Abstract
Complete mitochondrial genomes contain large and diverse datasets for species delineation. To better understand the divergence of the two morphologically indistinguishable weevil species in Curculionini, we first sequenced and compared their complete mitochondrial genomes. The complete mitochondrial genomes of Curculio chinensis and Curculio sp. were 19,713 bp with an A + T content of 76.61% and 19,216 bp with an A + T content of 76.85%, respectively. All 37 of the typical mitochondrial genes were determined in both species. The 13 protein sequences of the two species shared high homology (about 90%) except for ATP8 (73.08%). The differences in secondary structure of ATP8 were the number of possible proteins and nucleic acid binding sites. There were 22 and 15 mismatched base-pairs in the tRNA secondary structures from C. chinensis and Curculio sp., respectively. Maximum Likelihood and Bayesian analyses indicated that Curculio sp. is a novel species closely related to C. chinensis. The divergence time estimation suggests that Cryptorhynchinae and Curculionini lines diverged in the Cenozoic Period, while C. chinensis and Curculio sp. diverged at 6.7079 (95% CI 5–13) Mya. This study demonstrates the utility of using complete mitochondrial gene sets for phylogenetic analysis and enhances our understanding of the genetic basis for the evolution of the Curculionini.
Introduction
The camellia weevil is a host-specific predator of the seeds of Camellia oleifera Abel. and Camellia sinensis (L.) O. Ktze. (Theaceae) which have been widely cultivated as important economic trees in China, and it is often identified as Curculio chinensis Chevrolat1–4. The species exhibits low levels of regional diversity and low degrees of morphological differentiation. However, a few geographic populations are phylogenetically distinct, and show significant variation related to Camellia host isolation (C. oleifera and C. sinensis)4. Camellia weevils in species complexes are often difficult or impossible to identify using morphological characteristics of the larvae. It is impractical to identify camellia weevils by rearing larvae to adults because the larvae are long-lived and difficult to rear when removed from the seed1,2. Molecular identification has proven to be reliable and more effective5.
Mitochondria exist in plant and animal cells and are inherited maternally. They are associated with metabolism, life cycle, and apoptosis and are abundant in energy consuming tissues related to essential biological functions6,7. Genetic data from mitochondria are commonly used to study evolutionary relationships. Useful information for evolution studies continues to be found in mitochondrial gene markers8–11. Relatively high mutation rates and low recombination frequencies make mitochondrial genomes especially useful for evolutionary studies12–15. However, shorter mitochondrial gene sequences often do not provide adequate phylogenetic resolution16–18. For example, our previous phylogenetic analysis based on the cytochrome oxidase subunit I (COX1) gene revealed the presence of a novel Curculio sp. in China feeding on Camellia seed. Curculio sp. is closely related to C. chinensis. However, the information provided by COX1 alone did not provide sufficient support for naming the new species4. Fortunately, complete mitochondrial genomes have become available for the identification of animal species19. Also, mitochondrial genomes have been used for phylogenetic analyses in many species, and especially in recent DNA-barcoding studies15,20,21.
High-throughput sequencing methodologies such as whole genome sequencing (WGS) and next generation sequencing (NGS) provide complete mitochondrial genomes for phylogenetic analyses and the replacement of earlier markers such as COX1, COX2, and NAD genes. However, the ability to resolve closely-related lineages with mitochondrial genomes remains controversial7,13,15,22,23. Generally, a complete mitochondrial insect genome is a circular-DNA molecule comprising 15–21 kbp of DNA. It features a relatively conserved gene content including 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes, in addition to an A + T-rich region6,15,18,23,24. Insect mitochondrial genome gene sequences, and particularly protein coding genes, are relatively conserved8,9. The 13 PCGs of insect mitochondria are useful phylogenetic markers25,26 and have been used for phylogenetic analyses of species in the Diptera, Coleoptera and Lepidoptera7,15,22,24,27. In addition, insect mitochondrial tRNAs exhibit rearrangement phenomena28, while other genes rarely exhibit such rearrangements29. The transposition genotypes of tRNAs, in addition to the combinations of six tRNAs between the mitochondrial ND3 and ND5 genes provide additional informative characters23,30. Mitochondrial genomes are therefore excellent molecular markers for studies of species delineation, population genetics and evolution.
The increasing availability of mitochondrial genomes, combined with next-generation sequencing technologies31–33, have enabled detailed comparative phylogenetic analyses of many species complexes34,35 and the major taxonomic groups of several insect orders have undergone diagnostic revisions5,22,26,28,36. In this study, we obtained the complete mitochondrial genomes from C. chinensis and a novel Curculio sp., and compared them with the mitochondrial genomes of other Curculionid species. These analyses provided insight into their genome evolution as well as the phylogeny of the Curculionidae.
Results and Discussion
Features of the sequenced mitochondrial genomes
Coleoptera mitochondrial genomes have relatively simple structures and lack spacers and introns. With this tight gene arrangement, genetic rearrangement, inversions, and translocations occur infrequently during the mutation process6,37. The complete mitochondrial genome sequences of C. chinensis and the Curculio sp. were 19,713 bp and 19,216 long, respectively (GenBank accessions MG728094 and MG728095) (Table S1). The genomes of both species contained all 37 typical animal mitochondrial genes, including 13 protein-coding genes, 22 tRNA genes, and two rRNA genes (Fig. S1). The A + T-rich region generated reliable sequence data in both species which was high (>75%) compared to that found in other mitochondrial genomes sequenced using NGS15. No gene rearrangement was observed in either species compared with the putative ancestral and sibling superfamily arrangement (Fig. S1). This is consistent with the lack of rearrangement found in all sequenced Curculionini species8,11.
The mitochondrial genome of C. chinensis has intergenic spacers with lengths ranging from 1 to 103 bp in 24 different locations. Seven pairs of genes overlap with each other, with overlap lengths ranging from 1 to 17 bp. Eight pairs of genes are directly adjacent to one another, which include the pairs rrnL-trnV and trnV-rrnS (Table 1). The mitochondrial genome of Curculio sp. has intergenic spacer lengths ranging from 1 to 148 bp in 26 different locations. Eight pairs of genes overlap with each other, with overlap lengths ranging from 1 to 7 bp. Nine pairs of genes were directly adjacent one another, which included the pairs rrnL-trnV and trnV-rrnS (Table 2). In both species, the longest intergenic spacer was located between trnS2 and NAD1. The longest overlapping regions were located between trnF and NAD5 (Tables 1 and 2). The intergenic and overlapping regions of these two species were similar to the mitochondrial genomes of most other insects. Similarly, no gene rearrangement was found in either species compared to genomes from Coleoptera species that have experienced frequent gene rearrangement23,28.
