Autophagy is a process of lysosomal self-degradation of cellular components by forming autophagosomes. Autophagosome formation is an essential process in autophagy and is fine-tuned by various autophagy-related gene (ATG) products, including ATG5, ATG12, and ATG16.
KEYWORDS: 3′ UTR, RNA binding proteins, translational control
ABSTRACT
Autophagy is a process of lysosomal self-degradation of cellular components by forming autophagosomes. Autophagosome formation is an essential process in autophagy and is fine-tuned by various autophagy-related gene (ATG) products, including ATG5, ATG12, and ATG16. Although several reports have shown that numerous factors affect multiple levels of gene regulation to orchestrate cellular autophagy, the detailed mechanism of autophagosome formation still needs further investigation. In this study, we demonstrate that the RNA binding protein HuR (human antigen R) performs an essential function in autophagosome formation. We observe that HuR silencing leads to inhibition of autophagosome formation and autophagic flux in liver cells. Ribonucleoprotein immunoprecipitation (RIP) assay allows the identification of ATG5, ATG12, and ATG16 mRNAs as the direct targets of HuR. We further show that HuR mediates the translation of ATG5, ATG12, and ATG16 mRNAs by binding to their 3′ untranslated regions (UTRs). In addition, we show that HuR expression positively correlates with the levels of ATG5 and ATG12 in hepatocellular carcinoma (HCC) cells. Collectively, our results suggest that HuR functions as a pivotal regulator of autophagosome formation by enhancing the translation of ATG5, ATG12, and ATG16 mRNAs and that augmented expression of HuR and ATGs may participate in the malfunction of autophagy in HCC cells.
INTRODUCTION
Autophagy is a process of lysosomal self-degradation of cellular components by forming autophagosomes that are conserved in all eukaryotes (1, 2). This is a pivotal process to maintain cellular homeostasis (3). An autophagosome is a double-membrane vesicle that contains sequestered cytoplasmic cargos and transports them to lysosomes (4). Autophagosome formation is an essential step in autophagy and is fine-tuned by various autophagy-related gene (ATG) products, including ATG5, ATG12, and ATG16. During autophagosome formation, ATG5 is conjugated with ATG12 by ubiquitin-like conjugation systems and, further, forms a homodimer consisting of an ATG5-ATG12/ATG16 complex (5, 6). The complex is localized to autophagy-related membranes and mediates LC3 conversion (7–9). LC3 conversion is widely used as a marker of autophagosome formation (10). It has been reported that loss of ATG5, ATG12, or ATG16 results in a decrease of autophagosome formation, thereby impairing the autophagic process (11–13).
Autophagy is involved in various stress responses, and dysregulation of autophagy has been found in many diseases, including cardiac ischemia/reperfusion injury, Crohn’s disease, neurodegeneration, myopathy, and diabetes, functioning as the driving or exacerbating factor in the pathogenesis of the diseases (3). In addition, autophagy is implicated in cancer development, although it is still controversial whether autophagy promotes or suppresses the growth of cancer cells (14). Several reports have shown that autophagy is a prosurvival process in established cancer cells, and the inhibition of autophagy is one of the strategies for cancer therapy (14–17). Therefore, elucidation of the fine molecular mechanisms of autophagic processes is important in understanding the role of autophagy in the pathogenesis of several diseases. Several critical regulators have been identified, and their expression mechanisms were elucidated in various models (reviewed in references 4 and 18). Several microRNAs (miRNAs) have been reported to be pivotal regulators of autophagy (19–21). In this study, we investigated the regulatory mechanism of autophagosome formation at the RNA level.
HuR (human antigen R) (also known as HuA or ELAVL1) is a member of the Hu/ELAV (embryonic lethal abnormal vision)-like RNA binding protein (RBP) family containing three RNA recognition motifs (RRM) (22). HuR binds to AU-rich elements (ARE) in the untranslated regions (UTRs) of target mRNAs and regulates gene expression by affecting the stability or translation of target mRNAs (22, 23). HuR is ubiquitously expressed and has essential roles in immune response, angiogenesis, metastasis, and cancer development by regulating cell proliferation, migration, survival, death, and autophagy (24). HuR is known to promote cell growth and survival by increasing cancer-related genes, such as the Cox-2, hypoxia-inducible factor 1α (HIF-1α), and vascular endothelial growth factor (VEGF) genes, and augmented expression of HuR is related to cancer progression in some types of cancer (23, 25–27). Several efforts have been made to validate the potential of HuR as a molecular target for cancer therapy (28–30).
Here, we investigate the role of HuR in the regulation of autophagosome formation and show that HuR silencing reduces autophagosome formation and the autophagic flux of human liver cells. Along with previous studies showing SQSTM1/p62 regulation by HuR (31, 32), we identify ATG5, ATG12, and ATG16 mRNAs as novel targets of HuR and demonstrate augmented expression of ATG5, ATG12, ATG16, and HuR in hepatocellular carcinoma (HCC). Our results provide a molecular mechanism of autophagosome formation regulated by HuR and the potential of HuR targeting in cancer progression.
RESULTS
HuR regulates autophagosome formation and autophagic flux.
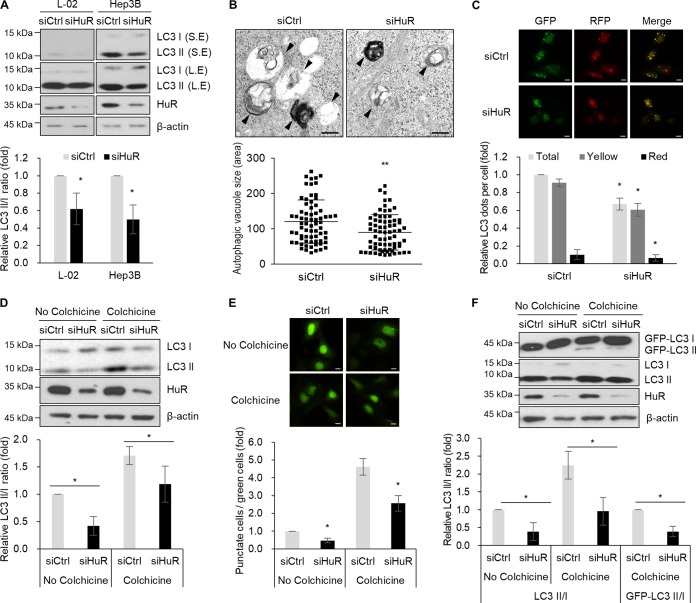
To understand the role of HuR in the regulation of autophagy, we investigated whether autophagosome formation is affected by downregulation of HuR in human liver cells, including L-02 and Hep3B cells. The LC3II/LC3I ratio was slightly, but consistently, reduced by HuR silencing in both L-02 and Hep3B cells (Fig. 1A). Electron microscopy images revealed that the sizes of autophagosomes and autolysosomes were reduced by downregulation of HuR (Fig. 1B). We also investigated autophagosome maturation after HuR downregulation using tandem fluorescence-tagged LC3 (33) and found that the total number of dots and the numbers of yellow dots and red dots were moderately decreased in HuR small interfering RNA (siRNA)-transfected cells (Fig. 1C). To further determine whether autophagic flux is affected by HuR downregulation, we investigated the LC3 turnover rate after treating cells with 0.4 μg/ml of colchicine, an inhibitor of autophagosome-lysosome fusion. Figure 1D shows that colchicine treatment increased LC3 conversion in Hep3B cells; however, HuR downregulation partially, but significantly, reduced colchicine-induced accumulation of autophagosomes. This result indicates that HuR silencing inhibited autophagosome formation and autophagic flux. Regulation of autophagosome formation by HuR was further examined by assessing the formation of green fluorescent protein (GFP) puncta in GFP-LC3-expressing U2OS cells (U2OS-GFP-LC3 cells). HuR downregulation resulted in a modest reduction in the number of GFP puncta-positive cells at the basal level, as well as after colchicine treatment (Fig. 1E). In addition, colchicine-induced accumulation of GFP-LC3II was also reduced by HuR silencing (Fig. 1F). These observations suggest that HuR has a role in the regulation of autophagosome formation and autophagic flux.
