Abstract
Stress granule formation is important for stress response in normal cells and could lead to chemotherapy resistance in cancer cells. Aberrant stress granule dynamics are also known to disrupt proteostasis, affect RNA metabolism, and contribute to neuronal cell death. Meanwhile, circadian abnormality is an aging-related risk factor for cancer and neurodegeneration. Whether stress granule dynamics are circadian regulated is entirely unknown. Here we show that the formation of stress granules varied by zeitgeber time in mouse liver. Moreover, altering circadian regulation by silencing the core circadian gene Bmal1 in a cell line expressing an endogenous GFP-tagged G3BP1 significantly increased stress granule dynamics, while the overexpression of Bmal1 decreased them. Surprisingly, increased stress granule dynamics and formation by transient decrease of BMAL1 coincided with increased resistance to stress-induced cell death. The circadian regulation of stress granules was mediated by oscillating eIF2α expression. At zeitgeber time when BMAL1 and eIF2α were at nadir, reduction of unphosphorylated eIF2α could significantly alter the ratio of phosphorylated/total eIF2α and quickly lead to increased formation of stress granules. Therefore, diurnal oscillating eIF2α connects the circadian cue to a cellular stress response mechanism that is vital for both neurodegeneration and cancer.
Introduction
The ability of cells to cope with environmental and cellular stress is vital for their flourishment and survival. Abnormal stress response is known to contribute to aging process and aging-related diseases such as cancer and neurodegenerative diseases1,2. By developing various stress response and anti-apoptotic mechanisms, cancer cells can proliferate in hostile microenvironment, such as hypoxia, or even chemotherapy drugs3. On the other hand, the inability of neurons to resist increased production of reactive oxygen species and endoplasmic reticulum (ER) stress either during normal aging or under pathogenic conditions will lead to neurodegenerative diseases4–6.
One of the cellular stress responses that have been intimately linked to stress resistance in cancer cells and the development of some neurodegenerative diseases is the formation of stress granules3,7–9. Stress granules are membrane-less cytoplasmic structures formed when translation initiation is inhibited during strong stress responses or viral infection10,11. They are composed of abundant messenger RNAs (mRNAs) stalled in translation initiation, RNA-binding proteins, and ribonucleoproteins. The formation of stress granules and the arrest of canonical translation could serve as a protective mechanism when the cellular resources are limited during stress10. While the translation of most constitutive proteins is suppressed, stress-induced mRNAs could be preferentially translated12. In tumor cells, stress granule induction promotes resistance to apoptosis in chemotherapy3. In neurons, the abnormal regulation of stress granules contributes to neurodegeneration.
Stress granules are dynamic structures characterized by constant exchange of protein components. The exchange rates of those components are different, with the proteins at the dense cores less dynamic13, and are affected by the interaction and local concentration of stress granule proteins14. Interestingly, a sizable portion of stress granule components are related to the pathogenesis of cancer and/or neurodegenerative diseases, particularly amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD)13. Proteins including FUS, TAF15, EWSR1, TDP43, TIA-1, VCP, and Ataxin-2 are not only genetically and/or pathologically related to ALS and FTD, but also involved in cancer development15–27. Furthermore, the most common cause of familial ALS and FTD, arginine-rich dipeptide repeats derived from C9orf72 hexanucleotide expansion repeats, could interact with stress granule components, affect the stress granule dynamics, and disrupt nucleocytoplasmic transport28–31.
Circadian rhythms are physiological and behavioral changes following an ~24 h cycle. The diurnal changes are governed by a molecular circadian clock, featuring two main feedback transcriptional–translational regulatory loops to direct the oscillating expression of target genes in an organ-specific manner32,33. The core circadian proteins BMAL1 and CLOCK transcriptionally activate Per, Cry, and Nr1d1/2, whose protein products, when accumulated, could negatively regulate the expression or transcriptional activity of BMAL134,35. The oscillating expression of these core circadian genes control the circadian activities. With aging, the circadian control is gradually weakened. There is a clear association between circadian dysregulation and various neurodegenerative diseases36,37. Sleep disturbance is frequently seen in patients with Alzheimer’s disease (AD) and Parkinson’s disease (PD)37–39, and abnormal circadian behaviors are detected in animal models of AD or PD before the onset of disease pathology40,41. In our recent study, we have observed sleep and circadian abnormalities before cognitive deficits in a FUS knock-in rodent model42. Therefore, circadian dysregulation could be a risk factor for neurodegenerative diseases. Whether circadian abnormalities may affect cancer risk is uncertain43, but DNA excision repair activity appears to be circadian regulated with peak activity at evening in mice44. In addition, chronotherapy in cancer has been practiced even though the underlying mechanisms are not entirely clear and the results were mixed43,45.
The generation of protective antioxidative enzymes, such as superoxide dismutase, shows diurnal rhythm46,47, and this is part of cellular defense system against weak stress insults from reactive oxygen species, the byproducts of normal cellular metabolism. Under ER stress or exogenous stress insults, stress granules will quickly appear. Whether the formation of stress granules could be regulated by circadian cues is entirely unknown. Even if there is such a regulation, as stress granule formation is a fast response to external stress, how would that be regulated by slow circadian signals? In this study, we have provided in vitro and in vivo evidence to demonstrate that stress granule formation is affected by circadian gene expression, and this regulation is due to the oscillation of total eIF2α. We have also unveiled a surprising result showing reduced expression of BMAL1 could protect cells against arsenite stress while promoting stress granule formation. Therefore, our results have suggested an efficient diurnal cellular protective mechanism to guard against increased stress in the active phase, and implied that circadian dysregulation may promote neurodegeneration due to aberrant stress response. In addition, our observations may provide a cellular mechanism to better understand cancer chronotherapy.
Results
To examine whether stress granule formation could be affected by the different time of the day in vivo, we intraperitoneally injected the mice with saline (control) or oxidative stressor sodium arsenite and harvested the liver tissues from circadian entrained mice at zeitgeber time 5 and 13 (ZT 5 and ZT 13). The differential expression of core circadian protein BMAL1 was validated (Supplementary Fig. 1a). The liver tissues were selected because liver is an organ with robust circadian-regulated gene expression48. Using stress granule markers PABP1 (Fig. 1a) and YB1 (Supplementary Fig. 1b), we have detected higher spontaneous and stress-induced stress granule formation at ZT 13 (Fig. 1a, b, Supplementary Fig. 1b, c). It is worth noting that although the spontaneously occurred stress granules are visible at ZT 13, they were much smaller than the ones formed in sodium arsenite-treated mice. This result suggested that the stress granules could be affected by circadian cues and are more easily formed at night in mice.
Fig. 1. Stress granules in mouse liver at zeitgeber time (ZT) 5 and ZT 13.