Table 1.
Annotation of the Curculio chinensis mitochondrial genome.
| Gene | Strand | Position | Length (bp) | Initiation codon | Stop codon | Anticodon | Intergenic nucleotide (bp) |
|---|---|---|---|---|---|---|---|
| trnI | N | 1–65 | 65 | GAT | 3,018 | ||
| trnQ | J | 3,084–3,152 | 69 | TTG | 1 | ||
| trnM | N | 3,154–3,221 | 68 | CAT | |||
| NAD2 | N | 3,222–4,232 | 1,011 | ATT | TAA | 14 | |
| trnW | N | 4,247–4,310 | 64 | TCA | −1 | ||
| trnC | J | 4,310–4,375 | 66 | GCA | 2 | ||
| trnY | J | 4,378–4,441 | 64 | GTA | −8 | ||
| COX1 | N | 4,434–5,978 | 1,545 | ATT | TAA | −5 | |
| trnL2 | N | 5,974–6,038 | 65 | TAA | |||
| COX2 | N | 6,039–6,722 | 684 | ATT | TAA | 1 | |
| trnK | N | 6,724–6,794 | 71 | CTT | 2 | ||
| trnD | N | 6,797–6,861 | 65 | GTC | |||
| ATP8 | N | 6,862–7,020 | 159 | ATT | TAA | −7 | |
| ATP6 | N | 7,014–7,688 | 675 | ATG | TAA | 4 | |
| COX3 | N | 7,693–8,480 | 788 | ATG | TA (A) | −1 | |
| trnG | N | 8,480–8,543 | 64 | TCC | |||
| NAD3 | N | 8,544–8,897 | 354 | ATT | TAG | −2 | |
| trnA | N | 8,896–8,962 | 67 | TGC | |||
| trnR | N | 8,963–9,024 | 62 | TCG | −2 | ||
| trnN | N | 9,023–9,086 | 64 | GTT | |||
| trnS1 | N | 9,087–9,153 | 67 | TCT | 7 | ||
| trnE | N | 9,161–9,224 | 64 | TTC | |||
| trnF | J | 9,225–9,289 | 65 | GAA | −17 | ||
| NAD5 | J | 9,273–11,000 | 1,728 | ATT | TAA | 9 | |
| trnH | J | 11,010–11,072 | 63 | GTG | −3 | ||
| NAD4 | J | 11,070–12,408 | 1,339 | ATG | T (AA) | −7 | |
| NAD4l | J | 12,402–12,695 | 294 | ATG | TAA | 2 | |
| trnT | N | 12,698–12,762 | 65 | TGT | |||
| trnP | J | 12,763–12,828 | 66 | TGG | 2 | ||
| NAD6 | N | 12,831–13,334 | 504 | ATT | TAA | ||
| COB | N | 13,335–14,474 | 1,140 | ATG | TAA | ||
| trnS2 | N | 14,475–14,541 | 67 | TGA | 103 | ||
| NAD1 | J | 14,645–15,571 | 927 | ATA | TAG | 25 | |
| trnL1 | J | 15,597–15,661 | 65 | TAG | 9 | ||
| rrnL | J | 15,671–16,944 | 1,274 | 22 | |||
| trnV | J | 16,967–17,032 | 66 | TAC | |||
| rrnS | J | 17,033–17,821 | 789 | 1,891 |
Table 2.
Annotation of the Curculio sp. mitochondrial genome.
| Feature | Strand | Position | Length (bp) | Initiation codon | Stop codon | Anticodon | Intergenic nucleotide (bp) |
|---|---|---|---|---|---|---|---|
| trnI | N | 1–65 | 65 | GAT | 2,007 | ||
| trnQ | J | 2,073–2,141 | 69 | TTG | 2 | ||
| trnM | N | 2,144–2,211 | 68 | CAT | |||
| NAD2 | N | 2,212–3,222 | 1,011 | ATG | TAA | 15 | |
| trnW | N | 3,238–3,301 | 64 | TCA | −1 | ||
| trnC | J | 3,301–3,366 | 66 | GCA | 2 | ||
| trnY | J | 3,369–3,432 | 64 | GTA | −8 | ||
| COX1 | N | 3,425–4,969 | 1,545 | ATT | TAA | −5 | |
| trnL2 | N | 4,965–5,029 | 65 | TAA | 21 | ||
| COX2 | N | 5,051–5,713 | 663 | ATG | TAA | 1 | |
| trnK | N | 5,715–5,785 | 71 | CTT | 2 | ||
| trnD | N | 5,788–5,851 | 64 | GTC | |||
| ATP8 | N | 5,852–6,010 | 159 | ATT | TAA | −7 | |
| ATP6 | N | 6,004–6,678 | 675 | ATG | TAA | 3 | |
| COX3 | N | 6,682–7,469 | 788 | ATG | TA (A) | −1 | |
| trnG | N | 7,469–7,536 | 68 | TCC | 6 | ||
| NAD3 | N | 7,543–7,890 | 348 | ATT | TAG | −2 | |
| trnA | N | 7,889–7,956 | 68 | TGC | |||
| trnR | N | 7,957–8,019 | 63 | TCG | −2 | ||
| trnN | N | 8,018–8,080 | 63 | GTT | |||
| trnS1 | N | 8,081–8,147 | 67 | TCT | 6 | ||
| trnE | N | 8,154–8,217 | 64 | TTC | |||
| trnF | J | 8,218–8,282 | 65 | GAA | −17 | ||
| NAD5 | J | 8,266–9,993 | 1,728 | ATT | TAA | 9 | |
| trnH | J | 10,003–10,065 | 63 | GTG | −3 | ||
| NAD4 | J | 10,063–11,401 | 1,339 | ATG | T (AA) | −7 | |
| NAD4l | J | 11,395–11,688 | 294 | ATG | TAA | 2 | |
| trnT | N | 11,691–11,755 | 65 | TGT | |||
| trnP | J | 11,756–11,820 | 65 | TGG | 2 | ||
| NAD6 | N | 11,823–12,326 | 504 | ATT | TAA | ||
| COB | N | 12,327–13,466 | 1,140 | ATG | TAA | ||
| trnS2 | N | 13,467–13,533 | 67 | TGA | 148 | ||
| NAD1 | J | 13,682–14,608 | 927 | ATA | TAG | 25 | |
| trnL1 | J | 14,634–14,698 | 65 | TAG | 5 | ||
| rrnL | J | 14,704–15,977 | 1,274 | 29 | |||
| trnV | J | 16,007–16,071 | 65 | TAC | |||
| rrnS | J | 16,072–16,856 | 785 | 2,359 |
Base composition
AT-skew, GC-skew, A + T content, and AT and GC asymmetries, are often used to assess the nucleotide-compositional differences of mitochondrial genomes38. The mitochondrial genomes of C. chinensis and the Curculio sp. were biased in nucleotide composition ((A + T)% > (G + C)%) across the whole genome, although the numbers of PCGs (n = 13) and rRNA genes (n = 22) were consistent with the genomes from other insects5,24. The A + T content of the whole genome was 76.61% for C. chinensis (40.16% A, 36.45% T, 9.89% G and 13.50% C) (Tables 3), and 77.08% for Curculio sp. (40.84% A, 36.24% T, 9.14% G and 13.79% C) (Table 4). The A + T content of all PCGs in C. chinensis ranged from 69.58% (COX1) to 83.38% (NAD6) (Table 3), and in Curculio sp. ranged from 69.84% (COX1) to 84.33% (NAD6) (Table 4). Most of the AT-skews of the two Curculio species were negative except COX2 and ATP8. Most of the GC-skews of both species were negative as well, indicating that the PCGs contained a higher percentage of T and C than A and G, as reported for most other insects22,38.
