Abstract
Metastatic disease remains one of the primary reasons for cancer-related death, yet the majority of in vitro cancer models focus on the primary tumor sites. Here we describe a metastasis-on-a-chip device, that houses multiple bioengineered 3D organoids, established by a 3D photopatterning technique employing extracellular matrix-derived hydrogel biomaterials. Specifically, cancer cells begin in a colorectal cancer (CRC) organoid, which resides in a single microfluidic chamber connected to multiple downstream chambers in which liver, lung, and endothelial constructs are housed. Under recirculating fluid flow, tumor cells grow in the primary site, eventually enter circulation, and can be tracked via fluorescent imaging. Importantly, we describe that in the current version of this platform, HCT116 CRC cells preferentially home to the liver and lung constructs; the corresponding organs of which CRC metastases arise the most in human patients. We believe that in subsequent studies this platform can be implemented to better understand the mechanisms underlying metastasis, perhaps resulting in identification of targets for intervention.
Keywords: Microfluidics, organ-on-a-chip, cancer, 3D in vitro model, tumor-on-a-chip
1. INTRODUCTION
Despite advances in how cancer patients are treated, metastasis of tumor cells from a primary site to target sites remains a significant hurdle in oncology and a leading cause of death in cancer patients. Metastasis from colon to liver is common in patients with colorectal cancer (CRC), leading to the 2nd highest number of cancer-related deaths in the U.S.1 Patients with hepatic metastatic disease have a poor prognosis, with 5-year survival rates below 5%.2 Despite treatment advances, metastasis is still not well understood, in particular i) the cause of initiation activation of tumor cell growth and metastasis, such as the specific signaling pathways, and ii) the role of the microenvironment in regulating these phenomenon.3, 4
Traditional in vitro 2D cultures have been the foundation of countless important scientific discoveries, but fail to recapitulate the in vivo 3D microenvironment of tissues and tumors.5 Cells grown in 2D experience different surface topography, stiffness, cell-cell/cell-matrix interactions, and media exposure compared to a 3D architecture. As such, maintenance under 2D conditions can substantially alter the genetic, molecular, and phenotypic properties of mammalian cells, producing experimental outcomes that may not be indicative of them in vivo.6, 7 For example, we demonstrated that, when grown in 2D culture, metastatic colon carcinoma cells exhibited an epithelial morphology and expression profile, and it was only when they were transitioned into a 3D liver organoid environment that they “switched” to adopt a mesenchymal and metastatic phenotype reflective of their in vivo origin.8, 9 Even animal models, such as patient-derived xenografts (PDX), which have become a gold standard in cancer biology have significant limitations. They have unsatisfactory take rates, and have only been successfully established using the most malignant of tumors. Moreover, they fail to incorporate human tumor stroma.10 Bioengineered 3D platforms using human patient-derived cells can better mimic the cell-cell, cell-ECM, and mechanical interactions of in vivo tissue, and are thus more suitable for mechanistic research and applications such as improving personalized medicine approaches.11, 12 Importantly, in recent years, there has been a rapid advance in the integration of 3D tissue constructs and organoids with microfluidic device architectures, resulting in a variety of tissue- and tumor-on-a-chip systems.13 These range from stand alone organoids, to single tissue-on-a-chip systems,14 to more complex multi-tissue organ-on-a-chip platforms.15
2. RESULTS AND DISCUSSION
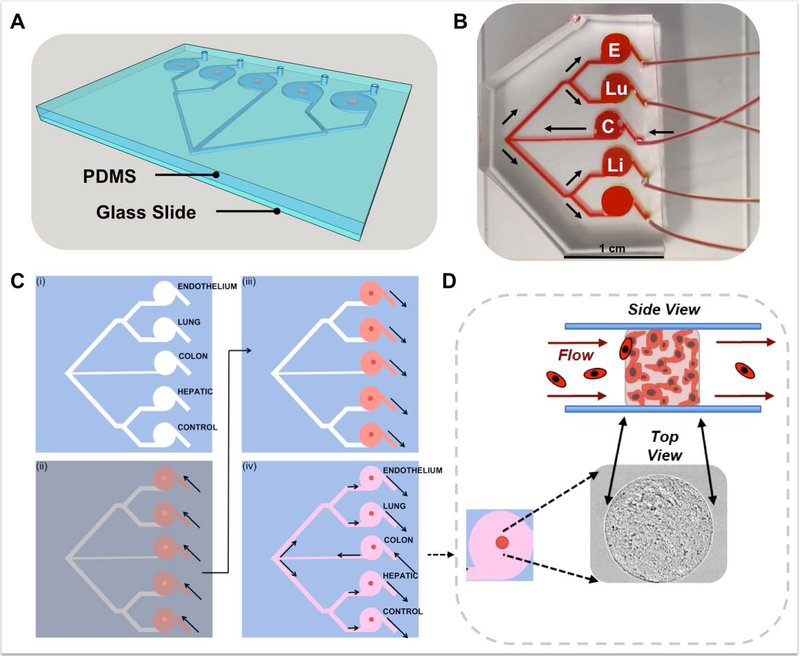
