Abstract
Many genes differentially expressed in brain tissue from human alcoholics and animals that have consumed large amounts of alcohol are components of the innate immune toll-like receptor (TLR) pathway. TLRs initiate inflammatory responses via two branches: (1) MyD88-dependent or (2) TRIF-dependent. All TLRs signal through MyD88 except TLR3. Prior work demonstrated a direct role for MyD88-dependent signaling in regulation of alcohol consumption. However, the role of TLR3 as a potential regulator of excessive alcohol drinking has not previously been investigated. To test the possibility TLR3 activation regulates alcohol consumption, we injected mice with the TLR3 agonist polyinosinic:polycytidylic acid (poly(I:C)) and tested alcohol consumption in an every-other-day two-bottle choice test. Poly(I:C) produced a persistent increase in alcohol intake that developed over several days. Repeated poly(I:C) and ethanol exposure altered innate immune transcript abundance; increased levels of TRIF-dependent pathway components correlated with increased alcohol consumption. Administration of poly(I:C) before exposure to alcohol did not alter alcohol intake, suggesting that poly(I:C) and ethanol must be present together to change drinking behavior. To determine which branch of TLR signaling mediates poly(I:C)-induced changes in drinking behavior, we tested either mice lacking MyD88 or mice administered a TLR3/dsRNA complex inhibitor. MyD88 null mutants showed poly(I:C)-induced increases in alcohol intake. In contrast, mice pretreated with a TLR3/dsRNA complex inhibitor reduced their alcohol intake, suggesting poly(I:C)-induced escalations in alcohol intake function are, at least partially, dependent on TLR3. Together, these results strongly suggest that TLR3-dependent signaling drives excessive alcohol drinking behavior.
Keywords: alcohol use disorder, toll-like receptors, cytokines, poly(I:C), ethanol, neuroimmune, drinking
1. Introduction
Alcohol consumption activates peripheral and central inflammatory pathways leading to increases in innate immune signaling (1–3). Inflammatory and immune-related signaling genes are differentially expressed in the frontal cortex of human alcoholics (4, 5), selectively-bred high drinking mice (6) and ethanol-exposed mice (7–10), indicating a role for innate immune signaling in regulating alcohol intake. Moreover, proinflammatory cytokines have been detected in the serum of human alcoholics and higher levels of cytokines correlated with alcohol craving (11). Taken together, this suggests that inflammatory and immune-related signaling may regulate alcohol craving and consumption. Behavioral validation studies confirmed that mice with null mutations in different immune-related genes drink less ethanol, whereas activation of innate immune signaling using lipopolysaccharide (LPS) increases ethanol intake (12, 13). Many of the innate immune genes implicated in these studies mediate their effects through activation of toll-like receptors (TLRs).
TLRs recognize a broad range of molecular motifs known as pattern-associated molecular patterns (PAMPs) to initiate innate immune responses (14). TLRs initiate inflammatory responses via two intracellular signaling transduction cascades: (1) Myeloid differentiation response gene 88 (MyD88)-dependent or (2) TIR-domain-containing adapter-inducing interferon β (TRIF)-dependent. Only TLR3 initiates inflammatory responses solely through the TRIF-dependent pathway.
Prior studies indicate a direct role for TLR signaling in the regulation of alcohol intake. Specifically, the field has focused on TLR4/MyD88-dependent signaling (12, 15–19). For instance, activation of TLR4 via LPS increased alcohol intake; whereas, siRNA inhibition of the downstream MyD88-dependent signaling component IKKβ decreased alcohol consumption (12, 19). However, despite evidence that TLR4/MyD88-dependent signaling is important for alcohol responses, it was recently found that TLR4 is not a critical determinant of excessive drinking in animal models (20, 21). Moreover, a recent study could not replicate that LPS pretreatment increases alcohol consumption in continuous two-bottle-choice or drinking-in-the-dark (22). This suggests that other innate immune pathways may be important in the regulation of excessive alcohol consumption. We hypothesize that TLR3/TRIF-dependent signaling may be a key determinant of excessive drinking.
Increased expression of TRIF-dependent pathway transcripts and proteins has been found in the frontal cortex and nucleus accumbens of male mice subjected to chronic voluntary alcohol consumption—the largest increases in TRIF-dependent pathway components were in the medial prefrontal cortex, which is why we chose this brain region for subsequent analysis (23). Increased expression of TRIF-dependent components has also been found in postmortem samples of cerebral cortex and basolateral amygdala from human alcoholics (24–26). TLR3 transcript abundance correlates with lifetime alcohol consumption in human alcoholics (25). Chronic ethanol exposure combined with TLR3 stimulation increases expression of proinflammatory cytokines in mouse frontal cortex (27). Inhibiting the downstream TRIF-signaling components IKKI and TBK1 reduces ethanol consumption in male mice, suggesting that an intact TRIF-dependent pathway in necessary for alcohol consumption (24). However, it is not known if activating TRIF-dependent signaling increases excessive drinking behavior.
In this study, we tested the hypothesis that activation of TLR3-dependent signaling increases alcohol intake. We activated TLR3-dependent signaling by administering the TLR3 agonist polyinosinic-polycytidylic acid (poly(I:C)) to C57BL/6J mice. We found that poly(I:C) increased ethanol intake through an interaction with ethanol that is, at least partially, dependent on TLR3. We also documented changes in innate immune transcripts after chronic poly(I:C) administration during chronic alcohol intake that may mediate increased alcohol consumption. These results establish TLR3-dependent signaling as an important regulator of alcohol consumption in male mice. Inhibiting TLR3-dependent signaling may be a novel strategy to reduce excessive alcohol drinking and the brain immune response to chronic ethanol exposure.
2. Materials and Methods
2.1. Mice.
Generation of Myd88 (B6.129P2(SJL)-Myd88tm1.1Defr/J, stock #009088) knockout (KO) mice was performed as described previously (28). Mutant strain purchased from Jackson Laboratories was backcrossed on a C57BL/6J genetic background more than 6 generations. Male C57BL/6J mice were originally purchased from Jackson Laboratories, Bar Harbor, ME at 8–10 weeks of age and bred to maintain our colony. Behavioral testing began when the mice were at least 2 months old, and mice were weighed every 4 days. All experiments were conducted in isolated behavioral testing rooms. The University of Texas at Austin Institutional Animal Care and Use Committee approved all experiments.
2.2. Drug administration
Poly(I:C) HMW obtained from Invivogen (San Diego, CA) was prepared as previously described (29). To determine the time course of poly(I:C), we injected poly(I:C) (5mg/kg) intraperitoneally (i.p.) and then sacrificed mice at 3, 24, and 48 hours post-injection before qRT-PCR analysis. The 5mg/kg dose was chosen to minimize the sickness response (29) and mitigate any adverse effect on voluntary ethanol consumption 24–36 h post-poly(I:C) injection. For ethanol drinking studies, poly(I:C) (2, 5, or 10mg/kg, i.p.) was administered every four days during no alcohol access periods. For the control injection, 0.9% saline (volume matched) was administered to control groups. Single use, sterile needles (27.5 gauge) were used. All injections we made between 8 am and 9 am to animals 8 −16 weeks of age.
TLR3/dsRNA complex inhibitor obtained from Sigma Aldrich (St. Louis, MO) was prepared for injection in 10% DMSO as previously described (30, 31). For qRT-PCR experiments, we injected 2mg inhibitor, waited 30 minutes and then injected poly(I:C) (5mg/kg, i.p.) and sacrificed mice at 3 hours post-injection. To test the effect of the inhibitor on poly(I:C)-induced escalations in alcohol consumption, mice were injected with poly(I:C) every four days, and underwent two-bottle choice every-other-day (EOD) drinking for a total of 36 days. After we verified that poly(I:C) increased alcohol intake, we injected 2mg of inhibitor, waited 30 minutes and then injected poly(I:C) (5mg/kg, i.p.) and allowed animals to undergo eight days EOD drinking with injections every four days. To test the effect of the inhibitor alone on alcohol consumption, the 2mg of inhibitor or saline (10% DMSO) was injected every four days for a total of 24 days during EOD drinking. To control for DMSO drug dilutions, saline (10% DMSO, volume matched) was administered to both the saline/saline and the saline/poly(I:C) groups.
2.3. Two-bottle choice every-other-day procedure
Intermittent EOD access to ethanol increases voluntary drinking in rats (32, 33) and mice (34–36). Mice were given EOD access to ethanol (15 or 20%) as previously described (12). The quantity of ethanol consumed was calculated as g/kg body weight/24 h. Total fluid intake was calculated as g/kg body weight/24h. For all EOD access drinking experiments, each group had ten animals, (n=10/group).
2.4. Preference for saccharin
Mice were tested for saccharin consumption using an EOD protocol in which one bottle contained water and the other contained saccharin solution. Mice were offered increasing concentrations of saccharin (0.0008 and 0.0016%). Each concentration was offered for a series of four poly(I:C) injections (16 days). Bottle positions were changed for each saccharin session. (n=10/group).
