Abstract
Prinsepia Royle (Rosaceae) is a genus native to China and the Himalayan region. In order to explain its current fragmented distribution pattern and to compare the impact of relatively recent climate changes on the genetic structure of Prinsepia species in different regions of China, a total of 66 populations and 617 individuals of four species of Prinsepia were genotyped, using three cpDNA markers. Meanwhile, phylogenetic reconstructions and divergence dating were conducted using the cpDNA haplotypes dataset and the nuclear ribosomal internal transcribed spacer (ITS) dataset, respectively. Ecological niche modeling (ENM) was performed to predict the potential distribution of each species of Prinsepia at present and during the Last Glacial Maximum. Both ITS and cpDNA gene trees support a north-south divergence of Prinsepia species in China. The divergence time of the northern and southern Clades occurred around the late Oligocene epoch. Combining the present distribution of Prinsepia species and their habitats, we inferred that the transition to a monsoon climate system in East Asia during the late Oligocene epoch, created a humid forest vegetation zone from central to East China, which potentially gave rise to the north-south divergence of Prinsepia species. Both regional climates and allopatric divergence may have played an important role in the speciation of P. sinensis and P. uniflora. P. sinensis had the lowest genetic diversity and a putative northward post-glacial colonization. The distribution range of P. uniflora was also extremely sensitive to interglacial-glacial cycles. P. utilis from southwestern China preserved more haplotypes than P. sinensis and P. uniflora due to its multiple and isolated refugia.
Keywords: arid belt, climatic barrier, East Asia, genetic differentiation, genetic diversity, monsoon systems
Introduction
East Asia harbors the most diverse temperate flora in the world, with a vast number (>600) of endemic genera (Wu and Wu, 1996; Manchester et al., 2009). Apart from the uplifts of the Qinghai-Tibet Plateau, the formation of the Asian monsoon system is also thought to have had a profound influence on the evolution and diversification of East Asian flora (Sun and Wang, 2005; Chen et al., 2018). The Asian monsoon system is comprised of the East Asian summer monsoon (EASM), the East Asian winter monsoon (EAWM), and the Indian summer monsoon (ISM) (Figure 1). Although the origin of the three components of the Asian monsoon system was asynchronous, all of them are considered to have existed at least by the late Oligocene epoch (Sun and Wang, 2005; Srivastava et al., 2012). It has been suggested that before the onset of the EASM, there was a broad belt of subtropical arid and semiarid vegetation, extending from east to northwest China (Guo et al., 2008). The eastern and central parts of this arid and semiarid vegetation zone were replaced by a humid forest vegetation zone with the establishment of the East Asian monsoon system, and the arid and semiarid zone became restricted to northwest China (Guo et al., 2008). Multiple changes to aridity or monsoon intensity have occurred in East Asia since then (Sun and Wang, 2005; Wang et al., 2008).
FIGURE 1.
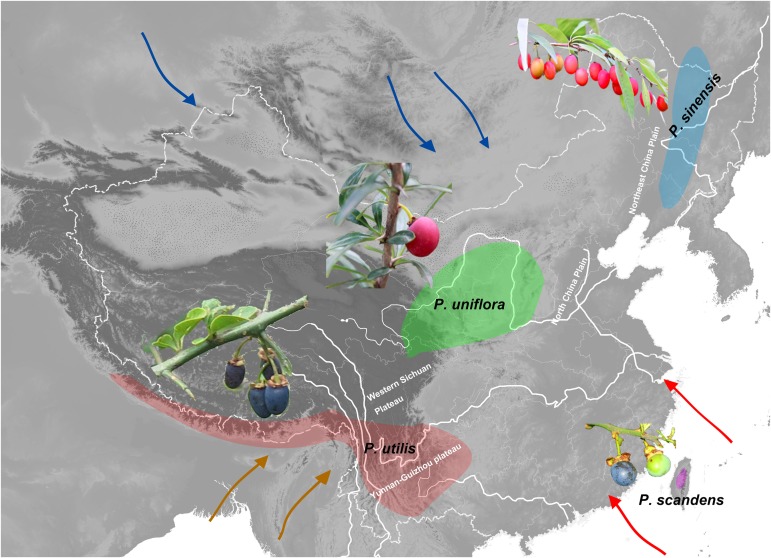
Approximate geographic distributions of the species of Prinsepia. Arrows of different colors indicate the East Asian summer monsoon (red), the East Asian winter monsoon (blue), and the Indian summer monsoon (green).
There are a few studies that focused on the origin of different vegetation types or ecosystems in East Asia as a result of East Asian monsoons (e.g., Kong et al., 2017; Yu et al., 2017). However, no studies have focused on the group of species that may have been present in the arid and semiarid vegetation zone in China during the Oligocene epoch. The east–west oriented arid belt is thought to have acted as a climate barrier to forest species between Northeast China and South China (Bai et al., 2016; Ye et al., 2017). Conversely, what is the fate of drought-tolerant or heliophilous plants distributed in this east–west oriented semiarid-arid belt, after the formation of the humid forest vegetation zone which was brought by the East Asian monsoon system?
Prinsepia Royle (Rosaceae) is native to China and the Himalayan region. Although the species of this genus exhibit a certain level of habitat divergence, they are all drought-tolerant shrubs, which have axillary spines and bear blossoms in early spring. This genus contains four species which are found in four separate areas (Figure 1; Gu and Bartholomew, 2003). Among them, P. uniflora Batalin is found in the semiarid region of north and northwest China, mainly on the Loess plateau (Figure 1). P. sinensis (Oliv.) Oliv. ex Bean is morphologically similar to P. uniflora, but this species is found in the eastern part of northeast China, an area more affected by the EASM (Figure 1). P. utilis Royle is found from southwest China to the Himalayan region (Figure 1) and the western Yunnan-Guizhou plateau is the core distribution area of this species. P. scandens Hayata, which only appears in the mountain ranges of mid-eastern Taiwan (Figure 1), is morphologically very similar to P. utilis.
