Abstract
Extracting RNA with high quality and integrity is crucial for molecular biology studies in eukaryotes. However, RNA isolation from cassava storage root raises a great concern because it contains large amounts of polysaccharides and polyphenol compounds. In the current study, four RNA extraction methods were evaluated for extracting RNA from cassava storage root. We found that the modified TM method (MTM) is timesaving and low-cost extraction method with high quality and quantities of RNA. The effectiveness of the improved method was assessed for qPCR analysis of four selected genes from total RNA of storage root. The improved protocol generated 4.18–5.94 µg RNA/g fresh weight. An A260/280 ratios of RNA samples are ranged from 2.14 to 2.17. The RIN values are ranged from 7.2 to 8.0. Importantly, isolated total RNA by MTM was successfully used for library construction and transcriptome sequencing. Therefore, we provide an efficient and low-cost method, MTM, for extracting high quality and quantities of RNA from cassava storage root.
Keywords: RNA isolation, Cassava storage root, High polysaccharides
Introduction
Cassava (Manihot esculenta), is an important staple food in the tropical regions (Rey and Vanderschuren 2017). Cassava originated in the Amazon River basin in the tropical regions of the Americas, is regarded as three major potato crops in addition with potatoes, sweet potatoes around the world (Morales 2007). Cassava is available for food, feed, and industrial exploitation (Patanun et al. 2013; Montagnac et al. 2009). Cassava storage root starch is one of the main raw materials for making starch in industry (Demiate and Kotovicz 2011; Zhu 2015). Sixty-five percent of cassava tuber production is consumed for human food in the world (Shanavas et al. 2011). Increasingly studies have been performed on cassava tuber. For example, Homeodomain proteins play a critical role in regulating various aspects of plant growth and development, which are differentially expressed during tuber development (Guo et al. 2014). Starch branching enzyme activity was correlated with the regulation of starch synthesis in the cassava storage root. Gene expressions of starch branching enzymes are induced by abscisic acid, glucose and glucose-1-P (Baguma et al. 2008). Genome-wide analysis reveals that the development of cassava storage root is associated with a phase change from cell division and accumulation of reserve substances (Yockteng et al. 2013). Activities of calcium-dependent protein kinases play a pivotal role in the signaling pathway of tuber initiation, and sulfite reductase might be involved in the regulation of tuber development (Wang et al. 2008). Moreover, major constraints facing production of cassava tubers is the rapid postharvest physiological deterioration (PPD), which is likely connected with rapid burst of reactive oxygen species accumulation (Bayoumi et al. 2010; Morante et al. 2010). High levels of phenolic acids, scopoletin, carotenoids, proteins, and augmented activities of guaiacol peroxidase and hydrogen peroxide in non-stored cassava roots can be used as potential biomarkers for cassava deterioration (Uarrota and Maraschin 2015). ATPases and peroxidases have an important role in the post-harvest deterioration of cassava tuber (Bunnik et al. 2013). On the contrary, exogenous melatonin could attenuate the postharvest physiological deterioration and reduce the accumulation of hydrogen peroxide (Ma et al. 2016). Therefore, works on the physiological as well as molecular aspects of cassava tuberisation and/or tube deterioration remain a great concern in cassava research.
In plant, obtaining high quality RNA is challenging for subsequent cDNA library construction, and molecular analysis such as real-time PCR (RT-PCR), cDNA synthesis and genetic analysis (Ogawa et al. 2003; Sim et al. 2013). Secondary metabolites such as polysaccharides and polyphenols could largely interfere with or degrade, or even coprecipitate with RNA during nucleotide purification process due to the similarity in physical and chemical properties between RNA and polysaccharide (Nonis et al. 2012; Gudenschwager et al. 2012; Ma and Yang 2011). Cassava storage root contains lots of polysaccharides and polyphenols, which resulted in hardly acquiring the high quantity and purity of RNAs for subsequent molecular analysis. Although several RNA extraction protocols have been developed for arabidopsis, rice, wheat, maize, sunflower and Jerusalem artichoke seeds, it may not well suitable for extracting total RNA from cassava tubers (Mornkham et al. 2013; Rayani and Dehghan Nayeri 2015; Xu et al. 2010; Baguma et al. 2008). In this study, four RNA extraction methods were evaluated for extraction RNA from cassava storage root, and modified TM method (MTM) is timesaving and low-cost extraction method that can obtain the high quality and quantities of RNA. Therefore, we provide an efficient and economical RNA extraction method for cassava storage root by comparing other three RNA extraction methods.