FIG 1.
Autophagosome formation is reduced by HuR downregulation. (A) L-02 and Hep3B cells were transfected with siCtrl and siHuR for 48 h, and the LC3 level was assessed by Western blotting analysis. S.E., short exposure; L.E., long exposure. The relative intensities of WB images are shown in the graph. (B) Hep3B cells were transfected with siCtrl and siHuR, and autophagosomes were observed by transmission electron microscopy. The arrowheads indicate the autophagosomes and autolysosomes. The sizes of autophagosomes were analyzed by measuring the areas of at least 70 autophagic vacuoles. Scale bars = 0.5 μm. **, P < 0.01. (C) After transfection of siRNAs, L-02 cells were further transfected with the mRFP-GFP-LC3 plasmid, and fluorescent LC3 dots from at least 300 cells were analyzed under a fluorescence microscope. Scale bars = 10 μm. The images are representative, and values are expressed as means and SD of the results from three independent experiments (Student's t test). *, P < 0.05. RFP, red fluorescent protein. (D) After transfection with siCtrl or siHuR, Hep3B cells were incubated with 0.4 μg/ml colchicine for 6 h, and the levels of LC3 and HuR were analyzed by Western blotting. The relative intensities of WB images are shown in the graph. *, P < 0.05. (E) After transfection with siCtrl or siHuR, U2OS cells stably expressing GFP-LC3 were incubated with 0.4 μg/ml colchicine for 6 h, and GFP-LC3 dots were analyzed under a fluorescence microscope. GFP-LC3 puncta were quantified as the ratio of GFP-LC3 punctate cells to total GFP-positive cells (at least 300 cells) under a fluorescence microscope. Scale bars = 10 μm. The data are representative of three independent experiments, and values are expressed as means and SD (Student's t test); *, P < 0.05. (F) GFP-LC3 or LC3 conversion in U2OS-GFP-LC3 cell lysates was analyzed by Western blotting; the relative intensities of WB images are shown in the graph. The images are representative of the results from three independent experiments.
HuR is associated with ATG5, ATG12, and ATG16 mRNAs.
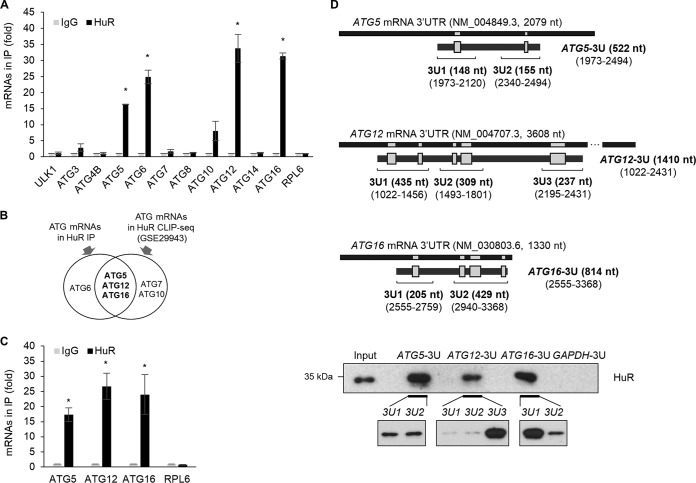
Based on our observation (Fig. 1), we hypothesized that HuR performs a role in regulating the expression of ATGs. To address this, HuR-containing ribonucleoprotein (RNP) particles were isolated by immunoprecipitation (IP) using HuR antibody, and RNP-associated mRNAs in the IP products were analyzed by reverse transcription-quantitative PCR (RT-qPCR) using specific primers (Table 1). The binding between HuR and a subset of ATG mRNAs, including ULK1, ATG3, ATG4B, ATG5, ATG6, ATG7, ATG8, ATG10, ATG12, ATG14, and ATG16 mRNAs, was assessed, and the results showed the enrichment of ATG5, ATG6, ATG10, ATG12, and ATG16 mRNAs in HuR IP products (Fig. 2A). In addition, we analyzed HuR photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) sequencing (CLIP-seq) data (GSE29943) on the UCSC Genome Browser (UCSC GB) (34) to investigate HuR binding sites at the 3′ UTR s of ATG5, ATG12, and ATG16 mRNAs (data not shown). Based on our experimental results and in silico analysis of CLIP-seq data, we found that ATG5, ATG12, and ATG16 mRNAs have putative HuR binding sites at their 3′ UTRs (Fig. 2B). The binding between HuR and the mRNAs was further investigated by ribonucleoprotein immunoprecipitation (RIP) and then RT-qPCR, and Fig. 2C shows that specific association of HuR with ATG5, ATG12, and ATG16 mRNAs was observed. To confirm the associations of HuR with ATG5, ATG12, and ATG16 mRNAs, we performed in vitro pulldown assays using biotin-labeled transcripts containing HuR binding sites (Fig. 2D, gray boxes) (ATG5-3U, ATG12-3U, and ATG16-3U), followed by Western blotting (WB) with HuR antibody. Figure 2D shows that HuR bound to the transcripts of ATG5-3U, ATG12-3U, and ATG16-3U but not to GAPDH-3U. To further validate HuR binding to each predicted region of the ATG mRNAs, the biotin pulldown assay was performed again using each fragment shown in Fig. 2D (ATG5-3U1, ATG5-3U2, ATG12-3U1, ATG12-3U2, ATG12-3U3, ATG16-3U1, and ATG16-3U2). HuR strongly bound to ATG5-3U1, ATG5-3U2, ATG12-3U3, and ATG16-3U1, while it moderately bound to ATG12-3U1, ATG12-3U2, and ATG16-3U2. These results suggest that HuR associated with the 3′ UTRs of ATG5, ATG12, and ATG16 mRNAs.
TABLE 1.
