Abstract
Demand for all-natural vanilla flavor is increasing, but its botanical source, Vanilla planifolia, faces critical challenges arising from a narrow germplasm base and supply limitations. Genomics tools are the key to overcoming these limitations by enabling advanced genetics and plant breeding for new cultivars with improved yield and quality. The objective of this work was to establish the genomic resources needed to facilitate analysis of diversity among Vanilla accessions and to provide a resource to analyze other Vanilla collections. A V. planifolia draft genome was assembled and used to identify 521,732 single nucleotide polymorphism (SNP) markers using Genotyping-By-Sequencing (GBS). The draft genome had a size of 2.20 Gb representing 97% of the estimated genome size. A filtered set of 5,082 SNPs was used to genotype a living collection of 112 Vanilla accessions from 23 species including native Florida species. Principal component analysis of the genetic distances, population structure, and the maternally inherited rbcL gene identified putative hybrids, misidentified accessions, significant diversity within V. planifolia, and evidence for 12 clusters that separate accessions by species. These results validate the efficiency of genomics-based tools to characterize and identify genetic diversity in Vanilla and provide a significant tool for genomics-assisted plant breeding.
Introduction
Vanilla is the second most valuable spice with increasing global demand1. Vanilla is an extract produced from the cured seed capsules (commonly referred to as “beans”) of the vining orchid Vanilla planifolia. The extract is used as a premium ingredient in ice cream, chocolate, perfumes, pharmaceuticals, and other products2. Vanilla beans have been used for their valuable aroma since pre-colonial times by early Mesoamericans, including its use to improve the flavor of chocolate by the Aztecs and Mayans3,4. Vanilla spread globally from Mesoamerica starting in the late 1500s including introduction into Europe in 1739 and domestically from Florida to Puerto Rico sometime before 19005,6. Today, clonal descendants, likely originating from these original introductions, are grown commercially. The result is an industry with heavy reliance on a limited genetic base. Improving the genetic diversity and horticultural performance of Vanilla is increasingly important as demand for natural ingredients like vanilla extract increases.
Historically, Vanilla production was limited to Mexico until supply constraints in the 1850s and 1860s pushed for expanded cultivation into other geographies. The demonstration of manual pollination in 1838 by Professor Charles Morren and an optimized, practical method in 1841 by a former slave named Edmond Albius enabled expanded commercial production beyond the native distribution7,8. Today, Madagascar dominates Vanilla production with Indonesia, Uganda, India, Comores, and Mexico all significantly contributing to global supply and with minor production in many other countries9. The United States imports more Vanilla beans than any other country (~1,500 to 2,000 metric tons annually), and produces high-value vanilla extract for export10. Cyclical supply deficits from major weather events, theft, poor quality beans, or geopolitical challenges have increased Vanilla bean price volatility. Recently, cured Vanilla beans were traded at over $600 per kilogram11. This price volatility can negatively impact growers, consumers, and the entire Vanilla supply chain.
Most vanilla extract comes from cured V. planifolia beans, but at least two other species including V. x tahitensis and V. pompona are also grown commercially on a more limited scale. The aroma profiles of each of these species vary, and these differences can be useful for various applications from specialty food ingredients to cosmetic uses. The standard of identity (CFR 169.175) for Vanilla bean extract includes only V. planifolia and V. x tahitensis for historical reasons7, though other species may have commercial relevance and favorable flavor profiles that could be used to improve the commercial species. Alternative sources of vanilla flavor include synthetic vanillin from paper and pulp mill byproducts, and petroleum-based synthesis2. Synthetic vanillin is important to meet the near insatiable demand for vanilla flavor, but industry trends based on consumer demands are increasingly favoring flavors from natural sources like the Vanilla bean.
Vanilla is pantropical, but the primary commercial species, V. planifolia, was first cultivated in Mesoamerica3. V. planifolia spread globally from Mesoamerica through the Caribbean islands and into Europe as early as 1510 with successful cultivation in England by 18078. Vanilla was then introduced into Africa (1852), India (1835), and into today’s major commercial regions of Reunion Island (1793) and Madagascar (1870)12. Since the early 1900s, commercial Vanilla production in the United States has been confined to Hawaii and Puerto Rico. Hawaii has naturalized Vanilla introductions originating from Mexico, Tahiti, Samoa, and Fiji with some cultivated for commercial production13. Puerto Rico has both native species and V. planifolia that was introduced from Florida and has been in cultivation since the early 1900s6. The Hawaiian Vanilla industry is now mostly tourism-based, and Puerto Rican Vanilla cultivation declined in the 1950s with only remnants of escaped plants surviving to this day. Today, growers and industry representatives are expressing increasing interest in domestic Vanilla cultivation including expansion into other suitable environments like south Florida where both native species and naturalized V. planifolia are already growing. The four native species in Florida include V. phaeantha (“leafy Vanilla”), V. barbellata (“worm vine orchid”), V. dilloniana (“Dillon’s Vanilla”), and V. mexicana (“Fuchs’ orchid”)14. These native species are endangered and exist in protected areas, except for V. dilloniana that is thought to be extirpated. The origin of the naturalized V. planifolia population in Florida is currently unknown.
There are over 100 species of Vanilla distributed approximately from latitudes 27°N to 27°S around the world. Many of these species are morphologically distinct for various characteristics including the presence or absence of leaves, aromatic or non-aromatic beans, climbing habit, variation in flower coloration, leaf size, and leaf shape to name a few. Many Vanilla species share similar vegetative traits and require a mature, flowering specimen in order to confirm identity. Additionally, morphological traits can vary with maturity and environmental conditions making species-level identification challenging. Molecular markers have been used to assess the diversity of Vanilla accessions from both herbarium specimens and natural populations. Such marker studies include isozymes15, RAPDs16–18, AFLPs4,19,20, microsatellites21–24, single gene sequences (usually plastid-derived)25–29, or some combination of the above30–35. Major limitations of these approaches include a lack of reproducibility, poor transferability between labs, high relative cost, and low information density. Furthermore, many of the common markers used to identify Vanilla species cannot be used to identify hybrids, or to assign relationships beyond species-level resolution. The development of genomics-based platforms should overcome these limitations and would enable marker-trait associations for plant breeding research needed to develop improved Vanilla cultivars.
Vanilla in general has not benefited from strategic plant breeding and the use of segregating populations to identify marker-trait associations. Literature on Vanilla plant breeding is limited with references to older Vanilla breeding programs12. Previous research has described the results of a wide cross between V. planifolia and V. aphylla resulting in four hybrid progeny that may display delayed flower wilting30,36. Vanilla hybrids cultivated in Costa Rica and natural hybrids discovered in Puerto Rico have also been characterized37,38. Additionally, an AFLP-based Vanilla genetic linkage map using a V. x tahitensis and V. pompona population has been reported20. Vanilla plant breeding could greatly be enhanced through the development and application of genomics-based molecular markers like single nucleotide polymorphisms (SNPs). Indeed, many authors have discussed the extremely narrow genetic diversity of commercially grown V. planifolia originating from a few foundational clones4,12,39–43. There is no technical or biological limitation that would prevent the breeding of improved Vanilla cultivars with improved yield and quality.
The objective of this research was to test the ability of genomics to analyze diversity within a living collection of Vanilla accessions. The outcomes of this research include the assembly of a draft Vanilla genome, the development of genomic resources to rapidly genotype Vanilla species, the identification of hybrid accessions, the assignment of species for unknown samples, and the identification of species-specific SNP markers that could aid in Vanilla identification. These resources will support the development of new Vanilla cultivars to meet evolving industry and consumer needs, and support a domestic Vanilla industry.
Results
Vanilla accessions included in this study
One goal of this study was to assemble a living collection of diverse Vanilla species needed to develop genomics-based tools for diversity analysis. The collection sourced accessions from botanical gardens, online vendors, private collectors, and native species. Accessions were maintained at the Tropical Research and Education Center of the University of Florida, in Homestead, FL. All 112 accessions included in this study, including six replicates as sequencing controls, selected for genotyping-by-sequencing (GBS) analysis are described in Table 1. Species assignments have been updated based on the results described in this study.
Table 1.
List of Vanilla accessions included in this study.