Because different plant genera differ in their evolutionary ages and niche evolution, their divergence or genetic structure should be affected by different geological processes and climatic events. From previous large-scale phylogenetic studies of Rosaceae, we supposed that the time of origin for Prinsepia pre-dated the onset of the East Asian monsoon system (Xiang et al., 2017; Zhang et al., 2017). In this study, we first wanted to elucidate the phylogenetic relationships among Prinsepia species, which exhibit a very particular fragmented distribution. The gap between the two northerly species and the two southerly species in this genus is exactly the core region of the temperate and subtropical forests in East Asia (Figure 1). We aimed to identify geological or climatic events behind the fragmented distribution within this genus. In addition, although quite a lot of phylogeographic studies have focused on plants in north and south China and their responses to the glacial-interglacial cycles of the Quaternary, phylogeographic studies of endemic genera of East Asia, containing several species, are rare. This research involved a phylogeographic study of all the species of Prinsepia and we aimed to compare the impact of the relatively recent environmental changes (Quaternary glaciations) on the genetic structure of each species in the different regions of China.
Materials and Methods
Plant Material
We sampled all the four species of Prinsepia, covering most of its distribution range in China. A total of 617 individuals from 66 populations were sampled, including 18 populations of P. uniflora, six populations of P. sinensis, 39 populations of P. utilis, and three populations of P. scandens. Two to fifteen individuals were collected from each population. The total numbers of individuals sampled of P. uniflora, P. sinensis, P. utilis, and P. scandens were 145, 61, 399, and 12, respectively. Silica-gel dried plant materials were all collected from the wild. The collection locations for each population are presented in Supplementary Appendix 1.
DNA Isolation, Amplification, and Sequencing
Total genomic DNA was extracted using a plant genomic DNA kit according to the manufacturer’s instructions (Tiangen, Beijing, China). We sequenced three intergenic spacer (IGS) regions of cpDNA (psbA-trnH, psbK-psbI, psbJ-petA) for phylogeographic analyses. Because the nuclear ribosomal internal transcribed spacer (ITS) region lacked variations within each species and between P. utilis and P. scandens, we sequenced five, four, six, and three samples for P. uniflora, P. sinensis, P. utilis, and P. scandens, respectively. The primer sequences and procedures for DNA amplification are listed in Table 1. All PCR products were sent to Sangon Biotech (Shanghai, China) for sequencing, and were sequenced using amplification primers. All sequences were submitted to GenBank (accession numbers: MK442012–MK442070).
Table 1.
The primer sequences and the corresponding references.
| Locus | PCR primers (5′–3′) | Reference |
|---|---|---|
| ITS | ITS4: TCCTCCGCTTATTGATATGC | White et al., 1990 |
| ITS5: GGAAGTAAAAGTCGTAACAAGG | ||
| PsbA-trnH | PsbA:GTTATGCATGAACGTAATGCTC | Sang et al., 1997 |
| trnH:CGCGCATGGTGGATTCACAAATC | ||
| PsbK-psbI | psbK: TTAGCCTTTGTTTGGCAAG | Lahaye et al., 2008 |
| psbI: AGAGTTTGAGAGTAAGCAT | ||
| psbJ-petA | psbJ: ATAGGTACTGTARCYGGTATT | Shaw et al., 2007 |
| petA: AACARTTYGARAAGGTTCAATT |
Phylogeographic Analyses of cpDNA
We combined three cpDNA fragments for the phylogeographic analyses. Fourteen indels (Gaps) were treated as a single mutation event and coded as substitutions A or T (a third base G or C will be used when three kinds of mutations coexisted). However, we deleted a 24 bp and a 65 bp region (aligned length) from the psbA-trnH data matrix, because they contained complex indel information and were too divergent to be aligned.
The number of cpDNA haplotypes (h), haplotype diversities (Hd) and nucleotide diversities (p) for each species were calculated using DNAsp V5.1 (Librado and Rozas, 2009). Based on the phylogenetic relationship, NETWORK v.5 (Fluxus Technology Ltd.) was used to generate the median-joining network for P. uniflora and P. sinensis, P. utilis and P. scandens, respectively. Permut 1.01 was used to compare Gst and Nst of P. uniflora and P. utilis, with 1000 permutations.
Divergence Dating
Bayesian phylogenetic reconstructions and divergence time estimations were conducted independently for concatenated cpDNA haplotypes and the ITS dataset, using Beast v1.8.4 (Drummond and Rambaut, 2007). We selected Exochorda serratifolia and Oemleria cerasiformis as outgroups for the cpDNA tree divergence time estimation, as used in previous large-scale phylogenetic studies of Rosaceae (Xiang et al., 2017; Zhang et al., 2017; Supplementary Appendix 2). The sequences for Sorbarieae and Kerrieae were also included in the ITS tree analysis (Supplementary Appendix 2). We used the original cpDNA sequences of all four species (without recoding and deleting two complex regions from psbA-trnH) to generate a haplotype file and these haplotypes were used for the divergence time estimation of cpDNA. The fossil records for Oemleria janhartfordae and Neviusia sp. from North America were used for calibration (DeVore et al., 2004; Benedict et al., 2011; Xiang et al., 2017). We used fossil constraint age, as provided by Xiang et al. (2017), and a standard deviation of 1.0 to calibrate the corresponding nodes. The MrModeltest 2.3 (Nylander, 2004) was used to determine the best-fitting nucleotide substitution model for the two datasets. The best-fit models for the ITS and cpDNA dataset were all GTR + G. An uncorrelated lognormal, relaxed clock model was applied to estimate rate change (Drummond et al., 2006). The birth-death process and Yule process were run independently as the tree priors. Marginal likelihood estimation, using path sampling and stepping-stone sampling, was used to perform model selection (Baele et al., 2012). Convergence of all parameters was checked using Tracer v1.7 (Rambaut et al., 2018). The analyses were run for 100 million generations and sampled every 1000 generations.
