Excitement grows for the advancement of science to improve human health outcomes in the emerging precision health era. For nurse scientists and nursing science, the opportunities of this era can be realized through careful attention to the philosophical and theoretical underpinnings of “precision science,” as well as the application of rigorous processes necessary to support the development of more accurate diagnostics and targeted treatments. Currently, there are multiple innovative initiatives that are important to nursing science, with specific overlaps in aims of scientific efforts, as well as in the population in which they are focused, to ultimately improve the care of patients. Consequently, members of an emerging pain and symptom science biomarker consortium, hereafter identified as the consortium, identified a number of scientific opportunities to develop additional insights into the clinical considerations related to pain, and utilized a collaborative framework to address the challenges for developing the next generation of precision symptom science across individuals and populations with pain. In this article, we outline several of the inherent challenges and opportunities for nursing science in the precision health era and detail the first stages of building a collaboration among nurse scientists with a focus on devising a shared theoretical perspective based on enhancements in our understanding of the biological factors associated with pain in a number of clinical conditions. We also describe several of the opportunities associated with building consortia that include NIH funded extramural and intramural scientists, as well as the foundational issues needed to allow for progress in addressing clinically relevant issues such as pain.
Commentary
The genesis and development of the consortium arose from discourse among colleagues, across academic institutions and disciplines, focused on understanding the common symptoms and pathways that we examine in a variety of different populations, including both human participants as well as animal models of pain and related symptoms. Individually, all of the investigators involved in the consortium have led studies focused on pain and other symptoms, however, have done so within the lens of their chosen patient population. Although this work has led to successful outcomes including novel interventions for managing symptoms, funded grant applications, and multiple publications, we realized that more scalable outcomes may require a different lens for analyzing biological and symptom measures collected from completed studies and/or designing prospective studies. In essence, we hypothesized that using existing data would allow for greater insights when combined through a collaborative network of investigators. This is also important for generating the data sources and sample sizes needed for complex symptom phenotypes across disease states and diverse populations in future research endeavors. Many nurse scientists were taught to focus intently and deeply on a specific scientific problem; however, we realized that a broader focus may also be important for contributing to the next generation of science, consistent with the goals and aspirations of the “precision” science era.
Symptom science is a prominent research focus for many extramural and intramural nurse scientists. The ultimate goals of this field of research are to be able to precisely identify individuals at risk for symptoms and develop targeted strategies to prevent or mitigate the severity of symptoms. However, most symptom science has been focused on population-specific symptoms, such as fatigue in cancer, acute and chronic pain, or multiple concurrent symptoms in a disease-bound model, with the notion that reducing heterogeneity will facilitate the interpretation of the resulting data. Less research has been done to examine symptoms in a disease-agnostic manner across conditions and populations, with the goal of identifying changes that are common to a variety of situations, suggesting that these alterations may drive the genesis/maintenance of the symptom(s).
For these reasons, we initially discussed prevalent symptoms across our populations of interest and decided to first focus on pain, a vexing, complex, and costly symptom on several levels. The direct medical costs and lost wages from pain are higher than those for cancer, heart disease, and diabetes combined (Pizzo, Clark, & Pokras, 2011). We then discussed the biological pathways that we have considered as priority areas in studying the transition from acute to chronic pain. Whether pain is classified as nociceptive, neuropathic, or inflammatory, there is evidence that there are similar cellular responses that contribute to sensitization, most notably through immune cell activation, suggesting that there may be an underlying common mechanism that contributes to multiple types of pain (Marchand, Perretti, & McMahon, 2005). The idea that the immune system is involved in a wide array of human diseases and symptoms is not new. However, the capabilities for tracing the biological signature of a symptom such as pain to a precise pathophysiological process involving the immune system may open the door to a deeper understanding of the transition from acute to chronic symptoms and, perhaps, novel interventional strategies. As we discussed our ideas with increasing excitement about working together in a group endeavor, we realized that before moving to the implementation phase of merging measures from completed studies and designing future studies, we needed to step back and consider how to ground our collaboration in the scientific paradigm of precision science. Here, we outline some of the issues that we have faced in the beginning stages of the consortium that may have implications for nurse scientists in developing collaborations to support the design and implementation of future studies grounded in precision science principles.
