Abstract
Cholesterol is an essential structural component of cellular membranes and precursor molecule for oxysterol, bile acid, and hormone synthesis. The study of intracellular cholesterol trafficking pathways has been limited in part due to a lack of suitable cholesterol analogues. Herein, we developed three novel diazirine alkyne cholesterol probes: LKM38, KK174, and KK175. We evaluated these probes as well as a previously described diazirine alkyne cholesterol analogue, trans-sterol, for their fidelity as cholesterol mimics and for study of cholesterol trafficking. LKM38 emerged as a promising cholesterol mimic because it both sustained the growth of cholesterol-auxotrophic cells and appropriately regulated key cholesterol homeostatic pathways. When presented as an ester in lipoprotein particles, LKM38 initially localized to the lysosome and subsequently trafficked to the plasma membrane and endoplasmic reticulum. LKM38 bound to diverse, established cholesterol binding proteins. Through a detailed characterization of the cellular behavior of a panel of diazirine alkyne probes using cell biological, biochemical trafficking assays and immunofluorescence approaches, we conclude that LKM38 can serve as a powerful tool for the study of cholesterol protein interactions and trafficking.
Keywords: lipids, lipoproteins, metabolism, biorthogonal probes, regulation
Cholesterol is an essential structural component of cellular membranes that regulates the membrane fluidity and permeability of phospholipid bilayers and the functions of integral membrane proteins. In addition, cholesterol serves as a precursor molecule for oxysterol, bile acid, and hormone synthesis. The distribution of cholesterol among cellular membranes is heterogeneous. The plasma membrane contains 60% to 70% of total cellular cholesterol at a concentration of ∼40 mol% (1, 2). In contrast, the ER and mitochondria are cholesterol-poor, with ∼1% of total cellular cholesterol in the ER, where its concentration is <5 mol% (1, 3). The low cholesterol content of the ER and its proximity to the cholesterol synthesis machinery facilitate sensitive monitoring of cellular cholesterol levels and tight regulation of uptake and synthesis. The ER-resident SREBP2 transcription factor serves as the primary regulatory axis for cholesterol homeostasis. When ER is cholesterol-depleted, membrane-bound SREBP2 traffics from the ER to the Golgi, where proteolytic processing liberates the soluble, mature transcription factor, which then translocates to the nucleus to upregulate genes involved in cholesterol synthesis and uptake. When the ER is cholesterol replete, SREBP2 is retained in the ER. The ER-resident protein ACAT converts excess sterol to cholesteryl esters for storage in lipid droplets (4). Excess cholesterol also promotes the conversion of cholesterol to oxysterols in the mitochondria, activating LXR, which upregulates the expression of genes involved in cholesterol efflux (5). The asymmetric distribution of cholesterol within and across different cellular membranes is critical in these regulatory mechanisms and persists despite the rapid flux of cholesterol between surface and internal pools (6).
Mammalian cells acquire cholesterol through the uptake of cholesterol and cholesteryl ester-laden lipoprotein particles by receptor-mediated endocytosis or through de novo synthesis in the ER (7). The bulk distribution of endocytosed, LDL-derived cholesterol and cholesteryl esters depends on the lysosomal proteins Niemann-Pick C (NPC) 1 and 2, as well as lysosomal acid lipase (LAL). LDL-derived cholesterol and cholesteryl esters are delivered to late endosomes/lysosomes. Under acidic conditions, LAL cleaves cholesteryl esters, liberating free cholesterol (7). Sequential “hydrophobic handoffs” are proposed to facilitate the transfer of cholesterol to the limiting lysosomal membrane, the first between the lipase and NPC2 and the second between NPC2 and NPC1. NPC2, a small soluble protein localized to the lysosomal lumen, binds cholesterol with the iso-octyl side chain buried deep within its binding pocket (8). By contrast, NPC1 is a large multidomain protein located in the limiting membrane of late endosome/lysosomes. Its luminal N-terminal domain binds the 3-hydroxyl group of cholesterol and is hypothesized to accept cholesterol from NPC2 (9–11). Mutations in either NPC1 or NPC2 result in dysregulated cholesterol homeostasis with the accumulation of unesterified cholesterol in the lysosomal compartment and impaired cholesterol reesterification (12).
The study of intracellular cholesterol trafficking pathways is central to understanding homeostatic pathways as well as disease pathogenesis. However, past approaches have been limited in part due to a lack of suitable cholesterol probes. Intrinsically fluorescent cholesterol analogues (dehydroergosterol and cholestatrienol) have weak fluorogenic properties. Bulky fluorophore conjugates (BODIPY- and NBD-cholesterol) fail to faithfully mimic cholesterol trafficking and membrane behavior (13, 14). Minimally modified diazirine alkyne cholesterol probes provide a new approach for the study of cholesterol trafficking. The alkyne functional group enables copper-catalyzed azide alkyne cycloaddition (“click chemistry”) of azide reporter molecules (e.g., fluorophores or biotin) after trafficking has taken place, while the diazirine group enables UV-activated cross-linking of probes to nearby biomolecules. A diazirine alkyne probe, trans-sterol, was recently used to identify the static cholesterol proteome (15). Several known cholesterol binding proteins such as SREBP cleavage-activating protein and HMG-CoA reductase were identified, highlighting the utility of this approach. Protein binding for two other diazirine alkyne cholesterol probes, LKM38 and KK174, has been shown to be sensitive to cholesterol competition (16, 17). Nonetheless, the extent to which these cholesterol probes mimic the trafficking characteristics of native cholesterol and its behavior with respect to the regulation of sterol homeostatic responses has yet to be determined.
Herein, we studied diazirine alkyne cholesterol probes LKM38, KK174, and trans-sterol probe, as well as a novel probe, KK175, for their fidelity as cholesterol mimics and use as dynamic cholesterol probes. Through detailed characterization of the cellular behavior of these diazirine alkyne probes, we identified new tools for the study of intracellular cholesterol trafficking.