In a previously described study, we demonstrated the utility of a 2-organoid metastasis-on-a-chip (MOC) platform. By providing circulating flow through the organoid system via microfluidics, we can achieve the dissemination of CRC cells from a colon organoid into circulation, after which metastatic cells can colonize a downstream liver organoid. This model was one of the first in vitro models of metastasis recapitulating migration from a 3D primary tissue to a 3D target tissue.9 This is notable and novel because phenotype of cells in the originating malignant tumors and metastases can vary significantly – for example, varying levels of invasiveness due to MMP secretion and stem cell-like genes16, 17 – making the ability to study both sites and microenvironments extremely important. Here we describe additional functionality of the system, expanding from a single downstream organoid site to 4 sites (Fig. 1A), although the actual number is arbitrary to fabricate. In this MOC system, movement of cells can be tracked from the primary site organoid, through circulation, and to 4 downstream potential sites of metastasis.
Figure 1. Metastasis-on-a-chip device design and in situ tumor and tissue construct biofabrication.
A) A 3D CAD visualization of the PDMS MOC channel and chamber system containing tumor and tissue constructs (orange) constructed from a soft lithography-molded PDMS component, sealed to a glass slide. B) Fluid flow overview within a generic MOC. Media is perfused into the device at the single inlet port into the CRC chamber (C) from the media reservoir from the micro-peristaltic pump. From the CRC chamber, the channels bifurcate twice, providing equal flow to the endothelial (E), lung (Lu), liver (Li), and gel control (not shown) constructs. C) In situ device biofabrication of tumor and tissue constructs is performed by a sequential photopatterning technique into a (i) clean MOC device. (ii) Hydrogel precursor solutions (dark red) containing encapsulated cells of each respective cell type is injected partially into each MOC device port, to fill the appropriate chamber. A photomask is placed over the device (grey), with apertures to define construct shape/size (bright red/orange). UV light pulses (~1 s) are used to photo-crosslink exposed regions into 3D constucts (dark red), after which uncrosslinked material (light red) is retracted or flushed from the devices. (iv) Cell culture media (pink) is introduced into devise through the CRC chamber inlet port. D) From a top view, within each MOC, constructs appear as circular units, that from a side view are cylindrical in nature. Constructs, comprised of cells in ECM hydrogels, exist under fluid flow, and have the capability to experience circulating cells either interact or pass by.
This updated MOC was designed to increase adherence to the physiological complexity of in vivo cancer progression, but in a simple, straightforward to operate platform. In the human body, there exist many potential sites for metastasis for many cancers. The platform described herein is an effort in modeling this complexity in a organ-on-a-chip microphysiological system (MPS), while recognizing that this is still an incomplete model of many other phenomena involved in metastasis (e.g. invasion through endothelium). In our previous MOC system, we modeled metastasis of CRC cells from gut to liver, the most prevalent site of metastasis in patients18. However, with the addition of multiple target tissue sites, we asked the question, is the preference for CRC metastasis to liver simply because the liver is the main site of lymphatic/vascular drainage from the gut (in other words, proximity), or is the microenvironment of the liver such that metastatic CRC cells are more able to colonize liver tissue in comparison to say lung tissue (seed and soil hypothesis)19, which is the next most common site of CRC metastasis. As such, our MOC device was designed to include a single perfusion inlet to a CRC construct, from which the flow bifurcated several times to 4 equidistant downstream chambers, 3 of which housed addition tissue constructs – liver, lung, and endothelial; with the fourth chamber to accommodate a hydrogel control (Fig. 1B). The endothelial construct was included as during metastasis, interactions between tumor cells and endothelium are important. We chose to employ a 2-point bifurcation design to ensure that all cells experience the same angle changes as they flow through the device, thus ensuring no bias towards any particular of the 4 end channels. This second generation MOC has now allowed us to probe the concept of proximity versus microenvironment preference. With all downstream organoids equal both in terms of distance and geometry from the CRC construct, would the cancer cells metastasize to the liver constructs preferentially?
