MicroRNA 155 (miR-155) is an oncomir, generated as a noncoding RNA from the BIC gene whose promoter activity is mainly controlled via activation protein 1 (AP-1) and NF-κB transcription factors. We found that the expression levels of miR-155 and programmed cell death 4 (Pdcd4) exhibit inverse relationships in tongue cancer cells (SAS and AWL) and tumor tissues compared to their relationships in normal FBM cells and normal tongue tissues, respectively.
KEYWORDS: AP-1, BIC, NF-κB, Pdcd4, apoptosis, miR-155, miR-155 sponge, tumor xenografts
ABSTRACT
MicroRNA 155 (miR-155) is an oncomir, generated as a noncoding RNA from the BIC gene whose promoter activity is mainly controlled via activation protein 1 (AP-1) and NF-κB transcription factors. We found that the expression levels of miR-155 and programmed cell death 4 (Pdcd4) exhibit inverse relationships in tongue cancer cells (SAS and AWL) and tumor tissues compared to their relationships in normal FBM cells and normal tongue tissues, respectively. In silico and in vitro studies with the 3′ untranslated region (UTR) of Pdcd4 via luciferase reporter assays, quantitative PCR (qPCR), and Western blotting showed that miR-155 directly targets Pdcd4 mRNA and blocks its expression. Ectopic expression of Pdcd4 or knockdown of miR-155 in tongue cancer cells predominantly reduces AP-1-dependent transcriptional activity of the BIC promoter and decreases miR-155 expression. In this study, we demonstrate that miR-155 expression is modulated by a feedback loop between Pdcd4, AP-1, and miR-155 which results in enhanced expression of miR-155 with a consequent progression of tongue tumorigenesis. Further, miR-155 knockdown increases apoptosis, arrests the cell cycle, regresses tumor size in xenograft nude mice, and reduces cell viability and colony formation in soft-agar and clonogenic assays. Thus, the restoration of Pdcd4 levels by the use of molecular manipulation such as using a miR-155 sponge has an essential role in the therapeutic intervention of cancers, including tongue cancer.
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs of 18 to 25 nucleotides in length involved in posttranscriptional gene regulation, mostly by binding to the 3′ untranslated region (UTR) of specific target messenger RNAs (mRNAs), causing mRNA degradation or translational repression (1). A single miRNA can regulate numerous target mRNAs, and, conversely, a single mRNA can be targeted by several miRNAs. miRNAs have a potential role to play in tumor development and sustenance (oncomirs) by downregulating tumor suppressor genes (2), but they can also act as tumor suppressors in a highly tissue-specific manner (3). The downregulation of tumor suppressor genes is essential for continuous proliferation and survival of cancer cells. One such essential tumor suppressor protein is programmed cell death 4 (Pdcd4), which is downregulated in various cancers of oral (4), breast (5), lung (6), liver (7), brain (8), and colon (9) tissues. Pdcd4 plays a vital role in regulating apoptosis, invasion, and metastasis (10–12) and is known to inhibit activation protein 1 (AP-1)-dependent transcription (13, 14). MicroRNA 155 (miR-155)-mediated downregulation of Pdcd4 was suggested earlier (15), and a recent study in lung cancer has claimed that miR-155 directly targets Pdcd4 and downregulates its expression in lung cancer cells (16) but did not provide convincing evidence for Pdcd4 3′ UTR-mediated downregulation as a mechanism. miR-155 was found to be overexpressed in oral (17), breast (18), tongue (19), pancreatic (20), prostatic (21), and thyroid cancers (22). miR-155 overexpression has also been implicated in enhanced cell proliferation, invasion, and metastasis (23) by downregulating the expression of tissue-specific target genes such as the APC (24), CDC73 (25), DET1 (26), FOXO3a (27), SMAD2 (28), and TP53INP1 (20) genes in many cancers. miR-155 is generated as a noncoding mRNA from the B-cell integration cluster (BIC) gene located on chromosome 21 whose promoter is mainly controlled via AP-1 (36 bp upstream of the start site) and NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells; 1,150 bp upstream from the start site) transcription factors present upstream of the core promoter region (29). AP-1, formed by the heterodimerization between members of the JUN and FOS protein families, is a critical regulator of cell proliferation, apoptosis, tumor invasiveness, angiogenesis, and other multiple hallmarks of cancer (30–32).