Ecological Niche Modeling
In order to study the distribution range shifts of each species from the Last Glacial Maximum (LGM; c. 21 kya) to the present, we carried out ecological niche modeling (ENM) using Maxent 3.2 (Phillips et al., 2006) with the default parameters. P. utilis and P. scandens were treated as a complex in this analysis because of their close phylogenetic relationship and similar habitat. The distribution data for each species were obtained from our collection and the herbarium records 2, 3 . After removing duplicates, we identified 18 sites for P. sinensis, 35 sites for P. uniflora and 68 sites for P. utilis and P. scandens (Supplementary Figure S1). The current and LGM climate data for 19 BIOCLIM variables were downloaded from the WorldClim database 4. We chose the highest resolution [30 arc-s (∼1 km)] for the current data and the Community Climate System Model 3.0 at 2.5 arc-min resolution for the LGM. Strongly correlated climatic variables, according to Pearson’s correlation coefficient (r > 0.9), were excluded (Supplementary Appendix 3). All models were run with 20 replicates in Maxent. Around 25% of the data in each run was randomly chosen for model testing and 75% was chosen for model training.
Climatic Data Analysis
In order to compare the difference in climate, between the distribution areas of adjacent species pairs (P. sinensis and P. uniflora; P. uniflora and P. utilis), we extracted the 19 BIOCLIM variables under current conditions (∼1960–1990) at the highest resolution [30 arc-s (∼1 km)] (Hijmans et al., 2005) using ArcGIS 10.25. The extracted locations were the same as the collection records in the ENM for P. sinensis and P. uniflora. However, we only used the distribution data for P. utilis on the Yunnan-Guizhou Plateau. We used a two-tailed t-test in Excel to compare the difference for each of the BIOCLIM variables at the sites of each pair of species.
Results
Bayesian Phylogenetic Analyses and Molecular Dating
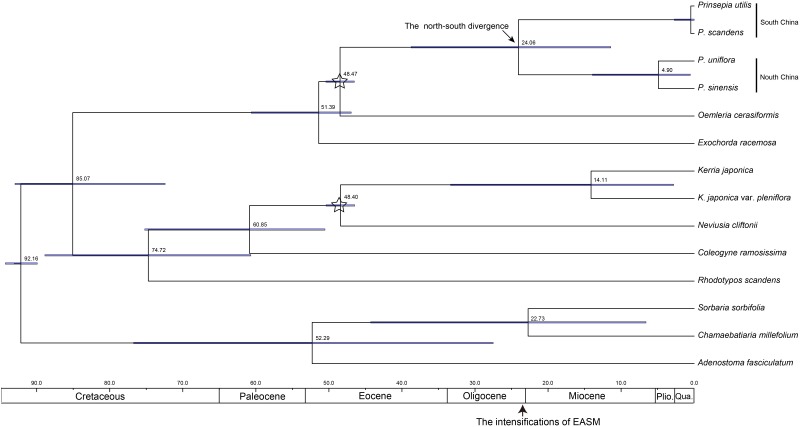
Both the ITS tree and the cpDNA tree supported a north-south divergence of Prinsepia species. P. uniflora and P. sinensis formed one clade, while P. utilis formed another monophyletic clade with P. scandens (Figure 2, 3). The populations of P. uniflora and P. sinensis each formed a monophyletic clade. In contrast, P. scandens did not form a monophyletic clade but was embedded in the clade of P. utilis (Figure 2, 3 and Supplementary Figure S2).
FIGURE 2.
The chronogram for Prinsepia derived from BEAST analysis of ITS sequences. Positions of the fossil calibrations are indicated by a five-pointed star. Divergence times are labeled on each node. Blue bars at nodes represent the 95% highest probability density (HPD) for the age of that node.
FIGURE 3.
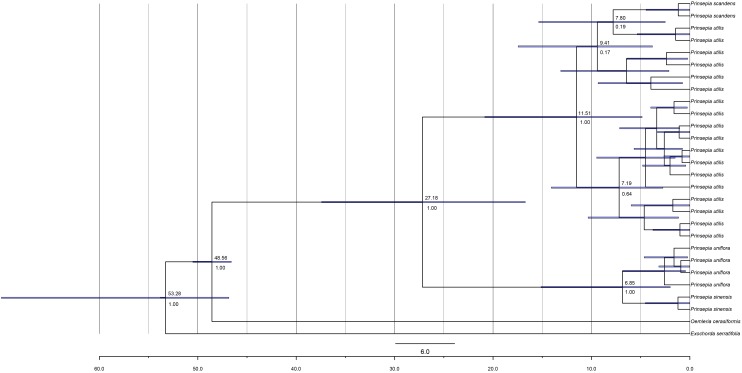
The chronogram of Prinsepia derived from BEAST analysis of combined cpDNA sequences.
The birth-death (BD) process was chosen as the best tree prior, based on the marginal likelihood estimation (MLE), using path sampling and stepping-stone sampling (MLEBD = −3041.00 and −3041.06, MLEYULE = −3041.86 and −3041.87 in ITS analysis; MLEBD = −3038.82 and −3038.82, MLEYULE = −3042.35 and −3042.40 in cpDNA analysis). The median origin time for Prinsepia was around 48.60 Mya, based on Oemleria, which is the existing genus most closely related to Prinsepia. The mean divergence time for the north and south clades of Prinsepia was 24.06 Mya (95% HPD = 11.41–38.75) in the ITS tree (Figure 2) and 27.18 Mya (95% HPD = 16.72–37.42) in the cpDNA tree (Figure 3). Within the North China clade, the mean divergence time for P. uniflora and P. sinensis was 4.90 Mya in the ITS tree and 6.85 Mya in the cpDNA tree (Figure 2, 3). Because of the lack of variation between P. utilis and P. scandens, in terms of their ITS sequences, we were unable to determine a divergence time for these two species. In the cpDNA tree, P. scandens did not, in fact, form a separate branch but was embedded within the clade of P. utilis.
Genetic Diversity and Genetic Structure
After recoding the indels and deleting two regions from the psbA-trnH matrix, we obtained a concatenated cpDNA dataset with a length of 1432 bp. The basic genetic data for the four Prinsepia species is shown in Table 2.
Table 2.