2.5. Brain and organ collection
For qRT-PCR experiments, brains were quickly harvested and the prefrontal cortex rapidly dissected before being snap frozen in liquid nitrogen. For immunohistochemistry experiments, mice were anesthetized with isoflourane, transcardially perfused with 0.9% saline until cleared of blood, and then perfused with freshly prepared 4% paraformaldehyde PFA in phosphate-buffered saline (PBS). Immediately thereafter, the brain and liver were removed and post fixed in 4% (PFA) at 4 °C for 24 h followed by cryoprotection for 24 h at 4 °C in 20% sucrose. Brains and livers were then placed in plastic molds containing optimum cutting temperature compound (OCT, VWR, Radnor, PA) and quickly frozen in isopentane on dry ice.
2.6. qRT-PCR
Total RNA was isolated, quantified and converted to cDNA as described (9). Relative quantification of mRNA levels was determined using BIORAD software as previously described (9, 10, 37). Gusb was selected as an endogenous control to normalize target gene mRNA levels. For all qRT-PCR, six animals were used per group, unless otherwise noted.
2.7. Immunohistochemistry
Thirty-micron sections from brain or liver were treated for immunohistochemistry, as described (37). Sections incubated with K1 (mouse α dsRNA, 1:200, Scicons, Hungary) and Alexa Fluor 488 (donkey α mouse, 1:1000, A21202, Invitrogen, Carlsbad, CA). Sections were mounted in 0.2% gelatin, dehydrated, and cover slipped with a DAPI (4′,6-diamidino-2-phenylindole)-containing mounting medium (Vector Labs, Burlingame, CA). For all immunohistochemistry experiments, four animals were used per group, unless otherwise noted.
2.8. Microscopy and quantitative image analysis
Identification of dsRNA positive cells was performed using a Zeiss Axiovert 200 M fluorescent light microscope for both brain and liver (Zeiss, Thornwood, NY) equipped with an Axiocam b/w camera as previously described for prefrontal cortex (37). Parameters used for image acquisition were identical across treatments. To quantify K1 synthetic dsRNA counts and fluorescence intensity in prefrontal cortex and liver, a single in-focus plane was acquired and analyzed as previously described using Image J (v1.49u, NIH) (37, 38).
2.9. Antibody controls
We tested the specificity of the primary antibody by injecting animals with a ssRNA agonist (R848, 200nmol, Invivogen, San Diego, CA), which should have no signal when using the dsRNA K1 antibody (Supplemental Figure 1A). We also tested the specificity of the secondary antibody (secondary control) by replacement of the primary antibody with only the serum of the appropriate species (Supplemental Figure 1B) or by replacement of both primary and secondary antibody with serum of the appropriate species (Supplemental Figure 1C). Furthermore, K1 has been widely used and its specificity has been previously validated (39–41).
2.10. Statistical analysis
Data are reported as mean ± SEM values, unless otherwise noted. The statistics software program GraphPad Prism (GraphPad Software, Inc., La Jolla, CA) was used to perform 2-way ANOVAs, Pearson correlations and Student’s t-tests. Drinking data were analyzed by repeated-measures 2-way ANOVA followed by Bonferroni post-hoc tests. Transcript abundance data were analyzed by two-way ANOVA followed by Tukey’s HSD post-hoc tests. The Pearson correlation (α = 0.05) was used to evaluate correlations between ethanol consumption and transcript abundance. Grubbs test (α = 0.05) was used to detect potential outliers. Student’s t-tests (two-tailed) were used to analyze raw qRT-PCR data, dsRNA immunopositive cell counts, and the TLR3/dsRNA inhibitor drinking experiment.
3. Results
3.1. Poly(I:C) rapidly increases innate immune transcript abundance in the prefrontal cortex.
Poly(I:C) induces a pro-inflammatory response in the central nervous system (27, 29, 42). We hypothesized that this proinflammatory response is due at least in part to increased TRIF-dependent pathway signaling, which would be reflected in increased abundance of pathway transcripts and transcriptional outputs. To capture changes over time, we measured the time course of poly(I:C) effects on prefrontal cortical transcripts for TRIF-and MyD88-dependent pathway members and for select cytokines and chemokines implicated in behavioral responses to ethanol (see (43) for review). Poly(I:C) (5mg/kg, i.p.) produced three distinct patterns of change in transcript levels: (1) genes dynamically changed by poly(I:C) across time, (2) genes persistently increased across time, and (3) genes unchanged by poly(I:C) (Figure 1, Supplemental Figure 2). Transcripts for the proinflammatory mediators Ifnβ1, Il1β, Ccl2, and Il6 showed a strong increase that peaked at 3 hours and returned to baseline by 48 hours [Ftreatment x time (2,30) = 18.52 (p<0.0001) for Ifnβ1, 5.54 (p<0.001) for Il1β, 14.59, (p<0.0001) for Ccl2, and 14.58 ( p<0.0001) for Il6]. In contrast, the transcript for Tlr4 was unique in that it decreased at 48 hours after poly(I:C) treatment [Ftreatment x time(2,30)=3.89, p=0.03]. Transcripts for Tlr3, Tlr2, Myd88, Ikkβ, and Ccl5 followed a different course and were elevated at all time points [Ftreatment(1,30)=108.7, (p<0.0001) for Tlr3, 70.8 (p<0.0001) for Tlr2, 44.07 (p<0.0001) for Myd88, 30.04 (p<0.0001) for Ikkβ and 242.3 (p<0.0001) for Ccl5]. In contrast to the downstream TRIF pathway member Ccl5, other TRIF-dependent pathway components (Ticam1, Ikkε, and Irf3) were unchanged by poly(I:C) treatment. These findings suggest that activation of TLR3 signaling not only increases levels of TRIF-dependent pathway outputs Ifnβ1 and Ccl5, but also of several innate immune genes that function outside of the TRIF-dependent pathway.
Figure 1: Poly(I:C) increases innate immune mRNA levels in prefrontal cortex.
Heat map showing transcript abundance at four time points, before and following administration of poly(I:C). The transcript levels are presented using fold-change values (Log2 format) normalized to endogenous control. The red and green colors indicate high and low transcript abundance, respectively. The scale representing the relative signal intensity values is shown on the right. Data were analyzed by hierarchical clustering, n=6 per group.
Poly(I:C) is a long dsRNA that can be recognized by either TLR3 or MDA5 (44–45). To determine whether the proinflammatory response after poly(I:C) was dependent on TLR3, we pretreated animals with a TLR3/dsRNA complex inhibitor (2mg, i.p.) followed by poly(I:C) (5mg/kg, i.p.) and measured prefrontal cortical transcripts for select inflammatory mediators implicated in TLR3 response to poly(I:C). TLR3/dsRNA complex inhibition reduced poly(I:C)-induced increases in Ifnβ1, Ccl5, Ccl2, Il6, and Il1β (Supplemental Figure 3). This suggests that the inflammatory response after poly(I:C) is, at least partially, mediated by TLR3.
Poly(I:C) is a polymer, therefore it might not reach the brain when administered by intraperitoneal injection. To investigate whether poly(I:C) is present in brain after systemic administration, we measured its presence in prefrontal cortex and liver using the K1 IgG2a monoclonal antibody, which detects synthetic dsRNA (39–41). Following administration of poly(I:C) (5mg/kg, i.p.), dsRNA-like immunoreactivity was detected in both prefrontal cortex and liver (Supplemental Figure 4). This finding suggests poly(I:C) does reach the brain after intraperitoneal administration, and direct activation of TLR3 in the central nervous system contributes to its effects.
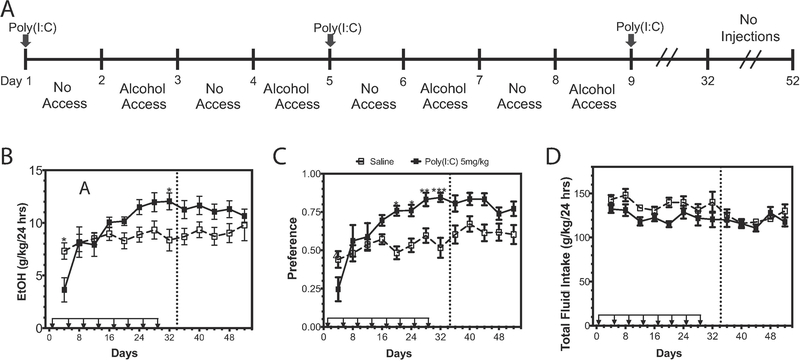
3.2. Poly(I:C) increases alcohol intake over time
Poly(I:C) can increase abundance of innate immune transcripts in brains of male mice, and other activators of innate immunity like LPS increase alcohol drinking (12), therefore we next tested the hypothesis that TLR3 activation increases alcohol intake. C57BL/6J male mice were administered poly(I:C) (5mg/kg, i.p.) every four days during an intermittent (EOD) consumption procedure (15% ethanol) (Figure 2A). Following the first injection of poly(I:C), there was a transient decrease in ethanol consumption, but after multiple injections, alcohol consumption increased [Figure 2B, Ftreatment x time (12, 216) = 3.68, p<0.0001]. A similar pattern was observed for ethanol preference [Figure 2C, Ftreatment x time (12, 216) = 4.77, p<0.0001]. Total fluid intake did not change during the course of the experiment [Figure 2D, p=0.26; Ftreatment(1,18)=4.22, p=0.055; Ftime(12,216)=4.05, p<0.0001; Ftreatment x time(12,216)=1.229].