In this study, we demonstrate that miR-155 is overexpressed in tongue cancer cells (SAS and AWL) and tongue tumor tissues compared to expression in normal FBM cells (a fetal buccal mucosal cell line) and normal tongue tissues, respectively. We hypothesized that miR-155-mediated targeting of the 3′ UTR of Pdcd4 mRNA results in the downregulation of Pdcd4 in tongue cancer and that this might account for the increased activation of AP-1-dependent transcription of the BIC promoter, which results in overexpression of miR-155. Here, we demonstrate for the first time that miR-155 directly targets Pdcd4 in tongue cancer cells and indirectly activates AP-1-dependent transcription of the BIC gene and, thus, promotes overexpression of miR-155 by an miR-155/Pdcd4/AP-1 positive feedback loop in tongue cancer.
RESULTS
The miR-155 expression level shows an inverse relation to that of its predicted target, Pdcd4, in tongue cancer cells and tissues.
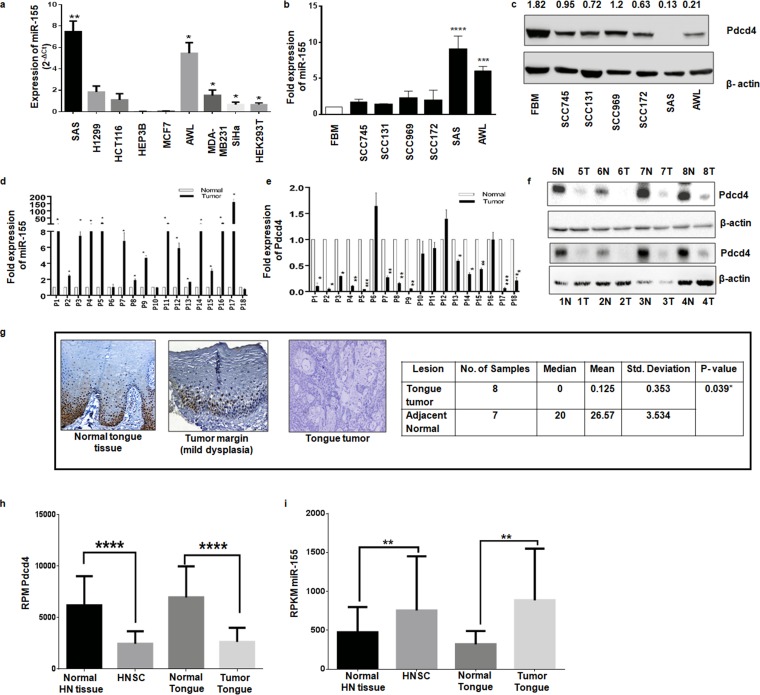
A panel of cancer cell lines of different tissue origins was screened for the endogenous expression of miR-155, and the results showed that miR-155 expression is significantly higher in tongue cancer cells (SAS and AWL) than in others tested (Fig. 1a). Next, we checked its expression in a panel of head and neck carcinoma cells and an immortalized fetal buccal mucosal cell line (FBM). We found that miR-155 expression was less in FBM cells and relatively high in head and neck cancer cells, with the highest levels being noticed in tongue cancer cells (SAS and AWL cells) (Fig. 1b), and, interestingly, that Pdcd4 is poorly expressed in tongue cancer cells (SAS and AWL cells) (Fig. 1c). In silico target prediction software predicted Pdcd4 as a target of miR-155, supported by the presence of an miR-155 seed sequence match (100%) from nucleotides 1774 to 1783 in the 3′ UTR of Pdcd4 with free energy (ΔG) of −21.4 kcal/mol (see Fig. S1B in the supplemental material). All of these results indicate that SAS and AWL cells show an inverse relationship for the expression levels of miR-155 and Pdcd4 (Fig. 1b and c), as often seen in miRNA target pairs in cancer cells (33).
FIG 1.