Materials and methods
Plant material
Cassava cultivars, ku50, was grown in the experimental fields of Institute of Tropical Agriculture and Forestry, Hainan University. The annual average temperature is around 24.3 °C, and the highest average temperature is about 28 °C, and the lowest average temperature is around 18 °C. One-year old cassava storage roots were harvested and immediately used.
Solution and reagents
RNA Extraction Buffer: 8 M LiCl, 2% (w/v) PVP40, 5% (v/v) β-mercaptoethanol. (adding before use). Solubilization Buffer: 1.4% (w/v) SDS, 0.075 M NaCl, 0.025 M EDTA, add 2% (v/v) β-mercaptoethanol just before use. Other reagents: Trizol® reagent (Invitrogen, USA), RNAplant Plus reagent (TIANGEN, Japan), chloroform, 3 M NaAc, pH 5.2, PVP40, absolute ethanol and liquid nitrogen, DNase I (Promega, USA), and isopropanol of analytic purity.
RNA isolation protocol
Total RNA was extracted from 0.3 g cassava storage root material with three independent replications. Before operation, all plastic materials were autoclaved, and then the glass material, mortar and pestle were baked for 3–6 h at 180 °C.
Three traditional extraction methods and one improved method were used: (a) Trizol method, extracting total RNA according to the instruction book of Trizol; (b) RNAplant Plus method, extracting total RNA following the direction of RNAplant Plus; (c) TM method, extracting total RNA according to the method reported by Tanupat Mornkham (Mornkham et al. 2013); (d) MTM method, extracting total RNA using the modified method based on TM method as followed.
Procedure of the total RNA extracted by the MTM method
PVP40 (polyvinylpolypyrrolidone) was added to the sample and ground in liquid nitrogen. Immediately transfer the sample to 1.5 ml enzyme-free tube and add 900 µl RNA extraction buffer, 45 µl β-mercaptoethanol, and 350 µl absolute ethanol, mixed by shaking, and incubate for 5 min at room temperature. Centrifuging at 5000 rpm for 3 min after mixing with 100 µl chloroform (high molecular weight impurities and lipids removed). Add 550 µl solubilization buffer and 550 µl chloroform to precipitate, vortex and centrifuge for 3 min at 5000 rpm at 4 °C. Then transfer the supernatant to a new 1.5 ml RNase-free centrifuge tube, add 1 ml Trizol and 200 µl chloroform, vortex and incubate for 3 min at room temperature. After centrifuging at 12,000 rpm, 4 °C for 15 min, transfer the supernatant to a new 1.5 ml RNase-free centrifuge tube, add 500 µl chloroform, vortex and centrifuge at 12,000 rpm for 15 min at 4 °C. The supernatant was then transferred to a fresh RNase-free centrifuge tube, and 500 µl isopropanol was added, vortex and incubated for 10 min at room temperature, and centrifuged at 12,000 rpm, 4 °C for 15 min. Discard the supernatant, add 1 ml 75% ethanol, gently shake and centrifuge at 12,000 rpm for 15 min at 4 °C, and then discard the supernatant. After centrifuging at 12,000 rpm, 4 °C for 2 min, the supernatant was discarded with a pipette tip. Add 50 µl DEPC water to the tubes after drying on the ice for 5 min, and then incubate in a 55 °C water bath for 5 min. Add 6 µl 10x DNase enzyme buffer, 4 µl DNase, incubated at 37 °C for 30 min. Add 240 µl DEPC water and 300 µl chloroform, vortex and centrifuged at 12,000 rpm, 4 °C for 10 min. Transfer the supernatant to a new tube, and add one-tenth volume of sodium acetate, three volumes of absolute ethanol, vortex and kept at − 80 °C for 1–2 h. Discard the supernatant after centrifuging at 12,000 rpm for 15 min at 4 °C. Add 1 ml 75% ethanol to wash the pellet, and add 30–50 µl DEPC water to dissolve the RNA.
