Abstract
As a follow-up to genome-wide association analysis of common variants associated with ovarian carcinoma (cancer), this study considers seven well-known ovarian cancer risk factors and their interactions with 28 genome-wide significant common genetic variants. The interaction analyses were based on data from 9,971 ovarian cancer cases and 15,566 controls from 17 case-control studies. Likelihood ratio and Wald tests for multiplicative interaction and for relative excess risk due to additive interaction were used. The top multiplicative interaction was noted between oral contraceptive pill (OCP) use (ever vs never) and rs13255292 (P-value = 3.48 × 10−4). Among women with the TT genotype for this variant, the odds ratio for OCP use was 0.53 (95% CI=0.46–0.60) compared to 0.71 (95%CI=0.66–0.77) for women with the CC genotype. When stratified by duration of OCP use, women with 1–5 years of OCP use exhibited differential protective benefit across genotypes. However, no interaction on either the multiplicative or additive scale was found to be statistically significant after multiple testing correction. The results suggest that OCP use may offer increased benefit for women who are carriers of the T allele in rs13255292. On the other hand, for women carrying the C allele in this variant, longer (5+ years) use of OCP may reduce the impact of carrying the risk allele of this SNP. Replication of this finding is needed. The study presents a comprehensive analytic framework for conducting gene-environment analysis in ovarian cancer.
Keywords: ovarian cancer, genetics, additive interaction, G × E
INTRODUCTION
Ovarian carcinoma (cancer) is a disease with high mortality; most women are diagnosed with advanced stage disease where five-year survival is less than 50% 1. Effective screening modalities have been elusive 2, and therefore primary prevention strategies remain the most promising avenue to minimize the incidence and mortality of ovarian cancer.
Several factors consistently associated with reduced or increased risk have been identified for ovarian cancer, including some that represent opportunities for chemoprevention or surgical intervention. Factors associated with reduced risk include oral contraceptive pill (OCP) 3 use aspirin use 4, tubal ligation 5, parity 3, salpingectomy 6–9 and bilateral salpingo-oophorectomy (BSO). Common germline genetic variation 10–20, first-degree family history of ovarian cancer 21, 22, menopausal hormone therapy use 23–25, greater body mass index (BMI) 26 and endometriosis 27 are risk factors for the disease. OCPs and aspirin use represent feasible chemoprevention strategies whereas salpingectomy is now recommended by many gynecologic societies as an ovarian cancer prevention approach for women seeking tubal sterilization, having a hysterectomy, or having other pelvic surgery.
Average lifetime risk of ovarian cancer diagnosis for women in the U.S. is 1.3% 28, but this number varies greatly depending on the composite exposure history of risk factors 29. Pearce et al. estimated the lifetime risk for women in the general population ranges from 0.35% (95%CI = 0.29% to 0.42%) to 8.8% (95%CI = 7.1% to 10.9%) depending on exposure history for six factors: OCP use, parity, tubal ligation, endometriosis, first degree family history of ovarian cancer and genetic risk score quintile 29.
However, these lifetime risk estimates were limited to six risk factors and did not consider their interaction with individual genetic variants identified through genome-wide association studies (GWAS) 28. The multiplicative scale is commonly used for gene-environment interaction (G × E) analysis. Additive interaction analysis has been suggested for case-control studies in many recent papers for a more mechanistic interpretation 30–34. Validity of a truly multiplicative model implies existence of additive interaction when the two factors under consideration have non-null main effects 35. Thus, failure to detect G × E interaction on multiplicative scale may imply there exists interaction on additive scale, but the ability to detect it depends on the sample size and the main and interaction effect sizes 35. We present here our efforts to evaluate both multiplicative and additive gene-environment interactions in ovarian cancer using data from the international Ovarian Cancer Association Consortium (OCAC) comprising 17 case-control studies.
We have included 28 common genetic variants previously associated with risk of ovarian cancer in genome-wide association analyses for our G × E analyses 36. Environmental factors included in our analysis are OCP use, parity, tubal ligation, breastfeeding, menopausal hormone therapy, usual adult BMI, and endometriosis. A small number of studies in OCAC had data available on aspirin use and thus we have not included this risk factor in our analysis here. Among our list of environmental factors, BMI, OCP use, tubal ligation, breastfeeding, and menopausal hormone therapy are of special interest because they are modifiable targets for prevention.
METHODS
Study Population
The OCAC is an international multidisciplinary consortium formed in 2005 (http://apps.ccge.medschl.cam.ac.uk/consortia/ocac/) with a goal of sharing data from worldwide ovarian cancer studies to establish reliable estimation of association between environmental and genetic factors related to risk of ovarian cancer 23, 37. Cases were defined as women with ovarian carcinoma (i.e., invasive epithelial ovarian cancers), fallopian tube cancer and primary peritoneal cancer. Controls were women without ovarian cancer and who had at least one ovary. For both cases and controls, individuals with prior cancers except non-melanoma skin cancers were excluded.
Genetic Association Analysis
In total, 28 single nucleotide polymorphisms (SNPs) previously identified through GWAS were included from 75 OCAC sites (Table 1). The first 26 SNPs were found to be significantly associated with either ovarian cancer overall or one or more histotypes 36. In addition, rs13255292 and rs10962643 were included because they were in the same region as two other significant SNPs but showed a strong independent association with ovarian cancer risk. The SNP at locus 15q26 (rs8037137), which was found to be genome-wide significant 13, was not included because not enough non-carriers were present in our analytic dataset for examining interactions. The genetic data included both genotyped and imputed variants (imputation being carried out using phase 2 Hapmap reference panel). More details regarding genotyping and imputation of the genetic data have been previously described 12, 17, 18, 20. The methods for analyzing the SNP data in the OCAC have also been described previously 12, 17, 18, 20. Briefly, logistic regression models were fit to examine the association between ovarian cancer and each genetic variant under an additive model (using risk allele dosage). The models were adjusted for ethnicity, genotyping panel and the leading principal components for each ethnicity. The summary results are shown in Table 1 and are also available through the OCAC website (http:/apps.ccge.medschl.cam.ac.uk/consortia/ocac/).
Table 1.
Odds ratios for marginal associations of 28 genetic susceptibility variants with ovarian cancer. Analysis used data with 26864 cases and 48034 controls from 75 study sites.