Devices were fabricated by soft lithography of a master mold with PDMS. The inlet and outlets of the negatively casted hemicylindrical microfluidics (Supplementary Fig. 1) PDMS layer were punched, followed by irreversible bonding to a glass slide; to form a sealed fluidic device. Using a micro-peristaltic pump, flow of media followed by ethanol through the devices was employed to qualitatively show that fluid flow appeared to reach each downstream organoid simultaneously (Fig. 2). Importantly, the fabrication method was designed to create hemicylindrical channels, to avoid trapping of circulating cells in angular features.
Figure 2. The MOC design supports consistent parallel chamber perfusion rates.
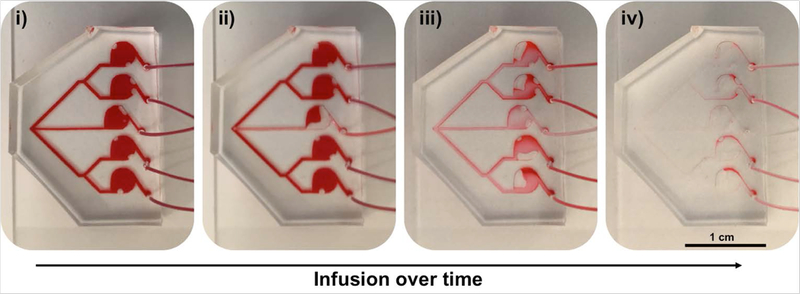
To demonstrate that fluid flow rates impact each parallel tissue construct chamber similarly, the MOC devices were filled with (i) DMEM cell culture media, further darkened with a food coloring dye for contrast. The media reservoir was replaced with a reservoir containing PBS, after which flow was initiated again, and (ii) PBS can be seen being infused into the inlet channel and CRC construct chamber. (iii) PBS inflow dilutes the DMEM/dye fluid, and reaches each parallel tissue construct chamber similarly, eventually (iv) taking the place of the majority of the DMEM/dye solution throughout the device.
Following device fabrication, tissue constructs were biofabricated by photopatterning-based cell encapsulation20 (Fig. 1C) using a hyaluronic acid (HA) and gelatin‐based hydrogel, commercially available as HyStem, that has been employed extensively in tissue engineering and regenerative medicine.21, 22 Examples include 3‐D culture,23, 24 tumor models,9, 25–27 bioprinting,28–32 and biofabrication of organoids for drug and toxicity screening.33–35 Recently, we modified this hydrogel system to provide extremely fast gelation kinetics,29 allowing for tissue construct fabrication with improved control over spatial polymerization, while maintaining established properties such as the pore size and elastic modulus of the original non-photocrosslinkable hydrogel. Few existing biomaterials mimic the native ECM and support the addition of bioactive factors, and are also highly adaptable to multiple biofabrication techniques. This system is comprised of naturally derived materials that are native to the body, and also supports the modular addition of additional ECM factors, such as cytokines and growth factors, if desired, via heparin-modulated binding,23, 28, 35, 36 thereby increasing its biomimetic properties compared to other photopolymerizable materials such as PEGDA and methacrylated gelatin. As described previously, the benefit with respect to the work described here is the capability of this hydrogel system to support on-demand photocrosslinking of discrete constructs in situ. This allows encapsulation of cells in 3D only upon UV light exposure, and limits hydrogel construct formation to only those regions exposed. This capability is not supported by more traditional gel materials with slow crosslinking kinetics, such as collagen Type I and Matrigel, or fast, but more difficult to control kinetics, such as alginate, making these materials less effective for biofabrication.37
Tissue construct photopatterning was performed in a stepwise fashion. Fig. 1C shows schematic of the MOC device and the biofabrication procedure with the four patterned tumor or tissue constructs, each formed in an individually addressable fluidic chamber. These constructs are mechanically robust even under significant flow (>30 μL/min), sustaining their photo-patterned shape. indicating that they are strongly adhered to the top and bottom of the chamber (PDMS and glass, respectively) through non-specific surface tension. Using this approach, in the sealed fluidic devices, the hydrogel‐based CRC, liver, lung, endothelial, and hydrogel control constructs were formed and then maintained under circulating flow (10 μL/min). Fig. 1D shows an enlarged top view of one of the tissue constructs within the MOC and a cartoon schematic illustrating conceptually the tissue construct under flow conditions and metastatic cancer cells traveling through the recirculating media with the capability to pass by or interact with the constructs.