MATERIALS AND METHODS
Chemicals and reagents
With the exception of McCoy’s medium (Gibco), sodium bicarbonate (Corning), and GlutaMAX (Promega), all cell culture reagents as well as methyl-β-cyclodextrin, CuSO4, tris-hydroxypropyltriazolylmethylamine (THPTA), ascorbic acid, BSA, SDS, glutaraldehyde, cholesterol oxidase, sphingomyelinase, nicotinic acid, O-[2-(1-azepanyl)ethyl]hydroxylamine dihydrocholoride, and PMSF were obtained from MilliporeSigma. Lipoprotein-deficient serum was prepared from FBS in-house using Cab-O-Sil (Supelco Analytical) (18). Lalistat (LALi) was a gift from the Maxfield Laboratory (compound 13) (19). Cholesterol was from Steraloids. Acetylated LDL (acLDL) was from Alfa Aesar. The commercially available deuterated compounds used in this study included d7-cholesteryl oleate (Avanti Polar Lipids), d5-cholesterol (Medical Isotopes, Inc.), and d5-cholestenone (CDN Isotopes). d17-Oleic acid (Cayman Chemical) and d4-cholesterol (Cambridge Isotope Laboratories) were used for the synthesis of d17-LKM38 oleate and d4-cholesteryl oleate standards, respectively. Additional reagents included methanol (JT Baker), chloroform (BDH VWR Analytical), and formic acid (Acros Organics).
Diazirine alkyne probes
The trans-sterol probe was made in-house using a previously published synthesis (15). The synthesis of LKM38 and KK174 have previously been reported (16, 17). The synthesis of KK175 and LKM38 oleate as well as LKM38-one, d17-LKM38 oleate, and d4-cholesteryl oleate standards are detailed in the supplemental Methods online. Diazirine alkyne probe stocks were prepared at 10 µM in ethanol unless otherwise stated.
Cell culture
U2OS cells expressing the human scavenger receptor A (SRA) were obtained from the Maxfield Laboratory (20). Chinese hamster ovary (CHO) cells expressing human influenza hemagglutinin (HA) epitope-tagged NPC2 were generated by the transduction of CHO cells with a retroviral construct expressing NPC2-HA (21, 22). Human skin fibroblasts were obtained from ATCC, and CYP27A1-deficient CTX-205-5 fibroblasts were a gift from Eran Leitersdorf (23). U2OS-SRA cells were cultured in McCoy’s medium containing 10% FBS, 1.2 g/l sodium bicarbonate, and 1 mg/ml G418. CHO cells were cultured in DMEM-F12 (1:1) containing 5% FBS, 0.5 mM sodium pyruvate, and 2 mM l-glutamine. Human fibroblasts were cultured in DMEM containing 10% FBS and 1× GlutaMAX. Lipoprotein-deficient medium (LPDM) contained lipoprotein-depleted FBS instead of regular serum. Starvation media were prepared by adding 20 µM lovastatin and 50 µM mevalonate to LPDM.
Cell viability assay
U2OS-SRA cells were seeded at 3,000 cells/well in 96-well plates overnight before being subjected to a 15 min pulse of 0.5% methyl-β-cyclodextrin in LPDM to deplete cellular cholesterol. Cells were then treated with starvation media containing 10 µM cholesterol, LKM38, KK174, KK175, trans-sterol, probe or vehicle. After 72 h, cell viability was assayed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Microscopy
U2OS-SRA cells were grown on coverslips at 1.8 × 105 cells/well in six-well dishes. Cells were incubated in LPDM containing LKM38 oleate reconstituted acLDL (24) with or without 10 µM LALi overnight. Media were spiked with 1 µM LysoTracker Red DND-99 (Thermo Fisher Scientific) 2 h before fixation for 10 min in 4% paraformaldehyde (Electron Microcopy Sciences). Coverslips were washed and then subjected to copper-catalyzed cycloaddition chemistry (click reaction). Cells were placed in 0.5 ml Tris-buffered saline; 20 µl 50 mM CuSO4, 20 µl 10 mM THPTA, 5 µl 10 mM fluorescein azide (Lumiprobe), and 20 µl 250 mM ascorbic acid were added sequentially to each well and then incubated for 30 min while rocking in the dark. This was immediately followed by a second, identical click reaction to maximize click efficiency. Coverslips were washed for 5 min each with PBS containing 40 mg/ml BSA, 0.5 M NaCl, and PBS before mounting on slides. Images were collected on a Nikon A1+ confocal microscope at 100× magnification (pixel size = 0.1 µm; Z-step size = 0.13 µm; optical resolution = 0.23 µm; optical sectioning = 0.42 µm). Colocalization quantification was performed using ImageJ.
Gene expression
U2OS-SRA cells or CTX-205-5 (CYP27A1-deficient) cells were plated at 4 × 105 or 2 × 105, respectively, in 6 cm dishes and grown overnight before incubation with starvation media containing 10 µM cholesterol, LKM38, KK174, KK175, trans-sterol, or vehicle (ethanol) for 14 h. After washing, RNA was collected in Trizol (Ambion) and prepared for quantitative PCR (qPCR) using the SuperScript III First-Strand Kit (Invitrogen). Relative RNA amount was determined by qPCR using PerfeCTa SYBR green FastMix (QuantaBio) and primers (IDT) to HMG-CoA synthase (HMGCS) (forward: GAT GTG GGA ATT GTT GCC CTT; reverse: ATT GTC TCT GTT CCA ACT TCC AG), ABCA1 (forward: TTC CCG CAT TAT CTG GAA AGC; reverse: CAA GGT CCA TTT CTT GGC TGT), and ribosomal protein lateral stalk subunit P0 (Rplp0) (forward: GGA GAC GGA TTA CAC CTT CCC; reverse: CAG CCA CAA AGG CAG ATG G). Relative quantification of target transcript abundance was calculated with the ddCT method using Rplp0 as an endogenous control on an ABI 7500 Fast Real-Time PCR system.