Figure 2: Poly(I:C) increases alcohol intake over time.
(A) C57BL/6J male mice were injected with saline or poly(I:C) (5mg/kg) every four days for a total of eight injections during two-bottle choice, every-other-day drinking (EOD,15%). (B) Ethanol (EtOH) intake (g/kg/24 h); (C) preference for EtOH; (D) total fluid intake. Arrows indicate days when animals received injections. Dashed line represents a “no injection” period during which animals continued EOD without injections. Data represented as mean ± s.e.m. (***p<0.0002, **p<0.0021, *p<0.05 for Bonferroni post-hoc tests, n=10 per group).
To determine whether the effect of poly(I:C) on alcohol intake was long lasting, we halted injections after day 29 and measured ethanol intake for another 5 ethanol drinking sessions. During this “no injection” period, there was still a significant main effect of poly(I:C) on ethanol preference [Figure 2C, Ftreatment (1,18)=6.78, p=0.018; Ftime (4,72)=4.37, p=0.0032; Ftreatment x time (4,72)=1.76, p=0.15] and a trend toward increased ethanol consumption [Figure 2B, Ftreatment (1,18)=3.21, p=0.08; Ftime (4,72)=0.61, p=0.65; Ftreatment x time (4,72)=1.286, p=0.28]. Together these results indicate that prior treatment with poly(I:C) can produce a persistent increase in ethanol preference over time.
3.3. Poly(I:C) increases alcohol intake across various doses
We investigated whether the effects of poly(I:C) on alcohol intake were dose-dependent. Mice were administered poly(I:C) (2mg/kg or 10mg/kg, i.p.) or saline every four days during EOD consumption of 15% ethanol for 36 days (Figure 3A). Similar to 5mg/kg poly(I:C), multiple injections of these doses were required to increase ethanol consumption [Figure 3B, Ftreatment x time (16,296)=2.86, p=0.0002]. A similar pattern was observed for ethanol preference [Figure 3C, Ftreatment x time (16,296)=2.45, p=0.001]. There was no change in total fluid intake [Figure 3D, Ftreatment (2,37)=0.50, p=0.61; Ftime (8,296)=9.63, p<0.0001; Ftreatment x time (16,296)=0.73, p=0.76]. There was also no significant difference between low dose poly(I:C) (2mg/kg) and high dose poly(I:C) (10mg/kg)-treated animals drinking 15% ethanol.
Figure 3: Poly(I:C) increases alcohol intake across multiple doses.
(A) C57BL/6J male mice were injected with saline, poly(I:C) (2mg/kg), or poly(I:C) (10mg/kg) every four days during a two-bottle choice every-other-day drinking procedure (EOD, 15% or 20%). (B) Ethanol (EtOH) intake (g/kg/24 h); (C) preference for EtOH; (D) total fluid intake. Dashed line represents a “no injection” period during which animals continued EOD without injections. Data represented as mean ± s.e.m. (****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 for Bonferroni post-hoc tests, n=10 per group).
These results suggest that poly(I:C) can increase alcohol intake across the three doses that we tested. However, because ethanol preference was nearly 1.0 when animals were provided 15% ethanol, we switched animals to 20% ethanol during a “no injection” period (beginning at day 38) to reduce ethanol preference and avoid a ceiling effect. During the 16-day “no injection” period the 10mg/kg poly(I:C) group retained increased ethanol consumption compared with saline-treated animals [Figure 3B, Ftreatment x time (8,148)=2.56, p=0.01]. There was a significant difference in consumption between the 10mg/kg poly(I:C) group and 2mg/kg poly(I:C) group only for the first day of the “no injection” period (p<0.01). Together this suggests that the effects of poly(I:C) on ethanol consumption when poly(I:C) is not repeatedly injected are partially dose-dependent.
We next investigated whether repeated poly(I:C) injections after a period without TLR3 activation would induce similar increases in ethanol drinking. After the “no injection” period, mice were again administered poly(I:C) (2mg/kg or 10mg/kg) or saline every four days during EOD consumption of 20% ethanol for 36 days (beginning at day 59). Both doses of poly(I:C) increased ethanol consumption and preference [Figure 3B ethanol intake: Ftreatment x time(18,333)=2.18, p=0.003); Figure 3C preference: Ftreatment x time(18,333)=1.58, p=0.06]. However, repeated injections of poly(I:C) did not continue to increase alcohol intake ad infinitum. Instead, alcohol intake plateaued over time, suggesting that there was an upper limit to how much poly(I:C) can increase drinking.
3.4. Poly(I:C) increases alcohol intake only during ethanol drinking.
We next investigated whether poly(I:C) alone was sufficient to increase alcohol intake. We administered poly(I:C) (5mg/kg, i.p.) or saline every four days for a total of eight injections before allowing mice to begin 20 days of EOD ethanol (15%) drinking (Figure 4A). Pretreatment with poly(I:C) did not increase ethanol consumption [Figure 4B, Ftreatment (1,20)=1.584, p=0.22; Ftime (4,80)=1.024, p=0.40; Ftreatment x time (4,80)=1.02, p=0.4], although there was a slight initial increase in ethanol preference [Figure 4C, Ftreatment x time(4,80)=3.015, p=0.02], suggesting that poly(I:C) pretreatment changed the pattern of escalation compared with saline-pretreated mice. Total fluid intake was unaffected by poly(I:C) pretreatment [Figure 4D, Ftreatment(1,20)=0.01, p=0.9; Ftime(4,80)=1.01, p=0.42; Ftreatment x time(4,80)=1.65, p=0.17]. Re-administration of poly(I:C) to these animals during access to ethanol increased ethanol consumption [Figure 4B, Ftreatment x time(7,140)=2.7, p=0.01] but did not change ethanol preference [Figure 4C, Ftreatment(1,20)=2.84, p=0.11; Ftime(7,140)=1.233, p=0.29; Ftreatment x time(7,140)=1.38, p=0.22]. Although there was a trend towards reduced total fluid consumption, this effect was not statistically significant [Figure 4D, Ftreatment(1,20)=3.778, p=0.06; Ftime(7,140)=2.55, p=0.01; Ftreatment x time(7, 140)=1.06, p=0.39]. These results suggest that the ability of poly(I:C) to increase alcohol intake depends on the presence of ethanol.
Figure 4: Pretreatment with poly(I:C) does not alter subsequent alcohol intake.
(A) C57BL/6J male mice were pretreated with saline or poly(I:C) (5mg/kg) every four days for a total of eight injections before beginning a two-bottle-choice every-other-day procedure (EOD,15%). (B) Ethanol (EtOH) intake (g/kg/24 h); (C) preference for EtOH; (C) total fluid intake. Dashed line represents when injections were resumed during the EOD procedure. Data represented as mean ± s.e.m. (**p<0.0021, *p<0.05 for Bonferroni post-hoc tests, n=10 per group).
3.5. Repeated poly(I:C) during ethanol drinking alters innate immune transcript abundance.
Innate immune response transcripts are increased in the brain after chronic exposure to ethanol or poly(I:C) and these increases are thought to drive ethanol consumption (1, 3, 13, 29, 46, 47). To test the hypothesis that poly(I:C)-induced changes in alcohol intake were due to activation of innate immune signaling, we measured changes in the abundance of transcripts for TRIF-and MyD88-dependent genes and for proinflammatory mediators in mice repeatedly administered 5mg/kg poly(I:C) every four days while consuming 15% ethanol in an EOD procedure (Supplemental Figure 5A). The values for each transcript, represented as the fold-change from the respective saline group, are shown as a heatmap in Figure 5A. Raw qRT-PCR data for each transcript are available in Supplemental Figure 5B. Poly(I:C) plus ethanol increased the abundance of transcripts for TRIF-dependent pathway genes (Tlr3, Ticam1, Ikkε, Irf3) and proinflammatory mediators (Ifnβ1, Ccl5, Ccl2, Il6, Il1β). In contrast, poly(I:C) plus ethanol decreased the abundance of MyD88-dependent pathway transcripts (Tlr4, Myd88, Ikkβ). Tlr2 and Ifnar1 transcripts were unchanged by treatment.
Figure 5: Chronic ethanol and poly(I:C) alter innate immune transcript abundance.
Male C57BL6/J mice underwent 36 days of two-bottle-choice, every-other-day drinking (EOD, 15%) with poly(I:C) (5mg/kg) injections every four days for a total of eight injections. Mice were euthanized and prefrontal cortex dissected 24 hours after the final drinking session on day 36. (A) Heat map of innate immune gene transcript abundance and correlations after both poly(I:C) and ethanol. The levels of transcripts are presented as fold-change values (Log2 format) normalized to respective saline-treated groups. The red and green colors indicate high and low transcript abundance or correlation strength. There were significant negative correlations between Myd88 (B) and amount of ethanol consumed and between Ikkβ (C) and amount of ethanol consumed in poly(I:C)-treated mice. There were significant positive correlations between Ticam1 (D) and amount of ethanol consumed and between Ikkε (E) and amount of ethanol consumed in poly(I:C)-treated mice. There was a significant negative correlation between Ifnb1 (F) and amount of ethanol consumed in poly(I:C)-treated mice. (****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 for Tukey post-hoc tests and correlations, n=6 per group).