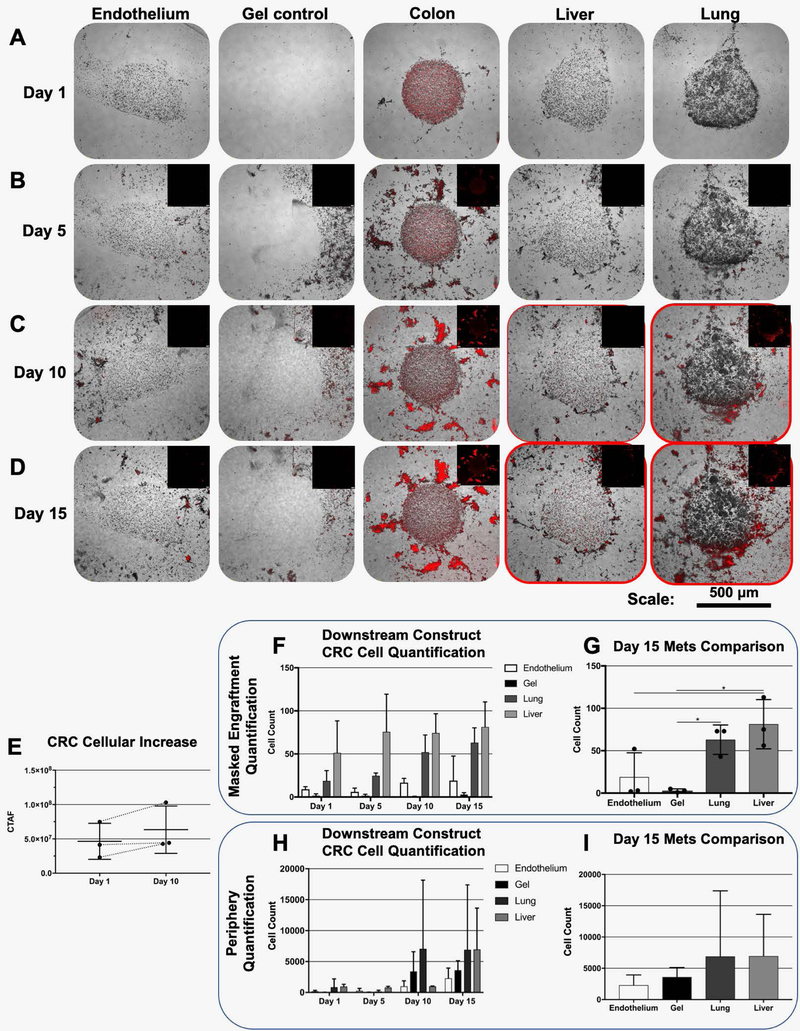
On day 1 of culture of one of the devices, the CRC tumor construct is shown as a clear cylindrical construct by standard phase microscopy within the MOC chip (Fig. 3A). This is notable, as the platform allows for in situ cell imaging, a feature that is crucial to subsequent tracking metastatic cells over time. At this early time point, viability was also verified by LIVE/DEAD staining and fluorescent imaging on chip. Shown is the same CRC tumor construct with little evidence of dead cells (red fluorescence), but rather a majority of viable cells comprising the construct (Fig. 3B). More importantly, the MOC system supports high viability across all constructs for over 2 weeks. On day 15 of culture, construct viability was visualized once more by LIVE/DEAD staining. Fluorescent imaging showed high levels of viability in CRC constructs (Fig. 3C), liver constructs (Fig. 3D), lung constructs (Fig. 3E), and endothelial constructs (Fig. 3F), verifying the ability to maintain 4 different 3D constructs under a common media.
Figure 3. Photopatterned MOC constructs exhibit high levels of cell viability and primary tumor site growth.
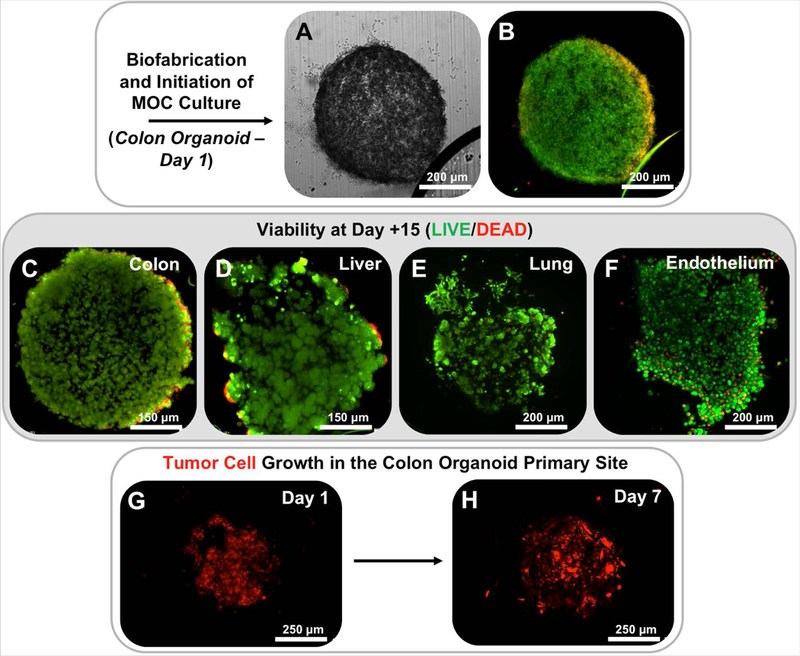
Following photopatterning, constructs are easily visualized in situ on-chip by A) phase microscopy and B) show high levels of cell viability as indicated by LIVE/DEAD staining and fluorescent imaging. Maintenance of viability continues through day 15 of culture for C) CRC, D) liver, E) lung, and F) endothelial constructs. Note – for viability stains, non-transfected HCT-116 cells were utilized. LIVE/DEAD: Green stain – calcein AM-stained viable cells; Red stain – ethidium homodimer-stained dead cell nuclei. G-H) Growth of RFP-labeled HCT116 cells over time in the primary site CRC tumor construct indicates tumor cell proliferation on chip.