Biochemical trafficking assays
For esterification and cholestenone assays, cells were plated at 6 × 104 cells/well in 12-well dishes. After overnight incubation, media were changed to LPDM supplemented with acLDL reconstituted with either LKM38 oleate or d7-cholesteryl oleate overnight in the presence of 10 µM LALi. Cells were chased in LALi-free media for 0–12 h before harvest. For esterification experiments, d9-oleate/BSA (25) was included during the chase period. For ester quantification, cellular lipids were extracted in 9:1 methanol-chloroform at −20°C. d17-LKM38 oleate or d4-cholesteryl oleate was added as an internal standard for LKM38 oleate or d7-cholesteryl oleate experiments, respectively. Cell lysate was collected by scraping. Supernatant was collected after centrifugation, dried under nitrogen, and resuspended in 9:1 methanol-chloroform. For the quantification of plasma membrane cholesterol, we used a cholesterol oxidase assay (26). In brief, cells were fixed in 1% glutaraldehyde and then exposed to 0.1 U/ml sphingomyelinase and 2 U/ml cholesterol oxidase to convert surface-accessible cholesterol or LKM38 to cholestenone or LKM38-one, respectively. Cellular lipids were extracted in 9:1 methanol-chloroform, with d5-cholestenone added as an internal standard. Extracts were dried under nitrogen, resuspended in 20 µl 0.05 M O-[2-(1-azepanyl)ethyl]hydroxylamine dihydrocholoride and 10 µl formic acid, and heated for 1 h at 50°C to derivatize cholestenones. LC/MS/MS was monitored in the positive mode for the ion transitions of interest (supplemental Table S1).
Immunoprecipitation of probe complexes
Following the incubation in LPDM overnight, cells were pulse-labeled for 1 h with 2.5 µM (for NPC1) or 10 µM [for NPC2, caveolin, and vesicle-associated membrane protein 7 (VAMP7)] LKM38/methyl-β-cyclodextrin complexes diluted in McCoy’s medium with 0, 10, or 100× cholesterol/methyl-β-cyclodextrin as a competitor. LKM38 and cholesterol methyl-β-cyclodextrin complexes were prepared in a ratio of 2.5 mM methyl-β-cyclodextrin/250 µM sterol and then sterile-filtered. The amount of cholesterol recovered in the cyclodextrin solution was determined by MS. Cells were washed with PBS before being subjected to UV cross-linking (366 nm) by UV Stratalinker 1800 (Stratagene) for 5 min on ice. Cells were collected by scraping in RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate) containing 1 mM PMSF and EDTA-free Protease Complete (Roche), sonicated, and incubated for 1 h while rotating at 4°C. Lysates were spun at 16,000 g for 10 min at 4°C, and supernatants were collected and analyzed for protein content by BCA (Thermo Fisher Scientific). Equal amounts of protein were added to Protein A Dynabeads (Novex) loaded with the appropriate antibody. After immunoprecipitation, beads were washed and subjected to click reaction. Beads were resuspended in 50 µl RIPA lysis buffer before the sequential addition of 1 µl 50 mM CuSO4, 1 µl 5 mM THPTA, 0.5 µl 10 mM rhodamine X azide (Lumiprobe), and 1 µl 250 mM ascorbic acid. The mixture was incubated for 30 min at 37°C while shaking at 1,100 rpm in the dark. Beads were washed in lysis buffer, suspended in 1× Laemmli sample buffer, and incubated for 10 min at 37°C before loading on 4% to 12% precast Bis-Tris gels (Invitrogen) for SDS-PAGE. Fluorescent images of gels were collected by a Typhoon scanner at 100 µm resolution. After imaging, gels were transferred to nitrocellulose and then probed for the appropriate target by Western blotting. Immunoprecipitation and Western blotting antibody conditions are provided in supplemental Table S2.
Fluorescent gel profiling
U2OS-SRA cells were treated with 10 μM probe (in ethanol) in culture media for 24 h. Media were changed to normal culture media, LPDM, or LPDM containing 100 μg/ml acLDL for 18 h. Cross-linking, click reaction, SDS-PAGE, and imaging were performed as described above, except gels were methanol-fixed before imaging. Parallel samples were prepared for Western blot analysis.
RESULTS
Characterization of diazirine alkyne probes
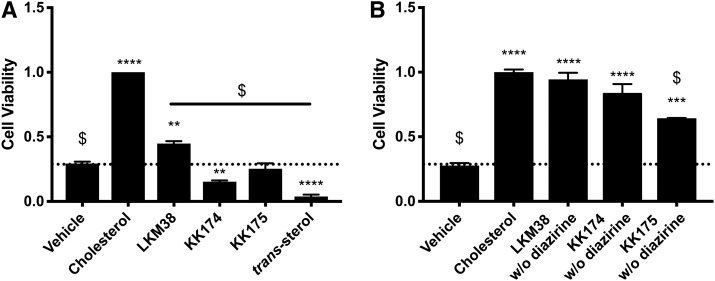
To develop tools for the study of cholesterol-protein interactions, we synthesized several novel cholesterol analogues. Each analogue contains a diazirine and alkyne group to enable UV cross-linking and copper-catalyzed cycloaddition, respectively. Probes were generated with diazirine and alkyne groups at different positions in an effort to survey the effect of these modifications on the behavior of the molecule (Fig. 1). We also evaluated a previously described diazirine alkyne cholesterol analogue, trans-sterol probe (15). In a stringent test of cholesterol mimicry, we examined the ability of diazirine alkyne probes to support the growth of cholesterol-auxotrophic cells (Fig. 2A). KK174 and trans-sterol were cytotoxic, while KK175 was cytostatic. By contrast, LKM38 partially supported auxotrophic cell growth, although not as robustly as cholesterol. Interestingly, alkyne-only probes supported growth at levels nearly equivalent to cholesterol, suggesting the diazirine group is the modification that limits the ability of the probes to support growth in the absence of cholesterol (Fig. 2B).
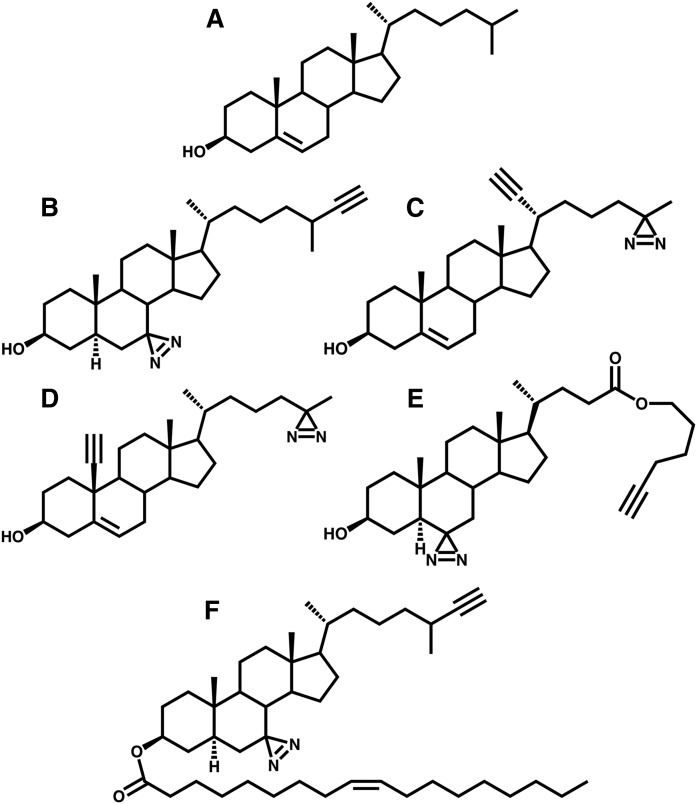
Fig. 1.
