Abstract
Asthma is the most common chronic disease in children. Inhaled corticosteroids (ICS) are the first-line treatment for asthma control, but up to one-third of children have a poor treatment response. The mechanism of ICS resistance is poorly understood, and the role of DNA methylation in ICS treatment response is not known. We examined the association between peripheral blood DNA methylation and ICS treatment response in 152 pediatric persistent asthmatics from the Childhood Asthma Management Program. Response to ICS was measured by the percentage change in forced expiratory volume in one second (FEV1) eight weeks after treatment initiation. The top CpG sites with a nominal P < 0.001 were correlated with gene expression using Pearson and partial correlations. In 152 subjects, mean age was 9.8 (± 2.0) years and median change in FEV1 after ICS initiation was 4.6% (± 10.4%). A total of 545 CpG sites were differentially methylated (nominal P < 0.05), and seven CpG sites had a nominal P < 0.001. Relative hypermethylation of cg20434811, cg02822723, cg14066280, cg27254601, and cg23913400 and relative hypomethylation of cg24937126 and cg24711626 were associated with an increase in FEV1 on ICS treatment. One CpG site was associated with gene expression. Relative hypermethylation of cg27254601 was associated with both an increase in FEV1 and BOLA2 expression (ρ = 0.25, P = 0.02). We identified a novel association between BOLA2 methylation, gene expression, and ICS response as measured by lung function. Pharmaco-epigenetics has the potential to detect treatment sensitivity in persistent childhood asthma.
Keywords: epigenetics, pharmacogenetics, DNA methylation, pediatric asthma, inhaled corticosteroids, lung function, FEV1
INTRODUCTION
Asthma is the most common chronic disease in infants and children[1]. Poorly controlled asthma leads to significant morbidity and mortality and is the most frequent cause of emergency department visits, hospitalizations, and school absenteeism in children[2]. The first-line treatment for asthma control is inhaled corticosteroids (ICS). However, up to one-third of children have a poor response to ICS[3]. The mechanism of ICS resistance is not well understood, and currently, no reliable methods exist to predict ICS sensitivity.
DNA methylation has been shown to regulate both asthma susceptibility and severity in addition to dexamethasone sensitivity and resistance[4–6]. A pilot study on DNA methylation and oral corticosteroid response found a nominal association between OTX2 promoter methylation in nasal epithelial cells and oral corticosteroid response in pediatric asthmatics[7]. The association between DNA methylation and ICS response has not yet been investigated.
To better understand the pharmaco-epigenomics of ICS sensitivity and resistance, we examined white blood cell DNA methylation in pediatric persistent asthmatics from the Childhood Asthma Management Program (CAMP). We identified differentially methylated CpG sites associated with ICS response as defined by improvement in lung function in the first eight weeks of treatment and correlated differential methylation to gene expression.
METHODS
CAMP was a four-year randomized controlled trial of three inhaled treatments for mild-to-moderate persistent childhood asthma[8]. Between December 1993 and September 1995, 1041 children aged 5 to 12 years were randomized to receive inhaled budesonide, nedocromil, or placebo. DNA methylation was measured in 554 subjects, of whom 163 subjects were on inhaled budesonide. All 163 subjects were of non-Hispanic white ethnicity, and 152 subjects remained after removal of sex mismatches and outliers. The study was approved by the institutional review boards of Brigham and Women’s Hospital and each of the CAMP study centers. Written informed consent with subject assent was provided by all parents.
Inhaled corticosteroid response
The outcome of interest was ICS response as measured by the percentage change in forced expiratory volume in one second (FEV1) eight weeks after initiating inhaled budesonide treatment. Longitudinal lung function measurements in the CAMP cohort enabled assessment of this outcome.
DNA methylation
DNA was extracted from whole blood using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) and bisulfite converted using the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA) followed by PCR amplification. DNA methylation was measured using the Infinium HumanMethylation27 BeadChip assay (Illumina, San Diego, CA), which quantified 27,578 CpG sites, and 17,078 CpG sites remained after removal of CpG sites on sex chromosomes, SNPs, indels, and repeats. Quantile normalization was performed, and DNA methylation beta values were converted to M values to correct for heteroscedasticity[9]. The percentages of white blood cell counts measured from whole blood were used to adjust for white blood cell composition in subsequent statistical models. Data analyses were performed using R software (version 3.4.0).
