Abstract
Lactobacillus plantarum is remarkably adaptable to diverse habitats and is widely used in food industry. In this study, the genome sequence of L. plantarum CAUH2 was analyzed and compared with other L. plantarum genome sequences. A comparison of the genome sequence of CAUH2 to L. plantarum ST-III reveals that the similarity of these two genomes reached up to 99% identity with 98% coverage, but the plasmid profiles of CAUH2 and ST-III are different. Notably, plasmid pCAUH203 in L. plantarum CAUH2 harbors seven genes involved in oxidative stress response, such as genes encoding thioredoxin-disulfide reductase, thioredoxin and DNA protection protein. Due to plasmid pCAUH203, the thioredoxin reductase activity of CAUH2 was 2.1-fold higher than that of ST-III. When exposed to 5 mM H2O2, this activity was further increased to 9.87 ± 1.60 mU per mg protein in CAUH2, which was 2.7-fold higher than that of ST-III, indicating that thioredoxin antioxidant system encoded by pCAUH203 might contribute to the H2O2 resistance. This hypothesis was further confirmed by survival assay under 10 mM H2O2 stress. The survival rate of CAUH2 was 12-fold higher than that of ST-III. Therefore, the complete genome sequencing of L. plantarum CAUH2 provides new insights into the molecular mechanism of its oxidative stress resistance.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1653-4) contains supplementary material, which is available to authorized users.
Keywords: L. plantarum CAUH2, Complete genome sequence, Plasmid, H2O2 resistance, Thioredoxin reductase
Introduction
Lactobacillus plantarum is a flexible, versatile and heterofermentative lactic acid bacteria (LAB) (Siezen et al. 2012). Among LAB, this species has one of the largest genomes and a particularly high proportion of regulatory genes, which allows it to survive in a variety of different environmental niches, such as raw or fermented plants, meat, dairy products and the gastro-intestinal tracts (GIT) of human and animals (Martino et al. 2016; Siezen et al. 2010). Consequently, L. plantarum is widely used as a starter culture in fermented food and feed industries (Leroy and De Vuyst 2004; Zhang et al. 2016). Moreover, some L. plantarum strains have been regarded as probiotics because of their beneficial effects, including improvement on the integrity of intestinal barrier, attenuation of gut disorder, as well as the modulation of gut microbiota and host immune response (Nieuwboer et al. 2016; Seddik et al. 2017). To better understand its application in industry, high-throughput sequencing was performed to explore the genome, diversity and evolution of L. plantarum in recent years.
So far, more than 300 L. plantarum strains have been sequenced, and the complete genome sequence of 92 L. plantarum strains can be found on NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/genomes/1108). A comparative genomic analysis of 54 L. plantarum strains revealed that there is a high degree of gene content variation among L. plantarum strains (Martino et al. 2016). Therefore, L. plantarum shows a wide phenotypic diversity with different capabilities of environmental adaptation and metabolic capacities (Siezen and van Hylckama Vlieg 2011). L. plantarum CAUH2 (previously named as LR1) was isolated from Szechuan Pickle, a traditionally fermented vegetable product in China. To gain new insight into genome details and explore its probiotic potential, the whole-genome sequence of L. plantarum CAUH2 was determined in this study.
Materials and methods
Bacterial strains, growth condition and DNA extraction
Lactobacillus plantarum CAUH2 and ST-III were cultured at 37 °C in De Man Rogosa Sharp (MRS) medium, respectively. Genomic DNA from L. plantarum CAUH2 was prepared using the genomic DNA Extraction Kit (Tiangen, Beijing, China) according to the manufacturer’s instruction. The lysozyme pretreatment was performed by adding lysozyme to TES buffer (50 mM Tris–HCl, 1 mM EDTA, 25% w/v sucrose; pH 8.0) to a final concentration of 30 mg per mL, and mixtures were incubated at 37 °C for 1 h (O’Sullivan D and Klaenhammer 1993).
Genome sequencing, assembly and annotation
The complete genome was sequenced with massively parallel sequencing Illumina technology. Two DNA libraries were constructed: a paired-end library with an insert size of 500 bp and a mate-pair library with an insert size of 5 kb. The 500 bp library was sequenced using an Illumina Miseq by PE300 strategy and the 5 kb library was sequenced using an Illumina Hiseq 2500 by PE100 strategy. A total of 821 MB reads with approximately 108-fold coverage were obtained and assembled by SOAPdenovo. Gene annotation was obtained by NCBI Prokaryotic Genome Annotation Pepline and Rapid Annotation Subsystem Technology (RAST server) (Aziz et al. 2008). The functions of the predicted protein-coding genes were annotated with the Clusters of Orthologous Groups (COG) database using the WebMGA. The genomic alignment was performed by Mauve software (Darling et al. 2004).