Table 3.
Base composition of the mitochondrial genome of Curculio chinensis.
| Region | A% | C% | G% | T% | A + T% | G + C% | AT skew | GC skew |
|---|---|---|---|---|---|---|---|---|
| Whole genome | 40.16 | 13.5 | 9.89 | 36.45 | 76.61 | 23.39 | 0.048 | −0.154 |
| NAD2 | 35.41 | 14.54 | 8.61 | 41.44 | 76.85 | 23.15 | −0.079 | −0.256 |
| COX1 | 32.62 | 16.83 | 13.59 | 36.96 | 69.58 | 30.42 | −0.062 | −0.106 |
| COX2 | 37.28 | 16.52 | 10.23 | 35.96 | 73.25 | 26.75 | 0.018 | −0.235 |
| ATP8 | 42.14 | 13.21 | 4.4 | 40.25 | 82.39 | 17.61 | 0.023 | −0.5 |
| ATP6 | 36.3 | 16.59 | 8.89 | 38.22 | 74.52 | 25.48 | −0.026 | −0.302 |
| COX3 | 36.42 | 16.62 | 11.42 | 35.53 | 71.95 | 28.05 | 0.012 | −0.186 |
| NAD3 | 36.72 | 12.43 | 8.19 | 42.66 | 79.38 | 20.62 | −0.075 | −0.205 |
| NAD5 | 29.75 | 8.1 | 13.08 | 49.07 | 78.82 | 21.18 | −0.245 | 0.235 |
| NAD4 | 28.75 | 8.14 | 12.47 | 50.63 | 79.39 | 20.61 | −0.276 | 0.21 |
| NAD4l | 29.25 | 5.1 | 13.95 | 51.7 | 80.95 | 19.05 | −0.277 | 0.464 |
| NAD6 | 39.09 | 8.53 | 6.75 | 45.63 | 84.72 | 15.28 | −0.077 | −0.117 |
| COB | 35.44 | 15.61 | 10.44 | 38.51 | 73.95 | 26.05 | −0.042 | −0.199 |
| NAD1 | 27.4 | 8.63 | 15.1 | 48.87 | 76.27 | 23.73 | −0.281 | 0.273 |
| rrnL | 36.81 | 6.36 | 13.34 | 43.49 | 80.3 | 19.7 | −0.083 | 0.355 |
| rrnS | 37.01 | 8.11 | 15.84 | 39.04 | 76.05 | 23.95 | −0.027 | 0.323 |
Table 4.
Base composition of the mitochondrial genome of Curculio sp.
| Region | A% | C% | G% | T% | A + T% | G + C% | AT skew | GC skew |
|---|---|---|---|---|---|---|---|---|
| Whole genome | 40.84 | 13.79 | 9.14 | 36.24 | 77.08 | 22.92 | 0.06 | −0.203 |
| NAD2 | 36.99 | 13.85 | 7.72 | 41.44 | 78.44 | 21.56 | −0.057 | −0.284 |
| COX1 | 32.04 | 16.31 | 13.85 | 37.8 | 69.84 | 30.16 | −0.082 | −0.082 |
| COX2 | 37.86 | 17.5 | 11.01 | 33.63 | 71.49 | 28.51 | 0.059 | −0.228 |
| ATP8 | 44.03 | 12.58 | 3.14 | 40.25 | 84.28 | 15.72 | 0.045 | −0.6 |
| ATP6 | 36.15 | 15.11 | 8.89 | 39.85 | 76 | 24 | −0.049 | −0.259 |
| COX3 | 36.17 | 15.61 | 11.55 | 36.68 | 72.84 | 27.16 | −0.007 | −0.15 |
| NAD3 | 34.48 | 15.23 | 8.05 | 42.24 | 76.72 | 23.28 | −0.101 | −0.309 |
| NAD5 | 29.11 | 8.97 | 13.43 | 48.5 | 77.6 | 22.4 | −0.25 | 0.199 |
| NAD4 | 29.05 | 9.19 | 12.32 | 49.44 | 78.49 | 21.51 | −0.26 | 0.146 |
| NAD4l | 29.25 | 5.78 | 13.27 | 51.7 | 80.95 | 19.05 | −0.277 | 0.393 |
| NAD6 | 39.88 | 8.53 | 7.14 | 44.44 | 84.33 | 15.67 | −0.054 | −0.089 |
| COB | 34.21 | 16.05 | 10.7 | 39.04 | 73.25 | 26.75 | −0.066 | −0.2 |
| NAD1 | 26.11 | 9.06 | 16.18 | 48.65 | 74.76 | 25.24 | −0.302 | 0.282 |
| rrnL | 35.79 | 6.36 | 13.81 | 44.03 | 79.83 | 20.17 | −0.103 | 0.37 |
| rrnS | 35.92 | 8.15 | 15.8 | 40.13 | 76.05 | 23.95 | −0.055 | 0.319 |
Protein-coding genes, codon usage, and protein conformance rates
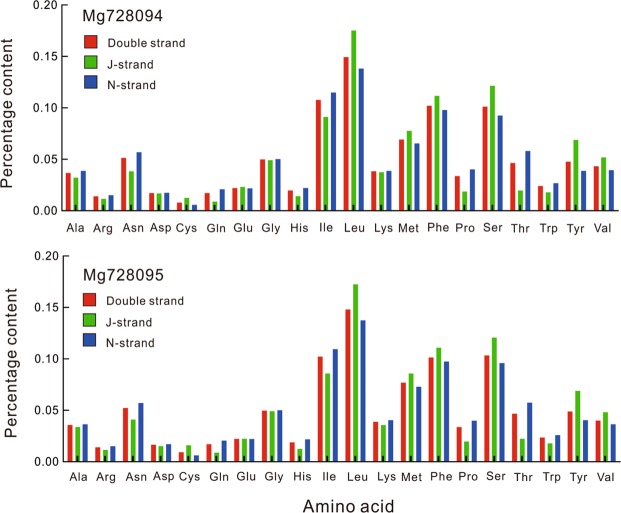
In the mitochondrial genomes of both C. chinensis and Curculio sp., nine of the 13 protein-coding genes were located on the majority strand (N-strand), while the other four protein-coding genes were located on the minority strand (J-strand) (Tables 1 and 2). In the C. chinensis mitochondrial genome, the total length of protein-coding genes was 11,148 bp, accounting for 56.55% of the whole genome. The total length of the protein-coding genes of Curculio sp. was 11,121 bp, accounting for 57.87% of the whole genome (Tables 1 and 2). PCGs contained Leu residues in the highest abundance, followed by Ile, Phe and Met. The four amino acids had the highest use frequency (Fig. 1), similar to other insect mitochondrial genomes36,39.