To identify potential targets that regulate poly(I:C)-induced escalations in drinking, we examined correlation between transcript abundance and average ethanol consumption. Several transcripts were significantly affected by poly(I:C) and alcohol exposure, but only five transcripts correlated with ethanol consumption. In the TRIF-dependent pathway, there was a significant positive correlation (r= 0.85, p=0.029) between Ticam1 transcript levels and ethanol intake in poly(I:C) treated-mice (Figure 5B). Transcript abundance for the downstream component Ikkε was also positively correlated (r=0.89, p=0.018) with ethanol intake (Figure 5C). Interestingly, there was a negative correlation between Myd88-dependent pathway components and ethanol intake (Figure 5D-E, Myd88, r= −0.82, p=0.046; Ikkβ, r=−0.85, p=0.035). There was also negative correlation (r= −0.94, p=0.0055) between type-1 interferon (Ifnβ) and alcohol intake. Together these data suggest that both the MyD88-and TRIF-dependent pathways may regulate poly(I:C)-induced escalations in alcohol intake.
3.6. Poly(I:C) increases alcohol intake independent of MyD88
Poly(I:C) can act via both MyD88-dependent and –independent mechanisms (42, 48). Moreover, male Myd88 global knockouts consume more ethanol than littermate controls (18). The strong negative correlation between poly(I:C)-induced escalations in ethanol intake and MyD88-dependent transcript abundance suggested that decreases in MyD88 may be necessary for poly(I:C)-induced increases in alcohol intake. If this were true, then administration of poly(I:C) should not increase alcohol intake in male Myd88 knockout mice. To test this hypothesis, we administered poly(I:C) (5mg/kg, i.p) every four days to Myd88 (−/−) knockout mice consuming 15% ethanol in an EOD procedure for 24 days. Poly(I:C) increased ethanol consumption [Figure 6A, Ftreatment (1,16)=10.99, p=0.0044; Ftime (5,80)=4.24, p=0.002; Ftreatment x time (5,80)=1.3, p=0.27] and preference [Figure 6B, Ftreatment(1,16)=14.58, p=0.001; Ftime(5,80)=4.86, p<0.001; Ftreatment x time(5,80)=1.73, p=0.14], but not total fluid intake [Figure 6C, Ftreatment x time=1.97, p=0.09]. These results suggest that poly(I:C) increases alcohol intake independent of MyD88.
Figure 6: Poly(I:C) increases alcohol intake independent of MyD88.
Mutant male mice (Myd88 −/−) were injected with saline or poly(I:C) (5mg/kg) every four days for a total of six injections during a two-bottle choice every-other-day drinking procedure (EOD, 15%). (A) Ethanol (EtOH) intake (g/kg/24 h); (B), preference for EtOH; (C) total fluid intake (n=10 per group).
3.7. Inhibition of the TLR3-dsRNA complex reduces poly(I:C)-induced increases in alcohol intake.
The strong positive correlation between poly(I:C)-induced escalations in alcohol intake and TRIF-dependent transcript abundance suggested that increases in TLR3-dependent signaling may be necessary for poly(I:C)-induced increases in alcohol intake. Additionally, as shown above, poly(I:C)-induced increases in proinflammatory transcripts are partially dependent on TLR3. If TLR3-dependent signaling regulates poly(I:C)-induced drinking behavior, then administration of a TLR3/dsRNA complex inhibitor should decrease alcohol intake in mice chronically administered poly(I:C). To test this hypothesis, we pretreated with saline/poly(I:C) (5mg/kg, i.p) every four days to male C57BL6/J mice consuming 15% ethanol in an EOD procedure for 36 days (Supplemental Figure 6). Once we verified poly(I:C) increased alcohol intake, we injected TLR3/dsRNA complex inhibitor (2mg, i.p.) then injected poly(I:C) (5mg/kg, i.p.) and allowed mice to undergo eight days EOD drinking. Mice treated with a TLR3/dsRNA complex inhibitor before poly(I:C) injection decreased ethanol consumption (Figure 7A). TLR3/dsRNA complex inhibition did not alter preference or total fluid intake in poly(I:C)-treated animals (Figure 7B-C). We further tested the hypothesis that TLR3 inhibition regulates alcohol intake by administering the TLR3/dsRNA inhibitor alone and monitoring drinking behavior. We found that the TLR3/dsRNA inhibitor alone reduced alcohol preference and slowed escalation of alcohol intake (Supplemental Figure 7). Together these results suggest that poly(I:C)-induced increases in alcohol intake are partially dependent on TLR3.
Figure 7: TLR3/dsRNA complex inhibitor reduces poly(I:C)-induced increases alcohol intake.
C57BL/6J male mice were injected with saline(10%DMSO) or poly(I:C) (5mg/kg) every four days for a total of nine injections during EOD (15%v/v). Once poly(I:C) increased alcohol intake above saline controls, we injected TLR3/dsRNA complex inhibitor (2mg, i.p.) then injected poly(I:C) (5mg/kg, i.p.) and allowed mice to undergo eight days EOD. (A) Ethanol (EtOH) intake (g/kg/24 h); (B) preference; (C) total fluid intake (g/kg/24h). There was a significant decrease in alcohol intake after each inhibitor pretreatment (Injection #1: t(16)=2.336, p=0.033; Injection #2, t(16)=2.83, p=0.013). Preference (Injection #1: t(16)=0.12, p=0.91; Injection #2, t(16)=0.64, p=0.52) and total fluid intake (Injection #1: t(16)=1.19, p=0.25; Injection #2, t(16)=1.96, p=0.07) were unaffected by treatment. Student’s t-tests (*p<0.05, **p<0.02 compared with saline/poly(I:C) group, n=10 per group).
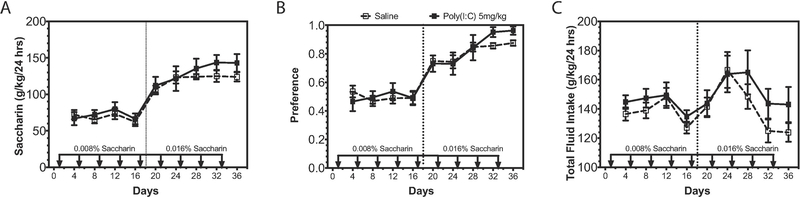
3.8. Poly(I:C) does not alter saccharin consumption
We also studied consumption of saccharin using an EOD procedure to determine whether altered sweet taste perception could account for changes in ethanol consumption. Poly(I:C) did not change EOD saccharin intake at either 0.0008% [Figure 8A, Ftreatment(1,12)=0.14, p=0.71; Ftime(3,36)=1.81, p=0.16; Ftreatment x time(3,36)=0.58, p=0.63] or 0.016% saccharin [Figure 8A, Ftreatment(1,12)=1.06, p=0.32; Ftime(4,48)=3.07, p=0.02; Ftreatment x time(4,48)=0.58, p=0.68]. Additionally, poly(I:C) did not change saccharin preference at either 0.0008% [Figure 8B, Ftreatment(1,12)=0.0001, p=0.98; Ftime(3,36)=0.24, p=0.86; Ftreatment x time(3,36)=0.91, p=0.44] or 0.0016% saccharin [Figure 8B, Ftreatment(1,12)=0.55, p=0.47; Ftime(4,48)=7.78, p<0.0001; Ftreatment x time(4,48)=0.83, p=0.51]. Total fluid intake was also unchanged by poly(I:C) [Figure 8C, Ftreatment(1,12)=1.14, p=0.31; Ftime(8,96)=5.55, p<0.0001; Ftreatment x time(8,96)=0.68, p=0.70]. These findings indicate that poly(I:C) does not change detection of sweet taste, suggesting that poly(I:C) does not increase alcohol intake by perturbing saccharin taste perception.
Figure 8: Poly(I:C) does not change taste perception.
C57BL/6J male mice were injected with saline or poly(I:C) (5mg/kg) every four days for a total of nine injections during a two-bottle choice every-other-day saccharin procedure (0.0008% or 0.016%). Dashed line represents change in saccharin concentration. (A) Saccharin consumption (g/kg/24 h); (B) preference for saccharin; (C) total fluid intake (g/kg/24 h) (n=10 per group).
4. Discussion
Our results reveal a novel role for TLR3-dependent signaling in regulation of alcohol intake. Activation of TLR3 by poly(I:C) increased transcript abundance of innate immune signaling components in prefrontal cortex. Repeated TLR3 activation, across multiple doses, increased alcohol intake over time. However, the effect of TLR3 activation on drinking was dependent on both repeated injections and the presence of ethanol, suggesting poly(I:C) is necessary for escalations in alcohol intake but not sufficient when administered alone. Poly(I:C) increased alcohol intake without altering saccharin taste preference, suggesting changes in consumption were not due to alterations in sweet taste preference. Ethanol and poly(I:C) exposure increased transcript abundance of both TRIF-and MyD88-dependent pathway components. TRIF-dependent pathway levels positively correlated with increased alcohol intake and MyD88-dependent pathway levels negatively correlated with alcohol intake. Poly(I:C) escalated alcohol intake in Myd88 knockout mice, supporting the hypothesis that increases in alcohol intake after poly(I:C) occur independently of MyD88. Inhibition of the TLR3/dsRNA complex reduced escalation of alcohol intake and poly(I:C)-induced escalations in alcohol intake, supporting the conclusion that increases in alcohol intake after poly(I:C) are partially dependent on TLR3/TRIF-dependent signaling. Together, these results suggest that activation of TLR3 signaling promotes alcohol consumption and therefore inhibition of TLR3 may be a potential therapeutic target for treating excessive drinking.