Genetic diversity assessment for four Prinsepia species using the concatenated cpDNA data set.
| Species | No. of samples | No. of haplotypes | Hd | π |
|---|---|---|---|---|
| Prinsepia sinensis | 61 | 2 | 0.064 | 0.00009 |
| Prinsepia utilis | 399 | 18 | 0.843 | 0.004 |
| Prinsepia uniflora | 145 | 6 | 0.775 | 0.00118 |
| Prinsepia scandens | 12 | 2 | 0.303 | 0.00021 |
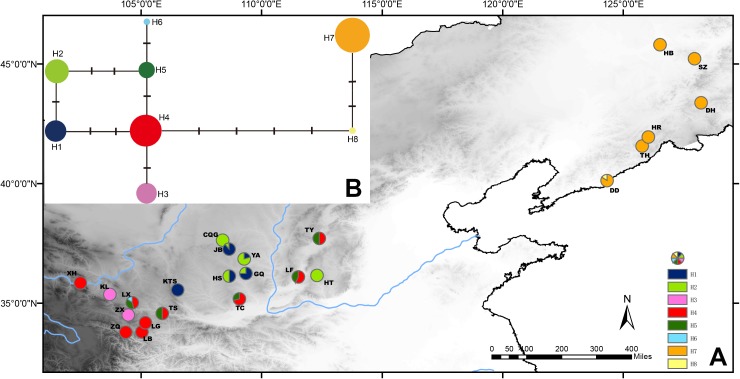
The population genetic structure of Prinsepia in north China is simple (Figure 4). Only two haplotypes were identified from 61 individuals of P. sinensis. All the populations of P. sinensis share an identical haplotype and only the Dandong (DD) population has an additional haplotype (Figure 4). Although six cpDNA haplotypes were identified from P. uniflora, these haplotypes were closely related (Figure 4). No significant phylogeographic structure was found for P. uniflora (Nst > Gst, p > 0.05).
FIGURE 4.
(A) Sampling localities and distribution of cpDNA haplotypes of P. uniflora and P. sinensis. (B) Median-joining network of the cpDNA haplotypes of P. uniflora and P. sinensis.
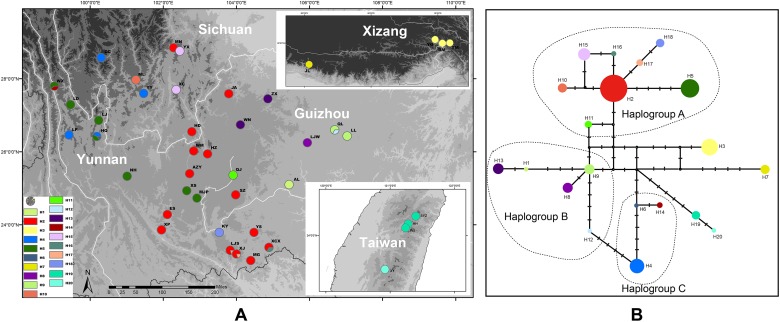
Twelve individuals of P. scandens were found to have two haplotypes (Figure 5), and these two haplotypes are closely related, and have seven nucleotide differences compared to the closest existing haplotype of P. utilis, according to the network analysis (Figure 5). The most closely related haplotype was found in the easternmost part of the distribution region of P. utilis in mainland China. A maximum of 18 haplotypes were identified from 399 individuals of P. utilis (Figure 5). P. utilis exhibited the highest haplotype diversity (0.843) and nucleotide diversity (0.004). Like the haplotypes from Taiwan, two haplotypes from Xizang and the other haplotypes of P. utilis were also connected by several hypothetical missing haplotypes (Figure 5). Permut did not reveal a significant phylogeographic structure for P. utilis (Nst > Gst, P > 0.05). However, a north-south oriented distribution pattern was found for several haplotypes in areas of the Hengduan mountains and Yunnan-Guizhou Plateau (Figure 5).
FIGURE 5.
(A) Sampling localities and distribution of cpDNA haplotypes of P. utilis and P. scandens. (B) Median-joining network of the cpDNA haplotypes of P. utilis and P. scandens.
Ecological Niche Modeling
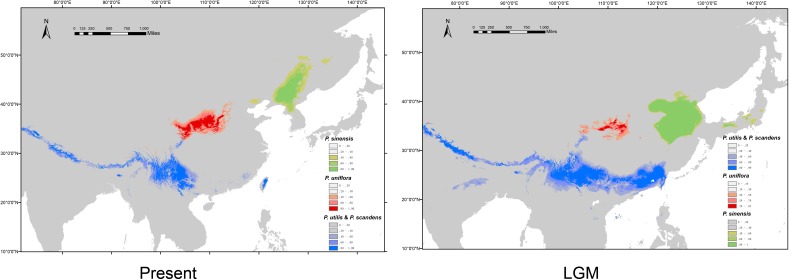
We obtained high AUC (the Area Under the Curve) values (>0.97) for each species and climate scenario, suggesting model performance of ENM are reliable. The predicted distribution range for P. sinensis during the LGM was more southerly than it is at present (Figure 6). P. uniflora had a more restricted distribution during the LGM than it does at present, and the range was also slightly south of the current distribution (Figure 6). The predicted species distribution during the LGM for P. utilis and P. scandens was significantly larger than the predicted species distributions under current conditions (Figure 6).
FIGURE 6.
Potential distribution probability of occurrence of the species of Prinsepia at present (1970–2000) and during the Last Glacial Maximum (LGM: c. 21 kya).
Climatic Data Analysis
The means of all but four (bio2, bio5, bio8, and bio10) of the BIOCLIM variables were significantly different (P < 0.01) when the sites of P. uniflora and P. sinensis were compared (Table 3). There were no statistically significant differences in mean values (P > 0.05) for three BIOCLIM variables (bio8, bio10, bio15) when comparing the ranges of P. uniflora and P. utilis (Table 3).
Table 3.
Results of t-tests on the 19 BioClim factors of adjacent species pairs.
| bio1 | bio2 | bio3 | bio4 | bio5 | bio6 | bio7 | bio8 | bio9 | bio10 | bio11 | bio12 | bio13 | bio14 | bio15 | bio16 | bio17 | bio18 | bio19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. utilis vs. P. uniflora | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.68 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.90 | 0.00 | 0.00 | 0.00 | 0.00 |
| P. uniflora vs. P. sinensis | 0.00 | 0.06 | 0.00 | 0.00 | 0.28 | 0.00 | 0.00 | 0.20 | 0.00 | 0.45 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
Significantly different: P < 0.05; P-values >0.05 were given in bold print. The Description of the bioclimatic variables can be found at the website (http://worldclim.org/bioclim).
