Abstract
Background
Beginning in December 2013, an epidemic of chikungunya virus (CHIKV) infection spread across the Caribbean and into virtually all countries in the Western hemisphere, with >2.4 million cases reported through the end of 2017.
Methods
We monitored a cohort of school children in rural Haiti from May 2014, through February 2015, for occurrence of acute undifferentiated febrile illness, with clinical and laboratory data available for 252 illness episodes.
Results
Our findings document passage of the major CHIKV epidemic between May and July 2014, with 82 laboratory-confirmed cases. Subsequent peaks of febrile illness were found to incorporate smaller outbreaks of dengue virus serotypes 1 and 4 and Zika virus, with identification of additional infections with Mayaro virus, enterovirus D68, and coronavirus NL63. CHIKV and dengue virus serotype 1 infections were more common in older children, with a complaint of arthralgia serving as a significant predictor for infection with CHIKV (odds ratio, 16.2; 95% confidence interval, 8.0–34.4; positive predictive value, 66%; negative predictive value, 80%).
Conclusions
Viral/arboviral infections were characterized by a pattern of recurrent outbreaks and case clusters, with the CHIKV epidemic representing just one of several arboviral agents moving through the population. Although clinical presentations of these agents are similar, arthralgias are highly suggestive of CHIKV infection.
Keywords: arbovirus, Haiti, chikungunya virus
Data were obtained for 252 Haitian children with acute febrile illness from May 2014 through February 2015. Findings document passage of the major 2014 Chikungunya virus epidemic (82 laboratory-confirmed cases), as well as ongoing outbreaks/cases of Dengue, Zika, and Mayaro.
The past few decades have been characterized by the emergence and geographic expansion of a series of arboviruses, including dengue virus serotypes 1–4 (DENV1–DENV4), chikungunya virus (CHIKV) [1], and Zika virus (ZIKV) [2]. In December 2013, CHIKV (Asian clade) was identified for the first time in the Western Hemisphere, on the Island of St Martin in the Caribbean [3]. From St Martin, an epidemic wave of CHIKV infection moved across the Caribbean and spread into virtually every country in the hemisphere, with >2.4 million cases reported to the Pan American Health Organization through the end of 2017 [4]. Although the CHIKV epidemic caused substantial morbidity, during the same time period (2013–2017) and in the same region >8.8 million dengue cases were reported to the Pan American Health Organization [5], together with >800000 suspected and confirmed ZIKV infections [5], underscoring the magnitude and diversity of arboviral infections within the population.
Moreira et al [6] have recently summarized results of studies on the epidemiology of acute febrile illness (AFI) in Latin America [6–11]. Although these studies have provided a basis for identifying the major pathogens responsible for febrile illness in the region, diagnostic criteria and approaches have differed widely, with no study having the ability to screen for previously unidentified pathogens; across all 17 studies evaluated by Moreira [6], the median number of pathogens tested per individual was 3.5. Studies have also tended to be cross-sectional, making it difficult to assess patterns of infection with different agents, data which are of potential importance in understanding drivers or facilitators for epidemic disease within the region.
Our group has maintained ongoing surveillance of a cohort of approximately 1250 children attending schools operated by the Christianville Foundation (a US-based nongovernmental organization) in a semirural region in Haiti close to Port-au-Prince [11]. In 2012–2013, acute undifferentiated febrile infection (ie, subjective fever in a child with no localizing signs or symptoms) was the third most common presenting complaint at the clinic serving the Christianville schools, with a rate of 235 cases per 1000 child-years of observation [11]. In these 2012–2013 studies, diagnostic testing of children with AFI was limited to screening for malaria (approximately 6% of patients) and possible typhoid fever. In 2014, anticipating the imminent onset of the CHIKV epidemic in Haiti, we expanded our evaluation to include collection of a blood sample for viral screening (including viral cultures) from case patients. Our goal was to diagnose and characterize the clinical and epidemiological features of AFI in these children, to understand temporal patterns of infection and explore differences in clinical presentation.
Our group has previously reported isolation of specific agents from individual members or small groups from this population [12–19], but we have not looked comprehensively at results or clinical presentations across an extended time period. The current report provides a comprehensive look at all virology results from May 2014 through February 2015, placing previous reports in temporal and phylogenic context; it also provides an in-depth analysis of clinical data from the CHIKV epidemic period.
