Mitogen-activated protein kinase 6/extracellular signal-regulated kinase 3 (MAPK6/ERK3) is an atypical member of the MAPKs. An essential role has been suggested by the perinatal lethal phenotype of ERK3 knockout mice carrying a lacZ insertion in exon 2 due to pulmonary dysfunction and by defects in function, activation, and positive selection of T cells.
KEYWORDS: MAPKAPK5/MK5, gene deletion, mitogen-activated protein kinases, protein kinase
ABSTRACT
Mitogen-activated protein kinase 6/extracellular signal-regulated kinase 3 (MAPK6/ERK3) is an atypical member of the MAPKs. An essential role has been suggested by the perinatal lethal phenotype of ERK3 knockout mice carrying a lacZ insertion in exon 2 due to pulmonary dysfunction and by defects in function, activation, and positive selection of T cells. To study the role of ERK3 in vivo, we generated mice carrying a conditional Erk3 allele with exon 3 flanked by loxP sites. Loss of ERK3 protein was validated after deletion of Erk3 in the female germ line using zona pellucida 3 (Zp3)-cre and a clear reduction of the protein kinase MK5 is detected, providing the first evidence for the existence of the ERK3/MK5 signaling complex in vivo. In contrast to the previously reported Erk3 knockout phenotype, these mice are viable and fertile and do not display pulmonary hypoplasia, acute respiratory failure, abnormal T-cell development, reduction of thymocyte numbers, or altered T-cell selection. Hence, ERK3 is dispensable for pulmonary and T-cell functions. The perinatal lethality and lung and T-cell defects of the previous ERK3 knockout mice are likely due to ERK3-unrelated effects of the inserted lacZ-neomycin resistance cassette. The knockout mouse of the closely related atypical MAPK ERK4/MAPK4 is also normal, suggesting redundant functions of both protein kinases.
INTRODUCTION
Mitogen-activated protein kinase (MAPK) cascades are conserved eukaryote signaling modules where the downstream effector kinases regulate cell proliferation, differentiation, and cell death by phosphorylation of protein substrates. MAPKs are also regulators in many physiological processes, including development and immune response. Multiple MAPKs were described in mammalian cells, which can be divided in five groups, the classical mitogen-responsive MAPKs (extracellular signal-regulated kinase 1 [ERK1] and ERK2), the stress-activated c-Jun N-terminal kinases (JNK1 to -3), p38MAPK (α, β, γ, and δ), the big MAPK ERK5, and the atypical MAPKs ERK3, ERK4, and ERK7 (1). MAPK activity is classically regulated by dual phosphorylation on a TXY motif in the activation loop of the kinase by MAPK kinases.
ERK3/MAPK6/p97MAPK and the closely related ERK4/MAPK4/p63MAPK are the only two MAPKs carrying long C-terminal extensions and lacking the dual TXY phosphorylation motif in the activation loop (2–4). Instead of the canonical dual-phosphorylation motif, ERK3 and ERK4 contain only a single phospho-acceptor serine (SEG motif) in the activation loop, which can be phosphorylated by p21-activated kinases in vitro and in transfected cells (5, 6). Apart from this, little is known about the mechanisms of regulation, substrate specificity, and physiological functions of atypical MAP kinases. In particular, mitogen and stress stimuli resulted in only weak phosphorylation at the SEG motif, and the only biologically relevant regulator of ERK3 and ERK4 identified so far is the MAP kinase-activated protein kinase MK5 (7–10). MK5 forms a complex with ERK3/4 and is phosphorylated at its activating site, T182, within this complex. In turn and still in the complex, ERK3 also autophosphorylates at various sites in its C-terminal extension. It is suggested that the ERK3/MK5 complex is involved in the regulation of dendrite morphology and septin function (11). However, it is not clear whether and how the productive complex formation between ERK3/4 and MK5 is regulated by extracellular stimuli or whether it just depends on their expression levels.
Additional functions described for ERK3 include its interaction with the cell cycle regulator Cdc14 (12), its contribution to meiotic spindle stability and metaphase-anaphase transition in mouse oocyte maturation (13), and its interaction with the steroid receptor coactivator 3 (SRC-3), an oncogenic protein overexpressed in multiple human cancers (14). ERK3 seemingly phosphorylates SRC-3 at S857 and regulates it interaction with the ETS- and SP1-type transcription factors (14, 15). Recently, the tyrosyl DNA phosphodiesterase 2 (TDP2), which repairs topoisomerase 2-linked DNA damage, was also described as a substrate of ERK3, and it was suggested that ERK3 phosphorylates TDP2 at S60 and stimulates its phosphodiesterase activity during the DNA damage response (16).
A major contribution to the understanding of the functions of ERK3 and ERK4 was the generation of constitutive knockout (KO) alleles of ERK3 (17) and ERK4 (18). Although both kinases are similar in structure and display similar molecular interactions, the phenotypes of both kinase knockouts differed significantly. While ERK4 knockout mice appeared normal, ERK3 knockout mice were not viable, displaying retarded intrauterine growth and pulmonary hypoplasia leading to acute perinatal respiratory failure (17). Furthermore, T-cell development, selection, and activation was impaired only in ERK3, but not in ERK4, knockout mice (19–21). The lack of phenocopy between both knockouts suggested distinct and nonredundant functions of the ERK3/MK5 and ERK4/MK5 signaling complexes.
The perinatal lethality of the constitutive ERK3 knockout mice limits the use of this mouse strain in disease models, which are mostly established for adult mice. To overcome this limitation, we generated a conditional allele where exon 3 of Erk3 is flanked by loxP sites. Here, we describe the unexpected finding that germ line deletion of exon 3 in mice causes the complete loss of ERK3 protein but does not lead to any of the phenotypes described for the ERK3 knockout mouse carrying the lacZ insertion. Mice lacking ERK3 protein are viable, allowing for the further analysis of ERK3 function in postnatal development and adult mice.
RESULTS
Conditional targeting of the ERK3 allele.
