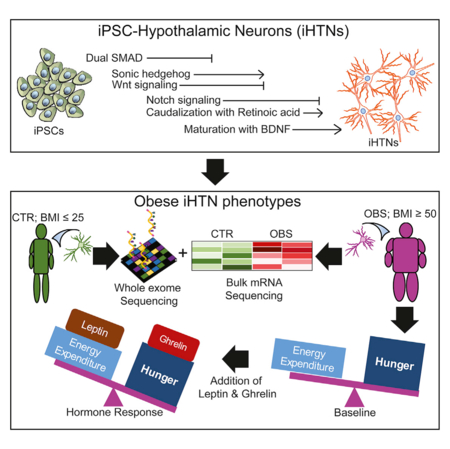
SUMMARY
The hypothalamus contains neurons that integrate hunger and satiety endocrine signals from the periphery and are implicated in the pathophysiology of obesity. The limited availability of human hypothalamic neurons hampers our understanding of obesity disease mechanisms. To address this, we generated human induced pluripotent stem cells (hiPSCs) from multiple normal body mass index (BMI; BMI ≤ 25) subjects and super-obese (OBS) donors (BMI ≥ 50) with polygenic coding variants in obesity-associated genes. We developed a method to reliably differentiate hiPSCs into hypothalamic-like neurons (iHTNs) capable of secreting orexigenic and anorexigenic neuropeptides. Transcriptomic profiling revealed that, although iHTNs maintain a fetal identity, they respond appropriately to metabolic hormones ghrelin and leptin. Notably, OBS iHTNs retained disease signatures and phenotypes of high BMI, exhibiting dysregulated respiratory function, ghrelin-leptin signaling, axonal guidance, glutamate receptors, and endoplasmic reticulum (ER) stress pathways. Thus, human iHTNs provide a powerful platform to study obesity and gene-environment interactions.
In Brief
Human hypothalamic neurons (HTNs) implicated in obesity have a limited availability. This study describes a reliable method for generating functional hormone-responsive HTNs from multiple normal and obese patient reprogrammed hiPSCs. Obese-patient-induced HTNs retained transcriptome profiles and functional phenotypes of high BMI, exhibiting aberrant obesity-related metabolic and respiratory pathways.
Graphical Abstract
INTRODUCTION
The hypothalamus, a region caudal to the thalamus and constituting about 0.3% of the adult human brain (MacLean, 1968), regulates the autonomic nervous and endocrine systems. The hypothalamus secretes various neurotransmitters, neuropeptides, and neurohormones that play critical roles in maintaining homeostatic processes in reproduction, stress, immune function, circadian rhythm, and energy metabolism (Utiger, 2017). Neurons of the arcuate nucleus (ARC) in the hypothalamus express pro-opiome-lanocortin (POMC), agouti-related peptide (AgRP), cocaine- and amphetamine-regulating transcript (CART), and neuropeptide Y (NPY), all of which respond to peripheral signaling hormones, including insulin, leptin, peptide YY (PYY), and ghrelin. By secreting neuropeptides such as NPY and α-melanocyte-stimulating hormone (α-MSH) (Wang et al., 2015), the hypothalamus in the CNS coordinates responses to hunger and satiety signals and determines food intake or expenditure.
Obesity is a global epidemic and a grave public health concern as the prevalence of obesity has more than doubled since 1970s and due to its comorbidity with several other cardiovascular risk factors, including high blood pressure and cholesterol levels, which often lead to coronary heart disease and stroke. Type 2 diabetes, sleep apnea, breathing problems, gallbladder disease, and certain cancers, are also linked to higher body mass index (BMI). With close to 200 genetic variants identified, in the majority of the population, obesity is polygenic where multiple genetic variants that are common and have small effects contribute to obesity susceptibility. Many of these genes regulate food intake and expenditure, energy metabolism, calorie burning efficiency, and how fat and sugar are metabolized resulting in body fat deposition. Given the role of the hypothalamus in feeding regulation, recent genome-wide association studies (GWASs) have also indicated a significant representation of CNS and hypothalamic processes in determining the BMI and regulation of obesity (Locke et al., 2015). Several of the BMI loci previously identified are located near genes expressed in the brain and CNS, specifically those involved in signaling pathways associated with clathrin-coated vesicles, decreased neurotransmitter release, glutamate signaling, synapse biology, and the generation of hormonal signals. Mutations in hypothalamic-specific genes, such as POMC and melanocortin 4 receptor (MC4R), in monogenic obesity syndromes also bolster the notion that the hypothalamus is a central coordinator of obesity and energy metabolism pathways in humans. Unfortunately, despite the great need for relevant neuronal models for obesity research, the relative inaccessibility of the hypothalamus tissue has precluded the collection of live cells from human subjects.
The use of human induced pluripotent stem cells (hiPSCs), however, obviates the need for direct collection by allowing the generation of hypothalamic-like neuronal cultures (iPSC-derived hypothalamic-like neurons [iHTNs]) from blood or fibroblasts of healthy and obese subjects to investigate the mechanisms of metabolic dysregulation impacting the CNS. To address this shortcoming, we employed hiPSCs generated from super-obese (BMI ≥ 50) and normal (BMI ≤ 25) subjects, and we differentiated them to hypothalamic-like neuronal cultures. Importantly, these iHTNs exhibited characteristics and functions of the human hypothalamic ARC, and they resembled phenotypically human hypothalamus tissue with responsiveness to exogenous hormonal signals, which was not demonstrated in previous studies that differentiated hiPSCs to hypothalamic-like neurons.
In this study, we identified that reliable generation of iHTNs required carefully timed and dose-dependent small molecule-assisted modulation of mothers against decapentaplegic (Smad) pathway inhibition, sonic hedgehog (Shh) activation, wingless-type MMTV integration site (Wnt) family, and Notch-signaling inhibition. Notably, we identified dysregulated obesity genes in differentiated iHTNs from super-obese patients with polygenic coding variants and that they retain the dysregulated obesogenic transcriptome-level profiles. This stem cell model demonstrates retention of obesogenic disease signatures in human iHTNs even after reprogramming to pluripotency, and it could serve as a key in determining early disease mechanisms in obesity at the level of the CNS. Addition of this platform to the arsenal of currently available obesity disease models may accelerate the development of therapeutics for intervention in patients with morbid obesity.
RESULTS
Generation of hiPSC-Derived Hypothalamic Neurons Using Directed Differentiation and Defined Media
Previous studies have reported the generation of hypothalamic-like neurons from human or mouse pluripotent stem cells with varying efficiencies (Merkle et al., 2015; Wang et al., 2015; Wataya et al., 2008) that involve undefined embryoid body differentiation methods and complex serum-based supplements for neuronal specification. Given that neuronal differentiation from hiPSC lines poses a challenge of inter- and intra-cell line variability, we devised a reliable and efficient method for generating iHTNs from hiPSC lines of multiple human donors in a completely chemically defined media without the use of any well-known neuronal supplements like N2 and B27, which consist of ~20 unique chemicals. Thus, we could easily vary concentrations of various components in our cell culture supplement (Table S2) to identify an optimal and reliable iHTN differentiation method that works across multiple hiPSC lines.
The method presented here for the directed generation of iHTNs from hiPSCs involves 3 stages: specification of neural ectoderm, followed by patterning toward ventral diencephalon, and finally maturation of hypothalamic neurons (Figure 1A). The neural ectoderm specification from hiPSCs was achieved by dual SMAD inhibition, followed by Shh activation and Wnt-signaling inhibition via a systematic combinatorial screen, which promoted the greatest percentage of hypothalamic progenitors based on the observation of NK2 homeobox 1- (Nkx2.1) and homeobox protein orthopedia- (Otp; hypothalamic neuron progenitor that specifies neuropeptidergic neurons) immunopositive cells at day 9 post-hiPSCs (Figures 1B and 1C). Since multiple morphogens, including Wnt, retinoic acid (RA), and insulin (Wataya et al., 2008), are critical in determining hypothalamic identity from hiPSCs, a combinatorial screening in 384-well plates was performed with varying doses of RA, insulin, and IWR-1-endo to efficiently determine the optimal dose for generation of Nkx2.1hi, Raxhi, and Sox1lo neuronal progenitors in this differentiation paradigm between days 2 and 9 of differentiation (Figures 1A, 1D, and 1E; Figures S2B and S2C). Neuronal progenitors of the ventral diencephalon express high levels of Nkx2.1 and Rax (retina and anterior neural fold homeobox, specific to retina and hypothalamus). It was observed that, in hiPSCs seeded in 384-well plates, the absence of insulin was detrimental to cell viability, as the least number of cells were observed in the condition receiving 0 μg/mL, while cells treated with 20 μg/mL exhibited the largest total cell numbers (Figure S2A). Absence of IWR-1-endo or low concentrations (1 μM) resulted in an Nkx2.1lo and Sox1hi cell population upon screening at day 9 of the protocol. At higher concentrations (10 μM) of IWR-1-endo, the largest number of Nkx2.1hi, Raxhi, and Sox1lo cells was observed. While there were higher levels of Raxhi cells at 100 μM IWR-1-endo, that condition showed low numbers of Nkx2.1+, and, furthermore, upon continuation of differentiation, it displayed low cell viability and decreased efficiency of iHTN formation. The absence of RA up to day 9 was also important for obtaining Nkx2.1hi and Raxhi cell populations. Based on this screening, 0 μM RA, 4 μg/mL insulin, and 10 μM IWR-1-endo was chosen as the most efficient combination for inducing early hypothalamic progenitors from hiPSCs between day 0 and day 9.
Figure 1. Combinatorial Screening for Forebrain Hypothalamic Neurons.
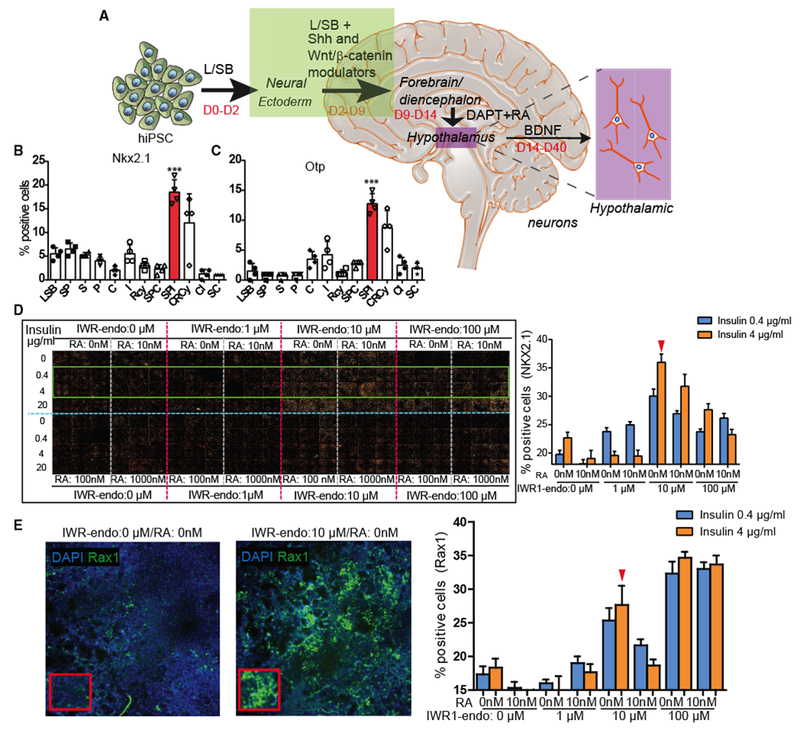
(A) A schematic of the iHTN direct differentiation protocol.
(B and C) L/SB + SPI combination showing the most (B) Nkx2.1 - and (C) Otp-positive progenitor cells. 4 independent experiments utilized 2 different cell lines and an average is represented. Small molecules utilized in (B) and (C) are as follows: L-LDN193189 (1 μM); SB, SB431542 (10 μM); S, Smoothened Agonist (1 μM); P, Purmorphamine (1 μM); I, IWR1-Endo (10 μM); RA, retinoic acid (0.01 μM); BDNF, brain-derived neurotrophic factor (10 ng/mL); C, CHIR99021 (3 μM); R, Robotnikinin (1 μM); and Cy, Cyclopamine (1 μM).
(D and E) The (D) montage and (E) image (left) and quantification (right) of large-scale combinatorial screen of varying doses of IWR1-endo, insulin, and retinoic acid (RA) in efficient differentiation of (D) Nkx2.1 + hypothalamic progenitors and (E) Rax+ hypothalamic progenitors in day 9 iHTN cultures. The green box in the montage (D) highlights the conditions represented in the bar graphs on the right. Red arrow in (D) denotes the dose combination selected for iHTN differentiation based on efficient Nkx2.1 and long-term neuropeptidergic neuron generation and cell survival. Red arrow in (E) shows that the selected combination shows efficient levels of Rax. 2 independent experiments each utilizing two cell lines were conducted and the average is represented. ***p < 0.001.
Error bars represent SEM.
iHTNs Closely Resemble Hypothalamic Neurons of the ARC
After directing the hiPSCs to hypothalamic progenitors, on day 9, the addition of 10 nM RA until day 14 for caudalization and the use of γ-secretase inhibitor DAPT to extend the G1/S phase and thereby exit cell cycle resulted in the largest populations of hypothalamic progenitors and neurons of the ARC. Furthermore, the addition of brain-derived neurotrophic factor (BDNF) until day 40 drove maturation of these neuronal progenitors into neurons that showed markers of hypothalamic ARC, which were capable of secreting neuropeptides. These iHTN progenitors could also be cryopreserved on day 20 of differentiation and reliably thawed (Table S5). Immunocytochemistry characterization of iHTNs obtained from our direct differentiation protocol revealed the presence of hypothalamus- and ARC-specific markers in multiple lines. These included Otp, Rax, NPY (a secreted neuropeptide of the orexigenic NPY neurons), CART (a neuropeptide produced by the anorexigenic CART neurons), α-MSH (a bioactive product of POMC-producing neurons), NPYR (NPY receptor Y2, receptor for NPY present), GhrR (ghrelin receptor, ghrelin response receptors present in ARC neurons), and MCH (melanin-concentrating hormone, orexigenic hypothalamic peptide found in hypothalamic MCH neurons) (Figure 2B). In addition, indicators of a diverse set of neuropeptidergic neurons that are present in the hypothalamic region were also observed, such as 5HT (serotonin), SST (somatostatin), NP II (neurophysin II), GABA (γ aminobutyric acid), TH (tyrosine hydroxylase), and CPE (carboxypeptidase E) (Figure 2B). The percentages of these neuronal subtypes derived from multiple hiPSC lines were scored as a percentage of DAPI+ total cells and quantified at day 40 of differentiation (Figure 2C).