MATERIALS AND METHODS
Patient Population/Specimen Collection
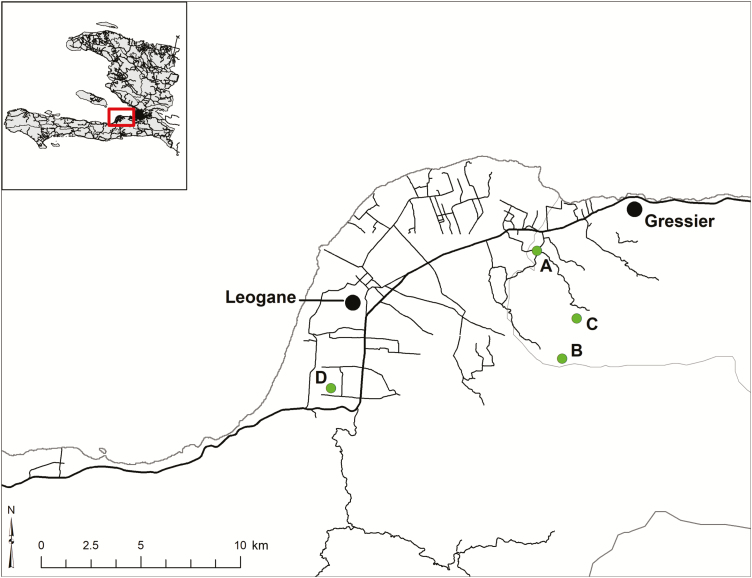
Blood samples were collected from schoolchildren with AFI seen at the Christianville School clinic beginning in May 2014; the current report summarizes results through February 2015. The clinic at the Christianville School is free and serves as the primary source of medical care for the approximately 1250 children at the 4 school campuses within the Christianville School system (Figure 1): this includes the main LaSalle/Gressier campus, housing kindergarten through grade13 (school A), and 3 small satellite elementary school campuses (schools B, C, and D).
Figure 1.
Map of study area. School locations are indicated by letters; the darker line represents the major highway through the region.
All schoolchildren who presented with AFI (defined based on a subjective history of fever and/or fever on presentation in a child with no obvious source of infection (ie, no respiratory symptoms, symptoms of urinary tract infection, or diagnostic criteria for malaria or typhoid) were included in this study [11]. All clinical data were collected and recorded by the clinic physician or nursing staff, with data extracted from clinical charts for analysis. The University of Florida and Haitian national institutional review boards approved all protocols, and written informed consent was obtained from parents or guardians of all study participants.
Laboratory Screening of Samples
Methods for sample collection and laboratory analysis have been reported elsewhere by our group [13–15, 19]. In brief: whole blood was collected in dipotassium ethylenediaminetetraacetic acid tubes (BD Vacutainer; Becton Dickinson), and plasma was extracted and frozen at −70°C. Samples were screened for CHIKV, DENV1-4, and ZIKV by means of real-time polymerase chain reaction (PCR) using standard procedures [19–21], with all virus isolations and RNA extractions for samples that might be CHIKV positive performed in a biosafety level 3 facility at the University of Florida Emerging Pathogens Institute in Gainesville.
Virus isolations from plasma specimens were attempted in Vero E6 cells (American Type Culture Collection CRL-1586). Cell cultures were maintained for up to 21 days after inoculation, or until CHIKV-induced cytopathic effects were observed in the cell monolayers. When cytopathic effects were observed throughout 50% of the monolayer, spent medium along with scraped cells were cryopreserved at −80°C for molecular testing. Total RNA extracted from the cell–spent media samples were subsequently tested by real-time reverse-transcription (RT) PCR [20–22], even if the corresponding plasma specimen had tested negative. In addition, spent medium from cultures displaying non-CHIKV cytopathic effects, which were DENV and ZIKV negative at real-time RT-PCR, were screened with a duplex RT-PCR for the viral RNAs of other alphaviruses and flaviviruses [23].
Analysis of Clinical and Demographic Data
Descriptive statistics were computed for the study population’s demographic characteristics and presentation of clinical symptoms by infection status. Demographic-specific attack rates (ARs) for AFI and for each pathogen were calculated by dividing the number of persons in the specific demographic category with the PCR-confirmed infection by the total number of persons in that category. Statistical significance in ARs was determined by comparing the 95% confidence intervals (CIs) between categories within each demographic variable; if the 95% CIs did not overlap, the categories were considered significantly different.
Forward and backward stepwise logistic regression models were employed for each pathogen-specific outcome (eg, presence of absence of CHIKV, referred to as “CHIKV model”) to narrow down the list of candidate predictors for use in the predictive models. The best-fit model for each pathogen outcome was selected so that adding or removing additional variables would not lower the Akaike information criterion. Only the selected variables that were informative (ie, had nonzero, noninfinity point estimates and associated CIs) were included in subsequent analysis.