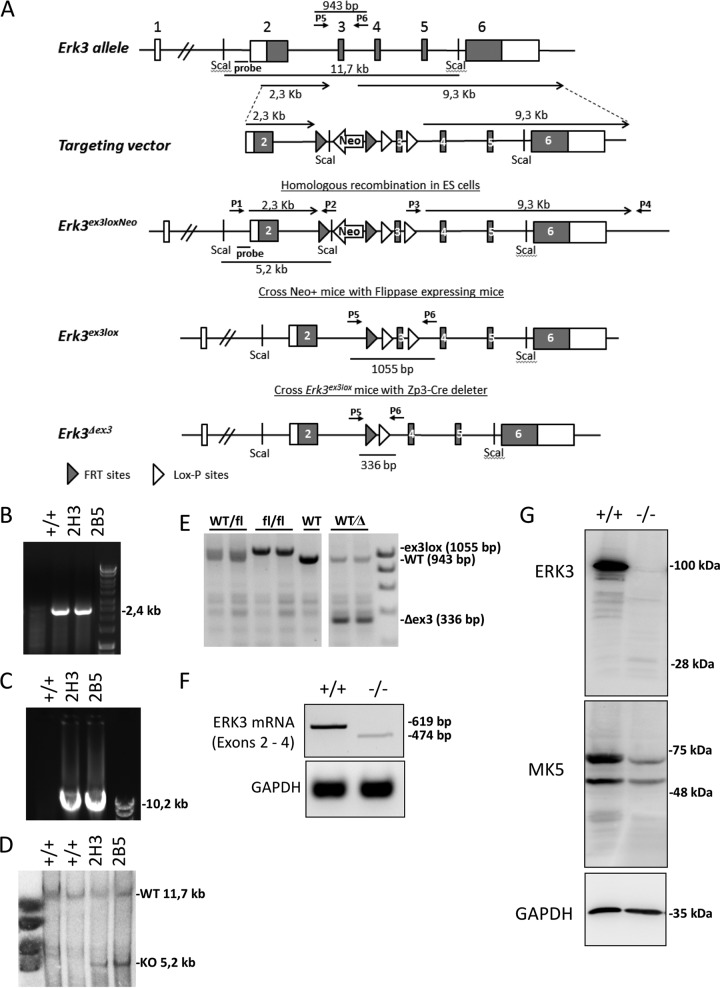
The murine Erk3 gene is comprised of 6 exons spanning 20 kb of genomic sequence (22). Exons 2 to 6 contain the open reading frame (ORF) sequence for ERK3. A targeting vector was designed to flank exon 3 of Erk3, coding for the activation loop and catalytic kinase subdomains VIII to X (amino acids 186 to 233), with loxP sites in embryonic stem (ES) cells (Erk3ex3lox) (Fig. 1A). The Neo selection cassette flanked by FLP recombination target (FRT) sites was inserted in intron 2. The correct integration of this vector was screened by PCR (Fig. 1B and C) and validated by Southern blot analysis (Fig. 1D) of genomic DNA. Germ line transmission was obtained with targeted Erk3ex3lox ES cell clone 2H3, and the neomycin resistance cassette was removed by breeding with Flp-deleter mice (23). Recombination was confirmed by PCR (Fig. 1E).
FIG 1.
Generation of the conditional ERK3 knockout mouse and deletion of ERK3 mRNA and protein. (A) Targeting strategy. (B to D) ES cell screening by PCR (B and C) and Southern hybridization (D). (B and C) PCR using primer combination P1/P2 and P3/P4 (shown in panel A) to detect Erk3ex3loxneo allele. (D) Southern blot analysis using probe (shown in panel A) and ScaI-digested DNA. (E) Detection of Flp- and Cre-mediated recombination by PCR using primer combinations (P5 and P6), leading to fragments as indicated in panel A. (F) ERK3 mRNA was amplified by PCR from total RNA of BMDMs using primers for exons 2 and 4. The ERK3 KO displays a single band weaker and smaller than that of the WT, indicating loss of exon 3. (G) Western blot analysis of total protein of BMDMs by an N-terminal-ERK3 antibody (53277; Abcam) and an MK5 antibody. Equal loading is demonstrated by GAPDH detection.
Germ line deletion of exon 3 of ERK3 leads to viable mice with the complete loss of ERK3 and reduced MK5 expression.
To validate that our targeting strategy results in a null allele, we first generated a germ line deletion of exon 3 (Erk3Δex3) by breeding mice with floxed exon 3 to Zp3-Cre mice that express CRE recombinase in oocytes (24), which eliminates ERK3 in all cells of the body, i.e., generates a constitutive knockout. Unexpectedly and in contrast to the previously described knockout (17), matings of heterozygous Erk3Δex3 mice gave rise to viable homozygous offspring at the expected Mendelian ratio (n = 105 of six crosses and 15 litters [Fig. 2A], 25 wild-type [WT], 45 heterozygotes, and 35 ERK3 KO mice; χ2 = 4.04 < 5.99 [P < 0.05]). PCR analysis of bone marrow-derived macrophages (BMDMs) demonstrated the absence of WT mRNA in Erk3Δe3x homozygotes and the presence of a reduced level of mRNA lacking exon 3 (Fig. 1F). Western blot analysis of BMDM lysates using various antibodies, including the N-terminus-specific one, which could detect the C-terminal truncations, revealed the lack of ERK3 in homozygous mutant mice (Fig. 1G). Detection of a very faint band at about 28 kDa could represent some remaining, rather instable protein fragment of only part of the catalytic domain (subdomains I to VII) encoded by exon 2 (amino acids 1 to 185). Deletion of exon 3 leads to a frameshift and immediate translation stop in potential alternatively spliced transcripts of the targeted allele. Interestingly, the protein level of the ERK3 interaction partner MK5 was also decreased in the ERK3 knockout (Fig. 1G).