Thioredoxin reductase activity assay
The thioredoxin reductase activity was measured with thioredoxin reductase activity assay kit (Abcam, Cambridge, MA, USA). L. plantarum overnight cultures were inoculated into 50 mL MRS medium. When cells reached an OD600nm of 0.8, 10 mL of each culture was collected and centrifuged at 6000×g for 2 min, then resuspended in the same volume MRS supplemented with or without 5 mM H2O2. After incubation for 30 min at 37 °C, cell pellets were collected by centrifugation, and then total protein was extracted by sonication with 0.1 M sodium phosphate buffer (pH 7.0). The concentration of total protein was quantified using the Qubit Protein Assay Kit and the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). The thioredoxin reductase activity was determined according to the manufacturer’s instruction, and showed as mU per mg protein. The results were obtained by three independent experiments.
Survival assay to H2O2 stress
To determine the tolerance to H2O2 stress, L. plantarum CAUH2 and ST-III overnight cultures were inoculated into 10 mL MRS medium, respectively. When cells reached an OD600nm of 0.8, 1 mL of each culture was collected and centrifuged at 6000×g for 2 min, then resuspended in the same volume MRS supplemented with 5 mM or 10 mM H2O2. After incubation for 30 min at 37 °C, the number of colony-forming units per milliliter (CFU/mL) was determined by plating tenfold serial dilutions on MRS agar and incubating at 37 °C for 16 h. Survival rate were calculated by dividing the number of CFU/mL after H2O2 challenge by the value of non-treated control. All results were obtained by at least three independent experiments with each performed in triplicate.
Statistical analysis
Data were analyzed using GraphPad Prism 6 software for Windows (GraphPad Software, Inc., La Jolla, CA, USA). When two groups were compared, an unpaired Student t test with Welch’s correction was used to calculate P values.
Nucleotide sequence accession numbers