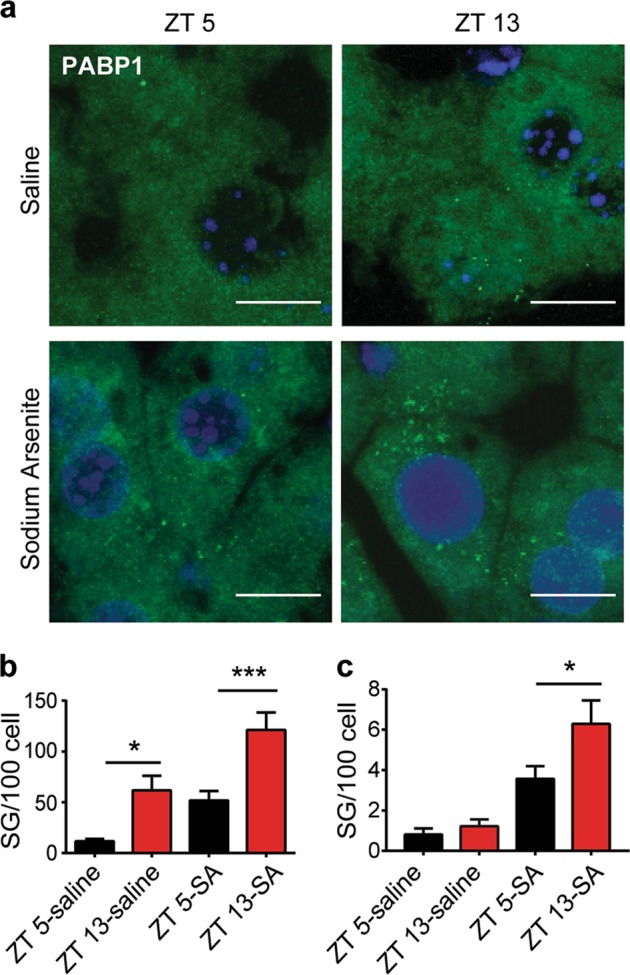
a The liver tissues from the wild-type (WT) mice harvested at ZT 5 and ZT 13 were labeled with anti-PABP1 (stress granule marker) antibody and revealed by immunofluorescence. Mice were intraperitoneally injected with saline or sodium arsenite (SA, 10 mg/kg) for 1 h before killing. Representative micrographs showing the presence of PABP1-positive stress granules (Scale bar = 10 μm). b, c Quantification of all visible PABP1 puncta (b) or large size puncta (>0.5 μm, c) respectively (n = 3 mice per group with 950–1000 cells examined per mouse. mean ± S.E.M.; two-way analysis of variance (ANOVA) with Tukey’s multiple comparison *P ≤ 0.05, ***P ≤ 0.001)
We then aimed to evaluate the circadian regulation on stress granule formation and dynamics. Overexpression of green fluorescent protein (GFP)-tagged stress granule marker G3BP1 in cell lines is a commonly adopted approach to study stress granule formation. However, we reasoned that exogenous expression of a key protein involved in stress granule assembly49 might not be the best approach to study the internal circadian regulation of stress granule formation. Therefore, we used CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) gene editing technique to insert a GFP tag in frame to the N terminus of the G3BP1 gene in the human neuroblastoma cell line SH-SY5Y (Fig. 2a, Supplementary Fig. 2a, b). Using this approach, we would retain the normal transcriptional and post-transcriptional regulation of the endogenous G3BP1. SH-SY5Y cells also demonstrated features of oscillating circadian gene expression (Supplementary Fig. 2c, d). The GFP-G3BP1 KI cells expressed G3BP1 in a cytoplasmic diffuse pattern and formed granules that colocalized with stress granule marker PABP1 upon stress (Fig. 2b), indicating the tagged G3BP1 behaved as expected during stress.
Fig. 2. Generation of GFP-G3BP1 knock-in (KI) cell line.
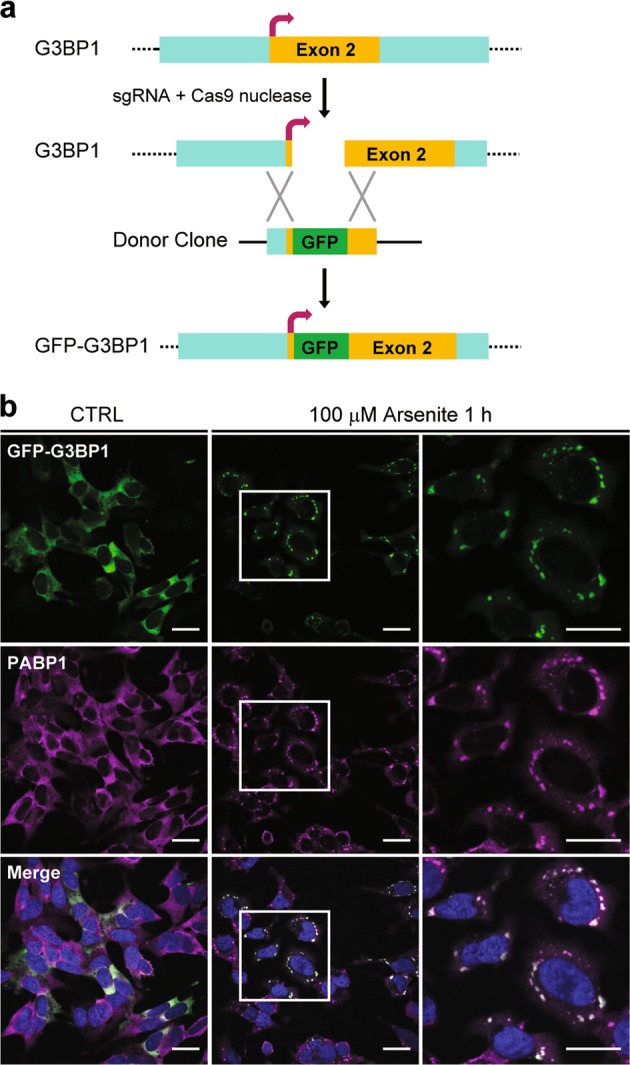
a Schematic representation of the strategy of tagging green fluorescent protein (GFP) at the N terminus of human G3BP1 using CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9). b Immunofluorescence confocal microscopy showing the co-localization of endogenous SG marker PABP1 with knock-in GFP-G3BP1. Stress granules were induced with 100 μM sodium arsenite for 1 h. The square areas in the middle panels were enlarged and shown in the right. Scale bar = 20 μm
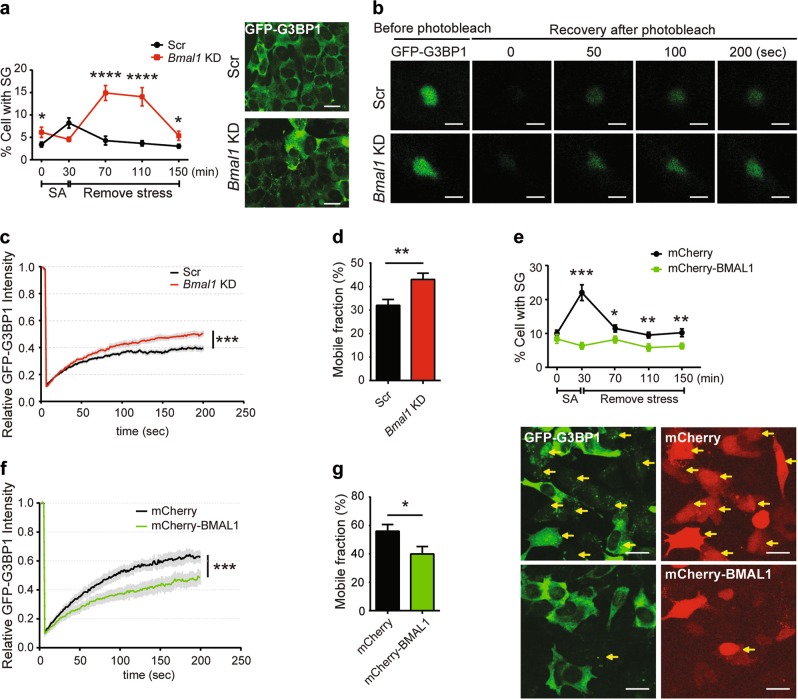
Bmal1 is a potent circadian regulator and single knockout (KO) of Bmal1 could cause arrhythmicity50. Furthermore, BMAL1 was expressed at low level at ZT 13 in mouse liver when stress granules increased (Fig. 1). Therefore, we chose to silence the expression of Bmal1 as a way to change circadian input and then evaluated stress granule formation. There was a slight increase of basal stress granule formed upon Bmal1 silencing (Fig. 3a). Upon transient oxidative stress shock with sodium arsenite for 30 min, the cells with lower BMAL1 expression showed a delayed but significant increased number of cells with stress granules (Fig. 3a). This observation was consistent with the higher number of stress granules formed in mouse liver at ZT 13 (Fig. 1). Next, we examined the change of stress granule dynamic using fluorescence recovery after photobleaching (FRAP) (Fig. 3b). Silencing of BMAL1 led to a statistically significant increase of stress granule dynamic (Fig. 3c) and mobile fraction (Fig. 3d). Interestingly, when we examined the stress granule dynamics in mouse embryonic fibroblasts (MEFs) derived from BMAL1 KO mice, we observed minor decrease of stress granule dynamics and no difference in stress granule formation when compared to the wild-type (WT) MEF (Supplementary Fig. 3a, b), suggesting that the BMAL1 KO cells have different stress response mechanism (see Discussion).