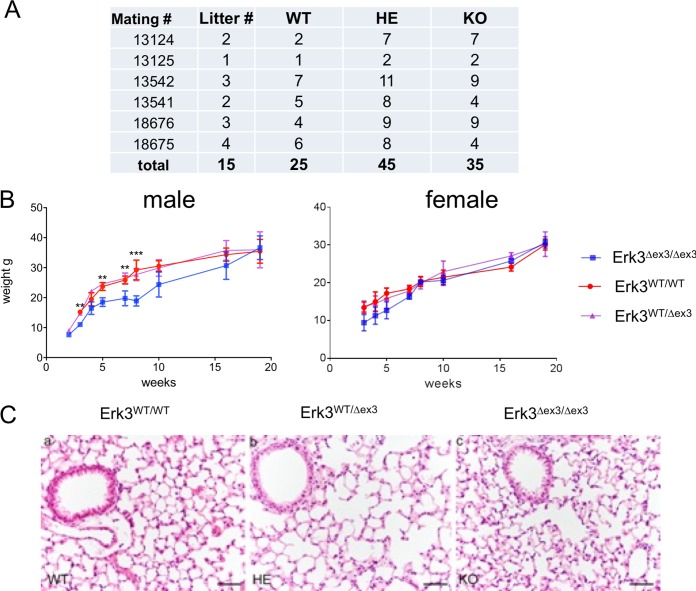
FIG 2.
Characterization of the viable ERK3 KO mice. (A) Mendelian ratio of the offspring of heterozygous (HE) Erk3Δex3 mice. Fifteen litters of six crosses (total n = 105) were analyzed. No statistically significant deviation from Mendelian ratio was detected (χ2 = 4.04 < 5.99, P < 0.05). (B) Slight and transient growth retardation of male but not female ERK3 KO mice (4 to 6 mice per group; **, P < 0.05; ***, P < 0.01). (C) Histopathology of the lungs from 35-day-old WT (a), ERK3+/− (b), and ERK3 KO (c) mice revealed no significant pathological alterations. Hematoxylin and eosin staining was used. Bars, 50 µm.
Early and transient growth retardation of Erk3Δex3/Δex3 mice.
We determined body weights of male and female Erk3Δex3/Δex3 mice and their littermates 3 to 20 weeks after birth. Male Erk3Δex3/Δex3 mice displayed significantly lower body weight 3 weeks after birth, indicating some early growth retardation (Fig. 2B). However, subsequently all Erk3Δex3/Δex3 mice gradually caught up in weight, developed normally, were apparently healthy, and became indistinguishable from wild-type and heterozygous littermates at an age of about 20 weeks. Examination of tissue sections stained with hematoxylin and eosin revealed no obvious abnormalities in the lungs of WT, Erk3+/Δex3, and Erk3Δex3/Δex3 mice (Fig. 2C). At present, several mice have already reached an age of 1 year or more, and several males and two females tested were fertile.
Loss of ERK3 does not impair T-cell development and proliferation.
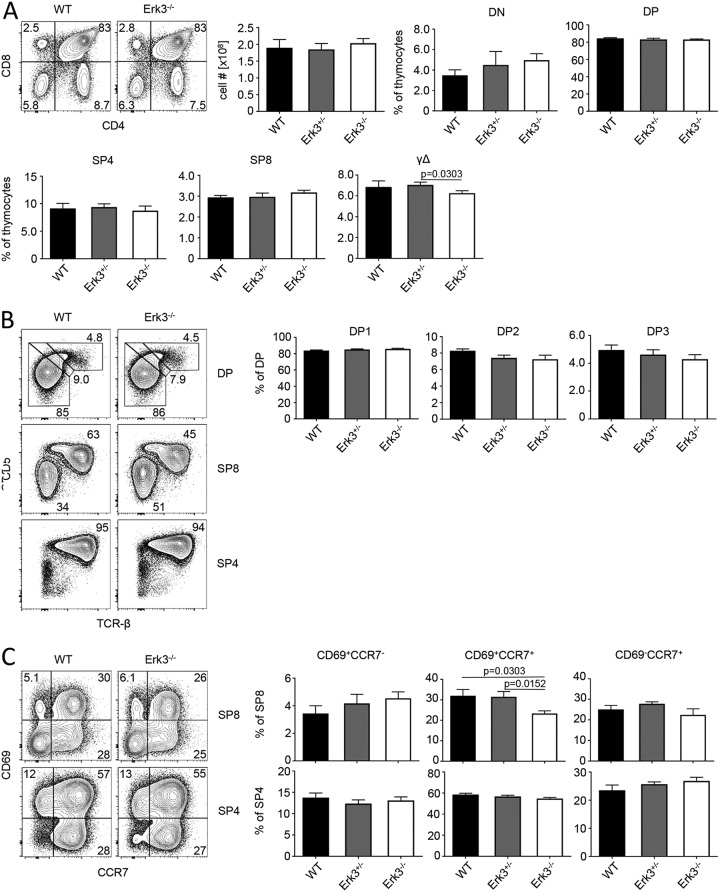
Because of the published effects of the previous ERK3 deletion on T-cell development, activation, and function (19–21), we analyzed T-cell development and function in our Erk3Δe3x/Δex3 mice. We first compared T-cell populations of the thymus of 5- to 6-week-old WT, ERK3+/−, and ERK3-deficient mice. We did not detect significant differences in total thymus cellularity. In addition, no differences were observed in the frequencies of any of the major thymocyte populations, CD4 and CD8 double-negative (DN), double-positive (DP), γδ T-cell receptor (TCR)-positive, and CD4 and CD8 single-positive (SP) cells (Fig. 3A). This is in contrast to the Erk3-lacZ allele, which strongly reduced numbers of thymocytes, most prominently within the CD4 and CD8 DP and CD8 SP subsets (19).
FIG 3.