Poly(I:C) produced transient activation of proinflammatory mediators in prefrontal cortex, with peak cytokine activation occurring at 3 hours post-injection—consistent with previous reports (27, 29, 49). However, poly(I:C) also increased transcript abundance of both TRIF-and MyD88-dependent components across a 48-hour period, suggesting that these longer lasting changes in innate immune signaling may drive drinking behavior. These poly(I:C)-induced changes in innate immune signaling were, at least partially, dependent on TLR3, because inhibition of the TLR3/dsRNA complex reduced activation of proinflammatory mediators. Staining for poly(I:C) presence in the liver and prefrontal cortex confirmed poly(I:C) can enter brain, which is consistent with poly(I:C) being able to disrupt the blood-brain barrier to permit direct brain access for dsRNA (50). This suggested that direct activation of TLR3 in the central nervous system contributes, at least partially, to the behavioral effects of poly(I:C). However, the greater amount of poly(I:C) signal detected in the liver raised the possibility that peripheral actions of poly(I:C) may play a role in how the innate immune system regulates drinking behavior and ethanol metabolism. Viral infections do alter liver metabolism (51). Activation of TLR3 by dsRNA induces natural killer cell accumulation and activation in the liver, leading to liver inflammation and injury (51). However, it has been shown that TLR3 activation with poly(I:C) ameliorates alcoholic liver injury via the stimulation of IL-10 production in hepatic stellate cells and Kupffer cells (52). Moreover, poly(I:C) does not alter alcohol dehydrogenase (ADH) expression in liver (52). Additionally, TLR3 knockout mice show no differences in ethanol metabolism when compared with littermate controls (data not shown). Therefore, if the absence of TLR3 does not affect ethanol metabolism, poly(I:C) does not effect ADH expression in liver and poly(I:C) treatment ameliorates alcoholic liver injury, we hypothesize that poly(I:C) is not changing ethanol metabolism. However, future studies will need to dissect the role of peripheral versus central TLR3 in the regulation of alcohol intake.
Repeated injections of poly(I:C) were required to increase alcohol intake, suggesting that long-lasting alterations in gene expression must occur to change behavior rather than transient innate immune changes due to acute TLR3 activation. All tested doses increased alcohol intake over time, indicating no dose-dependence of poly(I:C) on alcohol intake. For both the 5mg/kg and 10mg/kg doses, poly(I:C) elevated alcohol preference even when poly(I:C) injections were halted, consistent with a persistent change in innate immune signaling having occurred. However, alcohol intake did not continue to increase without continued poly(I:C) injections, indicating that poly(I:C) must be repeatedly injected to increase alcohol consumption. The requirement of repeated TLR3 activation suggests different and longer lasting signaling changes must occur to increase alcohol intake. A limitation of the current study is that a single time point was selected after poly(I:C) and ethanol consumption (24h), which provided a snapshot of how poly(I:C) changes gene expression. From these data we hypothesize that TRIF-dependent signaling may be necessary for escalations in alcohol intake along with longer-lasting changes in innate immune signaling pathways not investigated in this paper. Future studies will investigate the longer-lasting transcriptional changes necessary for poly(I:C)-induced increases in alcohol intake over time using RNA-sequencing in order to address how repeated TLR3 activation changes drinking behavior on the transcriptome level.
To determine if poly(I:C) alone is sufficient to increase alcohol intake we pretreated animals with poly(I:C) for eight injections before allowing animals to begin drinking. Pretreatment with poly(I:C) did not change alcohol intake, suggesting that poly(I:C) alone is not sufficient to change drinking behavior. Additionally after pretreatment, we demonstrate ethanol and poly(I:C) must be present together to increase alcohol intake, implying that ethanol adds a key component of neuroimmune activation that we have yet to discover. It has been hypothesized that alcohol-induced neuroinflammation directly contributes to the development of alcohol use disorders (53). Ethanol can activate TRIF-dependent and MyD88-dependent signaling as well as a wide array of proinflammatory mediators (2, 54). Others have shown that a variety of proinflammatory signals increases ethanol drinking and preference (13) and interfering with neuroinflammatory signaling reduces alcohol consumption (55, 56). Combined activation of TLRs alters the extent of cytokine production and can stimulate additive increases in the amount of cytokines produced, which could lead to greater increases in alcohol consumption over time (57–59). Thus, the combination of ethanol and poly(I:C) may be necessary to produce sufficient inflammatory signaling to change drinking behavior.
Repeated ethanol and poly(I:C) exposure altered innate immune transcript abundance in both TLR pathway branches. Additionally there was a robust increase in levels of proinflammatory mediators (Il6, Il1β, Ccl2, Ccl5) in poly(I:C)-treated mice, suggesting involvement of multiple innate immune signaling branches. When there is chronic activation of one immune signaling branch (e.g. TRIF-dependent), there can be compensatory downregulation of the other (e.g. MyD88-dependent) (60). This is thought to allow the innate immune system to facilitate simultaneous amplification of the appropriate level of protective responses during infection, attenuation of the damage inflicted by inflammation, and maintenance of homeostasis (61, 62). For example, poly(I:C) can downregulate TLR4 signaling via a TLR3-dependent mechanism to attenuate the immune response (63, 64). We considered whether TLR3 induced decreases in MyD88-dependent signaling could explain the increase in alcohol intake that we observed in mice treated with poly(I:C). In support of this hypothesis, knockout of Myd88 increases alcohol intake in male mice (20, 21). However, we found that poly(I:C) increased alcohol intake in Myd88 knockout mice—a limitation of this knockout study was that wildtype controls were not used in conjuction with Myd88 knockout mice. Taken together, this suggests that poly(I:C) increases alcohol intake through mechanisms independent of MyD88.
TLR3 activation can have either neuroprotective or neurodegenerative effects. For instance, poly(I:C) pretreatment exerts neuroprotective and anti-inflammatory effects in simulated cerebral ischemia models (65, 66). However, in a Parkinson’s model, poly(I:C) triggered nigrostriatal dopaminergic degeneration and microglial proliferation (67, 68). This suggests that when inflammation has already occured, TLR3 activation primes immune cells in brain for proinflammatory responses leading to enhanced neurodegeneration. We have previously shown EOD drinking enhances proinflammatory signaling (24), thus we hypothesized that poly(I:C) increases alcohol intake by enhancing inflammatory responses, specifically TLR3/TRIF-dependent responses. In support of this hypothesis we observed increases in TRIF-dependent pathway components after chronic poly(I:C) and ethanol, which positively correlated with alcohol intake. Furthermore, inhibition of the TLR3/dsRNA complex reduced drinking in poly(I:C)-treated animals, indicating that TLR3/TRIF-dependent signaling may promote excessive alcohol intake but other immune pathways may also mediate this effect. Global knockout of TLR3 reduces EOD drinking in male mice (Blednov unpublished observations) and pharmacological inhibition of the downstream TRIF-dependent pathway components IKKI and TBK1 decreases alcohol intake (24). TLR3 signaling may also have broader roles in regulating rewarding effects of drugs. For instance, TLR3 has been shown to modulate the behavioral effects of cocaine in mice and intra-nucleus accumbens injection of TLR3/dsRNA complex inhibitor significantly attenuated cocaine self-administration in mice (69). TLR3 activation also enhances morphine conditioned place preference acquisition (70). Therefore, poly(I:C) may increase alcohol intake by changing the rewarding properties of ethanol. Future studies will need to dissect if TLR3 activation changes ethanol sensitivity. Taken together, inhibition of TLR3 may be a valuable therapeutic target to reduce excessive alcohol consumption as well as treating other substance abuse disorders.
Although poly(I:C) is a synthetic dsRNA polymer that mimics viral infection, it should be noted that we are most likely not modeling viral infection in human alcoholics. In human alcoholics, chronic alcohol exposure over time interferes with the normal functioning of all aspects of the innate and adaptive immune response, including both cell-mediated and humoral responses (71). Most likely our model represents a heightened peripheral proinflammatory state, driven by TLR3 that leads to increases in alcohol intake. Since poly(I:C) injection results in more dsRNA signal in the liver compared with prefrontal cortex, we are likely activating more peripheral TLR3 resulting in an increase in proinflammatory mediators in the periphery that can cross the BBB and activate central neuroimmune pathways (such as IL-1β, IL-6, and IFNβ). Proinflammatory cytokines have been detected in the serum of human alcoholics and higher levels of cytokines correlates with alcohol craving (11). Therefore, we hypothesize that these proinflammatory mediators increase craving and increase alcohol intake over time. Future studies should address whether heightened viral load increases alcohol consumption in human populations, linking the idea that viral activation of TLR3 leads to escalation of alcohol intake over time.