| Identifier | Species | Source | Heterozygosity | BIC Group | Notes |
|---|---|---|---|---|---|
| AC101 | V. phaeantha | Online vendor | 0.0444 | 11 | 3 |
| AC102 | V. siamensis | Private collection | 0.0313 | 9 | |
| AC103 | V. pompona | Private collection | 0.0631 | 3 | |
| AC104 | V. pompona | Private collection | 0.0584 | 3 | 3 |
| AC105 | V. dilloniana and unknown | Online vendor | 0.0655 | 6 | 2 |
| AC106 | V. planifolia | Private collection | 0.0457 | 5 | 1, 3 |
| AC107 | V. pompona and V. phaeantha | USDA | 0.0600 | 3 | 2 |
| AC108 | V. planifolia and unknown | Online vendor | 0.0401 | 5 | 1, 2, 3 |
| AC109 | V. planifolia x V. phaeantha | Botanical Garden | 0.0498 | 11 | 2, 3 |
| AC110 | V. pompona | Private collection | 0.0627 | 3 | 1, 3 |
| AC111 | V. phalaenopsis | Botanical Garden | 0.0274 | 2 | |
| AC112 | V. madagascariensis | Botanical Garden | 0.0348 | 2 | |
| AC113 | V. planifolia | Botanical Garden | 0.0363 | 5 | |
| AC114 | V. insignis | Botanical Garden | 0.0331 | 10 | |
| AC115 | V. dilloniana | Botanical Garden | 0.0712 | 6 | 3 |
| AC116 | V. phaeantha | Botanical Garden | 0.0404 | 11 | |
| AC117 | V. planifolia | Online vendor | 0.0349 | 5 | |
| AC118 | V. imperialis | Botanical Garden | 0.0631 | 2 | 3 |
| AC119 | V. poitaei | Online vendor | 0.0409 | 6 | 3 |
| AC120 | V. planifolia | Botanical Garden | 0.0322 | 5 | |
| AC121 | V. aphylla | Online vendor | 0.0658 | 6 | |
| AC122 | V. dilloniana | Online vendor | 0.0699 | 6 | |
| AC123 | V. appendiculata | Botanical Garden | 0.0405 | 2 | 3 |
| AC124 | V. phaeantha | Botanical Garden | 0.0433 | 11 | 1, 3 |
| AC125 | V. imperialis | Botanical Garden | 0.0530 | 2 | |
| AC126 | V. planifolia | Naturalized species | 0.0346 | 5 | 3 |
| AC127 | V. roscheri | Botanical Garden | 0.0500 | 4 | 3 |
| AC128 | V. roscheri | Botanical Garden | 0.0370 | 2 | |
| AC129 | V. phaeantha and unknown | Botanical Garden | 0.0430 | 11 | 1, 2, 3 |
| AC130 | V. planifolia x V. pompona | Online vendor | 0.0543 | 5 | 2, 3 |
| AC131 | V. dilloniana | Online vendor | 0.0686 | 6 | |
| AC132 | V. pompona x V. phaeantha | Private collection | 0.0788 | 7 | 1, 2, 3 |
| AC133 | V. planifolia | Online vendor | 0.0363 | 5 | 1, 3 |
| AC134 | V. dilloniana | Online vendor | 0.0673 | 6 | 1, 3 |
| AC135 | V. phaeantha | Native Species | 0.0373 | 11 | |
| AC136 | V. phaeantha | Native Species | 0.0368 | 11 | |
| AC137 | V. phaeantha | Native Species | 0.0393 | 11 | |
| AC138 | V. phaeantha | Native Species | 0.0409 | 11 | |
| AC139 | V. barbellata | Private collection | 0.0428 | 4 | 3 |
| AC140 | V. barbellata | Private collection | 0.0422 | 6 | 1 |
| AC141 | V. roscheri | Botanical Garden | 0.0322 | 2 | |
| AC142 | V. pompona and V. odorata | Botanical Garden | 0.0423 | 8 | 1, 2, 3 |
| AC143 | V. palmarum and unknown | Botanical Garden | 0.0332 | 10 | 2 |
| AC144 | V. imperialis | Botanical Garden | 0.0562 | 2 | |
| AC145 | V. barbellata | Native Species | 0.0517 | 4 | |
| AC146 | V. barbellata | Native Species | 0.0529 | 4 | |
| AC147 | V. phaeantha x V. pompona | Native Species | 0.0557 | 7 | 2, 3 |
| AC148 | V. barbellata and unknown | Online vendor | 0.0288 | 9 | 2 |
| AC149 | V. barbellata | Native Species | 0.0503 | 4 | |
| AC150 | V. claviculata | Online vendor | 0.0454 | 4 | |
| AC151 | V. planifolia | Private collection | 0.0390 | 5 | |
| AC152 | V. barbellata | Private collection | 0.0459 | 4 | 1 |
| AC153 | V. pompona | Botanical Garden | 0.0584 | 3 | |
| AC154 | V. griffithii | Botanical Garden | 0.0358 | 9 | |
| AC155 | V. schwackeana | Botanical Garden | 0.0315 | 8 | |
| AC156 | V. pompona | Botanical Garden | 0.0557 | 3 | |
| AC157 | V. pompona and V. phaeantha | Private collection | 0.0823 | 7 | 2 |
| AC158 | V. pompona and V. odorata | Private collection | 0.0454 | 8 | 2, 3, 5 |
| AC159 | V. pompona | Private collection | 0.0638 | 3 | 5 |
| AC160 | V. planifolia | Private collection | 0.0397 | 5 | |
| AC161 | V. aphylla and unknown | Private collection | 0.0307 | 9 | 2 |
| AC162 | V. imperialis | Private collection | 0.0768 | 2 | |
| AC164 | V. planifolia | Private collection | 0.0362 | 5 | 5 |
| AC165 | V. pompona and V. odorata | Private collection | 0.0393 | 8 | 2, 3, 5 |
| AC166 | V. pompona | Private collection | 0.0573 | 3 | |
| AC167 | V. pompona | Private collection | 0.0569 | 3 | 5 |
| AC168 | V. planifolia | Private collection | 0.0352 | 5 | 3 |
| AC169 | V. pompona | Private collection | 0.0570 | 3 | |
| AC170 | V. planifolia x V. phaeantha | Private collection | 0.0502 | 11 | 2, 3, 5 |
| AC171 | V. planifolia | Botanical Garden | 0.0357 | 5 | |
| AC172 | V. imperialis | Botanical Garden | 0.0640 | 2 | |
| AC173 | V. planifolia | Private collection | 0.0360 | 5 | 3 |
| AC174 | V. planifolia | Private collection | 0.0385 | 5 | 1, 3 |
| AC175 | V. pompona and V. odorata | Private collection | 0.0612 | 3 | 1, 2, 3 |
| AC176 | V. planifolia | Online vendor | 0.0401 | 5 | |
| AC177 | V. odorata | Online vendor | 0.0342 | 1 | 3 |
| AC178 | V. planifolia | Online vendor | 0.0381 | 5 | 1, 3 |
| AC179 | V. pompona and unknown | Online vendor | 0.0492 | 3 | 2 |
| AC180 | V. planifolia | Online vendor | 0.0401 | 5 | |
| AC181 | V. planifolia | Online vendor | 0.0361 | 5 | 1, 3 |
| AC182 | V. dilloniana and unknown | Private collection | 0.0598 | 6 | 2 |
| AC183 | V. pompona | Private collection | 0.0553 | 3 | 3 |
| AC184 | V. planifolia | Private collection | 0.0365 | 5 | 3 |
| AC185 | V. planifolia | Private collection | 0.0368 | 5 | 3 |
| AC186 | V. planifolia | Private collection | 0.0405 | 5 | 3 |
| AC187 | V. planifolia | Private collection | 0.0378 | 5 | 4 (AC173) |
| AC188 | V. planifolia | Private collection | 0.0354 | 5 | 4 (AC174) |
| AC189 | V. pompona x V. phaeantha | Private collection | 0.0781 | 7 | 2, 4 (AC132) |
| AC190 | V. pompona | Private collection | 0.0574 | 3 | 4 (AC104) |
| AC191 | V. mexicana | Native Species | 0.0375 | 9 | 3 |
| AC192 | V. mexicana | Native Species | 0.0390 | 9 | |
| AC193 | V. barbellata | Private collection | 0.0486 | 4 | 3, 5 |
| AC194 | V. planifolia x V. phaeantha | Botanical Garden | 0.0517 | 11 | 2, 3 |
| AC195 | V. planifolia | Private collection | 0.0343 | 5 | 3 |
| AC196 | V. planifolia | Private collection | 0.0331 | 5 | 3 |
| AC197 | V. planifolia | Private collection | 0.0342 | 5 | 3 |
| AC198 | V. planifolia | Private collection | 0.0342 | 5 | |
| AC199 | V. planifolia | Private collection | 0.0398 | 5 | |
| AC200 | V. pompona | Private collection | 0.0643 | 3 | |
| AC201 | V. planifolia | Online vendor | 0.0365 | 5 | |
| AC202 | V. planifolia | Private collection | 0.0397 | 5 | 3, 5 |
| AC203 | V. planifolia | Private collection | 0.0383 | 5 | 3, 5 |
| AC204 | V. hartii | Private collection | 0.0376 | 10 | 3, 5 |
| AC205 | V. x tahitensis | Private collection | 0.0389 | 5 | 3, 5 |
| AC206 | V. x tahitensis | Private collection | 0.0367 | 5 | 3, 5 |
| AC207 | V. odorata | Private collection | 0.0426 | 1 | 3, 5 |
| AC208 | V. ensifolia | Private collection | 0.0305 | 8 | 3, 5 |
| AC209 | V. planifolia | Private collection | 0.0364 | 5 | 3, 5 |
| AC210 | V. odorata | Private collection | 0.0398 | 1 | 3, 5 |
| AC211 | V. odorata and unknown | Private collection | 0.0299 | 1 | 2, 3, 5 |
| AC212 | V. ensifolia | Private collection | 0.0328 | 8 | 3, 5 |
| AC213 | V. odorata | Private collection | 0.0369 | 1 | 3, 5 |
| AC214 | V. palmarum | Private collection | 0.0507 | 12 | 3, 5 |
| AC215 | V. palmarum | Private collection | 0.0513 | 12 | 3, 5 |
| AC216 | V. palmarum | Private collection | 0.0502 | 12 | 3, 5 |
| AC217 | V. palmarum | Private collection | 0.0496 | 12 | 3, 5 |
| AC218 | V. planifolia | Private collection | 0.0346 | 5 | 4 (AC173) |
| AC219 | V. planifolia | Private collection | 0.0345 | 5 | 4 (AC106) |
Each accession was given a unique identifier from AC101 to AC219 (accession AC163 was dropped due to poor library QC results). Shown are the species assignments based on rbcL sequencing (as available) and GBS data, source of the material, calculated heterozygosity based on GBS, clustering group assignment (BIC Group), and notes. Notes are coded as follows: (1) previously misclassified accession, (2) probable hybrid based on GBS results, (3) accessions with rbcL sequence data provided in this study, (4) duplicate samples (with duplicate sample ID shown in parentheses), and (5) previously unknown species assignment. Predicted parents of hybrids are shown in place of a species assignment with maternal parent shown first when known.