RNA assessment
RNA integrity was evaluated by the 28S, 18S and 5S rRNA bands from 1 µg total RNA in the 1.0% agarose gel electrophoresis. The gels were stained with ethidium bromide and visualized under UV light. The photographs were taken by Vilber Lourmat (VILBERLOURMAT, France), and the images were inverted in Adobe Photoshop (Adobe, USA). The absorbance ratios of total RNA extracted from all methods were evaluated at 260 nm/280 nm by Nanodrop 2000. The A260/A280 ratio was calculated to determine the purity of RNA sample. RIN values were determined on an Agilent 2100 Bioanalyzer using Expert software (Rev. B.02.08.SI648).
Reverse transcription and RT-qPCR analysis
After DNase treatment using RQ1 RNase-Free DNase (Promega, USA), 7 µl total RNA from cassava tubers were reverse transcribed in a 20 µL reaction using OligodT (2.5 µM) and digested with RNaseH according to the EasyScript® One-Step gDNA Removal and cDNA Synthesis Super Mix kit (TRANSGEN Tech, China). After the first strand cDNA synthesis, cDNA can then be used as a template for amplification by real-time PCR (RT-PCR). A PCR primer pairs were used for the amplification of the gene expression of cassava RSZ21b gene. The sequences were 5′-AAAAAGCAGGCTTAATGGCTCGGGTTTACG-3′ for the forward primer, and 5′-AGAAAGCTGGGTAATCTCCATTAGCATAAGGTG-3′ for the reverse primer. A 20 µl reaction volume was performed in a RNAase free tube using 10 µl 2 × PCR Mix, 1 µl cDNA template, 1 µl 10 µM forward primers, 1 µl 10 µM reverse primers and 7 µl DEPC water. The PCR cycles were programmed as followed: initial denaturation at 94 °C for 5 min followed by identical 35 cycles with 30 min denaturing at 94 °C, 30 s annealing at 56 °C, and 1 min elongation at 72 °C followed by a single final cycle of 12 °C. After the reaction, 5 µl assay mixture was used for agarose electrophoresis using 1 × TAE buffer following the common procedure. The remaining products were stored at − 20 °C for subsequent analysis. The quantitative RT-PCR (qPCR) experiments were performed in a LightCycler® 480 thermocycler (Roche, USA). The genes were ubiquitin and RSZ21b, which primer sequences were 5′-GCCTCCCAAGGTAGCTTTCA-3′ for the forward primer and 5′-GGTTAATGCAGGGCTCCACT-3′ for the reverse primer, 5′-TCGGAGAGGGATCTCGAAGA-3′ for the forward primer and 5′-GCATCCCTGCGATCATCAAAC-3′ for the reverse primer, respectively. The reaction was conducted with a final volume of 20 µl using the LightCycler® 480 SYBR Green I Master kit (Roche, USA) according to the manufacturer’s instructions. The qPCR condition was used as follows: denatured at 94 °C for 30 s; 45 cycles of 94 °C for 5 s, 60 °C for 15 s and 72 °C for 10 s, with melting and cooling. The PCR products were also analyzed on 1.0% agarose electrophoresis and visualized as described above.
Results and discussion
Methods of RNA isolation are devised for the basic principle of preventing degradation by extracting total RNA from cells/tissues in an environment inhibitory to RNases. Extracting RNA with high quality and integrity is prerequisite and crucial for carrying out molecular biology studies in plant. A large number of RNA extraction methods have been exploited or modified (Mornkham et al. 2013; Rayani and Dehghan Nayeri 2015; Xu et al. 2010; Baguma et al. 2008), however, plants tissues have large diverse in ingredient which require suitable approaches for different kinds of purpose. Meanwhile, the efficiency of certain method for extracting RNA is varied for distinct plants. Generally, it is difficult to extract high quality RNA from plants that rich in polysaccharides and polyphenols (Rayani and Dehghan Nayeri 2015). The physical and chemical properties of polysaccharides are similar to those of RNA that co-precipitation of polyphenols is easily occurs, and thus RNA quality is inevitably compromised (Mornkham et al. 2013). For this particular reason, RNA isolation from cassava storage root that contains high levels of polysaccharides, phenols, proteins and other secondary metabolites are difficult as it affects the RNA yield and RNA integration. Therefore, we aim to establish a simple, economical and efficient RNA extraction method to acquire high-quality RNA from cassava storage root t.