Diazirine alkyne cholesterol probes. Cholesterol (A), LKM38 (B), KK174 (C), KK175 (D), trans-sterol (E), and LKM38 oleate (F).
Fig. 2.
Growth of cholesterol-auxotrophic cells in the presence of diazirine alkyne probes. U2OS-SRA cells were pulsed for 15 min with 0.5% methyl-β-cyclodextrin in LPDM and treated for 72 h in starvation media containing 10 μM cholesterol, diazirine alkyne (A), or alkyne without diazirine (B) in ethanol. Cell viability was assessed using the CellTiter-Glo assay. Values are normalized to cholesterol-treated cells. Bars: mean + SEM. Each experiment was performed in quadruplicate, n = 3. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05 for compound versus vehicle, and $P < 0.001 for compound or vehicle versus cholesterol using one-way ANOVA followed by Dunnett’s test.
Effects of diazirine alkyne probes on the regulation of sterol homeostatic responses
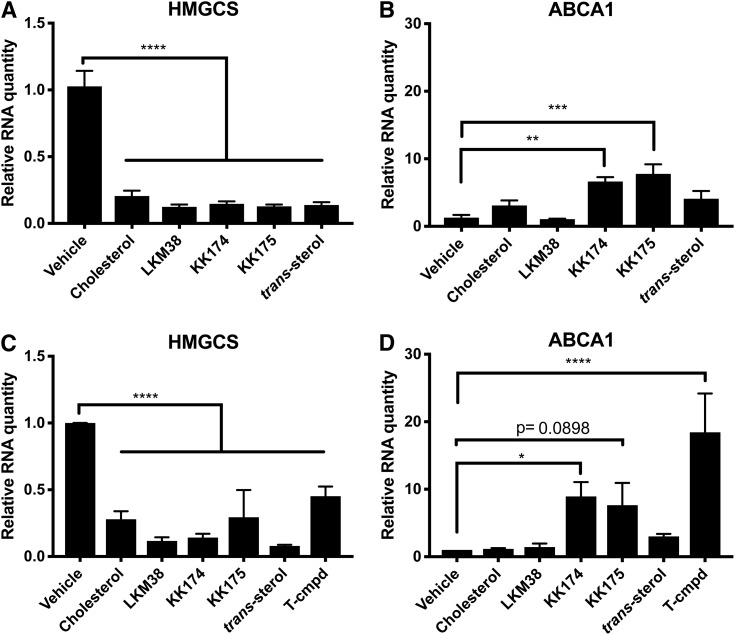
To assess the ability of diazirine alkyne probes to serve as authentic cholesterol mimics, we tested their capacity to regulate SREBP2 target gene expression, an important sterol homeostatic response that governs de novo cholesterol synthesis. Transcript levels of HMGCS, an SREBP2 target, were monitored after 14 h in the presence of cholesterol or diazirine alkyne probe as the only provided sterol. All diazirine alkyne probes suppressed HMGCS expression to a similar degree as cholesterol (Fig. 3A). To monitor for possible off-target oxysterol effects, the expression of ABCA1, an LXR target, was determined. Neither cholesterol, LKM38, nor trans-sterol significantly increased ABCA1 expression (Fig. 3B). In contrast, the diazirine alkyne probes KK174 and KK175 increased ABCA1 expression. When this experiment was performed in cells deficient in CYP27A1, a mitochondrial enzyme responsible for synthesizing the endogenous LXR ligand 27-hydroxycholesterol, similar suppression of HMGCS expression was observed (Fig. 3C). ABCA1 expression was significantly increased after KK174 treatment, and a trend was observed for KK175 (Fig. 3D). Although neither compound increased ABCA1 expression as robustly as the LXR agonist T0901317, these results indicate that these probes, in contrast to LKM38 and trans-sterol, directly activate LXR.
Fig. 3.
Diazirine alkyne probes regulate cholesterol homeostasis. U2OS-SRA cells (A, B) or CYP27A1-deficient human skin fibroblasts (C, D) were treated for 14 h in starvation media containing 10 μM probe or cholesterol in ethanol. HMGCS (A, C) and ABCA1 (B, D) mRNA expression was determined by qPCR relative to Rplp0. The LXR agonist T0901317 (T-cmpd) is shown as a positive control. Values are normalized to vehicle. Bars: mean + SEM, n = 5 (A, B) or n = 3 (C, D). ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05 for comparisons indicated using one-way ANOVA followed by Dunnett’s test.