Figure 2. Characterization of Neuropeptidergic Hypothalamic Neurons.
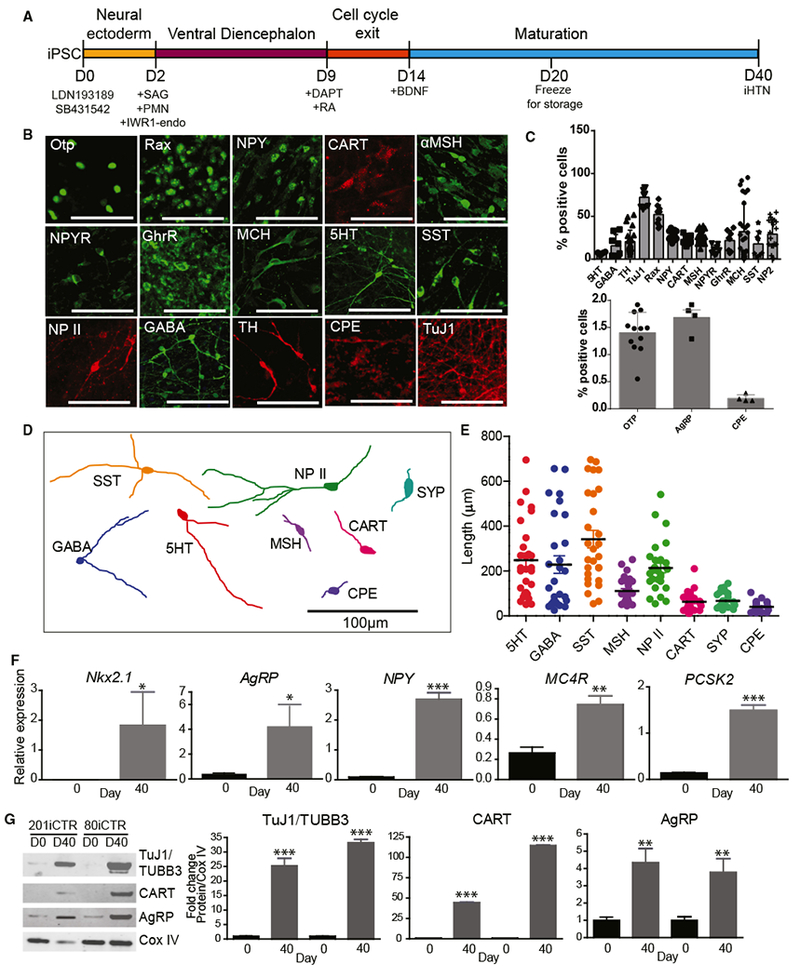
(A) Schematic of differentiation protocol for iHTNs.
(B) Immunocytochemical staining of iHTNs showing various neuronal, hypothalamic, and neuropeptidergic markers.
(C) Quantification of the various cell types contained in iHTN cultures (n = 8 lines).
(D) Representative cell traces of iHTNs expressing relevant markers show distinctive morphologies.
(E) Quantification of primary neurite length (n = 8 lines).
(F) qRT-PCR of hypothalamic and arcuate nucleus-specific genes showing significantly increased expression ofthe genes at day 40 of differentiation compared to day 0 (n = 3 lines).
(G) Representative immunoblots and quantified histograms showing increases in neuron numbers (TUBB3) and arcuate nucleus markers (CART and AgRP) in day 40 cultures compared to day 0 (n = 3 lines). *p < 0.05, **p < 0.01, ***p < 0.001. All statistical analysis was performed using unpaired t test.
Error bars represent SEM.
Tracing of the neurites was performed on the immunostained neuropeptidergic neurons to determine neurite length and morphology (Figure 2D). Neuroendocrine neurons expressing SST, 5HT, NP II, and GABA had relatively long neurites compared to neurons expressing ARC-specific markers, such as α-MSH, CART, CPE, and synaptophysin (SYP) (Figure 2E).
Targeted gene expression revealed that iHTNs expressed typical hypothalamic transcripts, including Nkx2.1 and ARC-specific genes such as AgRP, NPY, MC4R, and PCSK2, in day 40 neurons (Figure 2F). The gene expression results were further supported upon assessing protein levels in the day 40 iHTNs compared to the day 0 hiPSCs by immunoblotting. Day 40 iHTNs showed significantly elevated levels of β-III-tubulin, as well as neuropeptides CART (anorexigenic) and AgRP (orexigenic), relative to their day 0 hiPSC counterparts (Figure 2G). Together, these results suggest that the iHTNs produced by this improved method closely resemble many features of hypothalamic neurons of the ARC.
Transcriptomic Analysis Confirms Hypothalamic Identity of iHTNs
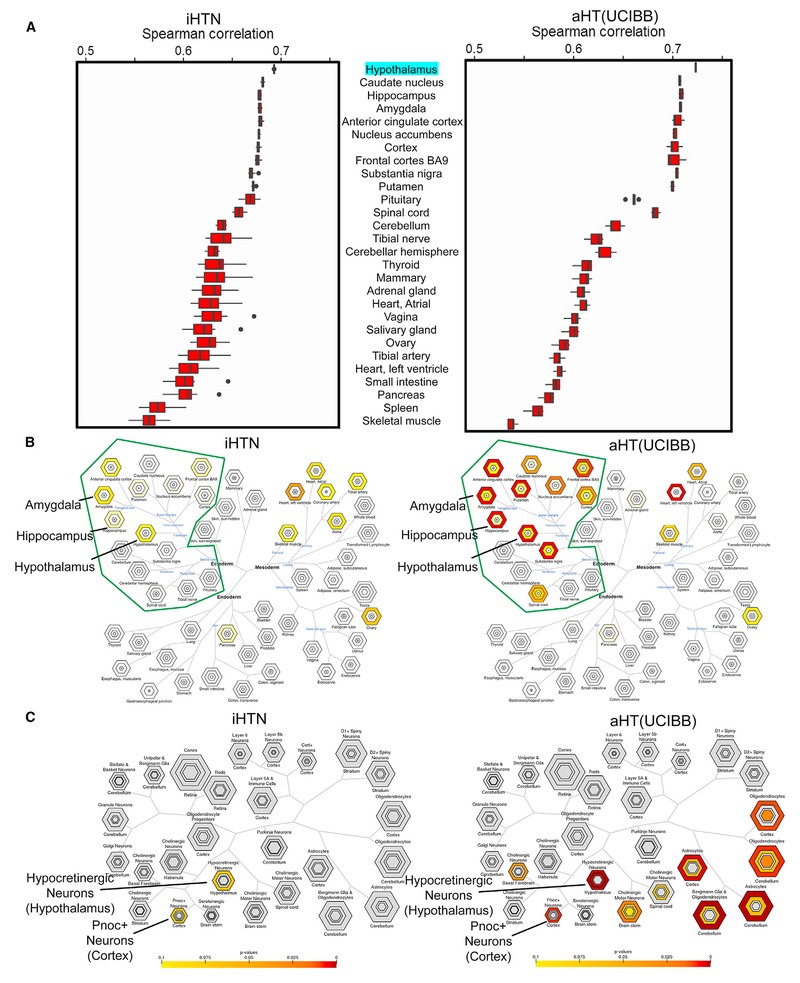
To determine the fidelity of iHTNs with the adult human hypothalamus, high-coverage (29.5 million reads on average) bulk mRNA sequencing transcriptomic comparisons of multiple day 40 iHTNs and adult post-mortem hypothalamus obtained from UCI Brain Bank (aHT-UCIBB) tissue were performed (Figure S4) (GEO: GSE95243). Spearman correlations were determined for iHTNs and aHT-UCIBB samples in comparison to the reference human hypothalamus gene expression values (transcripts per million [TPM]) obtained from the Genotype-Tissue Expression (GTEx) project (Lonsdale et al., 2013), and they are represented as boxplots (Figure 3A). When compared to all available tissue types in an unsupervised manner, transcriptomes of iHTNs scored similarly to transcriptomes of adult HT tissues. Most importantly, in both iHTNs and aHT-UCIBB tissues, the hypothalamus profile provided by GTEx scored the highest match. Adult HT samples scored 0.72 on average while our iHTN samples scored 0.69. In both cases, the next six closest matching tissues were other forebrain regions of the brain.
Figure 3. Characterization of Tissue and Cell Specificity of iHTNs.
(A) A boxplot showing Spearman correlations in iHTNs (n = 12) and post-mortem adult HT tissue (aHT[UCIBB]) (n = 6) compared to GTEx hypothalamus (aHT [GTEx]) gene expression.
(B) Tissue Specific Expression Analysis (TSEA) of iHTNs (left) and aHT(UCIBB) (right) using GTEx, showing that lab-generated iHTNs are brain related in origin (green boundary) and express forebrain- and hypothalamus-specific genes similar to adult hypothalamus tissue.
(C) Cell-Type Specific Expression Analysis (CSEA) of iHTNs (left) and aHT (right) using BrainAtlas, showing that lab-generated iHTNs comprise predominantly hypothalamic neurons in culture.
The top 500 genes found in the hiPSC-derived HT samples overlapped to a statistically significant degree with five regions of the brain, according to data available from GTEx (Figure 3B). The brain regions indicated included the hypothalamus as well as four regions proximal to the hypothalamus and bearing some functional similarities: hippocampus, putamen, amygdala, and anterior cingulate cortex. A few other tissue types also appeared in the tissue analysis, however, these appeared to be artifacts, as these non-neural tissue hits appeared in the top 500 genes transcribed in both aHT-UCIBB samples we analyzed, as well as the top 500 genes obtained directly from the original GTEx hypothalamus data (data not shown). The top 500 genes from the iHTNs and aHT-UCIBB tissue and the GTEx data (data not shown) were submitted to the Tissue Specific Expression Analysis (TSEA) tool to enrich for genes associated with the brain. These enriched sets were then submitted to the Cell-Type Specific Expression Analysis (CSEA) tool to be classified against brain regions drawn from BrainSpan: Atlas of the Developing Human Brain (Dougherty et al., 2010; Xu et al., 2014). Using the CSEA, the gene set from the iHTN samples strongly resembled hypocretinergic neurons of the hypothalamus as well as Pnoc+ neurons of the cortex (Figure 3C). Of note, upon CSEA of the human aHT-UCIBB samples, we found that, in addition to the hypocretinergic and Pnoc+ neurons, certain other related cell types were also detected, such as oligodendrocytes and astrocytes from cerebellum and cortex (Figure 3C). Since the aHT-UCIBB samples were obtained from a post-mortem human brain tissue, the glial cells present in the human brain tissue were likely responsible for the presence of non-neuronal cell profiles. However, iHTNs profiled in vitro are largely devoid of glial tissue, which explains the absence of oligodendrocyte and astrocyte signatures in the iHTNs.
iHTNs Exhibit Functional Characteristics and Responsiveness to Exogenous Peptides
We determined whether the iHTNs were electrically active and capable of firing trains of action potentials using Maestro microelectrode arrays (MEAs) (Figure 4A) in a 48-well-plate format (Axion Biosystems). A raster trace of each of the wells showed measurable firing by the iHTNs for the electrodes in contact with an active neuron (Figure 4B). Each horizontal row represented an electrode in the well. MEA measurements of iHTNs over time exhibited a steady increase in spontaneous electrical activity as the neuronal differentiation time of the culture increased, as indicated by the average spike numbers measured over a period of 10 min (Figure 4C). Average spike numbers for iHTNs steadily increased from day 17 until day 50, suggesting that the iHTNs matured over time and began expressing the complement of ion channels that support trains of spontaneous action potentials.
Figure 4. iHTNs Exhibit Electrical Activity and Response to Exogenous Peptides.
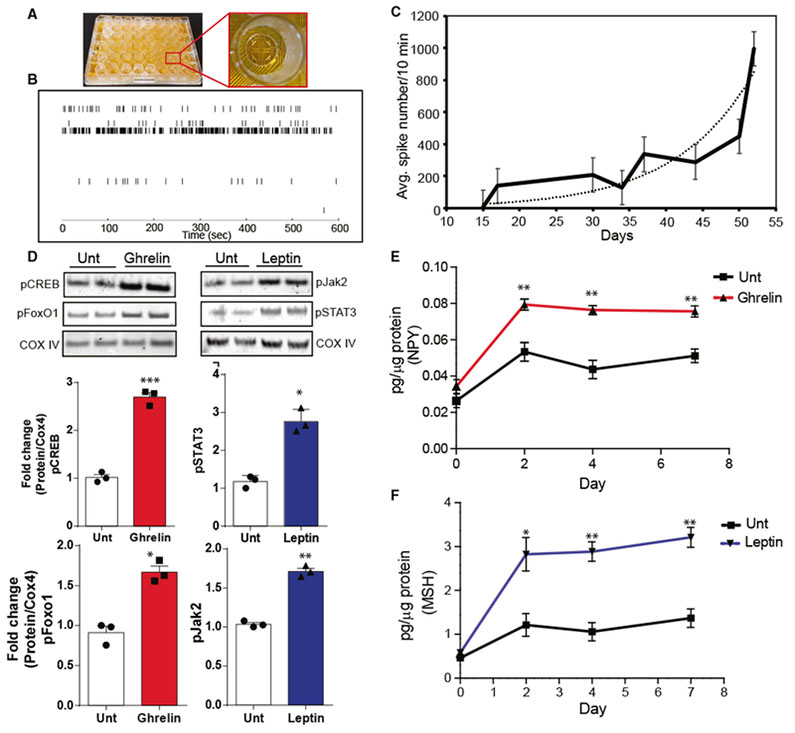
(A) An image of a 48-well microelectrode array (MEA) plate used for measuring neuronal electrophysiological activity over a 30-day period during iHTN differentiation and maturation.