Fig. 3. Altered BMAL1 expression affects stress granule (SG) formation and dynamics.
a Bmal1 silencing caused increased stress granule formation in GFP-G3BP1 knock-in (KI) cells exposed to sodium arsenite (SA) stress. Cells transfected with the scrambled or Bmal1 small interfering RNA (siRNA) were treated with 50 μM of SA for 0 or 30 min. After stress was removed, cells were kept in fresh medium for additional 40, 80, or 120 min before fixation for immunofluorescence microscopy analysis. The percentage of cells with stress granules under indicated condition was quantified (mean ± S.E.M.; n = 3 independent experiments, with 30 fields for each time point, at least 50 cells per field. *P ≤ 0.05, ****P ≤ 0.0001 by unpaired Student’s-t-test, Scale bar = 20 μm). Representative images show the SG formation 80 min after removing stress. b–d GFP-G3BP1 KI cells were transfected with Cy3-labeled scrambled siRNA (Scr) or Bmal1 siRNA and subsequently treated with 50 μM SA for 30 min. After stress was removed, green fluorescent protein (GFP)-positive stress granules in CY3-positive cells were analyzed by fluorescence recovery after photobleaching (FRAP). b Representative images show the stress granules before and after photobleaching at different time. Scale bar = 2 μm. c Signal intensity of GFP fluorescence of stress granule FRAP in Cy3-positive cells. The average fluorescence before photobleaching was designated as 1. d Mobile fraction calculated from the FRAP analysis in c (mean ± S.E.M.; n = 14-15 cells per sample, **P ≤ 0.01, ***P ≤ 0.001 by unpaired Student’s-t-test). e Overexpression of BMAL1 suppressed stress granule formation. GFP-G3BP1 KI cells expressing mCherry or mCherry-BMAL1 were stress shocked with 20 μM SA and fixed at indicated time for stress granule imaging and quantification (mean ± S.E.M.; n = 3 independent experiments, with 30 fields for each time point, at least 10–15 cells per field. *P ≤ 0.05, **P ≤ 0.01 ***P ≤ 0.001 by unpaired Student’s-t-test. Scale bar = 20 μm). Representative images show SG formation in cells treated with SA for 30 min. Yellow arrows indicate the SG status of representative mCherry-positive cells included in the quantification. f The signal intensity of GFP fluorescence of stress granule FRAP in mCherry-positive cells. g Mobile fraction calculated from the FRAP analysis in f (mean ± S.E.M.; n = 14–15 cells per sample, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 by unpaired Student’s-t-test)
To further validate the effects of BMAL1 on stress granule formation, we overexpressed mCherry-tagged BMAL1 in GFP-G3BP1 knock-in (KI) cells, and quantified the cells with stress granules. In contrast to cells with silenced BMAL1 expression, overexpression of BMAL1 suppressed the formation of stress granules upon stress (Fig. 3e). Furthermore, the stress granule dynamic was also significantly reduced (Fig. 3f), accompanied by decreased mobile fraction (Fig. 3g). To confirm these observations, we examined the regulation of stress granule dynamics in MEF cells derived from NR1D1/RevErbα-deficient mouse embryos. NR1D1/RevErbα is a negative regulator of BMAL1 expression51. Depletion of NR1D1/RevErbα led to the reduction of stress granule dynamics and mobile fraction (Supplementary Fig. 3c–e). Taken together, our results showed that the altered expression of core circadian regulators could affect stress granule formation and dynamics.
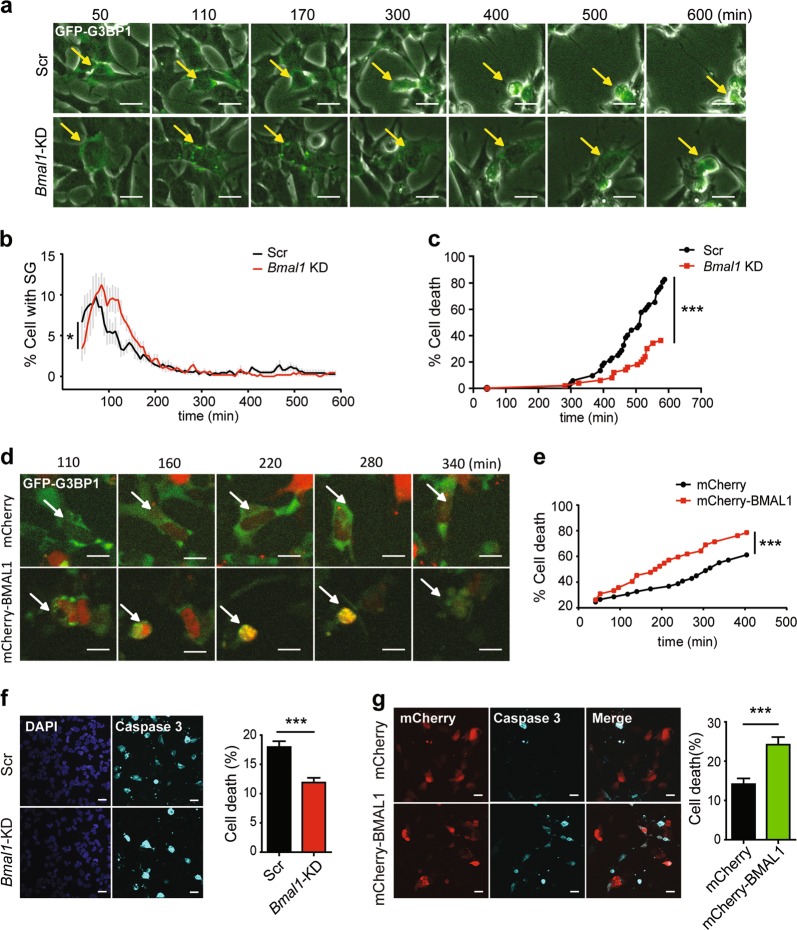
As stress granule formation is a pro-survival response during stress10, we evaluated the relationship of stress granule formation with cell death under stress when circadian cues are altered. We first used specific small interfering RNA (siRNA) to silence BMAL1 expression, and then treated GFP-G3BP1 KI cells with 20 μM sodium arsenite and recorded cell morphology and the appearance of stress granules using live imaging (fluorescence and bright field) for up to 600 min (Fig. 4a). The timing for stress granule appearance and the fate of these stress granule-positive cells were analyzed individually. In cells with silenced BMAL1, there was a delayed appearance of stress granules, but higher percentage of cells with stress granules (Fig. 4b), consistent with the patterns seen in cells exposed to transient stress shock (Fig. 3a). Interestingly, these stress granule-positive, BMAL1-silenced cells exhibited significant resistance to stress-induced cell death (Fig. 4c). In contrast, live imaging analysis of cells overexpressing mCherry-BMAL1 showed increased sensitivity to stress-induced cell death (Fig. 4d, e). To validate the effect of stress-induced cell death by altered BMAL1 expression, we examined activated caspase-3, an indicator for apoptotic cell death, in control or BMAL1-silenced cells, or in cells overexpressing BMAL1. While BMAL1 knockdown protected cells against stress-induced cell death (Fig. 4f), BMAL1 overexpression potentiated it (Fig. 4g). Therefore, transient low expression of BMAL1 facilitated the formation of stress granules, and protected cells against stress-induced cell death.