Primer sequences used in this study
| Purpose and primer designationa | Sequence (5′→3′)b |
Product size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| For qPCR (human) | |||
| ULK1 | GGCAAGTTCGAGTTCTCCCG | CGACCTCCAAATCGTGCTTCT | 97 |
| ATG3 | GATGGCGGATGGGTAGATACA | TCTTCACATAGTGCTGAGCAATC | 125 |
| ATG4B | GGTGTGGACAGATGATCTTTGC | CCAACTCCCATTTGCGCTATC | 172 |
| ATG5 | GTTTTGGGCCATCAATCGGAA | TCTCCTAGTGTGTGCAACTGT | 159 |
| ATG6 | AGGTTGAGAAAGGCGAGACA | AATTGTGAGGACACCCAAGC | 196 |
| ATG7 | ATGATCCCTGTAACTTAGCCCA | CACGGAAGCAAACAACTTCAAC | 114 |
| ATG8 | AACATGAGCGAGTTGGTCAAG | GCTCGTAGATGTCCGCGAT | 127 |
| ATG10 | TACGCAACAGGAACATCCAA | AACAACTGGCCCTACAATGC | 157 |
| ATG12 | TAGAGCGAACACGAACCATCC | CACTGCCAAAACACTCATAGAGA | 153 |
| ATG14 | GCGCCAAATGCGTTCAGAG | AGTCGGCTTAACCTTTCCTTCT | 91 |
| ATG16 | ATGCGCGGATTGTCTCAGGG | GTCCACTCATTACACATTGCTCT | 138 |
| HuR | AACTACGTGACCGCGAAGG | CGCCCAAACCGAGAGAACA | 194 |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG | 87 |
| β-Actin | GGACTTCGAGCAAGAGATGG | AGCACTGTGTTGGCGTACAG | 234 |
| RPL6 | AGATTACGGAGCAGCGCAAGATTG | GCAAACACAGATCGCAGGTAGCCC | 106 |
| For BPD assay (human) | |||
| ATG5-3U | CCAAGCTTCTAATACGACTCACTATAGGGAGACTGTGTTCTTTACACACTACACG | TGTGAAGAAATGTGCTATTAATCAGTG | 554 |
| ATG5-3U1 | ATG5-3U forward primer sequence | ACACTAGCTTGGAAGGAATGG | 181 |
| ATG5-3U2 | CCAAGCTTCTAATACGACTCACTATAGGGAGACAGACACGATCATGGTTTTAG | ATG5-3U reverse primer sequence | 186 |
| ATG12-3U | CCAAGCTTCTAATACGACTCACTATAGGGAGAAACATCTTCATTGTCAGTTCTCA | CAACAAGACCGAAACGCTG | 1,443 |
| ATG12-3U1 | ATG12-3U forward primer sequence | CCACAGTTATGTGATTGGGACT | 467 |
| ATG12-3U2 | CCAAGCTTCTAATACGACTCACTATAGGGAGAGATGAGTAAACCCAGTAGCATCTT | CAGGCTGGTCTCAAACTTGT | 341 |
| ATG12-3U3 | CCAAGCTTCTAATACGACTCACTATAGGGAGACCACTGAGGGTTCAGATGATAG | ATG12-3U reverse primer sequence | 269 |
| ATG16-3U | CCAAGCTTCTAATACGACTCACTATAGGGAGAACTCCCGAACTACAGACCC | TATTTACAATCAAGTTCTGTTGGCC | 846 |
| ATG16-3U1 | ATG16-3U forward primer sequence | TTCACACATCCTCTCCCACCTC | 237 |
| ATG16-3U2 | CCAAGCTTCTAATACGACTCACTATAGGGAGAGATGCCCTTGGTGCTTTAGTGC | ATG16-3U reverse primer sequence | 461 |
| GAPDH-3U | CCAAGCTTCTAATACGACTCACTATAGGGAGACCTCAACGACCACTTTGTCA | GGTTGAGCACAGGGTACTTTAT | 335 |
| For cloning (human) | |||
| ATG5-3U | aaaaAGATCTTAACTGTGTTCTTTACACACTACACG | aaaaGGTACCTGTGAAGAAATGTGCTATTAATCAGTG | 546 |
| ATG12-3U | aaaaAGATCTTAAAACATCTTCATTGTCAGTTCTCA | aaaaGGTACCCAACAAGACCGAAACGCTG | 1,435 |
| ATG16-3U | aaaaAGATCTTAAACTCCCGAACTACAGACCC | aaaaGGTACCTATTTACAATCAAGTTCTGTTGGCC | 839 |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Lowercase letters are extra nucleotides for efficient cleavage by restriction enzymes.
FIG 2.
Interaction between HuR and ATG5, ATG12, and ATG16 mRNAs. (A) The interactions between ATG mRNAs and HuR in Hep3B lysates were screened by RIP-qPCR analysis using anti-HuR or control IgG antibodies. RPL6 mRNA was used as a negative control. (B) Venn diagram of ATG mRNAs between HuR RIP screening and HuR CLIP-seq analysis (GSE29943). (C) Relative associations of ATG mRNAs with HuR compared to normal IgG were confirmed by RIP-qPCR. The data are expressed as means and SD of the results from three independent experiments (Student's t test); *, P < 0.05. (D) (Top) Schematics of 3′ UTRs of human ATG5, ATG12, and ATG16 mRNAs and their putative HuR binding sites (gray boxes; based on the CLIP-seq data [GSE29943]). Each biotinylated transcript containing putative HuR binding sites was synthesized for biotin pulldown analysis. (Bottom) Each biotinylated transcript was incubated with Hep3B cell lysates, and the interaction between HuR and each biotinylated transcript is shown by Western blot analysis using an anti-HuR antibody. Biotinylated GAPDH transcript was used as the negative control. The images are representative of the results from three independent experiments.
HuR silencing decreased the expressions of ATG5, ATG12, and ATG16.
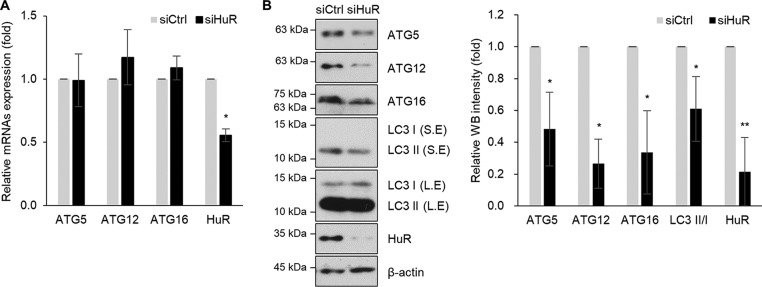
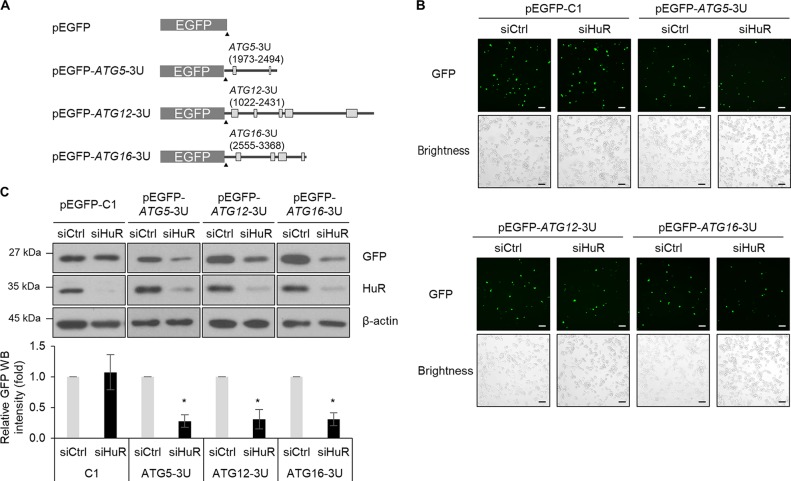
To investigate whether HuR affects the expressions of ATG5, ATG12, and ATG16, Hep3B cells were transiently transfected with siHuR or control siRNA, and the levels of mRNA and protein were assessed by RT-qPCR and Western blotting, respectively. No obvious changes in their mRNAs were found; however, HuR silencing downregulated the levels of ATG5, ATG12, and ATG16 proteins (Fig. 3). This was consistently observed in L-02 cells (data not shown). To further investigate HuR-mediated regulation of ATG5, ATG12, and ATG16, we performed enhanced green fluorescent protein (EGFP) reporter assays. EGFP reporter plasmids were constructed by inserting the 3′ UTR of each mRNA, as shown in Fig. 4A, and the expression of EGFP reporters after HuR downregulation was analyzed by fluorescence microscopy and Western blotting (Fig. 4B and C). HuR downregulation resulted in a reduction of expression of EGFP reporters containing 3′ UTRs of ATG5, ATG12, and ATG16 mRNAs but not control EGFP. Taken together, these results suggest that HuR regulated the expression of ATG5, ATG12, and ATG16 by binding to their 3′ UTRs.
FIG 3.
Expression of ATG5, ATG12, and ATG16 is regulated by HuR. Hep3B cells were transfected with siHuR or siCtrl for 48 h, and expression levels of ATG5, ATG12, and ATG16 were assessed by RT-qPCR and Western blotting. (A) The levels of ATG5, ATG12, ATG16, and HuR mRNAs were measured by RT-qPCR. The GAPDH mRNA level was used for normalization. (B) Endogenous ATG5, ATG12, ATG16, LC3, and HuR expression levels were analyzed by Western blotting, and the relative intensities of WB images are shown in the graph. β-Actin was used as a loading control. S.E., short exposure; L.E., long exposure. The data are representative of the results from three independent experiments, and values are expressed as means and SD (Student's t test). *, P < 0.05; **, P < 0.01.