Gene Expression
Gene expression data in CAMP were generated through Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE) Consortium[10, 11]. Gene expression was measured from whole blood using the Illumina HumanHT-12 v4 Expression BeadChip Kit (Illumina, San Diego, CA) in 448 subjects, and 107 subjects had both DNA methylation and gene expression data.
Statistical analyses
Differential DNA methylation was investigated using multivariable linear regression from the iCheck package[12]. Serum IgE level, eosinophil count, vitamin D level, history of atopy, parental history of asthma, and history of parental smoking were selected a priori as potential confounders. The univariate association between each potential confounder and ICS response was examined, and the statistically significant confounders were controlled for in the multivariable linear regression model. Age, sex, batch effect, clinic site, and white blood cell composition were also adjusted for in the multivariable model. The FDb.InfiniumMethylation.hg19 package was used to annotate the DNA methylation probes to the Genome Reference Consortium Human Build 37, including both genes in cis and trans[13]. For the top CpG sites that met a nominal P value < 0.001, the association between DNA methylation and gene expression was examined by using Pearson and partial correlations. In the partial correlation analysis, DNA methylation was adjusted for the same covariates described above, and gene expression was adjusted for age, sex, and batch effect.
RESULTS
In 152 subjects, the mean age was 9.8 (± SD 2.0) years, and the median change in FEV1 after ICS initiation was 4.6% (± IQR 10.4%) (Table 1). The majority of subjects were male (57.9%) and did not have a history of atopy, parental asthma, or parental smoking. A total of 545 CpG sites were differentially methylated (nominal P < 0.05) in association with an increase in FEV1 on ICS treatment (see Table, Supplemental Digital Content 1, http://links.lww.com/FPC/B340).
Table 1.
Baseline demographics for the Childhood Asthma Management Program subjects on inhaled corticosteroids (ICS) with DNA methylation data
| Study population (n = 152) |
|
|---|---|
| Age (mean ± SD) | 9.8 (± 2.0) |
| Age at asthma onset (median ± IQR) | 2.0 (± 3.0) |
| Sex | |
| Female | 64 (42.1%) |
| Male | 88 (57.9%) |
| FEV1 percentage predicted (mean ± SD) | 94.3 (± 13.1) |
| FEV1/FVC (absolute percentage) (mean ± SD) | 79.5 (± 8.5) |
| Percentage change in FEV1 eight weeks after ICS initiation (median ± IQR) | 4.6 (± 10.4) |
| History of atopy | 41 (27.0%) |
| History of parental asthma | 68 (46.9%) |
| History of parental smoking | 51 (33.8%) |
| Serum IgE level (ng/mL) (median ± IQR) | 394.0 (± 878.5) |
| Serum eosinophil count (cells/μL) (median ± IQR) | 400.0 (± 410.0) |
| Serum vitamin D level (ng/mL) (mean ± SD) | 43.3 (± 14.5) |
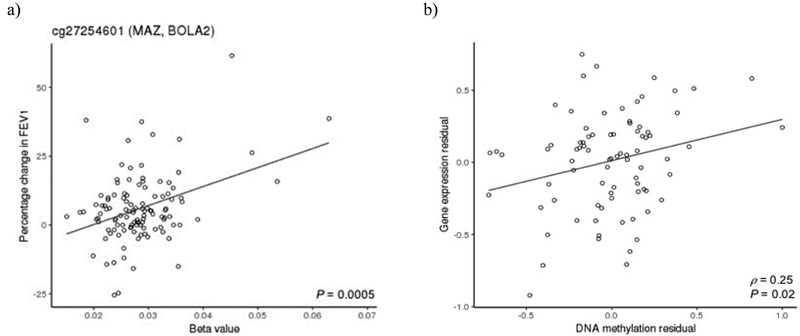
The association between DNA methylation and gene expression was examined for the top CpG sites (P < 0.001). One of the CpG sites (cg27254601) was associated with gene expression (Figure 1). Cg27254601 is located in the intronic region of BOLA2, a gene in cis on chromosome 16. Relative hypermethylation of cg27254601 was associated with increased BOLA2 expression (ρ = 0.25, P = 0.02).