Localization of lipoprotein-derived diazirine alkyne probe
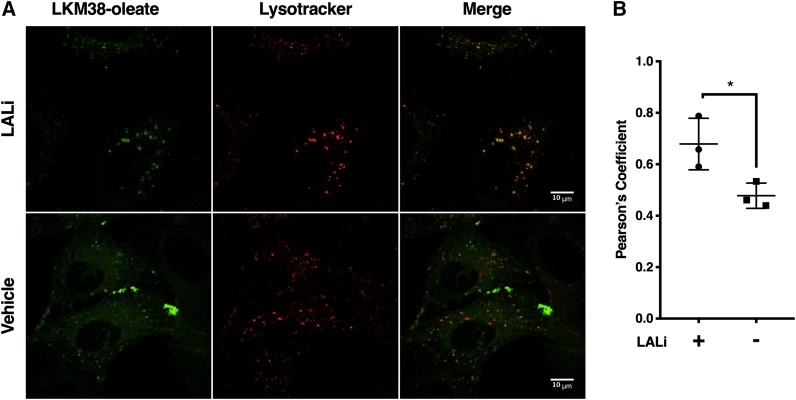
Based on the results in cholesterol auxotrophy and SREBP2 target gene expression assays, LKM38 was selected for further characterization. Cholesterol and cholesteryl ester-laden lipoprotein particles enter the cell through receptor-mediated endocytosis and then transit through the endocytic pathway to reach the lysosomal compartment. To assess whether diazirine alkyne probes trafficked in a similar manner, acLDL reconstituted with LKM38 oleate was delivered to U2OS-SRA cells. Click addition of azide-fluorophore allowed for the visualization of the cellular distribution of the probe. In the presence of an LAL inhibitor (LALi) that prevents the cleavage of cholesteryl esters to free cholesterol, punctate probe staining was observed (Fig. 4A). The probe signal colocalized with LysoTracker Red DND-99, a selective marker for acidic organelles, with a Pearson’s coefficient of 0.6788 (Fig. 4B). In the absence of LALi, both punctate and reticular staining patterns were observed, and LysoTracker colocalization was attenuated (Pearson’s coefficient: 0.4780). Thus, when provided as an ester in lipoprotein particles, LKM38 accumulates in lysosomes, and the action of LAL is required for trafficking to other cellular compartments, similar to native cholesterol.
Fig. 4.
LKM38 oleate localizes to the lysosomal compartment. U2OS-SRA cells were incubated in LPDM containing 50 μg/ml acLDL reconstituted with LKM38 oleate, labeled with lysotracker, and fluorescently labeled using copper-catalyzed azide alkyne cycloaddition of Alexa-488. A: Images collected by laser-scan confocal microscopy. B: Colocalization quantification. Pearson coefficients were calculated for 6–10 fields/condition/per experiment using ImageJ. Each data point represents the average Pearson coefficient for a biologically independent sample, n = 3. Bars: mean + SEM. *P < 0.05 using t-test.
Postlysosomal distribution of diazirine alkyne probe
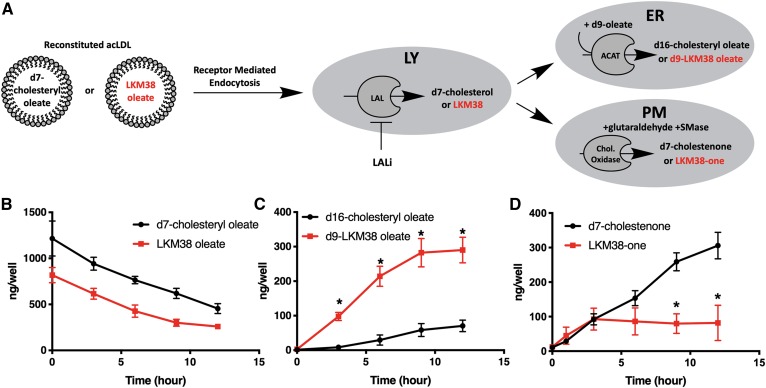
From the lysosome, cholesterol is distributed to other cellular compartments, including the ER and plasma membrane. To evaluate the fidelity of trafficking of the diazirine alkyne probe, the kinetics of probe arrival at these compartments was compared with deuterated cholesterol using LC/MS/MS-based biochemical trafficking assays. LKM38 oleate or d7-cholesteryl oleate was reconstituted in acLDL particles and delivered to U2OS-SRA cells in the presence of LALi (Fig. 5A). Cholesterol or probe metabolites were collected at several points after LALi washout. By supplying d9-oleate, the formation of a differentially deuterated reesterification product (d9-LKM38 oleate or d16-cholesteryl oleate) was used to monitor probe arrival at the ER. Arrival at the plasma membrane was determined using a cholesterol oxidase assay and quantification of oxidized derivatives (LKM38-one or d7-cholestenone). The difference in the amount of LKM38 oleate and d7-cholesteryl oleate loaded to cells was not statistically significant (Fig. 5B). After inhibitor washout, LAL cleaved LKM38 oleate and d7-cholesteryl oleate with similar kinetics. Re-esterification products were detected for both LKM38 and d7-cholesterol (Fig. 5C). Significantly more LKM38 was re-esterified relative to d7-cholesterol. At early time points, LKM38 and d7-cholesterol arrival at the plasma membrane was nearly identical; however, LKM38, in contrast to cholesterol, failed to continue to accumulate in the plasma membrane past 6 h (Fig. 5D). Together, these data suggest structural modifications present in the LKM38 diazirine alkyne probe do not affect sterol trafficking to the plasma membrane at short time points but do increase reesterification.
Fig. 5.
LKM38 traffics to the plasma membrane and ER compartments. A: U2OS-SRA cells were treated with LPDM containing 50 μg/ml acLDL reconstituted with d7-cholesteryl oleate or LKM38 oleate in the presence of LALi to load the lysosomal compartment and chased in LALi-free media for 0–12 h to allow sterol trafficking. B: Chase media contained d9-oleate/BSA to label newly synthesized esters. C: Cells were fixed and then treated with cholesterol oxidase before cellular lipids were extracted. d7-Cholesteryl oleate or LKM38 oleate (B), d16-cholesteryl oleate or d9-LKM38 oleate (C), and cholestenone or LKM38-one (D) were quantified by LC/MS/MS. Bars: mean + SEM, n = 3. *P < 0.01 using multiple t-tests with a false discovery rate of 1%.