Figure 1.
Percentage of each amino acid for proteins coded by PCGs in the two newly mitochondrial genomes of Curculio chinensis (MG728094) and Curculio sp. (MG728095).
In the mitochondrial genomes of both species, all PCGs start with the conventional initiation codons (ATN) as seen in other insects40. In C. chinensis PCGs, only one gene (NAD1) used ATA, seven used ATT, and five used ATG. In contrast, in the PCGs of Curculio sp., only one gene (NAD1) used ATA, while five and seven PCGs started with ATT and ATG, respectively (Tables 1 and 2). In both the C. chinensis and Curculio sp. mitochondrial genome, nine PCGS used TAA as the stop codon, and the NAD1 and NAD3 genes used TAG, while the COX3 and NAD4 genes used an incomplete stop codon T (Tables 1 and 2). The usage of incomplete stop codons in PCGs is common in invertebrate mitochondrial genomes5,41.
We calculated the homologous consistency of the 13 protein sequences of the two species as one group. Except for the ATP8 sequences that exhibited a value of 73.08%, the rest of the sequences had values of about 90% (Fig. 2A). Ratios of Ka/Ks values for each PCG in the two species showed that ATP8 had the largest ratio (0.3194) among all proteins (Fig. 2A). Two genes, ATP6 and ATP8 are the core subunits of Complex V, which consists of F0 and F1, and the two genes are directly involved in ATP synthesis42–44.
Figure 2.
Protein conformance of each protein coding gene (PCG) in the mitochondrial genomes of Curculio chinensis and Curculio sp. The Ka/Ks values of each PCG represented the ratios of non-synonymous substitutions (Ka) to synonymous substitutions (Ks) (A). ATP8 protein structure prediction (B). a: Protein sequence; b: Protein-protein binding; c: Secondary structure; d: Solvent accessibility; e: Transmembrane helix; f: Disordered region. (C) The predicted cellular compartment, mitochondrial membrane is highlighted in green in a schematic of a eukaryotic cell.
We further characterized the ATP8 proteins from both genomes and predicted their structures (Fig. 2B), since this was the most variable gene of the 13 PCGs. The ATP8 protein sequences in both species contained 52 amino acids. The C. chinensis ATP8 protein structure contained three possible protein binding sites and five possible nucleic acid binding sites (Fig. 2B). Across sites 9–29, there was a region that might produce a spiral structure (Fig. 2B). In the entire chain of the C. chinensis ATP8 protein, there were three disordered areas, two exposed regions, two buried regions, and one transmembrane helix region (Fig. 2B). The Curculio sp. ATP8 protein structure has four possible protein binding sites and four possible nucleic acid binding sites (Fig. 2B). Like the C. chinensis ATP8 protein structure, there was also a region that might produce a spiral structure across the sites comprising 9–29 (Fig. 2B). In the whole chain of the Curculio sp. ATP8 protein, there were two disordered areas, two exposed regions, three buried regions, and one transmembrane helix region (Fig. 2B). The SOPMA analysis of the ATP8 secondary structure revealed clear structural differentiation between the two species. The alpha helix represented 38.46% and 59.62% of the structures of C. chinensis and Curculio sp., respectively, while the extended strand regions were 13.08% and 11.54%, the beta turn regions were 9.62% and 1.92%, and the random coil accounted for 28.85% and 26.92%, respectively. Adaptive evolution of ATP synthase can occur among species living in different ecological niches39,44,45. Thus, we speculated that modifications in the sequence and conformation of ATP8 structures could affect the assembly and function of Complex V, and consequently modulate its ability to produce ATP in Curculio weevils.
Phylogenetic relationships and comparison of divergence times
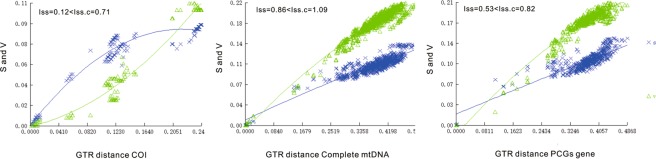
In previous studies, mitochondrial sequence length variation was low, resulting in minimal alignment ambiguity that was not investigated further23. We calculated saturation plots for COX1, complete mtDNA genomes, and the PCGs before we used these to build a phylogenetic tree. The plots showed uncorrected pairwise divergences in transitions (s) and transversions (v) against divergences calculated with the GTR model, and none of the three genes had reached saturation (Fig. 3).
Figure 3.
Saturation plots for (from left to right) COX1, the complete mtDNA genomes and PCGs. The plot showed uncorrected pairwise divergences in transitions (s) and transversions (v) against divergences calculated using the GTR model. Blue: transitions; Green: transversions.