Figure 1.
cg27254601 is associated with improved lung function on inhaled corticosteroids and BOLA2 expression
a) Relative hypermethylation of cg27254601 is associated with an improvement in forced expiratory volume in one second (FEV1) in pediatric asthmatics after initiating inhaled corticosteroids. b) Partial correlation between DNA methylation and gene expression controlling for percentage change in FEV1 after initiating inhaled corticosteroid therapy, age, sex, batch effect, clinic site, and lymphocyte counts (ρ = 0.25, P = 0.02).
DISCUSSION
ICS are the most effective treatment for asthma control, yet the markers and mechanisms of ICS resistance are poorly understood. We identified DNA methylation marks associated with improvements and decrements in lung function in childhood asthmatics on ICS. Moreover, one of the differentially methylated CpG sites was associated with gene expression, indicating a potential functional mechanism by which DNA methylation influences lung function. Relative hypermethylation of cg27254601 was associated with an increase in FEV1 and BOLA2 expression. Therefore, our results suggest a link between DNA methylation and ICS response as measured by lung function.
BOLA2 encodes the BolA Family Member 2 protein which complexes with monothiol glutaredoxin-3 to form a cellular iron-sulfur chaperone that aids in the maturation of iron-sulfur containing proteins[14]. BOLA2 is differentially expressed in airway epithelial cells and in asthmatic subjects[15–17]. Respiratory syncytial virus infection in human bronchial and small airway epithelial cells upregulates BOLA2 expression[15]. In addition, severe adult asthmatics have lower BOLA2 expression in peripheral blood compared to non-asthmatics[16]. Preliminary evidence also indicates an association between BOLA2 expression and childhood asthma in high air pollution regions[17]. We demonstrated that epigenetic regulation of BOLA2 is associated with ICS treatment response in pediatric asthmatics, and the mechanism linking BOLA2 expression and lung function in childhood asthma should be investigated in future studies.
DNA methylation of the glucocorticoid receptor gene (NR3C1) regulates glucocorticoid sensitivity in endothelial cells[6]. We did not find an association between NR3C1 methylation and ICS response, perhaps due to phenotypic and/or cell-type differences. In a study on the response to oral corticosteroids, nasal epithelial cell OTX2 is hypermethylated at baseline in pediatric asthmatic responders compared to non-responders[7]. Notably, we also found relative hypermethylation of OTX2 to be nominally associated with a good response to ICS, as evidenced by an improvement in FEV1 (standardized coefficient 2.123, P = 0.04) (see Table, Supplemental Digital Content 1, http://links.lww.com/FPC/B340). Currently, it is not known if inhaled corticosteroids administered locally change the DNA methylome in peripheral blood. Thus, our results in peripheral blood potentially replicate the OTX2 findings from the upper airway, and studies that simultaneously evaluate the DNA methylome from airway epithelium and peripheral blood are needed.
Repeated longitudinal measurements of lung function were obtained in CAMP, and studies with this phenotype combined with DNA methylation data are rare. Therefore, a suitable replication population does not currently exist. This study was limited by the relatively small sample size of ICS treated subjects and the use of the Illumina HumanMethylation27 assay. Our analyses focused on peripheral blood DNA methylation and gene expression because these measures are non-invasive and can be influenced by systemic asthma inflammation and ICS absorption. Cell type convolution was performed using white blood cells measured concurrently with DNA methylation[18]. Lymphocyte specific DNA methylation was not performed because asthma and ICS affect all lymphocyte types. Despite these limitations, we were able to detect novel DNA methylation associations and strengthen our findings with relevant gene expression data.
In summary, we identified a novel association between BOLA2 methylation, gene expression, and ICS response. Relative hypermethylation of BOLA2 was associated with increased gene expression and improvement in FEV1 in childhood asthmatics on ICS. We also report the potential validation of the association between OTX2 hypermethylation and corticosteroid response. Our results demonstrate the potential for DNA methylation to detect asthma treatment sensitivity and resistance. The underlying causes of DNA methylation and its downstream effects on ICS response are important future topics of research. Further understanding of pharmaco-epigenetics in asthma may identify new epigenetic biomarkers, mechanisms of treatment resistance, and treatments to complement current therapies.