The best-fit models for each pathogen (referred to as DENV1, DENV4, CHIKV, and CHIKV-ZIKV models) were then trained on a randomly selected 66% of the original data set, and validated against the remaining 34% of the data. This procedure was repeated 100 times for each pathogen model. Receiver operating characteristic (ROC) curves were generated for the average of the 100 replicates for each pathogen outcome. Predictive statistics including area under the ROC curve, positive predictive value (PPV), negative predictive value (NPV), sensitivity, and specificity were generated.
Finally, K-means clustering was applied to the data, to better observe how collections of clinical symptoms cluster with the presence and absence of each different pathogen. The variables included were binary, so that centroid values closer to 1 indicate that individuals classified into that cluster more often than not had that symptom. All data cleaning and analysis were carried out using R software, version 3.2.1 (R Core Team; 2014), and predictions were implemented using the forecast package [24].
RESULTS
Clinical and laboratory data were available for 252 children; we estimate that this represents approximately 95% of all children seen in the school clinic who met the criteria for AFI. We identified 223 AFI cases per 1000 child-years of observation, which is marginally less than the 235 cases per 1000 child-years that was observed in 2012–2013 [11].
Eighty-two children had a laboratory-confirmed diagnosis of CHIKV infection. Among these, 8 were infected with multiple viruses, including 6 with dual CHIKV-ZIKV, 1 with dual CHIKV-DENV2, and 1 with dual CHIKV and Mayaro virus (MAYV) infection (previously reported in [19]). Other infections identified included DENV1 (n = 24; 2 dual infections), DENV4 (n = 32), ZIKV (n = 9; 6 dual infections with CHIKV, as noted above, and 1 dual infection with DENV1 [13]), MAYV (n = 2; 1 dual infection with CHIKV, as noted above, and 1 dual infection with DENV1 [14]), coronavirus NL63 (n = 4 [15]), and enterovirus D68 (n = 1 [12]).
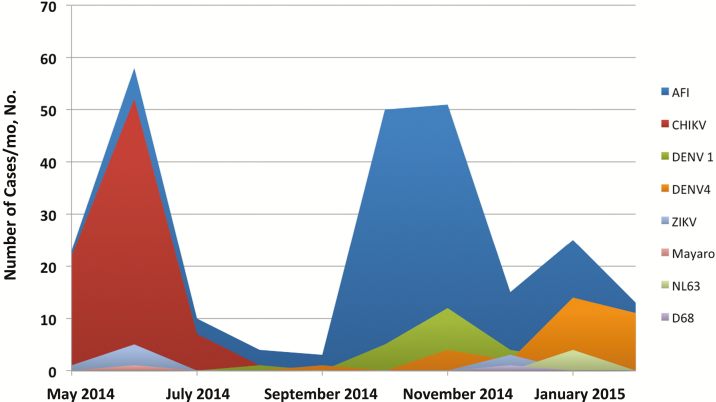
As shown in Figure 2, major AFI peaks were seen in June 2014, October and November 2014 and January 2015. CHIKV infections accounted for virtually all of the May–July AFI cases, consistent with the time that the CHIKV epidemic was known to be moving through Haiti. The staff at the school clinic initially assumed that the October–November AFI peaks reflected recurrence of CHIKV. However, at laboratory analysis we identified a cluster of DENV1 cases, followed by a small ZIKV cluster in December [14]. However, we were not able to find an cause for the majority of AFI cases in this time period, despite the use of viral culture techniques; work on identification of other possible etiologic agents for AFI cases in this time period continues. Finally, the January 2015 peak seems to have been due largely to an outbreak of DENV4 infection.
Figure 2.
Pathogen-specific surveillance over the course of the study period. Abbreviations: AFI, acute febrile illness; CHIKV, chikungunya virus; DENV1 and DENV4, dengue virus, serotypes 1 and 4; ZIKV, Zika virus.
Study population demographics, including age, sex, school location, and grade are presented in Table 1, along with demographic-specific ARs for AFI. Students in kindergarten had an AR of 39% (95% CI, 33%–44%), which was significantly greater than those in primary school students (15%; 12%–18%), and secondary school students (20%; 15%–26%). Kindergarteners also represented >45% of the students with AFI, but only 21% of the total cohort.
Table 1.