FIG 4.
Expression of ATG5, ATG12, and ATG16 is directly regulated by HuR via their 3′ UTRs. (A) Schematics of EGFP reporters. (B and C) Following sequential transfection with reporter plasmids for 24 h after knockdown of HuR in Hep3B cells, the EGFP fluorescence from at least 300 cells/experiment was observed under a fluorescence microscope (scale bars = 20 μm) (B) and the expression levels of GFP and HuR were analyzed by Western blotting. β-actin was used as a loading control. (C) The images are representative of the results from three independent experiments. The relative intensities of WB images are shown in the graph, and the values represent means and SD (Student's t test); *, P < 0.05.
HuR silencing decreases the translation of ATG5, ATG12, and ATG16 mRNAs.
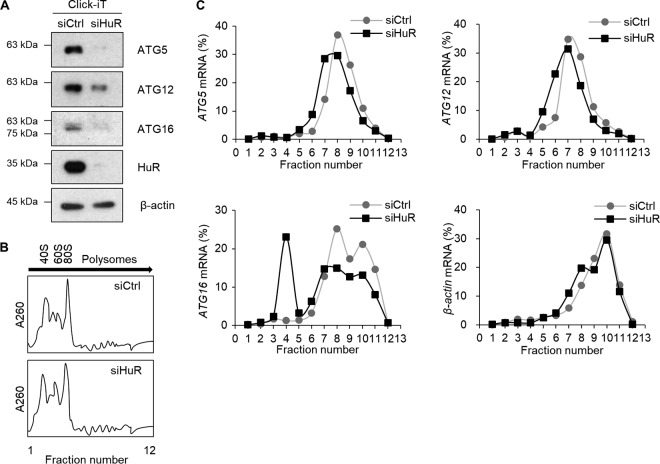
Because the levels of ATG5, ATG12, and ATG16 were reduced by HuR downregulation without obvious changes in their mRNA levels (Fig. 3), we further investigated whether the translation rates of those mRNAs were affected by HuR silencing in Hep3B cells. First, we analyzed newly synthesized proteins by tracing metabolically labeled proteins with l-azidohomoalanine (AHA), as described in Materials and Methods. De novo synthesis of ATG5, ATG12, and ATG16, assessed by Click-iT (Invitrogen) reaction followed by Western blotting, was reduced by HuR silencing (Fig. 5A). This result indicated that HuR plays a role in regulating the translation of ATG5, ATG12, and ATG16 mRNAs. In addition, we assessed the distribution of ATG5, ATG12, and ATG16 mRNAs on polyribosome (polysome) fractions as described previously (35, 36). For this, after transfection of HuR siRNA or control siRNA, cell lysates were fractionated using sucrose gradient ultracentrifugation, and the distribution of ATG5, ATG12, ATG16, and β-actin mRNAs from each fraction was assessed by RT-qPCR. Total RNA distributions in the two groups were similar (Fig. 5B); however, the distributions of polysome-associated ATG5, ATG12, and ATG16 mRNAs (found in fractions 6 to 9) were reduced in the siHuR-transfected group (Fig. 5C). These results indicate that HuR silencing downregulated the expression of ATG5, ATG12, and ATG16 by decreasing their translation.
FIG 5.
Translational regulation of ATG5, ATG12, and ATG16 mRNAs by HuR. (A) The amounts of nascent ATG5, ATG12, and ATG16 proteins changed by reduction of HuR in Hep3B cells were analyzed using the Click-iT system. After knockdown of HuR expression, the cells were preincubated for 1 h in methionine-free RPMI medium and then metabolically labeled for 4 h with the azide-containing methionine analog AHA. After incubation, cell lysates were prepared, and the newly synthesized AHA-incorporating proteins were cross-linked to alkyne-derivatized biotin by copper-catalyzed cycloaddition (Click-iT reaction). Biotin-conjugated newly synthesized proteins were collected by pulldown using streptavidin beads, followed by Western blotting. β-Actin was used as a loading control. (B) Hep3B cell lysates transfected with siHuR or siCtrl for 48 h were centrifuged through a 10 to 50% sucrose density gradient, and polysomes were fractionated according to size. (C) RNAs were isolated from each fraction from panel B, and the relative distribution of ATG5, ATG12, ATG16, and β-actin mRNAs on each fraction was analyzed by RT-qPCR. The data are representative of the results from three independent experiments.
Expression levels between HuR and ATGs were positively correlated in human HCC tissues.
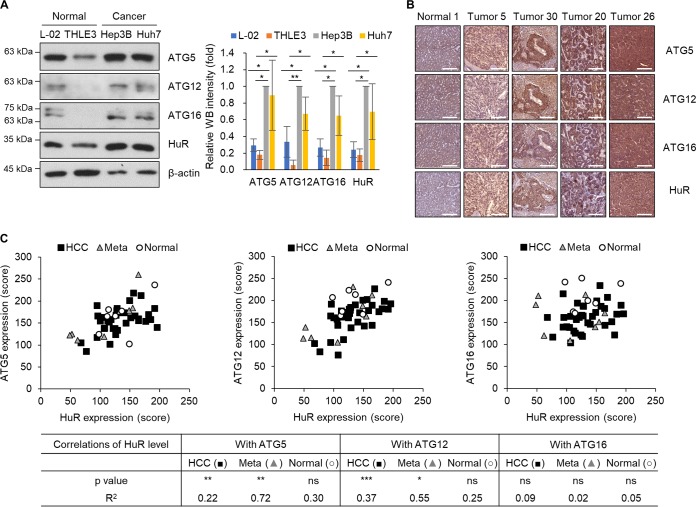
To investigate the correlation between HuR and ATGs, including ATG5, ATG12, and ATG16, in normal and cancer cells, we analyzed the expression of the proteins in human liver cell lines (Fig. 6A) and in human liver tissues from HCC tissue microarray (TMA) (Fig. 6B and Table 2). Western blotting revealed that the relative levels of HuR, ATG5, ATG12, and ATG16 in HCC cell lines (Hep3B and Huh7) were higher than those in immortalized hepatic cell lines (THLE3 and L-02) (Fig. 6A). In addition, the relative expression levels of HuR, ATG5, ATG12, and ATG16 were also studied by immunohistochemistry (IHC) using human HCC TMA containing a total of 50 cores, including the liver tissues from primary HCC (n = 35) and metastatic HCC (n = 8) and paired normal tissues (n = 7). The intensity of 3,3′-diaminobenzidine (DAB) signals of each tissue was measured, and the correlations between HuR and each ATG protein were further analyzed according to the guidelines (37); the results are shown in Fig. 6C. The ATG5 level was positively related to the HuR level in both groups of HCC tissues (primary HCC and metastatic HCC) (P < 0.01) but not in paired normal tissues. High correlation between ATG5 and HuR (R2 = 0.72) was observed in metastatic HCC tissues. The ATG12 level was also positively related to the HuR level in both groups of HCC tissues (P < 0.001 in the primary HCC group; P < 0.05 in the metastatic HCC group) but not in paired normal tissues. High correlation (R2 = 0.55) and moderate correlation (R2 = 0.37) between ATG12 and HuR were observed in primary HCC tissues and metastatic HCC tissues, respectively. However, the ATG16 level was not significantly corelated with the HuR level in this analysis. This result suggests that the HuR level is associated with ATG5 and ATG12 in the liver tissues from HCC patients, and differential expression of these proteins may be related to development of HCC.
FIG 6.
