Abstract
The oral microbiota is associated with oral diseases and digestive systemic diseases. Nevertheless, the causal relationship between them has not been completely elucidated, and colonisation of the gut by oral bacteria is not clear due to the limitations of existing research models. The aim of this study was to develop a human oral microbiota-associated (HOMA) mouse model and to investigate the ecological invasion into the gut. By transplanting human saliva into germ-free (GF) mice, a HOMA mouse model was first constructed. 16S rRNA gene sequencing was used to reveal the biogeography of oral bacteria along the cephalocaudal axis of the digestive tract. In the HOMA mice, 84.78% of the detected genus-level taxa were specific to the donor. Principal component analysis (PCA) revealed that the donor oral microbiota clustered with those of the HOMA mice and were distinct from those of specific pathogen-free (SPF) mice. In HOMA mice, OTU counts decreased from the stomach and small intestine to the distal gut. The distal gut was dominated by Streptococcus, Veillonella, Haemophilus, Fusobacterium, Trichococcus and Actinomyces. HOMA mice and human microbiota-associated (HMA) mice along with the GF mice were then cohoused. Microbial communities of cohoused mice clustered together and were significantly separated from those of HOMA mice and HMA mice. The Source Tracker analysis and network analysis revealed more significant ecological invasion from oral bacteria in the small intestines, compared to the distal gut, of cohoused mice. In conclusion, a HOMA mouse model was successfully established. By overcoming the physical and microbial barrier, oral bacteria colonised the gut and profiled the gut microbiota, especially in the small intestine.
Subject terms: Microbial ecology, Microbiome, Microbial ecology, Microbiome
A mouse model for human oral bacteria studies
Swabbing the mouths of germ-free mice with human saliva can provide an effective mouse model for studying the roles of human oral bacteria in mouth and gut diseases. The human oral microbiota-associated (HOMA) mice were developed by Lei Cheng of Sichuan University and colleagues in China and bred alone for 35 days. A significant proportion (85%) of the types of bacteria present in the human donors’ saliva successfully established themselves in the germ-free mice and colonised various parts of their guts. When the HOMA mice were co-bred with other types of mouse models, digestive tract bacteria transferred to them, preventing oral bacteria from significantly colonising the gut. The researchers conclude that a HOMA model was successfully established and could help overcome current limitations in investigations related to human oral bacteria and disease.
Introduction
Clinical trials have indicated that the oral microbiota is associated with dental caries and periodontitis,1–4 both of which give rise to an extensive loss of natural teeth in older people and are identified as public health problems worldwide.5 Accumulating evidence has even linked the human oral microbiota to oral cancer.6,7 In recent years, oral microecology dysbiosis has been proven to cause periodontitis4,8 and regarded as an indicator to predict early childhood caries (ECC).9 Thus, the oral microbiota has a key role in the initiation of oral diseases.
An increasing number of clinical research studies of the oral microbiota are being designed. However, the clinical investigations are usually restricted by complex conditions, including ethical issues. Regardless, a prospective cohort clinical study9 found that shifts in the microbiota preceded the manifestation of clinical symptoms of ECC. Unfortunately, most of the other studies were cross-sectional and could barely address whether the oral microbiota was the cause or effect in the development of oral diseases. In-vitro models also have limitations due to the abundant uncultivated phylotypes in the mouth.10 Animal models would have been considered a good choice to study the oral microbiota; however, the oral microbiota of mice, the most common experiment animal model, differs from that of humans. Therefore, a HOMA mouse model, with an oral microbiota similar to the human donors, must be established to reveal the cause-and-effect relationships between the oral microbiota and host pathologies, like the HMA mouse model.11,12
Not only oral diseases but also oral bacteria are linked to various digestive systemic diseases, including inflammatory bowel disease,13,14 colorectal cancer (CRC),15 pancreatic cancer,16,17 liver carcinoma18 and liver cirrhosis.19 Seedorf et al.20 demonstrated that mouth-derived bacteria such as Actinobacteria, Bacilli, Clostridia, Fusobacteria and Epsilonproteobacteria are able to overcome the host physical barrier and persist in the germ-free distal gut. Comparing the gut microbiome of patients suffering from liver cirrhosis with that of healthy control individuals, Qin et al.21 found that most (54%) of the patient-enriched faecal microbial species originated from the oral cavity, demonstrating that the oral microbiota had invaded the gut of patients with liver cirrhosis. These studies indicated that the oral microbiota influenced host health by invading and colonising the gut. The colonisation of oral microbiota in the gut is a key point to understand pathologic colonisation, facilitating studies of the pathogenic mechanisms of oral bacteria in systemic digestive diseases. However, invasion by oral microbiota by overcoming host physical barriers and gut microbiota barriers at various regions along the cephalocaudal axis of the gut is not well described.