miR-155 and Pdcd4 expression levels in cell lines and tongue tumors. (a) Endogenous expression of miR-155 in cancer cell lines of different tissue origins from three different passages calculated by the 2-ΔCT method (statistical analysis was performed by column statistics with a one-sample t test; n = 3). (b) Expression of miR-155 in FBM and different head and neck carcinoma cells normalized to that of U6 (n = 3). (c) Western blots for Pdcd4 and β-actin (as an internal control) in FBM and different head and neck carcinoma cells. (d) Expression of miR-155 normalized to that of U6 in normal and tumor tongue tissues (n = 3 as experimental triplicates). (e) Expression of Pdcd4 mRNA normalized to that of β-actin in normal and tumor tongue tissues (n = 3 as experimental triplicates). (f) Western blots for Pdcd4 and β-actin in tongue cancer tissue samples with adjacent normal tissues from patients 1 to 8. N, normal; T, tumor. (g) Representative immunohistochemistry images for Pdcd4 in normal tongue tissue, tumor margin (mild dysplasia), and tongue cancer tissue at ×20 magnification. (h) TCGA data were analyzed for the expression of Pdcd4 in head and neck and tongue cancers and compared with values in normal tissues. (i) TCGA data were analyzed for the expression of miR-155 in head and neck and tongue cancers and compared with values in normal tissues. Values are expressed as the means ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Since miR-155 and Pdcd4 expression levels in SAS and AWL cells show an inverse relationship, it was relevant to check if there is an inverse relationship in their expression levels in tongue cancer tissues. To this end, we analyzed miR-155 and Pdcd4 levels in 18 pairs of tongue cancer patient samples (adjacent normal and tumor). miR-155 expression was found to be higher in most of the tumor tissues (except in patient 6) than in their adjacent normal tissue sections when expression was analyzed by quantitative PCR (qPCR) (Fig. 1d). As expected, Pdcd4 mRNA levels were lower in tongue cancer tissues than in their adjacent normal tissues (Fig. 1e). The protein levels of Pdcd4 were detected by Western blotting in eight patients and by immunohistochemistry (IHC) in another 18 patients, and the results showed that Pdcd4 was barely identified in tumor samples compared to its expression in normal tissue (Fig. 1f and g). To emphasize this inverse correlation between miR-155 and Pdcd4 expression, we analyzed the data collected from The Cancer Genome Atlas (TCGA) for head and neck squamous carcinoma (HNSC) in general and for tongue cancer in particular. It was found that Pdcd4 and miR-155 expression shows an inverse relation in both HNSC and tongue cancers (Fig. 1h and i). These data indicated the existence of an inverse correlation in the expression levels of miR-155 and its target, Pdcd4, and prompted us to experimentally validate for the first time if Pdcd4 is a target of miR-155 in tongue cancer cells.
miR-155 negatively regulates Pdcd4 expression.
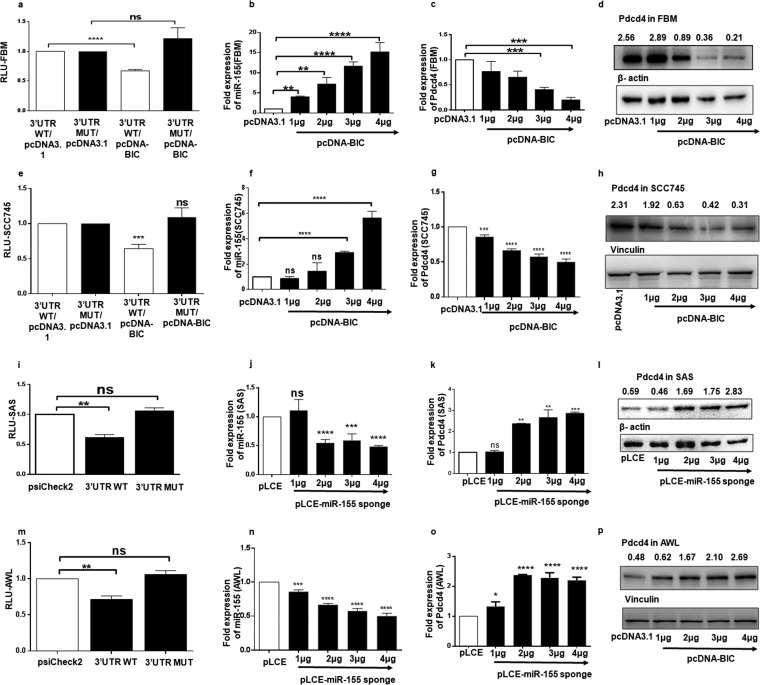
First, we cloned the wild-type (WT) 3′ UTR of Pdcd4 (nucleotides 1170 to 1913) and an miR-155 binding site mutant 3′ UTR (MUT) of Pdcd4 into psiCheck-2 (Fig. S2A). Since FBM and SCC745 cells express low and moderate levels of miR-155 (Fig. 1b), respectively, they were cotransfected with pcDNA-BIC expressing miR-155 and psiCheck-Pdcd4-WT or -MUT 3′ UTR. The normalized Renilla luciferase activity of the psiCheck-Pdcd4-WT 3′ UTR was significantly reduced upon ectopic expression of miR-155 in both FBM (Fig. 2a) and SCC745 cells (Fig. 2e), but there was not much change in that of psiCheck-Pdcd4-MUT 3′ UTR (Fig. 2a and e). Further, FBM and SCC745 cells were transfected with increasing amounts of pcDNA-BIC or pcDNA3.1(+) (1 μg to 4 μg) as a control, and a gradual increase in the expression levels of miR-155 over the control level was noticed using qPCR analysis (Fig. 2b and f). When cells were analyzed for the expression of Pdcd4 mRNA, there was a gradual decrease in the levels of Pdcd4 mRNA in FBM (Fig. 2c) and SCC745 (Fig. 2g) cells. The protein levels of Pdcd4 showed a gradual decrease with increases in the expression of miR-155 in FBM (Fig. 2d) and SCC745 (Fig. 2h) cells. These results suggest that overexpression of miR-155 has the potential to target Pdcd4 and downregulate its expression in FBM and SCC745 cells.