Correlation between ATG5, ATG12, ATG16, and HuR expression in human liver cell lines and liver tissues. (A) Whole-cell lysates were prepared from human hepatic cell lines (L-02 and THLE3) and human HCC cell lines (Hep3B and Huh7), and endogenous ATG5, ATG12, ATG16, and HuR expression levels in the whole-cell lysates were detected by Western blotting. The intensities of WB images from three independent experiments were analyzed and are shown in the graph. *, P < 0.05; **, P < 0.01. (B and C) ATG5, ATG12, ATG16, and HuR expression levels in liver tissues from HCC patitents were assessed by IHC. (B) Representative images of immunohistochemical detection of ATG5, ATG12, ATG16, and HuR in a human HCC TMA slide. Scale bars = 200 μm. (C) Correlation of HuR expression with ATG5, ATG12, or ATG16 presented in a scatter graph using the IHC stain score of each core (n = 50). HCC, primary HCC; Meta, metastatic HCC; Normal, paired normal tissues. The data represent P and R2 values of IHC analysis (37). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. 0 < R2 < 0.09, no correlation; 0.09 < R2 < 0.25, low correlation; 0.25 < R2 < 0.49, moderate correlation; 0.49 < R2 < 0.81, high correlation; 0.81 < R2 < 1.0, very high correlation.
TABLE 2.
Characteristics of liver tissues in tissue microarray
| Characteristic | Value |
|---|---|
| No. (%) of HCC patients | 38 (100) |
| Age (yr) | |
| Median | 56.5 |
| Range | 25–77 |
| Gender [no. (%) of patients] | |
| Male | 36 (94.74) |
| Female | 2 (5.26) |
| Stage [no. (%) of patients] | |
| I | 12 (31.58) |
| II | 11 (28.95) |
| III | 7 (18.42) |
| IV | 8 (21.05) |
| No. (%) of TMA cores | |
| Total | 50 (100) |
| Primary HCC tissue | 35 (70) |
| Paired metastatic tumor tissue from HCC | 5 (10) |
| Unpaired metastatic tumor tissue from HCC | 3 (6) |
| Paired normal liver tissue | 7 (14) |
DISCUSSION
Autophagy is a pivotal process to maintain cellular homeostasis in response to various stimuli, and tight regulation of autophagy is critical for the life of cells. Previous reports have shown that HuR affects autophagy activity by regulating SQSTM1/p62, ATG7, and ATG16, and we confirmed a regulatory function of HuR in autophagosome formation in this study (31, 32, 38). We observed that HuR silencing leads to the inhibition of autophagosome formation and autophagic flux in HCC Hep3B cells and human fetal hepatocyte cell line L-02 cells and identified ATG5, ATG12, and ATG16 mRNAs as direct targets of HuR. We showed that HuR mediates the translation of ATG5, ATG12, and ATG16 mRNAs by binding to their 3′ UTRs. We also showed that HuR expression positively correlates with the levels of ATG5 and ATG12 in HCC cells. Collectively, our results suggest that augmented expression of HuR may participate in the malfunction of autophagy in HCC cells by enhancing the translation of ATG5, ATG12, and ATG16 mRNAs.
HuR is involved in posttranscriptional gene regulation in eukaryotes and plays important roles in splicing, export, stability, localization, and translation by forming RNP particles (39). HuR affects a broad spectrum of cell physiology by regulating the metabolism of multiple target mRNAs (23, 26). In this study, we identified three different target mRNAs, ATG5, ATG12, and ATG16 mRNAs, as direct targets for HuR and demonstrated that HuR functions as a translational enhancer. Our results suggest that HuR performs a function in autophagy regulation by orchestrating autophagosome formation. Therefore, tight control of the HuR level in cells is critical for cellular homeostasis. Several studies have reported augmented expression of HuR in various types of cancer, including stomach, colon, kidney, thyroid, and lung cancer (25, 27, 40). It suggests that dysregulation of HuR contributes to cancer progression by regulating cell proliferation, migration, invasion, inflammation, and autophagy.
Several regulatory mechanisms governing HuR expression have been elucidated. The phospatidylinositol 3-kinase (PI3K)/AKT pathway is frequently mutated in several types of human cancer, and the PI3K/AKT/NF-κB axis resulted in the overexpression of HuR (40). The amount of HuR protein could be regulated by ubiquitination (41). Tumor suppressor esophageal-cancer-related gene 2 (ECRG2) promotes ubiquitnation-dependent degradation of HuR, and an inverse correlation between ECRG2 and HuR has been reported in esophageal and lung cancer (42, 43). In addition, several miRNAs, including miR-519, miR-125a, miR-16, miR-570-3p, and miR-219b-3p, have been identified as negative regulators of HuR (44–47). Interestingly, the levels of miR-519, miR-16, and miR-570-3p are downregulated in several cancer tissues and function as tumor suppressors (48–50). Further investigation would be needed to identify other regulators that affect HuR expression and to provide valuable information to understand the differential expression of HuR in various types of human diseases, including cancer.
Autophagy is a pivotal process for the survival of cells in response to various stimuli, such as starvation, hypoxia, and oxidative stress. Activation of autophagy at the basal level is essential in most cell types for the housekeeping role in organelle turnover and the removal of protein aggregates (1, 3). In certain types of cells, including neurons and hepatocytes, tight regulation of basal autophagy is critical for normal cellular function, and its dysregulation is implicated in the pathogenesis of human diseases, including cancer (51–53). Although the function of autophagy in HCC is still not fully understood, several reports have shown that changes in autophagy-regulatory genes, such as ATG5, ATG6, and ATG7, affect the development of HCC (54, 55). Basal autophagy is thought to play tumor-suppressive roles in hepatocytes; however, in established tumors, dysregulated autophagy contributes to cell survival, thereby promoting tumor growth (56). It has been reported that impaired autophagy induces the accumulation of tumor suppressors, including p53, p21, and p27, and it suppresses HCC development (57). Also, an autophagy inhibitor made HCC cells sensitive to anticancer drugs, such as sorafenib, and ATG5 depletion increased sorafenib-induced cell death (58, 59). These reports suggest that autophagy performs a protumor function in the development of HCC. In addition to these observations, several reports have shown that autophagy plays antitumor roles in HCC (reviewed in references 55 and 60). For example, autophagy deficiency resulted from the depletion of ATG5 and ATG7, and p62 accumulation promoted tumor growth in hepatocytes (61, 62). Unfortunately, the exact functions of autophagy in HCC are not fully elucidated and remain controversial. Further intensive investigations will be required to understand the role of autophagy in HCC development.
Tight regulation of ATG proteins is critical for the maintenance of basal autophagy that affects cellular homeostasis. Numerous studies have identified essential factors that regulate ATG expression and elucidated their regulatory mechanisms. Posttranslational modifications of ATG proteins, including phosphorylation, ubiquitination, SUMOylation, acetylation, and O-GlcNAc modification, are known to be involved in the dynamic regulation of autophagy (reviewed in references 63, to ,65). Transcription factors TFEB and FOXO function as key regulators of autophagy and lysosomal biogenesis by controlling the transcriptional networks of autophagy (65–67). Epigenetic events, such as chromatin modification by histone modification, have also been linked to autophagy regulation (68). In addition, noncoding RNAs (ncRNAs), including miRNAs and long noncoding RNAs (lncRNAs), are actively involved in the regulation of autophagy by affecting ATG expression (reviewed in references 21 and 69). RBPs, such as RCK-Dcp2, TDP-43, TIA-1, and HuD, are known to function as autophagy regulators (35, 70–72). In this study, we proposed HuR as a pivotal regulatory factor governing autophagosome formation and autophagic flux (Fig. 1). Although the comprehensive mechanism of autophagy regulation still needs to be updated, autophagy seems to be a very dynamic and complex process and requires diverse regulation at multiple levels of gene expression to maintain cellular homeostasis in response to various stimuli.