The complete genome sequence of Lactobacillus plantarum CAUH2 was deposited at GenBank under the accession number CP015126.1 and CP015127.2-CP015129.2. Lactobacillus plantarum CAUH2 strain was deposited at the China Center of Industrial Culture Collection under accession number CICC No. 23,941.
Results and discussion
Genome features of L. plantarum CAUH2
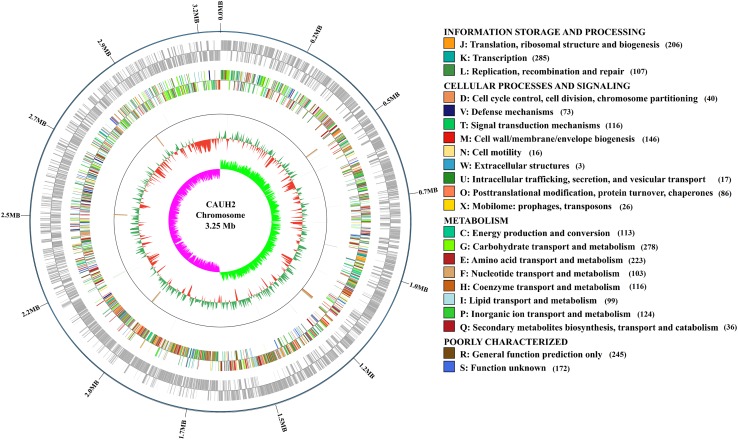
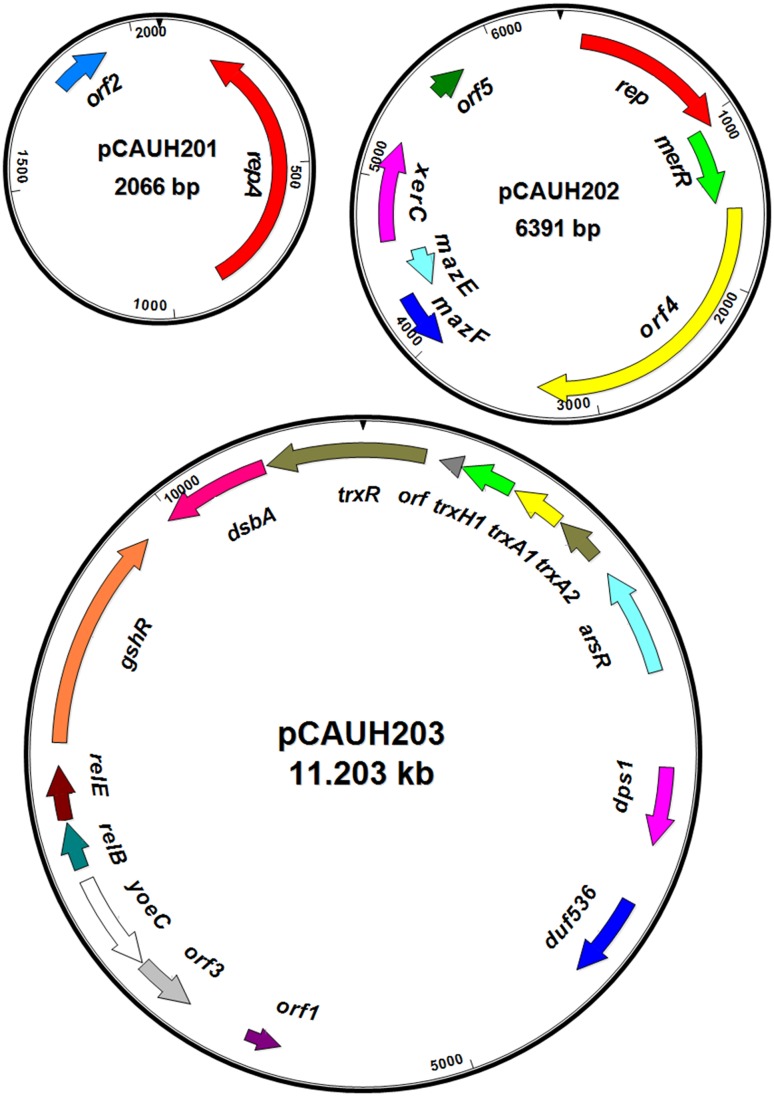
The complete genome of L. plantarum CAUH2 consists of a 3,254,946 bp circular chromosome with GC contents of 44.6% and three plasmids designated as pCAUH201 (2066 bp), pCAUH202 (6391 bp) and pCAUH203 (11,203 bp) (Figs. 1, 2). A total of 2986 protein-coding genes, 17 rRNA genes, 72 tRNAs and 83 pseudo genes were predicted in the chromosome. And, 2, 7 and 15 protein-coding genes were located on the plasmid pCAUH201, pCAUH202 and pCAUH203, respectively. The features of the complete genome sequence of L. plantarum CAUH2 are summarized in Table 1. A total of 2630 protein-coding genes were assigned to 22 COG categories (Tatusov et al. 2003). Among these genes, 1092 genes were associated with metabolism (41.52% proportion), including carbohydrate transport and metabolism, energy production and conversion, amino acid transport and metabolism (Fig. 1). The high number of genes involved in nutrients acquisition and utilization suggests the flexibility of CAUH2 in a large variety of niches.
Fig. 1.
Circular genome map of strain CAUH2. Genome map was drawn using Circos v0.64 (http://circos.ca/) (Krzywinski et al. 2009). Rings from the outermost to the center: (1) Scale marks of the genome; (2) protein-coding genes on the forward strand and reverse strand; (3) COG annotated coding sequences; (4) Non-coding RNA genes on the forward strand and reverse strand; (5) GC content; (6) GC skew
Fig. 2.
The physical maps of plasmid pCAUH201, pCAUH202 and pCAUH203. Plasmid map was generated by SeqBuilder program of Lasergene v7.1 (DNASTAR Inc, Madison, WI, USA). Closed arrows indicated open reading frames (orfs)
Table 1.