A draft genome for V. planifolia
A draft genome of accession AC173 was created to assist with read mapping and SNP calling. The estimated haploid genome size and heterozygosity were 1.13 Gb and 2.32%, respectively, based in the best model fit from GenomeScope (99.53% for 77 Kmer). The Kmer distribution did not show the two clear peaks that are usually present for an allotetraploid or hybrid. The genome size estimation using Kmers delivered a value that it is half of the values of previous estimations (1 C~2.26 Gb)42.
One paired end (insert size 300 bp) and three mate pair libraries (insert sizes 5, 8 and 10 Kb) were sequenced, delivering 45.90, 56.98, 53.04, and 47.36 Gb respectively (40X of paired ends and 139X of mate pairs). SOAPdenovo2 and Minia were used to assemble the sequencing data. For all of the assemblies, the SOAPdenovo2 assembly with a Kmer of 95 (VaplaK095A02) was preferred based on its assembly stats (see Methods for more details), and was selected for GBS read alignment. Gaps were filled in VaplaK095A02 to produce assembly Vapla0.1.1, and then contaminants and scaffolds less than 200 bp were filtered producing the final assembly Vapla0.1.4. Vapla0.1.4 is a highly fragmented assembly with a total size of 1.96 Gb (contigs) and 2.20 Gb (scaffolds) in agreement with published flow cytometry genome size estimations. Statistical results are summarized in Table 2.
Table 2.
Statistical summary for contigs and scaffolds in the VaplaK095A02 and Vapla0.1.4 assemblies. Publicly available RNA-Seq data was used to test the quality of the draft genome assemblies.
| Assembly | VaplaK095A02 | Vapla0.1.4 | ||
|---|---|---|---|---|
| Assembly Statistics | Contigs | Scaffolds | Contigs | Scaffolds |
| Total assembly size (Gb) | 1.20 | 2.34 | 1.96 | 2.20 |
| Total assembled sequences | 2,955,869 | 2,115,012 | 2,250,393 | 794,547 |
| Longest sequence length (Kb) | 54.03 | 626.54 | 129.70 | 630.91 |
| Average sequence length (Kb) | 0.40 | 1.12 | 0.87 | 2.77 |
| N50 index (sequences) | 445,497 | 11,865 | 47,789 | 9,596 |
| L50 length (Kb) | 0.60 | 41.10 | 9.62 | 53.33 |
| % RNA-Seq Mapped | NA | 87.1 ± 0.4 | NA | 96.7 ± 0.8 |
| % BUSCO Completed | NA | 8.6 | NA | 79.5 |
| % BUSCO Duplicated | NA | 1.2 | NA | 32.8 |
| % BUSCO Fragmented | NA | 16.7 | NA | 5 |
Two approaches were used to evaluate the completeness of the Vapla0.1.4 assembly gene space including (1) read mapping of three RNA-seq datasets from the National Center for Biotechnology Information (differentiated flower bud SRR1171644, placental laminae in mature pods of 6 months old Vanilla SRR150937444, and leaf SRR150935644), and (2) BUSCO45 analysis (Table 2). The high percentage of the reads of the different RNA-seq datasets that map to the Vapla0.1.4 reference indicates that the assembly draft may be capturing a high percentage of the gene space (>95%).
SNP analysis
The GBS library produced 124,085,946 reads with the expected barcode and cut site overhang (99.99% of 124,089,684 total Illumina reads). The TASSEL 3 GBS pipeline identified 11,711,559 unique tags from high quality barcoded reads, of which 6,643,190 (56.72%) aligned to the VaplaK095A02 draft Vanilla genome. A total of 521,732 unfiltered SNPs were produced, which have 0.93–15.39 mean read depth and 33.24–94.21% missing rate for the 118 samples (Supplementary Table S1). 5,082 SNPs remained for downstream analyses after filtering for maximum missing rate (<30%), minor allele frequency (>10%), minimum read depth (>10), maximum read depth (<1000), maximum heterozygosity rate (<20%), and linkage disequilibrium (<0.2). The SNP analysis results are shown in Supplementary Fig. S1.
Heterozygosity
The SNP analysis included calculations of heterozygosity for each accession as shown in Table 1. Heterozygosity as calculated by vcftools–het and ranged from 0.0274 for V. phalaenopsis AC111 to 0.0823 for AC157 from a private collection that is most likely a hybrid between V. phaeantha and V. pompona. Some species had an overall higher average rate of heterozygosity including V. dilloniana, V. imperialis, and V. pompona. Conversely, low average heterozygosity was calculated for V. planifolia, V. odorata, and V. x tahitensis. The native species ranged from lowest to highest heterozygosity for V. mexicana < V. phaeantha < V. barbellata < V. dilloniana, respectively.
GBS-based diversity analysis
Genetic diversity within the collection was assessed using the 5,082 filtered GBS SNPs. A plot of the first two principal components indicated that enough variation was captured within PCA1 (25.55%) and PCA2 (18.37%) to visually differentiate species (Fig. 1). Our PCA analysis grouped the accessions within distinct clusters representing the species V. pompona, V. planifolia, V. imperialis, V. odorata, V. palmarum, V. barbellata, V. dilloniana, V. phaeantha, and V. mexicana. Other species with only a few representatives clustered with V. odorata, V. barbellata, in a miscellaneous cluster, or were entirely separated as for the V. appendiculata (AC123) accession indicating that some accessions within this collection are genetically distinct. Exceptions to these trends were identified as probable hybrids as described below. Two probable V. x tahitensis accessions were located on the PCA plot between V. planifolia and V. odorata.
Figure 1.
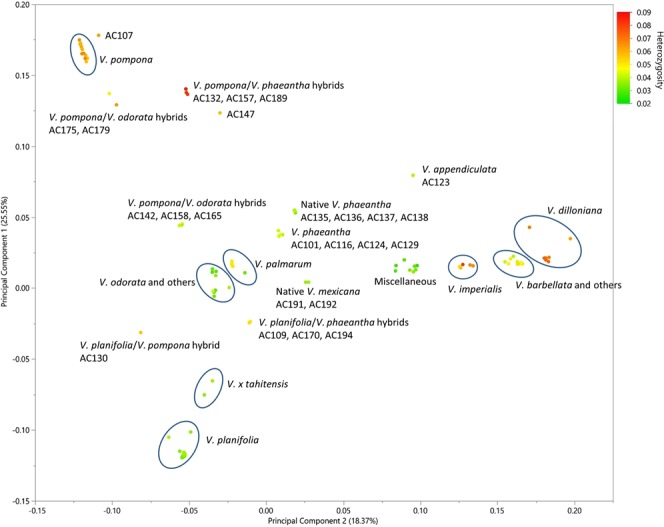
Plot of the first two principal components using 5,082 filtered SNPs. Individual accessions are shown by dots colored by heterozygosity rate with the highest heterozygosity shown in red and lowest shown in green. Species groupings are delineated by ovals surrounding groups of dots representing individual accessions.
Clustering and STRUCTURE analysis using filtered SNPs
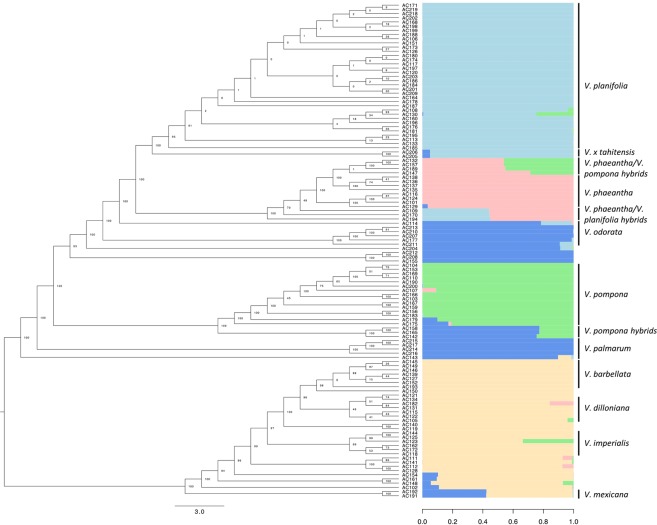
Clustering, phylogenetic, and STRUCTURE analyses46 of the filtered SNPs revealed distinct relationships among accessions (Fig. 2). Discriminant analysis of principal components DAPC using the top 40 principal components yielded evidence for 12 clusters (BIC Groups, Table 1, Supplementary Fig. S2). V. planifolia accessions had lower bootstrap support than other species and were all in cluster 5. A few accessions had higher bootstrap support for separation from the majority of V. planifolia and included AC195, AC113 (variegated accession), AC133, and AC185. V. x tahitensis accessions AC205 and AC206 are also part of the V. planifolia cluster as would be expected, while all V. odorata (five accessions) are in cluster 1. Cluster 3 was mostly V. pompona with a few potential V. pompona hybrids including AC107, AC179, and AC175 while other putative V. pompona hybrids formed cluster eight (AC158, AC165, AC142).
Figure 2.
Cladogram and genetic structure of 112 accessions and six replicates included in this study. Phylogenetic tree constructed using SNP data with bootstrap percentage for 1,000 replicates is shown. Bayesian clustering (STRUCTURE, K = 5) of 118 accessions is shown on the right portion of the figure. The x-axis quantifies cluster membership, and the y-axis represents the different accessions. The order and position of accessions on the y-axis are consistent with those in the phylogenetic tree. General species groupings are shown alongside the STRUCTURE plot. STRUCTURE colors were assigned according to a best fit of K = 5 from STURCTURE HARVESTER.