5. Conclusions
Altogether, this work has defined the role of TLR3 signaling in drinking behavior, defined the causal role and pathways necessary for TLR3-dependent increases in alcohol intake, and provides a potential pharmacotherapeutic target to improve treatment outcomes for excessive drinking. While, these studies suggest that increases in TLR3-dependent signaling exacerbate proinflammatory signaling resulting in increases in alcohol intake, we suggest targeted inhibition of TLR3 and its downstream mediators may represent novel treatment strategies for excessive drinking.
Supplementary Material
(A) Staining specificity for K1 antibody in prefrontal cortex and liver of C57BL/6J male mice injected with R848 (ssRNA TLR7/8 agonist). (B) Secondary controls in prefrontal cortex and liver of C57BL/6J male mice— primary antibody omitted, 10% donkey serum and donkey anti-mouse Alexa Flour 488 secondary. (C) Labeling staining controls in prefrontal cortex and liver of C57BL/6J male mice—primary antibody and secondary antibody omitted, 10% donkey serum only. Scale bar=50μm (n=4 per group).
The levels of transcripts are presented using fold-change values normalized to endogenous control, with six biological and three technical replicates. Data represented as fold change ± s.e.m (****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 compared with saline treatment at same time point; #p<0.05 compared with other poly(I:C) groups for Tukey post-hoc tests, n=6 per group).
C57BL/6J male mice were injected with poly(I:C) (5mg/kg). Mice were euthanized, prefrontal cortex dissected and liver removed three hours post-injection. (A-D) representative images of K1 dsRNA immunofluorescence, scale bars=100μm. (E) Corrected total cell fluorescence (CTCF) was increased after poly(I:C) injection in both brain and liver, though the increase was greater in the liver [Ftreatment(1,12)=665, p<0.0001; Forgan(1,12)=287, p<0.0001; Ftreatment x organ(1,12)=327, p<0.0001]. (F) As similar pattern was observed for measurements of integrated intensity [Ftreatment(1,12)=80.45, p<0.0001; Forgan(1,12)=122,4, p<0.0001; Ftreatment x organ(1,12)=31.91, p<0.0001]. (G) Poly(I:C) treatment increased the number of dsRNA immunopositive cells present in the prefrontal cortex (two-tailed Student’s t-test t(6)=25.73, ****p<0.0001). All data are expressed as mean ± s.e.m, n=4 per group.
(A) Drinking data for poly(I:C) (5mg/kg) EOD experiment used for qRT-PCR analysis. There was a main effect of poly(I:C) on ethanol intake [Ftreatment(1,18)=15.49, p=0.001; Ftime(7,126)=4.65, p=0.0001; Ftreatment x time(7,126)=1.433, p=0.19] and preference [Ftreatment(1,18)=13.54, p=0.001; Ftime(7,126)=5.09, p<0.0001; Ftreatment x time(7,126)=0.37, p=0.92]. There was no effect of poly(I:C) on total fluid intake [Ftreatment(1,18)=0.55, p=0.46; Ftime(7,126)=2.445, p=0.02; Ftreatment x time(7,126)=1.84, p=0.08]. Raw qRT-PCR data for 5mg/kg poly(I:C) EOD experiment. The levels of genes are presented as fold change ± s.e.m. All values were normalized to endogenous control. Data were analyzed by two-tailed, Student’s t-tests (****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 compared with saline group, n=7 per group).
C57BL/6J male mice were injected with saline (10%DMSO)/saline or saline(10%DMSO)/ poly(I:C) (5mg/kg) every four days for a total of nine injections during two-bottle choice, every-other-day drinking (EOD,15%), then poly(I:C) treated animals were pretreated with a TLR3/dsRNA complex inhibitor for eight days. (A) Ethanol (EtOH) intake (g/kg/24 h); (B) preference for EtOH; (C) total fluid intake (g/kg/24 h); (D-E) body weight (g). There was a main effect of poly(I:C) on ethanol [Ftreatment(1,17)=7.5, p=0.014; Ftime(8,136)=15.01, p<0.0001; Ftreatment x time(8,136)=2.29, p=0.02] and preference [Ftreatment(1,17)=3.626, p=0.07; Ftime(8,136)=15.89, p<0.0001; Ftreatment x time(8,136)=2.83, p=0.019]. Total fluid intake was not changed by poly(I:C) treatment [Ftreatment(1,17)=0.14, p=0.71; Ftime(8,136)=3.65, p=0.0007; Ftreatment x time- (8,136)=1.59, p=0.13]. There was a main effect of TLR3/dsRNA inhibitor pretreatment on poly(I:C)-induced ethanol consumption (Ftreatment(1,16)=7.25, p=0.01). There was no effect of TLR3/dsRNA inhibitor pretreatment on poly(I:C)-induced ethanol preference (Ftreatment(1,16)=0.16, p=0.69) or total fluid intake (Ftreatment(1,16)=3.35, p=0.08). There was no main effect of poly(I:C) or TLR3/dsRNA inhibitor on body weight [Ftreatment(1,18)=1.49, p=0.24; Ftime(10,180)=9.4, p<0.0001; Ftreatment x time(10,180)=2.69, p=0.004]. Arrows indicate days when animals received injections, the red arrow indicates switch to the TLR3/dsRNA inhibitor. Dashed line represents when saline(10%DMSO)/poly(I:C) treated animals were treated with inhibitor(10%DMSO)/poly(I:C). Data represented as mean ± s.e.m. (*p<0.05, significantly different from saline/saline group; #p<0.05, significantly different from saline/poly(I:C) group, Bonferroni post-hoc tests, n=10 per group).
The levels of transcripts are presented using fold-change values normalized to endogenous control, with six biological and three technical replicates. Values are presented as fold change ± SEM. (* represents significant difference from saline group, # represents significant difference from poly(I:C) group, ****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 compared with saline treatment at same time; #p<0.05 compared with other poly(I:C) groups, n=6 per group).
Male C57BL6/J mice were treated with either saline(10%DMSO) or TLR3/dsRNA complex inhibitor (2mg) and underwent 24 days of EOD-2BC (15%v/v ethanol). To determine how the inhibitor changed escalation of drinking behavior from baseline we subtracted subsequent behavioral drinking days from the first drinking day. (A) Inhibitor treatment trended toward decreased ethanol intake (Ftreatment(1,18)=3.49, p=0.07; Ftime(4,72)=5.57, p=0.0006; Ftreatment x time(4,72)=0.363, p=0.83). (B) Inhibitor treatment reduced escalation of alcohol preference (Ftreatment(1,18)=5.96, p=0.02; Ftime(4,72)=6.79, p=0.0001; Ftreatment x time(4,72)=1.149, p=0.34). (C) Inhibitor treatment modestly increased total fluid intake escalation (Ftreatment(1,18)=4.92, p=0.04; Ftime(4,72)=3.38, p=0.01; Ftreatment x time(4,72)=2.62, p=0.04). Data represented as mean ± s.e.m. (*p<0.05, significantly different from saline group, Bonferroni post-hoc tests, n=10 per group).
Highlights:
Activation of TLR3 via poly(I:C) increased alcohol intake.
Poly(I:C) and ethanol must be present together to change drinking behavior.
Increased alcohol intake due to poly(I:C) is independent of MYD88.
Increased alcohol intake due to poly(I:C) is dependent on TLR3.
Acknowledgments
Funding: This work was supported by the National Institutes of Health/National Institute of Alcohol Abuse and Alcoholism [U01 AA020926, P01 AA020683, AA013520, AA006399, AA025499].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors report no biomedical financial interests or potential conflicts of interest.