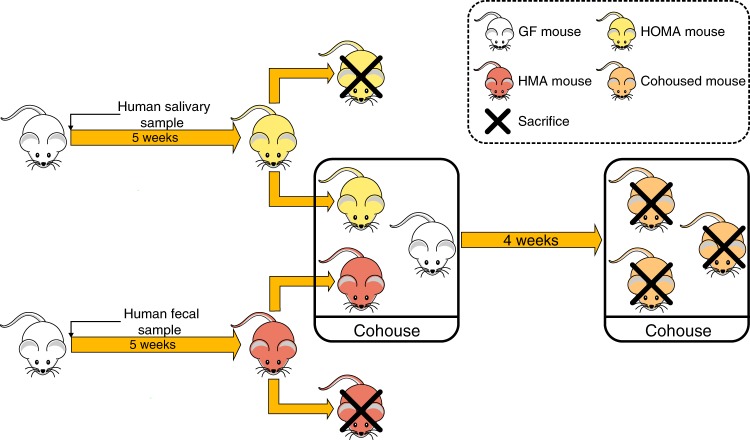
To develop the HOMA mouse model, we introduced the human salivary microbiota into GF mice and created a well-defined, representative animal model of the human oral microbial ecosystem. Using the HOMA mouse model, we investigated the colonisation of gut-selected oral bacteria along the longitudinal axis. Furthermore, we studied the competition of oral microbiota with the native gut microbiota in various regions of the gut and identified key bacteria during the ecological invasion, by cohousing HOMA mice, HMA mice and GF mice (Fig. 1).
Fig. 1.
Design of the human microbiota transplant and cohousing experiments
Results
The oral microbiota of the HOMA mouse model
The surveys of oral samples revealed the engraftment of the human oral microbiota: all bacterial phyla, classes, orders, 27 of 28 bacterial families, and 84.78% (39 of 46) of genus-level taxa were detected among the recipient mice. All seven genus-level taxa missed by the humanised mice exhibited a low abundance in the donor sample (0.21% on average). The oral microbiota of the donor was dominated by eleven genus-level taxa, with a high relative abundance (>1%), of which five, Veillonella, Fusobacterium, Streptococcus, Porphyromonas and Haemophilus, maintained a high abundance (>1% on average) among the recipient mice. The others were depleted to a low abundance among the recipient mice (Table S1).
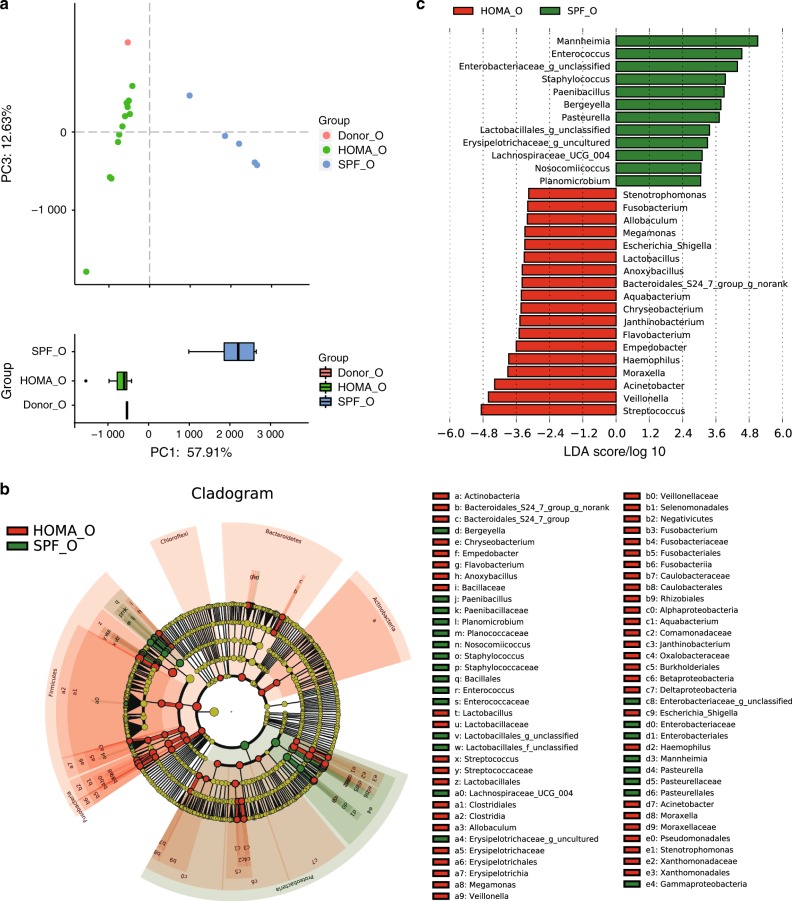
To further identify the advantages of the HOMA mouse model, we compared the oral microbiota of HOMA mice with SPF mice. PCA revealed that the donor oral microbiota clustered closely with the HOMA mouse but were distinct from SPF mouse microbiota, especially in PC1 (57.91%) (Fig. 2a). The oral microbiota of HOMA mice differed from that of SPF mice in taxonomic structure. Dominant genus-level taxa present in the donor saliva sample were significantly more abundant among HOMA mice than SPF mice, including Veillonella, Fusobacterium, Streptococcus and Haemophilus (Fig. 2b, c).