| SNP | Previously published best hita | Chr | Position | Risk Allele | Baseline Allele | RAF | ORb | P-valueb |
|---|---|---|---|---|---|---|---|---|
| rs12023270 | rs5872217015 | 1 | 38086578 | T | C | 0.264 | 1.08 (1.05,1.10) | 2.65×10−8 |
| chr2:111818658 | rs216510918 | 2 | 111818658 | C | A | 0.277 | 1.06 (1.04,1.09) | 2.03×10−6 |
| rs874898 | rs75259014 | 2 | 113974196 | C | G | 0.262 | 1.00 (0.98,1.03) | 7.36×10−1* |
| rs1562314 | rs71183014 | 2 | 177045560 | T | A | 0.638 | 1.10 (1.07,1.13) | 2.84×10−14 |
| rs11207182018 | 3 | 138849110 | allele 1 | G | 0.270 | 1.03 (1.00,1.06) | 5.17×10−2* | |
| chr3:156397692 | rs6227404117 | 3 | 156397692 | T | C | 0.048 | 1.47 (1.39,1.55) | 7.73×10−47* |
| rs987020718 | 3 | 190525516 | A | G | 0.666 | 1.05 (1.03,1.08) | 2.95×10−5 | |
| rs7705526 | rs1006969010 | 5 | 1285974 | A | C | 0.343 | 1.10 (1.07,1.12) | 5.52×10−14 |
| chr5:66121089 | rs55502517918 | 5 | 66121089 | allele2 | G | 0.526 | 1.03 (1.00,1.05) | 2.61×10−2* |
| chr8:82653644 | 8:8266881817 | 8 | 82653644 | G | A | 0.064 | 1.18 (1.12,1.23) | 3.25×10−12* |
| rs988665118 | 8 | 128817883 | G | A | 0.435 | 1.06 (1.03,1.08) | 2.89×10−6* | |
| rs1325529218 | NA | 8 | 129076573 | C | T | 0.700 | 1.07 (1.05,1.10) | 3.57×10−8* |
| rs10103314 | rs1400482 12 | 8 | 129560744 | A | C | 0.883 | 1.15 (1.11,1.20) | 5.76×10−15* |
| chr9:16915105 | rs1096269220 | 9 | 16915105 | C | G | 0.834 | 1.24 (1.20,1.28) | 4.54×10−41* |
| rs10962643 | NA | 9 | 16857403 | C | A | 0.699 | 1.17 (1.14,1.20) | 1.13×10−35* |
| rs320203 18 | 9 | 104943226 | C | A | 0.842 | 1.03 (1.00,1.06) | 5.21×10−2 | |
| chr9:136138765 15 | 9 | 136138765 | G | allele 3 | 0.176 | 1.12 (1.08,1.15) | 1.49×10−12* | |
| rs7084454 | rs14496237617 | 10 | 21821274 | A | G | 0.301 | 1.07 (1.05,1.10) | 3.32×10−8* |
| rs7902587 18 | 10 | 105694301 | T | C | 0.091 | 1.08 (1.03,1.12) | 4.54×10−4* | |
| chr12:121403724 | rs7953249 18 | 12 | 121403724 | A | G | 0.570 | 1.05 (1.03,1.07) | 2.58×10−5* |
| chr15:91531995 | rs8037137 13 | 15 | 91531995 | C | T | 0.829 | 1.08 (1.05,1.12) | 1.18×10−6* |
| rs11658063 | rs7405776 19 | 17 | 36103872 | G | C | 0.614 | 1.04 (1.02,1.07) | 2.98×10−4* |
| chr17:43552537 | rs1879586 17 | 17 | 43552537 | A | G | 0.164 | 1.12 (1.08,1.15) | 2.22×10−12* |
| rs7217120 | rs720782616 | 17 | 46484755 | C | T | 0.275 | 1.10 (1.07,1.13) | 8.69×10−13* |
| rs809824418 | 18 | 21405553 | G | A | 0.741 | 1.04 (1.01,1.07) | 4.23×10−3* | |
| rs4808075 11 | 19 | 17390291 | C | T | 0.268 | 1.13 (1.10,1.16) | 1.49×10−20* | |
| rs74597329 | rs68818714 | 19 | 39739155 | G | T | 0.301 | 1.02 (0.99,1.04) | 2.63×10−1 |
| rs6005807 18 | 22 | 28934313 | T | C | 0.095 | 1.09 (1.04,1.13) | 3.35×10−5* | |
Abbreviations: SNP, single-nucleotide polymorphism; RAF, risk allele frequency; Chr, chromosome; OR, odds ratio; allele1, GCCAGATTCAGAAT; allele2, GACACACAC; allele3, GCGCCCACCACTA.
If not specified, the previously published best hit is the same as the current best hit.
Logistic regression for ovarian cancer overall (regardless of histology), adjusted for ethnicity, study panel and leading principal components for each ethnicity (using a total of 47 principal components).
P-value > 0.01
Environmental Association Analysis
Environmental Variables (E):
A total of seven established environmental risk factors for ovarian cancer were of primary interest (Table 2), including four associated with decreased risk and three with increased risk for ovarian cancer or one specific histotype. These included: OCP use (measured as both ever/never and duration of OCP use (never users including <1 one year of use, 1-<5, 5+yr), tubal ligation (yes/no), breastfeeding (ever/never), parity (0, 1–2, 3+ full-term births (i.e., those lasting ≥6 months), type of menopausal hormone therapy use for more than 1 year after age 50 (never user, menopausal estrogen therapy only, any use of menopausal estrogen + progestin therapy), BMI (<25, 25-<30, 30+), and a history of endometriosis (yes/no).
Table 2.
Odds ratios for marginal associations of seven environmental risk factors with ovarian cancer risk with 13722 cases and 22975 controls from 19 study sites.
| Environmental risk factor | Before Imputationa |
After Imputationb |
||||
|---|---|---|---|---|---|---|
| Control | Case | Control | Case | ORc | P-valuec | |
| OCP use | ||||||
| Never | 0.347 | 0.444 | 0.351 | 0.452 | Ref | |
| Ever | 0.645 | 0.536 | 0.649 | 0.548 | 0.62 (0.59,0.66) | 5.24×10−73 |
| (missing) | 0.008 | 0.020 | ||||
| Duration of OCP use | ||||||
| Never users (including <1 year) | 0.425 | 0.542 | 0.430 | 0.554 | Ref | |
| 1−<5 year | 0.229 | 0.208 | 0.232 | 0.215 | 0.70 (0.66,0.74) | 8.23×10−32 |
| 5+ year | 0.332 | 0.222 | 0.338 | 0.231 | 0.48 (0.45,0.51) | 2.20×10−133 |
| (missing) | 0.014 | 0.028 | ||||
| Tubal ligation | ||||||
| No | 0.693 | 0.777 | 0.762 | 0.824 | Ref | |
| Yes | 0.208 | 0.160 | 0.238 | 0.176 | 0.73 (0.69,0.78) | 1.81×10−23 |
| (missing) | 0.098 | 0.063 | ||||
| Breastfeeding | ||||||
| No | 0.239 | 0.294 | 0.380 | 0.515 | Ref | |
| Yes | 0.532 | 0.410 | 0.620 | 0.485 | 0.76 (0.71,0.80) | 4.80×10−21 |
| (missing) | 0.229 | 0.296 | ||||
| Parity (number of full−term births) | ||||||
| 0 | 0.148 | 0.241 | 0.149 | 0.243 | Ref | |
| 1–2 | 0.487 | 0.434 | 0.489 | 0.438 | 0.59 (0.55,0.63) | 1.94×10−65 |
| 3+ | 0.359 | 0.315 | 0.362 | 0.319 | 0.50 (0.46,0.53) | 4.91×10−90 |
| (missing) | 0.006 | 0.011 | ||||
| Type of HT using more than 1 year after age 50 | ||||||
| Never use | 0.687 | 0.647 | 0.789 | 0.782 | Ref | |
| ET only | 0.060 | 0.075 | 0.066 | 0.084 | 1.22 (1.12,1.34) | 2.65×10−5 |
| Any EPT | 0.131 | 0.118 | 0.145 | 0.134 | 0.97 (0.90,1.04) | 3.55×10−1 |
| (missing) | 0.121 | 0.160 | ||||
| BMI | ||||||
| < 25 | 0.392 | 0.370 | 0.516 | 0.485 | Ref | |
| 25−<30 | 0.209 | 0.213 | 0.284 | 0.286 | 1.03 (0.98,1.09) | 2.55×10−1 |
| 30+ | 0.144 | 0.174 | 0.200 | 0.229 | 1.15 (1.08,1.22) | 6.11×10−6 |
| (missing) | 0.255 | 0.243 | ||||
| Endometriosis | ||||||
| No | 0.703 | 0.695 | 0.937 | 0.902 | Ref | |
| Yes | 0.047 | 0.076 | 0.063 | 0.098 | 1.60 (1.46,1.75) | 3.41×10−23 |
| (missing) | 0.250 | 0.230 | ||||
Abbreviations: OR, odds ratio; OCP, oral contraceptive pills; BMI, body mass index; HT, menopausal hormone therapy; ET, menopausal estrogen therapy; EPT, menopausal estrogen + progestin therapy; Ref, reference group.
Harmonized environmental data before imputation. Results of the complete cases analysis are provided in eTable 2.
Based on ten imputed E datasets.
Logistic regression model adjusted for reference age, race, education, family history, OCP use, tubal ligation, parity, BMI, endometriosis and study site.
Four other environmental variables were included in our analysis, as covariates: baseline age (<50, 50-<55, 55-<60, 60-<65, 65–70, 70+ years), race (non-Hispanic white, Hispanic White, Black, Other), education (less than high school, high school graduate, some college, college graduate) and first-degree family history of ovarian cancer (yes/no). In addition to these four covariates, study site, OCP use, tubal ligation, parity, BMI and endometriosis were also included in all models for the environmental association analysis and gene by environment interaction analysis.