ERK3 is largely dispensable for intrathymic T-cell development. (A) Representative flow cytometric analysis of thymi from WT and Erk3−/− mice stained with antibodies against CD4 and CD8α. Numbers in quadrants represent frequencies. Total cellularity of thymi from WT, ERK3+/−, and ERK3−/− mice. Statistical analysis of flow cytometric results from WT, ERK3+/−, and ERK3−/− mice. Cells were defined as DN (CD4− CD8α−), DP (CD4+ CD8α+), SP4 (CD4+ CD8α−), SP8 (CD4− CD8α+), and γδ T cells (CD4− CD8α− TCRγδ+). (B) Representative flow cytometric analysis of thymi from WT and ERK3−/− mice stained with antibodies against CD4, CD8α, TCRβ, and CD5. Numbers adjacent to gates represent frequencies. For statistical analysis of flow cytometric results, cells were defined as DP1 (TCRβlo CD5lo), DP2 (TCRβint CD5hi), and DP3 (TCRβhi CD5int) thymocytes. (C) Representative flow cytometric analysis of thymi from WT and ERK3−/− mice stained with antibodies against CD4, CD8α, CCR7, and CD69. Numbers in quadrants represent frequencies. For statistical analysis of flow cytometric results, cells were defined as described for panel A and as CD69+ CCR7−, CD69+ CCR7+, and CD69− CCR7+ thymocytes. (A to C) Pooled data of two independent experiments (n = 5 for WT, n = 6 for ERK3+/−, and n = 6 for ERK3−/−). Statistically significant differences with P values of <0.05 are indicated; P values of >0.05 are not shown.
It has been proposed that Erk3 contributes to positive selection of thymocytes (21). At steady state, DP thymocytes undergoing selection can be discriminated based on expression of the surface marker CD5 and TCRβ. Preselection DP cells are CD5lo TCRβlo (DP1), selecting DP cells are CD5hi TCRβint, and CD5hi TCRβhi cells (DP3) are precursors of CD8 SP thymocytes (25). We did not observe any alterations in ratios between the 3 DP thymocyte fractions, suggesting that positive selection in the thymus also is unaffected by loss of ERK3 (Fig. 3B). These data are consistent with normal frequencies of CD4 and CD8 SP cells in the absence of ERK3. In order to assess potential consequences of Erk3 deletion at the SP stages of T-cell development, we employed the same staining strategy. Again, we found no difference in surface phenotype in CD4 and CD8 SP cells (Fig. 3B). An alternative strategy to monitor thymocyte maturation is staining for the activation marker CD69 and chemokine receptor CCR7. Selection induces upregulation of CD69, followed by CCR7. CD69 is then rapidly downregulated, leaving a mature CD69− CCR7+ thymocyte population. We detected a statistically significant lower frequency of transitory CD69+ CCR7+ thymocytes within the CD8 lineage that had no impact on the frequency of fully mature CD69− CCR7+ CD8 SP cells in ERK3-deficient thymi (Fig. 3C). We detected no ERK3-dependent differences in frequencies of CD4 SP subsets.
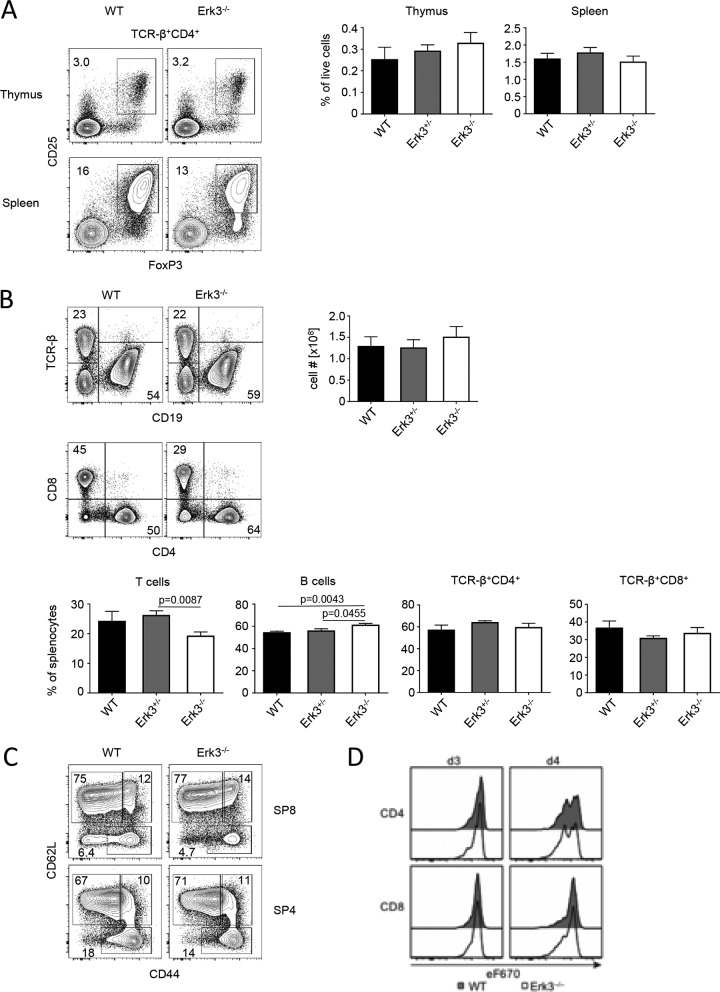
We next assessed development of Foxp3+ regulatory T cells (Treg) in the absence of functional ERK3. We found no Erk3-dependent differences in Treg cell frequencies in either thymus or spleen (Fig. 4A). The total cellularity of splenocytes was unaltered in the absence of ERK3 (Fig. 4B). Consistent with normal T-cell development in the thymus, we also did not detect any alteration in the ratio of T versus B cells in spleen, although we noted a marginal but statistically significant increase in the frequency of splenic B cells. In addition, we found no changes in the ratio between CD4+ and CD8+ T cells upon deletion of ERK3. Expression of CD44 and CD62L allows for discrimination of naive (CD44− CD62L+), central memory (CD44+ CD62L+), and effector memory (CD44+ CD62L−) T-cell subsets. We found no differences in the frequencies of naive, central memory, or effector T cells within the CD4 and CD8 T-cell compartments in spleen when comparing ERK3-sufficient to ERK3-deficient mice (Fig. 4C). These data suggest that loss of Erk3 expression does not result in aberrant T-cell activation at steady state. T cells from conventional ERK3 KO mice had reduced proliferation capacity upon unspecific TCR stimulation using anti-CD3 and anti-CD28 antibodies (4). Although T-cell activation was not altered in our newly generated ERK3 knockout mice at steady state, we compared proliferation capacity of WT and ERK3-deficient cells upon CD3/CD28 stimulation. In contrast to the conventional ERK3 KO, we could not observe any differences in T-cell proliferation, in either CD4 or CD8 T cells, for our new ERK3 KO (Fig. 4D). Taking these data together, we conclude that ERK3 is dispensable for intrathymic T-cell development, T-cell homeostasis, and T-cell proliferation.