Fig. 2.
Advancement of the HOMA mouse model. a PCA score plot of the oral microbiota of the human donor (Donor_O, red), HOMA mice (HOMA_O, green) and SPF mice (SPF_O, blue) at the genus level. b Taxonomic cladogram for HOMA mouse-enriched taxa (red) and SPF mouse-enriched taxa (green) obtained by LEfSe analysis of 16S sequences. c The HOMA mouse-enriched taxa are indicated by a negative LDA score (red), while the taxa enriched by SPF mice have a positive score (green). Taxa at the genus level with different abundances between groups and with an LDA score >3.0 are shown
Biogeography of the host gut-selected oral microbiota
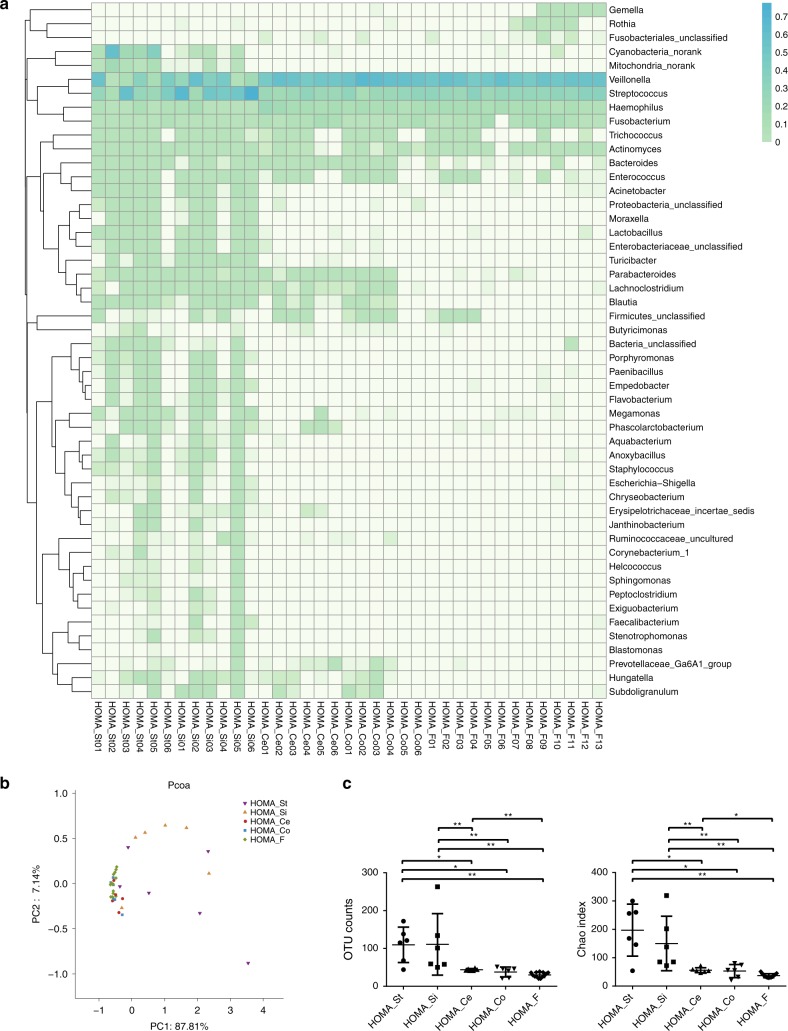
The 16S rRNA gene sequencing survey revealed that the oral bacteria colonised various segments of the gut. In the stomach, eighteen genus-level taxa were detected, with a relative abundance of more than 0.1% on average, eleven of which had a relative abundance exceeding 0.5% on average. In the small intestine, the relative abundances of 23 genus-level taxa exceeded 0.1% on average. Those with a relative abundance greater than 0.5% on average were Streptococcus, Veillonella, Haemophilus, Enterococcus, Fusobacterium, Acinetobacter, Enterobacteriaceae_unclassified, and Bacteroides. In the caecum, only six genus-level taxa were detected, with a relative abundance greater than 0.1% on average, including Veillonella, Streptococcus, Haemophilus, Fusobacterium, Bacteroides and Trichococcus. Genus-level taxa with a relative abundance greater than 0.1% in the colon were the same as those in the caecum. The main genus-level taxa in the whole gut were Streptococcus, Veillonella, Haemophilus, Fusobacterium, Trichococcus and Bacteroides (Fig. 3a, Table S2). All six main genus-level taxa in the gut were also the dominant genus-level taxa (>1%) in the mouth of the HOMA mouse (Table S1). Although the microbial communities colonising various regions shared some main bacteria, the differences among them were clear. Principal coordinates analysis (PCoA) showed that microbial communities present in the caecum, colon, and faeces clustered together and were distinct from those in the stomach and small intestine (Fig. 3b). OTU counts significantly decreased from the stomach and small intestine to the distal gut and from the caecum to faeces, as did the Chao index (Fig. 3c). Distal gut communities were depleted to a low diversity consortium. The relative abundances of Acinetobacter, Enterobacteriaceae_unclassified, Lactobacillus, Turicibacter, Proteobacteria_unclassified and Moraxella decreased from the stomach and small intestine to the distal gut and faeces. The relative abundances of Parabacteroides, Lachnoclostridium and Blautia decreased from the caecum and colon to the faeces (Fig. 3a). These results indicated that the oral bacteria were filtered out by the distal gut.