Protein labeling by the diazirine alkyne probe
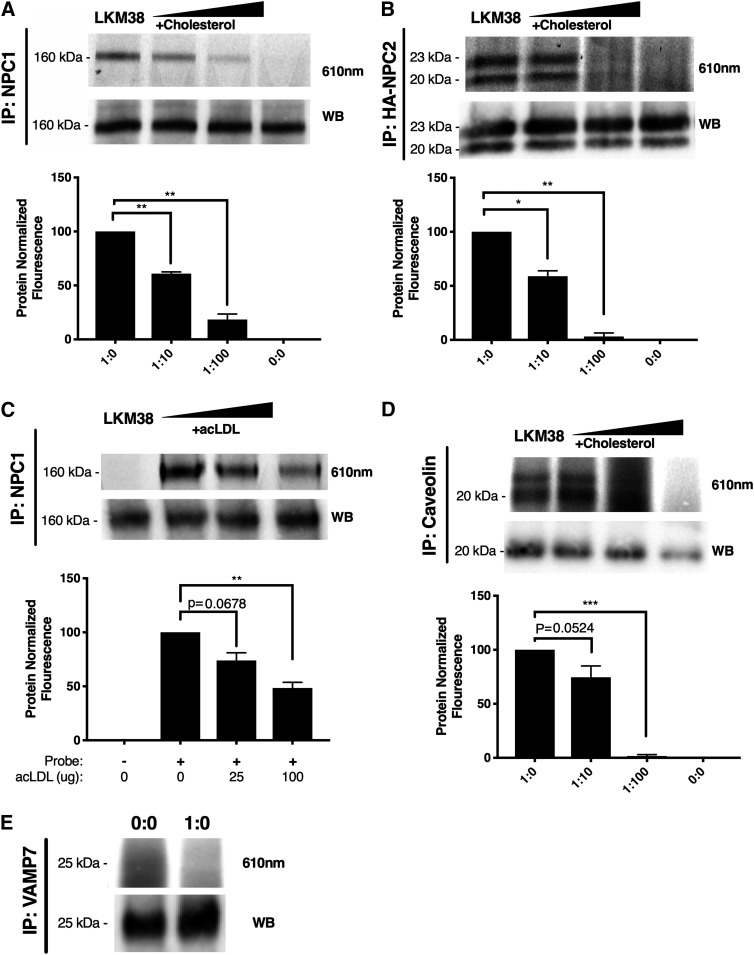
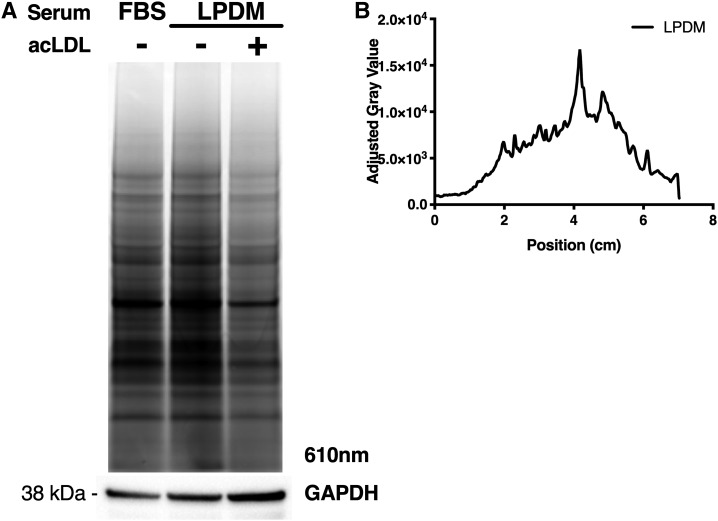
The diazirine modification to the sterol structure of LKM38 permits the cholesterol probe to be UV-cross-linked to molecules in close proximity. To test this functionality and to assay the probe interactome, we evaluated LKM38 labeling of structurally diverse, established cholesterol binding proteins, NPC1, NPC2, and caveolin. Following labeling, incubation with probe, and UV cross-linking, target proteins were immunoprecipitated from labeled lysates using the respective antibody, subjected to click addition of a fluorescein azide, separated by SDS-PAGE, and then imaged. LKM38 labeled endogenously expressed NPC1 and caveolin (Fig. 6A, C) and epitope-tagged NPC2 (Fig. 6B), as evidenced by the fluorescent signal at the appropriate molecular weight in the immunoprecipitates. When labeling was performed in the presence of increasing amounts of excess free cholesterol or in the presence of acLDL as a competitor, probe binding was diminished. Specific binding of LKM38 has previously been reported for the CRAC/CARC domain-containing protein SLC38A9 (17). Importantly, LKM38 failed to label VAMP7, an abundant transmembrane endolysosomal protein that has not been previously identified as interacting with cholesterol (Fig. 6D) (27). In order to obtain a broader view of the LKM38 interactome, total protein lysate was visualized after labeling with LKM38. Many fluorescent bands were identified, indicating that LKM38 has a large interactome (Fig. 7A). To test specificity, excess cholesterol, in the form of acLDL, was included after probe incubation. AcLDL incubation led to the reduced labeling of many of these species, suggesting these interactions are specific (Fig. 7B).
Fig. 6.
LKM38 specifically labels established cholesterol binding proteins. Normal human skin fibroblasts (A), NPC2-HA (B), or U2OS-SRA (C–E) cells were treated with 2.5 (A) or 10 μM (B–D) probe complexed to methyl-β-cyclodextrin without (1:0) or with 10× (1:10) or 100× (1:100) cholesterol or acLDL as competitor and cross-linked. Immunoprecipitated protein (IP) was clicked to rhodamine X azide before elution in sample buffer and separation by SDS-PAGE. Gels were imaged at 610 nm then transferred to nitrocellulose for Western blotting (WB) for NPC1 (160 kDa), NPC2 (23 and 20 kDa), caveolin (20 kDa), and VAMP7 (25 kDa). Bars: mean + SEM, n = 3. ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05 for comparisons indicated using paired two-tailed t-test.
Fig. 7.
Fluorescent gel profiling reveals specific labeling of proteins. Following treatment of U2OS-SRA cells with 10 μM probe in complete media, cells were incubated with complete media (FBS), LPDM, or LPDM containing 100 μg/ml acLDL. After 18 h, cells were cross-linked, lysed, and clicked to rhodamine X azide before separation by SDS-PAGE. A: Representative gel imaged at 610 nm, with parallel immunoblot for GAPDH. B: Gel profile quantified using ImageJ software. The graph quantifies the difference in the gray value between LPDM with and without acLDL as a measure of competition due to excess cholesterol.
DISCUSSION
Intracellular cholesterol trafficking is widely studied but remains poorly understood. Diazirine alkyne probes provide a new approach for the study of intracellular cholesterol distribution using a variety of techniques, including immunofluorescence microscopy, MS-based biochemical trafficking, and proteomics. In this study, we synthesized and characterized novel diazirine alkyne probes and investigated their fidelity as cholesterol mimics. Probes were designed with diazirine alkyne functional modifications located at different points on the sterol backbone to identify the least perturbing set of modifications. Novel probes were compared with the commercially available diazirine alkyne probe trans-sterol. Prior characterization of trans-sterol, LKM38, and KK174 has been limited to protein binding studies with nonphysiological probe delivery (16, 17). The results presented here are the first assessment of the ability of diazirine alkyne probes to support cell growth in the absence of cholesterol and to report on the interactions and trafficking itinerary of probe that is presented to cells in the context of lipoprotein particles.