Supplementary Material
Acknowledgments
Conflicts of Interest and Source of Funding: No conflicts of interest declared. Funding: NIH T32 AI007306, T32 HL007427, RC2 HL101543, R01 HL127332, R01 HL129935, U01 HL65899, P01 HL132825.
REFERENCES
- 1.Gold DR, Wright R. Population disparities in asthma. Annu Rev Public Health. 2005;26:89–113. [DOI] [PubMed] [Google Scholar]
- 2.Akinbami LJ, Moorman JE, Liu X. Asthma prevalence, health care use, and mortality: United States, 2005–2009. Natl Health Stat Report. 2011(32):1–14. [PubMed] [Google Scholar]
- 3.Szefler SJ, Martin RJ, King TS, Boushey HA, Cherniack RM, Chinchilli VM, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002;109(3):410–8. [DOI] [PubMed] [Google Scholar]
- 4.Xu CJ, Soderhall C, Bustamante M, Baiz N, Gruzieva O, Gehring U, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6(5):379–88. [DOI] [PubMed] [Google Scholar]
- 5.Gaffin JM, Raby BA, Petty CR, Hoffman EB, Baccarelli AA, Gold DR, et al. beta-2 adrenergic receptor gene methylation is associated with decreased asthma severity in inner-city schoolchildren: asthma and rhinitis. Clin Exp Allergy. 2014;44(5):681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mata-Greenwood E, Jackson PN, Pearce WJ, Zhang L. Endothelial glucocorticoid receptor promoter methylation according to dexamethasone sensitivity. J Mol Endocrinol. 2015;55(2):133–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Biagini Myers JM, Yadagiri VK, Ulm A, Chen X, Weirauch MT, et al. Nasal DNA methylation differentiates corticosteroid treatment response in pediatric asthma: A pilot study. PLOS ONE. 2017;12(10):e0186150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 9.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF, Jr., Liu AH, et al. Gene Expression Profiling in Blood Provides Reproducible Molecular Insights into Asthma Control. Am J Respir Crit Care Med. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raby B, Barnes K, Beaty TH, Bosco A, Carey VJ, Castro M, et al. Asthma Bridge: the Asthma Biorepository For Integrative Genomic Exploration. Am J Respir Crit Care Med. 2011;183:A6189. [Google Scholar]
- 12.Qiu W, Guo B, Anderson C, Klanderman B, Carey V, Raby B. iCheck: QC pipeline and data analysis tools for high-dimensional Illumina mRNA expression data. 2016:R package version 1.6.0. [Google Scholar]
- 13.Triche JT. FDb.InfiniumMethylation.hg19: annotation package for Illumina Infinium DNA methylation probes. 2014. R package version 2.2.0. [Google Scholar]
- 14.Frey AG, Palenchar DJ, Wildemann JD, Philpott CC. A Glutaredoxin.BolA Complex Serves as an Iron-Sulfur Cluster Chaperone for the Cytosolic Cluster Assembly Machinery. J Biol Chem. 2016;291(43):22344–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Jamaluddin M, Zhang Y, Sun H, Ivanciuc T, Garofalo RP, et al. Systematic Analysis of Cell-Type Differences in the Epithelial Secretome Reveals Insights into the Pathogenesis of Respiratory Syncytial Virus-Induced Lower Respiratory Tract Infections. J Immunol. 2017;198(8):3345–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigler J, Boedigheimer M, Schofield JPR, Skipp PJ, Corfield J, Rowe A, et al. A Severe Asthma Disease Signature from Gene Expression Profiling of Peripheral Blood from U-BIOPRED Cohorts. Am J Respir Crit Care Med. 2017;195(10):1311–20. [DOI] [PubMed] [Google Scholar]
- 17.Choi H, Song W, Sram R, Zhang B. Ambient polycyclic aromatic hydrocarbons, gene expression, DNA methylation, and childhood asthma [abstract] In: Abstracts of the 2016 ISEE; 2016 Sep 1–4; Rome, Italy: Research Triangle Park (NC): Environmental Health Perspectives; 2016. [Google Scholar]
- 18.Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017;9(4):539–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.