The quality of RNA extracted by traditional methods
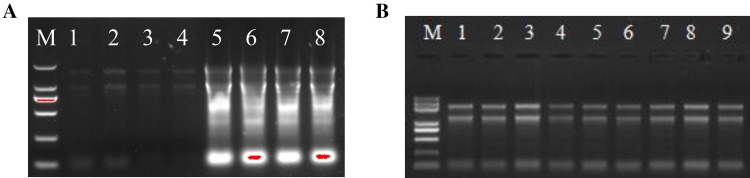
In general, extracting the total RNA from cassava storage root is expensive and unstable for commercial RNA isolation kit. We found that traditional Trizol method is not suitable for extracting RNA from cassava storage root as polysaccharides and polyphenols were combined together with extracts that formed in agglomerates which resulted in a less supernatant for subsequent step (Fig. 1). Additionally, our initial attempts using the other two existing methods, RNAplant Plus and TM method (Mornkham et al. 2013), to extract total RNA remain unsatisfactory from cassava storage root (Fig. 2; Table 1). When using the RNAplant Plus method, the higher RNA concentration is obtained (1714.1 ng/µl) and the ratio of OD 260/280 is 2.03. However, the RNA band is diffused in the 1% agarose gel electrophoresis. When using the TM method, the lower RNA concentration is obtained (29.6 to 58.7 ng/µl), however, the RNA band is not visible in the 1% agarose gel electrophoresis (Fig. 2), which is not suitable for subsequent analysis.
Fig. 1.
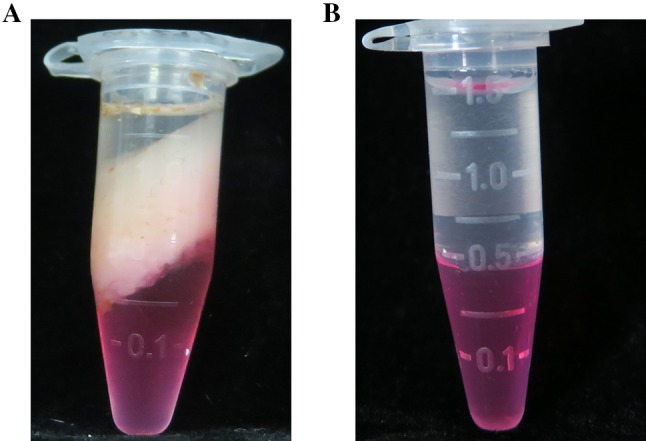
The agglomerates were formed by Trizol method but not with MTM method. a Trizol method: add 1 ml TRIzol and 200 µl chloroform into the grinding tuber, centrifuge at 12,000 rpm, 4 °C for 15 min after incubate for 3 min at room temperature; b MTM methods: grinding tuber is incubated with RNA extraction buffer, β-mercaptoethanol, and absolute ethanol, chloroform. After centrifuge, add 1 ml Trizol and 200 µl chloroform into supernatant, and centrifuge at 12,000 rpm, 4 °C for 15 min
Fig. 2.
Total RNAs extracting from cassava tubers were analyzed in the electrophoresis (1.0% agarose) by three methods (1 µg RNA per lane). a Lane M, DL2000 DNA maker, lane 1–4, RNA samples were isolated by TM method; lane 5–8, RNA samples were isolated by RNAplant Plus method; b lane M, DL2000 DNA maker, lane 1–9, RNA sample were isolated by MTM method
Table 1.