(B) A representative raster trace of iHTNs in a well measured on day 52. Each horizontal row represents an electrode in the well.
(C) Plot representing average neuronal spike number in the iHTNs measured from day 15to day 50 of iHTN development (n = 6 lines in 3 × 48 wells with each line seeded in 24 wells of a plate).
(D) Representative immunoblots and densitometry quantification in histograms showing signaling response of iHTNsto either untreated (Unt) control orexposed to exogenous ghrelin (left) and leptin (right). Activation of the respective pathway proteins is demonstrated via pCREB and pFoxO1 for ghrelin signaling and pSTAT3 and pJak2 for leptin signaling.
(E and F) ELISA measurements showing increased NPY secretion with ghrelin treatment (E) and increased α-MSH secretion with leptin treatment (F). *p < 0.05, **p < 0.01, ***p < 0.001. Data shown are representative of results from differentiated iHTNs in n = 3 independent experiments.
Error bars represent SEM.
One of the main functions of ARC hypothalamic neurons is to appropriately respond to hormones and neuropeptides both locally and peripherally. The responsiveness of iHTNs to exogenously added neuropeptides including, ghrelin (orexigenic) and leptin (anorexigenic), was tested by determining whether relevant downstream signaling pathways were activated. Upon the addition of ghrelin, there was an activation of the ghrelin-signaling pathway, measured by an increase in phosphorylation of CREB (pCREB; p < 0.001) and phosphorylation of FoxO1 (pFoxO1; p < 0.05) (Figure 4D). Upon the addition of leptin, there was an increase in the phosphorylation of Jak2 (pJak2; p < 0.05) and STAT3 (pSTAT3; p < 0.01), which indicates activation of the leptin-induced-signaling pathway (Figure 4D). In concordance with this behavior of iHTNs, ghrelin treatment also increased the release of orexigenic NPY into the culture medium (Figure 4E), while leptin treatment increased anorexigenic α-MSH release (Figure 4F). These data clearly suggest that the iHTNs were both electrically functional (as measured by their ability to fire spontaneously) and responsive to physiologically relevant neuropeptides necessary for the orexigenic and anorexigenic signaling typical of the hypothalamus. Taken together, these data suggest that this in vitro-directed differentiation protocol produces functional iHTNs.
Super-Obese-Derived iHTNs Show Signatures of Obesity
Disease modeling using hiPSCs has been of tremendous interest in recent years. Whether hiPSCs can model polygenic complex metabolic diseases such as obesity remains to be seen. In this study, we attempted to model obesity by reprogramming hiPSCs derived from five super-obese (OBS) individuals (BMI ≥ 50) (Figure 5A) to seven control (CTR) (BMI ≤ 25) subjects (Figure S1A) and then comparing the transcriptome signatures of the derived functional iHTNs. Given the severity of the obesity in the OBS donor group, we determined the underlying DNA level genetic variants by performing whole-exome DNA sequencing (WES) (Sequence Read Archive [SRA]: PRJNA416010) and identifying overlap with known obesity-related coding SNPs listed in the phenotype-gene relationships of the Online Mendelian Inheritance in Man (OMIM) obesity database and the Locke study (Locke et al., 2015). Obesity-related SNPs described here were coding variants with high-sequencing coverage (139.5×). All of the 5 OBS donor samples had 3 common variants, 2 on the adrenoceptor beta 2 (ADRB2) gene (rs1042714 and rs1042713) and 1 on Bardet-Biedl syndrome 2 (BBS2) gene (rs4784677) (Table 1). Four OBS donors shared a common variant in the ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) (rs1044498) and syndecan 3 (SDC3) (rs4949184) genes. Additional obesity-related coding variants were observed in leptin receptor (LEPR) and peroxisome proliferator-activated receptor gamma (PPARG) genes in up to two of the OBS donor hiPSCs.
Figure 5. Transcriptome Profiling of iHTNs from Super-Obese Patients Displays Molecular Signatures of Metabolic Disease and Obesity.
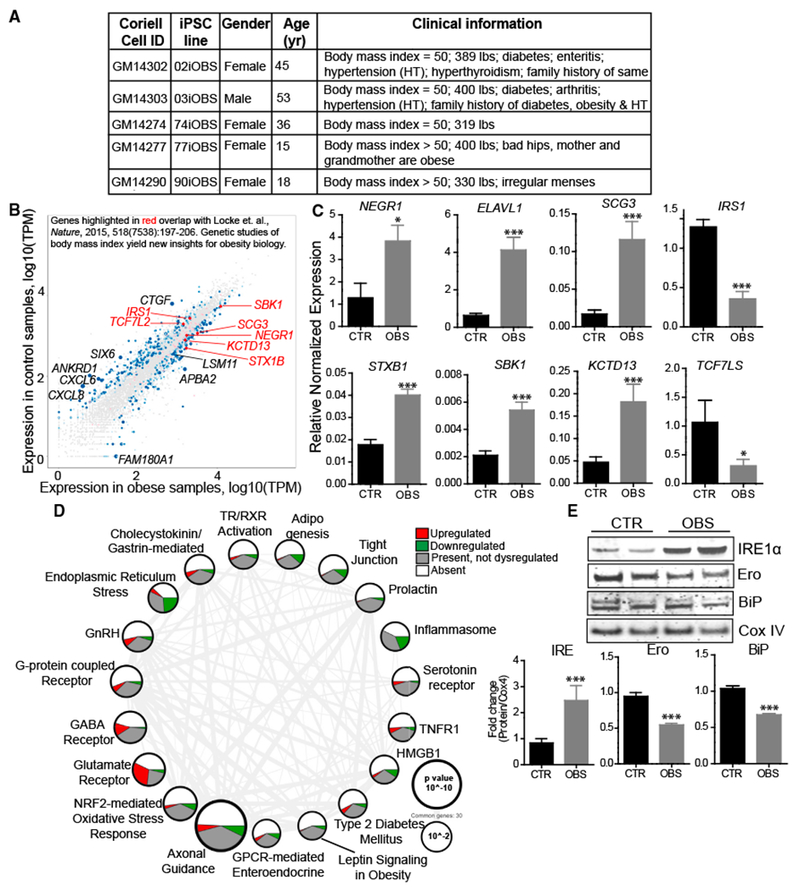
(A) Patient information on super-obese (OBS) patient-derived hiPSC lines.
(B) Scatterplot highlighting dysregulated expression of genes in iHTNs derived from normal control (BMI < 25) versus OBS patients (BMI > 25) based on RNA sequencing data and correlated with obesity-related gene identified in Locke et al. (2015) (n = 12 lines).
(C) qRT-PCR validation of genes identified from Locke et al. (2015) showing increased expression of NEGR1, ELAVL1, SCG3, STXB1, SBK1, and KCTD13 and decreased expression of IRS1 and TCF7LS in the OBS group compared to CTR (n = 10 lines).
(D) Ingenuity PathwayAnalysis showing differentially regulated pathways. Size ofeach node representsthe p value, while thethickness of lines connecting each node represents the number of common genes shared between the two pathways (n = 12 lines).
(E) Immunoblot validation ofendoplasmic reticulum (ER) stress pathway, where OBS iHTNs confirm ER stress activation with an increase in IRE1α and a decrease in Ero and BiP compared to CTR (n = 3 independent experiments consisting of one CTR and one OBS line each). *p < 0.05, ***p < 0.001.
Error bars represent SEM.
Table 1.
Coding SNPs Found in Exome-Sequenced Obesity hiPSC Lines
| Gene | SNP ID | Chromosome | Position | Base Change | Amino Acid Change | Alternative Base Frequency | Lines Containing | Homozygous|Heterozygous | OMIM |
|---|---|---|---|---|---|---|---|---|---|
| ADRB2 | rs1042714 | 5 | 148,206,473 | G > C | Glu > Gln | 0.796 | 5 | 5 | 0 | 601665 |
| rs1042713 | 5 | 148,206,440 | G > A | Gly > Arg | 0.476 | 5 | 2 | 3 | 601665 | |
| ENPP1 | rs1044498 | 6 | 132,172,368 | A > C | Lys > Gln | 0.342 | 4 | 2 | 2 | 601665/ 125853 |
| LEPR | rs1137101 | 1 | 66,058,513 | A > G | Gln > Arg | 0.584 | 2 | 1 | 1 | 614963 |
| rs1805094 | 1 | 66,075,952 | G > C | Lys > Asn | 0.142 | 2 | 0 | 2 | 614963 | |
| rs1137100 | 1 | 66,036,441 | A > G | Lys > Arg | 0.320 | 1 | 0 | 1 | 614963 | |
| PPARG | rs3856806 | 3 | 12,475,557 | C > T | His | 0.127 | 2 | 0 | 2 | 604367/ 125853 |
| SDC3 | rs2282440 | 1 | 31,347,320 | G > A | Asp > Asn | 0.161 | 1 | 1 | 2 | 601665 |
| rs4949184 | 1 | 31,347,399 | C > T | Val > Ile | 0.234 | 4 | 1 | 3 | 601665 | |
| rs2491132 | 1 | 31,349,647 | C > T | Thr > Ile | 0.076 | 1 | 0 | 1 | 601665 | |
| BBS2 | rs4784677 | 16 | 56,548,501 | C > T | Ser > Asn | 0.996 | 5 | 5 | 0 | 615981 |
The exome of each obese cell line was compared to the OMIM database of SNPs associated with obesity and with a list of potentially relevant SNPs described in Locke etal. (2015). SNPs from genes of relevance are shown above. Chromosome and position indicate the chromosome and locus of the SNP. Base change and amino acid change indicate the typical and alternative nucleotide and the resulting amino acid change during protein translation. Alternative base frequency is the frequency at which the mutation is observed in the world population. The lines containing column lists the number of lines in which the SNP was found and the zygosity column lists the number of lines in which the SNP was found on both chromosomes or on a single chromosome. The OMIM column contains a SNP entry in the Online Mendelian Inheritance in Man (OMIM) database.
With respect to iHTN differentiation efficiency, no differences in cell numbers expressing typical hypothalamic markers were observed between the CTR and OBS groups (Figure S3). Differential expression analysis of the bulk mRNA sequencing (mRNA-seq) transcriptome profiles of the two groups revealed dysregulation of relevant genes, which were identified based on a list provided by a combination of GWAS and Metabochip datasets meta-analysis of BMI data from 339,224 individuals (Locke et al., 2015). The dysregulated pathways and genes within BMI-associated loci identified by Locke et al. (2015) were enriched for expression in the brain and CNS, including clathrin-coated vesicles, glutamate signaling, and synapse biology.
We identified over half a dozen CNS-specific genes differentially regulated with a p value less than 0.05 in OBS iHTNs that overlapped with the Locke et al. (2015) study (Figure 5B). The relative expression levels of these transcripts were further validated in day 40 iHTNs, and we found that IRS1 and TCF7L2 were downregulated in the OBS group, while NEGR1, SCG3, SBK1, KCTD13, and STX1B were upregulated in the OBS group (Figure 5C) (Locke et al., 2015).
Ingenuity Pathway Analysis performed on all 657 significant differentially expressed genes (DEGs) between CTR and OBS iHTNs revealed that relevant signaling pathways regulating metabolism and energy homeostasis were dysregulated in the OBS group. These included axonal guidance signaling; glutamate receptor signaling; GABA receptor signaling; and G-protein-coupled receptor (GPCR) signaling that includes ghrelin, NPY, and CART receptors, all enriched in the CNS. Further, GPCR-mediated enteroendocrine signaling, endoplasmic reticulum (ER) stress and leptin signaling in obesity, type 2 diabetes mellitus, and adipogenesis were among the metabolic pathways that were perturbed in the OBS iHTNs. Finally, inflammasome, tumor necrosis factor receptor 1 (TNFR1), and high-mobility group box 1 (HMGB1) pathways were significantly enriched for DEGs, and they are known to be associated with obesity-related inflammatory processes in the brain (Figure 5D). To further confirm whether one of these dysregulated pathways manifests at the protein level, we performed immunoblotting on CTR and OBS iHTN lysates, and we evaluated proteins implicated in regulating ER stress specifically (IRE1α, Ero, and BiP). There was evidence of increased ER stress in OBS iHTNs, as indicated by the increased expression of IRE1α (ER transmembrane stress sensor protein) and decreased expression of proteins controlling folding, Ero and BiP (Figure 5E). Taken together, the WES and transcriptomic profiling of iHTNs point toward the possibility that the obesity-associated coding variants may contribute to these disease signatures even after reprogramming somatic cells.
OBS-Derived iHTNs Have Perturbations in Metabolism and Responses to Exogenous Neuropeptides
Next, the effect of obesity on iHTN mitochondrial respiratory function was determined by performing a mitochondrial stress test with the XFe24 Seahorse Extracellular Flux Analyzer. Interestingly, the oxygen consumption rate of iHTNs in the OBS group was significantly lower than the CTR group, pointing toward a decrease in basal mitochondrial respiration and oxygen consumption rate (OCR) in OBS iHTNs (Figure 6A). Additionally, the OBS iHTNs were more glycolytic than CTR iHTNs, as measured by a plot of the extracellular acidification rate (ECAR) versus OCR (Figure 6B). No differences were observed in the respiration rates in the iHTNs upon ghrelin or leptin addition compared to untreated condition (Figure S5).
Figure 6. Functional Comparison of Control and OBS-Derived iHTNs.