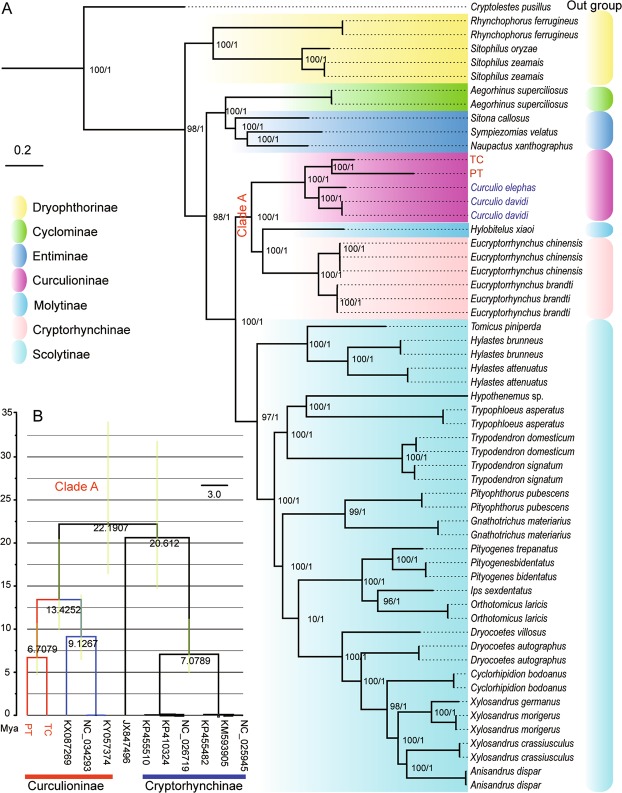
Maximum Likelihood and Bayesian phylogenetic trees were constructed based on the protein sequences of the 13 PCGs from the mitochondrial genomes of 57 Curculionid species (Fig. 4A). The results supported substantially our previous hypothesis that Curculio sp. is an undescribed species which was closely related to C. chinensis4. Our study showed that the Curculioninae fall into the diverse “CMC” clade (Curculioninae + Molytinae + Cryptorhynchinae), which is consistent with previous results32,46–50. Additionally, the genus Curculio is a typical member of Curculionini in Curculioninae, which showed close affinity with the subfamilies Molytinae and Cryptorhynchinae. Family-level studies have been used to estimate phylogenetic divergence times for Coleoptera using molecular data12,25,48,50. The studies suggest that the last common ancestor of Coleoptera occurred in the Permian period (253–297 Mya). The Cucujiformia species first occurred in the Triassic period (200–250 Mya). However, the Curculionidae might have first appeared in the Cretaceous period (60–150 Mya)12,48,50. Our data suggest that the Cryptorhynchinae + Molytinae and Curculioninae diverged at 22.1907 (95% credibility interval 16–35) Mya in the Cenozoic period (0–60 Mya), while C. chinensis and Curculio sp. diverged at 6.7079 (95% credibility interval 5–13) Mya (Fig. 4B). The divergence time between the two host plants, C. oleifera and C. sinensis was about 5–6 million years ago, which is consistent with the formation time of the earliest camellia fossils found in the tertiary stratum in Japan51. The geographic isolation of Camellia hosts might have played a role in the differentiation of camellia weevils.
Figure 4.
Maximum Likelihood and Bayesian phylogenetic tree based on protein sequences of 13 PCGs from the mitochondrial genomes of 57 species (A). Bootstrap supports of >90% for ML (upper) and posterior probabilities of >90% for BI (lower) were indicated around branches. PT: Curculio chinensis (MG728094) and TC: Curculio sp. (MG728095); The colors represent different subfamilies. Timescale for Clade A evolution and comparison of divergence times based on the 13 PCGs (B). The green horizontal bars represent 95% credibility intervals.
Transfer RNA and ribosomal RNA genes
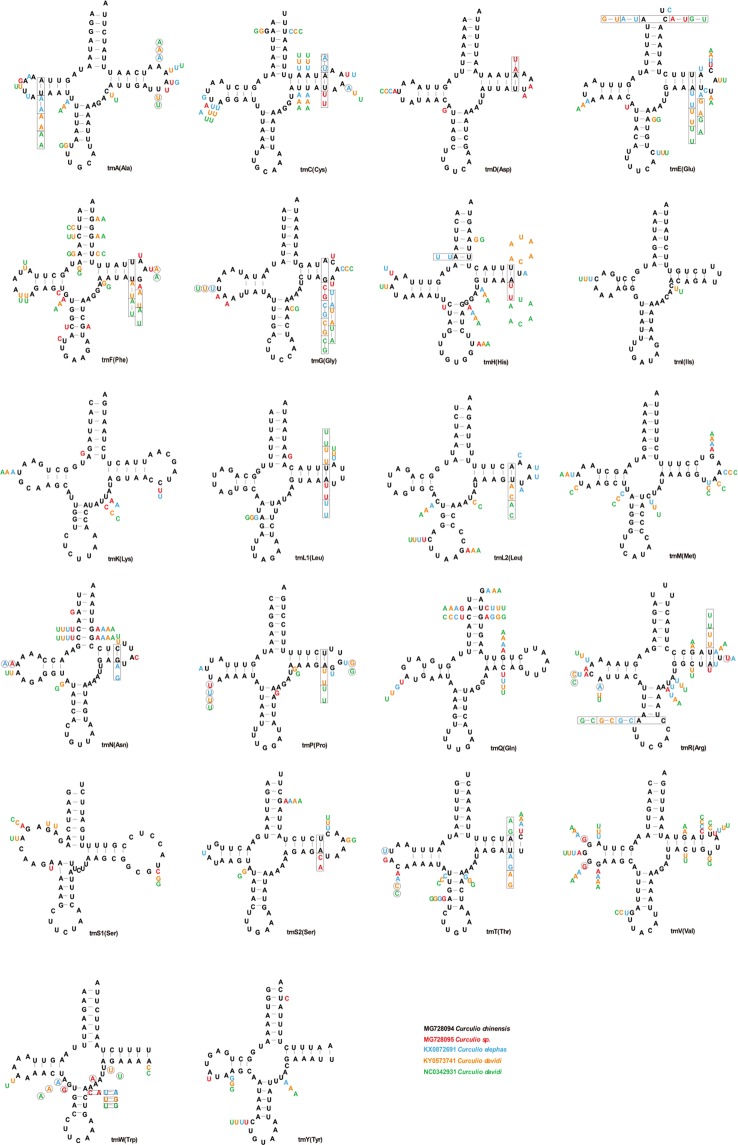
Consistent with the results of the phylogenetic relationships and comparison of estimated divergence times, all tRNA anticodons of the sequenced mitochondrial genomes of C. chinensis and Curculio sp. were identical to other Curculionini species (Tables 1 and 2). Of the 22 total tRNA genes, 14 are located on the N-strand and eight are located on the J-strand. Individual tRNAs of C. chinensis (MG728094) and Curculio sp. (MG728095) ranged from 63 bp (trnH) to 71 bp (trnK) in length. Secondary structure models of the tRNA genes from the two mitochondrial genomes were predicted using the Mitos WebServer (http://mitos.bioinf.uni-leipzig.de/). All tRNA genes from C. chinensis and Curculio sp. mitochondrial genomes fold into a canonical clover-leaf structure (Fig. 5).