Harmonization and Imputation of Environmental Data:
A brief description of environmental data harmonization across OCAC study sites is provided in eMethod 1 in the Supplementary Material. To optimize power and enhance the chance for discovery, we carried out multiple imputation of the environmental data. The maximal amount of data was used for imputation (see eMethod 1 and eFigure 1 in the Supplementary Material for details). A total of 19 studies comprising 13,722 cases and 22,975 controls with partially missing data were included for imputation. Of these 19 studies, 12 were from the US, 4 from Europe, 2 from Canada and 1 from Australia (see eTable 1 for a description of study sites). Further details for these 19 studies have been previously described (see Supplementary Material). The environmental variables included in our analysis were multiply imputed by chained equations (MICE) to produce ten imputed datasets. See details of imputation model in eMethod 2.1 in the Supplementary Material.
All analyses were performed on each of the ten imputed datasets, and coefficients/test statistics were properly combined to account for uncertainty due to imputation, following the recommended combination rule for multiply imputed datasets 38 (see details in eMethod 2.3 in the Supplementary Material). Our marginal environmental association analysis was based on combined inference from the ten imputed versions of this harmonized E data. Logistic regression models were used for evaluating marginal associations between the environmental risk factors with ovarian cancer after adjusting for covariate. The estimated ORs, their 95% CIs, as well as two-sided Wald tests after accounting for imputation uncertainty are presented in Table 2 along with summary statistics of complete cases before imputation. Full results of the complete cases analysis using logistic regression models are presented in eTable 2.
Gene by Environment Interaction Analysis
After marginal analysis of the genetic and environmental risk factors, we considered gene by environment (G × E) interaction analysis both on the multiplicative (odds ratio/relative risk) and the additive (relative excess risk due to interaction/absolute risk) scale 39. From the 19 studies with imputed environmental data, a subset of 17 case-control studies with 9,971 cases and 15,566 controls had available genetic data, thus G × E analyses were carried out on these 17 studies. Each imputed environmental dataset was merged with the genetic data for subsequent G × E analyses. Interaction analyses were then carried out separately on the ten imputed G × E datasets, and then all tests and coefficients reported were combined using appropriate multiple imputation combination rules 38.
For both multiplicative and additive interaction analysis, we started with global likelihood ratio tests (LRTs) for each G × E pair as several environmental factors had multiple categories resulting in tests for interactions with multiple degrees of freedom (df). These global joint tests, serving as a screening step for G × E interactions, were carried out for a total of 196 (7×28=196) G × E pairs. After the global tests, we then followed up on the suggestive interactions (with global test P-value < 0.2) and carried out a two-sided Wald test for interactions involving each separate category of an environmental risk factor.
For the k-th SNP , coded as a continuous allelic dosage, the j-th environmental risk factor , and a set of confounders/covariates , the basic fitted model for the probability of ovarian cancer of the i-th subject, namely, , is of the following form:
| [M1] |
where L = (levels of ) – 1, = (levels of ) – 1, and Q is the number of adjusted covariates.
Multiplicative Interaction Tests:
For testing the multiplicative interaction between and , we first used the global LRT with L degrees of freedom to test for the joint null hypothesis . If the global test P-value < 0.2, we further assessed the multiplicative interaction at each level of by using a Wald test with one degree of freedom for the null hypothesis for the l-th level.
Additive Interaction Tests:
Due to limitations of existing software (CGEN) 40 for testing additive interactions with continuous dosage data, we used the maximal probable genotype for imputed SNPs. We further conducted the LRTs with binary collapsing of SNPs assuming a dominant genetic susceptibility model (given the constraints in software) 31. For a given SNP and an environmental risk factor with L categories, a global LRT with L df was used for the following joint null hypothesis
where the regression coefficients are log odds ratio parameters described in model [M1]. This null hypothesis is based on a rare disease assumption 41, which is tenable for our study (lifetime risk of ovarian cancer in the US is approximately 1.3%) 42. If the global LRT P-value < 0.2, we further assessed the additive interaction at each level of through the relative excess risk due to interaction (RERI) 41. At the l-h level of , a Wald test with one degree of freedom (35) was used to test for the null hypothesis:
After the screening step, we further explored the structure of the most promising interactions (defined as global test P-value < 0.01). This was accomplished by exploring odds ratios corresponding to E in sub-groups defined by G (for the multiplicative interaction) or absolute risks for ovarian cancer in each configuration of the values of (G, E) (for the additive interaction). To better understand these two different scales of interaction, we also compared the observed joint ORs with the corresponding expected ORs under the multiplicative and the additive nulls.
To estimate sub-group specific absolute risk (AR) for each stratum defined by a given SNP and environmental risk factor, we need the relative risk and the joint distribution of and . The former was estimated from the fitted model [M1], and the latter was empirically estimated from the observed joint frequency of and in the control population (details in eMethod3 from the Supplementary Material). Table 4 presents the bootstrap confidence intervals for the estimated ARs and the risk differences (RDs) (see details in eMethod4 in the Supplementary Material). The results for G × E analysis are presented in Table 3 (multiplicative interaction), Table 4 (additive interaction) and eTable 5 (observed and expected joint OR under the two different nulls). All calculations were performed in the statistical software R 30, 40.
Table 4.
Absolute risks and risk differences stratified by levels of environmental risk factor and levels of genotype (for G-E pairs with global likelihood ratio test p-value < 0.01 on additive scale. Analysis used the G×E data with 9971 cases and 15566 controls from 17 study sites).
| SNPs | Environmental risk factor | N (cases/controls)a | Estimated ARs or RDs for E stratified by SNPs (95%CI)c | Global LRTd | Wald Teste | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| risk/baseline allele | variable | category | Genotype | Genotype | (df) | (df) | ||||
| rs11658063 G/C | Type of HT | CC | CG | GG | CC | CG | GG | |||
| Neither | 589/1142 | 2609/4518 | 3310/4956 | 1.27% (1.23%,1.32%) | 1.30% (1.28%,1.33%) | 1.33% (1.26%,1.40%) | Ref | Ref | ||
| ET only | 66/98 | 281/409 | 416/454 | 1.36% (1.15%,1.57%) | 1.63% (1.46%,1.79%) | 1.96% (1.59%,2.33%) | ||||
| RDb | 0.09% (−0.14%,0.31%) | 0.33% (0.15%,0.50%) | 0.63% (0.24%,1.02%) | 3.29×10−3(2) | 3.01×10−2(1) | |||||
| Any EPT | 105/207 | 498/952 | 606/1046 | 1.16% (1.04%,1.28%) | 1.21% (1.12%,1.30%) | 1.27% (1.09%,1.44%) | ||||
| RD | −0.12%(−0.26%,0.03%) | −0.09%(−0.20%,0.01%) | −0.06%(−0.26%,0.13%) | 7.04×10−1(1) | ||||||
| missing | 122/202 | 582/762 | 787/820 | |||||||
| rs9886651G/A | OCP use | AA | AG | GG | AA | AG | GG | |||
| Never | 1278/1718 | 2053/2502 | 900/1028 | 1.52% (1.42%,1.62%) | 1.70% (1.64%,1.76%) | 1.91% (1.77%,2.04%) | Ref | Ref | ||
| Ever | 1666/3105 | 2640/4978 | 1194/2072 | 1.07% (1.02%,1.12%) | 1.10% (1.07%,1.13%) | 1.14% (1.07%,1.21%) | ||||
| RD | −0.45%(−0.57%, −0.33%) | −0.60%(−0.69%, −0.51%) | −0.77%(−0.93%, −0.60%) | 5.32×10−3(2) | 9.90×10−3(1) | |||||
| missing | 70/47 | 113/79 | 57/37 | |||||||
Abbreviation: SNP, single-nucleotide polymorphism; AR, absolute risk; RD, risk difference; OCP, oral contraceptive pills; HT, menopausal hormone therapy; ET, menopausal estrogen therapy; EPT, menopausal estrogen + progestin therapy; Ref, reference group; df, degree of freedom.
Number of cases and controls were estimated from the original merged G×E data (before imputation) with 9971 cases and 15566 controls from 17 study sites, using maximal probable genotypes for imputed SNPs.
The risk difference corresponds to given category compared to the reference group, stratified by SNP.
ARs were estimated from logistic regression model by empirically estimated distribution of E and SNPs, while fixing all other covariates at their mode (determined from the original data).
LRT was performed for jointly testing additive interactions, assuming dominant effect model of SNPs (due to limitation of software).