FIG 2.
Targeting of 3′ UTR of Pdcd4 by miR-155 and the consequent changes in the expression of Pdcd4 in FBM, SAS, and AWL cells. (a and e) A dual-luciferase reporter assay was performed in FBM and SCC745 cells by cotransfecting them with the Pdcd4WT or MUT 3 UTR (100 ng) and pcDNA3.1 or pcDNA-BIC. These graphs represent normalized values (relative light units [RLU]) of Renilla/firefly luciferase activities (n = 3). (b and f) Expression of miR-155 normalized to that of U6 in FBM and SCC745 cells transfected with different amounts (1 μg to 4 μg) of pcDNA-BIC with pcDNA3.1 as a control (n = 3). (c and g) Expression of Pdcd4 mRNA in FBM and SCC745 cells transfected with different amounts (1 μg to 4 μg) of pcDNA-BIC with pcDNA3.1 as a control and normalized to that of β-actin mRNA (n = 3). (d and h) Western blots for Pdcd4 and β-actin in FBM cells and for Pdcd4 and vinculin in SCC745 cells transfected with different amounts (1 μg to 4 μg) of pcDNA-BIC, with pcDNA3.1 as a control. (i and m) Dual-luciferase reporter assay performed in SAS and AWL cells transfected with 100 ng of psiCheck-2, psiCheck-Pdcd4-WT 3′ UTR or psiCheck-Pdcd4-MUT 3′ UTR (n = 3). (j and n) Expression of miR-155 in SAS and AWL cells transfected with pLCE–miR-155 sponge plasmid (1 to 4 μg), with 4 μg of pLCE as a control (n = 3). (k and o) Expression of Pdcd4 mRNA in SAS and AWL cells transfected with pLCE–miR-155 sponge plasmid (1 to 4 μg) with 4 μg of pLCE as a control, normalized to that of β-actin (n = 3). (l and p) Western blots for Pdcd4 in SAS and AWL cells when transfected with 1 μg to 4 μg of pLCE–miR-155 sponge plasmid, with 4 μg of pLCE as a control; β-actin and vinculin were taken as internal controls, respectively. Values are expressed as the means ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant).
As SAS and AWL cells express high levels of miR-155 (Fig. 1a and b), they were transfected with psiCheck-2 and psiCheck-Pdcd4-WT or -MUT 3′ UTR; as expected, the Renilla luciferase activity showed a significant decrease with psiCheck-Pdcd4-WT 3′ UTR but not with the other two constructs (Fig. 2i and m). Next, we decreased the expression of miR-155 in SAS and AWL cells using miR-155 sponge constructs containing nine tandem repeats of miR-155 binding sites. Ectopic expression of pLCE–miR-155 sponge (1 to 4 μg) with pLCE as a control resulted in a decrease in endogenous levels of miR-155 (Fig. 2l and n), and Pdcd4 expressions increased at both the mRNA (Fig. 2k and o) and protein (Fig. 2h and p) levels. These results show that miR-155 regulates Pdcd4 expression by binding to its 3′ UTR in FBM, SCC745, SAS, and AWL cell lines.
The feedback loop between miR-155, Pdcd4, and AP-1 provides a mechanistic basis for the overexpression of miR-155 in tongue cancer.