Fig. 3.
Biogeography of gut-selected oral microbiota. a Heatmap of specimens showing the relative abundance of the main identified bacteria at the genus taxonomic level in each segment of HOMA mouse guts, including stomach (St), small intestine (Si), caecum (Ce), colon (Co) and faeces (F). b PCoA score plot of the microbiota from each segment of HOMA mouse guts based on unweighted UniFrac metrics. c The Kruskal–Wallis test was used to compare the difference between each segment of HOMA mouse guts in the OTU count and Chao index (*P < 0.05, **P < 0.01)
Ecological invasion by oral microbiota in the gut
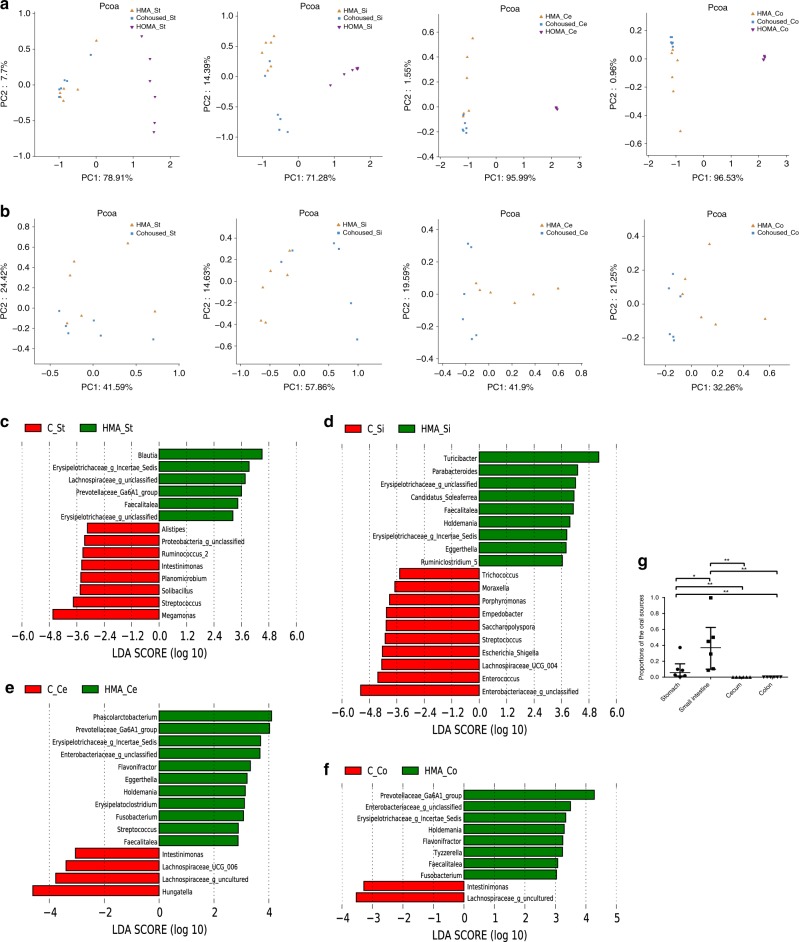
PCoA revealed that the microbial communities in every segment could not be distinguished by the original grouping 28 days after cohousing (Fig. 4a). Therefore, the gut microbiota of the cohoused mice could be regarded as an aggregate, regardless of the original mouse group. The microbial communities of cohoused mice were closely clustered with those of HMA mice and distinct from those of HOMA mice in every segment (Fig. 4a), suggesting that the oral microbiota was unable to challenge the dominant position of the gut microbiota in the gut. Interestingly, further analysis without HOMA mice showed that the microbial communities of cohoused mice could also be separated from HMA mice in every segment (Fig. 4b). These results indicated that although the oral microbiota was almost protected by the gut microbiota barrier, it reshaped the native gut microbiota. To further understand the effect of the oral microbiota on the community composition of the gut microbiota, LEfSe analysis was used. In the stomach, seven genus-level taxa were significantly increased from HMA mice to cohoused mice. One of the seven genus-level taxa was Streptococcus, which was the dominant genus (relative abundance > 1%) in the mouth of the HOMA mouse (Fig. 4c). In the small intestine, seven genus-level taxa were significantly increased from HMA mice to cohoused mice, six of which were dominant genera in the mouth of the HOMA mouse: Enterococcus, Streptococcus, Empedobacter, Porphyromonas, Moraxella and Trichococcus (Fig. 4d). In the distal gut, four genus-level taxa were significantly increased from HMA mice to cohoused mice. but none was the dominant genera in the mouth (Fig. 4e, f). Microbial Source Tracker was used to analyse the effects of cohousing on the flow of microbes between cage mates, which allowed us to determine whether the assembly processes were involved in shaping the communities. The results revealed significant ecological invasion by oral bacteria in the small intestine (Fig. 4g).