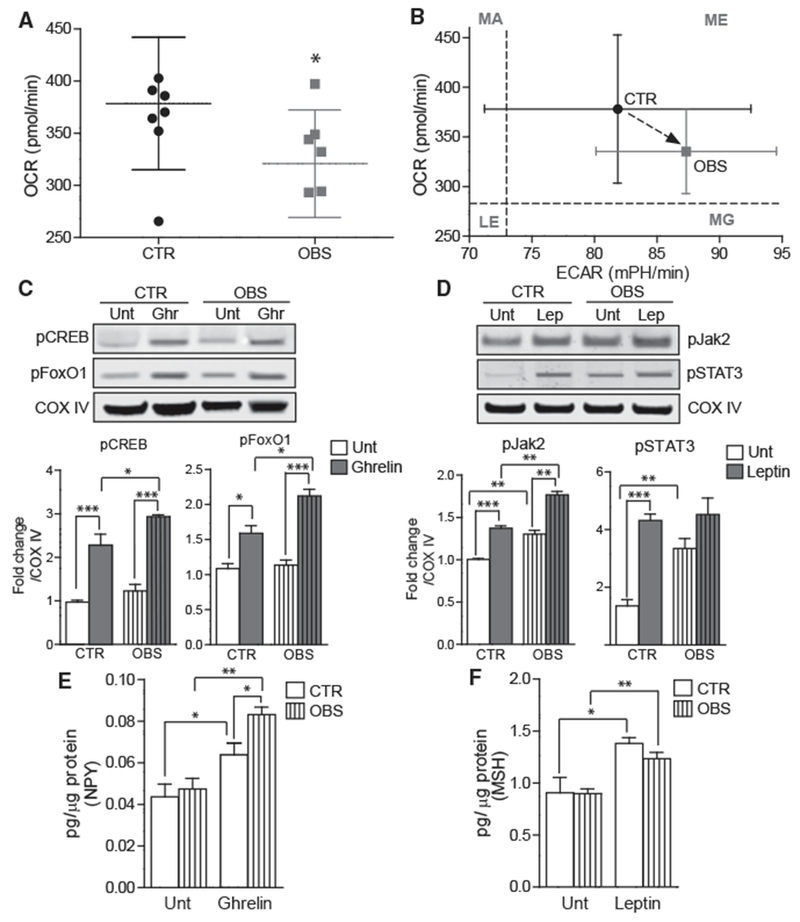
(A) Seahorse mitochondrial respirometry histogram showing decreased OCR in OBS iHTNs compared to control (CTR) iHTNs.
(B) Comparison of OCR versus extracellular acidification rate (ECAR, glycolysis measure) showing higher ECAR and lower OCR in OBS iHTNs. LE, less energetic; MA, more aerobic; MG, more glycolytic; ME, more energetic.
(C and D) Representative immunoblots showing activation of ghrelin pathway upon ghrelin treatment in iHTNs as measured by phosphorylation of CREB and FoxO1 (C) and moderate or no activation of leptin pathway upon leptin treatment as measured by phosphorylation of Jak2 and STAT3 (D).
(E) NPY secretion measured by ELISA showing increased response of OBS iHTNs to ghrelin exposure.
(F) α-MSH secretion measured by ELISA comparing baseline (Unt) to leptin treatment. Both CTR and OBS iHTNs were responsive to leptin exposure and increased α-MSH secretion; however, this measure was not significantly different between CTR and OBS groups. *p < 0.05, **p < 0.01, ***p < 0.001. Data shown are representative of results from differentiated iHTNs used in n = 3 independent experiments consisting of one CTR and one OBS line each.
Error bars represent SEM.
To investigate whether CTR or OBS iHTNs respond to hormonal signals similarly, the iHTNs derived from multiple hiPSC lines were exposed to ghrelin and leptin neuropeptides, and their differential responses were examined by measuring downstream signaling responses. Upon exposure to ghrelin, as described previously, the CTR iHTNs appropriately exhibited an increase in phosphorylation of CREB (pCREB) and FoxO1 (pFoxO1). The OBS iHTNs responded similarly, by increasing pCREB and pFoxO1 levels upon ghrelin exposure; however, the OBS iHTNs’ response was stronger relative to the levels of pCREB and pFoxO1 in ghrelin-treated CTR iHTNs. This increased signaling response to an orexigenic hormone such as ghrelin suggests that OBS iHTNs retained an innate increased food intake-signaling response in the presence of ghrelin (Figure 6C). Conversely, baseline leptin signaling (i.e., absent exogenous leptin treatment) was higher in OBS iHTNs relative to CTR iHTNs based on increased levels of pJak2 and STAT3 (pSTAT3). Upon exogenous leptin treatment, CTR iHTNs showed significantly increased pJak2 and pSTAT3, while OBS iHTNs only showed a significant increase in pJak2, but not in pSTAT3 (Figure 6D). These data suggest elevated baseline activation of the leptin-signaling pathway in the OBS iHTNs in the absence of exogenous signaling molecules.
With regard to neuropeptide secretion, orexigenic NPY was measured in the cell culture media following ghrelin treatment of iHTNs, and both CTR iHTNs and OBS iHTNs responded to ghrelin by secreting increased levels of NPY (Figure 6E). Although both groups responded to ghrelin treatment, it is noteworthy that OBS iHTNs again showed an exacerbated response to ghrelin treatment by secreting higher levels of NPY compared to CTR iHTNs. On the other hand, when leptin treatment was performed and secretion levels of anorexigenic α-MSH were assessed, we found that both iHTNs from CTR and OBS showed increased secretion of α-MSH at similar magnitudes (Figure 6F).
DISCUSSION
hiPSCs offer great promise in the field of disease modeling. Several neurodevelopmental (Chamberlain et al., 2010; Fuller et al., 2016; Sareen et al., 2012), neurodegenerative (Israel et al., 2012; Reinhardt et al., 2013; Yagi et al., 2011), and metabolic diseases (Gu et al., 2015; Wang et al., 2015) have been successfully modeled using hiPSCs. Given the seriousness of the global obesity epidemic and the alarming rise in the rate of childhood obesity, this study focused on determining whether we can successfully model aspects of CNS involvement in non-monogenic forms of obesity, which may have complex etiological origins. This study describes a reliable, controlled, efficient, and reproducible protocol for the generation of hypothalamic, neuropeptidergic neurons capable of secreting neurohormones, and it demonstrates that these neurons are responsive to exogenous hormone signals. The ultimate goal was to determine whether hypothalamic neuropeptidergic neurons derived from reprogrammed patient hiPSCs could faithfully capture signatures of complex obesity, by comparing differentiated hypothalamic neurons from hiPSCs of multiple normal BMI and OBS individuals with known coding variants in obesity loci. We observed that certain molecular and pathway-level signatures of human obesity can be recapitulated by diseased iHTNs derived from OBS patients, despite undergoing reprogramming and re-differentiation to hypothalamic neuronal cells. This study thus widens the application of hiPSCs in modeling CNS involvement in obesity as well as other metabolic diseases.
Differentiation of hypothalamic neuropeptidergic neurons has been previously described using mouse embryonic stem cells (ESCs) (Wataya et al., 2008), human ESCs (Merkle et al., 2015), and hiPSCs (Wang et al., 2015). Although these studies describe the generation of hypothalamic neurons at low differentiation efficiencies from a handful of PSC lines, in the context of metabolic diseases, it becomes necessary to verify the responsiveness of the in vitro-derived neuroendocrine neurons to various hormonal signals in order for them to be utilized as a valid model of metabolic signaling defects and testing strategies for therapeutic intervention. In this regard, we successfully demonstrated differentiation of multiple hiPSC lines from over 10 donors. Our results show that iHTN cultures are a mixed population of both orexigenic AgRP/NPY and anorexigenic POMC/CART neurons; hence, we rightly believed that they would respond well to both orexigenic (ghrelin) and anorexigenic (leptin) signals, and transcriptomic profiles showed the highest correlation with the RNA isolated from human brain hypothalamus.
In addition to the iHTN transcriptome profiles supporting the efficacy of our model to produce physiologically relevant hypothalamic neurons, this RNA-seq data also support the role of genetic loci that have been associated with obesity biology based on BMI (Locke et al., 2015). Specifically, several genetic loci for increased BMI were also differentially expressed in OBS iHTNs compared to CTR iHTNs. NEGR1 expression was increased in OBS, and this gene has been associated with synaptic function, glutamate signaling, and, most interestingly, extreme/early onset obesity. We also found TCF7L2 decreased in OBS compared to controls. TCF7L2 encodes a transcription factor of the same name, which has been implicated in type 2 diabetes (Jin and Liu, 2008), especially proglucagon (blood glucose-elevating prohormone) synthesis whereby activation of TCF7L2 leads to the repression of proglucagon synthesis. We found that TCF7L2 is lower in OBS individuals thereby possibly causing an increase in the production of proglucagon and risk of developing diabetes (Grant et al., 2006). IRS1 was also found to be downregulated in OBS, suggesting the dysregulation of yet another gene involved in the pathogenesis of type 2 diabetes (Kovacs et al., 2003). Among the other genes differentially regulated were KCTD13, a gene associated with sweet taste signaling, which is known to be a glucose sensor in hypothalamus (Ren et al., 2009), and SCG3 (secretogranin 3), encoding the SCG3 protein, which is a member of the neuroendocrine secretory protein family and may serve as precursor for biologically active endocrine peptides (Rong et al., 2002). Taken together, the genes that are differentially regulated in the OBS-derived iHTNs suggest there is a strong tendency toward dysregulation of endocrine and metabolic processes known to be abnormal in obese individuals.
Furthermore, pathway analysis of these DEGs between OBS iHTNs and CTR iHTNs revealed several dysregulated pathways relevant in obesity. For example, we found dysregulation of several endocrine-signaling pathways such as leptin signaling in obesity and cholecystokinin/Gastrin-mediated signaling, pointing toward a skewed response to these critical endocrine hormones, which are hallmarks of obesity (Chen et al., 2016; Lee et al., 2001). Additionally, many of the receptor-signaling pathways were also dysregulated, such as GPCR, GPCR-mediated enteroendocrine signaling, as well as glutamate receptor, GABA receptor, and serotonin receptor (also GPCRs) pathways, all of which are strongly involved in the neurotransmission and neuronal signaling.
The OBS donors contain polygenic obesity-related SNPs, suggesting these coding variants likely contribute to the obese phenotype exhibited by the iHTNs. The main SNPs were observed in obesity-related genes coding for LEPR, SDC3, ADRB2, ENPP1, PPARG, and BBS2. All 5 of the OBS hiPSCs contained SNPs in three regions: two variants of ADRB2 and one variant on the BBS2 gene. Both variants of the ADRB2 gene (rs1042713 and rs1042714) have been shown to be associated with obesity and/or BMI (Gjesing et al., 2007, 2009; Hayakawa et al., 2000). Although some studies did not find an association of these SNPs with obesity (Bengtsson et al., 2001; Pereira et al., 2003), the rs1042714-related 27Glu allele was shown to be significantly associated with obesity in Asians, Pacific Islanders, and Native Americans (Jalba et al., 2008). The BBS2 variant rs4784677 has also been associated with obesity (http://omin.org/entry/606151). This is a rare SNP for Bardet-Biedl Syndrome (BBS) and includes obesity as one of its clinical features. BBS in itself is a complex trait needing three mutant alleles for manifesting the phenotype. Hence, associating obesity with a mutation in the BBS2 gene cannot be done with confidence.
Besides these genes, SNPs in other obesity-related genes were observed in one or more of the obesity donors, but not in all. Several obesity-related SNPs in LEPR genes, such as rs1137101 (Furusawa et al., 2010), rs1805094 (Rojano-Rodriguez et al., 2016), and rs1137100 (Hastuti et al., 2016), were observed. PPARG SNP rs3856806 was observed in two samples, which have been associated with risk of obesity and type 2 diabetes (Lv et al., 2017). Two other obesity samples showed SNPs in one locus each for SDC3 gene, rs2491132 and rs2282440, both of which have been associated with obesity (Ha et al., 2006). Besides the above SNPs, four of our samples showed polymorphism in the ENPP1 gene (rs1044498), although some studies have failed to show a consistent association of this SNP with obesity (Grarup et al., 2006; Lyon et al., 2006; Zhao et al., 2011). We believe that the mutations found in the severe obesity donors used in this study provide a compelling explanation for the retained obesity phenotype exhibited by OBS hiPSC-derived iHTNs. Energy metabolism differed between the iHTNs from control versus OBS patients, consistent with the critical role of mitochondrial metabolism in ghrelin’s effects on NPY/AgRP neurons (Andrews et al., 2008). Future studies will explore the changes to mitochondrial metabolism in response to ghrelin between CTR and OBS iHTNs, particularly to examine UCP2 and free radical production, which are important to this pathway (Andrews et al., 2008, 2009; Toda and Diano, 2014).
Studies have shown the presence of increased levels of ghrelin in obesity (Cummings et al., 2002), but not much information is available on the effect of elevated ghrelin at a molecular level. We show that similar amounts of exogenously added ghrelin elicit an exaggerated effect in OBS iHTNs compared to CTR iHTNs. Thus, increased ghrelin levels in obese individuals, combined with hyper-responsive neurons, might result in exacerbated orexigenic signaling.
While baseline levels of CREB and FoxO1 were not different between CTR and OBS iHTNs, we found a significant increase in the baseline levels of pJak2 and pSTAT3 levels between the two groups, suggesting that at baseline there could be an increase in leptin levels, which is a hallmark of obesity (Scarpace and Zhang, 2007). In response to exogenous leptin, however, pJak2 and α-MSH secretion levels increased, but pSTAT3 did not. Unlike exogenous ghrelin exposure, OBS iHTNs did not show any exacerbated leptin-signaling response upon leptin exposure (Figure 7). These data suggest that OBS iHTNs have increased baseline leptin levels as well as a non-dramatic leptin signaling upon exposure to exogenous leptin, pointing toward a possible leptin resistance, which is also typical of obesity (Knight et al., 2010).
Figure 7. A Schematic Representation of the Hormonal Responses in CTR and OBS iHTNs in Baseline and Hormone-Treated Conditions.