The effect of HuR on LC3 conversion has been observed in several types of cells, including hepatic stellate cells, pancreatic ductal adenocarcinoma cells, renal tubular cells, and human embryonic kidney cells (38, 73–75). Here, we report HuR-mediated autophagosome formation in HCC cells and further demonstrate that HuR functions as the factor enhancing the expression of ATG5, ATG12, and ATG16, thereby influencing autophagic flux. We also observed positive correlations between HuR and some ATG proteins (ATG5 and ATG12) in the liver tissues from HCC patients. Although our results and a recent study show that HuR positively regulates ATG16 expression (38), a positive correlation between HuR and ATG16 was not observed in HCC tissues (Fig. 6). Particular miRNAs, including miR-519a, miR-374, miR-223, and miR-874, are known to target ATG16L1 mRNA in various types of cells (38, 76–78). Whether those miRNAs or other RBPs are involved in the regulation of ATG16 expression during cancer development needs to be determined in separate studies. Our results provide a regulatory mechanism of gene expression, including ATG5, ATG12, and ATG16 mRNAs, by HuR at the posttranscriptional level and demonstrate an essential role of HuR in autophagosome formation. Although we did not measure autophagy activity in HCC, further studies may provide the additional significance of HuR in autophagy dysregulation leading to cancer development.
MATERIALS AND METHODS
Cell culture, transfection of plasmids, and small interfering RNAs.
Human liver cells, including Hep3B (hepatocellular carcinoma), Huh7 (hepatocellular carcinoma), L-02 (immortal hepatic cell line), and THLE-3 (immortal hepatic cell line) cells were cultured in Dulbecco’s modified essential medium (DMEM) (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin at 37°C in 5% CO2. The U2OS cell line stably expressing GFP-LC3 was generated and maintained as described previously (79). EGFP reporters were cloned by inserting 3′ UTR fragments from the human ATG5, ATG12, and ATG16 mRNAs into pEGFP-C1 (BD Bioscience, Franklin Lakes, NJ). The tandem fluorescence-tagged LC3 (ptfLC3) plasmid was from Addgene (no. 21074). siRNAs (control siRNA [siCtrl; 5′-CCAUGACCAACUACGAUGA-3′] and HuR siRNA [siHuR; 5′-TGTGAAAGTGATCCGCGAC-3′]; Genolution Pharmaceuticals, Seoul, South Korea) or EGFP reporter plasmids were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Western blot analysis.
Whole-cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, and 1× protease inhibitor cocktail (Roche, Basel, Switzerland); separated by electrophoresis in SDS-containing polyacrylamide gels; and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). Incubation with primary antibodies against ATG5 (Abcam, Cambridge, MA; ab180327), ATG12 (Santa Cruz Biotechnology, Dallas, TX; sc-68884), ATG16 (Santa Cruz Biotechnology; sc-393274), LC3 (Novus, St. Charles, MO; nb600-1384), HuR (Santa Cruz Biotechnology; sc-5261), EGFP (Santa Cruz Biotechnology; sc-9996), or β-actin (Abcam; ab3280) was followed by incubation with the appropriate secondary antibodies conjugated with horseradish peroxidase (HRP) (Santa Cruz Biotechnology) and detection using enhanced luminescence (Bio-Rad, Hercules, CA). Additional antibody information is provided in Table 3. Images were analyzed with ImageJ software (80).
TABLE 3.
Antibody information
| Purpose and antibody | Manufacturer | Product no. | Dilution factor |
|---|---|---|---|
| For WB | |||
| ATG5 | Abcam | ab180327 | 1:1,000 |
| ATG12 | Santa Cruz Biotechnology | sc-68884 | 1:200 |
| ATG16 | Santa Cruz Biotechnology | sc-393274 | 1:100 |
| LC3 | Novus | nb600-1384 | 1:1,000 |
| HuR | Santa Cruz Biotechnology | sc-5261 | 1:1,000 |
| GFP | Santa Cruz Biotechnology | sc-9996 | 1:1,000 |
| β-Actin | Abcam | ab3280 | 1:10,000 |
| For IHC | |||
| ATG5 | Abcam | ab180327 | 1:100 |
| ATG12 | Santa Cruz Biotechnology | sc-68884 | 1:500 |
| ATG16 | Abgent | #AP1817b | 1:200 |
| HuR | Santa Cruz Biotechnology | sc-5261 | 1:100 |
RT-qPCR.
Total RNA was prepared from whole cells using TRIzol (Invitrogen), according to the manufacturer’s instructions. After RT using random hexamers and reverse transcriptase (Toyobo, Osaka, Japan), the abundance of transcripts was assessed by qPCR analysis using SYBR green PCR master mix (Kapa Biosystems, Wilmington, MA) and gene-specific primer sets (Table 1). RT-qPCR analysis was performed on a StepOne Plus system (Life Technologies, Carlsbad, CA).
RIP.
RIP analysis was performed using anti-HuR primary antibodies or control IgG (Santa Cruz Biotechnology) (35, 81). In brief, Hep3B cell lysates were incubated with anti-HuR or control IgG, and RNP complexes were precipitated using protein A-agarose beads for 2 h. After further incubation of RNP complexes with DNase I and proteinase K, RNAs were isolated from the complexes and analyzed by RT-qPCR using the primers listed in Table 1. Relative levels of HuR binding to ATG mRNAs in RIP products were normalized with GAPDH mRNA compared to normal IgG. RPL6 mRNA was used as a negative control as described previously (31).
Biotin pulldown analysis.
To synthesize biotinylated transcripts, PCR fragments were prepared using forward primers that contained the T7 RNA polymerase promoter sequence (T7; 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′). Table 1 lists the primers used to prepare biotinylated transcripts spanning the ATG5 (NM_004849.4), ATG12 (NM_004707.4), and ATG16 (NM_030803.7) mRNAs. After purification of the PCR products, biotinylated transcripts were synthesized using a MaxiScript T7 kit (Ambion, Waltham, MA) and biotin-CTP (Enzo Life Sciences, Farmingdale, NY). Whole-cell lysates (200 μg per sample) were incubated with 1 μg of purified biotinylated transcripts for 30 min at room temperature, and then complexes were isolated using streptavidin-coupled Dynabeads (Invitrogen). Proteins present in the pulldown material were studied by Western blot analysis as described previously (81).
Nascent protein synthesis analysis.
The quantities of nascent ATG5, ATG12, and ATG16 proteins were analyzed using a Click-iT system (Invitrogen) according to the manufacturer’s instructions. In brief, after knockdown of HuR expression using siHuR, cells preincubated (for 1 h) in methionine-free RPMI medium were labeled with the azide-containing methionine analog AHA for 4 h. Newly synthesized and AHA-incorporating proteins in cell lysates (500 μg) were cross-linked to alkyne-derivatized biotin by copper-catalyzed cycloaddition (Click-iT reaction). Biotin-conjugated proteins were isolaed using streptavidin beads and further analyzed by Western blotting.
Polysome analysis.
Forty-eight hours after transfection, Hep3B cells were preincubated with cycloheximide (100 μg/ml) for 15 min and lysed with polysome extraction buffer containing 50 mM MOPS (morpholinepropanesulfonic acid), 15 mM MgCl2, 150 mM NaCl, 100 μg/ml cycloheximide, 0.5% Triton X-100, 1 mg/ml heparin, 1× protease inhibitor cocktail, and RNase inhibitor, followed by centrifugation at 13,000 × g for 10 min. The lysates were further fractionated by ultracentrifugation through 10 to 50% linear sucrose gradients, as described previously (36, 81). RNAs from each fraction were isolated, and cDNA was synthesized as described above. Relative levels of ATG5, ATG12, ATG16, and β-actin mRNAs were analyzed by RT-qPCR using specific primers listed in Table 1.