1-df Wald test corresponds to the test individual RERI term (SNP = 2 vs SNP = 0, E = k vs E = reference group) is zero or not.
All models were estimated from logistic regression model with SNP, E variable, SNP E variable, assuming log-additive model (except for additive LRT which assumes dominant effect), using maximal probable genotypes for imputed SNPs, adjusted for reference age, race, education, family history, OCP use, tubal ligation, parity, BMI, endometriosis and study site and were performed on imputed datasets of G-E (9971 cases, 15566 controls) with proper pooling.
Table 3.
Odds ratios for marginal associations of seven environmental risk factors with ovarian cancer risk with 13722 cases and 22975 controls from 19 study sites.
| SNP | Environmental risk factor | N (cases/controls)a | Estimated ORb for E stratified by G (95%CI) | Globalc LRT | Waldd Test | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Risk/Baseline allele | Variable | Category | Genotype | Genotype | (df) | (df) | ||||
| rs13255292 C/T | OCP use | TT | TC | CC | TT | TC | CC | |||
| Never | 396/503 | 1758/2175 | 2077/2570 | Ref | Ref | Ref | ||||
| Ever | 446/1069 | 2286/4336 | 2768/4750 | 0.5 (0.46,0.60) | 0.61 (0.57,0.66) | 0.71 (0.66,0.77) | 3.48×10−4 (1) | 3.47×10−4 (1) | ||
| Missing | 24/15 | 96/56 | 120/96 | |||||||
| rs13255292 C/T | Duration of OCP use | TT | TC | CC | TT | TC | CC | |||
| < 1 yr | 451/636 | 2213/2670 | 2546/3145 | Ref | Ref | Ref | ||||
| 1−<5 yr | 171/362 | 854/1522 | 1082/1662 | 0.58 (0.50,0.69) | 0.68 (0.63,0.74) | 0.79 (0.72,0.87) | 7.26×10−3 (2) | 4.74×10−3 (1) | ||
| 5+ yr | 209/568 | 945/2269 | 1178/2470 | 0.43 (0.37,0.5) | 0.48 (0.44,0.52) | 0.53 (0.49,0.58) | 2.43×10−2 (1) | |||
| Missing | 35/21 | 128/106 | 159/135 | |||||||
| rs10962643 C/A | Parity (full term birth) | AA | AC | CC | AA | AC | CC | |||
| 0 | 230/220 | 940/940 | 1194/1080 | Ref | Ref | Ref | ||||
| 1–2 | 398/835 | 1741/3184 | 2202/3536 | 0.52 (0.44,0.61) | 0.56 (0.51,0.6) | 0.60 (0.54,0.66) | 7.52×10−3 (2) | 1.99×10−1 (1) | ||
| 3+ | 243/579 | 1242/2459 | 1664/2614 | 0.38 (0.32,0.46) | 0.46 (0.42,0.5) | 0.55 (0.49,0.61) | 2.86×10−3 (1) | |||
| Missing | 11/15 | 47/58 | 59/46 | |||||||
| chr9:16915105 C/G | Parity (full term birth) | GG | GC | CC | GG | GC | CC | |||
| 0 | 73/72 | 624/649 | 1667/1519 | Ref | Ref | Ref | ||||
| 1–2 | 111/300 | 1129/2285 | 3101/4970 | 0.46 (0.36,0.58) | 0.52 (0.47,0.59) | 0.60 (0.55,0.65) | 5.25×10−3 (2) | 5.10×10−2 (1) | ||
| 3+ | 70/220 | 749/1679 | 2330/3753 | 0.33 (0.26,0.43) | 0.42 (0.37,0.48) | 0.53 (0.48,0.58) | 1.25×10−3 (1) | |||
| missing | 2/7 | 37/36 | 78/76 | |||||||
Abbreviation: SNP, single-nucleotide polymorphism; OR, odds ratio; OCP, oral contraceptive pills; yr, year; Ref, reference group; df, degree of freedom, LRT, likelihood ratio test.
Number of cases and controls were estimated from the original merged GxE data (before imputation) with 9971 cases and 15566 controls from 17 study sites, using maximal probable genotypes for imputed SNPs.
ORs were estimated from the logistic regression model with SNP, E variable, SNP E variable.
LRT was performed for jointly testing multiplicative interactions.
Wald test for individual multiplicative interaction.
All models were estimated from the logistic regression model with SNP, E variable, SNP E variable, assuming log-additive model, using dosage data for imputed SNPs, adjusted for reference age, race, education, family history, OCP use, tubal ligation, parity, BMI, endometriosis and study site and were performed on imputed datasets of G-E (9971 cases, 15566 controls) with proper pooling.
RESULTS
The marginal G analysis was carried out on 26,864 cases and 48,034 controls and the results are shown in Table 1. These results are available through the OCAC website (http://apps.ccge.medschl.cam.ac.uk/consortia/ocac/). A total of 36,697 women with 13,722 ovarian cancer cases from 19 sites were included in the marginal E analysis using the imputed datasets. All seven environmental risk factors were associated with ovarian cancer in the expected direction (Table 2). OCP use for five or more years was associated with a 52% decrease in risk of ovarian cancer compared to never users (OR=0.48, 95%CI = 0.45 to 0.51). Tubal ligation (OR=0.73, 95%CI = 0.69 to 0.78) and breastfeeding (OR=0.76, 95%CI = 0.71 to 0.80) showed similar magnitudes of decreased risk. Also, having more than 3 children (versus none) was associated with a 50% (OR=0.5, 95%CI = 0.46 to 0.53) reduction in risk of ovarian cancer. Using menopausal estrogen therapy only for more than one year (OR=1.22, 95%CI = 1.12 to 1.34), being obese (OR=1.15, 95%CI = 1.08 to 1.22), and history of endometriosis (OR=1.60, 95%CI = 1.46 to 1.75) were all associated with increased risk of ovarian cancer. The inference remained robust before and after imputation (eTable 2.).
Gene by Environment Interaction Results
Global Likelihood Ratio Tests:
The global LRT essentially serves as a screening approach to identify a list of potentially interesting interactions. All interactions with global LRT P-value < 0.2 (40 on multiplicative scale and 41 on additive scale) are listed in eTable 3, while more detailed analysis of the top interactions, which showed the strongest significance (P-value < 0.01; 4 on multiplicative and 2 on additive scale), are shown in Table 3 and Table 4, respectively.
According to Global LRT results, the top interaction on the multiplicative scale was identified with the SNP rs13255292 and OCP use (ever and never use: P-value = 3.48 × 10−4; duration of use [<1 yr, 1–5 yr, 5+ yr]: P-value = 7.26 × 10−3) (Table 3). None of the observed interactions were significant based on a Bonferroni threshold of 0.05/(28 × 7)= 2.55 × 10−4.