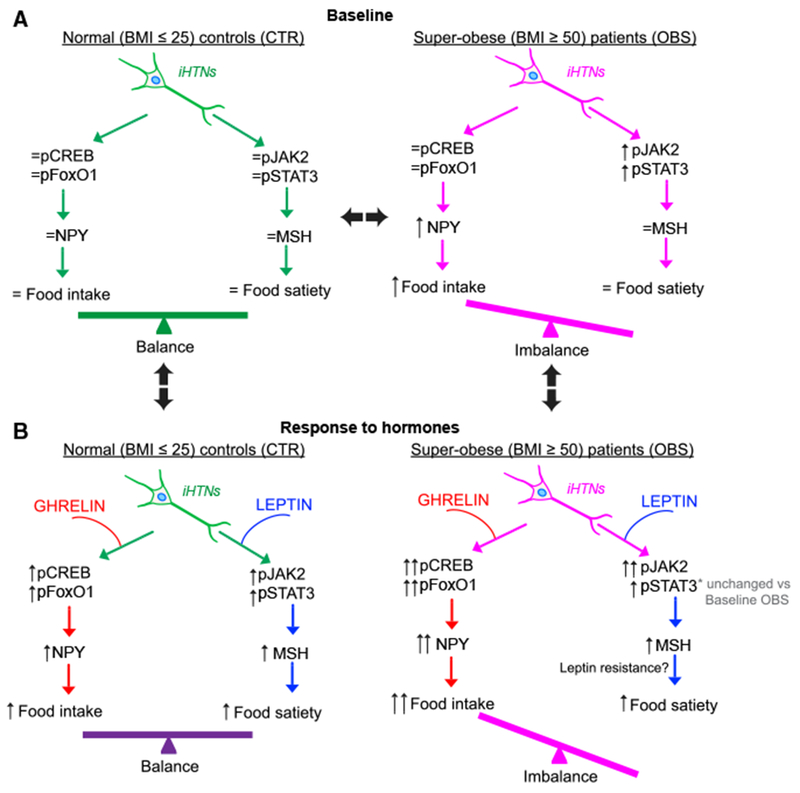
(A) At baseline, CTR iHTNs (left) show baseline levels of orexigenic (pCREB and pFoxO1) and anorexigenic pathway proteins (Jak2 and STAT3) and a baseline response to added hormones to maintain a balance between food intake (NPY) and energy expenditure (α-MSH). At baseline, OBS iHTNs (right) show an imbalance in food intake versus energy expenditure in that there is an activation of the orexigenic pathway while the anorexigenic pathway is not equally active.
(B) Upon treatment of relevant hormones such as ghrelin (orexigenic) and leptin (anorexigenic), the CTR iHTNs (left) maintain their balance between food intake and energy expenditure. Interestingly, OBS iHTNs (right) show a further imbalance in that there is a further increase in orexigenic signals upon ghrelin treatment, but leptin does not elicit an equally effective anorexigenic response. As a result, OBS iHTNs show exacerbated orexigenic signals as well as possible resistance to leptin.
In this study, the use of hiPSCs served as a unique platform for modeling obesity. The possibility to elucidate developmental and metabolic cues into the cause of obesity retrospectively has been made possible by this platform. Obtaining metabolically relevant tissues or cell types such as the hypothalamic neurons of the ARC from living individuals struggling with obesity would otherwise be impossible. hiPSC-based models offer great promise in dissecting the role of individual cell types in the onset of the disease, and they provide a platform of living cells that can be used for high-throughput drug screening. It is thus evident that polygenic obesity can be modeled using differentiated neurons derived from hiPSCs of donors afflicted with severe obesity. Although the patient lines had polygenic SNPs at few obesity loci, gene expression and molecular and metabolic profiling of the hypothalamic neurons revealed convergence of these DNA-level variants on specific obesogenic phenotypes in vitro. While this study particularly focused on obesity, there are several other disorders associated with hypothalamic dysfunction that could potentially be modeled and evaluated with this iHTN differentiation method, such as anorexia nervosa, alcoholism, diabetes mellitus and insipidus, anxiety disorders, as well as certain rare diseases hallmarked by neuroendocrine abnormalities. Likewise, many approved drugs used to treat patients with mental illness, such as antidepressants, antipsychotics, and mood stabilizers, can cause metabolic syndrome in some patients and ultimately lead to physical diseases of obesity, diabetes, and cardiovascular disease (Correll et al., 2015). Such patient-derived iHTN models may provide a platform to evaluate the mechanisms behind these unwanted side effects, and they could potentially be used to identify which patients are more susceptible to these unwanted outcomes.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Nkx2.1 | EMD Millipore | Cat# MAB5460 RRID: AB_571072 |
| OTP | GeneTex | Cat# GTX119601 RRID: AB_11164017 |
| SOX1 | R&D Systems | Cat# AF3369 RRID: AB_2239879 |
| Rax | ThermoFisher Scientific | Cat# PA5-11477 RRID: AB_2284916 |
| NPY | Millipore | Cat# AB9608 RRID: AB_2153720 |
| CART | Santa Cruz | Cat# sc-18068 RRID: AB_2068572 |
| α-MSH | Phoenix Pharmaceuticals | Cat# H-043-01 RRID: AB_10013604 |
| NPYR | Alomone Labs | Cat# ANR022 RRID: AB_2040032 |
| GhrR | Alomone Labs | Cat# AGR031 RRID: AB_2340976 |
| MCH | Sigma Aldrich | Cat# M8440 RRID: AB_260690 |
| Serotonin | Immunostar | Cat# 20080 RRID: AB_572263 |
| Somatostatin | Santa Cruz | Cat# sc-13099 RRID: AB_2195930 |
| Neurophysin II | Santa Cruz | Cat# sc-27093 RRID: AB_2061964 |
| GABA | Sigma-Aldrich | Cat# A2052 RRID: AB_477652 |
| Tyrosine Hydroxylase | Immunostar | Cat# 22941 RRID: AB_572268 |
| Carboxypeptidase E | R&D Systems | Cat# AF3587 RRID: AB_2083766 |
| β III Tubulin | Sigma-Aldrich | Cat# T8660 RRID: AB_477590 |
| COX IV | Cell Signaling Technology | Cat# 4850 RRID: AB_2085424 |
| pCREB | Cell Signaling Technology | Cat# 9198S RRID: AB_2561044 |
| CREB Total | Cell Signaling technology | Cat# 9197S RRID: AB_331277 |
| pFOXO1 | Cell Signaling technology | Cat# 9461S RRID: AB_329831 |
| FOXO1 Total | Cell Signaling Technology | Cat# 2880S RRID: AB_2106495 |
| pJAK2 | Cell Signaling Technology | Cat# 3776S RRID: AB_2617123 |
| JAK2 Total | Cell Signaling technology | Cat# 3230S RRID: AB_2128522 |
| pSTAT3 | Cell Signaling Technology | Cat# 9131S RRID: AB_331586 |
| STAT3 Total | Cell Signaling Technology | Cat# 9139S RRID: AB_331757 |
| SOX2 | Stemgent | Cat# 09-0024 RRID: AB_2195775 |
| TRA-1-81 | Stemgent | Cat# 09-0011 RRID: AB_1512171 |
| NANOG | Stemgent | Cat# 09-0020 RRID: AB_2298294 |
| TRA-1-60 | Stemgent | Cat# 09-0010 RRID: AB_1512170 |
| OCT4 | Stemgent | Cat# 09-0023 RRID: AB_2167689 |
| SSEA4 | Stemgent | Cat# 09-0006 RRID: AB_1512169 |
| Biological Samples | ||
| Brain sec. 1662; 30 year M | UC Irvine Brain Bank | N/A |
| Brain sec. 1838; 97 year cauc. F | UC Irvine Brain Bank | N/A |
| Brain sec. 1843; 78 year cauc. M | UC Irvine Brain Bank | N/A |
| Brain sec. 1919; 61 year cauc. M | UC Irvine Brain Bank | N/A |
| Brain sec. 2266; 53 year cauc. F | UC Irvine Brain Bank | N/A |
| Brain sec. 2884; 84 year cauc. M | UC Irvine Brain Bank | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Accutase | EMD Millipore | SCR005 |
| 0.2% Triton X-100 | BioRad | 1610407 |
| 3,3′,5,5′-tetra-methulbenzidine | Sigma-Aldrich | 860336-1G |
| 4% Paraformaldehyde | Electron Microscopy Sciences | 15714-S |
| 5% Donkey serum | EMD Millipore | 566460 |
| 5% Milk solution | BioRad | 1706404 |
| Antimycin A | abcam | ab141904 |
| Avidin conjugated Horseradish peroxidase | ThermoFisher | 29994 |
| BDNF | Miltenyi Biotec | 130-093-811 |
| Chloroform | Sigma-Aldrich | C2432-500ML |
| CryoStor | Stem Cell Technologies | 7930 |
| DAPT | Cayman | 13197 |
| FCCP carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone | Sigma-Aldrich | C2920-10MG |
| Ghrelin peptide | Cayman | 15072 |
| Hoechst | ThermoFisher | 62249 |
| IWR-endo | Cayman | 13659 |
| Laminin basement membrane | Sigma-Aldrich | L2020-1MG |
| LDN193189 | Cayman | 11802 |
| Leptin | Peprotech | 300-27 |
| Matrigel | Corning | 356253 |
| Oligomycin | Sigma-Aldrich | 75351-5MG |
| Purmorphamine | Tocris | 4551 |
| Pyruvate | Sigma-Aldrich | P5280-25G |
| Retinoic Acid | Cayman | 11017 |
| ROCK inhibitor Y-27632 (hydrochloride) | Cayman | 10005583 |
| Rotenone | Sigma-Aldrich | R8875-1G |
| SAG Smoothened agonist | Tocris | 4366 |
| SB431542 | Cayman | 13031 |
| Seahorse XF Base medium plus supplements | Agilent | 102353-100 |
| Streptomycin | abcam | ab143264 |
| Trizol | ThermoFisher | 15596026 |
| Tween-20 | Sigma-Aldrich | P9416-100ML |
| Critical Commercial Assays | ||
| Agencourt AMPure XP beads | Beckman Coulter Genomics | NC9959336 |
| Agilent high sensitivity DNA assay | Agilent | 5067-4626 |
| Agilent SureSelect Human Exon kit v4 | Agilent | 5190-8863 |
| Agilent Total RNA 6000 Nano Kit | Agilent | 5067-1511 |
| ELISA Kit for Alpha-Melanocyte Stimulating Hormone (aMSH) | Cloud-Clone | CEA239Hu |
| EMD Millipore Human Neuropeptide Y ELISA | EMD Millipore | EZHNPY-25K |
| GoScript Reverse Transcriptase | Promega | A5003 |
| Halt Protease Inhibitor Cocktail | ThermoScientific | 78430 |
| Illumina TruSeq stranded mRNA library | Illumina | 20020594 |
| Mammalian PER | ThermoScientific | 78501 |
| Microelectrode Array Plate, 48 wells | Axion | M768-KAP-48 |
| Nitrocellulose Membrane | BioRad | 1620115 |
| NuPAGE Novex Polyacrylaminde Gel | ThermoScientific | NP0322BOX |
| QIAGEN On-Column DNase | QIAGEN | 79254 |
| QIAGEN Rneasy Mini Kit | QIAGEN | 74104 |
| Seahorse culture plate | Agilent | 100777-004 |
| Streptavidin-coated Magnetic Beads | New England Biolabs | S1421S |
| Super-Script II Reverse Transcriptase | ThermoScientific | 18064014 |
| SYBR Green Master mix | Applied Biosystems | 4309155 |
| Agencourt AMPure XP beads | Beckman Coulter Genomics | NC9959336 |
| Agilent high sensitivity DNA assay | Agilent | 5067-4626 |
| Agilent SureSelect Human Exon kit v4 | Agilent | 5190-8863 |
| Agilent Total RNA 6000 Nano Kit | Agilent | 5067-1511 |
| ELISA Kit for Alpha-Melanocyte Stimulating Hormone (aMSH) | Cloud-Clone | CEA239Hu |
| EMD Millipore Human Neuropeptide Y ELISA | EMD Millipore | EZHNPY-25K |
| GoScript Reverse Transcriptase | Promega | A5003 |
| Halt Protease Inhibitor Cocktail | ThermoScientific | 78430 |
| Illumina TruSeq stranded mRNA library | Illumina | 20020594 |
| Mammalian PER | ThermoScientific | 78501 |
| Microelectrode Array Plate, 48 wells | Axion | M768-KAP-48 |
| Nitrocellulose Membrane | BioRad | 1620115 |
| NuPAGE Novex Polyacrylaminde Gel | ThermoScientific | NP0322BOX |
| QIAGEN On-Column DNase | QIAGEN | 79254 |
| QIAGEN Rneasy Mini Kit | QIAGEN | 74104 |
| Seahorse culture plate | Agilent | 100777-004 |
| Streptavidin-coated Magnetic Beads | New England Biolabs | S1421S |
| Super-Script II Reverse Transcriptase | ThermoScientific | 18064014 |
| SYBR Green Master mix | Applied Biosystems | 4309155 |
| Agencourt AMPure XP beads | Beckman Coulter Genomics | NC9959336 |
| Agilent high sensitivity DNA assay | Agilent | 5067-4626 |
| Agilent SureSelect Human Exon kit v4 | Agilent | 5190-8863 |
| Agilent Total RNA 6000 Nano Kit | Agilent | 5067-1511 |
| Pierce BCA Protein Assay Kit | ThermoScientific | 23225 |
| ELISA Kit for Alpha-Melanocyte Stimulating Hormone (aMSH) | Cloud-Clone | CEA239Hu |
| EMD Millipore Human Neuropeptide Y ELISA | EMD Millipore | EZHNPY-25K |
| GoScript Reverse Transcriptase | Promega | A5003 |
| Halt Protease Inhibitor Cocktail | ThermoScientific | 78430 |
| Illumina TruSeq stranded mRNA library | Illumina | 20020594 |
| Mammalian PER | ThermoScientific | 78501 |
| Microelectrode Array Plate, 48 wells | Axion | M768-KAP-48 |
| Nitrocellulose Membrane | BioRad | 1620115 |
| NuPAGE Novex Polyacrylaminde Gel | ThermoScientific | NP0322BOX |
| QIAGEN On-Column DNase | QIAGEN | 79254 |
| QIAGEN Rneasy Mini Kit | QIAGEN | 74104 |
| Seahorse culture plate | Agilent | 100777-004 |
| Streptavidin-coated Magnetic Beads | New England Biolabs | S1421S |
| Super-Script II Reverse Transcriptase | ThermoScientific | 18064014 |
| SYBR Green Master mix | Applied Biosystems | 4309155 |
| Deposited Data | ||
| RNA-seq data | NCBI: GEO | GEO: GSE95243 |
| Whole-exome sequence data | Sequence Read Archive | PRJNA416010 |
| Experimental Models: Cell Lines | ||
| iPSC line 02iCTR_nTn1 | CSMC Stem Cell Core | CS02iCTR_nTn1 |
| iPSC line 03iCTR_nTn1 | CSMC Stem Cell Core | CS03iCTR_nTn1 |
| iPSC line 25iCTR_n5 | CSMC Stem Cell Core | CS25iCTR_n5 |
| iPSC line 80iCTR_Tn3 | CSMC Stem Cell Core | CS80iCTR_Tn3 |
| iPSC line 87iCTR_n3 | CSMC Stem Cell Core | CS87iCTR_n3 |
| iPSC line 201iCTR_Tn6 | CSMC Stem Cell Core | CS201iCTR_Tn6 |
| iPSC line 02iOBS_n4 | CSMC Stem Cell Core | CS02iOBS_n4 |
| iPSC line 03iOBS_n3 | CSMC Stem Cell Core | CS03iOBS_n3 |
| iPSC line 04iOBS_n9 | CSMC Stem Cell Core | CS04iOBS_n9 |
| iPSC line 77iOBS_n3 | CSMC Stem Cell Core | CS77iOBS_n3 |
| iPSC line 90iOBS_n1 | CSMC Stem Cell Core | CS90iOBS_n1 |
| Oligonucleotides | ||
| See Table S6 | N/A | N/A |
| Software and Algorithms | ||
| 2100 Expert Software | Agilent | N/A |
| ANNOVAR release 2015Dev14 | Wang et al. 2015 | N/A |
| Cell Specific Enrichment Analysis | Xu et al., 2014 | N/A |
| DESeq2 | Bioconductor | N/A |
| Dougherty pSI Package for R | CRAN | N/A |
| Genome Analysis Tool Kit (GATK) v.3.5 | Broad Institute | N/A |
| GraphPad | PRISM | N/A |
| Integrated Pathway Analysis | QIAGEN | N/A |
| Maestro AxIS Software v.2.0.2.11 | Axion Biosystems | N/A |
| MetaExpress | Molecular Devices | N/A |
| R / Rstudio | Comprehensive R Archive Network | N/A |
| R Scripts used in this study | Github:AndrewRGross | N/A |
| RSEM v.1.2.20 | Li and Dewey, 2011 | N/A |
| STAR v2.5.0 | Dobin et al., 2013 | N/A |
| Tissue Specific Enrichment Analysis | Dougherty et al., 2010 | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dhruv Sareen.