References:
- 1.Robinson G, Most D, Ferguson LB, Mayfield J, Harris RA, Blednov YA (2014): Neuroimmune pathways in alcohol consumption: evidence from behavioral and genetic studies in rodents and humans. Int Rev Neurobiol 118:13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crews FT, Walter TJ, Coleman LG, Vetreno RP (2017): Toll-like receptor signaling and stages of addiction. Psychopharmacology (Berl) 234:1483–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montesinos J, Alfonso-Loeches S, Guerri C (2016): Impact of the Innate Immune Response in the Actions of Ethanol on the Central Nervous System. Alcohol Clin Exp Res 40:2260–2270. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. (2006): Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology 31:1574–1582. [DOI] [PubMed] [Google Scholar]
- 5.Okvist A, Johansson S, Kuzmin A, Bazov I, Merino-Martinez R, Ponomarev I, et al. (2007): Neuroadaptations in human chronic alcoholics: dysregulation of the NF-kappaB system. PLoS One 2:e930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. (2006): Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A 103:6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Harris RA, Ponomarev I (2011): Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol Clin Exp Res 35:659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, et al. (2013): Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics 14:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osterndorff-Kahanek E, Ponomarev I, Blednov YA, Harris RA (2013): Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS One 8:e59870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osterndorff-Kahanek EA, Becker HC, Lopez MF, Farris SP, Tiwari GR, Nunez YO, et al. (2015): Chronic ethanol exposure produces time-and brain region-dependent changes in gene coexpression networks. PLoS One 10:e0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclercq S, De Saeger C, Delzenne N, de Timary P, Starkel P (2014): Role of inflammatory pathways, blood mononuclear cells, and gut-derived bacterial products in alcohol dependence. Biol Psychiatry 76:725–733. [DOI] [PubMed] [Google Scholar]
- 12.Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA (2011): Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain Behav Immun 25 Suppl 1:S92–S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blednov YA, Ponomarev I, Geil C, Bergeson S, Koob GF, Harris RA (2012): Neuroimmune regulation of alcohol consumption: behavioral validation of genes obtained from genomic studies. Addict Biol 17:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda K, Akira S (2004): TLR signaling pathways. Semin Immunol 16:3–9. [DOI] [PubMed] [Google Scholar]
- 15.Alfonso-Loeches S, Pascual-Lucas M, Blanco AM, Sanchez-Vera I, Guerri C (2010): Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. J Neurosci 30:8285–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.June HL, Liu J, Warnock KT, Bell KA, Balan I, Bollino D, et al. (2015): CRF-amplified neuronal TLR4/MCP-1 signaling regulates alcohol self-administration. Neuropsychopharmacology 40:1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Yang AR, Kelly T, Puche A, Esoga C, June HL, et al. (2011): Binge alcohol drinking is associated with GABAA alpha2-regulated Toll-like receptor 4 (TLR4) expression in the central amygdala. Proc Natl Acad Sci U S A 108:4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blednov YA, Black M, Chernis J, Da Costa A, Mayfield J, Harris RA (2017): Ethanol Consumption in Mice Lacking CD14, TLR2, TLR4, or MyD88. Alcohol Clin Exp Res 41:516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truitt JM, Blednov YA, Benavidez JM, Black M, Ponomareva O, Law J, et al. (2016): Inhibition of IKKβ Reduces Ethanol Consumption in C57BL/6J Mice. eneuro (In Press). [DOI] [PMC free article] [PubMed]
- 20.Blednov YA, Black M, Benavidez JM, Da Costa A, Mayfield J, Harris RA (2017): Sedative and Motor Incoordination Effects of Ethanol in Mice Lacking CD14, TLR2, TLR4, or MyD88. Alcohol Clin Exp Res 41:531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris RA, Bajo M, Bell RL, Blednov YA, Varodayan FP, Truitt JM, et al. (2017): Genetic and Pharmacologic Manipulation of TLR4 Has Minimal Impact on Ethanol Consumption in Rodents. J Neurosci 37:1139–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lainiola M, Linden AM (2017): Alcohol intake in two different mouse drinking models after recovery from the lipopolysaccharide-induced sickness reaction. Alcohol 65:1–10. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy GM, Warden AS, Bridges CR, Blednov YA, Harris RA (2018): Chronic ethanol consumption: role of TLR3/TRIF-dependent signaling. Addict Biol 23:889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy GR, Warden A, Bridges C, Blednov, et al. (2017): Chronic ethanol consumption: Role of TLR3/TRIF-dependent signaling. Addiction Biology [DOI] [PMC free article] [PubMed]
- 25.Crews FT, Qin L, Sheedy D, Vetreno RP, Zou J (2013): High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biol Psychiatry 73:602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD (2012): Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 32:1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin L, Crews FT (2012): Chronic ethanol increases systemic TLR3 agonist-induced neuroinflammation and neurodegeneration. J Neuroinflammation 9:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou B, Reizis B, DeFranco AL (2008): Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity 29:272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham C, Campion S, Teeling J, Felton L, Perry VH (2007): The sickness behaviour and CNS inflammatory mediator profile induced by systemic challenge of mice with synthetic double-stranded RNA (poly I:C). Brain Behav Immun 21:490–502. [DOI] [PubMed] [Google Scholar]
- 30.Cheng K, Wang X, Yin H (2011): Small-molecule inhibitors of the TLR3/dsRNA complex. J Am Chem Soc 133:3764–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takemura N, Kawasaki T, Kunisawa J, Sato S, Lamichhane A, Kobiyama K, et al. (2014): Blockade of TLR3 protects mice from lethal radiation-induced gastrointestinal syndrome. Nat Commun 5:3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simms JA, Steensland P, Medina B, Abernathy KE, Chandler LJ, Wise R, et al. (2008): Intermittent access to 20% ethanol induces high ethanol consumption in Long-Evans and Wistar rats. Alcohol Clin Exp Res 32:1816–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wise RA (1973): Voluntary ethanol intake in rats following exposure to ethanol on various schedules. Psychopharmacologia 29:203–210. [DOI] [PubMed] [Google Scholar]
- 34.Melendez RI (2011): Intermittent (every-other-day) drinking induces rapid escalation of ethanol intake and preference in adolescent and adult C57BL/6J mice. Alcohol Clin Exp Res 35:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenwasser AM, Fixaris MC, Crabbe JC, Brooks PC, Ascheid S (2013): Escalation of intake under intermittent ethanol access in diverse mouse genotypes. Addict Biol 18:496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwa LS, Chu A, Levinson SA, Kayyali TM, DeBold JF, Miczek KA (2011): Persistent escalation of alcohol drinking in C57BL/6J mice with intermittent access to 20% ethanol. Alcohol Clin Exp Res 35:1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warden A, Truitt J, Merriman M, Ponomareva O, Jameson K, Ferguson LB, et al. (2016): Localization of PPAR isotypes in the adult mouse and human brain. Sci Rep 6:27618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A (2014): Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schönborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N (1991): Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res 19:2993–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukacs N (1997): Detection of sense:antisense duplexes by structure specific anti-RNA antibodies. In: Lichtenstein C, Nellen W, editors. Antisense Technology A Practical Approach Oxford: IRL Press, pp 281–295. [Google Scholar]
- 41.Lukács N (1994): Detection of virus infection in plants and differentiation between coexisting viruses by monoclonal antibodies to double-stranded RNA. J Virol Methods 47:255–272. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X, Levasseur PR, Michaelis KA, Burfeind KG, Marks DL (2016): A distinct brain pathway links viral RNA exposure to sickness behavior. Sci Rep 6:29885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayfield J, Harris RA (2017): The Neuroimmune Basis of Excessive Alcohol Consumption. Neuropsychopharmacology 42:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. [DOI] [PubMed] [Google Scholar]
- 45.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M (2006) Essential role of mda-5 in type I IFN responses to polyriboinosinic: polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A 103, 8459–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blednov YA, Bergeson SE, Walker D, Ferreira VM, Kuziel WA, Harris RA (2005): Perturbation of chemokine networks by gene deletion alters the reinforcing actions of ethanol. Behav Brain Res 165:110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crews FT, Vetreno RP (2016): Mechanisms of neuroimmune gene induction in alcoholism. Psychopharmacology (Berl) 233:1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001): Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. [DOI] [PubMed] [Google Scholar]
- 49.Field R, Campion S, Warren C, Murray C, Cunningham C (2010): Systemic challenge with the TLR3 agonist poly I:C induces amplified IFNalpha/beta and IL-1beta responses in the diseased brain and exacerbates chronic neurodegeneration. Brain Behav Immun 24:996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA (2004): Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med 10:1366–1373. [DOI] [PubMed] [Google Scholar]
- 51.Yin S, Gao B (2010): Toll-like receptor 3 in liver diseases. Gastroenterol Res Pract 2010. [DOI] [PMC free article] [PubMed]
- 52.Byun JS, Suh YG, Yi HS, Lee YS, Jeong WI (2013): Activation of toll-like receptor 3 attenuates alcoholic liver injury by stimulating Kupffer cells and stellate cells to produce interleukin-10 in mice. J Hepatol 58:342–349. [DOI] [PubMed] [Google Scholar]