Over time in culture, RFP‐labeled HCT116 cells comprising the CRC constructs proliferated in the primary tumor construct site, as evidenced by an increase in red fluorescence over time. Fig. 3G-H shows the red fluorescent signal in a CRC tumor construct increasing from day 1 to day 7. Primary site growth is often a precursor to malignant cells entering circulation18, 19, which we observed previously in our colon-to-liver MOC.9 In the current multi-target tissue platform, HCT116 proliferation is also verified in the CRC construct site by increased RFP signal (Fig. 4A-B). Moreover, by day 5 fluorescent tracking showed that HCT116 cells disseminated from the construct, into the periphery around the CRC construct, entering the circulating media flow (Fig. 4B). By day 10, RFP‐labeled cells were in circulation and able to home to the downstream target tissue construct sites (Fig. 4C) Initially, qualitative observations appeared to show metastasis predominantly the lung constructs, followed by the liver constructs, and finally the endothelial constructs where few tumor cells were observed (Fig. 4C-D, Supplementary Fig. 3, and Supplementary Fig. 4). However, if we employed a digital masking technique to remove cells outside of the construct borders in the images capture, and quantified cells that had engrafted in the constructs in an unbiased fashion (Supplementary Fig. 2), we see a different trend. First, we do observe an increase in CRC cell presence in the primary site (Fig. 4E). But more importantly, the number of engrafted cells in the lung and liver are actually not significantly different (Fig. 4F-G). However, the difference between localization around the periphery of the constructs versus engraftment is an important consideration. As described above, and in Fig. 4C-D, and Supplementary Fig. 3 and Supplementary Fig. 4, there do appear to be more HCT116 cells that localize around the periphery of the tissue constructs in the device chambers containing lung constructs. As such we ran the quantification method described above, but included peripheral cells and excluded the engrafted cells in the quantification by inverting the masking approach. As seen in Fig. 4H-I, there is a trend for HCT116 cells to predominantly localize around lung constructs on day 10, but this localization begins to even out between lung and liver constructs by day 15. Using the periphery quantification resulted in a large amount of variance between cell numbers, and as such the data in Fig. 4H-I was not statistically significant. Nevertheless, there does appear to be a difference between initial localization and long-term engraftment.
Figure 4. Metastasis tracking over time on multi-tissue construct MOC devices.
Tumor and tissue constructs are visible via phase microscopy and HCT116 CRC tumor cells are visualized by fluorescent imaging of RFP. Phase and fluorescent images are overlaid. A) Upon initiation of metastasis experiments, RFP+ cells are only visible at the CRC construct site. B) By day 5, RFP+ cells are seen having migrated out of the primary CRC construct site. C) On day 10, RFP+ cells are observed in significant numbers at lung construct sites, but are also present at liver construct sites, but less obviously, and D) this trend continues through day 15. E) Quantification of CRC cell presence in the primary organoid over time by total fluorescence (CTAF). F) Quantification of numbers of metastasized HCT116 cells engrafted in downstream constructs over time. G) Engrafted cells at day 15 with statistical analysis (* indicates p < 0.05). H) Quantification of numbers of HCT cells localized around the periphery of downstream constructs over time. I) Localized cells around construct peripheries at day 15.