Fig. 4. Reduced expression of BMAL1 protects against stress-induced cell apoptosis.
a Live-cell imaging of Bmal1-silenced GFP-G3BP1 knock-in (KI) cells exposed to 20 μM of sodium arsenite for up to 600 min (see Movie S1, 2). The same stress granule-generating cell in each group (Scr or Bmal1 knockdown (KD)) at different time was indicated by an arrow. Note the different timing of the apoptotic morphology (rounded cell shape and detachment from the dish) in cells transfected with Scr or Bmal1 small interfering RNA (siRNA). b Quantification of the percentage of cells with stress granules (mean ± S.E.M.; n = 500–600 cells per sample, *P ≤ 0.05 by two-way analysis of variance (ANOVA). Scale bar = 20 μm). c The death ratio of stress granule-positive cells (n = 50–100 cells per group, ***P ≤ 0.0001 by log-rank test). d Live-cell imaging of mCherry-BMAL1-expressing GFP-G3BP1 KI cells exposed to 20 μM of sodium arsenite (see Movie S3, 4). e The cell death ratio of mCherry- and stress granule-positive cell, ***P ≤ 0.0001 by log-rank test. f, g Caspase-3 activation in GFP-G3BP1 KI cells transfected with Bmal1 siRNA (f) or mCherry-BMAL1 expression plasmid (g). At 72 h after siRNA transfection, cells were treated with 50 μM SA for 12 h before analysis. For cells with mCherry-BMAL1 overexpression, they were treated 48 h post transfection with 20 μM SA for 7 h before analysis. Representative images of activated caspase-3 staining and the quantification of percentage of cell death as indicated by activated caspase-3 are shown for each condition (mean ± S.E.M.; n = 3 independent experiments, with 10 fields scored per experiment, each field >100 cells, ***P ≤ 0.001 by unpaired Student’s-t-test. Scale bar = 20 μm)
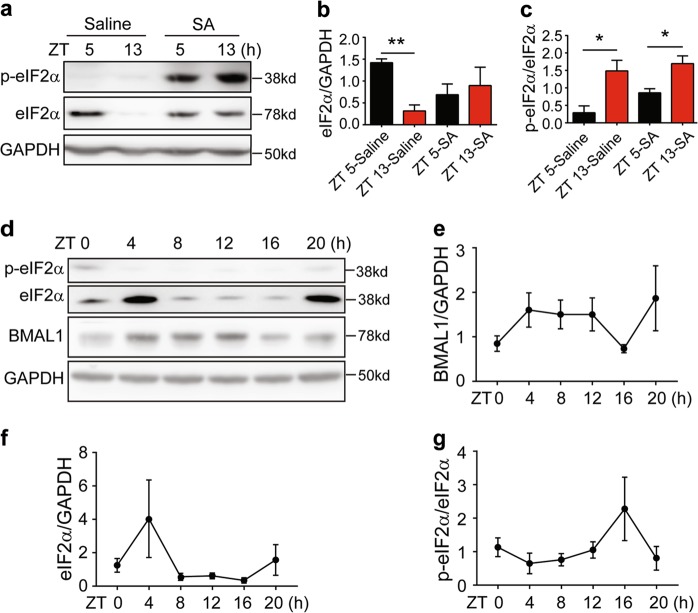
To investigate the mechanism that may account for the regulation of stress granules by circadian proteins, we examined whether it could be related to eIF2α, as eIF2α phosphorylation is a key signal to initiate the stress granule assembly12. We first examined the total and phosphorylated eIF2α in mouse liver collected at ZT 5 and ZT 13, with or without sodium arsenite exposure. At ZT 13, there was a significant drop of total eIF2α level at both the protein and mRNA levels (Fig. 5a, b, Supplementary Fig.4a), but the absolute phosphorylated eIF2α level remained stable or slightly increased (Fig. 5a). When exposed to arsenite stress, the mouse liver showed a clear increase of total and phosphorylated eIF2α (Fig. 5a). Interestingly, the ratio of phosphorylated eIF2α to total eIF2α was significantly elevated at ZT 13, even in the absence of stress insults (Fig. 5c), which may account for the higher basal stress granule levels at ZT 13 (Fig. 1a, b).
Fig. 5. Circadian oscillation of eIF2α and stress granules in mouse liver.