First, consortium members determined our broad focus on symptom science within the emerging “precision” initiatives. The multiple ways in which the term “precision” has been used to initiate a discussion of some important distinctions in the various uses of this term, with the biggest differences being between “precision medicine” and “precision population health/precision public health.” While presently there are multiple definitions, the original focus of precision medicine was aimed at large-scale methods for characterizing molecular measures (such as genomics, proteomics, metabolomics, microbiomics) across large samples and populations to enable “personalized” treatment to each individual through a deductive scientific process (Collins & Varmus, 2015). Precision population health is more broad and generalizable, joining biological with social and environmental determinants of health to develop precision approaches to interventions in individuals and populations (Khoury, Iademarco, & Riley, 2016). The definitions become important when distilling the theoretical as well as the empirical grounding of the consortium so that we share a common lens for examining pain and other distressing co-morbid symptoms in a manner that represents a comprehensive theoretical scientific view of the symptom.
Nurse scientists, given the focus on the environment that harkens back to Nightingale (Nightingale, 1992), have a theoretical perspective that includes factors more broad than the biological basis, and this perspective includes a focus on both individual and aggregate outcomes and prevention as well as treatment. Although the integration of laboratory and human research is beginning to collectively increase, understanding of the contribution of genome and other ‘omic variation to human disease is not yet fully established. Moreover there is less focus on environmental factors to understand risk, pathogenesis, prevention, or resistance to disease (Pickler, 2018). Given that humans undergo time-dependent changes that are influenced by genetic, environmental, and genotype-by-environment interactions, these multiple areas are not well-represented in model systems and present challenges. These complex relationships provide opportunities for nurse scientists, who work with individuals across the life-span, to collect data to more comprehensively understand the basis for health, and disease. Similarly, much of the current precision medicine initiative focuses on better understanding pathology of disease instead of possible salutary aspects of health. Likewise, other variables such as social determinants of health, exposures, and life-style habits that are known to impact health and health disparities have been less discussed than “omics” in the precision era, with less known about the interaction of social health determinants and “omics.” After many lively discussions, we agreed that there was a need to further refine our focus on precision symptom science with a perspective that includes not only the “omic” but also the social, societal, and environmental determinants of heath. The ultimate purpose of this initiative is to improve the health of patients through a more comprehensive understanding of the factors that interact to contribute to pain, with this evidence being essential to develop precision-based interventions to prevent, as well as treat pain.
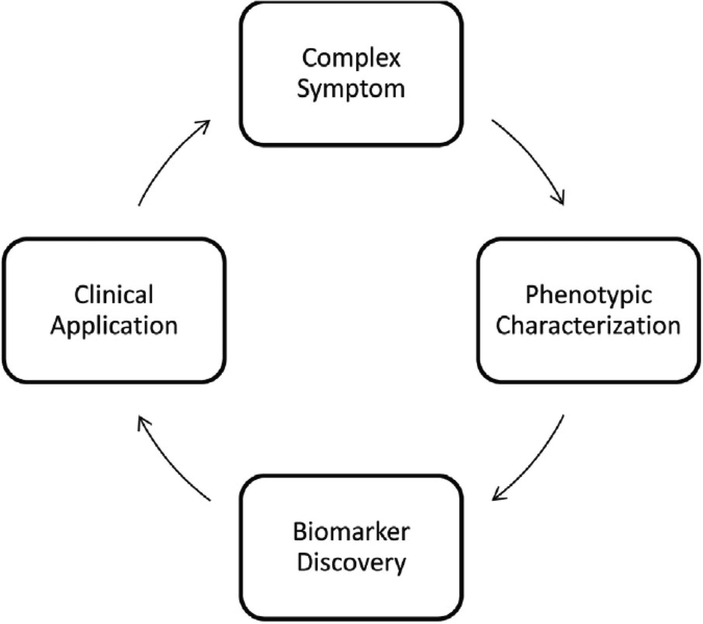
Next, we focused on choosing a conceptual model to ground our work. The National Institute of Nursing Research (NINR) developed the National Institutes of Health (NIH) Symptom Science Model (SSM) to guide this type of research (Cashion & Grady, 2015). The model begins by identifying a complex symptom, which is then characterized into a phenotype with biological and clinical data, followed by the application of genomic and other discovery methodologies to illuminate targets for therapeutic and clinical interventions (Figure 1). Our group chose this model for its parsimony, simplicity, and its direct relevance for our group’s research focus as it describes an investigative sequence to address the critical issue of pain. As we focused on the first step of our model, identifying a complex symptom, we encountered one of the vexing issues for symptom research: how to decide upon that definition the captures the components of that symptom, as well as a reliable and valid measure of how to determine the presence and severity of the symptom, which in our case is pain.
Figure 1.