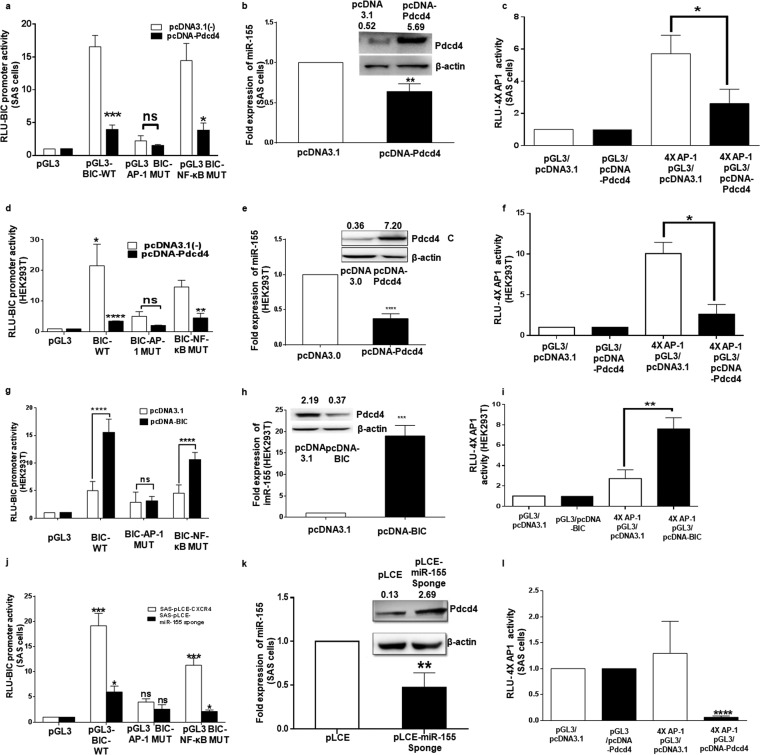
To understand whether BIC promoter regulation involves miR-155, Pdcd4, and AP-1, SAS and HEK293T cells were transfected with pGL-3 BIC promoter and pcDNA3.1(−) or pcDNA-Pdcd4. The luciferase activity of pGL3-BIC promoter was markedly reduced when it was cotransfected with pcDNA-Pdcd4 compared to the level with pcDNA3.1(−) (Fig. 3a and d). To validate the involvement of AP-1 or NF-κB transcription factors in BIC gene promoter regulation in these cells, we used promoter constructs mutated at the AP-1 or NF-κB binding sites together with pcDNA3.1(−) and pcDNA-Pdcd4. We observed that mutation of the conserved AP-1 binding site reduced BIC promoter activity, whereas mutation at the NF-κB binding site had less effect in both cell lines transfected with pcDNA3.1(−) (Fig. 3a and d). However, in pcDNA-Pdcd4 transfected cells, the effect was more significant in reducing the BIC promoter activity in the case of the NF-κB mutant (with WT AP-1) but not in the AP-1 mutant (with WT NF-κB) (Fig. 3a and d). Therefore, we conclude that Pdcd4 regulates BIC promoter primarily by AP-1-driven transcription in SAS and HEK293T cells (Fig. 3a) and SAS cells (Fig. 3d).
FIG 3.
The feedback loop between miR-155, Pdcd4, and AP-1 in SAS cells. (a and d) Dual-luciferase reporter activity of wild-type BIC promoter and BIC promoter mutated at the AP-1 or NF-κB binding site in the presence of pcDNA3.1(−) or pcDNA-Pdcd4 in SAS and HEK293T cells (n = 3). (b and e) Expression of miR-155 normalized to that of U6 upon overexpression of Pdcd4 in SAS and HEK293T cells (n = 3). Insets show Western blots of Pdcd4 and β-actin in SAS and HEK293T cells in the presence of pcDNA3.1(−) or pcDNA-Pdcd4. (c and f) AP-1–luciferase activity detected by 4× AP-1 binding sites cloned upstream of a pGL3-luciferase plasmid in the presence of pcDNA-Pdcd4 and pcDNA3.1(−) in SAS and HEK293T cells (n = 3). (g) Dual-luciferase reporter activity of wild-type BIC promoter and BIC promoter mutated at the AP-1 or NF-κB binding site in the presence of pcDNA3.1(−) or pcDNA-BIC in HEK293T cells (n = 3). (h) Fold expression of miR-155 upon ectopic expression of miR-155 in HEK293T cells (n = 3). The inset shows a Western blot for Pdcd4 and β-actin in HEK293T cells in the presence of pcDNA3.1 and pcDNA-BIC. (i) AP-1–luciferase activity detected by 4× AP-1 binding sites cloned upstream of the pGL3-luciferase plasmid in the presence of pcDNA-BIC and pcDNA3.1(+) in HEK293T cells (n = 3). (j) Luciferase activity of wild-type BIC promoter and BIC promoter separately mutated at the AP-1 or NF-κB binding site in the presence of pLCE and pLCE–miR-155 sponge in SAS cells (n = 3). (k) Expression of miR-155 normalized to that of U6 upon ectopic expression of pLCE and pLCE–miR-155 sponge in SAS cells (n = 3). The inset shows a Western blot for expression of Pdcd4 and β-actin in SAS cells upon ectopic expression of pLCE and pLCE–miR-155 sponge. (l) AP-1 activity detected by a 4× AP-1–luciferase construct in the presence of pLCE and pLCE–miR-155 sponge in SAS cells. Values are expressed as the means ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant).
Upon ectopic expression of Pdcd4 (Fig. 3b, inset) miR-155 levels decreased in SAS and HEK293T cells (Fig. 3b and e). To validate the regulation of AP-1 transcription by Pdcd4, we cotransfected SAS cells and HEK293T with pGL3 basic or a luciferase reporter construct containing four AP-1 binding sites (4× AP-1–luciferase) with pcDNA-Pdcd4 or pcDNA3.1(−), and this decreased the AP-1–luciferase activity over that of corresponding controls in the presence of Pdcd4 in both SAS (Fig. 3c) and HEK293T cells (Fig. 3f).