Features of Lactobacillus plantarum CAUH2 genome
| Attributes | Chromosome | Plasmid | ||
|---|---|---|---|---|
| pCAUH201 | pCAUH202 | pCAUH203 | ||
| Genome size (bp) | 3,254,946 | 2066 | 6391 | 11,203 |
| GC content (%) | 44.6 | 37.8 | 33.7 | 37.0 |
| 5S rRNAs | 6 | 0 | 0 | 0 |
| 16S rRNAs | 6 | 0 | 0 | 0 |
| 23S rRNAs | 5 | 0 | 0 | 0 |
| tRNAs | 72 | 0 | 0 | 0 |
| Other RNAs | 4 | 0 | 0 | 0 |
| Total genes | 3162 | 3 | 7 | 16 |
| Protein-coding genes | 2986 | 2 | 7 | 15 |
| Genes assigned to COGs | 2630 | 1 | 5 | 10 |
According to NCBI genome neighbor report, the closest genome sequence of L. plantarum CAUH2 is that of L. plantarum ST-III (99.9003% gapped identity) among the 92 complete L. plantarum genomes (Wang et al. 2011). The average nucleotide sequence identity (ANI) further showed that the similarity between L. plantarum CAUH2 genome and ST-III genome reached up to 99% identity with 98% coverage. Moreover, genomic alignment between strains CAUH2 and ST-III showed that the chromosome sequences were highly identical, and there was no genomic rearrangement (Fig. S1). However, there is only one theta-type plasmid pST-III (53,560 bp) in L. plantarum ST-III (Chen et al. 2012). The plasmid pST-III harbors genes encoding K+-transport system, glycine/betaine/carnitine ABC transporters and inorganic ion transporters, which play an important role in hyperosmotic resistance (Chen et al. 2012). In L. plantarum CAUH2, the plasmid pCAUH201 is a 2066-bp cryptic plasmid which has been proven to replicate via the rolling circle replication mechanism in our previous study (Li et al. 2009). Bioinformatics analysis further revealed that the plasmid pCAUH202 harbors seven genes encoding replication initiation protein (Rep), transcriptional regulator (MerR), site-specific integrase (XerC), type II toxin–antitoxin system and two hypothetical proteins. Notably, the largest plasmid pCAUH203 (11,203 bp) harbors 15 protein-coding genes (Fig. 2 and Table S1), and 7 genes were predicted to be involved in oxidative stress response, including trxR encoding thioredoxin-disulfide reductase, trxH1, trxA1 and trxA2 encoding thioredoxin, dps1 encoding DNA protection protein, gshR encoding NAD(P)/FAD-dependent oxidoreductase and dsbA encoding DsbA family oxidoreductase. Among these oxidative stress-associated proteins, thioredoxin reductase (TrxR) and thioredoxin (TrxH1, TrxA1 and TrxA2) could constitute a thioredoxin antioxidant system, which was predicted to play an important role in the oxidative stress response of L. plantarum CAUH2.
Thioredoxin reductase activity of CAUH2 and ST-III
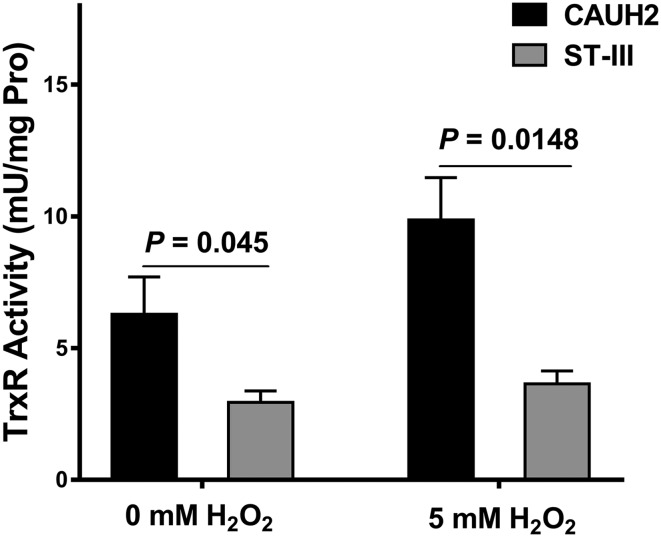
To investigate whether the plasmid coding thioredoxin antioxidant system can confer L. plantarum CAUH2-enhanced thioredoxin reductase activity, the thioredoxin reductase assay was performed using 5,5′-dithio-bis(2-nitrobenzoic acid)-based method (Arnér and Holmgren 2001). In L. plantarum CAUH2, the thioredoxin reductase activity was 6.28 ± 1.42 mU per mg protein without H2O2 treatment, which was 2.1-fold higher than that of L. plantarum ST-III (P < 0.05, Fig. 3). When exposed to 5 mM H2O2, thioredoxin reductase activity was further increased to 9.87 ± 1.60 mU per mg protein in CAUH2, which was 2.7-fold higher than that of ST-III. These results indicated that thioredoxin reductase could be induced by H2O2, which is in accordance with previous studies (Serrano et al. 2007). In bacteria, thioredoxin, thioredoxin reductase and NADPH constitute the thioredoxin antioxidant system, which is a key antioxidant system against the oxidative stress (Lu and Holmgren 2014). The reduced form of thioredoxin serves as a hydrogen donor for thiol-dependent peroxidases to remove intracellular reactive oxygen species (ROS) or reduces ribonucleotide reductase, methionine sulfone reductase and other redox-sensitive transcription factors under oxidative stress (Arnér and Holmgren 2000). Then, the oxidized thioredoxin is converted to reduced form by thioredoxin reductase using NADPH as a cofactor. Thus, the activity of thioredoxin reductase is very important for thioredoxin antioxidant system (Serrano et al. 2007). Therefore, the thioredoxin antioxidant system encoded by plasmid pCAUH203 may confer strain CAUH2 enhanced tolerance to oxidative stress than its closest relative L. plantarum ST-III.