Population Demographics and Demographic-specific Attack Rates
| Variablea | Patients, No. (%) | AFI AR (95% CI) | |
|---|---|---|---|
| AFI Population (n = 252) | Total Population (N = 1390) | ||
| Sex | |||
| Female | 120 (47.6) | 705 (50.72) | 0.17 (.14–.20) |
| Male | 132 (52.4) | 685 (49.28) | 0.19 (.16–.22) |
| Schoolb | |||
| School A | 180 (71.4) | 558 (40.14) | 0.32 (.28–.36) |
| School B | 13 (5.2) | 217 (15.61) | 0.06 (.03–.09) |
| School C | 47 (18.7) | 126 (9.06) | 0.38 (.29–.47) |
| School D | 12 (4.8) | 231 (16.62) | 0.06 (.03–.09) |
| Gradeb | |||
| Kindergarten | 114 (45.2) | 296 (21.29) | 0.35 (.30–.41) |
| Primary | 96 (38.1) | 648 (46.62) | 0.14 (.11–.16) |
| Secondary | 42 (16.7) | 207 (14.89) | 0.19 (.14–.25) |
Abbreviations: AFI, acute febrile illness; AR, attack rate; CI, confidence interval.
aThe mean age in the AFI population was 7.8 years (standard deviation, 4.5). The mean age for the school-age population was 8.84 years.
bStatistically significant differences between ≥1 categories in this variable.
Pathogen-specific ARs along with their associated 95% CIs are presented in Table 2 by demographic categories. Of note, students in school A (AR, 9%; 95% CI, 7%–11%) had a significantly higher AR for CHIKV than those in school D (1%; 0%–2%).
Table 2.
Pathogen-specific Demographics and Attack Rates
| Variablea | Cohort, No. |
DENV1 Only (n = 24) | DENV4 Only (n = 32) | CHIKV Only (n = 74) | Dual CHIKV-ZIKV (n = 6) | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. (%) | AR (95% CI), % | No. (%) | AR (95% CI), % | No. (%) | AR (95% CI), % | No. (%) | AR (95% CI), % | ||
| Sex | |||||||||
| Female | 705 | 11 (45.8) | 0.02 (.01–.02) | 17 (53.1) | 0.02 (.01–.03) | 39 (52.7) | 0.06 (.04–.07) | 4 (66.7) | 0.01 (.0–.01) |
| Male | 685 | 13 (54.2) | 0.02 (.01–.03) | 15 (46.9) | 0.02 (.01–.03) | 35 (47.3) | 0.05 (.03–.07) | 2 (33.3) | 0.0 (.0–.01) |
| School | |||||||||
| School A | 736 | 19 (79.2) | 0.03 (.02–.04) | 22 (68.8) | 0.03 (.02–.04) | 64 (86.5) | 0.09 (.07–.11) | 5 (83.3) | 0.01 (.0–.01) |
| School B | 230 | 1 (4.2) | 0.0 (.0–.01) | 0 (0.0) | 0.0 (.0–.0) | 0 (0.0) | 0 (.0–.0) | 0 (0.0) | 0.0 (.0–.0) |
| School C | 174 | 3 (12.5) | 0.02 (.0–.04) | 8 (25.0) | 0.05 (.01–.08) | 8 (10.8) | 0.02 (.01–.8) | 1 (16.7) | 0.01 (.0–.2) |
| School D | 244 | 0 (0.0) | 0.0 (.0–.0) | 2 (6.3) | 0.01 (.0–.02) | 2 (2.6) | 0.01 (.0–.02) | 0 (0.0) | 0.0 (.0–.0) |
| Grade | |||||||||
| Kindergarten | 401 | 9 (37.5) | 0.02 (.01–.04) | 13 (40.6) | 0.03 (.02–.05) | 17 (23.0) | 0.04 (.02–.06) | 3 (50.0) | 0.01 (.0–.02) |
| Primary | 736 | 11 (45.8) | 0.01 (.01–.02) | 11 (34.4) | 0.01 (.01–.02) | 38 (51.4) | 0.05 (.03–.07) | 2 (33.3) | 0.0 (.0–.01) |
| Secondary | 247 | 4 (16.7) | 0.02 (.0–.03) | 8 (25.0) | 0.03 (.01–.05) | 19 (25.7) | 0.08 (.04–.11) | 1 (16.7) | 0.0 (.0–.01) |
Abbreviations: AR, attack rate; CHIKV, chikungunya virus; CI, confidence interval; DENV1 and DENV4, dengue virus, serotypes 1 and 4; ZIKV, Zika virus.
aThe mean (standard deviation) age was 8.3 (4.5) years for DENV1 only, 8.1 (4.7) years for DENV4 only, 9.6 (4.4) years for CHIKV only, 7.5 (5.4) years for dual CHIKV-ZIKV infection, and 8.8 (3.9) years for the school age population as a whole.