- 53.Crews FT, Zou J, Qin L (2011): Induction of innate immune genes in brain create the neurobiology of addiction. Brain Behav Immun 25 Suppl 1:S4–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warden A, Erickson E, Robinson G, Harris RA, Mayfield RD (2016): The neuroimmune transcriptome and alcohol dependence: potential for targeted therapies. Pharmacogenomics 17:2081–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agrawal RG, Hewetson A, George CM, Syapin PJ, Bergeson SE (2011): Minocycline reduces ethanol drinking. Brain Behav Immun 25 Suppl 1:S165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Truitt JM, Blednov YA, Benavidez JM, Black M, Ponomareva O, Law J, et al. (2016): Inhibition of IKKβ Reduces Ethanol Consumption in C57BL/6J Mice. eneuro [DOI] [PMC free article] [PubMed]
- 57.Liu B, Liu Q, Yang L, Palaniappan SK, Bahar I, Thiagarajan PS, et al. (2016): Innate immune memory and homeostasis may be conferred through crosstalk between the TLR3 and TLR7 pathways. Sci Signal 9:ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenberger K, Derkow K, Dembny P, Krüger C, Schott E, Lehnardt S (2014): The impact of single and pairwise Toll-like receptor activation on neuroinflammation and neurodegeneration. J Neuroinflammation 11:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krummen M, Balkow S, Shen L, Heinz S, Loquai C, Probst HC, et al. (2010): Release of IL-12 by dendritic cells activated by TLR ligation is dependent on MyD88 signaling, whereas TRIF signaling is indispensable for TLR synergy. J Leukoc Biol 88:189–199. [DOI] [PubMed] [Google Scholar]
- 60.Johnson AC, Li X, Pearlman E (2008): MyD88 functions as a negative regulator of TLR3/TRIF-induced corneal inflammation by inhibiting activation of c-Jun N-terminal kinase. J Biol Chem 283:3988–3996. [DOI] [PubMed] [Google Scholar]
- 61.Kawai T, Akira S (2011): Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. [DOI] [PubMed] [Google Scholar]
- 62.Tan RS, Ho B, Leung BP, Ding JL (2014): TLR cross-talk confers specificity to innate immunity. Int Rev Immunol 33:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang PF, Fang H, Chen J, Lin S, Liu Y, Xiong XY, et al. (2014): Polyinosinic-polycytidylic acid has therapeutic effects against cerebral ischemia/reperfusion injury through the downregulation of TLR4 signaling via TLR3. J Immunol 192:4783–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan LN, Zhu W, Li C, Xu XL, Guo LJ, Lu Q (2012): Toll-like receptor 3 agonist Poly I:C protects against simulated cerebral ischemia in vitro and in vivo. Acta Pharmacol Sin 33:1246–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan LN, Zhu W, Li Y, Xu XL, Guo LJ, Lu Q, et al. (2014): Astrocytic Toll-like receptor 3 is associated with ischemic preconditioning-induced protection against brain ischemia in rodents. PLoS One 9:e99526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X, Ha T, Lu C, Lam F, Liu L, Schweitzer J, et al. (2015): Poly (I:C) therapy decreases cerebral ischaemia/reperfusion injury via TLR3-mediated prevention of Fas/FADD interaction. J Cell Mol Med 19:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deleidi M, Hallett PJ, Koprich JB, Chung CY, Isacson O (2010): The Toll-like receptor-3 agonist polyinosinic:polycytidylic acid triggers nigrostriatal dopaminergic degeneration. J Neurosci 30:16091–16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bobyn J, Mangano EN, Gandhi A, Nelson E, Moloney K, Clarke M, et al. (2012): Viral-toxin interactions and Parkinson’s disease: poly I:C priming enhanced the neurodegenerative effects of paraquat. J Neuroinflammation 9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu R, Bu Q, Fu D, Shao X, Jiang L, Guo W, et al. (2018): Toll-like receptor 3 modulates the behavioral effects of cocaine in mice. J Neuroinflammation 15:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Chen S, Liu H, Lu L, Zhai H (2010): Synthetic double-stranded RNA polyinosinic-polycytidylic acid augments morphine-induced conditioned place preference in rats. Behav Pharmacol 21:369–373. [DOI] [PubMed] [Google Scholar]
- 71.Pasala S, Barr T, Messaoudi I (2015): Impact of Alcohol Abuse on the Adaptive Immune System. Alcohol Res 37:185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Staining specificity for K1 antibody in prefrontal cortex and liver of C57BL/6J male mice injected with R848 (ssRNA TLR7/8 agonist). (B) Secondary controls in prefrontal cortex and liver of C57BL/6J male mice— primary antibody omitted, 10% donkey serum and donkey anti-mouse Alexa Flour 488 secondary. (C) Labeling staining controls in prefrontal cortex and liver of C57BL/6J male mice—primary antibody and secondary antibody omitted, 10% donkey serum only. Scale bar=50μm (n=4 per group).
The levels of transcripts are presented using fold-change values normalized to endogenous control, with six biological and three technical replicates. Data represented as fold change ± s.e.m (****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 compared with saline treatment at same time point; #p<0.05 compared with other poly(I:C) groups for Tukey post-hoc tests, n=6 per group).
C57BL/6J male mice were injected with poly(I:C) (5mg/kg). Mice were euthanized, prefrontal cortex dissected and liver removed three hours post-injection. (A-D) representative images of K1 dsRNA immunofluorescence, scale bars=100μm. (E) Corrected total cell fluorescence (CTCF) was increased after poly(I:C) injection in both brain and liver, though the increase was greater in the liver [Ftreatment(1,12)=665, p<0.0001; Forgan(1,12)=287, p<0.0001; Ftreatment x organ(1,12)=327, p<0.0001]. (F) As similar pattern was observed for measurements of integrated intensity [Ftreatment(1,12)=80.45, p<0.0001; Forgan(1,12)=122,4, p<0.0001; Ftreatment x organ(1,12)=31.91, p<0.0001]. (G) Poly(I:C) treatment increased the number of dsRNA immunopositive cells present in the prefrontal cortex (two-tailed Student’s t-test t(6)=25.73, ****p<0.0001). All data are expressed as mean ± s.e.m, n=4 per group.
(A) Drinking data for poly(I:C) (5mg/kg) EOD experiment used for qRT-PCR analysis. There was a main effect of poly(I:C) on ethanol intake [Ftreatment(1,18)=15.49, p=0.001; Ftime(7,126)=4.65, p=0.0001; Ftreatment x time(7,126)=1.433, p=0.19] and preference [Ftreatment(1,18)=13.54, p=0.001; Ftime(7,126)=5.09, p<0.0001; Ftreatment x time(7,126)=0.37, p=0.92]. There was no effect of poly(I:C) on total fluid intake [Ftreatment(1,18)=0.55, p=0.46; Ftime(7,126)=2.445, p=0.02; Ftreatment x time(7,126)=1.84, p=0.08]. Raw qRT-PCR data for 5mg/kg poly(I:C) EOD experiment. The levels of genes are presented as fold change ± s.e.m. All values were normalized to endogenous control. Data were analyzed by two-tailed, Student’s t-tests (****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 compared with saline group, n=7 per group).
C57BL/6J male mice were injected with saline (10%DMSO)/saline or saline(10%DMSO)/ poly(I:C) (5mg/kg) every four days for a total of nine injections during two-bottle choice, every-other-day drinking (EOD,15%), then poly(I:C) treated animals were pretreated with a TLR3/dsRNA complex inhibitor for eight days. (A) Ethanol (EtOH) intake (g/kg/24 h); (B) preference for EtOH; (C) total fluid intake (g/kg/24 h); (D-E) body weight (g). There was a main effect of poly(I:C) on ethanol [Ftreatment(1,17)=7.5, p=0.014; Ftime(8,136)=15.01, p<0.0001; Ftreatment x time(8,136)=2.29, p=0.02] and preference [Ftreatment(1,17)=3.626, p=0.07; Ftime(8,136)=15.89, p<0.0001; Ftreatment x time(8,136)=2.83, p=0.019]. Total fluid intake was not changed by poly(I:C) treatment [Ftreatment(1,17)=0.14, p=0.71; Ftime(8,136)=3.65, p=0.0007; Ftreatment x time- (8,136)=1.59, p=0.13]. There was a main effect of TLR3/dsRNA inhibitor pretreatment on poly(I:C)-induced ethanol consumption (Ftreatment(1,16)=7.25, p=0.01). There was no effect of TLR3/dsRNA inhibitor pretreatment on poly(I:C)-induced ethanol preference (Ftreatment(1,16)=0.16, p=0.69) or total fluid intake (Ftreatment(1,16)=3.35, p=0.08). There was no main effect of poly(I:C) or TLR3/dsRNA inhibitor on body weight [Ftreatment(1,18)=1.49, p=0.24; Ftime(10,180)=9.4, p<0.0001; Ftreatment x time(10,180)=2.69, p=0.004]. Arrows indicate days when animals received injections, the red arrow indicates switch to the TLR3/dsRNA inhibitor. Dashed line represents when saline(10%DMSO)/poly(I:C) treated animals were treated with inhibitor(10%DMSO)/poly(I:C). Data represented as mean ± s.e.m. (*p<0.05, significantly different from saline/saline group; #p<0.05, significantly different from saline/poly(I:C) group, Bonferroni post-hoc tests, n=10 per group).
The levels of transcripts are presented using fold-change values normalized to endogenous control, with six biological and three technical replicates. Values are presented as fold change ± SEM. (* represents significant difference from saline group, # represents significant difference from poly(I:C) group, ****p<0.0001, ***p<0.0002, **p<0.0021, *p<0.05 compared with saline treatment at same time; #p<0.05 compared with other poly(I:C) groups, n=6 per group).
Male C57BL6/J mice were treated with either saline(10%DMSO) or TLR3/dsRNA complex inhibitor (2mg) and underwent 24 days of EOD-2BC (15%v/v ethanol). To determine how the inhibitor changed escalation of drinking behavior from baseline we subtracted subsequent behavioral drinking days from the first drinking day. (A) Inhibitor treatment trended toward decreased ethanol intake (Ftreatment(1,18)=3.49, p=0.07; Ftime(4,72)=5.57, p=0.0006; Ftreatment x time(4,72)=0.363, p=0.83). (B) Inhibitor treatment reduced escalation of alcohol preference (Ftreatment(1,18)=5.96, p=0.02; Ftime(4,72)=6.79, p=0.0001; Ftreatment x time(4,72)=1.149, p=0.34). (C) Inhibitor treatment modestly increased total fluid intake escalation (Ftreatment(1,18)=4.92, p=0.04; Ftime(4,72)=3.38, p=0.01; Ftreatment x time(4,72)=2.62, p=0.04). Data represented as mean ± s.e.m. (*p<0.05, significantly different from saline group, Bonferroni post-hoc tests, n=10 per group).