Discussion
The North-South Divergence of Prinsepia Shaped by the EASM and the Fragmented Distribution of Prinsepia in North and South China
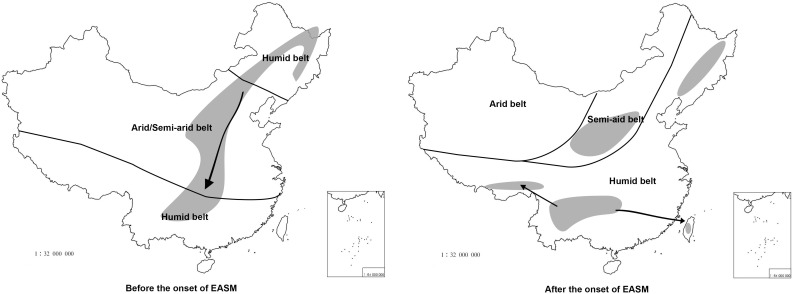
Oemleria is a monotypic genus endemic to western North America; this is the existing genus most closely related to Prinsepia (Xiang et al., 2017). Because the reliable fossil of Oemleria has been dated to late in the early Eocene epoch (49.42 ± 0.54 Ma) (Benedict et al., 2011), the origin time for Prinsepia should not follow after this time, if no other group had become extinct. All the related genera of Prinsepia, including Oemleria, Exochorda Lindl., Neviusia A. Gray, Kerria Candolle, Rhodotypos Siebold and Zuccarini, and Coleogyne Torrey, are all temperate taxa, which are distributed in East Asia or North America. These existing temperate taxa or their ancestors in East Asia or North America had a more northerly distribution in the Paleogene epoch, i.e., Arcto-Tertiary Flora (Milne and Abbott, 2002), so Prinsepia probably diverged from Oemleria in a high or middle-high latitude area and spread southward gradually along the mountain chains of China (Figure 7).
FIGURE 7.
The putative distribution of Prinsepia before and after the onset of the EASM. The climate zones follow Guo et al. (2008). Thick black lines represent boundaries of different climatic zones and arrows represent dispersal routes.
All four Prinsepia species have axillary spines, indicating that the ancestral species of Prinsepia may have grown in semi-arid areas. P. uniflora is a drought-tolerant shrub that grows in the semiarid zone of northwest China. Although P. utilis exhibited a certain degree of habitat divergence from P. uniflora (Table 3), it still cannot grow in humid and dense forests. P. utilis tends to grow in full sun and well-drained areas and blooms in the driest season (early spring). Descendent species are likely to occupy more or less the same ecological niches as their ancestor. From the current habitats of P. uniflora and P. utilis, which are found on opposite sides of central China, we infer that the ancestral species of Prinsepia was likely to have occurred (or survived) in the semiarid vegetation zone of central China during the Oligocene epoch (Figure 7).
The divergence time (median) of the northern clade and the southern clade of Prinsepia was the late Oligocene epoch in both cpDNA and ITS trees, which is very close to the onset or intensification of the EASM (Sun and Wang, 2005). With the influence of the East Asian monsoon circulation system from the late Oligocene epoch onward, eastern, and central China transformed into a humid forest vegetation zone (Guo et al., 2008; Yu et al., 2017). From the current distribution of P. utilis and P. uniflora we can see that although the mountainous environment of the eastern intervening area between these two species can provide suitable altitudes for Prinsepia plants, this region is exactly the core region of the temperate and subtropical forests of East Asia, which does not contain a large area of suitable habitat for Prinsepia species. The western intervening area, where Prinsepia does not occur now, is the eastern part of the Qinghai-Tibet Plateau, i.e., the Western Sichuan Plateau (Figure 1). Although the result of the ENM showed that the eastern margin of the Western Sichuan Plateau has conditions suitable for P. utilis under both current and LGM climate models, P. utilis actually only has a northern presence in southwest Sichuan (Figure 1, 7). Therefore, the ancestral species of Prinsepia, which gave rise to the north-south divergence of this genus, was more likely found in the mountain chains of central China rather than western Sichuan (Figure 7).
After the establishment of the EASM during the Oligocene epoch, the dry belt within China migrated northward (Guo et al., 2008; Figure 7). P. uniflora is mainly found in the semiarid zone of north and northwest China and this species is also the most drought-tolerant of all the members of the genus (Gu and Bartholomew, 2003). Although the Western Yunnan-Guizhou Plateau is located at the meeting point of the Indian and East Asia summer monsoons (Figure 1) and the forest vegetation in this area is well-developed, this region is famous for its winter and spring drought, as well as plentiful sunlight. More importantly, this region has a great variety of landscapes, providing various habitats for different plants. Many drought-tolerant or heliophilous shrubs can be found here and many drought-tolerant temperate genera have been found to have evolved distinct species in this region, for example Cotinus nana (Min and Barfod, 2008), Ostryopsis nobilis (Li and Skvortsov, 1999), and Pistacia weinmanniifolia (Min and Barfod, 2008).
If niche conservatism contributed greatly to the initial north-south divergence of Prinsepia, local adaptation may have had an influence on the subsequent speciation of P. sinensis and P. uniflora. From the climate data analysis, we found that all the bioclimatic variables related to precipitation, showed significant differences between the sites where P. sinensis and P. uniflora were found. In contrast, four bioclimatic variables relating to temperature, especially the temperature of growing season (the wettest Quarter and warmest Quarter), showed no significant difference between these two species (Table 3). P. sinensis is found in the eastern part of northeast China, which is more influenced by the EASM. The divergence time of P. sinensis and P. uniflora was from the late Miocene to the Pliocene. It has been suggested that the EASM intensified in the late Miocene–Pliocene (Ao et al., 2016), which may relate to the divergence of P. sinensis and P. uniflora. An alternative scenario is that ecological divergence was behind the allopatric speciation of these two species. Allospecies always differ in their ecological niches to some extent, reflecting adaptations to different climatic regimes or different competitive abilities (Rundell and Price, 2009). The Northeast China Plain and North China Plain may have played an important role in the obstruction of gene flow between these two species during different geological periods (Figure 1). And from the results of ENM, although the predicted distribution of P. sinensis was much more southerly in the LGM than its current distribution, it is still confined to the eastern area and has experienced increasing isolation from P. uniflora.