The proportion of positive cases of each pathogen with specific symptoms is presented in Table 3. By definition, all children in the study had a history of fever; those with a measured temperature of >37°C when seen in the clinic were included in the category of “fever at visit.”
Table 3.
Prevalence of Symptoms by Pathogen
| Symptom | Patients, No. (%) | ||||
|---|---|---|---|---|---|
| DENV1 Only (n = 24) | DENV4 Only (n = 32) | Any DENV (n = 56) | CHIKV Only (n = 74) | Dual CHIKV-ZIKV (n = 6) | |
| Fever at visit | 13 (54.1) | 9 (28.1) | 22 (39.3) | 29 (39.2) | 2 (33.3) |
| Headache | 9 (37.5) | 14 (43.8) | 23 (41.1) | 29 (39.2) | 1 (16.7) |
| Abdomen pain | 9 (37.5) | 16 (50.0) | 25 (44.6)a | 19 (25.7)a | 1 (16.7) |
| Myalgia | 6 (25.0) | 1 (3.1) | 7 (12.5)b | 43 (58.1)b | 4 (66.7) |
| Arthralgia | 5 (20.8) | 0 (0.0) | 5 (8.9)b | 47 (63.5)b | 4 (66.7) |
| Rash | 0 (0.0) | 2 (6.3) | 2 (3.6) | 3 (4.1) | 0 (0.0) |
Abbreviations: CHIKV, chikungunya virus; DENV, dengue virus; DENV1 and DENV4, DENV serotypes 1 and 4; ZIKV, Zika virus.
a P < .05.
b P < .001.
Compared with patients with CHIKV, a significantly greater proportion of patients who tested positive for “any DENV” experienced abdominal pain, whereas patients with CHIKV had significantly higher rates of arthralgia and myalgia than those with any DENV. Supplementary Table 1 presents the localization of arthralgias by infection outcome for patients with confirmed CHIKV only and dual CHIKV-ZIKV infection: arthralgias in CHIKV-only patients were most common in the knee (48.9%), wrist (44.7%), and ankle (36.2%).
Table 4 shows the results from the 4 stepwise logistic regression models that were performed for variable selection for each of the 4 outcomes of interest. Of note, no single variable was included in every model. Individuals aged 10–14 year or older had significantly greater odds of CHIKV infection than 3–4-year-olds (odds ratio, 3.6; 95% CI, 1.4–9.8). In addition, patients with AFI who had arthralgia had 16.2-fold increased odds of CHIKV infection (95% CI, 8.0–34.4) compared with patients with AFI who did not present with arthralgia, after adjusting for age category and sex. Although the number of cases was small, the presence of myalgia was significantly associated with increased odds of dual CHIKV-ZIKV infection (odds ratio, 6.5; 95% CI, 1.2–47.6), compared with the absence of myalgia
Table 4.
Stepwise Logistic Regression for Each Pathogen
| Variable | OR (95% CI) | |||
|---|---|---|---|---|
| DENV1 Only | DENV4 Only | CHIKV Only | CHIKV-ZIKV Dual Infection | |
| Sex | ||||
| Male | …a | … | 0.6 (.3–1.2) | … |
| Female | … | … | Reference | … |
| Age category | ||||
| 3–4 y | … | … | Reference | … |
| 5–9 y | … | … | 1.43 (.6–3.6) | … |
| 10–14 y | … | … | 3.6 (1.4–9.8) | … |
| 15–20 y | … | … | 2.4 (.7–8.5) | … |
| Fever at visit | 2.1 (.9–4.9) | - | … | … |
| Headache | … | 2.5 (1.1–5.5) | … | … |
| Myalgia | … | … | … | 6.5 (1.2–47.6) |
| Abdominal pain | … | … | … | … |
| Arthralgia | … | … | 16.2 (8.0–34.4) | … |
Abbreviations: CHIKV, chikungunya virus; CI, confidence interval; DENV1 and DENV4, dengue virus, serotypes 1 and 4; OR, odds ratio; ZIKV, Zika virus.
aEllipses indicate that the variable was not selected for inclusion in the stepwise model for the outcome of interest.