Wald Tests for Multiplicative interactions:
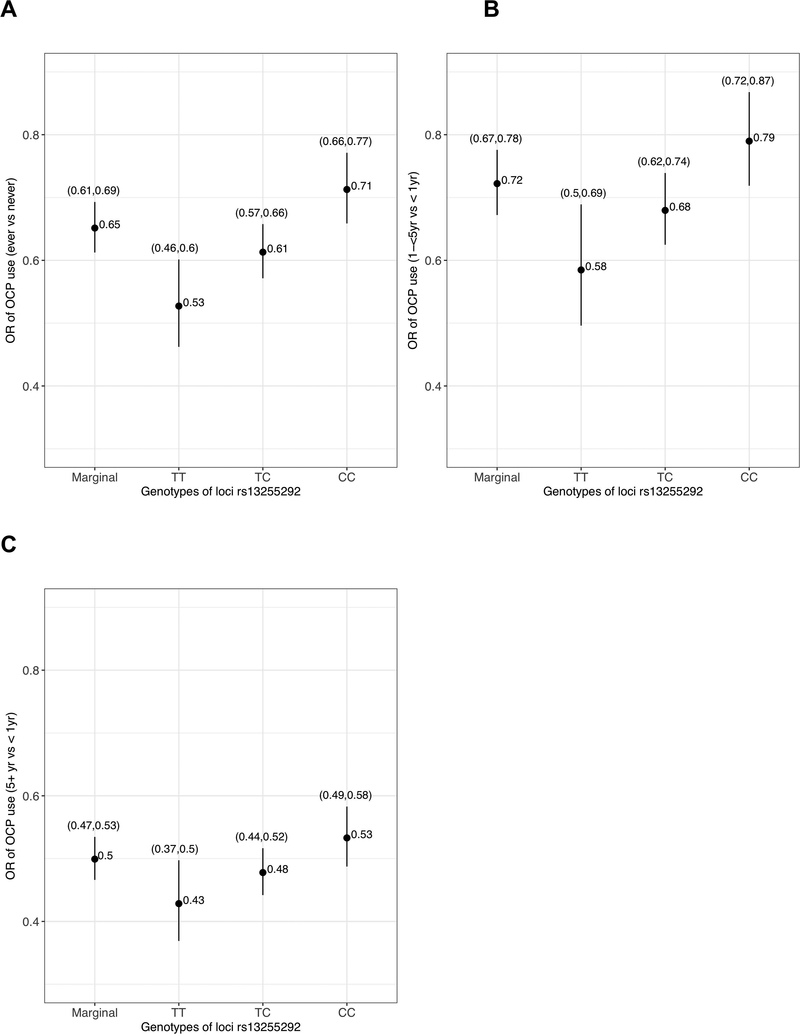
For the most promising multiplicative interactions reported in Table 3 we carried out an in-depth analysis to better understand the structure of interactions by estimating the ORs (with accompanying Wald CIs and tests) corresponding to E in strata defined by G. For example, the OR for OCP use among women with the TT genotype for rs13255292 is estimated to be 0.53 (95%CI = 0.46 to 0.60), whereas for the CC genotype the estimated OR is 0.71 (95%CI = 0.66 to 0.77) suggesting a stronger protective effect of OCP use among TT genotypes (Table 3, Figure 1A).
Figure 1A–1C.
ORs of oral contraceptive (OCP) use, marginally, or stratified by number of risk allele of rs13255292. The ORs were calculated from a logistic regression model assuming log-additive effect of SNPs. (A) OR of OCP (ever vs never) (B) OR of 1 to 5 years of OCP use (vs < 1 year) (B) OR of more than 5 years of OCP use (vs < 1 year).
When OCP use was further stratified by duration, we observed an interesting pattern in its interaction with rs13255292. The estimated OR corresponding to 1–5 year of OCP use vs < 1 year use in the TT genotype group was 0.58 (95%CI = 0.50 to 0.69) compared to an OR of 0.79 (95%CI = 0.72 to 0.87) among women with CC genotype, showing effect modification by the risk allele (C) of rs13255292 (Table 3, Figure 1B). This is akin to the result with ever/never user. However, the OR corresponding to 5+ years of OCP use vs < 1 year of use for the TT genotype group was 0.43 (95%CI = 0.37 to 0.50) and for the CC genotype was 0.53 (95%CI = 0.49 to 0.58) (Table 3, Figure 1C). With overlapping confidence intervals, there is no significant difference in the odds ratios for long-term OCP users across genotype sub-groups. Table 3 shows that the P-value of the Wald test for interaction of rs13255292 and 1–5 years of OCP use (vs < 1 yr) was lower (P-value = 4.74 × 10−3), when compared to the P-value for interaction of the same variant with 5+ years of OCP use (vs < 1 yr) (P-value = 2.43 × 10−2).
Wald Test for Additive interaction/RERI:
For the most statistically significant additive interactions in Table 4, we estimated the sub-group specific absolute risks (ARs) and risk differences (RDs) in each E by G stratum. For example, for the strongest additive interaction based on the global likelihood ratio tests in Table 4, there was suggestive evidence that rs11658063 modified the effect of menopausal estrogen therapy use, compared to never use of menopausal hormone therapy (P-value = 3.01 × 10−2). Among women with the GG genotype, never users of menopausal hormone therapy had an estimated AR of 1.33% (95%CI =1.26% to 1.40%) while women who used menopausal estrogen therapy had an estimated AR of 1.96% (95%CI = 1.59% to 2.33%), leading to an absolute risk increase of 0.63% (95%CI = 0.24% to 1.02%) (Table 4, eFigure 2).
For women with the CC genotype, the estimated AR was 1.27% (95%CI = 1.23% to 1.32%) for never receiving menopausal hormone therapy and 1.36% (95%CI = 1.15% to 1.57%) for receiving menopausal estrogen only therapy. This implies virtually no increased risk from taking menopausal estrogen only therapy among women with the CC genotype (95%CI = −0.14% to 0.31%; Table 4, eFigure 2). The results on the additive interactions were in general weaker in terms of the strength of P-values.
DISCUSSION
We have conducted a comprehensive multiplicative and additive interaction analysis of previously identified common genetic variants and environmental factors unequivocally associated with ovarian cancer risk. We observed six suggestive interactions (with P-value < 0.01), four on the multiplicative scale and two on the additive scale. The lack of statistical significance of interactions after multiple testing correction from a large collection of data and well-curated studies enable us to conclude that it is unlikely that there are substantive interactions with single variants and environmental factors regardless of the choice of scale. This is consistent with what has been observed for other cancers. One may argue that the Bonferroni threshold for multiple comparisons is likely to be conservative for this set of correlated environmental factors, but the general pattern of findings remains consistent with smaller magnitude of interaction effect sizes. However, there are several interesting findings from this analysis that may be worthwhile to follow-up in future G × E studies of ovarian cancer.
Mechanistic Insight:
In addition to guiding targeted prevention strategies, G × E analysis has the potential to provide mechanistic insight into the complex multifactorial structure of the underlying biological pathway. One issue complicating observed gene-environment interactions of even confirmed susceptibility loci is that the true casual alleles and the biological impact of the variants are unknown. Our top interaction is between OCP use and rs13255292. This variant lies in the 8q24 region which harbors several risk loci for ovarian cancer 18 and other cancers 43, 44. The SNP is in the PVT1 gene which interacts with the oncogene MYC 45. MYC has long been reported to be at least in part under hormonal control 46, 47 thus an interaction with OCP use is plausible. Conversely, our top additive interaction is between menopausal estrogen use and rs11658063 which falls in HNF1B. To our knowledge there is no relationship between HNF1B and hormones thus underscoring the difficulty of understanding these gene-environment interactions given our limited understanding of the function of the variants and even more broadly the biological role of the genes.
Exposure Pathways and Potential for Targeted Prevention:
The strongest interactions are observed with OCP use or menopausal estrogen use which are modifiable exposures. Our most promising finding is the potential interaction between SNP rs13255292 and OCP use. This finding, if replicated could potentially lead to improved understanding of exposure pathways.
Analytic Architecture and the Choice of Scale for Measuring Interaction:
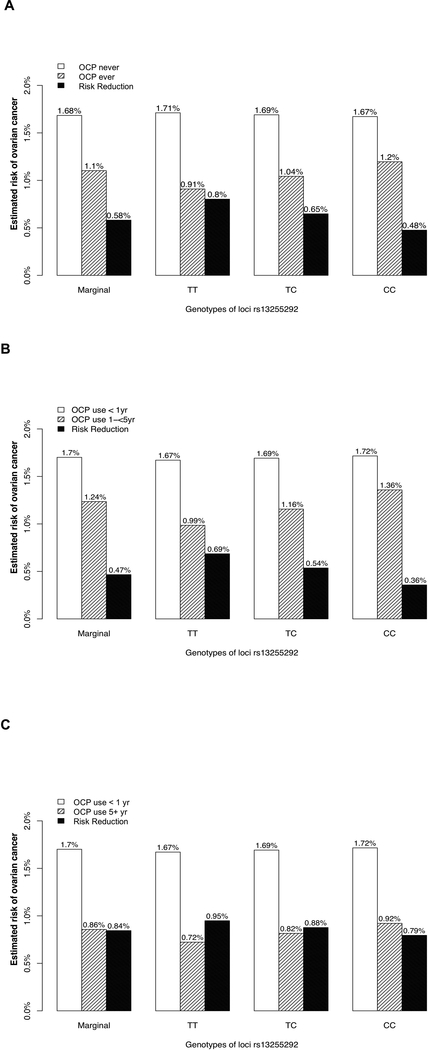
We present a comprehensive analytical framework to carry out post-GWAS G × E analysis on both multiplicative and additive scale. Our framework starting with data harmonization and imputation followed by Global likelihood ratio tests and single df Wald tests provides a principled analytic architecture for such analysis. Our analysis reiterates the well-known fact that testing the additive and multiplicative nulls are very similar when the marginal associations are weak but could depart when both marginal associations are large in magnitude and the sample size is finite. In eTable 5, we present observed joint odds ratios for strata defined by G and E along with the expected odds ratios under the multiplicative null and the additive null. We use our top hit rs13255292 and OCP use (ever versus never) and length of OCP use (<1yr, 1-<5 yrs, 5+ yrs) as an illustration. One can note that the expected ORs are fairly close under both models. However, their estimated departure from the observed joint OR is more pronounced for the 1-<5 yrs sub-group when compared to 5+ yrs, explaining the suggestive evidence for rejecting the null.