dhruv.sareen@cshs.org
Dhruv Sareen
Cedars-Sinai Medical Center
8700 Beverly Blvd.
Los Angeles, CA 90048
AHSP 8418
EXPERIMENTAL MODEL AND SUBJECT DETAILS
iPSC Model
iPSCs were obtained from Cedars-Sinai’s iPSC core and cultured in essential 8 (E8) or mTeSR medium prior to differentiation. Twelve lines were differentiated into hypothalamic neurons: seven lines from healthy individuals and five from individuals exhibiting extreme obesity. The sample number, N, here refers to the 12 iPSC lines studied, and was selected to use all five available cell lines from obese individuals, as well as an equivalent number of controls.
Adult Post-Mortem Brain Sections
Hypothalamic tissue from 6 post-mortem individuals was obtained as a control. The number of samples was selected to match the number of control and obese-patient-derived iPSC cell lines.
Subject Details
Obese iPSC lines included five females and two males. Control iPSC lines included three males and three females. Brain sections came from four males and two females. The gender of subjects was mixed to make use of available obese-patient-derived iPSC lines. Sex-specific analyses were performed but discerned no difference in results. With regards to sample size, obtained 5 OBS iPSC lines and hence matched that with 5 CTR iPSC lines. However, since we wished to include lines derived from a variety of parent cell types, we included 7 CTR iPSC lines overall.
| Subject | Type | Disease Condition | Sex | Age | Race | Parent cell type |
|---|---|---|---|---|---|---|
| 02iCTR_n1 | iPSC line | Non-obese | M | 51 | Unknown | PBMCs |
| 03iCTR_n1 | iPSC line | Non-obese | M | 34 | Unknown | PBMCs |
| 25iCTR_n2 | iPSC line | Non-obese | M | 76 | Unknown | Fibroblasts |
| 80iCTR_n3 | iPSC line | Non-obese | F | 84 | Unknown | PBMCs |
| 87iCTR_n3 | iPSC line | Non-obese | F | Unknown | Hispanic | Lymphoblastoid |
| 201iCTR_n6 | iPSC line | Non-obese | F | Unknown | Unknown | PBMCs |
| 688iCTR_n5 | iPSC line | Non-obese | M | Unknown | Unknown | Lymphoblastoid |
| 02iOBS_n4 | iPSC line | Obese | F | 45 | Caucasian | Lymphoblastoid |
| 03iOBS_n3 | iPSC line | Obese | M | 53 | Caucasian | Lymphoblastoid |
| 74iOBS_n9 | iPSC line | Obese | F | 36 | Caucasian | Lymphoblastoid |
| 77iOBS_n3 | iPSC line | Obese | F | 15 | Black | Lymphoblastoid |
| 90iOBS_n1 | iPSC line | Obese | F | 18 | Hispanic | Lymphoblastoid |
| 1662 | Brain sec. | Non-obese | M | 30 | Unknown | N/A |
| 1838 | Brain sec. | Non-obese | F | 97 | Caucasian | N/A |
| 1843 | Brain sec. | Non-obese | M | 78 | Caucasian | N/A |
| 1919 | Brain sec. | Non-obese | M | 61 | Caucasian | N/A |
| 2266 | Brain sec. | Non-obese | F | 53 | Caucasian | N/A |
| 2884 | Brain sec. | Non-obese | M | 84 | Caucasian | N/A |
METHOD DETAILS
Ethics Statement and Reprogramming to iPSCs
Human PBMCs were obtained from whole blood draws of healthy volunteers at were Cedars-Sinai under the auspices of the Cedars-Sinai Medical Center Institutional Review Board (IRB) approved protocol Pro00028662. The reprogramming of iPSC cell lines and differentiation protocols in the present study were carried out in accordance with the guidelines approved by Stem Cell Research Oversight committee (SCRO) and IRB, under the auspices of IRB-SCRO Protocols Pro00032834 (iPSC Core Repository and Stem Cell Program) and Pro00036896 (Sareen Stem Cell Program). Reprogramming and characterization of PBMCs and fibroblasts from skin punch biopsies and lymphoblastoid cell lines (LCL) was carried out as previously described (Barrett et al., 2014). We employed one clone each of 7 CTR lines and 5 OBS subjects.
Briefly reprogramming was performed with either the Cell Line Nucleofector Kit C (VACA-1004, Lonza) or B cell Nucleofector Kit (VPA-1001, Lonza) using 1.5 μg of each episomal plasmid (Addgene) expressing 7 factors: OCT4, SOX2, KLF4, L-MYC, LIN28, SV40LT and p53 shRNA (pEP4 E02S ET2K, pCXLE-hOCT3/4-shp53-F, pCXLE-hUL, and pCXLE-hSK). Patient derived cells (1 × 106 cells per nucleofection) were harvested, centrifuged at 1500rpm for 5 minutes, re-suspended in Nucleofector® Solution with the E-010 program. These nucleofected cells were plated on feeder-independent BD Matrigel growth factor-reduced Matrix (Corning/BD Biosciences, #354230) and cultures maintained at 20% O2 during the process. Cells were gradually transitioned to reprogramming media (RM) by adding 1 mL RM to the original respective culture media daily for the next 3 days to aid in attachment. Reprogramming media comprises of DMEM/F12, 1% NEAA, 1% GlutaMax, 1% N2, 2% B27, 0.5% Antibiotic-Antimycotic (GIBCO #15240-062), 0.1 μM β-mercaptoethanol, 100ng/ml bFGF (Peprotech), 1:1000 (~;1000 units) hLIF (Millipore, #LIF1010), 0.5 μM PD0325901 (Cayman Chemicals, #13034), 3 μM CHIR99021 (Tocris, #4423), 10 μM HA-100 (Santa Cruz Biotech, #203072), and 0.5 μM A-83-01 (Tocris, #2939). The cells were then maintained in RM 15 days with fresh media every other day. They were then gradually changed to chemically-defined mTeSR®1 medium between 17-20 days post-nucleofection. Individual iPSC colonies appeared between day 25-32 and those with best morphology were mechanically isolated, transferred onto 12-well plates with fresh Matrigel for further expansion and banking (Figure S1).
iPSC-derived hypothalamic neuron differentiation (iHTN)
For differentiation into iHTNs, iPSCs were accutase-treated and plated as single cells in 6-well Matrigel-coated plates at a density of approx. 1×106 cells/well in E8 medium with ROCK-inhibitorY27632 (10 μM; Stemgent). The next day iHTN differentiation was initiated by neuroectoderm differentiation by dual SMAD inhibition using LDN193189 (1 μM, Cayman) and SB431542 (10 μM, Cayman) and this treatment is carried on for 48 hours in an in-house neural differentiation medium (Table S2). The neural ectoderm specification from hiPSCs was achieved by dual SMAD inhibition with LDN193189, a BMP type I receptor inhibitor of activin receptor-like kinases (ALKs) ALK2 and ALK3, and SB431542, a transforming growth factor-beta 1 (TGF-β1) of ALK-4, −5 and −7 (Chambers et al., 2009). This was followed by Sonic hedgehog activation by Smoothened agonist SAG (1 μM, Tocris) and purmorphamine (PMN, 1 μM, Tocris) and Wnt signaling inhibition using IWR-endo (10 μM, Cayman) from Day 2 to day 9 to direct the cells toward ventral diencephalon with regular media change every 2 days. Shh activation augments a ventral diencephalon forebrain cell fate in the CNS (Bragina et al., 2010; Hu et al., 2016), while activation of Wnt/β-catenin signaling promotes a more posterior brain identity in the CNS. Subsequently, on day 2 after neural conversion of PSCs small molecule pathway modulators of sonic hedgehog (Shh) [smoothened agonist (SAG), purmorphamine (PMN), robotnikinin (R), and cyclopamine (Cy)] and Wnt//β-catenin signaling [CHIR99021 and IWR-1-endo] were screened in different permutations between day 2 and 8 of differentiation to identify a combination that augmented ventral diencephalon hypothalamic cell identity during post-neural ectoderm specification. The combination of Shh signaling – activated by treatment with SAG and PMN – and inhibition of Wnt/β-catenin signaling – via the addition of Wnt/β-catenin pathway blocker IWR-1-endo – promoted the greatest percent of hypothalamic progenitors, based on the observation of NK2 homeobox 1 (Nkx2.1) and homeobox protein orthopedia (Otp; hypothalamic neuron progenitor that specify neuropeptidergic neurons) immunopositive cells at day 9 post-iPSCs. Day 9 to day 14 the cells are slowly made to exit cell cycle using DAPT (10 μM, Cayman) in the presence of caudalizing agent retinoic acid (0.01 μM, Cayman). On day 14, the cells were treated with Accutase and re-plated onto laminin-coated plates in the presence of maturation medium containing brain-derived neurotrophic factor BDNF (10 ng/mL, Miltenyi) and maintained until day 40. Upon assessing for hypothalamic markers we detected Otp, Rax, NPY (neuropeptide Y; a secreted neuropeptide of the orexigenic NPY neurons), CART (cocaine amphetamine regulated transcript; a neuropeptide produced by the anorexigenic CART neurons), α-MSH (α-melanocyte stimulating hormone; a bioactive product of POMC producing neurons), NPYR (neuropeptide Y receptor Y2; receptor for NPY present), GhrR (ghrelin receptor; ghrelin response receptors present in ARC neurons) and MCH (melanin concentrating hormone; orexigenic hypothalamic peptide found in hypothalamic MCH neurons) (Figure 2B). For freezing, Day 20 neurons were kept frozen in CryoStor® (Stem cell technologies) and thawed when needed and cultured for a further 20 days with an average of 70 ± 9% viability (Table S5).
Adult hypothalamic tissue (aHT-UCIBB) processing
Brain Tissue
Frozen brain tissue was obtained from 6 autopsy donors, 4 males and 2 females, from the University of California, Irvine Brain Bank (UCIBB). Brains used in this study were obtained after informed consent of next-of-kin and with approval from the University of California, Irvine Institutional Review Board (IRB). Autopsy donors were primarily of Caucasian ethnicity, were 67 ± 24 (mean ± SD) years of age at the time of death, had an agonal factor status of 0, had their brains collected after a short postmortem interval (PMI) (13.3 ± 8.3 hr), and had no reported medical history of obesity. Approximately 0.6-0.75cm3 of the hypothalamic region was micro-dissected from each subject for these studies.
RNA Extractions and Quantification
Frozen brain tissue was manually homogenized with a plastic pestle in a 1.5ml microfuge tube, in the presence of 0.5ml Trizol, and was allowed to incubate at room temperature for 5min. The pestle was rinsed with an additional 0.5ml Trizol, for a total volume of 1ml. 0.2ml of chloroform was then added to the solution, which was vigorously shaken and allowed to incubate at room temperature for an additional 2min. Phase separation was performed by centrifugation (14,000×g, 4°C, 20min), and the aqueous phase containing the RNA was removed by pipette into a new RNase-free tube and was processed further using a column-based RNA extraction kit. RNA processing/cleanup was performed using the QIAGEN RNeasy Mini Kit, including the optional 15min incubation with the QIAGEN On-Column DNase, all according to the manufacturer’s instructions. RNA was eluted with 60 μL RNase-free water. RNA was quantified and qualified using the Agilent Total RNA 6000 Nano Kit and the Agilent 2100 Bioanalyzer and 2100 Expert Software. RNA Integrity Numbers (RIN) ranged between 5.3-8.1, with an average RIN of 6.6.