To gain further insights into the regulation of the BIC promoter by a feedback loop formed by miR-155/Pdcd4/AP-1, we cotransfected HEK293T cells with pGL3-BIC and pcDNA-BIC or pcDNA3.1(+), and we found a significant increase in luciferase activity of BIC promoter over that of the corresponding controls, while the BIC–AP-1 mutant was not much affected in the presence of miR-155 (Fig. 3g). The BIC–NF-κB mutant shows reduced activity in the presence of miR-155, indicating the involvement of the AP-1 site which is in native form in this construct (Fig. 3g). Overexpression of miR-155 was confirmed by qPCR (Fig. 3h), and Pdcd4 levels were quantified by Western blotting (inset). To validate the regulation of AP-1 transcription by miR-155, we cotransfected HEK293T cells with pGL3 basic or the 4× AP-1–luciferase construct with pcDNA-BIC or pcDNA3.1(+), and this increased the AP-1–luciferase activity over that of corresponding controls in the presence of miR-155 in both groups of HEK293T cells (Fig. 3i).
As SAS cells have higher expression of miR-155, we cotransfected them with the pGL3-BIC and pLCE or the pLCE–miR-155 sponge, and it was found that BIC promoter activity was reduced in the presence of the miR-155 sponge, which results in higher expression of Pdcd4 (Fig. 3j). The reduced expression of miR-155 expression was confirmed by miR-155-specific stem-loop qPCR (Fig. 3k). The upregulation of Pdcd4 was confirmed by Western blotting on ectopic expression of the miR-155 sponge (Fig. 3k, inset). The AP-1–luciferase activity decreased upon cotransfection of the 4× AP-1–luciferase construct with a pLCE–miR-155 sponge compared to that with pLCE (Fig. 3l). All of this experimental evidence indicates the existence of a feedback loop operating in SAS cells, which may be responsible for high expression of miR-155 in these cells.
miR-155 targets Pdcd4 whereas the miR-155 sponge restores the Pdcd4 levels inhibiting the tumorigenic properties of SAS cells.
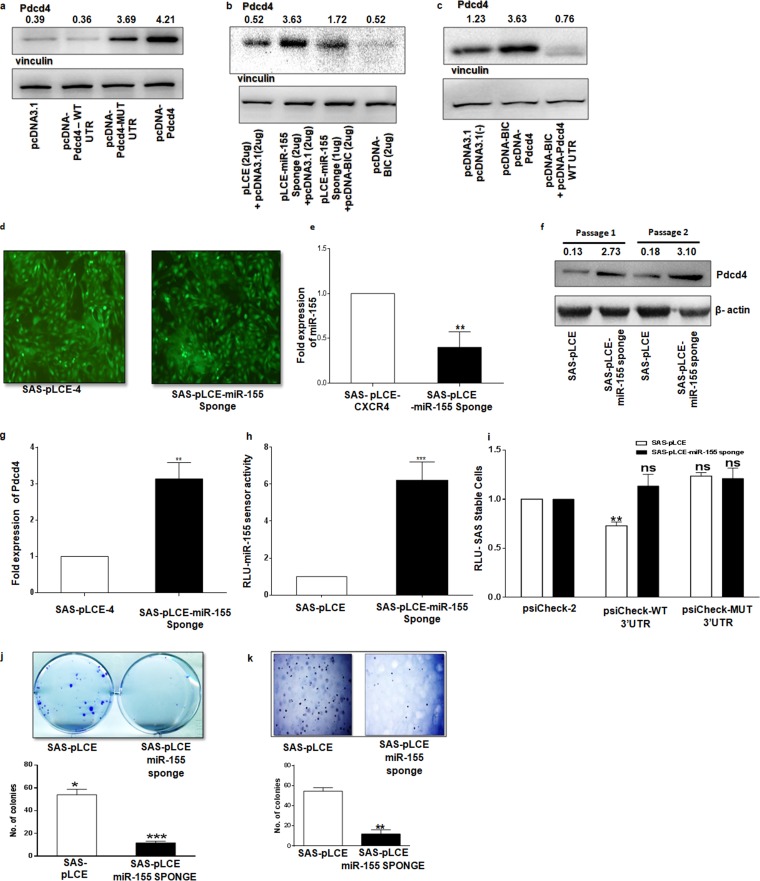
To gain more mechanistic insights and establish that miR-155 acts through Pdcd4, SAS cells were transfected with pcDNA-Pdcd4-WT 3′ UTR, pcDNA-Pdcd4-MUT 3′ UTR, and pcDNA-Pdcd4 (only the open reading frame [ORF] was cloned). It was found that pcDNA-Pdcd4-WT 3′ UTR shows downregulation compared to expression of pcDNA-Pdcd4-MUT 3′ UTR and pcDNA-Pdcd4 (Fig. 4a). A rescue experiment was performed in SAS cells in which the miR-155 sponge and pcDNA-BIC were cotransfected; the results clearly show that miR-155 inhibition with the sponge increased Pdcd4 expression and that reexpression of miR-155 decreased its levels (Fig. 4b). We also cotransfected SAS cells with pcDNA3.1 or pcDNA-BIC together with pcDNA-Pdcd4 and pcDNA-Pdcd4-WT 3′ UTR and found that overexpression of miR-155 through pcDNA-BIC together with pcDNA-Pdcd4-WT 3′ UTR downregulates Pdcd4 expression while no such effect was found with pcDNA-Pdcd4 (Fig. 4c). These results point out that miR-155 acts on Pdcd4 expression via the miR-155 binding site and that this is an essential regulatory mechanism for miR-155 expression in SAS cells.