The National Institute of Nursing Research Symptom Science Model. Adapted from “the National institutes of health/national institutes of nursing research intramural research program and the development of The National institutes of health symptom science model” by A. K. Cashion & P. A. Grady, 2015, Nursing Outlook, 63(4), p. 485. Copyright 2015 by the National institutes of nursing research.
First, we compared the different methods we used to rigorously phenotype pain across studies of patients with differing types of pain. We each shared a list of the measures that were used and selected common data elements (CDEs) to compare across conditions. CDEs are standardized instruments that can be used across studies to measure variables of interest. Symptom CDEs have been developed through the PROMIS initiative, with several recommended for use in NINR-funded studies, including anxiety, depression, fatigue, cognitive impairment and pain (Eckardt et al., 2017). While CDEs allow investigators to compare symptom characteristics across different populations, they may lack specificity in detecting important disease-specific symptom phenotypes (Cohen, Thompson, Yates, Zimmerman, & Pullen, 2015). Several investigators on our team also used quantitative sensory testing, and we compared our protocols to ensure that all procedures were carried out consistently across the studies. Since our first goal was to harmonize our datasets across different populations, which were collected independently, we needed to compromise by using the instruments that were most often used, and then normalize the scales to enable comparisons (such as visual analogue scales measured from 0 to 10, 1–10 or 1–100). Realizing that our group will have the opportunity to choose shared measures for future research, we tackled additional harmonization issues and complexities in the phenotyping of pain, consistent with the first part of the NIH-SSM model.
Our group also encountered some difficulty in standardizing other data, including demographic information across studies (Redeker et al., 2015). For example, one issue that is frequently overlooked in the precision era is refining and defining the “demographic” variables of race, ethnicity, sex and gender. Measures of ancestry, race, and ethnicity may be used interchangeably, although the connotations of each are quite different and have important implications for research, particularly in the “post-genomic” era (Yudell, Roberts, DeSalle, & Tishkoff, 2016). Race is generally self-reported based on many factors or assigned by an investigator based on the individual’s appearance. Ethnicity is one’s state of belonging to a social group that has common national or cultural values and traditions. Although typically very little information on ethnicity is collected in research studies, it can be very important information for understanding the shared influence of lifestyle, diet, and geographic location of primary residence (Diez Roux, 2001). The routine collection of computationally derived genetic ancestry (now quantifiable using genotyping arrays or sequencing) along with self-reported ancestry could be considered for refining the ancestry construct (Fujimura & Rajagopalan, 2011; Landry, Ali, Williams, Rehm, & Bonham, 2018), particularly for research focused on founder mutations that occur disproportionally in homogenous groups, such as mutations in BRCA1- and BRCA2 genes (Rebbeck & Sankar, 2005). Without further focus on the race/ethnicity and ancestry contribution to health, it is likely that a continuation of the disproportionate study of European cohorts will continue to limit the generalizability of findings to specific population groups. Similarly, the constructs of sex and gender need to be addressed more comprehensively, as most studies include these as dichotomous variables. Sex can be determined by many characteristics beyond self-report, such as the number and type of sex chromosomes, external genitalia and/or internal reproductive anatomy, sex hormones, or type of gonads (Gonzales & Ehrenfeld, 2018). In addition, sex may change over an individual’s lifetime, and this needs to be collected as data and better connected to clinical outcomes (Ainsworth, 2015). Lastly, gender involves many variables that are also not fixed, and additional data to better understand the relationship of gender-related difference in clinical outcomes is warranted. Identifying ways to develop shared understandings and precise definitions of these important demographic variables is crucial.
After we clarified our commitment to further discussions about demographic variables, our group moved to a discussion of the pros and cons of a variety of omics measures that may be used to better understand the biological factors associated with pain. With advances in technology, multiple biological measures are now possible using relatively small amounts of stored specimens. Genome-wide association studies (GWAS) require large sample sizes (at least 1,000 subjects) and rare variants are difficult to study, whereas gene expression studies or epigenetic studies (methylation) use smaller cohorts (e.g., 30–50 cases/controls) and convey information that relates to the function of the gene, which has been linked to clinical symptoms. In support of this, gene expression studies in blood have been used successfully to identify biomarkers of susceptibility and/or severity for migraine headache, epilepsy, liver disease and neurodegenerative disorders (Zhao et al., 2017). All of the investigators in the consortium have collected blood as part of their protocol and acknowledged that their informed consent process included language regarding their participants’ willingness to allow their data to be used for additional purposes, such as for exploring different mechanisms of pain through the sharing of data within the consortium. Regarding specimen selection, in a peripheral blood specimen, white blood cells are the most valuable RNA-containing cells but are outnumbered by red blood cells approximately 1000:1 and ex vivo degradation can occur rapidly with RNA compared to the more stable structure of DNA. Therefore, developing a standardized protocol for the extraction of RNA, checking the sample quantity and quality, and performing the RNA sequencing was essential. We also acknowledged that technical protocols and instrumentation differ from lab to lab, thereby introducing variation that could confound the interpretation of data analyses. Thus, we decided to have all of the samples processed in one laboratory to reduce processing and batch variability.