The variables selected for each pathogen model were then used in additional logistic regression models for each outcome that were trained on a randomly selected 66.7% of the data and validated against the remaining 33.3%. This process was repeated 100 times, and prediction metrics are presented in Supplementary Table 2. Of note, all models had 0% PPV, except for the CHIKV model (the model that predicted presence or absence of CHIKV based on the covariate [arthralgia] selected for inclusion from Table 4). The CHIKV model had a mean PPV of 66% (standard deviation [SD], 0.06; interquartile range [IQR], 0.62–0.70) and mean NPV of 80% (SD, 0.04; IQR, 0.78–0.83) across all 100 validation runs (Supplementary Table 2). The mean area under the ROC curve was 0.80 (SD, 0.05; IQR, 0.7–0.83; Supplementary Figure 1). In an additional model that predicted the binary CHIKV outcome as a function of presence of arthralgia without any demographic covariates, the PPV was 56% (SD, 0.05; IQR, 0.53–0.60; data not shown).
Results from the K-means cluster analysis for all symptoms with 4 clusters are shown in Supplementary Figure 2, and the cluster assignments by participant infection status are presented in Supplementary Table 3.
All patients in cluster 1 experienced arthralgia, leg pain, and ankle pain, and >75% experienced myalgia. Twenty-eight of the CHIKV cases were grouped into cluster 3, which also contained the vast majority of “no pathogen identified” patients and all with DENV4; patients in this cluster experienced low rates of all symptoms except abdominal pain (approximately 40%) and headache (approximately 25%). Eighteen of the 23 patients grouped into cluster 4 had CHIKV only. Patients in this cluster experienced a wide variety of clinical symptoms, including very high rates of arthralgia, leg pain, knee pain, and myalgia.
DISCUSSION
CHIKV was first identified in Tanzania in 1952, with subsequent association with sporadic cases of disease and localized outbreaks in Africa and parts of Asia [3]. In 2004, there was a reemergence of the virus, initially in Kenya, with a series of subsequent epidemics in and around the Indian Ocean. In December 2013, CHIKV infections were confirmed for the first time in the western hemisphere [4, 25]. It is clear from our study, and other longitudinal studies [7, 26], that as the epidemic moved through islands and countries it was the dominant cause of AFI for a period of 2–4 months. However, in both longitudinal and cross-sectional studies [7, 26, 27], what also emerges is a more complex picture of a mélange of arboviral infections, with frequent coinfections and cases and case clusters attributable to other viruses surrounding the CHIKV epidemic peak. In Haiti, in contrast to other studies that relied on serologic assays, we performed actual viral cultures on a majority of case patients; findings further underscored the extent of coinfections [19], and resulted in the identification of “unexpected” viruses, such as ZIKV and MAYV. It remains to be seen what impact this mix of infections has on host susceptibility and disease occurrence and severity [28, 29].
As has been noted in multiple studies [8, 22, 23, 30–32], it can be difficult, particularly in endemic regions, to identify an infecting arbovirus based solely on clinical presentation. The current study made use of a school cohort, and consequently ages were concentrated between 3 years and 20 years, reflecting attendance in prekindergarten through “grade 13” (an additional year of school generally taken as part of the secondary education cycle). As noted above, our clinical staff was convinced that the AFI peak seen in October and November 2014 (Figure 2) reflected a “recurrence” of CHIKV; in actuality, it built on a DENV1 case cluster, with some ZIKV mixed in at the end, and a still unidentified presumptive pathogen responsible for the majority of cases. Although we recognize the difficulties inherent in making clinical arboviral diagnoses, our data reinforce the frequency with which arthralgias are seen in CHIKV cases (with knees, wrists, and ankles most commonly affected, in keeping with findings in studies in other locales [9, 30]).
Within our AFI study population, age, sex, and arthralgias were predictive of CHIKV infection, with a PPV of 66% (Supplementary Table 2). The link with age is not immediately clear, but we hypothesize that it reflects lack of immunity to CHIKV among all population age groups for this emergent epidemic, in contrast to the likelihood of immunity to other endemic agents, such as DENV, among older students. Obviously, the PPV will drop with low-level endemicity, compared with epidemic CHIKV; nonetheless, arthralgias seem to be useful markers for initial recognition of a possible CHIKV outbreak and targeting of the diagnostic workup.
This study is subject to some limitations. Samples were collected through a syndromic surveillance program whereby children who presented to the school clinic with a history of fever and/or had a fever at the time of the clinic visit (but no other localizing signs or symptoms) had blood collected and tested for viral infections. Given that viremia is relatively transient in some of these infections, it is possible that children with a history of fever, but no fever when seen in the clinic, were past the period of viremia, resulting in an underestimation of actual case numbers. Different rates of asymptomatic and subclinical infection for the various pathogens may also have resulted in differential case detection, potentially biasing our results.