The putative hybrid accessions and native Vanilla species were included in multiple clusters. The known V. planifolia x V. pompona hybrid AC130 was part of cluster 5 with the V. planifolia accessions. Hybrids between V. phaeantha and V. pompona formed cluster 7 with a few V. pompona and V. phaeantha accessions. Native and procured V. phaeantha accessions along with a group of V. phaeantha/V. pompona hybrids formed cluster 11. Cluster 4 included the native V. barbellata species and V. claviculata, and Cluster 6 included V. dilloniana and V. poitaei. Cluster 9 included V. griffithii, V. aphylla, V siamensis, and a V. barbellata accession along with the native V. mexicana accessions.
Species identification using rbcL sequencing
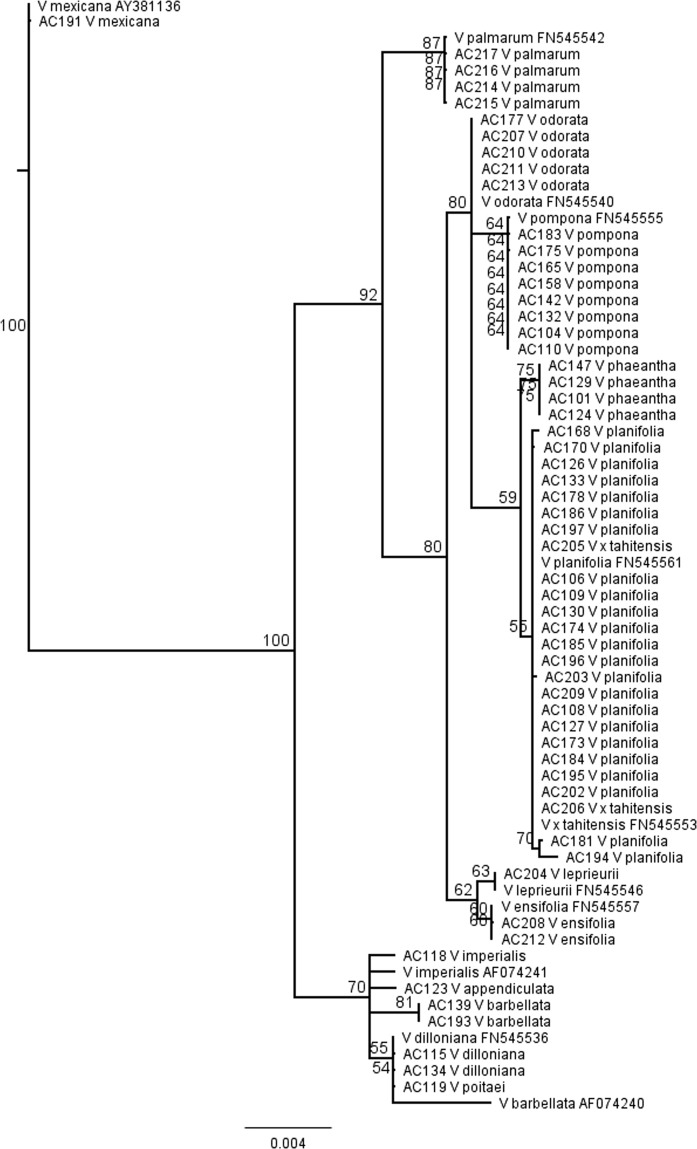
Single gene sequencing of Vanilla species is useful for identifying accessions at the species level47, because these sequences can be compared to publicly available data at bioinformatics repositories including the National Center for Biotechnology Information (ncbi.nlm.nih.gov). Single gene sequencing is also useful in the identification of maternal parents of potential hybrids, because many of the single gene sequences for genotyping Vanilla are from plastid-derived targets. Fifty-seven accessions were selected for single gene sequencing of the rbcL locus (Fig. 3). The results from this analysis are in close agreement with previously published sequences except for V. barbellata AF074240 where the reference sequence is more similar to V. dilloniana accessions than the V. barbellata accessions in this study. No rbcL reference sequences were available for V. phaeantha, V. appendiculata, or V. poitaei and were therefore not included in this analysis. GBS and rbcL sequencing results indicate that native V. mexicana is distinct from all other accessions.
Figure 3.
Phylogenetic tree based on ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) partial locus sequencing for selected accessions in the study. Included are published rbcL sequences obtained from NCBI as indicated by accessions IDs. Bootstrap values for 100 clustering replicates are shown.
Diversity within V. planifolia
V. planifolia is the major commercial species for the genus, and thus diversity within this species is especially valuable for future plant breeding research. SNPs specifically selected for V. planifolia analysis would be expected to vary from the 5,082 SNPs selected to analyze diversity among many species based on the various filtering criteria imposed. Diversity within V. planifolia was therefore analyzed separately from the other species. The results are shown for 27 accessions in Fig. 4. There were 565 SNPs identified and used for this analysis. V. planifolia accessions AC181 and AC185 are more distantly related than the other V. planifolia accessions and were excluded to improve the resolution of the remaining accessions in Fig. 4.
Figure 4.
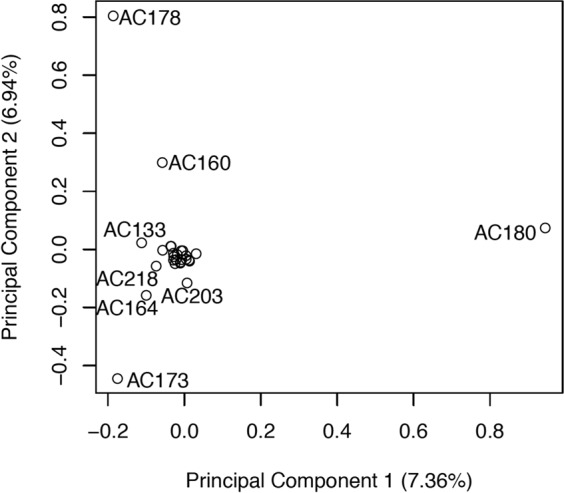
PCA plot showing diversity within 27 V. planifolia accessions using 565 filtered SNPs. Diverse accessions AC108, AC181, and AC185 were excluded to improve resolution of the remaining accessions. Distinct accessions are labeled by accession number.
Fst analysis
Fst reflects the population differentiation due to genetic structure. High Fst values indicate a considerable degree of differentiation among populations. Distinct groups were identified from Fst analysis of the Vanilla collection (Fig. 5). One group includes V. phaeantha, V. schwackeana, V. ensifolia, V. odorata, V. insignis, V. pompona, V. palmarum, and V. hartii which share moderate pairwise Fst values. A second group includes V. aphylla, V. roscheri, V. barbellata, V. claviculata, V. dilloniana, V. poitaei, V. griffithii, V. siamensis, V. imperialis, V. appendiculata, V. madagascariensis, and V. phalaenopsis which also share low to moderate pairwise Fst values. These two groups share lower Fst, indicating that lower genetic differences exist among accessions in each group. Additionally, V. planifolia and V. x tahitensis share moderate pairwise Fst (0.704), meaning that these two have somewhat dissimilar genetic structure. The phylogenetic tree and STRUCTURE analysis also supported the similar grouping of V. planifolia and V. x tahitensis.
Figure 5.
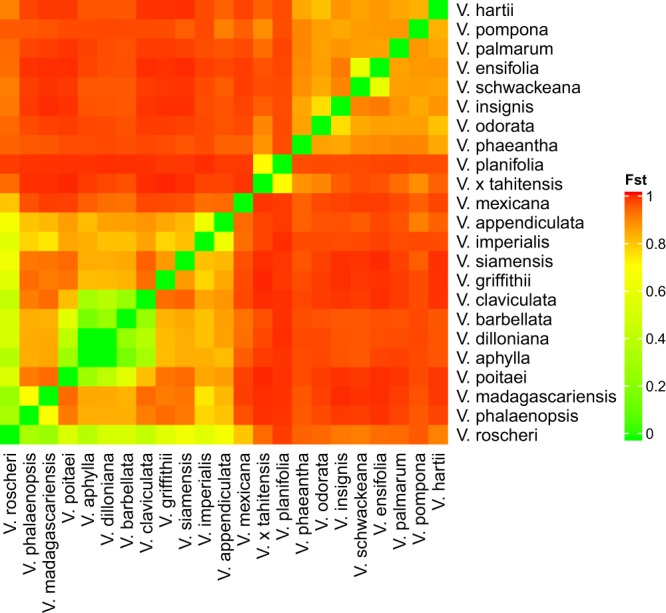
Heatmap illustrating pairwise Fst values among 23 Vanilla species. High Fst values are shown in red and low Fst values are shown in green.
Fst analysis also separated most of the leafless species by low Fst values when compared to each other and high values when compared to V. planifolia. This pattern includes accessions for V. aphylla, V. roscheri, V. barbellata, V. claviculata, V. dilloniana, and V. poitaei. The exceptions include V. claviculata compared to V. poitaei, and V. madagascariensis that is leafless yet has more moderate Fst values compared to the other leafless species.