In addition to the scientific considerations and measurement issues, the logistical and bureaucratic details associated with developing a partnership among scientists at multiple institutions is an ongoing and important component of the consortium building process. To develop a partnership, there were a number of practical considerations to address, including: “ownership” of ideas in the context of merging and sharing of data, the use biospecimens and resources, and the development of ways to meet the needs of individuals and group. We held thoughtful discussions regarding authorship, use of data, data sharing, and intellectual property potential and generated an agreement that all parties found acceptable. We followed the established protocols from each institution and put into place material transfer and data use agreements to operationalize the collaboration. Funding issues will be addressed with input from each investigator’s academic home, in collaboration with the consortium, in an iterative manner. These processes and practical issues were not trivial and need to always be carefully considered. The downstream ramifications for the principal investigators, trainees, and institutions can be significant. Negotiation of the responsibility for analyses of the data and development of manuscripts was undertaken within the consortium to reflect the contribution of each author in the order of authors within the resulting publications, and these considerations were also made with the intent to allow individuals to further develop expertise related to symptom science for career development (DeVon, Rice, Pickler, Krause-Parello, & Richmond, 2016).
Conclusion
There are multiple challenges and opportunities for nurse scientists who work with individuals across the life-span in the “precision” era. The careful consideration of how the underlying tenets of nursing science fit into “precision medicine” and “precision health” initiatives is important for individuals and groups who are designing research to harness the potentialities, and to appreciate the challenges in gaining a better understanding of the multiple influences on health, disease risk, and disease outcomes. For the evolution of nursing science, understanding not only the evolution of scientific discovery but also newer models of collaboration will be necessary to lead the next generation of symptom science discovery and translation. The emerging paradigm of precision science requires different strategies for obtaining data sources and the sample sizes necessary to study complex symptom phenotypes across disease states and diverse populations. The integration of diverse information from molecular pathways, biologic processes, and behavioral models is critical to predict and treat disease, improve survival, manage symptoms and enhance quality of life. This symbiosis of differing, but complementary knowledge is necessary to ensure nursing science improves health through its translation into practical clinical applications. Further attention to the inherent challenges and opportunities for nursing science in the precision science era, with a focus on the language and mores of scientific discovery is needed for the potential of nursing research to be fully realized. It is our hope that this commentary will provide a roadmap, or template, to aid future groups to build collaborations among nurse scientists.
Table 1.
Glossary of Terms
| Terms | Definition |
|---|---|
| Precision Medicine | Large-scale methods for characterizing molecular measures (such as genomics, proteomics, metabolomics, microbiomics) across large samples and populations to enable “personalized” treatment to each individual through a deductive scientific process.1 |
| Precision Population Health/Precision Public Health | Joining biological factors with social and environmental determinants of health to develop precision approaches to interventions in individuals and populations.2 |
| Precision Science | Application of rigorous processes necessary to support the development of more accurate diagnostics and targeted treatments. |
| Precision Symptom Science | A perspective that includes not only the “omic” but also the social, societal, and environmental determinants of heath. The ultimate purpose of this initiative is to improve the health of patients through a more comprehensive understanding of the factors that interact to contribute to pain, with this evidence being essential to develop precision-based interventions to prevent, as well as treat pain. |
| Symptom Science | Goal is to be able to precisely identify individuals at risk for symptoms and develop targeted strategies to prevent or mitigate the severity of symptoms. A perspective that includes not only the “omic” but also the social, societal, and environmental determinants of heath. |
Acknowledgement:
The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Nursing Research (NINR) or the National Institutes of Health (NIH).
Footnotes
The authors have no conflicts of interest to report.
Contributor Information
Susan G. Dorsey, University of Maryland School of Nursing, Baltimore, MD, USA.
Mari A. Griffioen, University of Delaware School of Nursing, Newark, DE, USA.
Cynthia L. Renn, University of Maryland School of Nursing, Baltimore, MD, USA.