FIG 4.
ERK3 is largely dispensable for T-cell homeostasis and proliferation. (A) Treg cell numbers in thymus and spleen. Dot plots show representative flow cytometric analysis of thymi and spleen from WT and ERK3−/− mice stained with antibodies against CD4, TCRβ, Foxp3, and CD25. Numbers adjacent to gates represent frequencies. Graphs display statistical analysis of flow cytometric results from WT, ERK3+/−, and ERK3−/− mice. (B) Representative flow cytometric analysis of spleen from WT and ERK3−/− mice stained with antibodies against CD19, TCRβ, CD4, and CD8. Numbers in quadrants represent frequencies. Total cellularity of spleen from WT, ERK3+/−, and ERK3−/− mice. For statistical analysis of flow cytometric results from WT, ERK3+/−, and ERK3−/− mice, cells were defined as T cells (TCRβ+), B cells (CD19+), SP4 (TCRβ+ CD4+), and SP8 (TCRβ+ CD8+). (C) Representative flow cytometric analysis of SP4 and SP8 splenic T cells from WT and ERK3−/− mice stained with antibodies against CD4, CD8, CD44, and CD62L to identify naive (CD44− CD62L+), central memory (CD44+ CD62L+), and effector memory (CD44+ CD62L−) T-cell subsets. Pooled data from two independent experiments were used for panels A to C (n = 5 for WT, n = 6 for ERK3+/−, and n = 6 for ERK3−/−). Statistically significant differences with P values of <0.05 are indicated; P values of >0.05 are not shown. (D) Flow cytometric analysis of CD4 (upper) and CD8 (lower). T-cell proliferation from WT and ERK3–/– mice after 3 and 4 days of culture. Shown are representative contour plots of experiments with n = 2 mice per genotype.
DISCUSSION
Here, we report the generation and analysis of a novel Erk3 null allele and show that deletion of ERK3 is not essential for viability, pulmonary function, or T-cell development, in contrast to previously described mice carrying a lacZ insertion into exon 2 of Erk3 (17, 19–21). The reason for these differences likely lies in the different targeting strategies used to disrupt Erk3. The constitutive ERK3 knockout was generated by insertion of lacZ in frame with the translation start of exon 2 of the ERK3 gene 12 codons downstream of the initiation codon and a neomycin resistance cassette downstream of lacZ. The conditional allele reported here removes exon 3 of Erk3 and leaves only a single FRT and a single loxP site in intron 2. The insertion of a lacZ-neomycin resistance cassette often altered transcription of genes in the flanking DNA around the targeted gene and, accordingly, the phenotype of the mutation (26–29). This may be due to the loss or disruption of intragenic regulatory elements, the constitutive promoter driving the neomycin gene, removal of insulating DNA in the targeted alleles, and local silencing due to disruption of normal chromatin organization by the exogenous construct (26). The detailed transcriptome analysis of 29 targeted alleles in mice revealed that downregulated genes flanking the targets were rather equally distributed 5′ and 3′ of the target, and the median distance from the target was 34 kb (26). In this regard, it is interesting that on chromosome 9 the gene for an essential subunit of RNA polymerase II, leo1, is 40 kb 3′ to the Erk3 gene. Hence, one can speculate that downregulation of essential genes, such as leo1, severely compromises cell viability and contributes to the previously reported Erk3 mutant phenotype. In contrast to effects of insertion cassettes and to the best of our knowledge, no effects of insertions of single loxP and FRT sites into intronic sequences have been described. Therefore, a likely explanation for the perinatal lethality and lung and T-cell defects observed in the previous ERK3 knockout mice is ERK3-unrelated effects of the inserted lacZ-neomycin resistance cassette.
We and others have described the ERK3/MK5 and ERK4/MK5 signaling modules (7, 9, 10, 30). Deletion of MK5 and ERK4 in mice resulted in some mild growth retardation (31, 32) and no significant phenotype at all (18), respectively. In contrast, disruption of Erk3 had a lethal phenotype (17). So far, it was difficult to understand that deletion of different components of these modules do not mutually phenocopy but display completely different phenotypes. This also questioned the physiological relevance of these signaling modules. However, the phenotype of slight and transient male growth retardation described for deletion of exon 3 of Erk3 here phenocopies the MK5 knockout and reconciles the genetic analysis of these signaling complexes with their existence in a specific signaling module. Furthermore, the observation that the level of the interaction partner MK5 is significantly reduced in the Erk3Δex3/Δex3 BMDMs strongly supports the existence of the ERK3/MK5 signaling complex in vivo. Possibly the remaining MK5 in our Erk3Δex3/Δex3 mice is stabilized by its interaction with ERK4.
So far, the differences between the phenotypes of the conventional knockouts of ERK3 and ERK4 were explained by specific nonredundant functions of these closely related atypical MAPKs (18), which contrasts with the similarities in structure and molecular interactions of both kinases (5, 33, 34). The viability and fertility of mice with deletion of exon 3 of Erk3 is much more similar to the phenotype of the conventional ERK4 knockout. Hence, one cannot exclude a redundant function of both atypical MAPKs, which could be revealed in the future by generation of the exon 3 ERK3/ERK4 double knockout mouse.
MATERIALS AND METHODS
Generation of ERK3 (MAPK6) conditional knockout mice.