a–c Protein expression of eIF2α and p-eIF2α in the liver tissues of the wild-type mice at zeitgeber time (ZT) 5 and ZT 13. Mice were treated with saline or sodium arsenite (10 mg/kg) for 1 h before tissue harvest. Values in (b, c) represented mean ± S.E.M.; n = 3 independent experiments, *P ≤ 0.05, **P ≤ 0.01 by unpaired Student’s-t-test. d–g Protein levels of BMAL1, eIF2α, and p-eIF2α at indicated zeitgeber times. f Representative immunoblots showing the expression levels of eIF2α, p-eIF2α, BMAL1, and GAPDH. e–g Quantification of relative expression of indicated proteins (mean ± S.E.M.; N = 5 mice per time point)
The significant difference of total eIF2α also suggested a possibility that eIF2α could be circadian regulated. We analyzed published proteomic and transcriptome dataset from mouse liver52 and found that the expression of eIF2α was indeed oscillating (Supplementary Fig. 4b, c), and the oscillation would be shifted in CRY1/2 KO mice53 or weakened in BMAL1 KO mice54 (Supplementary Fig. 4d, e). In addition, eIF2α expression also oscillates in baboons, although with a peak around ZT 12–16 in the brain (Supplementary Fig. 4f, g)55. To thoroughly examine the change of eIF2α at various circadian time, we collected liver and brain tissues from entrained mice every 4 h (n = 5 mice at each time point) and determined the expression of eIF2α and BMAL1 (Fig. 5d) as well as the spontaneous stress granule formation at each time point (Supplementary Fig. 5a, b). The BMAL1 protein showed oscillating expression as expected, with relatively lower expression at ZT 16–20 (Fig. 5e), which also coincided with increased abundance of stress granules (Supplementary Fig. 5a, b). Consistent with our analysis of proteomic and transcriptome data (Supplementary Fig. 4b–g), the oscillation of total eIF2α was observed in liver (Fig. 5d, f), cortex, and hypothalamus (Supplementary Fig. 5c, d). Remarkably, the relative expression of phosphorylated eIF2α in the liver also showed an oscillating pattern, but in the opposite phase as the total eIF2α (Fig. 5g). It is worth noting that the absolute level of phosphorylated eIF2α did not vary significantly between ZT 4 and ZT 16 (Fig. 5d), even though the total eIF2α expression was at peak and trough, respectively. Therefore, at ZT 12–16 when the total eIF2α was at trough, even minor phosphorylation of eIF2α would quickly reduce the pool of unphosphorylated eIF2α, increase the ratio between phosphorylated and total eIF2α, and cause translation arrest and stress granule formation.
To directly test whether reduced pool of unphosphorylated eIF2α could contribute to increased stress granule formation in cells with low expression of BMAL1, we first transiently silenced the BMAL1 expression using siRNA, and then stress shocked SH-SY5Y cells with sodium arsenite. While the total eIF2α slightly decreased, the phosphorylated eIF2α clearly increased (Fig. 6a–c), suggesting reduced abundance of unphosphorylated eIF2α. We then transiently transfected mCherry-tagged eIF2α in GFP-G3BP1 KI cells pretreated with BMAL1 or control siRNA, and examined the stress granule formation after stress shock by immunofluorescence microscopy. eIF2α expression significantly suppressed the formation of stress granules (Fig. 6d, e). Therefore, decreased pool of unphosphorylated eIF2α in cells with low BMAL1 expression contributed to the formation of stress granules.
Fig. 6. Reduced BMAL1 expression increased the ratio of P-eIF2α to total eIF2α and the number of stress granules.
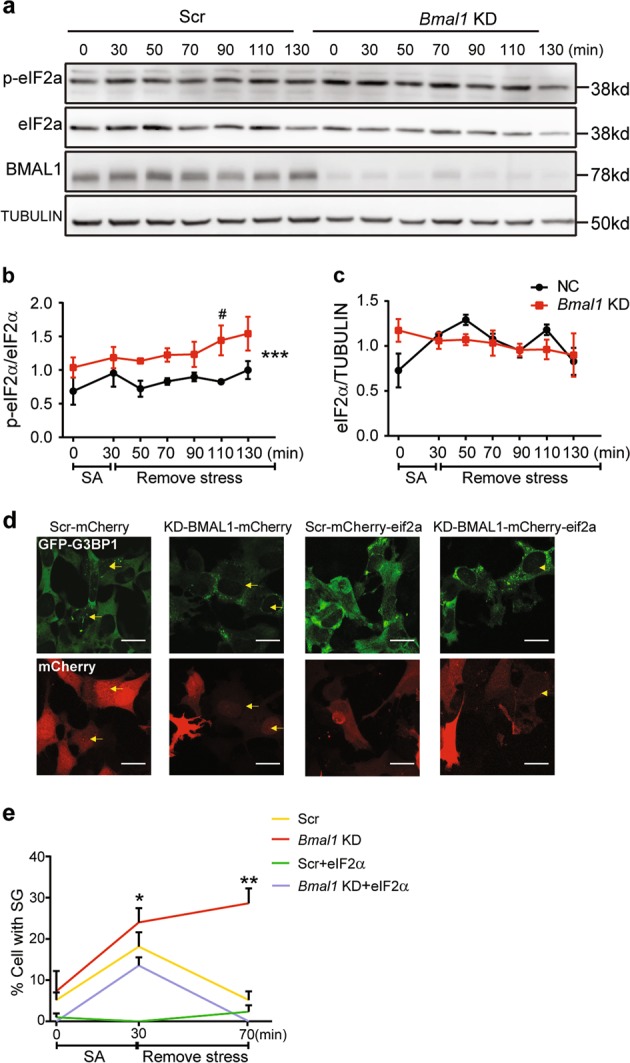
a–c Protein levels of p-eIF2α, eIF2α, and BMAL1 at indicated time points after stress shock in cells with reduced BMAL1 expression. Representative immunoblots are shown in a, and the quantification of p-eIF2α and eIF2α are presented in b and c, respectively (mean ± S.E.M.; n = 3 independent experiments, two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test, * represents the P value of two-way ANOVA among 50–130 min ***P ≤ 0.001; # represents the P value of Sidak’s multiple comparison, #P ≤ 0.05). d, e Expression of total eIF2α blocked the increased stress granule formation by BMAL1 silencing. mCherry-eIF2α or mCherry was transfected in GFP-G3BP1 knock-in (KI) cells with control or small interfering RNA (siRNA) targeting BMAL1, and the number of stress granules formed was analyzed after stress shock. Representative images are shown in d, and the quantification is presented in e (mean ± S.E.M.; n = 150–200 cells per sample, *P ≤ 0.05, **P ≤ 0.01 by unpaired Student’s t-test between KD-BMAL1 and KD-BMAL1-eIF2α. Scale bar = 20 μm). Yellow arrows indicate mCherry and SG double-positive cells
Discussion
In this study, we have provided evidence to show that stress granule formation and dynamics are affected by circadian cues, and this regulation is caused by oscillating eIF2α expression. Furthermore, we have discovered that altering the circadian gene expression by decreasing the expression of BMAL1 could promote stress granule formation and protect arsenite stress-induced apoptosis. Therefore, our findings have shed new light on the circadian influence on the cellular defense mechanisms against stress insults and could help us better understand the circadian influence on neurodegeneration and cancer.
The regulation of stress granule formation by phosphorylation of the translation initiation protein eIF2α is well documented56; however, the diurnal oscillation of eIF2α has never been reported. Stress granules are quickly assembled when cells are exposed to sudden increase of environmental and cellular stress57. In contrast, the rhythmic changes in physiology and behaviors governed by circadian clock usually happen at much slower pace. The oscillating eIF2α expression happens to seamlessly connect the circadian rhythm and stress granule formation. We have found that in mouse liver the eIF2α expression troughs when the BMAL1 expression is at nadir at early night time, also an active phase for nocturnal animals with increased lipid peroxidation46,58. Phosphorylation of eIF2α under stress condition at night would act as a fast-acting signaling event to further reduce the small pool of unphosphorylated eIF2α that is required for translational initiation, thus causing translation arrest and stress granule formation. On the other hand, the same stress insult at day time (ZT 5) in mouse would cause a much smaller impact on stress granule formation. This regulation provides an efficient cellular mechanism to manage increased stress during active phase for nocturnal animals. Interestingly, the peak expression of BMAL1 and eIF2α in primates are between ZT 12 and 16, about anti-phase to that in mice55 (Supplementary Fig. 4f, g). Therefore, the regulation of stress granules by oscillating eIF2α could be a conserved mechanism in mammals to cope with increased stress during active phase.