Immunofluorescence
Cells that were subject to immunofluorescence were first fixed using 4% paraformaldehyde (PFA) for 20 minutes and subsequently washed with PBS. After blocking the cells with 5% donkey serum (Millipore) with 0.2% Triton X-100 (Bio-rad) in PBS for a minimum of 2 hours, the cells were then treated with an appropriate concentration of relevant primary antibody combinations (1:250) overnight at 4°C. After thorough washing using PBS with 0.1% Tween-20, the cells are then treated with appropriate species-specific Alexa Fluor-conjugated secondary antibody combinations for 45 minutes (1:500). Hoechst stains were used to mark the nuclei and the cells were then visualized using appropriate fluorescent filters using ImageXpress Micro XLS (Molecular devices).
Quantitative PCR
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) and RNA (2 μg) was first DNase treated and reverse transcribed to cDNA with oligo(dT) using the Promega Reverse Transcriptase System (Promega). Reactions were performed in three replicates using SYBR Green master mix (Applied Biosystems) using primer sequences specific to each gene on a CFX384 Real Time system (Biorad). Each PCR cycle consisted of 95°C for 10 minutes, 95°C30 s – >58°C for 60 s, for 40 cycles, and 72°C for 5 minutes. Genes of interest were normalized to RPL13A.
Immunoblots
Cell pellets were collected and lysed (mammalian PER, Thermo scientific + 1× protease inhibitor cocktail, Thermo Scientific) and samples were prepared after protein quantification. We loaded about 15 μg protein per lane of a polyacrylamide gel (NuPAGE Novex 4%–12% Bis-Tris Protein Gels). Once the gels were resolved, they were transferred onto nitrocellulose membrane and subsequently blocked in 5% milk solution for a minimum of 2 hours. This was followed by a one-step i-Bind process which treated the membrane with primary antibody, washing and secondary antibody steps (Life technologies). We employed Li-Cor® IRDye secondary antibodies (680 and 800 wavelength infrared dyes) and detection of bands was carried out in a Li-Cor ODyssey CLx imager (Li-Cor).
RNA sequencing
Library construction was performed using the Illumina TruSeq Stranded mRNA library preparation kit. Total RNA samples were assessed for concentration using the Nanodrop 8000 (Thermo Scientific) and quality using the 2100 Bioanalyzer (Agilent). One microgram of total RNA per sample was used for poly-A mRNA selection using streptavidin-coated magnetic beads. cDNA was synthesized from enriched and fragmented RNA using reverse transcriptase (Super-Script II, Invitrogen) and random primers. The cDNA was further converted into double-stranded DNA, and the resulting dsDNA was enriched with PCR for library preparation. The PCR-amplified library was purified using Agencourt AMPure XP beads (Beckman Coulter Genomics). The concentration of the amplified library was measured with a NanoDrop spectrophotometer and an aliquot of the library was resolved on an Agilent 2100 Bioanalyzer. Sample libraries are multiplexed and sequenced on a NextSeq 500 platform (Illumina) using 75bp single-end sequencing. On average, 29.5 million reads were generated from each sample.
α-MSH ELISA
We employed the Cloud-Clone Corp. Enzyme-Linked Immunosorbent Assay (ELISA) kit to measure secreted levels of α-MSH from our cell supernatants. This competitive inhibition enzyme immunoassay technique uses a monoclonal α-MSH antibody pre-coated microplate which launches a competitive inhibition reaction between biotin-labeled α-MSH and the unlabeled α-MSH from standard and samples. An avidin conjugated Horseradish peroxidase helps identify the unbound sites within each sample and the substrate mediated color development is inversely proportional to the concentration of α-MSH in the sample. A log-log graph was used to quantify the amount of α-MSH present in each sample.
NPY ELISA
We employed the EMD Millipore Human Neuropeptide Y ELISA kit to measure secreted NPY levels. This is a sandwich ELISA which captures NPY in the sample by antihuman NPY IgG and immobilizes the resulting complex to wells of a microtiter plate coated with pre-titered amount of anchor antibodies. The captured NPY then binds to a second biotinylated antibody to NPY followed by conjugation of horseradish peroxidase to the immobilized biotinylated antibodies. NPY is quantified by monitoring horseradish peroxidase activities in the presence of the substrate 3,3′,5,5′-tetra-methylbenzidine. The enzyme activity is measured spectrophotometrically by the increased absorbency at 450 nm. The increase in absorbency is directly proportional to the amount of captured NPY.
Metabolic Phenotyping and Seahorse Respirometry Assay
The Seahorse XFe24 Extracellular Flux Analyzer (Seahorse Biosciences) was used to perform mitochondrial stress tests and obtain real-time measurements of oxygen consumption rate (OCR) in cells. iFGEs and iHTNS treated with or without EDCs were seeded in a 24-well Seahorse culture plate at a density of 10,000-15,000 cells/well on Day 14 of differentiation and allowed to differentiate on the plate thereon until Day 40 when the assay was performed. For analysis of OCR, culture media was first exchanged for Seahorse XF Base media (supplemented to 10mM Glucose, 1mM Pyruvate, and 1mM Glutamine, pH 7.4) and cells were allowed to equilibrate for 1 hour at 37°C in a non-CO2 incubator before performing the assay. Chemical reagents targeted to specific functions of the mitochondria (Sigma) were used at final concentrations as follows: 1 μM Oligomycin - an ATP synthase inhibitor, 1 μM (FCCP) carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone - an uncoupling agent, and a mixture of 0.5 μM antimycin A - a cytochrome C reductase inhibitor and 0.5 μM rotenone - a complex I inhibitor. Results were normalized to protein concentration determined by BCA assay (Thermo Scientific).
MEA measurements
We employed a 48-well microelectrode array (MEA) plate (Figure 4A) for measurement of neuronal firing in iHTNs. MEA plates were precoated with Laminin and day 14 iHTN progenitors were single-cell dissociated and plated where they were maintained in iHTN maturation medium. Each day since plating cells i.e day 14, measurements were obtained for 10 minutes spans until day 60. The recording conditions were at 37°C using the standard neural setting (Axion Biosystems Maestro Axis software version 2.0.2.11). Recorded spike number per electrode were averaged after disregarding noisy electrodes from analysis.
Exogenous peptide treatments
The neurons were treated with exogenous ghrelin (10nM) and leptin (100ng/ml) on days 33, 35 and 37 of iHTN differentiation and respective conditioned media was collected on days 35, 37 and 40, i.e., day 2,4 and 7 of treatment.
Whole Exome Sequencing: Library preparation and sequencing
Sequencing libraries were generated using Agilent SureSelect Human All Exon V4 kit (Agilent Technologies, CA, USA) which targeted 20,965 genes and 334,378 exons, following manufacturer’s recommendations. Briefly, a total amount of 1 μg genomic DNA per sample was fragmented by a M200 shearing system (Covaris, Massachusetts, USA) to generate 180-280bp fragments. After end-repair, A-tailing, ligation, PCR enrichment and hybridization with biotin labeled probes, library DNA were captured by magnetic beads with streptomycin, followed by indexing PCR. PCR Products were purified using AMPureXP system (Beckman Coulter, Beverly, USA) and quantified using Agilent high sensitivity DNA assay on an Agilent Bioanalyzer 2100 system. Sequencing libraries were loaded on a HiSeq X Ten (Illumina, San Diego, CA) with paired-end 2×150bp sequencing setting to obtain an average sequencing depth of ~43.5M reads. The sequence data have been deposited in the Sequence Read Archive (SRA) with the accession code PRJNA416010.
QUANTIFICATION AND STATISTICAL ANALYSIS
Neuron tracing and neurite length measurement
Cell quantification and morphological analysis was performed on images using MetaExpress. The number of cell types for a specific marker was determined as a percentage of Hoechst positive nuclei. For neuronal tracing Neurolucida was employed and the analysis of neurite length and the distance spanned by the primary neurite (tip of neurite to cell body) was measured in microns.
Data analysis for RNA seq
Raw reads obtained from RNA-Seq was aligned to the transcriptome using STAR (version 2.5.0) (Dobin et al., 2013)/ RSEM (version 1.2.20) (Li and Dewey, 2011) with default parameters, using a custom human GRCh38 transcriptome reference downloaded from https://www.gencodegenes.org, containing all protein coding and long non-coding RNA genes based on human GENCODE version 23 annotation. Differential expression was calculated using the DESeq2 module in R using expression counts. Gene expression for all other observations were normalized by sequencing depth to Transcripts per Million reads (TPM) (Tables S3 and S4). All R scripts used in analysis are available on Github at https://github.com/Sareen-Lab/HT-Neurons-in-Obesity. Raw reads were made available on NCBI’s GEO portal at accession number GSE95243.
Spearmen Correlation
The Spearman correlation is a measure of the similarity of the rank of two lists: lists of identical order produce a score of 1 and lists with no relationship in order produce a score of 0. Expression data was compared to references from the Genotype-Tissue Expression (GTEx) project database. The highest expressed 10,000 genes from each sample and reference were compared using a Spearman correlation in R using the rcorr function. Distant references were filtered out of the expression data for visual clarity. The resulting distance matrix was then exported as a boxplot. The entire R script used to generate these tables and the resulting heatmap can be found on Github at https://github.com/Sareen-Lab/HT-Neurons-in-Obesity/.
Bioinformatics (TSEA, CSEA)
We assessed the similarity of our samples to several human tissues and cell types using the Dougherty pSI tissue identification method (Dougherty et al., 2010; Xu et al., 2014) to identify tissue identity exhibited by the iHTNs. This method predicts a sample’s closest tissue identity by calculating the probability of each gene in each set occurring outside of a given tissue. The similarity between a sample and a set of reference cell or tissue is scored types based on the number of highly enriched genes in the sample which have been identified as having a statistically significant association with a given reference (Dougherty et al., 2010). First, a reference expression set is used to assign to each gene a Specificity Index based on the probability of a gene occurring outside of a given tissue (pSI). Since they represent a probability, they are abbreviated as pSI, and increase in significance as they get smaller. This method is performed by first scoring each gene in an expression dataset using the specificity index calculation. These scores represent the probability of a given gene occurring outside of a given tissue by chance. The output of the specificity index calculation is a specificity index table with the same dimensions as the original expression table, but in which expression values have been replaced with pSI values. Then, the pSI values for each gene in a queried set are used to predict the probability of such a set occurring by chance in each of the tissues included in the original reference set.
Scoring all genes in an expression table allows us to assemble list of genes with significant specificity index scores for each reference tissue included in the original expression table. These gene lists represent the genes which have been identified as uniquely representative of a given reference tissue. We can then compare the highest expressed genes in a sample against the gene lists for each reference using another algorithm developed along with the specificity index calculation. This comparison is repeated for each reference four times across four different stringency levels to determine the similarity of the sample against large gene lists of moderate specificity scores as well as small, tightly restrictive gene lists containing genes with very small specificity scores.
In subsequent publications, Dougherty et al. displayed the results of this comparison as a set of nested hexagons which display the comparison algorithm’s confidence in the color of the hexagons. This analysis can be performed instantly by submitting highly expressed genes to a form on the Dougherty lab website. The Dougherty lab website allows users to compare their samples to the Genotype-Transcription Expression project (GTEx) 2013 dataset, which they label as the Tissue Specific Expression Analysis (TSEA) and several transcriptomes taken from the BrainSpan Atlas of Developing Human Brain which they label as the Cell Specific Expression Analysis (CSEA). To compare our sample to the currently available GTEx dataset, we downloaded the updated data and repeated their pSI calculation using the R package pSI. We then compared the genes with the top 1000 median expression values among our iPSC derived neurons and the genes with the top 1000 median expression values among the adult hypothalamus brain sections we sequenced in R to the new pSI table we’d generated and plotted it in a manner like theirs using Cytoscape in Figure 3. The pSI table we generated and the template files for generating the associated hexagonal bullseye plots in cytoscape are included in supplemental data.
Additionally, we submitted our top expressed genes from both iPSC hypothalamus and brain hypothalamus to TSEA and CSEA. CSEA could not detect a signal from either of these gene lists or from the GTEx hypothalamus gene list, so we instead submitted each of these gene lists to TSEA and then submitted those genes which TSEA associated with the brain into CSEA.
Pathway Analysis using Integrated Pathway Analysis
Pathways which differed in their mean expression between the obese and control samples were identified using Integrated Pathway Analysis (IPA, QIAGEN). The submitted dataset included 8000 genes, as this was the maximum number of rows supported by IPA under our settings. These 8000 genes were selected by ranking all genes based on the p value associated with their differential expression. Fold changes were only included for the 1498 genes with a p value less than 0.1. The remainder were reassigned a log fold change as zero so that IPA would recognize their presence but conservatively assume no difference in expression between obese and control samples. The pathways detected and the fraction of genes with up, down, or indeterminate regulation were formatted in R and plotted in Cytoscape.
Each node in Figure 5D represents a pathway IPA predicted to be dysregulated. The size of the node correlates to the p value IPA assigned to the pathway. The pathways are arranged clockwise in ascending p value. The fraction of genes in the pathway which were found to be upregulated in our obese samples is reflected in the size of the red wedge. Downregulated genes are represented by the green wedge and genes which were detected but not conclusively up or downregulated are represented in gray. The number of genes common to any pair of nodes is reflected in the thickness of the line connecting those nodes.