Furthermore, the spectrum of clinical illness in patients who did experience symptomatic infections may be variable, which could influence care seeking. For example, because of the link between CHIKV infection and arthralgias, patients with CHIKV may have been more likely to seek medical care [33]. In addition, the clinical symptom data used in this analysis were not collected in a questionnaire format. Rather, symptom presence or absence was elicited from patients on presentation to the clinic as part of a routine medical history. Thus, it is possible that less severe (or perceived less severe) symptoms were not reported by the patient and thus incorrectly marked as “absent” in the medical records.
Despite these limitations, our data provide a unique picture of the shifting causes of AFI in Haiti, and they highlight the apparent endemicity of a number of arboviral species. Interesting questions regarding viral origins are also raised by our identification of a cluster of ZIKV/CHIKV dual infections in May and June of 2014, at the very beginning of what subsequently became the ZIKV epidemic; as previously reported by our group, CHIKV and ZIKV strains from coinfected patients clustered monophyletically in their respective phylogeny, with molecular clock analysis suggesting that the 2 clades were introduced into the local population within overlapping time frames [19].
As in any study of arboviruses, there is also a need to link findings with entomologic data. White et al [34] have recently published results of mosquito studies within this same region and time period, which demonstrated exclusive carriage of CHIKV by Aedes aegypti; however, in studies conducted in 2016, we isolated a new “American” subgroup of African lineage CHIKV from Aedes albopictus in this same area of Haiti, further highlighting the dynamic flow of strains (and vectors) across time. Clearly, ongoing surveillance is essential within this setting, both to monitor occurrence and transmission pathways of known pathogens, such as CHIKV, DENV, ZIKV, and MAYV, and to identify new and emergent viruses and viral clades, which may, in turn, form the basis for future epidemics.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the students and teachers at the Christianville School system, and staff members within the clinic, for their interest and willingness to participate in these studies.
Financial support. This work was supported by the US Department of Health and Human Services (grants R01 AI126357-01S1 to J. G. M. and Models of Infectious Disease Agent Study U01 GM110721-01 to D. A. T. C.) and the National Institutes of Health (grants to D. A. T. C.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.GBD 2013 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990–2013: quantifying the epidemiological transition. Lancet 2015; 386:2145–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campos GS, Bandeira AC, Sardi SI. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis 2015; 21:1885–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yactayo S, Staples JE, Millot V, Cibrelus L, Ramon-Pardo P. Epidemiology of chikungunya in the Americas. J Infect Dis 2016; 214:441–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geographic spread of chikungunya in the Americas December 2013–December 2017 Available at: http://ais.paho.org/phip/viz/ed_chikungunya_amro.asp. Accessed 23 February 2018.
- 5. Pan American Health Organization, World Health Organization. Dengue: PAHO/WHO data, maps and statistics Available at: http://www.paho.org/hq/index.php?option=com_topics&view=readall&cid=3273&Itemid=0. Accessed 22 February 2018.
- 6. Moreira J, Bressan CS, Brasil P, Siqueira AM. Epidemiology of acute febrile illness in Latin America. Clin Microbiol Infect 2018; 24:827–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharp TM, Ryff KR, Alvarado L, et al. Surveillance for chikungunya and dengue during the first year of chikungunya virus circulation in Puerto Rico. J Infect Dis 2016; 214:475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alva-Urcia C, Aguilar-Luis MA, Palomares-Reyes C, et al. Emerging and reemerging arboviruses: a new threat in Eastern Peru. PLoS One 2017; 12:e0187897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galán-Huerta KA, Martínez-Landeros E, Delgado-Gallegos JL, et al. Molecular and clinical characterization of chikungunya virus infections in Southeast Mexico. Viruses 2018; 10:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sahadeo N, Mohammed H, Allicock OM, et al. Molecular characterisation of chikungunya virus infections in Trinidad and comparison of clinical and laboratory features with dengue and other acute febrile cases. PLoS Negl Trop Dis 2015; 9:e0004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beau De Rochars VEM, Alam MT, Telisma T, et al. Spectrum of outpatient illness in a school-based cohort in Haiti, with a focus on diarrheal pathogens. Am J Trop Med Hyg 2015; 92:752–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. ElBadry M, Lednicky J, Cella E, et al. Isolation of an enterovirus D68 from blood from a child with pneumonia in rural Haiti: close phylogenetic linkage with New York strain. Pediatr Infect Dis J 2016; 35:1048–50. [DOI] [PubMed] [Google Scholar]