Importantly, the hydrogel control constructs show RFP-labeled cells around the periphery of the constructs, but no significant engraftment and growth compared to those of lung and liver constructs. Proliferation of tumor cells at the metastatic sites appears to be indicated by images taken at day 15 (Fig. 4D). Remarkably, despite randomly shuffling the particular on-chip locations of the target tissue constructs, the HCT116 cells overwhelmingly have a preference for “metastasizing” to the liver and lung constructs. This is shown onboard additional MOC systems, two of which are highlighted in Supplementary Fig. 3 and Supplementary Fig. 4. It should be noted that the particular locations of each tissue construct type in these devices were randomized to ensure that the physical design of the MOC device did not bias metastasis to a particular tissue type.
These results raise several interesting points. First, despite these constructs being simplistic versions of actual in vivo tissues, it suggests that, indeed, proximity of a potential metastatic site may have some precedence over microenvironment; or if not precedence, can be a significant factor. However, additional mechanistic studies are needed to make any claims with respect to this hypothesis. Accordingly, in future studies, we plan to 1) modulate the cellular compositions of the target tissue constructs, including stroma and immune cells, and in the case of the tumor constructs, include these components as the tumor microenvironment surrounding the primary tumor site, 2) incorporate endothelial barriers around each tumor and tissue construct, potentially derived from tissue-specific endothelial cells, through which tumor cells would need to intravasate/extravasate, and 3) improve the mimicry of our hydrogel biomaterial to native tissue ECM. All of these directions will serve to create tissue constructs more indicative of their in vivo counterparts, and thus improve the clinical relevance of the MOC platform. With a more physiologically accurate system such as this, we can then begin additional receptor-based signaling blockade manipulations to better understand the particular pathways at play during metastatic events, and how tumor cells exert a preference in metastasizing to particular sites. Second, it is important to note that this MOC platform can distinguish preferences of metastatic cells to home and colonize different tissues/tissue constructs. One of our next initiatives will be to employ patient tumor biospecimen-derived cells to generate the primary site tumor constructs. Likewise, we are exploring the potential to also employ primary cells from patients to biofabricate the “healthy” tissue constructs. Such systems could be used diagnostically and prognostically, to determine therapies that may minimize the risk of metastatic disease, and predict potential tissues that may be most susceptible to developing metastatic legions. Such a platform technology could have great impact in both basic science research and clinical applications.
3. MATERIALS AND METHODS
3.1. Cell culture
Human CRC cells (HCT‐116, transfected previously with red fluorescent protein [RFP]), human hepatoma cells (HepG2), and human lung epithelial cells (A549) were expanded in 2‐D on tissue culture plastic in 15 cm tissue‐treated dishes until 90% confluence with Dulbecco’s Minimum Essential Medium (DMEM, Sigma, St. Louis, MO), containing 10% fetal bovine serum (FBS, Hyclone, Logan, UT). Human endothelial cells (HUVEC) were expanded similarly, but using Endothelial Growth Medium (EGM-10, Lonza, Walkerstown, MD), containing 10% FBS, and supplements included by the manufacturer. Cells were detached from the substrate with Trypsin/EDTA (Hyclone) and resuspended in media before use in further studies.
3.2. MOC device fabrication
MOCs were fabricated by conventional soft lithography,38–40 with modifications specific to the style of devices required for our studies.20 A 200 μm thick rectangular positive master mold was fabricated by photolithography through a photomask into SU-8 2050 (MicroChem, Newton, MA). A secondary 150 μm radius hemicylindrical positive master mold was fabricated in baked SU-8 2050 by soft lithography of a 50 μm curved PDMS-membrane covered PDMS replica of the rectangular mold. A 10:1 mixture of polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning, Midland, MI) with its curing agent was mixed and degassed before being poured directly onto the hemicylindrical mold and cured at 60 °C for 60 min. Following curing, the devices were removed from the mold and the respective inlet and outlets were punched out with a blunt needle. The embossed surface of the PDMS replica and the top surface of a clean glass slide were cleaned and N2 plasma oxidized (Harrick Plasma, Ithaca, NY), after which the PDMS component was bounded to the glass slide, irreversibly sealing the device. The devices were sterilized by UV light prior to use. Fluidic connections were made using PTFE tubing and 2-Stop PVC 0.51mm ID tubing (Cole-Parmer, Vernon Hills, IL). The PTFE tubing connected the 4 outlets of each device to individual PVC tubing lengths that drained to a single media reservoir. An additional line of tubing recirculated the media of the reservoir via a MP2 Precision micro-peristaltic pump (Elemental Scientific, Inc., Omaha, NE) to the single inlet per device, forming closed circuits (Fig. 1B).