Electron microscopy.
For transmission electron microscopy (TEM) observation, cells were fixed with 1% osmium tetroxide and embedded in Epon 812. Ultrathin sections were observed with a transmission electron microscope (JEM 1010; JEOL, Japan). Autophagic vacuole size was measured using ImageJ software (n = 70).
TMA and IHC.
Paraffin-embedded HCC-related TMA slides, including HCC cores (n = 35), paired metastatic tumor tissue cores from HCC (n = 5), unpaired metastatic tumor tissue cores from HCC (n = 3), and paired normal liver tissue cores (n = 7) from human donors, were purchased from Super Bio Chips (Seoul, South Korea). The slides were deparaffinized, rehydrated, and incubated with EDTA buffer (1 mM; pH 8.0) at 95°C for antigen retrieval. All steps of IHC staining were performed according to the manufacturer’s instructions (Immune Bio Science Corp., Mukilteo, WA). After blocking with blocking solution, the slides were incubated with anti-HuR (Santa Cruz Biotechnology; sc-5261), ATG5 (Abcam; ab108327), ATG12 (Santa Cruz Biotechnology; sc-68884), or ATG16 (Abgent, San Diego, CA; AP1817b) antibodies, respectively, at 4°C overnight and further incubated with a horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. DAB was applied for color development. Antibody information is provided in Table 3. The strength of positivity was analyzed with a panoramic MIDI slide scanner system (3D Histech Ltd., Budapest, Hungary). The stain intensity score of each core was calculated using ImageJ and IHC profiler programs. This study was approved by the Institutional Review Board (IRB) at the Catholic University of Korea, College of Medicine (IRB approval number MC17SESI0110) and carried out in accordance with the Declaration of Helsinki.
Statistical analysis.
Data are expressed as means and standard deviations (SD) of three independent experiments. The statistical significance of the data was analyzed by Student’s t test using GraphPad Prism 5.0 software. Correlation data were analyzed by the Pearson correlation method using GraphPad Prism 5.0 software.
ACKNOWLEDGMENTS
This work is supported by National Research Foundation of Korea (NRF) grants funded by the government of Korea (2017R1A2B2009381 and 2012R1A5A2047939).
REFERENCES
- 1.Glick D, Barth S, Macleod KF. 2010. Autophagy: cellular and molecular mechanisms. J Pathol 221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadwell K, Stappenbeck TS, Virgin HW. 2009. Role of autophagy and autophagy genes in inflammatory bowel disease. Curr Top Microbiol Immunol 335:141–167. doi: 10.1007/978-3-642-00302-8_7. [DOI] [PubMed] [Google Scholar]
- 3.Murrow L, Debnath J. 2013. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol 8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. 1998. A protein conjugation system essential for autophagy. Nature 395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N, Noda T, Ohsumi Y. 1999. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J 18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. 2008. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19:2092–2100. doi: 10.1091/mbc.e07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 9.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malhotra R, Warne JP, Salas E, Xu AW, Debnath J. 2015. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy 11:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 14.White E. 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maes H, Rubio N, Garg AD, Agostinis P. 2013. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med 19:428–446. doi: 10.1016/j.molmed.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Hu Q, Shen HM. 2016. Pharmacological inhibitors of autophagy as novel cancer therapeutic agents. Pharmacol Res 105:164–175. doi: 10.1016/j.phrs.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 17.Lei Y, Zhang D, Yu J, Dong H, Zhang J, Yang S. 2017. Targeting autophagy in cancer stem cells as an anticancer therapy. Cancer Lett 393:33–39. doi: 10.1016/j.canlet.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC. 2016. Mammalian autophagy: how does it work?. Annu Rev Biochem 85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 19.Frankel LB, Lund AH. 2012. MicroRNA regulation of autophagy. Carcinogenesis 33:2018–2025. doi: 10.1093/carcin/bgs266. [DOI] [PubMed] [Google Scholar]
- 20.Zhai H, Fesler A, Ju J. 2013. MicroRNA: a third dimension in autophagy. Cell Cycle 12:246–250. doi: 10.4161/cc.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. 2017. Autophagy-regulating microRNAs and cancer. Front Oncol 7:65. doi: 10.3389/fonc.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinman MN, Lou H. 2008. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdelmohsen K, Gorospe M. 2010. Posttranscriptional regulation of cancer traits by HuR. WIREs RNA 1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srikantan S, Gorospe M. 2012. HuR function in disease. Front Biosci 17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. 2003. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Wang B, Bi J, Zhang C. 2011. Cytoplasmic HuR expression correlates with angiogenesis, lymphangiogenesis, and poor outcome in lung cancer. Med Oncol 28:577. doi: 10.1007/s12032-010-9734-6. [DOI] [PubMed] [Google Scholar]
- 27.Miyata Y, Watanabe S, Sagara Y, Mitsunari K, Matsuo T, Ohba K, Sakai H. 2013. High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS One 8:e59095. doi: 10.1371/journal.pone.0059095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotta-Loizou I, Vasilopoulos SN, Coutts RH, Theocharis S. 2016. Current evidence and future perspectives on HuR and breast cancer development, prognosis, and treatment. Neoplasia 18:674–688. doi: 10.1016/j.neo.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimbo M, Blanco FF, Huang YH, Telonis AG, Screnci BA, Cosma GL, Alexeev V, Gonye GE, Yeo CJ, Sawicki JA, Winter JM, Brody JR. 2015. Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget 6:27312–27331. doi: 10.18632/oncotarget.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, Padhye S, Xu L, Weir SJ, Anant S, Meisner-Kober N, Brody JR, Dixon DA. 2016. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget 7:74043–74058. doi: 10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, Rilla K, Akhtar S, Provenzani A, D’Agostino VG, Govoni S, Pascale A, Agostini H, Petrovski G, Salminen A, Kaarniranta K. 2013. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One 8:e69563. doi: 10.1371/journal.pone.0069563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchesi N, Thongon N, Pascale A, Provenzani A, Koskela A, Korhonen E, Smedowski A, Govoni S, Kauppinen A, Kaarniranta K, Amadio M. 2018. Autophagy stimulus promotes early HuR protein activation and p62/SQSTM1 protein synthesis in ARPE-19 cells by triggering Erk1/2, p38(MAPK), and JNK kinase pathways. Oxid Med Cell Longev 2018:4956080. doi: 10.1155/2018/4956080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura S, Noda T, Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 34.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. 2011. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Kim C, Kim W, Lee H, Ji E, Choe YJ, Martindale JL, Akamatsu W, Okano H, Kim HS, Nam SW, Gorospe M, Lee EK. 2014. The RNA-binding protein HuD regulates autophagosome formation in pancreatic beta cells by promoting autophagy-related gene 5 expression. J Biol Chem 289:112–121. doi: 10.1074/jbc.M113.474700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KM, Cho H, Choi K, Kim J, Kim BW, Ko YG, Jang SK, Kim YK. 2009. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev 23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukaka MM. 2012. Statistics corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med J 24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 38.Palanisamy K, Tsai TH, Yu TM, Sun KT, Yu SH, Lin FY, Wang IK, Li CY. 2018. RNA-binding protein, human antigen R regulates hypoxia-induced autophagy by targeting ATG7/ATG16L1 expressions and autophagosome formation. J Cell Physiol doi: 10.1002/jcp.27502. [DOI] [PubMed] [Google Scholar]