Identification and analysis of Single Nucleotide Variants
The raw reads passed the quality control were aligned by BWA (version 0.7.8) to the human reference genome GRCh37, with an average coverage of 139.5X coverage on targeted regions. The Genome Analysis Toolkit (GATK) version 3.5 was used for variants calling according to GATK Best Practices recommendations (DePristo et al., 2011). Variants were filtered against public databases and annotated with ANNOVAR (release 2015Dev14) (Wang et al., 2010). A total of 48 genes potentially associated with obesity were summarized from OMIM Catalog (Term #601665, “Obesity”) (Hamosh et al., 2002) as well as recent studies (Hendricks et al., 2017). Mutations with 1) mapping coverage more than 30 independent reads, 2) predicted to affect protein function and 3) located within exon regions of obesity related genes were considered as significant candidates and summarized in Tables 1 and S1.
Statistical Analysis of Protein Expression
Experiments were performed using four independent donor-derived iPSC-derived hypothalamic neuroendocrine cultures (iHTNs). Multiple differentiations and treatments performed in independent experiments and are stated in each figure legend. All data are represented as mean ± s.e.m. p values < 0.05 were considered significant – * p < 0.05, ** p < 0.01 and *** p < 0.001. Statistical analyses were performed on GraphPad Prism using student’s unpaired t test (with or without Welch’s correction) or one-way Analysis of variance (ANOVA) and Bonferroni post-test for multiple comparisons.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is GEO: GSE95243. The accession number for the whole genome sequencing data reported in this paper is Sequence Read Archive: PRJNA416010. Any other data are available upon request. All software used is available either commercially or as freeware. All custom codes are available on GitHub at https://github.com/Sareen-Lab/HT-Neurons-in-Obesity/.
Supplementary Material
Highlights.
Obesity-associated coding variants are detected in super-obese hiPSCs
Super-obese iHTNs retain dysregulated obesogenic disease signatures
Impaired respiratory function and ghrelin-leptin signaling exist in super-obese iHTNs
Ghrelin and leptin exacerbate orexigenic response of super-obese iHTNs
ACKNOWLEDGMENTS
We would like to acknowledge Rebecca Hernandez for her contribution in conducting part ofthe hormone treatment experiments, ELISA, and immunocytochemistry (ICC). We would also like to acknowledge the donors that provided biospecimens tissue (blood, LCLs, and fibroblasts) for hiPSC generation, as well as the autopsy donors and their families for providing tissue to the UCI Brain Bank, without whom these studies would not have been possible. This work was supported by Cedars-Sinai institutional programmatic funds (D.S.) and in part by the NIH National Center for Advancing Translational Science (NCATS) UCLA CTSI grant UL1TR000124. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and six tables and can be found with this article online at https://doi.org/10.1016/j.stem.2018.03.009.
DECLARATION OF INTERESTS
A provisional patent application 62/354,04 describing methods and compositions for differentiation of iPSCs has been filed in the United States Patent and Trademark Office. Apart from the patent filing, the authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Andrews ZB, Liu ZW, Walllingford N, Erion DM, Borok E, Friedman JM, Tscho¨ p MH, Shanabrough M, Cline G, Shulman GI, et al. (2008). UCP2 mediates ghrelin’s action on NPY/AgRP neurons by lowering free radicals. Nature 454, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews ZB, Erion D, Beiler R, Liu Z-W, Abizaid A, Zigman J, Elsworth JD, Savitt JM, DiMarchi R, Tschoep M, et al. (2009). Ghrelin promotes and protects nigrostriatal dopamine function via a UCP2-dependent mitochondrial mechanism. J. Neurosci. 29, 14057–14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R, Ornelas L, Yeager N, Mandefro B, Sahabian A, Lenaeus L, Targan SR, Svendsen CN, and Sareen D (2014). Reliable generation of induced pluripotent stem cells from human lymphoblastoid cell lines. Stem Cells Transl. Med. 3, 1429–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson K, Orho-Melander M, Melander O, Lindblad U, Ranstam J, Råstam L, and Groop L (2001). Beta(2)-adrenergic receptor gene variation and hypertension in subjects with type 2 diabetes. Hypertension 37, 1303–1308. [DOI] [PubMed] [Google Scholar]
- Bragina O, Sergejeva S, Serg M, Zarkovsky T, Maloverjan A, Kogerman P, and Zarkovsky A (2010). Smoothened agonist augments proliferation and survival of neural cells. Neurosci. Lett. 482, 81–85. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Chen P-F, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, and Lalande M (2010). Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc. Natl. Acad. Sci. USA 107, 17668–17673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, and Studer L (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chen L, Sanseau P, Freudenberg JM, and Rajpal DK (2016). Significant obesity-associated gene expression changes occur in the stomach but not intestines in obese mice. Physiol. Rep 4, e12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll CU, Detraux J, De Lepeleire J, and De Hert M (2015). Effects of antipsychotics, antidepressants and mood stabilizers on risk for physical diseases in people with schizophrenia, depression and bipolar disorder. World Psychiatry 14, 119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DE, Clement K, Purnell JQ, Vaisse C, Foster KE, Frayo RS, Schwartz MW, Basdevant A, and Weigle DS (2002). Elevated plasma ghrelin levels in Prader Willi syndrome. Nat. Med 8, 643–644. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011). A frame-work for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Schmidt EF, Nakajima M, and Heintz N (2010). Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 38, 4218–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller HR, Mandefro B, Shirran SL, Gross AR, Kaus AS, Botting CH, Morris GE, and Sareen D (2016). Spinal Muscular Atrophy Patient iPSC-Derived Motor Neurons Have Reduced Expression of Proteins Important in Neuronal Development. Front. Cell. Neurosci 9, 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Naka I, Yamauchi T, Natsuhara K, Kimura R, Nakazawa M, Ishida T, Inaoka T, Matsumura Y, Ataka Y, et al. (2010). The Q223R polymorphism in LEPR is associated with obesity in Pacific Islanders. Hum. Genet 127, 287–294. [DOI] [PubMed] [Google Scholar]
- Gjesing AP, Andersen G, Burgdorf KS, Borch-Johnsen K, Jørgensen T, Hansen T, and Pedersen O (2007). Studies of the associations between functional β2-adrenergic receptor variants and obesity, hypertension and type 2 diabetes in 7,808 white subjects. Diabetologia 50, 563–568. [DOI] [PubMed] [Google Scholar]
- Gjesing AP, Sparsø T, Borch-Johnsen K, Jørgensen T, Pedersen O, Hansen T, and Olsen NV (2009). No consistent effect of ADRB2 haplotypes on obesity, hypertension and quantitative traits of body fatness and blood pressure among 6,514 adult Danes. PLoS ONE 4, e7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SFA, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. (2006). Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet 38, 320–323. [DOI] [PubMed] [Google Scholar]
- Grarup N, Urhammer SA, Ek J, Albrechtsen A, Gl€umer C, Borch-Johnsen K, Jørgensen T, Hansen T, and Pedersen O (2006). Studies of the relationship between the ENPP1 K121Q polymorphism and type 2 diabetes, insulin resistance and obesity in 7,333 Danish white subjects. Diabetologia 49, 2097–2104. [DOI] [PubMed] [Google Scholar]
- Gu M, Mordwinkin NM, Kooreman NG, Lee J, Wu H, Hu S, Churko JM, Diecke S, Burridge PW, He C, et al. (2015). Pravastatin reverses obesity-induced dysfunction of induced pluripotent stem cell-derived endothelial cells via a nitric oxide-dependent mechanism. Eur. Heart J 36, 806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha E, Kim M-J, Choi B-K, Rho J-J, Oh D-J, Rho T-H, Kim K-H, Lee HJ, Shin D-H, Yim SV, et al. (2006). Positive association of obesity with single nucleotide polymorphisms of syndecan 3 in the Korean population. J. Clin. Endocrinol. Metab 91, 5095–5099. [DOI] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, and McKusick VA (2002). Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 30, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastuti P, Zukhrufia I, Padwaswari MH, Nuraini A, and Sadewa AH (2016). Polymorphism in leptin receptor gene was associated with obesity in Yogyakarta, Indonesia. Egypt. J. Med. Hum. Genet 17, 271–276. [Google Scholar]
- Hayakawa T, Nagai Y, Kahara T, Yamashita H, Takamura T, Abe T, Nomura G, and Kobayashi K (2000). Gln27Glu and Arg16Gly polymorphisms of the beta2-adrenergic receptor gene are not associated with obesity in Japanese men. Metabolism 49, 1215–1218. [DOI] [PubMed] [Google Scholar]
- Hendricks AE, Bochukova EG, Marenne G, Keogh JM, Atanassova N, Bounds R, Wheeler E, Mistry V, Henning E, Korner A, et al. ; Understanding Society Scientific Group; EPIC-CVD Consortium; UK10K Consortium (2017). Rare Variant Analysis of Human and Rodent Obesity Genes in Individuals with Severe Childhood Obesity. Sci. Rep 7, 4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Qu ZY, Cao SY, Li Q, Ma L, Krencik R, Xu M, and Liu Y (2016). Directed differentiation of basal forebrain cholinergic neurons from human pluripotent stem cells. J. Neurosci. Methods 266, 42–49. [DOI] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, et al. (2012). Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 482, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalba MS, Rhoads GG, and Demissie K (2008). Association of codon 16 and codon 27 beta 2-adrenergic receptor gene polymorphisms with obesity: a meta-analysis. Obesity (Silver Spring) 16, 2096–2106. [DOI] [PubMed] [Google Scholar]
- Jin T, and Liu L (2008). The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol. Endocrinol. 22, 2383–2392. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Hannan KS, Greenberg ML, and Friedman JM (2010). Hyperleptinemia is required for the development of leptin resistance. PLoS ONE 5, e11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs P, Hanson RL, Lee Y-H, Yang X, Kobes S, Permana PA, Bogardus C, and Baier LJ (2003). The role of insulin receptor substrate-1 gene (IRS1) in type 2 diabetes in Pima Indians. Diabetes 52, 3005–3009. [DOI] [PubMed] [Google Scholar]
- Lee JH, Reed DR, and Price RA (2001). Leptin resistance is associated with extreme obesity and aggregates in families. Int. J. Obes. Relat. Metab. Disord 25, 1471–1473. [DOI] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. ; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium (2015). Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. ; GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Zhang L, Sun J, Cai Z, Gu Q, Zhang R, and Shan A (2017). Interaction between peroxisome proliferator-activated receptor gamma polymorphism and obesity on type 2 diabetes in a Chinese Han population. Diabetol. Metab. Syndr 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon HN, Florez JC, Bersaglieri T, Saxena R, Winckler W, Almgren P, Lindblad U, Tuomi T, Gaudet D, Zhu X, et al. (2006). Common variants in the ENPP1 gene are not reproducibly associated with diabetes or obesity. Diabetes 55, 3180–3184. [DOI] [PubMed] [Google Scholar]
- MacLean PD (1968). The human brain in figures and tables: A quantitative handbook, by S. M. Blinkov and I. I. Glezer. Science 161, 454–455. [Google Scholar]
- Merkle FT, Maroof A, Wataya T, Sasai Y, Studer L, Eggan K, and Schier AF (2015). Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development 142, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AC, Floriano MS, Mota GFA, Cunha RS, Herkenhoff FL, Mill JG, and Krieger JE (2003). Beta2 adrenoceptor functional gene variants, obesity, and blood pressure level interactions in the general population. Hypertension 42, 685–692. [DOI] [PubMed] [Google Scholar]
- Reinhardt P, Schmid B, Burbulla LF, Scho¨ ndorf DC, Wagner L, Glatza M, Höing S, Hargus G, Heck SA, Dhingra A, et al. (2013). Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell 12, 354–367. [DOI] [PubMed] [Google Scholar]
- Ren X, Zhou L, Terwilliger R, Newton SS, and de Araujo IE (2009). Sweet taste signaling functions as a hypothalamic glucose sensor. Front. Integr. Neurosci 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojano-Rodriguez ME, Beristain-Hernandez JL, Zavaleta-Villa B, Maravilla P, Romero-Valdovinos M, and Olivo-Diaz A (2016). Leptin receptor gene polymorphisms and morbid obesity in Mexican patients. Hereditas 153, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YP, Liu F, Zeng LC, Ma WJ, Wei DZ, and Han ZG (2002). Cloning and characterization of a novel human secretory protein: secretogranin III. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 34, 411–417. [PubMed] [Google Scholar]
- Sareen D, Ebert AD, Heins BM, McGivern JV, Ornelas L, and Svendsen CN (2012). Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS ONE 7, e39113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpace PJ, and Zhang Y (2007). Elevated leptin: consequence or cause of obesity? Front. Biosci 12, 3531–3544. [DOI] [PubMed] [Google Scholar]
- Toda C, and Diano S (2014). Mitochondrial UCP2 in the central regulation of metabolism. Best Pract. Res. Clin. Endocrinol. Metab 28, 757–764. [DOI] [PubMed] [Google Scholar]
- Utiger R (2017). Hypothalamus (Encycl. Brittanica, Inc.). [Google Scholar]
- Wang K, Li M, and Hakonarson H (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Meece K, Williams DJ, Lo KA, Zimmer M, Heinrich G, Martin Carli J, Leduc CA, Sun L, Zeltser LM, et al. (2015). Differentiation of hypothalamic-like neurons from human pluripotent stem cells. J. Clin. Invest 125, 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wataya T, Ando S, Muguruma K, Ikeda H, Watanabe K, Eiraku M, Kawada M, Takahashi J, Hashimoto N, and Sasai Y (2008). Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc. Natl. Acad. Sci. USA 105, 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, and Dougherty JD (2014). Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J. Neurosci. 34, 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, and Suzuki N (2011). Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum. Mol. Genet. 20, 4530–4539. [DOI] [PubMed] [Google Scholar]
- Zhao T, Liu Z, Zhang D, Liu Y, Yang Y, Zhou D, Chen Z, Yu L, Zhang Z, Feng G, et al. (2011). The ENPP1 K121Q polymorphism is not associated with type 2 diabetes or obesity in the Chinese Han population. J. Hum. Genet. 56, 12–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is GEO: GSE95243. The accession number for the whole genome sequencing data reported in this paper is Sequence Read Archive: PRJNA416010. Any other data are available upon request. All software used is available either commercially or as freeware. All custom codes are available on GitHub at https://github.com/Sareen-Lab/HT-Neurons-in-Obesity/.