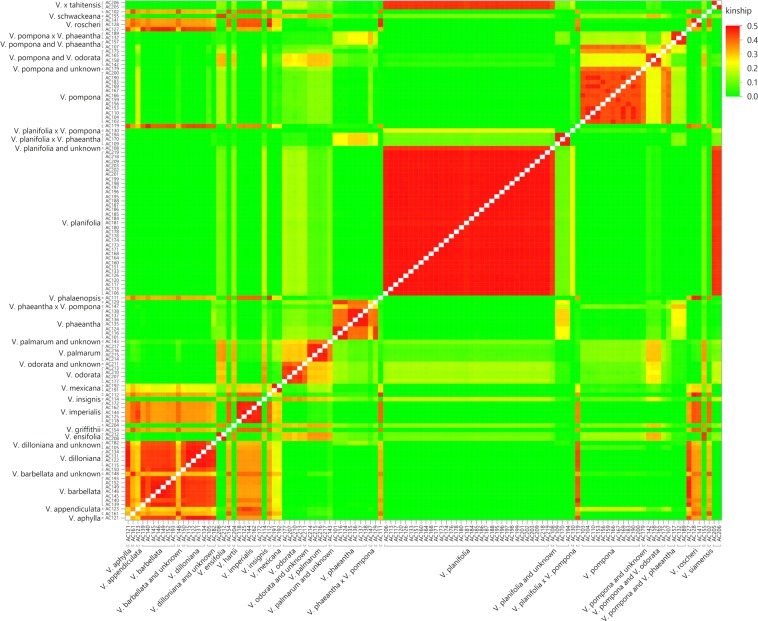
Kinship analysis
Kinship analysis was conducted to test the relationship among individuals (Fig. 6). As expected, the species with multiple accessions (for example, V. planifolia and V. pompona) showed high kinship values among individuals of their respective species. V. x tahitensis has high kinship with V. planifolia as expected, and moderate kinship with V. odorata. Probable hybrid accessions can be visually identified by species assignment and contrasting kinship values compared to the majority of accessions in that species. The most diverse species including the leafless species tend to have higher kinship values with each other than to leafy species including V. planifolia and V. pompona.
Figure 6.
Kinship heatmap of 112 accessions and six replicates in this study. Accessions are grouped by species as assigned by rbcL sequencing (as available) and by GBS results. Lower kinship values are shown in green, moderate values in yellow, and higher values are shown in red.
Identification of probable hybrids
Genomics-based analysis of the Vanilla collection enabled the identification of probable hybrids. One accession, AC130, is a V. planifolia (maternal parent) x V. pompona (paternal parent) hybrid confirmed by sequencing both the partial rbcL gene and by cloning and sequencing ITS amplicons from this accession (Supplementary Fig. S3). The hybrid nature of this accession is confirmed as shown in the STRUCTURE analysis including roughly three quarters V. planifolia alleles and one quarter V. pompona alleles.
Other potential hybrids were also identified by interpreting the combined results from the clustering and STRUCTURE analyses. These included accessions AC132 and AC157 that were received as V. odorata and V. pompona, respectively, but both contain markers for V. pompona and V. phaeantha. The maternal parent of AC132 was confirmed to be V. pompona by rbcL sequencing in agreement with the GBS results. AC189 is a duplicate sample of AC132 and the GBS results are consistent between the two samples. Both of the preceding accessions were from a single source, and could actually be cuttings from one original sample. The GBS results for AC147 provide support that this accession is a hybrid between V. phaeantha and V. pompona, but in contrast to AC132 and AC157, the maternal parent of AC147 was V. phaeantha as confirmed by rbcL sequencing. AC147 is unique as a probable hybrid because it was collected in southern Florida on protected land. Accessions AC132 and AC157 are separated on the PCA plot, and are unlikely to be from the same original clone.
AC109, AC170, and AC194 are all probable hybrids between V. planifolia and V. phaeantha. Each has V. planifolia as the maternal parent as confirmed by rbcL sequencing, are similarly located on the PCA plot, and have higher average heterozygosity (0.051) than V. planifolia (0.036). Each of these three accessions came from a different source (one private collector and two different botanical gardens). V. pompona was shown as the maternal parent of putative hybrids AC142 (sourced from a botanical garden), AC158 and AC165 (private collection), and AC175 (a separate private collection) yet all showed hybrid characteristics from the STRUCTURE analysis potentially with V. odorata in their ancestry. AC175 was part of cluster 3 while AC142, AC158, and AC165 formed cluster 8 with V. ensifolia and V. hartii. All potential hybrids are noted in Table 1.
Misclassified accessions
There were 15 obviously misclassified accessions in this study, but only species with multiple accessions could be used to create consensus species assignments based on GBS and single gene sequencing. AC140 and AC152 were received as V. dilloniana, but are actually V. barbellata. AC181, AC133, and AC178 were received as V. imperialis, V. mexicana, and V. pompona, respectively, and are V. planifolia. AC132, AC110, AC142, and AC175 were received as V. odorata, V. planifolia, V. planifolia, and V. planifolia, respectively, and are V. pompona. AC134 was received as V. phaeantha and is V. dilloniana. AC124 and AC129 were received from two different botanical gardens as V. x tahitensis and are actually both V. phaeantha. AC106, AC108, and AC175 were also received as V. x tahitensis, but in the absence of V. odorata alleles were reclassified as V. planifolia according to their genotypes.
Other accessions may be misclassified, but lack sufficient supporting information to confidently reassign a species designation. For example, AC123 that was received as V. appendiculata, but single gene sequencing and GBS results both closely match V. imperialis. This could be biologically relevant, or an incorrect species assignment. Overall, the misclassified accessions were not limited to a single germplasm source, but were received from multiple botanical gardens, online vendors, and private collections.
Species-specific SNPs
Species-specific, diagnostic molecular markers would be advantageous for quickly identifying species in new collections, and also for confirming hybrid progeny when breeding. The greatest limitations to validating species-specific SNPs include sampling enough diversity within a species and across relevant species to obtain high confidence for marker specificity. Towards developing species-specific SNPs, the 521,732 SNPs from this study were screened for those that could be species-specific. The results are reported in Supplementary Data S1. V. planifolia had 1,611 species-specific SNPs, and V. pompona had 3,230. The other species had values ranging from 227 for V. aphylla to 6,187 identified for V. palmarum.
Discussion
The primary objective of this study was to examine the utility of genomics-based diversity analysis to characterize a living Vanilla collection. The collection included 112 accessions from 23 species obtained from botanical gardens, private collections, online vendors, and collected from natural areas in southern Florida as part of conservation research. We developed a draft Vanilla genome as a reference for GBS analysis of the living collection. GBS yielded 5,082 filtered SNPs resulting in the largest genomics dataset for Vanilla to date, and the first application of genomics-based diversity analysis in this genus. The increased resolution among accessions due to increased marker numbers and reduced cost per data point accelerated the discovery of potential hybrids, identified misclassified accessions, and demonstrated the suitability of these methods to analyze diversity across and within Vanilla species.
A draft Vanilla genome for diversity analysis
We created a draft V. planifolia draft genome assembly to facilitate read mapping and SNP calling. The assembly proved its utility for the development of genetic markers and the study of the population structure in a biodiversity panel in spite of being highly fragmented. The V. planifolia genome size was previously reported as 2.26 +/− 0.05 Gb using flow cytometry48, being twice the genome size that was estimated by Kmer distributions. This could indicate that either previous flow cytometry values overestimated genome size, or that V. planifolia is an autopolyploid (2n = 4x = 24) with a haploid genome size of ~2.26 Gb. Using flow cytometry to estimate the genome size of orchid species is especially challenging due to endoreduplication48.
Leveraging the draft genome simplified SNP calling, but also presented a few limitations. For example, the number of mapped reads is expected to decrease as diversity increases possibly leading to the exclusion of SNPs that could be relevant to studies with other Vanilla species. Still, having obtained over 5,000 filtered SNPs for diversity analysis across the 23 species was more than sufficient to meet the study objectives. In the future, the results from the draft genome could be further developed to generate a V. planifolia reference genome that would capture a greater proportion of the genic space, and would enable gene discovery.
Diversity analysis
The level of similarity among V. planifolia accessions, even those collected from disparate sources, is not surprising considering the general consensus that genetic diversity in V. planifolia is limited. This is probably due to the ease of propagation by cuttings and the worldwide distribution of a few foundational clones. Still, evidence of diversity among V. planifolia accessions was found in this study and includes a few accessions with only limited source information. All results from rbcL sequencing matched expectations from the published literature except for misclassified accessions29. Some V. planifolia accessions were genetically distinct. For example, AC133 was originally labeled as V. mexicana, but was confirmed to be V. planifolia by rbcL sequencing and was distinguished from the majority of the V. planifolia accessions based on the GBS results. AC185 was obtained from a commercial grower and described as a plant with unique morphology. The GBS results support the hypothesis that this accession is genetically distinct compared to other V. planifolia accessions including other types from the same source (AC184 and AC186). These results suggest that expanded sampling using genomics-based analysis methods could uncover hidden diversity even within V. planifolia. Uncovering genomics-based diversity could be a particularly useful tool when selecting parents for a Vanilla breeding program.
V. x tahitensis is a commercial species that can be sold with the “vanilla bean” label in the US and European markets. While V. planifolia commands much more market share, V. x tahitensis has a unique flavor profile and interesting agronomic characteristics including non-dehiscence49,50. It is therefore an important species within the genus and warrants additional genomics-based research. Previous work has shown a close genetic relationship between the V. planifolia and V. x tahitensis4,16,18,33, but definitive data (especially at the genomics level) supporting the origins of V. x tahitensis is still lacking. V. x tahitensis is not easily differentiated from V. planifolia using single gene sequencing as the two are closely related, but use of genomics-based molecular markers should be able to efficiently identify alleles from both V. planifolia and V. odorata in V. x tahitensis.