At least based on the results of molecular analyses, P. scandens may not be an independent species, but may be just an evolutionary lineage of P. utilis. The morphological characters of P. scandens are also very similar to P. utilis. A total of 50 genera of seed plants exhibit a disjunct distribution between the island of Taiwan and southwest mainland China (Ye et al., 2012). The divergence time between P. scandens and the populations of P. utilis could not be determined from the cpDNA estimation because the corresponding node was only weakly supported. Meanwhile, no variation was found between the ITS sequences of P. scandens and P. utilis from southwest China. Although it is difficult to explain why such a huge discrepancy existed between the mutation rate of the ITS region and cpDNA fragments, we are inclined to believe that the divergence time for P. scandens and P. utilis is relatively recent. P. scandens may originate from a dispersal event from southwest China. From the ENM analysis, we find that the suitable ranges during the LGM for P. utilis and P. scandens were larger than under current conditions and these two species may have had contact with each other during the LGM. This result is similar to some cold tolerant plants which have disjunct distributions between southwest China and Taiwan (Gao et al., 2015; Niu et al., 2017). P. scandens is found in areas of high elevation in Taiwan. The climate of south China became cold and dry during the LGM, thus providing a large area of suitable habitats for P. utilis and P. scandens.
Genetic Diversity and Geographic Structure of Prinsepia Species in North and South China
Prinsepia sinensis has only two haplotypes based on three cpDNA fragments in our study. One of these was shared by all six populations and the other haplotype was only found in the DD population, which is located in the southernmost part of the species’ range (Figure 4). Although phylogeographical analyses of some species have supported the suggestion that potential refugia existed in northeast China during glacial periods (Qiu et al., 2011; Zhao et al., 2013), P. sinensis probably retreated south area at this time, as suggested by its unique haplotype in northern areas and the ENM result; this situation is similar to the post-glacial recolonization of European biota (Hewitt, 1999).
A relatively high level of genetic diversity was observed in P. uniflora, which may be the result of the broad west to east range of this species. Perhaps because no large geographical barriers exist in this region, the haplotypes of P. uniflora have relatively close relationships with each other and no phylogeographical structure was found. P. uniflora mainly grows on the Loess plateau and prefers a semiarid habitat and its distribution during the LGM was much smaller than its current range, according to the ENM result (Figure 6). This suggests that although the drought resistant plants on the Loess plateau were able to retreat to southern areas during the LGM, they may still not have been able to cross climatic barriers such as the Ta-pa Mountains, where precipitation is plentiful and there are well-developed forests because of the EASM.
Southwest China is not only well-known for its high species richness, but is also characterized by the high degree of genetic diversity that it hosts (Sun et al., 2017). This high diversity can be attributed to a combination of long-term isolation between different mountain systems and environmentally stable habitats during the Pleistocene epoch (Qu et al., 2014). P. utilis has a higher haplotype diversity than the two northern species and a north-south oriented distribution pattern was found for several haplotypes (H2 and H5) (Figure 5), which may be closely related to the north-to-south orientation of the mountains in southwest China, and historical river separation (Zhang et al., 2011). The north-south features of southwest China created barriers to gene flow, as demonstrated for many groups (Liu et al., 2013; Feng et al., 2016; Luo et al., 2017).
Prinsepia utilis not only has the highest genetic diversity, but was also found to exhibit large genetic differentiation across the whole distribution range. The two haplotypes from Xizang were connected with the haplotypes from YGP by several hypothetical missing haplotypes (Figure 5B). P. utilis is distributed in a long and narrow area along the Himalayas. Many species with such a distribution have been found to exhibit allopatric divergence from conspecific populations in the Hengduan Mountains region (Jia et al., 2011; Li et al., 2011), because gene flows between them can be easily blocked by north-south oriented mountains or allopatric fragmentation caused by climate oscillations. Although no significant phylogeographic structure was found for P. utilis in our Permut analysis, the haplotypes of P. utilis in the Hengduan Mountains and YGP can be divided into three haplogroups according to their phylogenetic relationship and geographical distribution (Figure 5B). Several haplotypes (H2, H5, H10–11, H15–18) from southwest Sichuan, western Yunnan and central areas of Yunnan constituted a haplogroup (haplogroup A). An ancestral haplotype (H2) can be identified in this haplogroup, and this haplotype also has the widest distribution. The other haplotypes were all derived from this haplotype and showed fewer variations from it. In addition, the derived haplotypes were all found in regions adjacent to the range of haplotype H2. Perhaps because this region is the most suitable habitat for P. utilis according the ENM result, the ancestral haplotype experienced historical range expansion and subsequent isolation, which created the new haplotypes. The haplotypes from eastern Yunnan and Guizhou (H1, H8–9, and H12–13) formed haplogroup B. Haplogroup C contained three haplotypes form western Yunnan (H4, H6, and H14). Haplogroup B was found to have a close relationship to both haplogroups A and C, and haplogroup C showed a closer relationship to haplogroup B. Haplogroups B and C are geographically isolated, and the intervening areas are occupied by haplotypes from haplogroup A. Given these facts, we believe that the current phylogeographic structure of P. utilis can be attributed to the allopatric divergence of the ancestral lineage (i.e., haplogroups B, C, the haplotypes from Xizang and P. scandens), and regional range expansion of the renascent lineage (haplogroup A). Meanwhile, secondary contacts between the derived lineage and the remnant lineage were also found in two populations of P. utilis in northwest Yunnan (HQ and WX) (Figure 5A).