We discussed the multiplicative interaction results for rs13255292 and OCP use in the previous section. We now explore the structure of additive interaction for this G × E result (Figure 2A-2C). Marginally, without including any genetic information, from a pure environmental association analysis we observed a relationship between duration of OCP use and risk reduction for ovarian cancer. For 1–5 years of OCP use (vs <1 year) the estimated absolute risk difference was 0.47% (95%CI = 0.37% to 0.56%), while the estimated absolute risk difference for long-term use of OCPs (5+ year vs <1 year) was 0.84% (95%CI = 0.77% to 0.92%) (Figure 2B-2C, eTable 4), in agreement with previous findings that longer duration of OCP use is associated with larger risk reduction in ovarian cancer 3. However, when stratified by rs13255292 genotype, we observed an interesting pattern. Among individuals with TT genotype, the corresponding absolute risk difference estimate for 1–5 year of OCP use (vs <1 year) was 0.69% (95%CI = 0.49% to 0.88%), whereas among individuals with CC genotypes the corresponding risk reduction estimate was 0.36% (95%CI = 0.22% to 0.50%), implying potential effect modification by the C allele at locus rs13255292 (P-value = 1.12 × 10−2) (Figure 2B, eTable 4). In contrast, the absolute risk difference is estimated at 0.95% (95%CI = 0.78% to 1.12%) for women with TT genotype and at 0.79% (95%CI = 0.69% to 0.90%) in women with CC genotype. This indicates that longer OC use is associated with greater risk reduction overall and the risk reduction might be even greater for women with the TT genotype than those with the CC genotype. From Figure 2B-2C we observe the interplay between “nature vs nurture” with risk due to germline genetic mutations offset by long-term use of a modifiable protective factor. This analysis also highlights the benefit of measuring duration of exposure as opposed to a coarse indicator of ever/never use.
Figure 2A–2C.
Estimated absolute risk (AR) of ovarian cancer given OCP use and number of copies of C allele, among non-Hispanic white college graduates aged below 50 with no family history of ovarian cancer, BMI below 25, no tubal ligation, no endometriosis, with one child. The ARs were calculated from a logistic regression model assuming log-additive effect of SNPs while all covariates fixed at their most frequent level as described above. (A) ARs stratified by OCP (ever vs never) and genotype (B) ARs stratified by 1 to 5 years of OCP use (vs < 1 year) and genotype (F) ARs stratified by more than 5 years of OCP use (vs < 1 year) and genotype. Risk differences were also reported as the solid black bar.
Prior work in G × E for ovarian cancer has focused solely on multiplicative interactions. We previously reported no departures from a multiplicative model with the first six risk loci identified through GWAS with a reduced set of exposures 3. Follow-up work identified an interaction with menopausal estrogen therapy use and rs10069690 in the TERT gene 48, but that finding was not replicated in the present analysis which included a larger set of studies. Fridley and colleagues have reported on G × E taking a candidate gene approach with several promising findings 49. There are several studies in other cancers examining G × E on the multiplicative scale with limited success in identifying interactions, but to our knowledge, only prostate cancer and bladder cancer have been studied on the additive scale. In prostate cancer, suggestive additive interactions between vitamin D, confirmed genetic variants and risk have been identified 50. In bladder cancer, additive interaction has been explored between confirmed genetic loci and smoking with risk of disease 31. In this work the authors were able to demonstrate that the absolute risk of bladder cancer for current smokers varied from 2.9% to 9.9% based on the polygenetic risk score quartile. These results are similar to our findings on the additive scale with absolute risk differing based on genetics and hormone therapy use; an interesting next step for our work is to consider the polygenetic risk score for all of these confirmed ovarian cancer susceptibility alleles.
There are several limitations of the current analysis. Though we considered both multiplicative and additive interactions, the logistic model in (M1) is linear in covariates and exposures. We ignored potential non-linearity and exposure x exposure as well as exposure x covariate interactions. Similarly, we ignored any higher order interactions. A completely non-parametric machine learning approach, based on a recursive partition of the predictor space may avoid misspecification of the model, but would lack interpretability from an epidemiologic and public health perspective. We also acknowledge that this exploration of interaction is purely statistical, a more causal interpretation in a biological sense will require functional validation. One may also want to explore G × E interaction with loci that are not significant at genome-wide threshold but are significant at a less stringent threshold or even conduct genome-wide G × E scans.
The associations between ovarian cancer risk and some of the variants included here were limited to specific histotypes of ovarian cancer, however we have only presented results for all epithelial ovarian cancers combined. Developing histotype-specific risk stratification approaches is not feasible because for any given histotype the absolute risk is unlikely to ever reach an actionable threshold on a population level. In addition, risk reducing strategies are the same across histotypes and thus there is little benefit to considering histotype specific results from a precision prevention perspective. Heterogeneous associations between environmental risk factors and ovarian cancer risk by histology has previously been well characterized 3, 23, 27. There is value in understanding histotype associations for disease etiology and mechanisms and this will be the focus of future work.
The analyses presented here offer insight into potential biological mechanisms, opportunities for ovarian cancer risk stratification, and approaches to studying gene-environment interactions. Ideally, replication for the six promising findings would be undertaken, but this is challenging with ovarian cancer given that most studies with the relevant data are included here. Functional studies for the regions harboring our most promising findings are underway and it is possible that the association described here may help inform those investigations 51. Also, gene-environment interaction analyses can also be used to identify novel genetic associations 51 and thus a deeper evaluation of variants that are still borderline significant, but do not exactly achieve a genome-wide threshold is warranted for subsequent G × E analysis. Of particular interest will be to conduct risk stratification and risk prediction analysis using a summative polygenic risk score and to conduct an agnostic genome-wide search for G × E interaction. Despite the limitations the comprehensive framework of data harmonization, imputation, screening test followed by characterization of effect and risk estimates that has been used in this analysis can serve as a robust model for future gene-environment interaction analyses.
Supplementary Material
Novelty and Impact:
Our paper conducts gene x environment interaction analysis on both additive and multiplicative scales using data from 9,971 ovarian cancer (OC) cases and 15,566 controls. Seven OC risk factors are considered with 28 variants identified from previous GWAS. The top interaction was between oral contraceptive pill (OCP) use (ever vs never) and rs13255292 (P-value=3.48×10−4). The protective benefit of OCP use differs by genotype suggesting that prevention strategies need tailoring to an individual’s genotypic profile.
ACKNOWLEDGEMENTS
The AOCS also acknowledges the cooperation of the participating institutions in Australia and acknowledges the contribution of the study nurses, research assistants and all clinical and scientific collaborators to the study. The complete AOCS Study Group can be found at www.aocstudy.org. We would like to thank all of the women who participated in these research programs. The cooperation of the 32 Connecticut hospitals, including Stamford Hospital, in allowing patient access, is gratefully acknowledged. This study was approved by the State of Connecticut Department of Public Health Human Investigation Committee. Certain data used in this study were obtained from the Connecticut Tumor Registry in the Connecticut Department of Public Health. The authors assume full responsibility for analyses and interpretation of these data. The German Ovarian Cancer Study (GER) thank Ursula Eilber for competent technical assistance. The NHS/NHSII studies thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. We particularly thank I. Jacobs, M.Widschwendter, E. Wozniak, A. Ryan, J. Ford and N. Balogun for their contribution to the study. We also thank Harvey Risch for contributing data for this analysis. Usha Menon has shares in Abcodia Ltd., which has an interest in early cancer detection.