The V. planifolia and V. odorata hybrid origin hypothesis of V. x tahitensis could easily be tested using a genomics-based approach33. Accessions AC205 and AC206 are probably true V. x tahitensis clones based on PCS, STRUCTURE, and the current hybrid origin theory. The STRUCTURE results for these accessions show a high level of V. planifolia ancestry and only a low level of V. odorata ancestry. Both of these accessions were donated from a private collection as unknown species. The accessions were artificially propagated from material most likely collected in Belize where V. x tahitensis could be endemic51. Further genomics research is needed to characterize additional V. x tahitensis accessions in order to identify its origins and unlock the traits that make this hybrid unique from agronomic and sensory perspectives.
Native Vanilla species
The four native Florida Vanilla species V. phaeantha, V. barbellata, V. dilloniana, and V. mexicana are all endangered. Conservation of these native species including collection, propagation, and reintroduction can be aided by diversity analysis. The native V. phaeantha species formed its own branch when clustering using the GBS data. This suggests some level of genetic diversity compared to V. phaeantha from other geographies. The identification of a potential hybrid between V. phaeantha and V. pompona (AC147) in a natural setting was unexpected. There are no known established populations of V. pompona in southern Florida, but V. pompona is common in private collections. The origin of this hybrid could exemplify the ease with which Vanilla species hybridize and establish in natural areas. This would suggest that hybrids like V. x tahitensis and AC147 could be more common than currently presumed.
V. barbellata and V. dilloniana are difficult to identify morphologically due to their shared leafless morphology and overlapping growing regions. GBS-based species identification could be a powerful tool for distinguishing these species as shown in this study. In contrast, V. mexicana appears to be morphologically distinct among Vanilla species, and also by single gene and GBS analysis. Future conservation work for each of these species should focus on identifying and maintaining as much diversity as can be captured prior to habitat destruction. The results from this study show the potential of GBS to support conservation efforts for these species.
Analysis of hybrids and misidentified accessions
We identified 21 potential hybrids out of 112 total accessions (18.8%). The actual occurrence of hybrid accessions is probably underestimated in other collections, because the majority of common molecular markers used in Vanilla are from plastid-derived genes. This common strategy would only identify the maternal parent of a potential hybrid. The value of the GBS approach to characterize diversity in a Vanilla collection has also been shown to putatively identify the paternal parent of hybrid accessions more efficiently and cost effectively than other methods.
Approximately 13% of accessions in this study were confirmed to be misidentified based on consensus, and this is probably an underestimate. The consensus approach can identify the species of unknown accessions as long as verified species are included in the database. Some species in the collection are represented by one or a few accessions, thereby reducing confidence in assigning and re-assigning species identities. Missing data can also cause artifacts in the clustering analysis. Therefore, the results for accessions with limited sample numbers should be cautiously interpreted. The genomics-based characterization of Vanilla collections, including those at botanical gardens, would greatly benefit the Vanilla community, especially when sharing germplasm.
Future work
Species-specific molecular markers would be useful for identifying unknown Vanilla accessions, and for confirming parentage of interspecific hybrid progeny. Additionally, a fewer number of informative SNPs could be used to differentiate accessions at reduced cost. Unknown and misclassified accessions are common in Vanilla, because species can look very similar and morphological traits (including leaf shape) can vary during development and across environments. Flower morphology often supports a species designation, but flowering can be an infrequent and unreliable event. One limitation of our approach for identifying diagnostic markers relies on the use of the V. planifolia draft genome for read mapping and SNP calling. While this approach has several advantages, it is expected that diverse species will have fewer mapped reads and therefore fewer SNPs called. This could favor SNP calls across conserved loci and result in the identification of fewer species-specific markers for distantly related accessions. Ultimately, species-specific SNPs like those included in Supplementary Data S1 will have to be identified and validated based on specific research objectives.
In conclusion, the development of a draft V. planifolia genome and genotyping a living collection of Vanilla species enabled diversity analysis, hybrid identification, and the designation of species assignments. The benefits of using GBS SNP data compared to other molecular markers include transferability between labs, ease of marker development, reduced genotyping costs, and increased reproducibility. Results from this work can easily be expanded to other Vanilla collections. Anticipated benefits would support breeding programs and the creation of improved Vanilla cultivars needed to meet cyclical supply challenges and prepare Vanilla to enter a modern era of cultivar development.
Methods
Vanilla plant accessions
Vanilla cuttings were obtained from domestic botanical gardens, local orchid enthusiasts, online vendors, and international collaborators to establish a living collection at the University of Florida’s Tropical Research and Education Center in Homestead, Florida. The collection comprises 112 accessions from 23 species, and also included accessions with unknown species designations. The list of Vanilla accessions used in this study is summarized in Table 1.
Native Florida Vanilla species were collected from southern Florida county and state parks under research permits working with park biologists following state regulations. V. barbellata and V. phaeantha were propagated by cuttings. V. mexicana leaves were sampled directly from different individuals in the natural setting, because this species is difficult to propagate by cuttings. V. dilloniana is extremely rare if not extinct in natural areas, and was therefore obtained through online vendors and local orchid enthusiasts. Collection sites cannot be disclosed due to permitting restrictions instituted to protect endangered species.
Plant maintenance
Cuttings were established on raised beds with 16 cm of mulch in a shade house under 50% filtered light. Beds were 18 m long and 1.8 m wide. Individual trellises were spaced 1.2 m apart and included a central post, a column of wire mesh as a climbing substrate, and horizontal wooden post for vine support at 1.2 m from the ground. Supplemental irrigation was supplied through drip lines during the dry season. Average temperatures in Homestead, Florida vary between 18 °C in the winter months to 27 °C in the summer. Average rainfall is ~4.4 cm per month in the dry season (November to March), and up to ~20.6 cm on average per month during the peak of the rainy season (June to September) (Florida Automated Weather Network, http://fawn.ifas.ufl.edu)52.
DNA extraction
DNA extraction was performed using a modified CTAB method. 1 gram (fresh weight) of mature leaf tissue was ground to a fine power in liquid nitrogen, mixed with 3 ml of DNA extraction buffer (2% CTAB, 1.4 M NaCl, 100 mM Tris, 2 mM EDTA, 1% β-mercaptoethanol, pH 8.0) in a 15 ml conical tube, vortexed to completely disperse the tissue, and then incubated at 65 °C for 10 minutes. Samples were then cooled to room temperature for 3–5 minutes and 3 ml chloroform:isoamyl alcohol (24:1 v/v) was added to each sample, vortexed, and centrifuged for 10 minutes at 18,000 × g. The supernatant was transferred to a fresh tube to precipitate DNA using two volumes of 95% ethanol. Samples were centrifuged at 10,000 × g for 10 minutes to pellet DNA, aspirated, washed once with one ml of 70% ethanol, centrifuged at 10,000 × g for 2 minutes, aspirated, and finally resuspended in 75 µl molecular biology grade water.
A draft V. planifolia genome
A draft V. planifolia genome was created to facilitate read mapping and SNP calling. V. planifolia accession AC173 was selected for sequencing, and is a clone of the same accession used for a previously published transcriptome44. DNA was extracted as above, and submitted to the Duke GCB Sequencing Core for library preparation, and sequenced on two lanes of an Illumina HiSeq 4000 (paired-end 150 bp). Libraries with four insert sizes including mate-pairs (300 bp, 5–7 kb, 8–10 kb, and 10–12 kb) were used to create the draft genome. Adapters, low quality extremes (qscore < 30) and reads shorter than 50 bp were removed using Fastq-mcf from the Ea-utils package version 1.1.2-53753. PCR duplications were filtered using Prinseq version 0.20.454. Reads were corrected using Musket version 1.155. Genome size and heterozygosity were estimated analyzing the Kmer distribution56 using Jellyfish version 2.2.657 and GenomeScope (web accessed on 2018-02-07)58 for the Kmers 31, 55, and 77. The reads were assembled using two different assemblers: Minia version 3, git commit efef7c759, and SOAPdenovo2 version r24060. The assemblers were run with the following list of different kmers: 17, 21, 25, 31, 39, 47, 51, 53, 55, 57, 59, 61, 63, 71, 79, 87, 95, 103, 119 and 123. The assemblies were evaluated based on minimum difference between assembly size and genome estimated size, longest contig sequence, N90/L90, and N50/L50. The assembly with the highest rank across these parameters was selected for scaffolding and gap filling with the SOAPdenovo2 package version r24060.
The evaluation of the completeness of the assembly was performed using two approaches. First, three different RNA-seq datasets were downloaded from the NCBI SRA database including (1) SRR1171644: differentiated flower bud, (2) SRR1509374: placental laminae in six weeks initial pods, and (3) SRR1509356: leaves. Reads were processed with Fastq-mcf as described previously. Reads were mapped to the assembly version VaplaK095A02 using Hisat2 version 2.1.061 with the default parameters. Second, BUSCO version 3.0.245 was run on the assembly VaplaK095A02 with the default parameters.
Assemblies were run on the Cascades server at the Advance Research Computer center (https://www.arc.vt.edu/computing/cascades/) using a Largemem node. Assembly gene space evaluations were run on a Ubuntu server (Linux 4.4.0-97) with 64 threads, 128 Gb of RAM memory and 2 Tb of hard drive.
Sequencing the Vanilla collection
Library preparation and sequencing for Genotyping-By-Sequencing (GBS) was performed at the University of Minnesota Genomics Center on a NextSeq 1 × 150 bp using 300–744 bp size selection with dual restriction enzymes BamHI and NsiI. A pilot study using eight Vanilla accessions found that 1 M reads per sample was sufficient to obtain an informative number of SNPs (data not shown). Therefore, 1 M reads per sample was selected for the 118 samples in this study including 112 accessions and 6 duplicates as sequencing controls.