Yields and purity of total RNA isolated from cassava tuber by three traditional methods
| Method | Concentration (ng/μl) | A260 (Abs) | A280 (Abs) | 260/280 | 260/230 | Factor |
|---|---|---|---|---|---|---|
| TM | 29.6 | 0.741 | 0.375 | 1.98 | 1.38 | 40.00 |
| TM | 56.9 | 1.423 | 0.733 | 1.94 | 1.72 | 40.00 |
| TM | 30.4 | 0.760 | 0.402 | 1.89 | 0.93 | 40.00 |
| TM | 58.7 | 1.467 | 0.774 | 1.90 | 1.31 | 40.00 |
| RNAplant Plus | 969.8 | 24.244 | 11.924 | 1.961 | 2.18 | 40.00 |
| RNAplant Plus | 582.4 | 14.559 | 7.485 | 1.974 | 1.94 | 40.00 |
| KNAplant Plus | 1714.1 | 42.853 | 21.051 | 1.95 | 2.11 | 40.00 |
| RNAplant Plus | 1303.4 | 32.586 | 16.084 | 2.034 | 2.10 | 40.00 |
| MTM | 34.3 | 0.857 | 0.394 | 2.17 | 2.01 | 40.00 |
| MTM | 37.2 | 0.929 | 0.435 | 2.14 | 1.97 | 40.00 |
| MTM | 57 | 1.425 | 0.664 | 2.15 | 1.99 | 40.00 |
| MTM | 51.8 | 1.294 | 0.606 | 2.14 | 1.94 | 40.00 |
The quality of RNA samples isolated from cassava tubers by MTM method is great
Considering the high levels of starch and lipid present existed in the cassava storage root, we modified the TM methods by adjusting the amounts of reagents and adding extra important steps. The optimal modified part is that we used RNA extraction reagent Trizol and chloroform with two times volume in the MTM method when compared with TM methods. Large amounts of Trizol regent used in this study could efficiently lyses the tissue, and the RNA enzyme inhibitor prevents RNA degradation. Massive chloroform can efficiently remove high molecular weight materials and lipids, inhibit the activity of RNAase, extract phenol from the aqueous phase, separate the organic and inorganic phases, and allow RNA to enter the aqueous phase. Importantly, all centrifugation steps are performed at 4 °C to prevent RNA degradation. The success of total RNA isolation should be verified by assessing the quantity, quality and integrity of RNA. The concentration of RNA isolated by MTM method is about 34.3–57 ng/µl, which is less different from TM method, however, in the 1.0% agarose gel electrophoresis, it was showed that ribosomal RNA bands of 28S, 18S and 5S were intact and bright, demonstrating that the total RNA isolated by MTM method was not degraded (Fig. 2; Table 1). Moreover, the yields of total RNA (µg/g fresh weight) isolated from the different cassava storage roots are shown in Table 2, and the yields of total RNA extracting from the cassava storage roots ranged from 4.18 to 5.94 µg/g, which is optimal for downstream applications.
Table 2.