Fig. 4.
The shift in microbial composition after cohousing. a PCoA score plot of the microbiota from each gut segment of HOMA mice, HMA mice and cohoused mice. b PCoA score plot of the microbiota from each gut segment from HMA mice and cohoused mice. c HMA mouse-enriched genus-level taxa in the stomach are indicated by a positive LDA score (green), while the cohoused (C) mouse-enriched taxa have a negative LDA score (red). d The HMA mouse-enriched genus-level taxa in the small intestine are indicated by a positive LDA score (green), while the cohoused mouse-enriched taxa have a negative LDA score (red). e The HMA mouse-enriched genus-level taxa in the caecum are indicated by a positive LDA score (green), while the cohoused mouse-enriched taxa have a negative LDA score (red). f The HMA mouse-enriched genus-level taxa in the colon are indicated by a positive LDA score (green), while the cohoused mouse-enriched taxa have a negative LDA score (red). g Microbial Source Tracker analysis showed the proportions of the different sources present in the microbiota of the cohoused mice in each gut segment. The Kruskal–Wallis test was used to compare the proportions of the oral sources present in each gut segment of the cohoused mice (*P < 0.05, **P < 0.01)
Porphyromonas competed for colonisation with the small intestinal microbiota
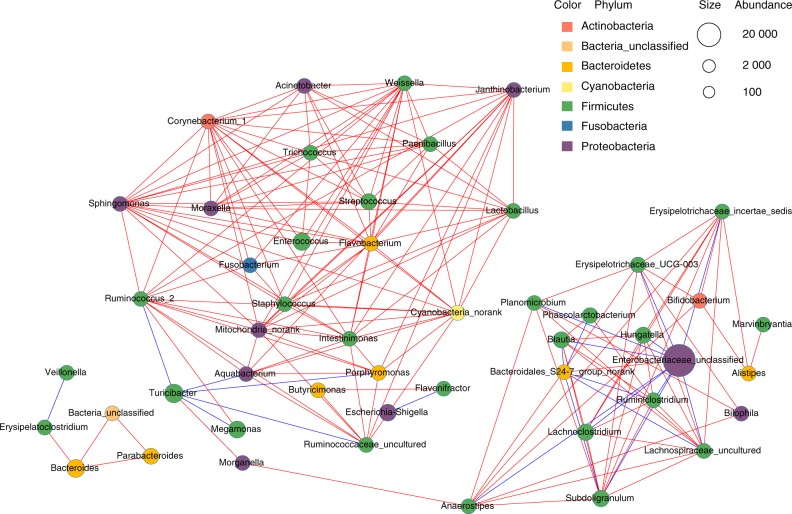
To further study the functional positions of oral bacteria in the microbial community colonising the small intestine, the co-occurrence network of the top 50 abundant genus-level taxa was used. Porphyromonas was found to correlate negatively with Turicibacter (Fig. 5). Before invasion by the oral microbiota, Turicibacter was the most dominant genus in the small intestine with the highest relative abundance (40.40% on average). Following invasion by the oral microbiota, the relative abundance of Porphyromonas increased significantly, and the abundance of Turicibacter decreased to 8.79% on average (Fig. 4d, Fig. S1). Moreover, Porphyromonas was found to correlate positively with these genera dominating the mouth of the HOMA mouse, including Streptococcus, Enterococcus, Acinetobacter, Moraxella, Trichococcus, Fusobacterium, Flavobacterium and Lactobacillus (Fig. 5, Table S1). These results suggested that Porphyromonas, as common oral bacteria, had a key role in competing for colonisation with the native main genus in the small intestine.
Fig. 5.