SNP calling
The Tassel 3 GBS pipeline62 was used to call SNPs from the sequenced GBS library using a reference draft V. planifolia genome. Quality filtering was performed primarily using build-in functions in VCFtools63. The minor allele frequency filter was set to 0.1 and minimum locus coverage was set to retain SNPs that are covered in at least 70% of the individuals. The minimum read depth and maximum read depth were set to 10 and 1000, respectively. The heterozygosity rate was set to 0.2 to eliminate SNPs with high amount of heterozygous calls resulting from alignment of paralogs and duplicate sequences. The SNPs only existing in one species were identified as species-specific SNPs from the total 521,732 SNPs.
Phylogenetic analysis
The filtered VCF files were converted to Phylip format by concatenating the SNPs with PGDSpider v.2.0.9.064 with IUPAC ambiguity codes for polymorphic data. The maximum likelihood (ML) phylogenetic tree with 1000 rapid bootstrap inference was constructed by using RAxML v.8.2.1265. The analysis was run using an ascertainment bias correction (ASC) for the data set containing only concatenated informative SNP positions, and a general time-reversible substitution model accounting for among-site rate heterogeneity (ASC_GTRGAMMA model). The RAxML results were graphically visualized in FigTree v.1.4.3 (downloaded from http://tree.bio.ed.ac.uk/software/figtree/).
Population STRUCTURE
The population structure was investigated to identify clusters of genetically related individuals using the Bayesian clustering method implemented in STRUCTURE, v.2.3.446. Ten independent STRUCTURE runs were performed for each of K = 2–16 (K = number of genetic clusters) with a burn-in of 10000 and 20000 iterations. The number of clusters that best fit the observed genotype data was inferred by examining the average and standard deviation (SD) of the natural log probability of each model66 using the online version of STRUCTURE HARVESTER67. Additionally, the R package SNPRelate68 was used to perform principal component analysis (PCA) and the R package adegenet69 was used to perform discriminant analysis of principal components (DAPC). Loci were trimmed to linkage disequilibrium (LD < 0.2) using the command “snpgdsLDpruning”. DAPC was performed on all individual genotypes by using the successive K-means approach, implemented by the find.clusters function, to identify the optimal number of groups based on Bayesian Information Criterion (BIC).
To investigate the diversity of V. planifolia further, the V. planifolia only PCA was performed on 27 accessions where three diverse V. planifolia lines (AC108, AC181, and AC185) identified from structure analysis were excluded. Pairwise Fst were calculated with the Weir and Cockerham formula70 with the SNPRelate package68.
rbcL sequencing
Partial sequencing of the rbcL gene26 was preferentially used to confirm species identifications of selected accessions and to identify the maternal parent of probable hybrids. Primers rbcL forward 5′ CTTCACAAGCAGCAGCTAGTTC 3′ and rbcL reverse 5′ ATGTCACCACAAACAGAAAC 3′ were used to amplify an ~1,300 bp fragment of the rbcL gene in 25 µl reactions using Phusion High-Fidelity polymerase (F531, Thermo Scientific) according to the manufacturer’s instructions. PCR amplification was conducted using an Applied Biosystems SimpliAmp Thermal Cycler with the following program: initial denaturation 5 minutes at 95 °C followed by 30 amplification cycles of 30 seconds at 95 °C, 30 seconds at 55 °C, and 60 seconds at 72 °C, with a final 10 minute extension at 72 °C. Sanger Sequencing was conducted at GENEWIZ (GENEWIZ, South Plainfield, NJ) using rbcL forward as the sequencing primer. Sequences were verified against published Vanilla rbcL sequences from NCBI (ncbi.nlm.nih.gov) using the default settings for MUSCLE alignment71 and Geneious Tree Builder within the Geneious Software package (Geneious version 11.1, Newark, NJ). Selected NCBI sequences included V. mexicana (AY381136), V. dilloniana (FN545536), V. barbellata (AF074240), V. imperialis (AF074241), V. palmarum (FN545542), V. leprieurii (FN545546), V. ensifolia (FN545557), V. x tahitensis (FN545553), V. planifolia (FN545561), V. pompona (FN545555), and V. odorata (FN545540).
Accession AC130 was suspected of being a V. planifolia x V. pompona hybrid based on the prevalence of this genotype where it was sourced in Costa Rica37. Its hybrid status was confirmed by PCR amplification (as above), cloning (TOPO TA Cloning Kit, Thermo Fisher), and sequencing (as above) the ITS partial locus using forward primer 5′ TATGCTTAAAYTCAGCGGGT 3′ and reverse 5′ AACAAGGTTTCCGTAGGTGA 3′ with T7 as the sequencing primer34,37.
Supplementary information
Acknowledgements
Funding for this research was provided by the University of Florida Dean for Research. We also acknowledge Advanced Research Computing (ARC) for use of the CASCADES supercomputing resource at Virginia Tech.
Author Contributions
E.B. and A.C. conceived and designed the study with critical input provided by Y.H., M.R. and A.B. Y.H. and A.C. wrote the main manuscript text. A.B. created the genome assembly and related analyses. Y.H. and M.R. conducted the SNP analyses. M.B. was responsible for DNA extractions and plant curation and maintenance. All authors revised and reviewed the final manuscript.
Data Availability
rbcL and ITS sequences for the accessions in this study are deposited at NCBI under the Genbank IDs listed in Supplementary Table S3. The Whole Genome Shotgun project including raw reads and the draft genome have been deposited at DDBJ/ENA/GenBank under BioProject PRJNA507095. The genome version described in this paper is SDXO01000000. The GBS datasets are deposited at NCBI under BioProject PRJNA507246.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elias Bassil, Email: ebassil@ufl.edu.
Alan H. Chambers, Email: ac@ufl.edu
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40144-1.
References
- 1.Gallage, N. J. & Møller, B. L. Vanilla: The most polular flavour in Biotechnology of Natural Products (eds Schwab, W., Lange, B. M. & Wüst, M.) 3–24 (Springer, 2018).
- 2.Gallage NJ, Møller BL. Vanillin–bioconversion and bioengineering of the most popular plant flavor and its de novo biosynthesis in the vanilla orchid. Mol. Plant. 2015;8:40–57. doi: 10.1016/j.molp.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Bruman H. The culture history of Mexican vanilla. Hisp. Amer. Hist.Rev. 1948;28:360–376. doi: 10.2307/2507753. [DOI] [Google Scholar]
- 4.Lubinsky P, Bory S, Hernandez J, Kim SC, Gomez-Pompa A. Origins and dispersal of cultivated vanilla (Vanilla planifolia jacks. [Orchidaceae]) Econ. Bot. 2008;62:127–138. doi: 10.1007/s12231-008-9014-y. [DOI] [Google Scholar]
- 5.Fouche JG, Jouve L. Vanilla planifolia: history, botany and culture in Reunion island. Agronomie. 1999;19:689–703. doi: 10.1051/agro:19990804. [DOI] [Google Scholar]
- 6.Childers, N. F. Vanilla culture in Puerto Rico. Vol. 28 (US Department ofAgriculture, 1948).
- 7.Berenstein N. Making a global sensation: Vanilla flavor, synthetic chemistry, and the meanings of purity. Hist. Sci. 2016;54:399–424. doi: 10.1177/0073275316681802. [DOI] [PubMed] [Google Scholar]
- 8.Childers, N., Cibes, H. & Hernandez-Medina, E. Vanilla-the orchid of commerce in The Orchids (ed. Withner, C.) 477–508 (The Ronald Press Company, 1959).
- 9.Divakaran, M., Nirmal-Babu, K. & Grisoni, M. Biotechnological applications in vanilla in Vanilla (eds Odoux, E. and Grisoni, M.) 51–73 (CRC Press, 2010).
- 10.FAO. Vanilla: Post-harvest Operations (2009).
- 11.Pilling, D. The real price of Madagascar’s vanilla boom. FInancial Times, https://www.ft.com/content/02042190-65bc-11e8-90c2-9563a0613e56 (June 5th, 2018).
- 12.Bory S, Grisoni M, Duval MF, Besse P. Biodiversity and preservation of vanilla: present state of knowledge. Genet. Resourc. Crop Evol. 2008;55:551–571. doi: 10.1007/s10722-007-9260-3. [DOI] [Google Scholar]
- 13.Conter, F. E. Vanilla cultivation in Hawaii. Press bulletin (Hawaii Agricultural Experiment Station); no. 6 (1903).
- 14.Chambers, A. H. Establishing Vanilla Production and a Vanilla Breeding Program in the Southern United States. In Handbook of Vanilla Science and Technology (eds Havkin-Frenkel, D. & Belanger F. C.) 165–180 (John Wileys & Sons Ltd, 2018).