Yields and purity of concentrated total RNA isolated from cassava tuber by MTM method
| Sample name | Concentration (ng/μl) | Volume (μl) | Yield (μg) | OD260/280 | OD260/230 | 25S/18S | RIN |
|---|---|---|---|---|---|---|---|
| 1 | 310 | 19 | 5.89 | 2.013 | 2.039 | 1.2 | 8 |
| 2 | 300 | 19 | 5.7 | 2 | 2.055 | 1.4 | 8 |
| 3 | 294 | 20 | 5.88 | 1.96 | 1.96 | 1.2 | 7.8 |
| 4 | 306 | 19 | 5.81 | 1.962 | 1.987 | 1.4 | 7.2 |
| 5 | 297 | 20 | 5.94 | 1.961 | 1.961 | 1.5 | 7.4 |
| 6 | 294 | 19 | 5.59 | 1.974 | 1.948 | 1.4 | 7.3 |
| 7 | 220 | 19 | 4.18 | 1.95 | 1.918 | 1.3 | 7.4 |
| 8 | 240 | 19 | 4.56 | 2.034 | 1.905 | 1.4 | 7.4 |
| 9 | 238 | 19 | 4.52 | 2.017 | 1.983 | 1.4 | 7.2 |
RNA in downstream applications
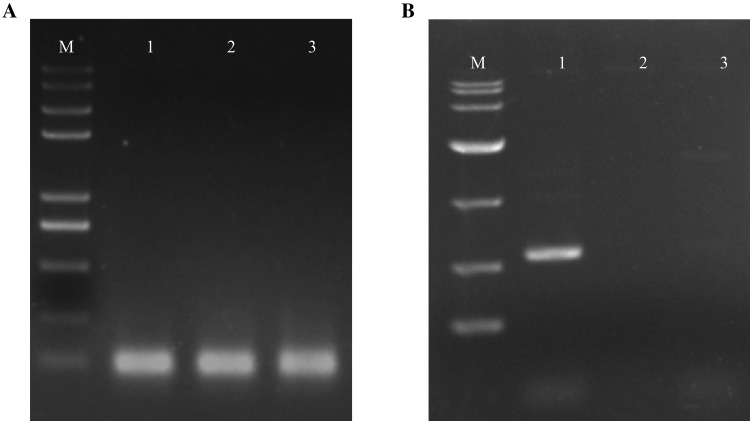
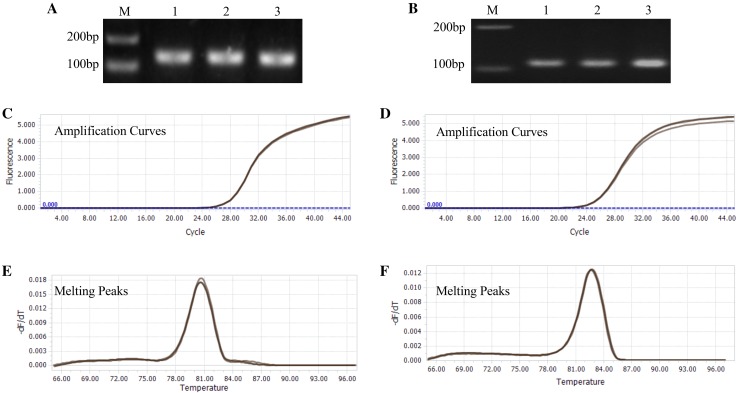
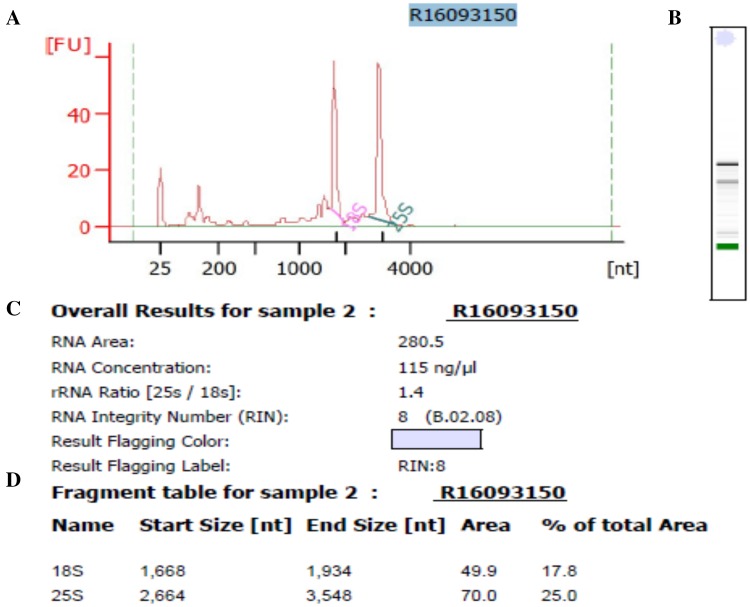
The quality of isolated total RNA from MTM method was confirmed in several ways. First, the Nanodrop 2000 result shows that the A260/280 ratios of RNA samples ranged between 2.14 and 2.17, and the A260/230 ratios ranged between 1.94 and 2.01, indicating that the RNA samples are free of contaminated proteins, DNA, polyphenolics and organic solvents (Table 1). Second, RNA samples were successfully reverse transcribed and the target gene was amplified from cDNA by PCR. For the ubiquitin gene, objective bands were observed in size of approximately 120 bp in the 1% agarose gel for three methods (Fig. 3a). For the RSZ21b gene, objective band was observed in size of approximately 600 bp in the 1% agarose gel for MTM method. Unexpectedly, none of objective bands were obtained from TM method and RNAplant Plus, respectively (Fig. 3b), indicating that large bands could be acquired by MTM methods. Third, qPCR was used to amplify the ubiquitin and RSZ21b genes to evaluate the integrity of the isolated RNA using the MMY method. The target bands with expected sizes of the ubiquitin and RSZ21b genes were observed (Fig. 4a, b). The values of qPCR cycle thresholds (Ct) were ranged from 24 to 30 cycles (Fig. 4c, d), and the melting curve was specific, with a single peak occurring at about 81 °C, 83 °C for ubiquitin and RSZ21b, respectively (Fig. 4e, f). Finally, RNA samples were concentrated and analyzed by the Agilent 2100 Bioanalyzer. The RIN values are ranged from 7.2 to 8, which is optimal to have good results for the next-generation sequencing library construction and analysis. A representative electropherogram of total RNA and the generated gel images showed the clear peaks of rRNAs and intact RNA (Fig. 5).