Although eIF2α oscillation and phosphorylation appear to be the main driver of circadian control of stress granules, other mechanisms may also contribute to the circadian regulation of stress granules. Based on our analysis of stress granule proteome13 and circadian-regulated transcriptome59, the expression of a score of stress granule component proteins, such as FUS, TDP43, and CRIBP, are under circadian control. Given that the stress granule assembly and dynamics are heavily influenced by protein-protein interaction14, the abundance of these proteins at various circadian time may also affect stress granules. In addition, the stress granule assembly and dynamics are affected by the presence of various ATPase, such as valosin-containing protein (VCP), in the granules14,60 and depend on the adenosine triphosphate (ATP)13,14. Given that ATP production could also be affected by circadian regulation of mitochondrial biogenesis61,62, it would be of interest to evaluate whether ATP abundance may contribute to the timing and sensitivity of stress granule formation.
One surprising finding from our study is the protective effect of low BMAL1 expression during stress response. BMAL1 KO mice have significant disrupted circadian rhythm and metabolism63,64 and have shortened life span with early aging pathologies65. In another study, BMAL1 KO mice show synaptic degeneration and increased neuronal oxidative damage66. However, transiently reduced BMAL1 expression promotes the stress granule formation and was protective in our study. Our observations are consistent with a recent paper showing the anti-apoptotic effect of reduced BMAL1 expression during unfolded protein response67. We have further demonstrated the difference in the stress response between cells with BMAL1 knock-out background and transient reduction (Supplementary Fig. 3a, b; Fig. 3a, c). Therefore, unlike the harmful effects of long-lasting BMAL1 depletion, daily oscillation of BMAL1 and eIF2α could increase the sensitivity to stress insults during the active dark phase in mice and could transiently stop protein translation by forming protective stress granules. It is not surprising for multifunctional proteins such as BMAL1 to exhibit divergent effects on cell survival and death based on the duration and abundance of their expression. For example, transient induction of the wild-type p53 is pro-survival while accumulation of p53 leads to apoptosis68. It will be of interest to examine the stress granule formation in detail in BMAL1 KO mice during the aging process to fully assess the role of BMAL1 in stress response and cell death.
As circadian rhythm is known to decay with aging, our study may provide a new cellular mechanism to account for the abnormal stress granule assembly, disassembly, and dynamics that could contribute to increased risks for cancer, viral infection, and neurodegenerative diseases in aged population. Remarkably, a group of proteins implicated in the pathogenesis of ALS and/or FTD are components of stress granules and could even participate in the assembly or disassembly of stress granules18,28,60,69,70, thus further strengthening the link between stress granule formation and neurodegenerative diseases. Interestingly, some mutations that cause neurodegenerative diseases could lead to altered circadian regulation by causing neuronal loss in brain regions affecting circadian and sleep71,72, or directly participating in circadian regulation. With time, they could start a feedforward cycle to affect stress response and accelerate the development of neurodegenerative diseases. Consistent with this idea, our recent studies have shown that ALS/FTD-associated FUS is a circadian regulator73 and that F521C KI rats exhibit circadian abnormalities as early symptoms42.
Chronotherapy has been practiced to improve the efficacy of anti-cancer drugs by modulating their pharmacokinetics and pharmacodynamics45,74. DNA repair and apoptosis, which some anti-cancer drugs target, are circadian regulated43,74. Together with these pathways, stress granules could be part of the circadian-regulated components contributing to the molecular cellular mechanisms in chronotherapy in cancer. Stress granule formation promotes cancer survival and resistance to chemotherapy agents3. Mutant Kras enhances cancer cell survival by stimulating stress granules75. Conversely, suppressing stress granules could increase the drug sensitivity in Hela cells76. Consistent with our findings showing increased stress granule formation in the evening, a recent study has reported that chemotherapy agent cisplatin causes less damage and side effects in the wild-type mice when the drug was delivered at that time77. Therefore, our current study will prompt the future investigation to decipher the circadian regulation of stress granules in cancer cells and help to optimize chronotherapy regimen to maximize the drug efficacy in cancer cells while minimizing the side effects in normal cells by considering the timing of stress granule formation.
Methods
Animals
All animal works were performed in accordance with the regulations by the Animal Care and Use Committee of Institute of Neuroscience, Chinese Academy of Science. Mice were killed under anesthesia, and all efforts were made to minimize mice suffering.
The 7–8-week-old wild-type C57BL/6J male mice were purchased from SLAC (Shanghai Laboratory Animal Center) and were maintained on a 12 h light:12 h dark (lights on at 7 a.m.) throughout the study. Mice were housed in the facility for 2 weeks before experiments. Food and water were available ad libitum. At ZT 4 (zeitgeber time 4, means 4 h after lights on) and ZT 12, the mice received intraperitoneal injection of saline or sodium arsenite (10 mg/kg) and the liver tissues were harvested at ZT 5 and ZT 13 for immunofluorescence and western blot. For the experiment to evaluate eIF2α circadian oscillation and stress granule formation oscillation, mice were single-housed for at least 2 weeks prior to experimental use, and 5 mice were harvested for small part of liver, cortex, and hypothalamus at each time point every 4 h (starting at zeitgeber time 0) for western blot. The mouse livers were then perfused through hepatic portal vein with 4% paraformaldehyde for subsequent immunofluorescence labeling.
Antibodies and drugs
The primary antibodies used are: mouse anti-eIF2α (sc-133132 Santa Cruz Biotechnology, 1:1000 for western blot); rabbit anti-phospho-eIF2α (9721 Cell Signaling Technology, 1:1000 for western blot); rabbit anti-BMAL1(14020 Cell Signaling Technology, 1:1000 for western blot); mouse anti-GAPDH (60004 Proteintech, 1:1000 for western blot); mouse anti-β-tubulin (M20005, Abmart; 1:3000 for western blot); rabbit anti-PABP1 (ab2060, Abcam, 1:100 for immunofluorescence), rabbit anti-YB1 (ab76149, Abcam, 1:200 for immunofluorescence); and rabbit anti-cleaved-caspase-3 (9661 Cell Signaling Technology, 1:200 for immunofluorescence). The drug used was sodium arsenite (S7400-100G, Sigma, 100 mg/kg for mouse and 20 or 50 μM for cells).
Plasmids and siRNA
The plasmids, mCherry-tagged BMAL1 and eIF2α, were generated by PCR using human complementary DNA (cDNA) library and cloned into the EGFP-C1 vector (Invitrogen), with EGFP changed to mCherry.
The siRNA constructs targeting the human BMAL1 were synthesized at Genepharma with the following sequences:
BMAL1-996 (sense, 5’-CCUCAACUACAGCCAGAAUTT-3’; antisense, 5’-AUUCUGGCUGUAGUUGAGGTT-3’);
BMAL1-1565 (sense, 5’-GCACAUCGUGUUAUGAAUATT-3’; antisense, 5’-UAUUCAUAACACGAUGUGCTT-3’);
BMAL1-12247 (sense, 5’-GCCUUCAGUAAAGGUUGAATT-3’; antisense, 5’-UUCAACCUUUACUGAAGGCTT-3’).