- 15.Nielsen LR, Siegismund HR. Interspecific differentiation and hybridization in Vanilla species (Orchidaceae) Heredity. 1999;83:560–567. doi: 10.1038/sj.hdy.6885880. [DOI] [PubMed] [Google Scholar]
- 16.Besse P, et al. RAPD genetic diversity in cultivated vanilla: Vanilla planifolia, and relationships with V-tahitensis and V-pompona. Plant Sci. 2004;167:379–385. doi: 10.1016/j.plantsci.2004.04.007. [DOI] [Google Scholar]
- 17.Minoo D, et al. Genetic variations and interrelationships in Vanilla planifolia and few related species as expressed by RAPD polymorphism. Genet. Resourc. Crop Evol. 2008;55:459–470. doi: 10.1007/s10722-007-9252-3. [DOI] [Google Scholar]
- 18.Schlüter PM, Arenas MAS, Harris SA. Genetic variation in Vanilla planifolia (Orchidaceae) Econ. Bot. 2007;61:328. doi: 10.1663/0013-0001(2007)61[328:GVIVPO]2.0.CO;2. [DOI] [Google Scholar]
- 19.Bory S, et al. Patterns of introduction and diversification of Vanilla planifolia (Orchidaceae) in Reunion Island (Indian Ocean) Am. J. Bot. 2008;95:805–815. doi: 10.3732/ajb.2007332. [DOI] [PubMed] [Google Scholar]
- 20.Lepers-Andrzejewski S, Causse S, Caromel B, Wong M, Dron M. Genetic linkage map and diversity analysis of Tahitian vanilla (Vanilla × tahitensis, Orchidaceae) Crop Sci. 2012;52:795–806. [Google Scholar]
- 21.Ramos-Castellá AL, et al. Evaluation of molecular variability in germplasm of vanilla (Vanilla planifolia G. Jackson in Andrews) in Southeast Mexico: implications for genetic improvement and conservation. Plant Genet. Resourc. 2017;15:310–320. doi: 10.1017/S1479262115000660. [DOI] [Google Scholar]
- 22.Bory S, et al. Development of microsatellite markers in cultivated vanilla: Polymorphism and transferability to other vanilla species. Sci. Hort. 2008;115:420–425. doi: 10.1016/j.scienta.2007.10.020. [DOI] [Google Scholar]
- 23.Perez VB, et al. Molecular and microclimatic characterization of two plantations of Vanilla planifolia (Jacks ex Andrews) with divergent backgrounds of premature fruit abortion. Sci. Hort. 2016;212:240–250. doi: 10.1016/j.scienta.2016.10.002. [DOI] [Google Scholar]
- 24.Gigant, R. L. et al. Microsatellite markers confirm self-pollination and autogamy in wild populations of Vanilla mexicana Mill.(syn. V. inodora) (Orchidaceae) in the Island of Guadeloupe in Microsatellite Markers (ed. Abdurakhmonov, I.) 529–592 (InTech, 2016).
- 25.Soto Arenas MA, Dressler RL. A revision of the Mexican and Central American species of Vanilla plumier ex Miller with a characterization of their ITS region of the nuclear ribosomal DNA. Lank. Inter. J. Orchid. 2010;9:285–354. [Google Scholar]
- 26.Cameron KM. Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol. Phyl. Evol. 2004;31:1157–1180. doi: 10.1016/j.ympev.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Bouetard A, et al. Evidence of transoceanic dispersion of the genus Vanilla based on plastid DNA phylogenetic analysis. Mol. Phylo. Evol. 2010;55:621–630. doi: 10.1016/j.ympev.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 28.Cameron KM. & Carmen Molina, M. Photosystem II gene sequences of psbB and psbC clarify the phylogenetic position of Vanilla (Vanilloideae, Orchidaceae) Cladistics. 2006;22:239–248. doi: 10.1111/j.1096-0031.2006.00102.x. [DOI] [Google Scholar]
- 29.Cameron KM, et al. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot. 1999;86:208–224. doi: 10.2307/2656938. [DOI] [PubMed] [Google Scholar]
- 30.Divakaran M, Babu KN, Ravindran PN, Peter KV. Interspecific hybridization in vanilla and molecular characterization of hybrids and selfed progenies using RAPD and AFLP markers. Sci. Hort. 2006;108:414–422. doi: 10.1016/j.scienta.2006.02.018. [DOI] [Google Scholar]
- 31.Verma PC, et al. The extent of genetic diversity among Vanilla species: comparative results for RAPD and ISSR. Ind. Crop Prod. 2009;29:581–589. doi: 10.1016/j.indcrop.2008.11.006. [DOI] [Google Scholar]
- 32.Sreedhar R, Venkatachalam L, Roohie K, Bhagyalakshmi N. Molecular analyses of Vanilla planifolia cultivated in India using RAPD and ISSR markers. Orchid Sci. Biotech. 2007;1:29–33. [Google Scholar]
- 33.Lubinsky P, et al. Neotropical roots of a Polynesian spice: the hybrid origin of Tahitian vanilla, Vanilla tahitensis (Orchidaceae) Amer. J. Bot. 2008;95:1040–1047. doi: 10.3732/ajb.0800067. [DOI] [PubMed] [Google Scholar]
- 34.Cameron KM. On the value of nuclear and mitochondrial gene sequences for reconstructing the phylogeny of vanilloid orchids (Vanilloideae, Orchidaceae) Ann. Bot. 2009;104:377–385. doi: 10.1093/aob/mcp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villanueva-Viramontes S, et al. Wild Vanilla planifolia and its relatives in the Mexican Yucatan Peninsula: Systematic analyses with ISSR and ITS. Bot. Sci. 2017;95:169–187. doi: 10.17129/botsci.668. [DOI] [Google Scholar]
- 36.Nissar VM, Hrideek T, Kuruvilla K, Madhusoodanan K, Thomas J. Studies on pollination, inter specific hybridization and fruit development in vanilla. J. Plant. Crop. 2006;34:167. [Google Scholar]
- 37.Belanger, F. C. & Havkin-Frenkel, D. Molecular analysis of a Vanilla hybrid cultivated in Costa Rica in(eds In Handbook of Vanilla Science and Technology (eds Havkin-Frenkel, D. & Belanger F. C.) 256–265 (John Wileys & Sons Ltd, 2018).
- 38.Nielsen LR. Natural hybridization between Vanilla claviculata (W. Wright) Sw. and V. barbellata Rchb. f.(Orchidaceae): genetic, morphological, and pollination experimental data. Bot. J. Linn. Soc. 2000;133:285–302. doi: 10.1111/j.1095-8339.2000.tb01547.x. [DOI] [Google Scholar]
- 39.Soto Arenas, M. A. Filogeografía y recursos genéticos de las vainillas de México. Instituto Chinoin AC. Informe final SNIB-Conabio, proyecto J101 (1999).
- 40.Lubinsky, P. Conservation of wild vanilla in First international congress on the future of Vanilla business. Princeton N.J. Novemeber 11–12 (2003).
- 41.Ecott, T. Vanilla: Travels in search of the Luscious Substance (Penguin UK, 2005).
- 42.Correll DS. Vanilla-Its botany, history, cultivation and economic import. Econ. Bot. 1953;7:291–358. doi: 10.1007/BF02930810. [DOI] [Google Scholar]
- 43.Bouriquet, G. Le vanillier et la vanille dans le monde (Paul Lechevalier, 1954).
- 44.Rao X, et al. A deep transcriptomic analysis of pod development in the vanilla orchid (Vanilla planifolia) Bmc Genom. 2014;15:964. doi: 10.1186/1471-2164-15-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 46.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cameron, K. Vanilla orchids: Natural history and cultivation (Timber Press, 2012).
- 48.Travnicek P, et al. Challenges of flow-cytometric estimation of nuclear genome size in orchids, a plant group with both whole-genome and progressively partial endoreplication. Cytom Part A. 2015;87a:958–966. doi: 10.1002/cyto.a.22681. [DOI] [PubMed] [Google Scholar]
- 49.Lapeyre-Montes, F., Conejero, G., Verdeil, J.-L. & Odoux, E. Anatomy and biochemistry of vanilla bean development in Vanilla Medicinal and Aromatic Plants - Industrial Profiles (eds Odoux, E. & Grisoni, M.) 149–172 (CRC Press, 2010).
- 50.Brunschwig, C., Collard, F.X., Lepers-Andrzejewski, S. & Raharivelomanana, P. Tahitian Vanilla (Vanilla × tahitensis): A Vanilla Species with unique features in Active Ingredients from Aromatic and Medicinal Plants (ed. El-Shemy, H.) Ch. 3; 10.5772/66621 (InTech, 2017).
- 51.Gretzinger, N. & Dean, D. Vanilla production in the context of culture, economics, and ecology of Belize in Handbook of Vanilla Science and Technology, (eds Havkin-Frenkel, D. & Belanger, F. C.) 50–68 (John Wileys & Sons Ltd, 2018).
- 52.Lusher WR, Jackson JL, Morgan KT. The Florida automated weather network: ten years of providing weather information to Florida growers. Proc. Florida St. Hort. Soc. 2008;121:69–74. [Google Scholar]
- 53.Aronesty E. Comparison of sequencing utility programs. The Open Bioinformatics Journal. 2013;7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 54.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu YC, Schroder J, Schmidt B. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics. 2013;29:308–315. doi: 10.1093/bioinformatics/bts690. [DOI] [PubMed] [Google Scholar]
- 56.Li RQ, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vurture GW, et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chikhi R, Rizk G. Space-efficient and exact de Bruijn graph representation based on a Bloom filter. Algorithm Mol. Biol. 2013;8:22. doi: 10.1186/1748-7188-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo RB, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217x-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, Landmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Meth. 2015;12:357–U121. doi: 10.1038/Nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glaubitz JC, et al. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. Plos One. 2014;9:e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lischer HEL, Excoffier L. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics. 2012;28:298–299. doi: 10.1093/bioinformatics/btr642. [DOI] [PubMed] [Google Scholar]
- 65.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 67.Earl DA, Vonholdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resourc. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 68.Zheng XW, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC genetics. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population-structure. Evol. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 71.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nuc. Acid. Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
rbcL and ITS sequences for the accessions in this study are deposited at NCBI under the Genbank IDs listed in Supplementary Table S3. The Whole Genome Shotgun project including raw reads and the draft genome have been deposited at DDBJ/ENA/GenBank under BioProject PRJNA507095. The genome version described in this paper is SDXO01000000. The GBS datasets are deposited at NCBI under BioProject PRJNA507246.