptani H, Reyes TM, Hawrylycz M, Abel T, Nickl-Jockschat T, 2018. Linking spatial gene expression patterns to sex-specific brain structural changes on a mouse model of 16p11.2 hemideletion. Transl. Psychiatry 8, 109 10.1038/s41398-018-0157-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JKY, Lerch JP, Doering LC, Foster JA, Ellegood J, 2016. Regional brain volumes changes in adult male FMR1-KO mouse on the FVB strain. Neuroscience 318, 12–21. 10.1016/j.neuroscience.2016.01.021 [DOI] [PubMed] [Google Scholar]
- Lee M, Martin GE, Berry-kravis E, Losh M, 2016. A developmental, longitudinal investigation of autism phenotypic profiles in fragile X syndrome. J. Neurodev. Disord 1–10. 10.1186/s11689-0169179-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans A, Jones DK, 2009. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn. Reson. Med 61, 1336–1349. 10.1002/mrm.21890 [DOI] [PubMed] [Google Scholar]
- Li D, Karnath H-O, Xu X, 2017. Candidate Biomarkers in Children with Autism Spectrum Disorder: A Review of MRI Studies. Neurosci. Bull 33, 219–237. 10.1007/s12264-017-0118-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta L. de la T., Huang J, Lowe JK, Blencowe BJ, Horvath S, Geschwind DH, 2016. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427. 10.1038/nature20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumple A, Mcmurray M, Johns J, Lauder J, Makam P, Radcliffe M, Oguz I, 2013. 3-Dimensional Diffusion Tensor Imaging (DTI) Atlas of the Rat Brain. PLoS One 8, e67334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Lisa I, Sonnenblick, Rudie JD, Ghahremani D, Mumford JA, Russell A, Poldrack, Dapretto M, Geschwind DH, Susan Y, 2010. Altered Functional Connectivity in Frontal Lobe Circuits Is Associated with Variation in the Autism Risk Gene CNTNAP2 Ashley. Sci. Transl. Med 2 10.1126/scitranslmed.3001344.Altered [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, 2013. The Laboratory Rat: Relating Its Age With Human’s. Int. J. Prev. Med 4, 624–630. [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN, 2010. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci 11, 490–502. 10.1038/nrn2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Alloway KD, 2013. Rat whisker motor cortex is subdivided into sensory-input and motor-output areas. Front. Neural Circuits 7, 4 10.3389/fncir.2013.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Nichols TE, 2009. Threshold-free cluster enhancement: Addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 44, 83–98. 10.1016/j.neuroimage.2008.03.061 [DOI] [PubMed] [Google Scholar]
- Südhof TC, 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911. 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora D, Gomez AM, Michaud J-F, Yam PT, Charron F, Scheiffele P, 2017. Cellular Functions of the Autism Risk Factor PTCHD1 in Mice. J. Neurosci 37, 11993–12005. 10.1523/JNEUROSCI.1393-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderver A, Tonduti D, Kahn I, 2014. Characteristic Brain Magnetic Resonance Imaging Pattern in Patients With Macrocephaly and PTEN Mutations. Am J Med Genet A. 164A, 627–633. 10.1017/S0954579414000868.Child-evoked [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos AN, Lett TAP, Lerch JP, Tiwari AK, Ameis SH, Rajji TK, Müller DJ, Mulsant BH, Kennedy JL, 2011. Neurexin-1 and frontal lobe white matter: An overlapping intermediate phenotype for schizophrenia and autism spectrum disorders. PLoS One 6 10.1371/journal.pone.0020982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Liu Z, Zhang H, Escolar ML, Gilmore JH, Gouttard S, Fillard P, Maltbie E, Gerig G, Styner M, 2011. DTI registration in atlas based fiber analysis of infantile Krabbe disease. Neuroimage 55, 1577–1586. 10.1016/j.neuroimage.2011.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein M, Ben-Sira L, Levy Y, Zachor DA, Itzhak E. Ben, Artzi M, Tarrasch R, Eksteine PM, Hendler T, Bashat D. Ben, 2011. Abnormal white matter integrity in young children with autism. Hum. Brain Mapp 32, 534–543. 10.1002/hbm.21042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witteveen JS, Willemsen MH, Dombroski TCD, van Bakel NHM, Nillesen WM, van Hulten JA, Jansen EJR, Verkaik D, Veenstra-Knol HE, van Ravenswaaij-Arts CMA, Wassink-Ruiter JSK, Vincent M, David A, Le Caignec C, Schieving J, Gilissen C, Foulds N, Rump P, Strom T, Cremer K, Zink AM, Engels H, de Munnik SA, Visser JE, Brunner HG, Martens GJM, Pfundt R, Kleefstra T, Kolk SM, 2016. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet 48, 877–887. 10.1038/ng.3619 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.