Cy3-labeled BMAL1 siRNA (sense, 5’-GCACAUCGUGUUAUGAAUATT-3’; antisense, 5’-UAUUCAUAACACGAUGUGCTT-3’);
Cy3-labeled Scramble siRNA (sense, 5’-UUCUCCGAACGUGUCACGUTT-3’; antisense, 5’-ACGUGACACGUUCGGAGAATT-3’);
Scramble siRNA (sense, 5’-UUCUCCGAACGUGUCACGUTT-3’; antisense, 5’-ACGUGACACGUUCGGAGAATT-3’).
For Bmal1 siRNA transfection, total of 5 μL of above constructs were mixed at 1:1:1 ratio for cells cultured in 3.5 cm dishes.
Western blot
The proteins from mouse tissues or cultured cells protein were extracted with RIPA lysis buffer (150 mM NaCl, 50 mM Tris (pH = 8.0), 1% NP40, 1% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche) as needed. Proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the protein bands were visualized using Bio-Rad western ECL substrate kit. The band intensity in immunoblots was determined by Bio-Rad Quantity One software.
Cell culture and transfection
MEFs were collected from E13.5 embryos using pregnant mice from Nr1d1 KO mice or BMAL1 KO mice mating pairs. E13.5 embryos were eviscerated. Embryos’ epidermis was cut into piece then digested into single cell with Tyrisin (Tyrisin-EDTA 0.25% phenol red Thermo Fisher). Large cell lumps were removed through cell strainer (40 μm nylon FALCON).
MEFs and SH-SY5Y cells were cultured at 37 °C in 5% CO2 in Dulbecco’s modified Eagle’s medium (Invitrogen), supplemented with 10% fetal bovine serum (Invitrogen) and antibiotics (penicillin and streptomycin, HyClone, SV30010). Cells were transfected as needed using Lipofectamine 2000 reagent (Invitrogen) or Lipofectamine RNA-MAX (Invitrogen). The total amount of plasmid DNA or siRNA was adjusted to 5 μg per 3.5 mm dish. Cells were harvested at 48 or 72 h (siRNA) post transfection for western blotting.
Quantitative reverse transcriptase PCR
For real-time PCR, total RNAs were extracted from cells using Trizol. The full-length cDNA library was constructed by reverse transcription PCR using PrimeScriptTM RT Master Mix Perfect Realtime (RR036A, Takara). Quantitative reverse transcriptase PCR was performed using iQTM SYBR@ Green Supermix (1708882, Bio-Rad) with Bio-Rad CFX Connect real-time PCR system.
Generation of GFP-G3BP1 KI SH-SY5Y cell line by CRISPR/cas9
The genomic sequences surrounding the coding region of human G3BP1 N-terminal (±200 bp) were analyzed for potential CRISPR/Cas9 cleavage sites, and the CRISPR design tool (http://crispr.mit.edu/) was used to design single-guide RNA (sgRNA). The five sgRNAs with high predicted score were synthesized and subcloned into the pX330 vector (Addgene) containing the flanking sgRNA sequences and a codon-optimized Cas9.
For selecting the sgRNA leading to optimal Cas9 cleavage, sgRNA plasmid which also expresses mCherry was transfected in human neuroblastoma cell line SH-SY5Y (ATCC). After 48 h, transfected cells were dissociated with trypsin and sorted using flow cytometry to identify mCherry-positive cells. T7 endonuclease I digestion patterns of PCR-amplified genomic DNA from these cells were analyzed to determine the optimal sgRNA that can lead to highest cleavage efficiency. Donor sequences containing GFP and 20 bp flanking G3BP1 sequences were cloned to donor vector (pX85, Addgene). To facilitate efficient homologous recombination, the selected sgRNA target sequences were fused to the G3BP1 homologous arms as previously described78 At 24 h after transfection of donor and sgRNA plasmids (1:1), the SH-SY5Y cells were selected for mCherry-positive cells using flow cytometry. After 1–2 weeks, GFP-positive cells were sorted and selected for expansion and functional validation using immunocytochemistry.
Fluorescence recovery after photobleaching
GFP-G3BP1 KI cells were plated in 25 mm glass dish for 24 h. Cells were then transfected with plasmids or siRNAs. After 48 or 72 h of incubation, cells were treated with sodium arsenite for 30 min to induce stress granules. The stress granules in siRNA-positive (CY3) or mCherry-positive cells were photobleached and GFP intensity was measured before and after bleaching. For FRAP experiments in MEF cells, WT and Nr1d1 KO or BMAL1 KO MEF cells were transfected with GFP-G3BP1 plasmids. After 48 or 72 h of incubation, cells were treated with sodium arsenite to induce stress granules. Images were taken with an inverted laser scanning confocal microscope (Nikon A1R) with 60× TIRF oil immersion lens equipped with a humidified 5% CO2 incubator at 37 °C with an environment-controlled chamber.
Immunofluorescence and imaging
SH-SY5Y cells were cultured on glass coverslips. Cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.25% Triton X-100 for 10 min, blocked with 3% bovine serum albumin/phosphate-buffered saline (PBS) (Blocking buffer) for 30 min. The cells were then incubated with specific primary antibodies for 12 h at 4 °C, followed by incubation with appropriate Alexa Fluor-conjugated secondary antibodies (Donkey anti-Mouse/Rabbit/Goat-488/546/647, Invitrogen; 1:500) for 2 h at room temperature and mounted with a drop of Vectashield mounting solution (94010, Vector Laboratories, Inc.).
Mouse liver was perfused and fixed with 4% paraformaldehyde for 24 h and then transferred to 30% sucrose for 48 h. At least 3 frozen sections (8 μm) of each tissue were mounted on glass slide. After permeabilization with 0.25% Triton X-100/PBS for 1h, slides were soaked into 95 °C antigen retrieval solution (10 mM sodium citrate, 0.05% Tween 20, pH = 6.0) for 20 min and then transferred into water for 10 min at room temperature, followed by blocking with Blocking buffer for 30 min. To detect the formation of stress granules in all cell types in the liver tissues, frozen sections were stained with anti-PABP1 or YB1 antibody, and fluorescence-conjugated secondary antibodies. We chose >0.5 μm cut-off as a more stringent standard to identify large stress granules. Stress granule diameter was measured using Fiji software. Images were taken with an inverted laser scanning confocal microscope (Nikon A1R) with 60× TIRF oil immersion lens.
Live-cell imaging and hazard analysis
Live-cell imaging experiments were performed on Olympus FV10I use 60× water objective lens. GFP-G3BP1 KI cells were plated in 29 mm dish with glass bottom. Before imaging, the medium was changed to FluoroBrite medium containing sodium arsenite. During imaging, cells were maintained in a humidified 5% CO2 incubator at 37 °C with an environment-controlled chamber. Multi-positions and Z-stacking were used during the imaging. During imaging, 488 and 546 nm laser was used with laser intensity set below 10% to avoid phototoxicity. Time-lapse images were collected at 6 min interval for 400–600 min and the images were analyzed with Fiji software.
Statistical analyses
For data analysis, the results were presented as mean ± S.E.M., with statistical significance analyzed using Student’s t-test in all the experiments. Two-way analysis of variance (ANOVA) was used for analyzing stress granule formation with time in mouse liver, live-cell imaging, eif2α, and phosphorylated eIF2α level in sodium arsenite-treated SH-SY-5Y cells (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; GraphPad, Prism 5).