Fig. 3.
The thioredoxin reductase was measured in L. plantarum CAUH2 and ST-III and showed as activity (mU) per mg protein of sample tested. Data are reported as mean ± standard deviation (SD) from at least three independent experiments and analyzed by an unpaired, two-tailed Student t test. The significant difference was indicated by P value < 0.05
Survival assay of CAUH2 and ST-III under H2O2 stress
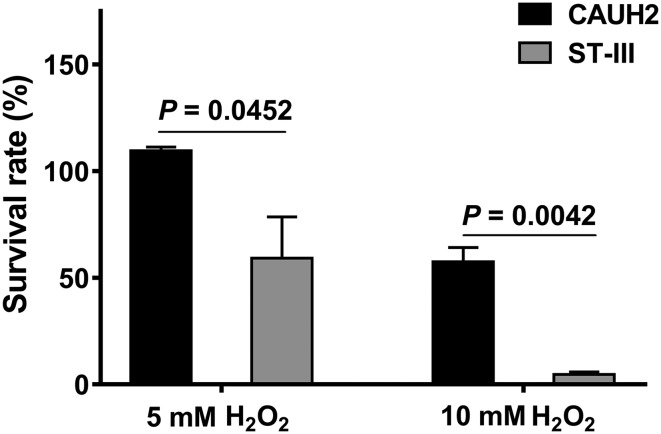
As the difference between CAUH2 and ST-III chromosome was less than 1%, and genes in these regions mainly encoded transposases, prophage proteins and hypothetical proteins, the oxidative stress tolerance of these two strains was compared to investigate the role of the plasmid pCAUH203. L. plantarum CAUH2 and ST-III were treated with 5 mM and 10 mM H2O2 treatment, respectively. The results showed that the viability of L. plantarum CAUH2 was not affected by 5 mM H2O2, whereas the survival rate of ST-III was decreased to average 59% (Fig. 4). When exposed to 10 mM H2O2, the survival rate of CAUH2 was average 57%, which was 12-fold higher than that of ST-III (P < 0.05, Fig. 4). These results further suggested that the plasmid pCAUH203 contributed to the enhanced H2O2 resistance for L. plantarum CAUH2. Generally, L. plantarum suffers from unavoidable oxidative stress caused by reactive oxygen species (ROS) during manufacturing or consumption. ROS, including the superoxide anion, hydrogen peroxide, the hydroxyl radical and organic hydroperoxides, can cause significant damage to various cellular structures such as DNA, proteins and cell membranes (Touati 2000). Therefore, tolerance to oxidative stress is crucial for L. plantarum to survive and further exert its beneficial effects (Corcoran et al. 2008; Mills et al. 2011). L. plantarum ST-III has been applied as a probiotic strain in commercial yogurt products (Bright Dairy & Food Co., Ltd., Shanghai, China) for many years. Compared with ST-III, L. plantarum CAUH2 showed higher tolerance to oxidative stress. Therefore, CAUH2 could also be a potential probiotic strain applied in food and feed industries. In addition, no antibiotic resistance genes were found in the novel plasmid pCAUH203, suggesting that this plasmid could be used to improve the oxidative stress tolerance of other probiotic strains via conjugative transfer (Grohmann et al. 2003; Ito et al. 2009).
Fig. 4.