Multiple readouts of cholesterol’s function were used to compare the ability of the different probes to mimic cellular functions of cholesterol. SREBP-regulated responses rely primarily on the behavior of the sterol in the ER membrane and can serve as sensitive readouts of cholesterol function in this organelle. LKM38 and trans-sterol similarly suppressed transcription of an SREBP target gene in the absence of off-target oxysterol transcriptional effects. We also tested for the ability of probes to support the growth of cholesterol-auxotrophic cells, a more rigorous test of cholesterol mimicry that relies on the ability of the probe to serve as the sole sterol source in the cell and assume all of the cellular functions of cholesterol. Of the diazirine alkyne probes evaluated, only LKM38 was able to sustain the growth of cholesterol-auxotrophic cells, and trans-sterol was cytotoxic in this assay. We hypothesize that the position of the diazirine on the trans-sterol probe and/or side-chain modifications may have prevented sterol-protein interactions required for cellular functions beyond the SREBP system. Based on these findings, LKM38 was selected for further evaluation as a cholesterol trafficking mimic.
Biochemical trafficking experiments allowed for the characterization of the postlysosomal trafficking of probe. Esterification of the LKM38 probe was more robust than for d7-cholesterol. Increased esterification of LKM38 could be an indication of increased delivery to the ER compartment or increased ACAT activity, possibly due to increased membrane availability (28, 29). Trafficking of LKM38 to the plasma membrane was essentially identical to native cholesterol trafficking to this compartment at early time points. Divergence at longer time points could be due to increased availability in the plasma membrane or an inability of LKM38 to recycle back to this compartment. Together, these data indicate that LKM38 is a useful probe for studying the postlysosomal cholesterol trafficking to the plasma membrane, but a bias of LKM38 toward lipid droplets should be considered when using this probe as a postlysosomal cholesterol trafficking tool. Increased association of a commonly used fluorescent cholesterol conjugate, BODIPY-cholesterol, with lipid droplets has also been observed (30).
Alkyne modification to the backbone structure of diazirine alkyne probes provides a highly flexible approach for the study of molecular distribution by fluorescence microscopy. The alkyne group is a minor modification that does not greatly influence the character of the molecule, as evidenced by the ability of a variety of alkyne cholesterol probes to support the growth of cholesterol-auxotrophic cells to nearly the same degree as cholesterol. Unlike the intrinsic fluorescent cholesterol probes, such as dehydroergosterol and cholestatrienol, which have low quantum yields, the click chemistry-compatible alkyne group of diazirine alkyne probes allows for the addition of a strong, photostable fluorescent azide (of which there are many commercially available) to be added after trafficking has taken place. This approach is compatible with fixed cell imaging to visualize static cholesterol distribution, similar to the prior use of a C19-alkyne cholesterol (31). By using a targeted lipoprotein delivery system and the alkyne functionality of diazirine alkyne probes, we were able to monitor the dynamic redistribution of cholesterol from the lysosome to the rest of the cell using confocal microscopy.
The cross-linking functionality of diazirine alkyne probes enables the study of probe-protein interactions, which are not possible with BODIPY-cholesterol and dehydroegosterol. We demonstrated binding for LKM38 to NPC1, NPC2, and caveolin and have previously reported binding to the CRAC/CARC domain-containing protein SLC38A9 (17). These interactions are specific, as each was sensitive to cholesterol competition. These proteins represent four structurally distinct cholesterol-binding motifs. The ability of LKM38 to bind specifically to structurally diverse proteins with distinct cholesterol binding motifs has the potential to expand the interactome accessible for study using this technique. While these experiments were based on the fluorescent detection of labeled targets, click addition of different azide handles allows for the enrichment of labeled proteins for MS-based protein identification. Such an approach was used for trans-sterol, a commercially available diazirine alkyne cholesterol probe (15); however, trans-sterol did not identify NPC2 or label NPC1 in a cholesterol-specific manner. Our data suggest that the structure of LKM38 has the potential to serve as a more sensitive and biologically relevant cholesterol probe.
In summary, we have characterized the authenticity of diazirine alkyne probes as cholesterol mimics using a variety of cell biological approaches. LKM38 emerged as a high-fidelity cholesterol mimic through its ability to sustain growth in auxotrophic cells and appropriately regulate cholesterol homeostasis. Moreover, through biochemical trafficking assays and immunofluorescence microscopy, we showed that lysosomally derived LKM38 is distributed to the plasma membrane and ER, providing evidence that this probe is suitable for trafficking studies. Finally, we demonstrated specific cholesterol-protein interactions between LKM38 and known cholesterol binding proteins. Taken together, these findings validate LKM38 as a flexible tool for the study of intracellular cholesterol trafficking.
Supplementary Material
Acknowledgments
The authors thank Hideji Fujiwara for advice regarding cholestenone derivatization.