Figure 5.
Comparison on the secondary structure of tRNA genes in Curculionini mitochondrial genomes. The secondary structures were drawn from tRNA genes of Curculio chinensis. Variations at each site in other four species of Curculionini were indicated near the corresponding nucleotide. Each species was marked by a unique color as shown in the legend at the bottom right of the figure.
The dihydrouridine (DHU) arm of all the tRNAs was a large loop, instead of the conserved stem-and-loop structure, which is consistent with typical metazoan mitochondrial genomes40. While the amino acid acceptor stem was conserved across 7 bp in all tRNA genes, the anticodon loops exhibited differences. trnH and trnR were conserved across 8 bp, while the rest of the 20 tRNAs were conserved across 7 bp (Fig. 5). The DHU arms in the tRNAs from C. chinensis and Curculio sp. were 0 to 4 bp long. The AC arms were 4 to 5 bp long, and the TΨC arms varied in length from 3 to 5 bp. The variable loops ranged from 4 to 8 bp. We also compared the variation in stem regions of tRNA genes among five other Curculionini species (Fig. 5). Among the 22 tRNA genes, trnI was the most conserved, and lacked nucleotide variation in stem regions between C. chinensis and Curculio sp. The rest of the tRNAs exhibited between 1–10 site mutations. The trnF had the highest number of site mutations on stem regions (10 sites), followed by trnV (7 sites) (Fig. 5). Among the 22 tRNA genes, there was no nucleotide variation in the stem regions between Curculio davidi (KY053741) and Curculio davidi (NC034931) (Fig. 5). Curculio elephas (KY0872691) had the most nucleotide variation in stem regions compared with C. chinensis (Fig. 5). Base pairs other than canonical A-Us and C-Gs were occasionally used in C. chinensis and Curculio sp. tRNAs, which is based on predicted tRNA secondary structures (Fig. 5). We found 22 and 15 mismatched base pairs in the tRNAs from C. chinensis and Curculio sp., respectively. Among the 22 mismatched base pairs in C. chinensis, three were U-U pairs, located in the amino acid acceptor stems and anticodon arm stems. The others were A–C pairs located in the amino acid acceptor stem. Curculio sp. had four U-U pairs t located in the TΨC stems (Fig. 5).
Methods
Sample collection and dna extraction
Camellia weevil samples were collected from Tengchong County in Yunnan Province, China. The field collected samples were initially placed in 100% ethyl alcohol and stored at −80 °C prior to DNA extraction. Total genomic DNA was extracted separately from the whole body of individual samples using a DNeasy tissue kit (Qiagen, Hilden, Germany). Voucher DNA was deposited in the entomological collections of the Research Institute of Subtropical Forestry, Chinese Academy of Forestry.
Mitochondrial genome sequencing and assembly
The mitochondrial genome sequences were obtained by next-generation sequencing. Prior to library construction, the DNA was quantified by Qubit 3.0 (Invitrogen, Life technologies, Carlsbad, CA, USA)5. The library (Lib. Type: PE400; Lib. Insert Size: 400 bp) with two indexes was constructed using the Illumina TruSeq@ DNA PCR-Free HT Kit and sequenced by Shanghai Personal Biotechnology CO., Ltd (Shanghai, China) using Illumina Miseq with the strategy of 251 bp paired-ends by paired sequencing mode. Raw sequence reads were generated on the Illumina Miseq sequencing platform in FASTQ format, and read quality was evaluated using the FastQC software package (http://www.bioinformatics.babraham.ac.uk/projects/fastqc)52. Reads containing ambiguous nucleotides and reads with an average quality value lower than Q30 were excluded from further analyses. The high-quality second-generation sequencing data were assembled de novo to generate contig and scaffold sequences using the A5-miseq v.2015052253 and SPAdes v.3.9.054 assembly pipelines. According to the sequencing depth extraction sequence of the splicing sequence, the high sequencing depth was blastn with the NT library in NCBI (BLAST v2.2.31+) and compared with the mitochondrial sequence of each splicing result. The mitochondrial splicing results were combined using the Mummer v.3.1 software to integrate splicing results. Linear analysis was used to determine the positions between contigs and fill gaps between contigs using the Pilon v.1.18 software package55. The results were then corrected to obtain the final mitochondrial genome sequences.
Mitochondrial genome annotation
The complete set of linear contigs was uploaded to the MITOS web page server (http://mitos2.bioinf.uni-leipzig.de/) for functional annotation56. The optional setting for ‘Genetic Code’ was selected as 05-verterbrate, and the remaining settings were set according to the default parameters. The circular mitogenomes of both samples were visualized using the Organellar Genome Draw web server tool (http://ogdraw.mpimp-golm.mpg.de/)57. The sequin file generated from MITOS was edited and submitted to NCBI according to the ORF Finder results (NCBI GenBank accession number MG728094, MG728095).
Comparative analysis of mitochondrial genomes
The mitochondrial genomes of five Curculionini species, including the two newly sequenced Curculio genomes, were compared. Gene arrangement, base composition, and PCG codon usage features were analyzed. We also analyzed base compositional differences based on the secondary structures of tRNA genes among the mitochondrial genomes of the five species. The AT- and GC-skew were calculated using the following formulas: AT-skew = (A% − T%)/(A% + T%) and GC-skew = (G% − C%)/(G% + C%)38. Intergenic spacers and overlapping regions between genes were manually counted. The rate of protein conformance among the 13 PCGs was analyzed using DNAMAN. Ka and Ks substitution values were calculated using the DNaSP V5.1058. Multiple protein structure prediction web servers (https://www.predictprotein.org/ and http://www.prabi.fr/)59 were used to predict the secondary structure of the ATP8 protein: The amino acid composition and coding sequence composition of the protein, combined with regional, screw, spiral transmembrane regions, and other irregular regions were analyzed. The protein loci of potential exposure areas and hidden areas were also predicted60,61.
Phylogenetic analyses
Substitution saturation of different genes was tested in DAMBE5 using the GTR substitution model as a reference62,63. The best model of evolution for all genes and protein sequences was the GTR + I + G model, as determined by the jModelTest software package64. For the phylogenetic analysis, 55 published mitochondrial genomes were downloaded from NCBI as references and used along with the two Curculio sequenced mitochondrial genomes (Table S1).