Conclusion
This study revealed that the humid belt in central-eastern China which was created by the EASM in the late Oligocene epoch might have acted as a climatic barrier for drought-tolerant plants. After a north-south divergence of Prinsepia, drastic genetic divergence occurred within the northern or southern clade. From our phylogeographic results, we infer that the distribution ranges of the plants in north China seem to have been highly impacted by the glacial interglacial cycles, resulting in this region hosting lower genetic diversity than southwest China. In contrast, southwest China retained more haplotypes due to its multiple and independent refugia, with the genetic divergence closely related to the high mountain and deep valley systems and the environmental conditions in southwest China.
Author Contributions
HS and BT conceived the study. BT, ZW, and XM collected the samples. BT completed the experiments. XM and BT analyzed the data. XM was the lead writer. BT, ZW, and HS corrected the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Xinxin Zhu for providing the photography of P. scandens. We also thank Zhiqiang Lu for his help in material collection.
Funding. This study was supported by the Major Program of the National Natural Science Foundation of China (31590823 to HS), the National Key R&D Program of China (2017YFC0505200 to HS), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203 to HS), theNational Science Foundation of China-Yunnan Joint Fund (U1802232 to HS), and the International Program of Chinese Academy of Sciences (151853KYSB2016021 to HS).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00128/full#supplementary-material
References
- Ao H., Roberts A. P., Dekkers M. J., Liu X. D., Rohling E. J., Shi Z. G., et al. (2016). Late miocene–pliocene asian monsoon intensification linked to antarctic ice-sheet growth. Earth Planet. Sci. Lett. 444 75–87. 10.1016/j.epsl.2016.03.028 [DOI] [Google Scholar]
- Baele G., Lemey P., Bedford T., Rambaut A., Suchard M. A., Alekseyenko A. V. (2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 29 2157–2167. 10.1093/molbev/mss084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai W. N., Wang W. T., Zhang D. Y. (2016). Phylogeographic breaks within asian butternuts indicate the existence of a phytogeographic divide in East Asia. New Phytol. 209 1757–1772. 10.1111/nph.13711 [DOI] [PubMed] [Google Scholar]
- Benedict J. C., DeVore M. L., Pigg K. B. (2011). Prunus and Oemleria (Rosaceae) flowers from the late early eocene republic flora of northwestern Washington State, USA. Int. J. Plant Sci. 172 948–958. 10.1086/660880 [DOI] [Google Scholar]
- Chen Y. S., Deng T., Zhou Z., Sun H. (2018). Is the East Asian flora ancient or not? Natl. Sci. Rev. 5 920–932. 10.1093/nsr/nwx156 [DOI] [Google Scholar]
- DeVore M. L., Moore S. M., Pigg K. B., Wehr W. C. (2004). Fossil Neviusia leaves (Rosaceae: Kerriae) from the lower-middle eocene of southern british columbia. Rhodora 106 197–209. [Google Scholar]
- Drummond A. J., Ho S. Y., Phillips M. J., Rambaut A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. 10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Zhao Q., Xu J., Qin J., Yang Z. L. (2016). Drainage isolation and climate change-driven population expansion shape the genetic structures of Tuber indicum complex in the hengduan mountains region. Sci. Rep. 6:21811. 10.1038/srep21811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. D., Zhang Y., Gao X. F., Zhu Z. M. (2015). Pleistocene glaciations, demographic expansion and subsequent isolation promoted morphological heterogeneity: a phylogeographic study of the alpine Rosa sericea complex (Rosaceae). Sci. Rep. 5:11698. 10.1038/srep11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C. Z., Bartholomew B. (2003). “Prinsepia Royle,” in Flora of China Vol. 9 eds Wu Z. Y., Raven P. H. (St. Louis, MO: Missouri Botanical Garden Press; ), 389–391. [Google Scholar]
- Guo Z. T., Sun B., Zhang Z. S., Peng S. Z., Xiao G. Q., Ge J. Y., et al. (2008). A major reorganization of asian climate by the early miocene. Clim. Past 4 153–174. 10.5194/cp-4-153-2008 [DOI] [Google Scholar]
- Hewitt G. M. (1999). Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 68 87–112. 10.1111/j.1095-8312.1999.tb01160.x [DOI] [Google Scholar]
- Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25 1965–1978. 10.1038/sdata.2018.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D. R., Liu T. L., Wang L. Y., Zhou D. W., Liu J. Q. (2011). Evolutionary history of an alpine shrub Hippophae tibetana (Elaeagnaceae): allopatric divergence and regional expansion. Biol. J. Linn. Soc. 102 37–50. 10.1111/j.1095-8312.2010.01553.x [DOI] [Google Scholar]
- Kong H. H., Condamine F. L., Harris A. J., Chen J. L., Pan B., Moller M., et al. (2017). Both temperature fluctuations and east asian monsoons have driven plant diversification in the karst ecosystems from southern China. Mol. Ecol. 26 6414–6429. 10.1111/mec.14367 [DOI] [PubMed] [Google Scholar]
- Lahaye R. R. Y., Savolainen V., Duthoit S., Maurin O., Bank M. V. D. (2008). A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger national park (South Africa) as a model system. Nat. Precedings [Preprint]. Available at: http://precedings.nature.com/documents/1896/version/1 (accessed May 10, 2015). [Google Scholar]
- Li P. C., Skvortsov A. K. (1999). “Ostryopsis Decaisne,” in Flora of China Vol. 4 eds Wu Z. Y., Raven P. H. (St. Louis, MO: Missouri Botanical Garden Press; ), 289. [Google Scholar]
- Li Y., Zhai S. N., Qiu Y. X., Guo Y. P., Ge X. J., Comes H. P. (2011). Glacial survival east and west of the ’Mekong-Salween Divide’ in the himalaya-hengduan mountains region as revealed by AFLPs and cpDNA sequence variation in Sinopodophyllum hexandrum (Berberidaceae). Mol. Phylogenet. Evol. 59 412–424. 10.1016/j.ympev.2011.01.009 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Liu J., Moller M., Provan J., Gao L. M., Poudel R. C., Li D. Z. (2013). Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytol. 199 1093–1108. 10.1111/nph.12336 [DOI] [PubMed] [Google Scholar]