Fig. 3.
Amplified cDNAs by RT-PCR were analyzed in the agarose gel electrophoresis. a Lane M represents DL10000 DNA marker; lane 1–3 represents RT-PCR product of ubiquitin; b lane M represents DL2000 DNA marker; lane 1–3 represents RT-PCR product of RSZ21b. Lane 1, MTM method; lane 2, TM method; lane 3, RNAplant Plus method
Fig. 4.
qPCR analysis of four genes in cassava storage root. a, b RT-PCR amplification of the ubiquitin gene (left) and the RSZ21b gene (Right) from the templates obtained in the 1st-stranded cDNA from cassava tubers, M represents DL2000 DNA marker; c, d amplification curves of the PCR products for ubiquitin gene (left) and the RSZ21b gene (Right); e, f melting peaks of the PCR products for the amplified ubiquitin gene (left) and the RSZ21b gene (Right)
Fig. 5.
Bioanalyzer results of total RNA isolated by the MTM method. RNA isolations were obtained from cassava tubers and run on an Agilent 2100 Bioanalyzer. a Electropherogram, the y axis represents fluorescence units, and the x axis represents second; b represented gel image by Bioanalyzer analysis; c overall results for isolated RNA samples. The RIN value was assigned by the 2100 expert software (Rev. B.02.08.SI648); d results of rRNA analysis
Conclusion
In this study, we provided an improved method, MTM, for RNA extraction with high-quality and -quantity from cassava storage root that contains the high levels of starch and lipids. The RNA obtained by MTM method can be used for various downstream applications including cDNA synthesis, RT-PCR, qPCR and RNA-seq. In conclusion, the method reported here is useful for RNA extraction from cassava storage root that rich in lipids, polyphenolics and other secondary metabolites.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2018YFD1000500), National Natural Science Foundation of China (31670250), the startup funding of Hainan University (KYQD1562) to ZW, and project of cultivating talents for outstanding undergraduate students in crop science of Hainan University.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Baguma Y, Sun C, Boren M, Olsson H, Rosenqvist S, Mutisya J, Rubaihayo PR, Jansson C. Sugar-mediated semidian oscillation of gene expression in the cassava storage root regulates starch synthesis. Plant Signal Behav. 2008;3(7):439–445. doi: 10.4161/psb.3.7.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayoumi SAL, Rowan MG, Beeching JR, Blagbrough IS. Constituents and secondary metabolite natural products in fresh and deteriorated cassava roots. Phytochemistry. 2010;71(5–6):598–604. doi: 10.1016/j.phytochem.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Bunnik EM, Chung DW, Hamilton M, Ponts N, Saraf A, Prudhomme J, Florens L, Le Roch KG. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum. Genome Biol. 2013;14(11):R128. doi: 10.1186/gb-2013-14-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demiate IM, Kotovicz V. Cassava starch in the Brazilian food industry. Cienc Tecnol Alime. 2011;31(2):388–397. doi: 10.1590/S0101-20612011000200017. [DOI] [Google Scholar]
- Gudenschwager O, Gonzalez-Aguero M, Defilippi BG. A general method for high-quality RNA isolation from metabolite-rich fruits. S Afr J Bot. 2012;83:186–192. doi: 10.1016/j.sajb.2012.08.004. [DOI] [Google Scholar]
- Guo D, Li HL, Tang X, Peng SQ. Cassava (Manihot esculenta Krantz) genome harbors KNOX genes differentially expressed during storage root development. Genet Mol Res. 2014;13(4):10714–10726. doi: 10.4238/2014.December.18.13. [DOI] [PubMed] [Google Scholar]
- Ma XB, Yang J. An optimized preparation method to obtain high-quality RNA from dry sunflower seeds. Genet Mol Res. 2011;10(1):160–168. doi: 10.4238/vol10-1gmr979. [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhang T, Zhang P, Wang ZY. Melatonin attenuates postharvest physiological deterioration of cassava storage roots. J Pineal Res. 2016;60(4):424–434. doi: 10.1111/jpi.12325. [DOI] [PubMed] [Google Scholar]