FINANCIAL SUPPORT
AUS (GCT, PMW): The Australian Ovarian Cancer Study was supported by the U.S. Army Medical Research and Materiel Command under DAMD17–01-1–0729, the Cancer Councils of New South Wales, Victoria, Queensland, South Australia and Tasmania and The Cancer Foundation of Western Australia (Multi-State Applications 182, 191 and 211) and the National Health and Medical Research Council of Australia (NHMRC; ID400413, ID400281, 199600 and 1043134). The Australian Ovarian Cancer Study gratefully acknowledges additional support from Ovarian Cancer Australia and the Peter MacCallum Foundation; CON (HAR): National Institutes of Health (R01-CA063678, R01-CA074850; R01-CA080742); DOV (HRH, JAD, MAR): National Institutes of Health R01-CA112523, R01-CA87538 and K22 CA193860; GER (JCC, RTF): German Federal Ministry of Education and Research, Programme of Clinical Biomedical Research (01 GB 9401) and the German Cancer Research Center (DKFZ); HAW (MTG, PJT): U.S. National Institutes of Health (R01-CA58598, N01-CN-55424 and N01-PC-67001); HOP (FM, RBN): DOD: DAMD17–02-1–0669 and NCI: K07-CA080668, R01-CA95023, P50-CA159981; NIH/National Center for Research Resources/General Clinical Research Center grant MO1-RR000056; MAL (AJ, SKK): Funding for this study was provided by research grant R01- CA61107 from the National Cancer Institute, Bethesda, Md; research grant 94 222 52 from the Danish Cancer Society, Copenhagen, Denmark; and the Mermaid I project; MAY (ELG, SJW): National Institutes of Health (R01-CA122443, P30-CA15083, P50-CA136393); Mayo Foundation; Minnesota Ovarian Cancer Alliance; Fred C. and Katherine B. Andersen Foundation; NCO (AB, JMS): National Institutes of Health (R01-CA76016) and the Department of Defense (DAMD17–02-1–0666); NEC (DWC, KLT): National Institutes of Health R01-CA54419 and P50-CA105009 and Department of Defense W81XWH-10–1-02802; NHS (SST): UM1 CA186107, P01 CA87969, R01 CA49449, R01-CA67262, UM1 CA176726; NJO (EVB, SHO): National Cancer Institute (NIH-K07 CA095666, R01-CA83918, NIH-K22-CA138563, P30-CA072720, and P30-CA008748) and the Cancer Institute of New Jersey; OVA (ABW, LSC, NDL): This work was supported by Canadian Institutes of Health Research grant (MOP-86727) and by NIH/NCI 1 R01CA160669–01A1; POL (BT, NW): Intramural Research Program of the National Cancer Institute; SON (HAR): National Health Research and Development Program, Health Canada, grant 6613–1415-53; STA (ASW, WS): NIH grants U01 CA71966 and U01 CA69417; UCI (HAC, AZ): NIH R01-CA058860, and the Lon V Smith Foundation grant LVS-39420; UKO (UM, SAG): The UKOPS study was funded by The Eve Appeal (The Oak Foundation) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre; USC (AWL, AHW, CLP, MCP): P01CA17054, P30CA14089, R01CA61132, N01PC67010, R03CA113148, R03CA115195, N01CN025403, T32 ES013678, and California Cancer Research Program (00–01389V-20170, 2II0200).
This work was supported by the National Cancer Institute, US National Institutes of Health (grant R01 CA076016, P30 CA04), and the National Cancer Institute’s Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative (grant U19-CA148112). and the National Cancer Institute (Grant P30 CA046592 for CLP and BM). Lastly, this work was also supported by the National Science Foundation (grant NSF DMS 1406712 BM) and the National Institute of Environmental Health Sciences (NIH ES 20811 BM).
The Collaborative Oncological Gene Environment Study is funded through the European Commission’s Seventh Framework Programme (agreement 223175 HEALTH F2 2009–223175). The Ovarian Cancer Association Consortium is supported by a grant from the ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (grant PPD/RPCI.074)
Abbreviations:
- AR
absolute risk
- BMI
body mass index
- BSO
bilateral salpingo-oophorectomy
- CI
confidence interval
- df
degrees of freedom
- G × E
gene-environment interaction
- GWAS
genome-wide association study
- LRT
likelihood ratio test
- OCAC
Ovarian Cancer Association Consortium
- OCP
oral contraceptive pill
- OR
odds ratio
- RD
risk difference
- SNP
single nucleotide polymorphism
REFERENCES
- 1.Society AC. Cancer Facts & Figures 2017. American Cancer Society; 2017. [Google Scholar]
- 2.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, Amso NN, Apostolidou S, Benjamin E, Cruickshank D, Crump DN, Davies SK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet (London, England) 2016;387:945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce CL, Rossing MA, Lee AW, Ness RB, Webb PM, for Australian Cancer S, Australian Ovarian Cancer Study G, Chenevix-Trench G, Jordan SM, Stram DA, Chang-Claude J, Hein R, et al. Combined and interactive effects of environmental and GWAS-identified risk factors in ovarian cancer. Cancer Epidemiol Biomarkers Prev 2013;22:880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trabert B, Ness RB, Lo-Ciganic WH, Murphy MA, Goode EL, Poole EM, Brinton LA, Webb PM, Nagle CM, Jordan SJ, Australian Ovarian Cancer Study Group ACS, Risch HA, et al. Aspirin, nonaspirin nonsteroidal anti-inflammatory drug, and acetaminophen use and risk of invasive epithelial ovarian cancer: a pooled analysis in the Ovarian Cancer Association Consortium. J Natl Cancer Inst 2014;106:djt431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sieh W, Salvador S, McGuire V, Weber RP, Terry KL, Rossing MA, Risch H, Wu AH, Webb PM, Moysich K, Doherty JA, Felberg A, et al. Tubal ligation and risk of ovarian cancer subtypes: a pooled analysis of case-control studies. Int J Epidemiol 2013;42:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falconer H, Yin L, Gronberg H, Altman D. Ovarian cancer risk after salpingectomy: a nationwide population-based study. J Natl Cancer Inst 2015;107. [DOI] [PubMed] [Google Scholar]
- 7.Lessard-Anderson CR, Handlogten KS, Molitor RJ, Dowdy SC, Cliby WA, Weaver AL, Sauver JS, Bakkum-Gamez JN. Effect of tubal sterilization technique on risk of serous epithelial ovarian and primary peritoneal carcinoma. Gynecol Oncol 2014;135:423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madsen C, Baandrup L, Dehlendorff C, Kjaer SK. Tubal ligation and salpingectomy and the risk of epithelial ovarian cancer and borderline ovarian tumors: a nationwide case-control study. Acta Obstet Gynecol Scand 2015;94:86–94. [DOI] [PubMed] [Google Scholar]
- 9.Yoon SH, Kim SN, Shim SH, Kang SB, Lee SJ. Bilateral salpingectomy can reduce the risk of ovarian cancer in the general population: A meta-analysis. Eur J Cancer 2016;55:38–46. [DOI] [PubMed] [Google Scholar]
- 10.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nature genetics 2013;45:371–84, 84e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY, Weidhaas J, Paik D, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nature genetics 2010;42:880–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, Widschwendter M, Vierkant RA, Larson MC, Kjaer SK, Birrer MJ, Berchuck A, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nature genetics 2010;42:874–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kar SP, Beesley J, Amin Al Olama A, Michailidou K, Tyrer J, Kote-Jarai Z, Lawrenson K, Lindstrom S, Ramus SJ, Thompson DJ, Kibel AS, Dansonka-Mieszkowska A, et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer discovery 2016;6:1052–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelemen LE, Lawrenson K, Tyrer J, Li Q, Lee JM, Seo JH, Phelan CM, Beesley J, Chen X, Spindler TJ, Aben KK, Anton-Culver H, et al. Genome-wide significant risk associations for mucinous ovarian carcinoma. Nature genetics 2015;47:888–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, Spindler TJ, Lin YG, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nature genetics 2015;47:164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Permuth-Wey J, Lawrenson K, Shen HC, Velkova A, Tyrer JP, Chen Z, Lin HY, Chen YA, Tsai YY, Qu X, Ramus SJ, Karevan R, et al. Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nature communications 2013;4:1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nature genetics 2013;45:362–70, 70e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar SP, Lawrenson K, Winham SJ, Dennis J, Pirie A, Riggan MJ, Chornokur G, Earp MA, Lyra PC Jr., et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nature genetics 2017;49:680–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H, Fridley BL, Song H, Lawrenson K, Cunningham JM, Ramus SJ, Cicek MS, Tyrer J, Stram D, Larson MC, Kobel M, Ziogas A, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nature communications 2013;4:1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, Anton-Culver H, Chang-Claude J, Cramer DW, DiCioccio R, Dork T, Goode EL, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nature genetics 2009;41:996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auranen A, Pukkala E, Makinen J, Sankila R, Grenman S, Salmi T. [Cancer incidence in the first-degree relatives of ovarian cancer patients]. Duodecim; laaketieteellinen aikakauskirja 1997;113:46–50. [PubMed] [Google Scholar]