- Luo D., Xu B., Li Z. M., Sun H. (2017). The ‘Ward Line–Mekong–Salween Divide’ is an important floristic boundary between the eastern himalaya and hengduan mountains: evidence from the phylogeographical structure of subnival herbs Marmoritis complanatum (Lamiaceae). Biol. J. Linn. Soc. 185 482–496. 10.1093/botlinnean/box067 [DOI] [Google Scholar]
- Manchester S. R., Chen Z. D., Lu A. M., Uemura K. (2009). Eastern Asian endemic seed plant genera and their paleogeographic history throughout the northern hemisphere. J. Syst. Evol. 47 1–42. 10.1111/j.1759-6831.2009.00001.x [DOI] [Google Scholar]
- Milne R. I., Abbott R. J. (2002). The origin and evolution of tertiary relict floras. Adv. Bot. Res. 38 281–314. 10.1016/S0065-2296(02)38033-9 [DOI] [Google Scholar]
- Min T. L., Barfod A. (2008). “Anacardiaceae,” in Flora of China Vol. 11 eds Wu Z. Y., Raven P. H. (St. Louis, MO: Missouri Botanical Garden Press; ), 335–345. [Google Scholar]
- Niu Y. T., Ye J. F., Zhang J. L., Wan J. Z., Yang T., Wei X. X., et al. (2017). Long-distance dispersal or postglacial contraction? insights into disjunction between himalaya–hengduan mountains and taiwan in a cold-adapted herbaceous genus. Triplostegia Ecol. Evol. 8 1131–1146. 10.1002/ece3.3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J. A. A. (2004). MrModeltest v2. Program distributed by the author. Genet. Soc. Am. 123 597–601. [Google Scholar]
- Phillips S. J., Anderson R. P., Schapire R. E. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Model. 190 231–259. 10.1016/j.ecolmodel.2005.03.026 [DOI] [Google Scholar]
- Qiu Y. X., Fu C. X., Comes H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59 225–244. 10.1016/j.ympev.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Qu Y. H., Ericson P. G. P., Quan Q., Song G., Zhang R. Y., Gao B., et al. (2014). Long-term isolation and stability explain high genetic diversity in the Eastern Himalaya. Mol. Ecol. 23 705–720. 10.1111/mec.12619 [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell R. J., Price T. D. (2009). Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol. Evol. 24394–399. 10.1016/j.tree.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Sang T., Crawford D. J., Stuessy T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84 1120–1136. 10.2307/2446155 [DOI] [PubMed] [Google Scholar]
- Shaw J., Lickey E. B., Schilling E. E., Small R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94 275–288. 10.3732/ajb.94.3.275 [DOI] [PubMed] [Google Scholar]
- Srivastava G., Spicer R. A., Spicer T. E. V., Yang J., Kumar M., Mehrotra R., et al. (2012). Megaflora and palaeoclimate of a late oligocene tropical delta, makum coalfield, Assam: evidence for the early development of the South Asia Monsoon. Palaeogeogr. Palaeoclimatol. 342-343 130–142. 10.1016/j.palaeo.2012.05.002 [DOI] [Google Scholar]
- Sun H., Zhang J., Deng T., Boufford D. E. (2017). Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers. 39 161–166. 10.1016/j.pld.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang P. (2005). How old is the Asian monsoon system?—palaeobotanical records from China. Palaeogeogr. Palaeoclimatol. 222 181–222. 10.1016/j.palaeo.2005.03.005 [DOI] [Google Scholar]
- Wang Y., Cheng H., Edwards R. L., Kong X., Shao X., Chen S., et al. (2008). Millennial- and orbital-scale changes in the East Asian monsoon over the past 224,000 years. Nature 451 1090–1093. 10.1038/nature06692 [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. San Diego, CA: Academic Press. [Google Scholar]
- Wu Z. Y., Wu S. G. (1996). ““A proposal for a new floristic kingdom (realm)–the Asiatic kingdom, its delineation and characteristics,”,” in Floristic Characteristics and Diversity of East Asian Plants, eds Zhang A. L., Wu S. G. (Beijing: China Higher Education; ), 3–42. [Google Scholar]
- Xiang Y., Huang C. H., Hu Y., Wen J., Li S., Yi T., et al. (2017). Evolution of rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 34 262–281. 10.1093/molbev/msw242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. F., Chen Z. D., Bing L., Qin H. N., Yang Y. (2012). Disjunct distribution of vascular plants between southwestern area and Taiwan area in China. Biodivers. Sci. 20 482–494. [Google Scholar]
- Ye J. W., Bai W. N., Bao L., Wang T. M., Wang H. F., Ge J. P. (2017). Sharp genetic discontinuity in the aridity-sensitive Lindera obtusiloba (Lauraceae): solid evidence supporting the Tertiary floral subdivision in East Asia. J. Biogeogr. 44 2082–2095. 10.1111/jbi.13020 [DOI] [Google Scholar]
- Yu X. Q., Gao L. M., Soltis D. E., Soltis P. S., Yang J. B., Fang L., et al. (2017). Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215 1235–1248. 10.1111/nph.14683 [DOI] [PubMed] [Google Scholar]
- Zhang S. D., Jin J. J., Chen S. Y., Chase M. W., Soltis D. E., Li H. T., et al. (2017). Diversification of rosaceae since the late cretaceous based on plastid phylogenomics. New Phytol. 214 1355–1367. 10.1111/nph.14461 [DOI] [PubMed] [Google Scholar]
- Zhang T. C., Comes H. P., Sun H. (2011). Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern sino-himalayan region and its correlation with historical river capture events. Mol. Phylogenet. Evol. 60 1–12. 10.1016/j.ympev.2011.04.009 [DOI] [PubMed] [Google Scholar]
- Zhao C., Wang C. B., Ma X. G., Liang Q. L., He X. J. (2013). Phylogeographic analysis of a temperate-deciduous forest restricted plant (Bupleurum longiradiatum Turcz.) reveals two refuge areas in China with subsequent refugial isolation promoting speciation. Mol. Phylogenet. Evol. 68 628–643. 10.1016/j.ympev.2013.04.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.