- 39.Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 40.Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha TK, Byun DS, Chae KS, Lee BH, Chun HS, Lee KY, Kim HJ, Chi SG. 2008. NF-kappaB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis. Gastroenterology 135:2030–2042. doi: 10.1053/j.gastro.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, Gorospe M. 2009. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J 28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucchesi C, Sheikh MS, Huang Y. 2016. Negative regulation of RNA-binding protein HuR by tumor-suppressor ECRG2. Oncogene 35:2565–2573. doi: 10.1038/onc.2015.339. [DOI] [PubMed] [Google Scholar]
- 43.Cui Y, Bi M, Su T, Liu H, Lu SH. 2010. Molecular cloning and characterization of a novel esophageal cancer related gene. Int J Oncol 37:1521–1528. [DOI] [PubMed] [Google Scholar]
- 44.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. 2008. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci U S A 105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu F, Zhang X, Lei Y, Liu X, Liu Z, Tong T, Wang W. 2010. Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J Cell Biochem 111:727–734. doi: 10.1002/jcb.22762. [DOI] [PubMed] [Google Scholar]
- 46.Roff AN, Craig TJ, August A, Stellato C, Ishmael FT. 2014. MicroRNA-570-3p regulates HuR and cytokine expression in airway epithelial cells. Am J Clin Exp Immunol 3:68–83. [PMC free article] [PubMed] [Google Scholar]
- 47.Guo J, Li M, Meng X, Sui J, Dou L, Tang W, Huang X, Man Y, Wang S, Li J. 2014. MiR-291b-3p induces apoptosis in liver cell line NCTC1469 by reducing the level of RNA-binding protein HuR. Cell Physiol Biochem 33:810–822. doi: 10.1159/000358654. [DOI] [PubMed] [Google Scholar]
- 48.Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, Cabo R, Gorospe M. 2010. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 9:1354–1359. doi: 10.4161/cc.9.7.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D'Angelo D, Palmieri D, Mussnich P, Roche M, Wierinckx A, Raverot G, Fedele M, Croce CM, Trouillas J, Fusco A. 2012. Altered microRNA expression profile in human pituitary GH adenomas: down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J Clin Endocrinol Metab 97:E1128–E1138. doi: 10.1210/jc.2011-3482. [DOI] [PubMed] [Google Scholar]
- 51.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 53.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. 2010. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang Y, Lin J, Tsung A. 2012. Manipulation of autophagy by MIR375 generates antitumor effects in liver cancer. Autophagy 8:1833–1834. doi: 10.4161/auto.21796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, Liao JZ, He XX, Li PY. 2017. The role of autophagy in hepatocellular carcinoma: friend or foe. Oncotarget 8:57707–57722. doi: 10.18632/oncotarget.17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun K, Guo XL, Zhao QD, Jing YY, Kou XR, Xie XQ, Zhou Y, Cai N, Gao L, Zhao X, Zhang SS, Song JR, Li D, Deng WJ, Li R, Wu MC, Wei LX. 2013. Paradoxical role of autophagy in the dysplastic and tumor-forming stages of hepatocarcinoma development in rats. Cell Death Dis 4:e501. doi: 10.1038/cddis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, Ou JH. 2015. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ 22:1025–1034. doi: 10.1038/cdd.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J, SHARP Investigators Study Group. 2008. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 59.Yuan H, Li AJ, Ma SL, Cui LJ, Wu B, Yin L, Wu MC. 2014. Inhibition of autophagy significantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J Gastroenterol 20:4953–4962. doi: 10.3748/wjg.v20.i17.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dash S, Chava S, Chandra PK, Aydin Y, Balart LA, Wu T. 2016. Autophagy in hepatocellular carcinomas: from pathophysiology to therapeutic response. Hepat Med 8:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kessler SM, Laggai S, Barghash A, Schultheiss CS, Lederer E, Artl M, Helms V, Haybaeck J, Kiemer AK. 2015. IMP2/p62 induces genomic instability and an aggressive hepatocellular carcinoma phenotype. Cell Death Dis 6:e1894. doi: 10.1038/cddis.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. 2015. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11:28–45. doi: 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J. 2015. Regulation of autophagy by protein post-translational modification. Lab Invest 95:14–25. doi: 10.1038/labinvest.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Botti-Millet J, Nascimbeni AC, Dupont N, Morel E, Codogno P. 2016. Fine-tuning autophagy: from transcriptional to posttranslational regulation. Am J Physiol Cell Physiol 311:C351. doi: 10.1152/ajpcell.00129.2016. [DOI] [PubMed] [Google Scholar]
- 66.Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. 2013. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol 23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 67.Fullgrabe J, Klionsky DJ, Joseph B. 2014. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 68.Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M. 2015. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 11:867–880. doi: 10.1080/15548627.2015.1034410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang L, Wang H, Shen Q, Feng L, Jin H. 2017. Long non-coding RNAs involved in autophagy regulation. Cell Death Dis 8:e3073. doi: 10.1038/cddis.2017.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu G, McQuiston T, Bernard A, Park YD, Qiu J, Vural A, Zhang N, Waterman SR, Blewett NH, Myers TG, Maraia RJ, Kehrl JH, Uzel G, Klionsky DJ, Williamson PR. 2015. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol 17:930–942. doi: 10.1038/ncb3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bose JK, Huang CC, Shen CK. 2011. Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem 286:44441–44448. doi: 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sánchez-Jiménez C, Izquierdo JM. 2013. T-cell intracellular antigen (TIA)-proteins deficiency in murine embryonic fibroblasts alters cell cycle progression and induces autophagy. PLoS One 8:e75127. doi: 10.1371/journal.pone.0075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan C, Chen J, Chen N. 2016. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep 6:22640. doi: 10.1038/srep22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen W, Zhang L, Zhou ZQ, Ren YQ, Sun LN, Man YL, Ma ZW, Wang ZK. 2018. Effects of long non-coding RNA LINC00963 on renal interstitial fibrosis and oxidative stress of rats with chronic renal failure via the Foxo signaling pathway. Cell Physiol Biochem 46:815–828. doi: 10.1159/000488739. [DOI] [PubMed] [Google Scholar]
- 75.Carrascoso I, Alcalde J, Sanchez-Jimenez C, Gonzalez-Sanchez P, Izquierdo JM. 2017. T-cell intracellular antigens and Hu antigen R antagonistically modulate mitochondrial activity and dynamics by regulating optic atrophy 1 gene expression. Mol Cell Biol 37:e00174-17. doi: 10.1128/MCB.00174-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Y, Guerrero-Preston R, Ratovitski EA. 2012. Phospho-DeltaNp63alpha-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle 11:1247–1259. doi: 10.4161/cc.11.6.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Zhou D, Ren Y, Zhang Z, Guo X, Ma M, Xue Z, Lv J, Liu H, Xi Q, Jia L, Zhang L, Liu Y, Zhang Q, Yan J, Da Y, Gao F, Yue J, Yao Z, Zhang R. 2018. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy 14:1–15. doi: 10.1080/15548627.2018.1522467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang H, Tang J, Zhang L, Bu Y, Zhang X. 2018. miR-874 regulates multiple-drug resistance in gastric cancer by targeting ATG16L1. Int J Oncol 53:2769–2779. doi: 10.3892/ijo.2018.4593. [DOI] [PubMed] [Google Scholar]
- 79.Choe YJ, Ha TJ, Ko KW, Lee SY, Shin SJ, Kim HS. 2012. Anthocyanins in the black soybean (Glycine max L.) protect U2OS cells from apoptosis by inducing autophagy via the activation of adenosyl monophosphate-dependent protein kinase. Oncol Rep 28:2049–2056. doi: 10.3892/or.2012.2034. [DOI] [PubMed] [Google Scholar]
- 80.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee EK, Kim W, Tominaga K, Martindale JL, Yang X, Subaran SS, Carlson OD, Mercken EM, Kulkarni RN, Akamatsu W, Okano H, Perrone-Bizzozero NI, de Cabo R, Egan JM, Gorospe M. 2012. RNA-binding protein HuD controls insulin translation. Mol Cell 45:826–835. doi: 10.1016/j.molcel.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]