FIG 4.
miR-155-mediated effect on Pdcd4 in SAS cells and its consequent effect on in vitro oncogenic properties. (a) Western blot for Pdcd4 and vinculin in SAS cells upon transfection with 500 ng of pcDNA3.1, pcDNA-Pdcd4-WT 3′ UTR, pcDNA-Pdcd4-MUT 3′ UTR, and pcDNA-Pdcd4 (ORF only). (b) Western blot for Pdcd4 and vinculin in SAS cells when cotransfected pLCE–miR-155 sponge/pcDNA-BIC in gradual increments, with pLCE/pcDNA3.1 as a control. (c) Western blots for Pdcd4 and vinculin in SAS cells upon cotransfection of pcDNA-Pdcd4 (ORF only)/pcDNA-BIC, pcDNA-Pdcd4-WT 3′ UTR/pcDNA-BIC and their respective controls with pcDNA3.1(+)(−). (d) SAS-pLCE and SAS–pLCE-miR-155 sponge-sorted cells were individually transduced by lentivirus expressing GFP (acting as a probe to sort the positive cells). (e) Expression of miR-155 in SAS-pLCE and SAS–pLCE–miR-155 sponge cells normalized to that of U6 (n = 3). (f) Western blot of Pdcd4 and β-actin in SAS-pLCE and SAS–pLCE–miR-155 sponge cells with two different cell passages. (g) Expression of Pdcd4 in SAS-pLCE and SAS–pLCE–miR-155 sponge cell, normalized to that of U6 (n = 3). (h) Dual-luciferase reporter assay performed with pGL-3X complementary binding sites for an miR-155/miR-155 sensor plasmid (n = 3). (i) A dual-luciferase assay was performed in SAS-pLCE and SAS–pLCE–miR-155 sponge cells transfected with 100 ng of psiCheck-2, psiCheck-Pdcd4-WT 3′ UTR or psiCheck-Pdcd4-MUT 3′ UTR (n = 3). (j) Representative images of a clonogenic assay with SAS-pLCE and SAS–pLCE–miR-155 sponge cells. The graph shows the mean number of colonies from three separate experiments (n = 3). (k) Representative images of soft-agar assay with SAS-pLCE and SAS–pLCE–miR-155 sponge cells, and the graph is a representation of a number of colonies from three separate experiments (n = 3). Values are expressed as the means ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant).
Further, we generated stable clones of SAS cells that express the miR-155 sponge using lentiviruses. The virus titer was optimized for efficient transduction, and positively transduced cells were enriched by cell sorting in a FACSAria III using green fluorescent protein (GFP) as a marker (Fig. 4d). Real-time PCR for miR-155 shows significant downregulation of miR-155 expression in SAS–pLCE–miR-155 sponge compared to the level with SAS-pLCE cells (Fig. 4e). Pdcd4 is upregulated both at translational and transcriptional levels in SAS–pLCE–miR-155 sponge cells compared to the level in SAS-pLCE cells (Fig. 4f and g). To further confirm the effectiveness of the sponge, we used an miR-155 luciferase reporter sensor system having three copies (3×) of the exact binding sequence for miR-155 and found that the luciferase activity was almost 7-fold higher in pLCE–miR-155 sponge stable cells than in SAS-pLCE cells (Fig. 4h). Furthermore, SAS-pLCE cells and SAS–pLCE–miR-155 sponge cells were transfected with psiCheck-2 or psiCheck-Pdcd4-WT or -MUT 3′ UTR. As expected, the Renilla luciferase activity showed a significant decrease with psiCheck-Pdcd4-WT in SAS-pLCE cells but not in SAS–pLCE–miR-155 sponge cells and the psiCheck-Pdcd4-MUT 3′ UTR (Fig. 4i). All of this experimental evidence clearly shows that miR-155 directly targets Pdcd4 in SAS cells and that depletion of miR-155 upregulates the expression of Pdcd4.