Ann K. Cashion, National Institute of Nursing Research, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, USA.
Luana Colloca, University of Maryland School of Nursing, Baltimore, MD, USA.
Colleen K. Jackson-Cook, Virginia Commonwealth University School of Medicine, Richmond, VA, USA.
Jessica Gill, National Institute of Nursing Research, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, USA.
Wendy Henderson, National Institutes of Nursing Research, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, USA.
Hyungsuk Kim, National Institute of Nursing Research, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, USA.
Paule V. Joseph, National Institute of Nursing Research, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, USA.
Leorey Saligan, National Institute of Nursing Research, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, USA.
Angela R. Starkweather, University of Connecticut School of Nursing, Storrs, CT, USA.
Debra Lyon, University of Florida Gainesville College of Nursing, Gainesville, FL, USA.
References
- Ainsworth C (2015). Sex redefined. Nature, 518(7539), 288. [DOI] [PubMed] [Google Scholar]
- Cashion AK, & Grady PA (2015). The National institutes of health/national institutes of nursing research intramural research program and the development of The National institutes of health symptom science model. Nursing outlook, 63(4), 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MZ, Thompson CB, Yates B, Zimmerman L, & Pullen CH (2015). Implementing common data elements across studies to advance research. Nursing outlook, 63(2), 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, & Varmus H (2015). A new initiative on precision medicine. New England Journal of Medicine, 372(9), 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVon HA, Rice M, Pickler RH, Krause-Parello CA, & Richmond TS (2016). Setting nursing science priorities to meet contemporary health care needs. Nursing outlook, 64(4), 399–401. doi: 10.1016/j.outlook.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Diez Roux AV (2001). Investigating neighborhood and area effects on health. American journal of public health, 91(11), 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt P, Culley JM, Corwin E, Richmond T, Dougherty C, Pickler RH, … DeVon HA (2017). National nursing science priorities: Creating a shared vision. Nursing outlook, 65(6), 726–736. [DOI] [PubMed] [Google Scholar]
- Fujimura JH, & Rajagopalan R (2011). Different differences: The use of ‘genetic ancestry’versus race in biomedical human genetic research. Social Studies of Science, 41(1), 5–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales G, & Ehrenfeld JM (2018). Sex is not gender and why it matters for population health. British Journal of Anaesthesia, 120(5), 1130–1131. doi: 10.1016/j.bja.2018.01.030 [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Iademarco MF, & Riley WT (2016). Precision Public Health for the Era of Precision Medicine. American journal of preventive medicine, 50(3), 398–401. doi: 10.1016/j.amepre.2015.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Ali N, Williams DR, Rehm HL, & Bonham VL (2018). Lack of diversity in genomic databases is a barrier to translating precision medicine research into practice. Health Affairs, 37(5), 780–785. [DOI] [PubMed] [Google Scholar]
- Marchand F, Perretti M, & McMahon SB (2005). Role of the immune system in chronic pain. Nature Reviews Neuroscience, 6(7), 521. [DOI] [PubMed] [Google Scholar]
- Nightingale F (1992). Notes on nursing: What it is, and what it is not: Lippincott Williams & Wilkins. [Google Scholar]
- Pickler RH (2018). Precision’s Promise. Nursing Research, 67(4), 271–272. doi: 10.1097/nnr.0000000000000295 [DOI] [PubMed] [Google Scholar]
- Pizzo PA, Clark NM, & Pokras OC (2011). Relieving Pain in America : A Blueprint for Transforming Prevention, Care, Education, and Research. Washington, D.C.: National Academies Press; Retrieved from www.nap.edu [PubMed] [Google Scholar]
- Rebbeck TR, & Sankar P (2005). Ethnicity, ancestry, and race in molecular epidemiologic research. Cancer Epidemiology and Prevention Biomarkers, 14(11), 2467–2471. [DOI] [PubMed] [Google Scholar]
- Redeker NS, Anderson R, Bakken S, Corwin E, Docherty S, Dorsey SG, … Pullen C (2015). Advancing symptom science through use of common data elements. Journal of Nursing Scholarship, 47(5), 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudell M, Roberts D, DeSalle R, & Tishkoff S (2016). Taking race out of human genetics. Science, 351(6273), 564–565. [DOI] [PubMed] [Google Scholar]
- Zhao W, Beers DR, Hooten KG, Sieglaff DH, Zhang A, Kalyana-Sundaram S, … Sathe GM (2017). Characterization of gene expression phenotype in amyotrophic lateral sclerosis monocytes. JAMA neurology, 74(6), 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]