The co-occurrence network was generated from the small intestinal microbiota of the cohoused mice. Different coloured edges represent a positive (red) and a negative (blue) correlation, respectively. Each node represents a genus-level taxon, and the size of each node is proportional to the abundance. The colour of the nodes indicates their classification at the phylum level
Discussion
In the past years, cumulative research data have implied a tight association between dysbiosis of the oral microbiota and diseases.3,6,7,16,22 However, it has been difficult to verify the contribution of the oral microbiota to diseases via clinical studies due to their limitations. The lack of understanding of the effect and pathogenic mechanism of dysbiotic oral microbiota manifests in a great gap between the large amount of data and clinical applications.23 Thus, for oral microbiota investigations, the establishment of a HOMA mouse model can have an important role in translational medicine, similar to the HMA mouse model. In the present study, 84.78% (39 of 46) of the genus-level taxa were from donor saliva, similar to the HMA mouse model receiving 11 of 12 bacterial classes, and 88% (58 of 66) of the genus-level taxa were human.12 Additionally, in subsequent study, we inoculated the contents of another two donor salivary glands into GF mice and obtained similar results.24 Additionally, the HOMA mouse was a better representative for the donor than traditional SPF mice (Fig. 2a). Therefore, it is not difficult to conclude that the HOMA mouse model was established successfully. Currently, the HMA mouse is an ideal model to study the role of the disease-associated gut microbiome.11 In future, we believe that the HOMA mouse model could be used to investigate the effect of a dysbiotic oral microbiota on oral diseases, such as dental caries, periodontics and oral cancer. In addition to oral disease, the HOMA mouse model will be applied to verify whether the oral microbiota is associated with some digestive systemic diseases.
In most previous studies, the faecal microbiota was collected to represent the gut microbiota; however, some researchers have had different opinions and have suggested to divide the digestive tract into different sections to study the gut microbiota.25 By collecting ileostomy samples from humans, Zoetendal et al.26 found that the small intestine was enriched with Streptococcus sp. and Escherichia coli. Interestingly, in the present analysis, invasion by oral bacteria into the small intestine increased the relative abundance of Streptococcus and Enterobacteriaceae (Fig. 4d). Furthermore, in the small intestines of the cohoused mice, nearly 40% of the taxa were from oral microbial communities, which reshaped the community composition in the small intestine of the HMA mouse (Fig. 4g). Thus, especially in the small intestine, the oral microbiota had an important role in building the integrated gut microbiota.
In the present study, oral bacteria overcame the host physical barrier and colonised the gut in HOMA mice (Fig. 3a, Table S2). However, in cohoused mice, the oral bacteria showed minimal colonisation of the gut, especially the distal gut (Fig. 4g). This result is consistent with a previous study,20 in which all the distal guts of HMA mice cohoused with mice with the microbiota from soil or zebrafish were dominated by caecum-derived microbiota at 7 days after cohousing. These results indicated that gut microbiota has an important role as a barrier in resisting the foreign bacteria from mouth. This resistance might due to greater acceptability in the gut of the gut microbiota than the oral microbiota, and the creation of a more stable microenvironment by the gut microbiota to resist foreign bacteria. However, the microbiota barrier of the gut was not consistently indestructible, especially in the small intestine, where six of seven increasing genus-level taxa in the cohoused mice were dominant genera in the mouth of the HOMA mouse, including Porphyromonas (Fig. 4d). As a key oral genus to overcome the gut microbiota barrier, Porphyromonas was tightly associated with these genera that dominated the mouth of the HOMA mouse, but it correlated negatively with Turicibacter, the most dominant genus in the small intestine of HMA mice. Prior to invasion by oral microbiota, the relative abundance of Turicibacter in other regions of the gut was lower than that in the small intestine (Fig. S1), which might explain why more oral bacteria invaded the small intestine instead of the other regions. The small intestine is responsible for the majority of substance transformation27 and is covered by a thinner mucin layer than the distal gut.28 Thus, the small intestinal microbiota more effectively impacts digestive systemic health, suggesting that ecological invasion in the small intestine by Porphyromonas had a marked effect on digestive systemic health. For example, oral administration of Porphyromonas gingivalis, belonging to Porphyromonas, has been confirmed to induce gut microbiota dysbiosis and impair mucosal barrier function, leading to the dissemination of Enterobacteria to the liver.29,30
Another interesting phenomenon is revealed by the barrier function of the gut microbiota. Fusobacterium overcame the physical barrier and became the dominant genus in the gut of the HOMA mouse. However, after receiving the gut microbiota by cohousing, the abundance of Fusobacterium decreased markedly, and even the gut microbiota barrier was partly overcome by oral microbiota in the small intestine. Fusobacterium was still stopped by the microbiota barrier, but the resistance to Fusobacterium was supported by the gut microbiota from a healthy donor here. Those individuals suffering CRC fail to resist Fusobacterium.15,31,32 The accumulating Fusobacterium nucleatum overcome the defective gut microbiota barrier from the CRC patient and further promote tumour development.33–35 In conclusion, resistance from various gut microbial communities is a key point to understand the effect of oral microbiota on gut microbiota and digestive systemic health, and it should be investigated in future studies.