Phylogenetic analyses incorporated both the Bayesian inference method (BI) using the program MRBAYES version 3.15265. Maximum Likelihood (ML) methods used the CIPRES server RAxML online (www.phylo.org). We used a Bayesian framework based on the PCG data to estimate the divergence times of clades using the BEAST v.1.6.1 software package. The substitution model (GTR + I + G) was also used for these analyses, as determined to best model the data by Jmodeltest64. The analysis was conducted with an expansion growth model and an uncorrelated lognormal relaxed clock, with a proposed insect molecular clock. Rates of nucleotide substitution were 10−2 subs/s/my/l for each mitochondrial protein-coding gene. The mean rate was 1.115 while the lower rate was 0.747 and the upper rate was 1.52313. Markov chains were analyzed three times for 500,000,000 generations, with sampling every 1,000 generations. The Tracer v.1.5 software package was used to verify the posterior distribution and the effective sample sizes (ESSs) from the MCMC output to ensure that the values were greater than 200. The chain analysis process was tested three times to ensure data stability. The Tree Annotator v1.7.5 component within the BEAST package66 was used to summarize a burn-in of 25% trees after the stationary chain likelihood values were established. The phylogenetic trees were viewed and edited using the FigTree software package.
Supplementary information
Acknowledgements
We thank Yin Xie and Cheng-lian Xin for helping with camellia weevil collection. This work was supported by the Forestry Industry Research Special Funds for Public Nonprofit Project (grant no. 201304403), the External Cooperation Program of Chinese Academy of Forestry and Zhejiang Province (No. 2016SY03).
Author Contributions
S.K.Z., J.P.S. and W.Z. contributed fieldwork. S.K.Z., Y.N.L. and H.P. performed the laboratory experiments and analyzed the data. Y.D.W. and H.J.W. coordinated the study and participated in conceptual design and manuscript preparation. S.K.Z. and J.P.S. performed most of the work for conceptual design and manuscript preparation.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jin-ping Shu, Email: shu_jinping001@163.com.
Yang-dong Wang, Email: Wyd11111@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-39895-8.
References
- 1.Shu JP, et al. Preliminary analysis on the causes of pre-harvest fruit drop in Camellia oleifera. China Plant Protection. 2013;33:9–14. [Google Scholar]
- 2.Li H, et al. Correlation between damage of Curculio chinensis and fruit traits of Camellia meiocarp. Scientia Silvae Sinicae. 2014;50:151–155. [Google Scholar]
- 3.Li MM, et al. Relationship between Curculio chinensis damage and physical characteristics of cones among Camellia oleifera varieties. Forest Research. 2017;30:232–237. [Google Scholar]
- 4.Zhang SK, et al. Genetic diversity in the camellia weevil, Curculio chinensis Chevrolat (Coleptera: Curculionidae) and inferences for the impact of host plant and human activity. Entomol. Sci. 2018;21:447–460. doi: 10.1111/ens.12329. [DOI] [Google Scholar]
- 5.Chen PY, et al. Next-generation sequencing of two mitochondrial genomes from family pompilidae (hymenoptera: vespoidea) reveal novel patterns of gene arrangement. Int. J. Mol. Sci. 2016;17:1641. doi: 10.3390/ijms17101641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moraes CT, et al. Mitochondrial DNA structure and function. Int. Rev. Neurobiol. 2002;53:3–23. doi: 10.1016/S0074-7742(02)53002-6. [DOI] [PubMed] [Google Scholar]
- 7.Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 8.Junqueira AC, et al. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae) Gene. 2004;339:7–15. doi: 10.1016/j.gene.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 9.Junqueira AC, et al. Large-scale mitogenomics enables insights into Schizophora (Diptera) radiation and population diversity. Sci. Rep. 2016;6:21762. doi: 10.1038/srep21762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron SL, O’Donoghue PJ, Adlard RD. Four new species of Macropodinium (Ciliophora: Litostomatea) from Australian Wallabies and Pademelons. J. Eukaryot. Microbiol. 2010;48:542–555. doi: 10.1111/j.1550-7408.2001.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 11.Li H, et al. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae) Plos One. 2012;7:e29419. doi: 10.1371/journal.pone.0029419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt T, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 13.Pons J, et al. Nucleotide substitution rates for the full set of mitochondrial protein-coding genes in Coleoptera. Mol. Phylogenet. Evol. 2010;56:796–807. doi: 10.1016/j.ympev.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Nie RE, Yang XK. Research progress in mitochondrial genomes of Coleoptera. Acta Entomologica Sinica. 2014;57:860–868. [Google Scholar]
- 15.Chen R, et al. DNA barcoding reveals a mysterious high species diversity of conifer-feeding aphids in the mountains of southwest China. Sci. Rep. 2016;6:20123. doi: 10.1038/srep20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich M, Muqim N. Sequence and phylogenetic analysis of the complete mitochondrial genome of the flour beetle Tribolium castanaeum. Mol. Phylogenet. Evol. 2003;26:502–512. doi: 10.1016/S1055-7903(02)00335-4. [DOI] [PubMed] [Google Scholar]
- 17.Fenn JD, Cameron SL, Whiting MF. The complete mitochondrial genome sequence of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Mol. Biol. 2007;16:239–252. doi: 10.1111/j.1365-2583.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- 18.Cameron SL. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014;39:400–411. doi: 10.1111/syen.12071. [DOI] [Google Scholar]
- 19.Salvato P, et al. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae) Bmc Genomics. 2008;9:331. doi: 10.1186/1471-2164-9-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanford E, Kelly MW. Local adaptation in marine invertebrates. Ann Rev Mar Sci. 2011;3:509–535. doi: 10.1146/annurev-marine-120709-142756. [DOI] [PubMed] [Google Scholar]
- 21.Saitoh T, et al. DNA barcoding reveals 24 distinct lineages as cryptic bird species candidates in and around the Japanese Archipelago. Mol. Ecol. Resour. 2015;15:177. doi: 10.1111/1755-0998.12282. [DOI] [PubMed] [Google Scholar]
- 22.Pu DQ, et al. Mitochondrial genomes of the hoverflies Episyrphus balteatus and Eupeodes corollae (Diptera: Syrphidae), with a phylogenetic analysis of Muscomorpha. Sci. Rep. 2017;7:44300. doi: 10.1038/srep44300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie RE, et al. The phylogeny of Galerucinae (Coleoptera: Chrysomelidae) and the performance of mitochondrial genomes in phylogenetic inference compared to nuclear rRNA genes. Cladistics. 2018;33:113–130. doi: 10.1111/cla.12196. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Xue D, Han H. The complete mitochondrial genome of Biston panterinaria (Lepidoptera: Geometridae), with phylogenetic utility of mitochondrial genome in the Lepidoptera. Gene. 2013;515:349–358. doi: 10.1016/j.gene.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 25.Mckenna DD, et al. The beetle tree of life reveals that Coleoptera survived end-Permian mass extinction to diversify during the Cretaceous terrestrial revolution. Syst. Entomol. 2015;40:835–880. doi: 10.1111/syen.12132. [DOI] [Google Scholar]