- Montagnac JA, Davis CR, Tanumihardjo SA. Nutritional value of cassava for use as a staple food and recent advances for improvement. Compr Rev Food Sci Food Saf. 2009;8(3):181–194. doi: 10.1111/j.1541-4337.2009.00077.x. [DOI] [PubMed] [Google Scholar]
- Morales FJ. Tropical whitefly IPM Project. Adv Virus Res. 2007;69:249–311. doi: 10.1016/S0065-3527(06)69006-4. [DOI] [PubMed] [Google Scholar]
- Morante N, Sanchez T, Ceballos H, Calle F, Perez JC, Egesi C, Cuambe CE, Escobar AF, Ortiz D, Chavez AL, Fregene M. Tolerance to Postharvest physiological deterioration in cassava roots. Crop Sci. 2010;50(4):1333–1338. doi: 10.2135/cropsci2009.11.0666. [DOI] [Google Scholar]
- Mornkham T, Wangsomnuk PP, Fu YB, Wangsomnuk P, Jogloy S, Patanothai A. Extractions of high quality RNA from the seeds of Jerusalem artichoke and other plant species with high levels of starch and lipid. Plants (Basel) 2013;2(2):302–316. doi: 10.3390/plants2020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonis A, Vezzaro A, Ruperti B. Evaluation of RNA extraction methods and identification of putative reference genes for real-time quantitative polymerase chain reaction expression studies on olive (Olea europaea L.) fruits. J Agric Food Chem. 2012;60(27):6855–6865. doi: 10.1021/jf300419w. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15(7):1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patanun O, Lertpanyasampatha M, Sojikul P, Viboonjun U, Narangajavana J. Computational identification of MicroRNAs and their targets in cassava (Manihot esculenta Crantz.) Mol Biotechnol. 2013;53(3):257–269. doi: 10.1007/s12033-012-9521-z. [DOI] [PubMed] [Google Scholar]
- Rayani A, Dehghan Nayeri F. An improved method for extraction of high-quality total RNA from oil seeds. Biotechnol Lett. 2015;37(4):927–933. doi: 10.1007/s10529-014-1752-6. [DOI] [PubMed] [Google Scholar]
- Rey C, Vanderschuren H. Cassava mosaic and brown streak diseases: current perspectives and beyond. Annu Rev Virol. 2017;4(1):429–452. doi: 10.1146/annurev-virology-101416-041913. [DOI] [PubMed] [Google Scholar]
- Shanavas S, Padmaja G, Moorthy SN, Sajeev MS, Sheriff JT. Process optimization for bioethanol production from cassava starch using novel eco-friendly enzymes. Biomass Bioenerg. 2011;35(2):901–909. doi: 10.1016/j.biombioe.2010.11.004. [DOI] [Google Scholar]
- Sim MC, Ho CL, Phang SM. A simple and effective method for RNA isolation and cDNA library construction from the brown seaweed Sargassum polycystum (Fucales, Phaeophyceae) J Appl Phycol. 2013;25(5):1277–1285. doi: 10.1007/s10811-013-9980-z. [DOI] [Google Scholar]
- Uarrota VG, Maraschin M. Metabolomic, enzymatic, and histochemical analyzes of cassava roots during postharvest physiological deterioration. BMC Res Notes. 2015;8:648. doi: 10.1186/s13104-015-1580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tian W, Li Y. Development of an efficient protocol of RNA isolation from recalcitrant tree tissues. Mol Biotechnol. 2008;38(1):57–64. doi: 10.1007/s12033-007-0073-6. [DOI] [PubMed] [Google Scholar]
- Xu J, Aileni M, Abbagani S, Zhang P. A reliable and efficient method for total RNA Isolation from various members of spurge family (Euphorbiaceae) Phytochem Anal. 2010;21(5):395–398. doi: 10.1002/pca.1205. [DOI] [PubMed] [Google Scholar]
- Yockteng R, Almeida AM, Yee S, Andre T, Hill C, Specht CD. A method for extracting high-quality RNA from diverse plants for next-generation sequencing and gene expression analyses. Appl Plant Sci. 2013 doi: 10.3732/apps.1300070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F. Composition, structure, physicochemical properties, and modifications of cassava starch. Carbohydr Polym. 2015;122:456–480. doi: 10.1016/j.carbpol.2014.10.063. [DOI] [PubMed] [Google Scholar]