- 22.Stratton JF, Pharoah P, Smith SK, Easton D, Ponder BA. A systematic review and meta-analysis of family history and risk of ovarian cancer. British journal of obstetrics and gynaecology 1998;105:493–9. [DOI] [PubMed] [Google Scholar]
- 23.Lee AW, Ness RB, Roman LD, Terry KL, Schildkraut JM, Chang-Claude J, Doherty JA, Menon U, Cramer DW, Gayther SA, Risch H, Gentry-Maharaj A, et al. Association Between Menopausal Estrogen-Only Therapy and Ovarian Carcinoma Risk. Obstetrics and gynecology 2016;127:828–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearce CL, Chung K, Pike MC, Wu AH. Increased ovarian cancer risk associated with menopausal estrogen therapy is reduced by adding a progestin. Cancer 2009;115:531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collaborative Group On Epidemiological Studies Of Ovarian C, Beral V, Gaitskell K, Hermon C, Moser K, Reeves G, Peto R. Menopausal hormone use and ovarian cancer risk: individual participant meta-analysis of 52 epidemiological studies. Lancet (London, England) 2015;385:1835–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen CM, Nagle CM, Whiteman DC, Ness R, Pearce CL, Pike MC, Rossing MA, Terry KL, Wu AH, Australian Cancer S, Australian Ovarian Cancer Study G, Risch HA, et al. Obesity and risk of ovarian cancer subtypes: evidence from the Ovarian Cancer Association Consortium. Endocr Relat Cancer 2013;20:251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce CL, Templeman C, Rossing MA, Lee A, Near AM, Webb PM, Nagle CM, Doherty JA, Cushing-Haugen KL, Wicklund KG, Chang-Claude J, Hein R, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol 2012;13:385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SEER Cancer Statistics Review 1975–2014, based on November 2016 SEER data submission, posted to the SEER web site, April 2017.
- 29.Pearce CL, Stram DO, Ness RB, Stram DA, Roman LD, Templeman C, Lee AW, Menon U, Fasching PA, McAlpine JN, Doherty JA, Modugno F, et al. Population Distribution of Lifetime Risk of Ovarian Cancer in the United States. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2015;24:671–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Lee S, Lee AW, Wu AH, Bandera EV, Jensen A, Anne Rossing M, Moysich KB, Chang-Claude J, Doherty J, Gentry-Maharaj A, Kiemeney L, et al. Robust Tests for Additive Gene-Environment Interaction in Case-Control Studies Using Gene-Environment Independence. Am J Epidemiol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Closas M, Rothman N, Figueroa JD, Prokunina-Olsson L, Han SS, Baris D, Jacobs EJ, Malats N, De Vivo I, Albanes D, Purdue MP, Sharma S, et al. Common genetic polymorphisms modify the effect of smoking on absolute risk of bladder cancer. Cancer Res 2013;73:2211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knol MJ, VanderWeele TJ, Groenwold RH, Klungel OH, Rovers MM, Grobbee DE. Estimating measures of interaction on an additive scale for preventive exposures. Eur J Epidemiol 2011;26:433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G, Mukherjee B, Lee S, Lee AW, Wu AH, Bandera EV, Jensen A, Rossing MA, Moysich KB, Chang-Claude J, Doherty JA, Gentry-Maharaj A, et al. Robust Tests for Additive Gene-Environment Interaction in Case-Control Studies Using Gene-Environment Independence. Am J Epidemiol 2018;187:366–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanderWeele TJ, Vansteelandt S. A weighting approach to causal effects and additive interaction in case-control studies: marginal structural linear odds models. Am J Epidemiol 2011;174:1197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.VanderWeele TJ. Sample Size and Power Calculations for Additive Interactions. Epidemiol Methods 2012;1:159–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolton KL, Ganda C, Berchuck A, Pharaoh PD, Gayther SA. Role of common genetic variants in ovarian cancer susceptibility and outcome: progress to date from the Ovarian Cancer Association Consortium (OCAC). Journal of internal medicine 2012;271:366–78. [DOI] [PubMed] [Google Scholar]
- 37.R L, D R. Chapter 10: Bayes and Multiple Imputation Statistical Analysis With Missing Data, 2nd ed. NJ: John Wiley & Sons, 2002. [Google Scholar]
- 38.Nickels S, Truong T, Hein R, Stevens K, Buck K, Behrens S, Eilber U, Schmidt M, Haberle L, Vrieling A, Gaudet M, Figueroa J, et al. Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS genetics 2013;9:e1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharjee SCN, Han S and Wheeler W. CGEN: An R package for analysis of case-control studies in genetic epidemiology, 2012.
- 40.Han SS, Rosenberg PS, Garcia-Closas M, Figueroa JD, Silverman D, Chanock SJ, Rothman N, Chatterjee N. Likelihood ratio test for detecting gene (G)-environment (E) interactions under an additive risk model exploiting G-E independence for case-control data. Am J Epidemiol 2012;176:1060–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.SEER. Cancer Stat Facts: Ovarian Cancer. In: SEER, ed., vol. 2017: National Cancer Institute. [Google Scholar]
- 42.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature genetics 2007;39:638–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J, Zhang Y, Zheng W, Michailidou K, Ghoussaini M, Bolla MK, Wang Q, Dennis J, Lush M, Milne RL, Shu XO, Beesley J, et al. Fine-scale mapping of 8q24 locus identifies multiple independent risk variants for breast cancer. Int J Cancer 2016;139:1303–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng YY, Bagchi A. The PVT1-MYC duet in cancer. Mol Cell Oncol 2015;2:e974467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science 2002;295:2465–8. [DOI] [PubMed] [Google Scholar]
- 46.Wang C, Mayer JA, Mazumdar A, Fertuck K, Kim H, Brown M, Brown PH. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol Endocrinol 2011;25:1527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee AW, Bomkamp A, Bandera EV, Jensen A, Ramus SJ, Goodman MT, Rossing MA, Modugno F, Moysich KB, Chang-Claude J, Rudolph A, Gentry-Maharaj A, et al. A splicing variant of TERT identified by GWAS interacts with menopausal estrogen therapy in risk of ovarian cancer. Int J Cancer 2016;139:2646–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usset JL, Raghavan R, Tyrer JP, McGuire V, Sieh W, Webb P, Chang-Claude J, Rudolph A, Anton-Culver H, Berchuck A, Brinton L, Cunningham JM, et al. Assessment of Multifactor Gene-Environment Interactions and Ovarian Cancer Risk: Candidate Genes, Obesity, and Hormone-Related Risk Factors. Cancer Epidemiol Biomarkers Prev 2016;25:780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimitrakopoulou VI, Travis RC, Shui IM, Mondul A, Albanes D, Virtamo J, Agudo A, Boeing H, Bueno-de-Mesquita HB, Gunter MJ, Johansson M, Khaw KT, et al. Interactions Between Genome-Wide Significant Genetic Variants and Circulating Concentrations of 25-Hydroxyvitamin D in Relation to Prostate Cancer Risk in the National Cancer Institute BPC3. Am J Epidemiol 2017;185:452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAllister K, Mechanic LE, Amos C, Aschard H, Blair IA, Chatterjee N, Conti D, Gauderman WJ, Hsu L, Hutter CM, Jankowska MM, Kerr J, et al. Current Challenges and New Opportunities for Gene-Environment Interaction Studies of Complex Diseases. Am J Epidemiol 2017;186:753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.