- 26.Kômoto N, Yukuhiro K, Tomita S. Novel gene rearrangements in the mitochondrial genome of a webspinner, Aposthonia japonica (Insecta: Embioptera) Genome. 2012;55:222–233. doi: 10.1139/g2012-007. [DOI] [PubMed] [Google Scholar]
- 27.Adina M, et al. Mitochondrial genome sequence and gene order of Sipunculus nudus give additional support for an inclusion of Sipuncula into Annelida. Bmc Genomics. 2009;10:27. doi: 10.1186/1471-2164-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timmermans MJ, Vogler AP. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea) Mol. Phylogenet. Evol. 2012;63:299–304. doi: 10.1016/j.ympev.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 29.Dowton M, Castro LR, Austin AD. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invertebr. Syst. 2002;16:345–356. doi: 10.1071/IS02003. [DOI] [Google Scholar]
- 30.Cramptonplatt A, et al. Soup to tree: the phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Mol. Biol. Evol. 2015;32:2302–2316. doi: 10.1093/molbev/msv111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, et al. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. Gigascience. 2013;2:4. doi: 10.1186/2047-217X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillett CP, et al. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea) Mol. Biol. Evol. 2014;31:2223–2237. doi: 10.1093/molbev/msu154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang M, et al. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014;42:e166. doi: 10.1093/nar/gku917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron SL, et al. Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera) Bmc Genomics. 2011;12:394. doi: 10.1186/1471-2164-12-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowton M, et al. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol. Biol. Evol. 2009;26:1607–1617. doi: 10.1093/molbev/msp072. [DOI] [PubMed] [Google Scholar]
- 36.Wei SJ, Li Q, Achterberg KV, Chen XX. Two mitochondrial genomes from the families Bethylidae and Mutillidae: independent rearrangement of protein-coding genes and higher-level phylogeny of the Hymenoptera. Mol. Phylogenet. Evol. 2014;77:1–10. doi: 10.1016/j.ympev.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassanin A, Léger N, Deutsch J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005;54:277–298. doi: 10.1080/10635150590947843. [DOI] [PubMed] [Google Scholar]
- 39.Peng Q, et al. Mitogenomic analysis of the genus Pseudois: evidence of adaptive evolution of morphological variation in the ATP synthase genes. Mitochondrion. 2012;12:500–505. doi: 10.1016/j.mito.2012.07.107. [DOI] [PubMed] [Google Scholar]
- 40.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 1992;141:173–216. doi: 10.1016/S0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 41.Masta SE, Boore JL. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol. Biol. Evol. 2004;21:893–902. doi: 10.1093/molbev/msh096. [DOI] [PubMed] [Google Scholar]
- 42.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida M, Muneyuki E, Hisabori T. ATP synthase - a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 44.Zhang QL, et al. Gene sequence variations and expression patterns of mitochondrial genes are associated with the adaptive evolution of two Gynaephora species (Lepidoptera: Lymantriinae) living in different high-elevation environments. Gene. 2017;610:148–155. doi: 10.1016/j.gene.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Gu P, et al. Evidence of adaptive evolution of alpine pheasants to high-altitude environment from mitogenomic perspective. Mitochondrial DNA Part A. 2016;27:455–462. doi: 10.3109/19401736.2014.900667. [DOI] [PubMed] [Google Scholar]
- 46.Andújar C, et al. Phylogenetic community ecology of soil biodiversity using mitochondrial metagenomics. Mol. Ecol. 2015;24:3603–3617. doi: 10.1111/mec.13195. [DOI] [PubMed] [Google Scholar]
- 47.Timmermans MJTN, et al. Family-level sampling of mitochondrial genomes in Coleoptera: compositional heterogeneity and phylogenetics. Genome Biol. Evol. 2015;8:161–175. doi: 10.1093/gbe/evv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang SQ, et al. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commun. 2018;9:205. doi: 10.1038/s41467-017-02644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunter NL, Oberprieler RG, Cameron SL. Molecular phylogenetics of Australian weevils (Coleoptera: Curculionoidea): exploring relationships in a hyperdiverse lineage through comparison of independent analyses. Austral Entomol. 2016;55:217–233. doi: 10.1111/aen.12173. [DOI] [Google Scholar]
- 50.Shin S, et al. Phylogenomic data yield new and robust insights into the phylogeny and evolution of weevils. Mol. Biol. Evol. 2018;35(4):823–836. doi: 10.1093/molbev/msx324. [DOI] [PubMed] [Google Scholar]
- 51.Zhao SZ, Gong C, Xing WN. Comparative genomic and phylogenetic analysis of Camellia oleifera and C. sinensis. South China Forestry. Science. 2013;4:1–5. [Google Scholar]
- 52.Andrews, S. FastQC: a quality control tool for high throughput sequence data. (Babraham Institute: Cambridge, UK). http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 53.Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina Miseq data. Bioinformatics. 2014;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 54.Bankevich A, et al. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J. Mol. Cell Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. Plos One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernt M, et al. Mitos: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 57.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 58.Librado P, Rozas R. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 59.Yachdav G, et al. PredictProtein-an open resource for online prediction of, protein structural and functional features. Nucleic Acids Res. 2014;42:W337–W343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tokuno M, et al. Long-term stored hemoglobin-vesicles, a cellular type of hemoglobin-based oxygen carrier, has resuscitative effects comparable to that for fresh red blood cells in a rat model with massive hemorrhage without post-transfusion lung injury. Plos One. 2016;11:e0165557. doi: 10.1371/journal.pone.0165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Müller K, Fedosov DA, Gompper G. Understanding particle margination in blood flow – a step toward optimized drug delivery systems. Med. Eng. Phys. 2016;38:2–10. doi: 10.1016/j.medengphy.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 62.Xia X. Dambe5: a comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013;30:1720–1728. doi: 10.1093/molbev/mst064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia X, Xie Z. Dambe: software package for data analysis in molecular biology and evolution. J. Hered. 2001;30:1720–1728. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 64.Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 65.Ronquist FR, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 66.Drummond AJ, et al. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.