Survival of L. plantarum CAUH2 and ST-III after hydrogen peroxide treatment. Survival was calculated by dividing the number of CFU/mL after H2O2 challenge by the value of non-treated control. Data are reported as mean ± SD from at least three independent experiments and analyzed by an unpaired, two-tailed Student’s t test. The significant difference was indicated by P value < 0.05
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (contract No.31701571).
Author contributions
ZZ, FR and YH designed research; ZZ, YY, JW and GW performed research; YY and GW contributed new reagents or analytic tools; ZZ, FR and YH analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Arnér ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Arnér ES, Holmgren A (2001) Measurement of thioredoxin and thioredoxin reductase. Curr Protoc Toxicol, Chap. 7:Unit 7.4. [DOI] [PubMed]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ai L, Zhou F, Ren J, Sun K, Zhang H, Chen W, Guo B. Complete nucleotide sequence of plasmid pST-III from Lactobacillus plantarum ST-III. Plasmid. 2012;67:236–244. doi: 10.1016/j.plasmid.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Corcoran BM, Stanton C, Fitzgerald G, Ross RP. Life under stress: the probiotic stress response and how it may be manipulated. Curr Pharm Des. 2008;14:1382–1399. doi: 10.2174/138161208784480225. [DOI] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kawai Y, Arakawa K, Honme Y, Sasaki T, Saito T. Conjugative plasmid from Lactobacillus gasseri LA39 that carries genes for production of and immunity to the circular bacteriocin gassericin A. Appl Environ Microbiol. 2009;75:6340–6351. doi: 10.1128/AEM.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy F, De Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Tech. 2004;15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- Li R, Zhai Z, Yin S, Huang Y, Wang Q, Luo Y, Hao Y. Characterization of a rolling-circle replication plasmid pLR1 from Lactobacillus plantarum LR1. Curr Microbiol. 2009;58:106–111. doi: 10.1007/s00284-008-9280-z. [DOI] [PubMed] [Google Scholar]
- Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- Martino ME, Bayjanov JR, Caffrey BE, Wels M, Joncour P, Hughes S, Gillet B, Kleerebezem M, van Hijum SA, Leulier F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ Microbiol. 2016;18:4974–4989. doi: 10.1111/1462-2920.13455. [DOI] [PubMed] [Google Scholar]
- Mills S, Stanton C, Fitzgerald GF, Ross RP. Enhancing the stress responses of probiotics for a lifestyle from gut to product and back again. Microb Cell Fact. 2011;10(Suppl 1):S19. doi: 10.1186/1475-2859-10-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwboer M, Hemert S, Claassen E, Vos WM. Lactobacillus plantarum WCFS1 and its host interaction: a dozen years after the genome. Microb Biotechnol. 2016;9:452–465. doi: 10.1111/1751-7915.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan DJ, Klaenhammer TR. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddik HA, Bendali F, Gancel F, Fliss I, Spano G, Drider D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob Proteins. 2017;9:111–122. doi: 10.1007/s12602-017-9264-z. [DOI] [PubMed] [Google Scholar]
- Serrano LM, Molenaar D, Wels M, Teusink B, Bron PA, Vos WMD, Smid EJ. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb Cell Fact. 2007;6:29. doi: 10.1186/1475-2859-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, van Hylckama Vlieg JE. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb Cell Fact. 2011;10(Suppl 1):S3. doi: 10.1186/1475-2859-10-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HT, Rademaker JL, Starrenburg MJ, Kleerebezem M, Molenaar D, van Hylckama Vlieg JE. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol. 2010;12:758–773. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- Siezen RJ, Francke C, Renckens B, Boekhorst J, Wels M, Kleerebezem M, van Hijum SA. Complete resequencing and reannotation of the Lactobacillus plantarum WCFS1 genome. J Bacteriol. 2012;194:195–196. doi: 10.1128/JB.06275-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinf. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen C, Ai L, Zhou F, Zhou Z, Wang L, Zhang H, Chen W, Guo B. Complete genome sequence of the probiotic Lactobacillus plantarum ST-III. J Bacteriol. 2011;193:313–314. doi: 10.1128/JB.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Xu D, Zhao XH, Song Y, Liu YL, Li HN. Biodegradation of two organophosphorus pesticides in whole corn silage as affected by the cultured Lactobacillus plantarum. 3 Biotech. 2016;6:73. doi: 10.1007/s13205-016-0364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.