Footnotes
Abbreviations:
- acLDL
- acetylated LDL
- CHO
- Chinese hamster ovary
- HA
- human influenza hemagglutinin
- HMGCS
- HMG-CoA synthase
- LAL
- lysosomal acid lipase
- LALi
- lalistat
- LPDM
- lipoprotein-deficient media
- NPC
- Niemann-Pick C
- qPCR
- quantitative PCR
- Rplp0
- ribosomal protein lateral stalk subunit P0
- SRA
- scavenger receptor A
- THPTA
- tris-hydroxypropyltriazolylmethylamine
- VAMP7
- vesicle-associated membrane protein 7
This work was supported by National Institutes of Health Grants R01 HL067773 (D.F.C., D.S.O., J.E.S.), T32 HL134635 (M.F.), and F31 HL142167 (M.F.) and the Taylor Family Institute for Innovative Psychiatric Research (D.F.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Wüstner D., and Solanko K.. 2015. How cholesterol interacts with proteins and lipids during its intracellular transport. Biochim. Biophys. Acta. 1848: 1908–1926. [DOI] [PubMed] [Google Scholar]
- 2.Das A., Brown M. S., Anderson D. D., Goldstein J. L., and Radhakrishnan A.. 2014. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 3: e02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan A., Goldstein J. L., McDonald J. G., and Brown M. S.. 2008. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein J. L., DeBose-Boyd R. A., and Brown M. S.. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. [DOI] [PubMed] [Google Scholar]
- 5.Hong C., and Tontonoz P.. 2014. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug Discov. 13: 433–444. [DOI] [PubMed] [Google Scholar]
- 6.Lange Y., Ye J., Rigney M., and Steck T. L.. 1999. Regulation of endoplasmic reticulum cholesterol by plasma membrane cholesterol. J. Lipid Res. 40: 2264–2270. [PubMed] [Google Scholar]
- 7.Brown M. S., Dana S. E., and Goldstein J. L.. 1975. Receptor-dependent hydrolysis of cholesteryl esters contained in plasma low density lipoprotein. Proc. Natl. Acad. Sci. USA. 72: 2925–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu S., Benoff B., Liou H. L., Lobel P., and Stock A. M.. 2007. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J. Biol. Chem. 282: 23525–23531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., and Infante R. E.. 2009. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 137: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Infante R. E., Radhakrishnan A., Abi-Mosleh L., Kinch L. N., Wang M. L., Grishin N. V., Goldstein J. L., and Brown M. S.. 2008. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J. Biol. Chem. 283: 1064–1075. [DOI] [PubMed] [Google Scholar]
- 11.Deffieu M. S., and Pfeffer S. R.. 2011. Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc. Natl. Acad. Sci. USA. 108: 18932–18936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanier M. T., and Millat G.. 2003. Niemann-Pick disease type C. Clin. Genet. 64: 269–281. [DOI] [PubMed] [Google Scholar]
- 13.Maxfield F. R., and Wustner D.. 2012. Analysis of cholesterol trafficking with fluorescent probes. Methods Cell Biol. 108: 367–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sezgin E., Can F. B., Schneider F., Clausen M. P., Galiani S., Stanly T. A., Waithe D., Colaco A., Honigmann A., Wustner D., et al. . 2016. A comparative study on fluorescent cholesterol analogs as versatile cellular reporters. J. Lipid Res. 57: 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulce J. J., Cognetta A. B., Niphakis M. J., Tully S. E., and Cravatt B. F.. 2013. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods. 10: 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budelier M. M., Cheng W. W. L., Bergdoll L., Chen Z. W., Janetka J. W., Abramson J., Krishnan K., Mydock-McGrane L., Covey D. F., Whitelegge J. P., et al. . 2017. Photoaffinity labeling with cholesterol analogues precisely maps a cholesterol-binding site in voltage-dependent anion channel-1. J. Biol. Chem. 292: 9294–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellano B. M., Thelen A. M., Moldavski O., Feltes M., van der Welle R. E., Mydock-McGrane L., Jiang X., van Eijkeren R. J., Davis O. B., Louie S. M., et al. . 2017. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 355: 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas D., Morgenthaler J., Lacbawan F., Long B., Runz H., Garbade S. F., Zschocke J., Kelley R. I., Okun J. G., Hoffmann G. F., et al. . 2007. Abnormal sterol metabolism in holoprosencephaly: studies in cultured lymphoblasts. J. Med. Genet. 44: 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenbaum A. I., Cosner C. C., Mariani C. J., Maxfield F. R., Wiest O., and Helquist P.. 2010. Thiadiazole carbamates: potent inhibitors of lysosomal acid lipase and potential Niemann-Pick type C disease therapeutics. J. Med. Chem. 53: 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pipalia N. H., Subramanian K., Mao S., Ralph H., Hutt D. M., Scott S. M., Balch W. E., and Maxfield F. R.. 2017. Histone deacetylase inhibitors correct the cholesterol storage defect in most Niemann-Pick C1 mutant cells. J. Lipid Res. 58: 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ory D. S., Neugeboren B. A., and Mulligan R. C.. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 93: 11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brookheart R. T., Michel C. I., Listenberger L. L., Ory D. S., and Schaffer J. E.. 2009. The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. J. Biol. Chem. 284: 7446–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitersdorf E., Safadi R., Meiner V., Reshef A., Bjorkhem I., Friedlander Y., Morkos S., and Berginer V. M.. 1994. Cerebrotendinous xanthomatosis in the Israeli Druze: molecular genetics and phenotypic characteristics. Am. J. Hum. Genet. 55: 907–915. [PMC free article] [PubMed] [Google Scholar]
- 24.Krieger M. 1986. Reconstitution of the hydrophobic core of low-density lipoprotein. Methods Enzymol. 128: 608–613. [DOI] [PubMed] [Google Scholar]
- 25.Pugach E. K., Feltes M., Kaufman R. J., Ory D. S., and Bang A. G.. 2018. High-content screen for modifiers of Niemann-Pick type C disease in patient cells. Hum. Mol. Genet. 27: 2101–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millard E. E., Srivastava K., Traub L. M., Schaffer J. E., and Ory D. S.. 2000. Niemann-pick type C1 (NPC1) overexpression alters cellular cholesterol homeostasis. J. Biol. Chem. 275: 38445–38451. [DOI] [PubMed] [Google Scholar]
- 27.Ohgami N., Ko D. C., Thomas M., Scott M. P., Chang C. C., and Chang T. Y.. 2004. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 101: 12473–12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen B. N., Bielska A. A., Lee T., Daily M. D., Covey D. F., Schlesinger P. H., Baker N. A., and Ory D. S.. 2013. The structural basis of cholesterol accessibility in membranes. Biophys. J. 105: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bielska A. A., Olsen B. N., Gale S. E., Mydock-McGrane L., Krishnan K., Baker N. A., Schlesinger P. H., Covey D. F., and Ory D. S.. 2014. Side-chain oxysterols modulate cholesterol accessibility through membrane remodeling. Biochemistry. 53: 3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wüstner D., Solanko L., Sokol E., Garvik O., Li Z., Bittman R., Korte T., and Herrmann A.. 2011. Quantitative assessment of sterol traffic in living cells by dual labeling with dehydroergosterol and BODIPY-cholesterol. Chem. Phys. Lipids. 164: 221–235. [DOI] [PubMed] [Google Scholar]
- 31.Jao C. Y., Nedelcu D., Lopez L. V., Samarakoon T. N., Welti R., and Salic A.. 2015. Bioorthogonal probes for imaging sterols in cells. ChemBioChem. 16: 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.