Supplementary information
Acknowledgements
This work is supported by the National Natural Science Foundation of China grant (81771425) to J.X.
Authors' contributions
R.W. designed and performed most of the experiments; X.J., P.B. and M.Q assisted in various experiments; J.X. conceived the project, designed experiments, analyzed results, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G. Raschellà
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1471-y).
References
- 1.Dues DJ, et al. Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways. Aging. 2016;8:777–795. doi: 10.18632/aging.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid. Redox Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 5.Hetz C, Saxena S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017;13:477–491. doi: 10.1038/nrneurol.2017.99. [DOI] [PubMed] [Google Scholar]
- 6.Morris G, et al. The endoplasmic reticulum stress response in neuroprogressive diseases: emerging pathophysiological role and translational implications. Mol. Neurobiol. 2018;55:8765–8787. doi: 10.1007/s12035-018-1028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maziuk B, Ballance HI, Wolozin B. Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front. Mol. Neurosci. 2017;10:89. doi: 10.3389/fnmol.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ash PE, Vanderweyde TE, Youmans KL, Apicco DJ, Wolozin B. Pathological stress granules in Alzheimer’s disease. Brain Res. 2014;1584:52–58. doi: 10.1016/j.brainres.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggieri A, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:VSTROE>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain S, et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ticozzi N, et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156B:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- 16.Couthouis J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor BS, et al. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenzie IR, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–816 e809. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 20.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairns NJ, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am. J. Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JO, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 25.Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- 26.Huynh DP, Figueroa K, Hoang N, Pulst SM. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- 27.Delattre O, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee KH, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788 e717. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K, et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173:958–971 e917. doi: 10.1016/j.cell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi JS. Molecular components of the circadian clock in mammals. Diabetes Obes. Metab. 2015;17:6–11. doi: 10.1111/dom.12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shearman LP, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 36.Wulff K, Gatti S, Wettstein JG, Foster RG. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 2010;11:589–599. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- 37.Musiek ES, Holtzman DM. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science. 2016;354:1004–1008. doi: 10.1126/science.aah4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorpy MJ, Adler CH. Parkinson’s disease and sleep. Neurol. Clin. 2005;23:1187–1208. doi: 10.1016/j.ncl.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Harper DG, et al. Disturbance of endogenous circadian rhythm in aging and Alzheimer disease. Am. J. Geriatr. Psychiatry. 2005;13:359–368. doi: 10.1097/00019442-200505000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Sethi M, et al. Increased fragmentation of sleep-wake cycles in the 5XFAD mouse model of Alzheimer’s disease. Neuroscience. 2015;290:80–89. doi: 10.1016/j.neuroscience.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kudo T, Loh DH, Truong D, Wu Y, Colwell CS. Circadian dysfunction in a mouse model of Parkinson’s disease. Exp. Neurol. 2011;232:66–75. doi: 10.1016/j.expneurol.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Zhang T, et al. Sleep and circadian abnormalities precede cognitive deficits in R521C FUS knockin rats. Neurobiol. Aging. 2018;72:159–170. doi: 10.1016/j.neurobiolaging.2018.08.025. [DOI] [PubMed] [Google Scholar]
- 43.Sancar A, et al. Circadian clock, cancer, and chemotherapy. Biochemistry. 2015;54:110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl. Acad. Sci. USA. 2009;106:2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu. Rev. Pharmacol. Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 46.Wilking M, Ndiaye M, Mukhtar H, Ahmad N. Circadian rhythm connections to oxidative stress: implications for human health. Antioxid. Redox Signal. 2013;19:192–208. doi: 10.1089/ars.2012.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putker M, O’Neill JS. Reciprocal control of the circadian clock and cellular redox state - a critical appraisal. Mol. Cells. 2016;39:6–19. doi: 10.14348/molcells.2016.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tourriere H, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Sun Y, et al. MOP3, a component of the molecular clock, regulates the development of B cells. Immunology. 2006;119:451–460. doi: 10.1111/j.1365-2567.2006.02456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, et al. A proteomics landscape of circadian clock in mouse liver. Nat. Commun. 2018;9:1553. doi: 10.1038/s41467-018-03898-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauvoisin D, et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang G, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 2016;8:324ra316. doi: 10.1126/scitranslmed.aad3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mure LS, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018;359:pii: eaao0318. doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 57.Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. eLife. 2016;5:pii: e18413. doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diaz-Munoz M, Hernandez-Munoz R, Suarez J, Chagoya de Sanchez V. Day-night cycle of lipid peroxidation in rat cerebral cortex and their relationship to the glutathione cycle and superoxide dismutase activity. Neuroscience. 1985;16:859–863. doi: 10.1016/0306-4522(85)90100-9. [DOI] [PubMed] [Google Scholar]
- 59.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang T, et al. Pathogenic mutations in the valosin-containing protein/p97(VCP) N-domain inhibit the SUMOylation of VCP and lead to impaired stress response. J. Biol. Chem. 2016;291:14373–14384. doi: 10.1074/jbc.M116.729343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 62.Jacobi D, et al. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 2015;22:709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Musiek ES, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J. Clin. Invest. 2013;123:5389–5400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bu Y, et al. A PERK-miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat. Cell Biol. 2018;20:104–115. doi: 10.1038/s41556-017-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim E, Giese A, Deppert W. Wild-type p53 in cancer cells: when a guardian turns into a blackguard. Biochem. Pharmacol. 2009;77:11–20. doi: 10.1016/j.bcp.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 69.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baron DM, et al. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol. Neurodegener. 2013;8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Musiek ES, Xiong DD, Holtzman DM. Sleep, circadian rhythms, and the pathogenesis of Alzheimer disease. Exp. Mol. Med. 2015;47:e148. doi: 10.1038/emm.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou JN, Hofman MA, Swaab DF. VIP neurons in the human SCN in relation to sex, age, and Alzheimer’s disease. Neurobiol. Aging. 1995;16:571–576. doi: 10.1016/0197-4580(95)00043-E. [DOI] [PubMed] [Google Scholar]
- 73.Jiang X, et al. Neurodegeneration-associated FUS is a novel regulator of circadian gene expression. Transl. Neurodegener. 2018;7:24. doi: 10.1186/s40035-018-0131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ozturk N, Ozturk D, Kavakli IH, Okyar A. Molecular aspects of circadian pharmacology and relevance for cancer chronotherapy. Int. J. Mol. Sci. 2017;18:pii: E2168. doi: 10.3390/ijms18102168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grabocka E, Bar-Sagi D. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell. 2016;167:1803–1813 e1812. doi: 10.1016/j.cell.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Timalsina S, et al. Chemical compounds that suppress hypoxia-induced stress granule formation enhance cancer drug sensitivity of human cervical cancer HeLa cells. J. Biochem. 2018;164:381–391. doi: 10.1093/jb/mvy062. [DOI] [PubMed] [Google Scholar]
- 77.Dakup PP, et al. The circadian clock regulates cisplatin-induced toxicity and tumor regression in melanoma mouse and human models. Oncotarget. 2018;9:14524–14538. doi: 10.18632/oncotarget.24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao X, et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017;27:801–814. doi: 10.1038/cr.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.