Erk3ex3lox mice (Mapk6tm1Mgl) were generated as indicated in Fig. 1A. Briefly, the targeting vector, containing lox sites flanking exon 3 of the mouse Erk3 gene and FRT sites flanking the neomycin cassette, was linearized with AsiSI and electroporated in 129Ola ES cells. Two positive clones (2H3 and 2B5) were obtained by PCR screen, and homologous recombination was confirmed by Southern blot analysis. DNA samples were digested with ScaI and probed with a 5′ external probe (PCR product that amplified the 846-bp genomic fragments 5′probe-ERK3-FW, 5′-GTACAGACATGCCTGTACTCATGC-3′, and 5′probe-ERK3-RC, 5′-CTATGCTAACCGACTTAACATGGGAC-3′). Positive clones were injected into blastocysts for the generation of chimeric mice. Agouti germ line pups were derived from the mating of chimeric male mice, obtained following the blastocyst injection of Erk3-targeted ES cell clone 2H3, with C57BL/6 females. The resulting Erk3ex3loxNeo mice were crossed with C57BL/6-(C3)-Tg(Pgk1-FLPo)10Sykr/J flippase-expressing mice (23) to delete the neomycin cassette retaining the loxP-flanked (floxed) exon 3, leading to Erk3ex3lox mice. Subsequent Cre recombinase expression then catalyzes exon 3 excision, resulting in an additional frameshift mutation downstream of this exon. For generation of oocyte-specific knockout animals, Erk3 homozygous floxed mice were crossed with B6-Zp3CretmTgCre (24). Erk3wt/ex3lox; Zp3Cre mice were bred to generate Erk3wt/Δex3 mice. Erk3wt/Δex3 mice were crossed, and littermates of different resulting genotypes (Erk3wt/wt [+/+], Erk3wt/Δex3 [+/−], and Erk3Δex3/Δex3 [−/−]) were analyzed. All mice were maintained at the animal facility of the Hannover Medical School under individually ventilated cage (IVC) conditions with free access to food and water. Mice were handled according to European guidelines (2010/63/EU) as well as the German Animal Welfare Act. All animal experiments were approved by the Lower Saxony State Office for Consumer Protection and Food Safety (file Z2017/47).
DNA isolation and genotyping.
Tail biopsy specimens, cells, and colonies were digested overnight at 55°C in lysis buffer (50 mM Tris-Cl [pH 8.0], 100 mM EDTA, 100 mM NaCl, and 1% SDS) containing proteinase K (0.5 mg/ml). For tissue samples, proteins were salted out with extra NaCl. DNA was precipitated with isopropanol, washed with 70% ethanol, and dissolved in water. Genotyping PCR was performed with Hotstar Taq (Qiagen) with extra Mg2+ under standard conditions. The primers used were the following: primer 1, ERK3-1-genotyping-FW, CCGTTTGAGTTTCTTGAGTG; primer 2, ERK3-3-genotyping-RV, CGTGGTATCGTTATGCG (primer 1 and 2 combination amplifies a 2.4-kb fragment in the case of homology recombination [short-arm integration]); primer 3, long-arm FW, CAGCTTTTGTTCCCTTTAGTGCTCGAC; primer 4, long-arm RC, AGGACTCCTACATCCTGAGCTACCTCTCTAG (primer 3 and 4 combination amplifies a 10.2-kb fragment in the case of homology recombination [long arm integration]); primer 5, ERK3_1-target-seq.-FW, TGGACAGAGCACTGGAAG; and primer 6, ERK3-loxP-RC, CTTAAGACAGGAGTGTGGATC (primer 5 and 6 combination amplifies 943 bp of WT, 1,055 bp of exon 3 floxed, or 336 bp of exon 3 deleted fragment).
PCRs were separated on 2% agarose gels and images were acquired using the INTAS gel documentation system.
Cell culture.
To generate BMDMs, bone marrow cells were flushed from the femurs of mice. Cells were cultured on 10-cm dishes in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, penicillin-streptomycin, and 50 ng/ml recombinant macrophage colony-stimulating factor (M-CSF) (Wyeth, Boston, MA) under humidified conditions with 5% CO2 at 37°C for 7 days.
Analysis of ERK3 mRNA expression.
Total RNA was isolated from BMDM of ERK3+/+ and ERK3−/− mice. RNA was purified using the Extractme total RNA extraction kit (BioScience) according to the manufacturer's instructions. cDNA from 500 ng RNA was synthesized using the first-strand cDNA synthesis kit (Fermentas/Thermo) in combination with random hexamer primers. Sense and antisense oligonucleotides (ERK3-mRNA-FW, 5′-TGGACAGAGCACTGGAAG-3′; ERK3-mRNA-RC, 5′-CTTAAGACAGGAGTGTGGATC-3′) specific for Erk3 were used to amplify a 619-bp ERK3 mRNA fragment spanning exons 2 to 4 from ERK3+/+ or a 474-bp fragment lacking the 145 bases of exon 3 from ERK3−/− cells. Amplification of a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA fragment was used as a loading control (GAPDH-fw, 5′-CATGGCCTTCCGTGTTCCTA-3′; GAPDH-rc, 5′-CCTGCTTCACCACCTTCTTGAT-3′).
SDS-PAGE, Western blotting, and antibodies.
Protein extracts were prepared by direct lysis of the cells in the culture plate with 2× Laemmli’s SDS sample buffer. Protein lysates were separated by SDS-PAGE on 7.5% to 16% gradient gels and transferred by semidry blotting to Hybond ECL nitrocellulose membranes (GE Healthcare). Primary antibodies used were anti-ERK3 (EP1720Y; ab53277) from Abcam, MK5 (HPA015515) from Atlas antibody, and GAPDH from Millipore. Secondary horseradish peroxidase-conjugated antibodies (Santa Cruz Biotechnologies) were used. Antigen-antibody complexes were detected with enhanced chemiluminescence (ECL) detection solution (solution A, 1.2 mM luminol in 0.1 M Tris-HCl [pH 8.6]; solution B, 6.7 mM p-coumaric acid in dimethyl sulfoxide; 35% H2O2 solution; ratio, 3333:333:1) using an LAS-3000 luminescent image analyzer (Fujifilm).
Histology.
Mice were euthanized individually with carbon dioxide in a standard mouse IVC. Lungs were harvested and inflation fixed with 10% neutral buffered formalin after euthanasia. Lung was cut at different levels, processed through a gradient of alcohols and xylene, and embedded in paraffin. For histological examination, 2- to 3-μm-thick sections were cut and stained with hematoxylin and eosin.
Flow cytometry and cell sorting.