Overall, we first established a HOMA mouse model, which copied the oral microbiota of the human donor. Using this animal model, we found that both physical and microbiota barriers filtrated the oral microbiota in the digestive tract. Additionally, the oral microbiota invaded and profiled the gut microbiota, especially in the small intestine. Oral Porphyromonas was the key bacterial species competing with the small intestinal microbiota.
Materials and methods
Sample collection from humans
The study was authorised by the Ethical Committee of Sichuan University (WCHSIRB-D-2016-070). The saliva was collected using a sterilised tube from an adult donor with natural dentition without periodontitis or active caries and without the use of antibiotics in the previous 3 months. The donor was required not to brush teeth for 24 h and abstain from food/drink intake for 2 h prior to donating saliva. Faeces were collected from the same person using a sterilised sealable plastic bag. A portion of the saliva and faeces were sent to the lab and inoculated into GF mice within 30 min. The rest was stored immediately at –80 °C.
Animal husbandry
The animal experimentation protocols were approved by the Ethical Committee of Sichuan University (WCHSIRB-D-2016-118) and the Third Military Medical University. Six-week-old GF male Kunming mice were maintained in the Experimental Animal Research Center at the Third Military Medical University. All GF mice were bred in plastic gnotobiotic isolators, where the temperature and humidity were maintained at 20–26 ℃ and 40%–70%, respectively. They were fed a standard diet (GB-T14924.3-2001) sterilised by 60 co gamma radiation. Thirteen-week-old SPF mice were also maintained in the Experimental Animal Research Center. They were fed in the barrier housing facility.
Establishment of the HOMA and HMA mouse models
To establish the HOMA mouse model, swabs dipped in 200 μL fresh saliva from the male donor were used to seed oral microbiota in the GF mice (n = 13) by swabbing without anaesthesia. Swabbing was performed only once. The HMA mouse model was developed as previously described.36 The faeces were resuspended in 10 mL sterile potassium phosphate buffer (0.1 mol·L−1, pH 7.2). Eight GF mice were inoculated by intragastric gavage with 1 mL human faeces suspension each, and 2-mL aliquots were spread on the fur. HOMA mice and HMA mice were bred in separated plastic gnotobiotic isolators. After 35 days, oral microbial samples were collected from the HOMA mice with swabs. The oral microbial samples from SPF mice were collected in the same way. Faeces of HOMA mice were also collected. Six of thirteen HOMA mice and six of eight HMA mice were subsequently killed randomly, and the contents of the stomach, small intestine, caecum and colon were collected. All the samples were immediately stored at −80 °C.
Cohousing experiment
Two HOMA mice and two HMA mice was transferred to a new germ-free plastic isolator containing two GF mice (Fig. 1). These six mice were then distributed into two triads, each of which included a HOMA mouse, a HMA mouse and a GF mouse housed in one cage, by which the animals could exchange components of their microbiota. After 28 days, the cohoused mice were killed, and the contents of the stomach, small intestine, caecum and colon were collected. All these samples were immediately stored at −80 °C.
16S rRNA gene sequencing
The samples were processed by Shanghai Majorbio Bio-Pharm Technology Co., Ltd (Shanghai, China). Total DNA was extracted, amplified and sequenced according to standard procedures.37,38 Briefly, microbial DNA was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer’s protocol. The DNA concentration was assessed using a Nanodrop (Thermo Scientific), and the quality was determined by agarose gel electrophoresis. Bacterial 16S rRNA gene sequences spanning the variable regions V4-V5 were amplified using the primer 515F_907R. The amplicons were then extracted from 2% agarose gels and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) and quantified by QuantiFluor™-ST (Promega, U.S.). Purified amplicons were pooled in equimolar amounts and subjected to paired-end sequencing (2 × 300) on an Illumina MiSeq platform.
Bioinformatics and statistical analysis
Raw fastq files were demultiplexed and quality-filtered by QIIME (version 1.9.1).39 Operational taxonomic units (OTUs) were clustered with a 97% similarity cut-off using UPARSE (version 7.1). The taxonomy of each 16S rRNA gene sequence was analysed using the RDP Classifier (http://rdp.cme.msu.edu/) against the SILVA rRNA database (http://www.arb-silva.de) with a confidence threshold of 70%. After the elimination of interference sequence, alpha diversity estimator calculations were performed using Mothur v.1.30.2. Phylogenetic beta diversity measures, such as unweighted UniFrac distance metrics analysis, was determined using the representative sequences of OTUs for each sample, and PCA and PCoA were conducted according to the distance matrices. LEfSe analysis (linear discriminant analysis [LDA] coupled to effect size measurements) was conducted to calculate bacteria with significant difference in relative abundance between the groups. Using a normalised relative abundance matrix, LEfSe showed taxa with significantly different abundances, and LDA estimated the effect size of the feature.37,40 In this study, a P value threshold of 0.05 (Wilcoxon rank-sum test) and an effect size threshold of 3 were used for all bacteria discussed. Microbial Source Tracker analysis was performed using the Source Tracker package based on Bayesian inference.20,41 The co-occurrence network of the top 50 abundant genus-level taxa was inferred based on the Spearman correlation matrix with a strict P-value threshold (P < 0.05) and a high correlation value (r > 0.6) to filter strong correlations. The combined result was exported to Cytoscape V.3.2.1.37
The data were subjected to nonparametric Kruskal–Wallis analysis. Differences were considered significant when P < 0.05. SPSS21.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
Data availability
The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP116564).