3.3. Tissue and tumor construct photopatterning-based biofabrication
ECM-based HA/gelatin hydrogels were formed using HyStem-HP (ESI-BIO, Alameda, CA). The thiolated HA component (Heprasil) and the thiolated gelatin component (Gelin-S) were dissolved in water containing 0.1% w/v of the photoinitiator 4-(2-hydroxyethoxy)phenyl-(2-propyl)ketone (Sigma St. Louis, MO) to make 1% w/v solutions. The polyethylene glycol diacrylate crosslinker (Extralink, ESI-BIO) was dissolved in the photoinitiator solution to make a 2% w/v solution. Heprasil, Gelin-S, and Extralink were then mixed in a 2:2:1 ratio by volume. The resulting solution used to resuspend cells at a cell density of 10 million cells/mL for the liver, lung and endothelial organoids, and 20 million cells/mL for the colon. The hydrogel precursor–cell mixtures were introduced into each of the device channels closest to the corresponding chamber for that tissue type using the inlet/outlet ports (Fig. 1C(ii)). The construct position was then defined in each chamber by UV exposure (365 nm, 18 W/cm2) for 1 s through a printed transparency photomask with apertures of 500 μm diameter to initiate a thiol–ene crosslinking reaction, resulting in cell encapsulation within the patterned hydrogel regions of approximately 500 μm diameter (Fig. 1C(iii)). The remaining unreacted precursor–cell mixture was flushed from the device using PBS, leaving cylindrical tumor or tissue constructs within each chamber of the device, supported by media infusion through the primary inlet port (Fig. 1C(iv)). The same photopatterning technique was carried out to fabricate a cell-free hydrogel-only construct as a control. The results described here are performed in triplicate using three separate fluidic devices in which tissue construct locations were randomized. Viability was verified with LIVE/DEAD staining (ThermoFisher, Waltham, MA) and images where captured by creating maximum projections of 120 μm Z-stacks using a TCS LSI macro-confocal microscope (Leica Microsystems, Wetzlar, Germany).
3.4. Device operation and tumor metastasis tracking
For each MOC device, medium flow from a 4-mL media reservoir (custom-modified cryotube) was introduced by the micro-peristaltic pump into the primary inlet port at a flow rate of 10 µL/min, and recirculated. For integrated organoid studies, DMEM-10 was supplemented with EGM-2 (2% FBS rather than 10%) in a 3:1 ratio, to better support endothelial constructs. During MOC system culture, constructs and presence of fluorescent RFP‐labeled tumor foci were documented over time at each construct site by inverted microscopy (Leica IX83). Composite images were taken of the primary construct on days 1, 5, 10, and 15, in which images of each construct were captured with light microscopy and epifluorescence at 594 nm, after which overlays were created to visualize both gross tumor and tissue construct shape, and presence of RFP-labeled HCT116 cells. These images were employed to analyze the metastatic progression in each tissue construct type. Supplementary Fig. 2 describes the quantification methodology used to determine the number of cells engrafted at each site.
Supplementary Material
ACKNOWLEDGEMENTS
AS acknowledges funding from the Wake Forest Institute for Regenerative Medicine Promoting Discoveries Pilot Program, and the Wake Forest Clinical and Translational Science Institute Open Pilot Program under NIH CTSA UL1 TR001420.
Funding information
The Wake Forest Institute for Regenerative Medicine Promoting Discoveries Pilot Program; The Wake Forest Clinical and Translational Science Institute Open Pilot Program under NIH CTSA UL1 TR001420.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Xu T et al. Hybrid printing of mechanically and biologically improved constructs for cartilage tissue engineering applications. Biofabrication 5, 015001 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Young M, Ordonez L & Clarke AR What are the best routes to effectively model human colorectal cancer? Molecular oncology 7, 178–189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fidler IJ, Kim SJ & Langley RR The role of the organ microenvironment in the biology and therapy of cancer metastasis. J Cell Biochem 101, 927–936 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Langley RR & Fidler IJ Tumor cell-organ microenvironment interactions in the pathogenesis of cancer metastasis. Endocr Rev 28, 297–321 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Kunz-Schughart LA, Freyer JP, Hofstaedter F & Ebner R The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J Biomol Screen 9, 273–285 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Ho WJ et al. Incorporation of multicellular spheroids into 3-D polymeric scaffolds provides an improved tumor model for screening anticancer drugs. Cancer science 101, 2637–2643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drewitz M et al. Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol J 6, 1488–1496 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Skardal A, Devarasetty M, Rodman C, Atala A & Soker S Liver-Tumor Hybrid Organoids for Modeling Tumor Growth and Drug Response In Vitro. Ann Biomed Eng (2015). [DOI] [PMC free article] [PubMed]