Monoclonal antibodies specific for CCR7 (4B12), CD4 (GK1.5), CD5 (53-7.3), CD8α (53-6.7), CD19 (6D5), CD25 (PC61.5), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3), Foxp3 (MF23), TCRβ (H57-597), and TCRγδ (GL3) were AmCyan, brilliant violet 510 (BV510), BV421, Pacific blue (PB), eFluor450, fluorescein isothiocyanate (FITC), Alexa Fluor 488, Alexa Fluor 647, phycoerythrin (PE), peridinin chlorophyll protein-Cy5.5 (PerCP-Cy5.5), PE-Cy7, allophycocyanin (APC), and APC-Cy7 and were purchased from eBioscience, BD Biosciences, or BioLegend. Cells were acquired using a BD FACSCanto II, and data were analyzed using FlowJo software (Tree Star). Discrimination of dead cells was performed using either the Zombie Aqua fixable viability kit or 7-amino-actinomycin D according to the manufacturer’s instructions, and doublets were excluded.
Cell preparations.
Thymus and spleen were crushed through a 70-µm cell strainer (Corning) to obtain single-cell suspensions. For spleen, red blood cells (RBCs) were lysed using Qiagen RBC lysis solution according to the manufacturer’s instructions.
Statistical analysis.
All analyses were performed using GraphPad Prism software (version 7). Data are represented as means plus or minus standard errors of the means. Analysis of significance between 3 groups of mice was performed using one-way analysis of variance followed by Tukey’s test.
Analysis of T-cell proliferation.
Splenic and peripheral lymph node single-cell suspensions were pooled and stained with cell proliferation dye eFluor670 (eBioscience). Staining was performed at a cell concentration of 1 × 107 cells/ml in phosphate-buffered saline with 1.25 µM eFluor670 for 10 min at 37°C. For in vitro activation, 0.15 × 106 labeled cells in 200 µl medium (RPMI 1640 supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, 1.75 µl β-mercaptoethanol/500 ml medium, and 100 U/ml freshly added recombinant human interleukin-2 [rhIL-2]) were seeded into coated round-bottom 96-well plates. Plates were coated overnight at 4°C with anti-CD3 (clone 17.A2; final concentration of 0.5 µg/ml) and anti-CD28 (clone 37.51; final concentration of 1 µg/ml) in a volume of 100 µl per well. After 48 h of incubation at 37°C with 5% CO2, cells were split 1:2 into noncoated 96-well plates and supplemented with fresh rhIL-2. Seventy-two and 96 h after the start of culture, cells were harvested and stained with anti-CD4 (clone RM4-5; BioLegend) and anti-CD8b (clone RMCD8; homemade) antibodies, and proliferation was analyzed by flow cytometry. For live-dead discrimination, 4′,6-diamidino-2-phenylindole was used.
ACKNOWLEDGMENT
The work of M.G. is supported by Deutsche Forschungsgemeinschaft.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/MCB.00527-18.
REFERENCES
- 1.Cargnello M, Roux PP. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, DePinho RA, Panayotatos N, Cobb MH, Yancopoulos GD. 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65:663–675. [DOI] [PubMed] [Google Scholar]
- 3.Zhu AX, Zhao Y, Moller DE, Flier JS. 1994. Cloning and characterization of p97MAPK, a novel human homolog of rat ERK-3. Mol Cell Biol 14:8202–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez FA, Raden DL, Rigby MR, Davis RJ. 1992. Heterogeneous expression of four MAP kinase isoforms in human tissues. FEBS Lett 304:170–178. [DOI] [PubMed] [Google Scholar]
- 5.Déléris P, Trost M, Topisirovic I, Tanguay P-L, Borden KLB, Thibault P, Meloche S. 2011. Activation loop phosphorylation of ERK3/ERK4 by group I p21-activated kinases (PAKs) defines a novel PAK-ERK3/4-MAPK-activated protein kinase 5 signaling pathway. J Biol Chem 286:6470–6478. doi: 10.1074/jbc.M110.181529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.la Mota-Peynado De A, Chernoff J, Beeser A. 2011. Identification of the atypical MAPK Erk3 as a novel substrate for p21-activated kinase (Pak) activity. J Biol Chem 286:13603–13611. doi: 10.1074/jbc.M110.181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seternes O-M, Mikalsen T, Johansen B, Michaelsen E, Armstrong CG, Morrice NA, Turgeon B, Meloche S, Moens U, Keyse SM. 2004. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J 23:4780–4791. doi: 10.1038/sj.emboj.7600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perander M, Aberg E, Johansen B, Dreyer B, Guldvik IJ, Outzen H, Keyse SM, Seternes O-M. 2008. The Ser(186) phospho-acceptor site within ERK4 is essential for its ability to interact with and activate PRAK/MK5. Biochem J 411:613–622. doi: 10.1042/BJ20071369. [DOI] [PubMed] [Google Scholar]
- 9.Kant S, Schumacher S, Singh MK, Kispert A, Kotlyarov A, Gaestel M. 2006. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J Biol Chem 281:35511–35519. doi: 10.1074/jbc.M606693200. [DOI] [PubMed] [Google Scholar]
- 10.Schumacher S, Laass K, Kant S, Shi Y, Visel A, Gruber AD, Kotlyarov A, Gaestel M. 2004. Scaffolding by ERK3 regulates MK5 in development. EMBO J 23:4770–4779. doi: 10.1038/sj.emboj.7600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand F, Schumacher S, Kant S, Menon MB, Simon R, Turgeon B, Britsch S, Meloche S, Gaestel M, Kotlyarov A. 2012. The extracellular signal-regulated kinase 3 (mitogen-activated protein kinase 6 [MAPK6])-MAPK-activated protein kinase 5 signaling complex regulates septin function and dendrite morphology. Mol Cell Biol 32:2467–2478. doi: 10.1128/MCB.06633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen CA, Bartek J, Jensen S. 2008. A functional link between the human cell cycle-regulatory phosphatase Cdc14A and the atypical mitogen-activated kinase Erk3. Cell Cycle 7:325–334. doi: 10.4161/cc.7.3.5354. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Ou X-H, Wang Z-B, Xiong B, Tong J-S, Wei L, Li M, Yuan J, Ouyang Y-C, Hou Y, Schatten H, Sun Q-Y. 2010. ERK3 is required for metaphase-anaphase transition in mouse oocyte meiosis. PLoS One 5:e13074–e13078. doi: 10.1371/journal.pone.0013074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, Solis LM, Wistuba II, Tsai SY, Tsai M-J, O'Malley BW. 2012. ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Investig 122:1869–1880. doi: 10.1172/JCI61492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Bian K, Vallabhaneni S, Zhang B, Wu R-C, O'Malley BW, Long W. 2014. ERK3 promotes endothelial cell functions by upregulating SRC-3/SP1-mediated VEGFR2 expression. J Cell Physiol 229:1529–1537. doi: 10.1002/jcp.24596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bian K, Muppani NR, Elkhadragy L, Wang W, Zhang C, Chen T, Jung S, Seternes O-M, Long W. 2016. ERK3 regulates TDP2-mediated DNA damage response and chemoresistance in lung cancer cells. Oncotarget 7:6665–6675. doi: 10.18632/oncotarget.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klinger S, Turgeon B, Lévesque K, Wood GA, Aagaard-Tillery KM, Meloche S. 2009. Loss of Erk3 function in mice leads to intrauterine growth restriction, pulmonary immaturity, and neonatal lethality. Proc Natl Acad Sci U S A 106:16710–16715. doi: 10.1073/pnas.0900919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rousseau J, Klinger S, Rachalski A, Turgeon B, Déléris P, Vigneault E, Poirier-Héon J-F, Davoli MA, Mechawar N, Mestikawy El S, Cermakian N, Meloche S. 2010. Targeted inactivation of Mapk4 in mice reveals specific nonredundant functions of Erk3/Erk4 subfamily mitogen-activated protein kinases. Mol Cell Biol 30:5752–5763. doi: 10.1128/MCB.01147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquis M, Daudelin JF, Boulet S, Sirois J, Crain K, Mathien S, Turgeon B, Rousseau J, Meloche S, Labrecque N. 2014. The catalytic activity of the mitogen-activated protein kinase extracellular signal-regulated kinase 3 is required to sustain CD4+ CD8+ thymocyte survival. Mol Cell Biol 34:3374–3387. doi: 10.1128/MCB.01701-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquis M, Boulet S, Mathien S, Rousseau J, Thébault P, Daudelin J-F, Rooney J, Turgeon B, Beauchamp C, Meloche S, Labrecque N. 2014. The non-classical MAP kinase ERK3 controls T cell activation. PLoS One 9:e86681. doi: 10.1371/journal.pone.0086681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirois J, Daudelin J-F, Boulet S, Marquis M, Meloche S, Labrecque N. 2015. The atypical MAPK ERK3 controls positive selection of thymocytes. Immunology 145:161–169. doi: 10.1111/imm.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turgeon B, Lang BF, Meloche S. 2002. The protein kinase ERK3 is encoded by a single functional gene: genomic analysis of the ERK3 gene family. Genomics 80:673–680. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, Wang C, Sun H, LeRoith D, Yakar S. 2009. High-efficient FLPo deleter mice in C57BL/6J background. PLoS One 4:e8054. doi: 10.1371/journal.pone.0008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewandoski M, Wassarman KM, Martin GR. 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol 7:148–151. [DOI] [PubMed] [Google Scholar]
- 25.Saini M, Sinclair C, Marshall D, Tolaini M, Sakaguchi S, Seddon B. 2010. Regulation of Zap70 expression during thymocyte development enables temporal separation of CD4 and CD8 repertoire selection at different signaling thresholds. Sci Signal 3:ra23–ra23. doi: 10.1126/scisignal.2000702. [DOI] [PubMed] [Google Scholar]
- 26.West DB, Engelhard EK, Adkisson M, Nava AJ, Kirov JV, Cipollone A, Willis B, Rapp J, de Jong PJ, Lloyd KC. 2016. Transcriptome analysis of targeted mouse mutations reveals the topography of local changes in gene expression. PLoS Genet 12:e1005691-19. doi: 10.1371/journal.pgen.1005691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaul A, Köster M, Neuhaus H, Braun T. 2000. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell 102:17–19. [DOI] [PubMed] [Google Scholar]
- 28.Braun T, Rudnicki MA, Arnold HH, Jaenisch R. 1992. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell 71:369–382. [DOI] [PubMed] [Google Scholar]
- 29.Wang B, Wang N, Whitehurst CE, She J, Chen J, Terhorst C. 1999. T lymphocyte development in the absence of CD3 epsilon or CD3 gamma delta epsilon zeta. J Immunol 162:88–94. [PubMed] [Google Scholar]
- 30.Aberg E, Perander M, Johansen B, Julien C, Meloche S, Keyse SM, Seternes O-M. 2006. Regulation of MAPK-activated protein kinase 5 activity and subcellular localization by the atypical MAPK ERK4/MAPK4. J Biol Chem 281:35499–35510. doi: 10.1074/jbc.M606225200. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Kotlyarov A, Laabeta K, Gruber AD, Butt E, Marcus K, Meyer HE, Friedrich A, Volk H-D, Gaestel M. 2003. Elimination of protein kinase MK5/PRAK activity by targeted homologous recombination. Mol Cell Biol 23:7732–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronkina N, Johansen C, Bohlmann L, Lafera J, Menon MB, Tiedje C, Laaß K, Turk BE, Iversen L, Kotlyarov A, Gaestel M. 2015. Comparative analysis of two gene-targeting approaches challenges the tumor-suppressive role of the protein kinase MK5/PRAK. PLoS One 10:e0136138. doi: 10.1371/journal.pone.0136138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aberg E, Torgersen KM, Johansen B, Keyse SM, Perander M, Seternes O-M. 2009. Docking of PRAK/MK5 to the atypical MAPKs ERK3 and ERK4 defines a novel MAPK interaction motif. J Biol Chem 284:19392–19401. doi: 10.1074/jbc.M109.023283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Déléris P, Rousseau J, Coulombe P, Rodier G, Tanguay P-L, Meloche S. 2008. Activation loop phosphorylation of the atypical MAP kinases ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J Cell Physiol 217:778–788. doi: 10.1002/jcp.21560. [DOI] [PubMed] [Google Scholar]