Supplementary information
Acknowledgements
This study was supported by the National Key Research and Development Program of China 2016YFC1102700 (X.Z.); National Natural Science Foundation of China grant 81372889 (L.C.), 81370906 (W.H.), 81600858 (B.R.) and 81430011 (X.Z.); Youth Grant of the Science and Technology Department of Sichuan Province, China 2017JQ0028 (L.C.); and National Basic Research Program of China 973 Program 2013CB532406 (W.H).
Author contributions
L.C., X.Z. and H.W. conceived and designed the experiments; B.L., J.Y., B.Z., X.P., W.L., B.R. and M.L. performed the experiments; B.L., Y.G. and J.Z. analysed the data; B.L. and Y.G. wrote the manuscript; H.W. and L.C. revised the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Hong Wei, Phone: +8613808388737, Email: weihong63528@163.com.
Xuedong Zhou, Phone: +8602885501439, Email: zhouxd@scu.edu.cn.
Supplementary material
The online version of this article (10.1038/s41368-018-0043-9) contains supplementary material, which is available to authorised users.
References
- 1.Johansson I, Witkowska E, Kaveh B, Holgerson PL, Tanner AC. The microbiome in populations with a low and high prevalence of caries. J. Dent. Res. 2016;95:80–86. doi: 10.1177/0022034515609554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belda-Ferre P, et al. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 2014;8:1879–1891. doi: 10.1038/ismej.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao E, et al. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe. 2017;22:120–128.e124. doi: 10.1016/j.chom.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen PE, Ogawa H. Prevention of dental caries through the use of fluoride--the WHO approach. Community Dent. Health. 2016;33:66–68. [PubMed] [Google Scholar]
- 6.Schmidt BL, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE. 2014;9:e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pushalkar S, et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12:144. doi: 10.1186/1471-2180-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajishengallis G, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teng F, et al. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe. 2015;18:296–306. doi: 10.1016/j.chom.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Dewhirst FE, et al. The human oral microbiome. J. Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrieta MC, Walter J, Finlay BB. Human microbiota-associated mice: a model with challenges. Cell Host Microbe. 2016;19:575–578. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1:6–14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strauss J, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 14.Ismail Y, et al. Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PLoS ONE. 2012;7:e38217. doi: 10.1371/journal.pone.0038217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostic AD, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrell JJ, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan X, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120–127. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu H, et al. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci. Rep. 2016;6:33142. doi: 10.1038/srep33142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bajaj JS, et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260–1271. doi: 10.1002/hep.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seedorf H, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin N, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 22.Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 2011;10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mankoff SP, Brander C, Ferrone S, Marincola FM. Lost in translation: obstacles to translational medicine. J. Transl. Med. 2004;2:14. doi: 10.1186/1479-5876-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, et al. Research on oral microbiota of monozygotic twins with discordant caries experience - in vitro and in vivo study. Sci. Rep. 2018;8:7267. doi: 10.1038/s41598-018-25636-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoetendal EG, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarker SA, Ahmed T, Brussow H. Hunger and microbiology: is a low gastric acid-induced bacterial overgrowth in the small intestine a contributor to malnutrition in developing countries? Microb. Biotechnol. 2017;10:1025–1030. doi: 10.1111/1751-7915.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: connecting spatial organization to function. Cell Host Microbe. 2017;21:433–442. doi: 10.1016/j.chom.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimatsu K, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci. Rep. 2014;4:4828. doi: 10.1038/srep04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima M, et al. Oral administration of P. gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of enterobacteria to the liver. PLoS ONE. 2015;10:e0134234. doi: 10.1371/journal.pone.0134234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gur C, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu T, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563.e516. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng B, et al. Effects of age and strain on the microbiota colonization in an infant human flora-associated mouse model. Curr. Microbiol. 2013;67:313–321. doi: 10.1007/s00284-013-0360-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang AH, et al. Human colorectal mucosal microbiota correlates with its host niche physiology revealed by endomicroscopy. Sci. Rep. 2016;6:21952. doi: 10.1038/srep21952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, et al. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci. Rep. 2015;5:15220. doi: 10.1038/srep15220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling Z, et al. Alterations in the fecal microbiota of patients with HIV-1 infection: an observational study in a Chinese population. Sci. Rep. 2016;6:30673. doi: 10.1038/srep30673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knights D, et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP116564).