- 9.Skardal A, Devarasetty M, Forsythe S, Atala A & Soker S A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol Bioeng 113, 2020–2032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrolecki LE et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev 35, 547–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devarasetty M, Mazzocchi AR & Skardal A Application of bioengineered 3D tissue and tumor organoids in drug development and precision medicine: current and future. BioDrugs In press (2018). [DOI] [PMC free article] [PubMed]
- 12.Mazzocchi AR, Soker S & Skardal A in Tumor Organoids, Vol. In Press (eds. Soker S & Skardal A) (Springer Nature, Berlin, Germany; 2017). [Google Scholar]
- 13.Esch EW, Bahinski A & Huh D Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 14, 248–260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell MJ, Jain RK & Langer R Engineering and physical sciences in oncology: challenges and opportunities. Nat Rev Cancer 17, 659–675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skardal A et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep 7, 8837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karakiulakis G et al. Increased type IV collagen-degrading activity in metastases originating from primary tumors of the human colon. Invasion & metastasis 17, 158–168 (1997). [PubMed] [Google Scholar]
- 17.Franci C et al. Biomarkers of residual disease, disseminated tumor cells, and metastases in the MMTV-PyMT breast cancer model. PLoS One 8, e58183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villeneuve PJ & Sundaresan RS Surgical management of colorectal lung metastasis. Clin Colon Rectal Surg 22, 233–241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fidler IJ The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 3, 453–458 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Skardal A, Devarasetty M, Soker S & Hall AR In situ patterned micro 3D liver constructs for parallel toxicology testing in a fluidic device. Biofabrication 7, 031001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burdick JA & Prestwich GD Hyaluronic acid hydrogels for biomedical applications. Adv Mater 23, H41–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serban MA & Skardal A Hyaluronan chemistries for three-dimensional matrix applications. Matrix Biol (2018). [DOI] [PubMed]
- 23.Skardal A et al. Tissue specific synthetic ECM hydrogels for 3-D in vitro maintenance of hepatocyte function. Biomaterials 33, 4565–4575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Skardal A & Prestwich GD Engineered extracellular matrices with cleavable crosslinkers for cell expansion and easy cell recovery. Biomaterials 29, 4521–4531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devarasetty M, Wang E, Soker S & Skardal A Mesenchymal stem cells support growth and organization of host-liver colorectal-tumor organoids and possibly resistance to chemotherapy. Biofabrication 9, 021002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazzocchi AR, Rajan SAP, Votanopoulos KI, Hall AR & Skardal A In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Sci Rep 8, 2886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skardal A, Devarasetty M, Rodman C, Atala A & Soker S Liver-Tumor Hybrid Organoids for Modeling Tumor Growth and Drug Response In Vitro. Ann Biomed Eng 43, 2361–2373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skardal A et al. A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomater 25, 24–34 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Skardal A et al. Bioprinting Cellularized Constructs Using a Tissue-specific Hydrogel Bioink. J Vis Exp (2016). [DOI] [PMC free article] [PubMed]
- 30.Skardal A, Zhang J, McCoard L, Oottamasathien S & Prestwich GD Dynamically crosslinked gold nanoparticle - hyaluronan hydrogels. Adv Mater 22, 4736–4740 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Skardal A et al. Photocrosslinkable hyaluronan-gelatin hydrogels for two-step bioprinting. Tissue Eng Part A 16, 2675–2685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skardal A, Zhang J & Prestwich GD Bioprinting vessel-like constructs using hyaluronan hydrogels crosslinked with tetrahedral polyethylene glycol tetracrylates. Biomaterials 31, 6173–6181 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Devarasetty M et al. Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids. Biosensors (Basel) 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forsythe SD et al. Environmental toxin screening using human-derived 3D bioengineered liver and cardiac organoids. Frontiers in Public Health In press (2018). [DOI] [PMC free article] [PubMed]
- 35.Skardal A et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Scientific Reports 7, 8837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skardal A et al. A tunable hydrogel system for long-term release of cell-secreted cytokines and bioprinted in situ wound cell delivery. J Biomed Mater Res B Appl Biomater (2016). [DOI] [PMC free article] [PubMed]
- 37.Caliari SR & Burdick JA A practical guide to hydrogels for cell culture. Nat Methods 13, 405–414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald JC & Whitesides GM Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc Chem Res 35, 491–499 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Qin D, Xia Y & Whitesides GM Soft lithography for micro- and nanoscale patterning. Nat Protoc 5, 491–502 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Kang E, Shin SJ, Lee KH & Lee SH Novel PDMS cylindrical channels that generate coaxial flow, and application to fabrication of microfibers and particles. Lab Chip 10, 1856–1861 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.