- 13. Lednicky J, Beau De Rochars VM, El Badry M, et al. Zika virus outbreak in Haiti in 2014: molecular and clinical data. PLoS Negl Trop Dis 2016; 10:e0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lednicky J, De Rochars VMB, Elbadry M, et al. Mayaro virus in child with acute gebrile illness, Haiti, 2015. Emerg Infect Dis 2016; 22:2000–02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beau De Rochars VM, Lednicky J, White S, et al. Isolation of coronavirus NL63 from blood from children in tural Haiti: phylogenetic similarities with recent isolates from Malaysia. Am J Trop Med Hyg 2017; 96:144–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White SK, Morris JG, Elbadry MA, Beau De Rochars VM, Okech BA, Lednicky JA. Complete genome sequences of chikungunya viruses isolated from plasma specimens collected from Haitians in 2014. Genome Announc 2017; 5:e00148–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elbadry M, White S, Loeb J, et al. Complete genomic sequence of dengue virus 1, isolated from plasma collected from a Haitian child in 2014. Genome Announc 2017; 5:e00331–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elbadry MA, White SK, Loeb JC, et al. Complete genomic sequence of dengue virus serotype 4 isolated from plasma collected from a Haitian child in 2014. Genome Announc 2017; 5:e01160–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White S, Mayian C, El Badry M, et al. Detection and phylogenetic characterization of arbovirus dual-infections among persons during a chikungunya fever outbreak, Haiti, 2014. PLoS Negl Trop Dis 12: e0006505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lanciotti RS, Kosoy OL, Laven JJ, et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 2007; 13:764–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santiago GA, Vergne E, Quiles Y, et al. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis 2013; 7:e2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lanciotti RS, Kosoy OL, Laven JJ, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 2008; 14:1232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Morais Bronzoni RV, Baleotti FG, Nogueira RMR, Nunes M, Figueiredo LTM. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian aphaviruses and flaviviruses. J Clin Microbiol 2005; 43:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hyndman R. Forecast: forecasting functions for time series and linear models. 2016. Available at: http://github.com/robjhyndman/forecast. Accessed April 2017. [Google Scholar]
- 25. Cassadou S, Boucau S, Petit-Sinturel M, Huc P, Leparc-Goffart I, Ledrans M. Emergence of chikungunya fever on the French side of Saint Martin island, October to December 2013. Euro Surveill Bull 2014; 19. pii: 20752. [DOI] [PubMed] [Google Scholar]
- 26. Tomashek KM, Lorenzi OD, Andújar-Pérez DA, et al. Clinical and epidemiologic characteristics of dengue and other etiologic agents among patients with acute febrile illness, Puerto Rico, 2012–2015. PLoS Negl Trop Dis 2017; 11:e0005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carrillo-Hernández MY, Ruiz-Saenz J, Villamizar LJ, Gómez-Rangel SY, Martínez-Gutierrez M. Co-circulation and simultaneous co-infection of dengue, chikungunya, and Zika viruses in patients with febrile syndrome at the Colombian-Venezuelan border. BMC Infect Dis 2018; 18:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katzelnick LC, Gresh L, Halloran ME, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science 2017; 358:929–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Halai UA, Nielsen-Saines K, Moreira ML, et al. Maternal Zika virus disease severity, virus load, prior dengue antibodies, and their relationship to birth outcomes. Clin Infect Dis 2017; 65:877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Danis-Lozano R, Díaz-González EE, Trujillo-Murillo KDC, et al. Clinical characterization of acute and convalescent illness of confirmed chikungunya cases from Chiapas, S. Mexico: a cross sectional study. PLoS One 2017; 12:e0186923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magalhaes T, Braga C, Cordeiro MT, et al. Zika virus displacement by a chikungunya outbreak in Recife, Brazil. PLoS Negl Trop Dis 2017; 11:e0006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sasmono RT, Perkasa A, Yohan B, et al. Chikungunya detection during dengue outbreak in Sumatra, Indonesia: clinical manifestations and virological profile. Am J Trop Med Hyg 2017; 97:1393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paniz-Mondolfi AE, Rodriguez-Morales AJ, Blohm G, Marquez M, Villamil-Gomez WE. ChikDenMaZika syndrome: the challenge of diagnosing arboviral infections in the midst of concurrent epidemics. Ann Clin Microbiol Antimicrob 2016; 15:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. White SK, Mavian C, Salemi M, et al. A new ‘American’ subgroup of African-lineage chikungunya virus detected in and isolated from mosquitoes collected in Haiti, 2016. PLoS One 2018; 13:e0196857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.