A clonogenic or colony formation assay is an in vitro cell survival assay designed to measure the ability of a single cell to grow into a colony (34). There was a drastic decrease in the capacity of SAS–pLCE–miR-155 sponge single cells to develop into colonies compared to that of SAS-pLCE-stable cells (Fig. 4j). Also, tumor cells have a propensity to grow in an anchorage-independent manner and hence can form colonies in a semisolid medium such as soft agar (35). The anchorage-independent growth of the stable pLCE and pLCE–miR-155 sponge cells was analyzed by a soft-agar assay in six-well tissue culture plates for 24 days. The colony formation capability on soft agar decreased in pLCE–miR-155 sponge cells compared to that in SAS-pLCE cells (Fig. 4k). These results suggest that reducing the miR-155 levels in SAS cells reduces their survival and the ability of a single cell form a colony on soft agar (34). There was a drastic decrease in the capacity of SAS–pLCE–miR-155 sponge single cells to develop into colonies compared to that of SAS-pLCE stable cells (Fig. 4j).
The miR-155 sponge inhibits cell viability, arrests the cell cycle, and induces apoptosis in SAS cells.
Since Pdcd4 is known to be upregulated upon induction of apoptosis (36), it was of interest to check the changes in cellular apoptosis, cell cycle, and cell viability. First, we monitored cell viability at different time points by MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] assay and found that SAS–pLCE–miR-155 sponge cells, but not the SAS-pLCE cells, exhibited a gradual decrease in viability with an increase in time (Fig. 5a). Next, cell cycle analysis (by propidium iodide) of SAS–pLCE–miR-155 sponge cells showed an increase in the number of cells in sub-G0 and G2/M phases of the cell cycle compared with the numbers in SAS-pLCE cells (Fig. 5b). An increased percentage of annexin V-stained cells was found upon downregulation of miR-155 in SAS cells (Fig. 5c). As Pdcd4 is among the first few proteins upregulated during the process of apoptosis, it was thus of interest to analyze essential proapoptotic proteins such as cleaved caspase 3, cleaved caspase 9, and cleaved poly(ADP-ribose) polymerase (PARP), They were found upregulated in SAS–pLCE–miR-155 sponge-transfected cells compared to levels in SAS-pLCE cells (Fig. 5d). We also analyzed the expression of Bcl2 and Bax and found that Bax, but not Bcl2, was upregulated in SAS–pLCE–miR-155 sponge cells compared to levels in SAS-pLCE cells (Fig. 5e). Taken together, the above results show cell cycle arrest at sub-G0 and G2/M phases together with an increased number of annexin V-positive cells in SAS–pLCE–miR-155 sponge cells compared to levels in SAS-pLCE cells. Also, increased expression levels of cleaved caspase 3, cleaved caspase 9, cleaved PARP, and Bax indicate that miR-155 depletion in SAS cells leads to a decrease in cell viability and the induction of cell cycle arrest and apoptosis.
FIG 5.
Effect of miR-155 inhibition on cell viability, cell cycle, and apoptosis. (a) Effect of miR-155 depletion on cell viability as measured by MTT assay (n = 3). (b) Cell cycle analysis by propidium iodide staining of SAS-pLCE and SAS–pLCE–miR-155 sponge cells. Each bar in the graph represents the percentage of cells present in a particular phase of cell cycle (n = 3). (c) Effect of miR-155 downregulation on cellular apoptosis detected by annexin V staining (n = 3). (d) Western blots for cleaved PARP, cleaved caspase 9, cleaved caspase 3, Pdcd4, and vinculin (as an internal control). The GFP level acts as an indirect indicator that miR-155 is effectively inhibited by a miR-155 sponge. (e) Western blots for Bcl2, Bax, Pdcd4, and vinculin (acting as an internal control). Values are expressed as the means ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
miR-155 knockdown reduces xenograft formation in nude mice.
Consistent with the above in vitro results, nude mice injected with 5 × 106 SAS–pLCE–miR-155 sponge cells showed a significant reduction in tumor burden over that in SAS-pLCE cells. The growth kinetics recorded at six different time points postinjection and the tumor volumes (in cubic millimeters) measured are shown in Fig. 6a. Tumor weights were also measured at the end of the study; the weights for the xenografts generated by SAS–pLCE–miR-155 sponge cells were significantly lower than those for xenografts generated by SAS-pLCE cells (Fig. 6b). The expression of miR-155 was demonstrated by qPCR of the RNA collected from tumor xenografts generated by SAS–pLCE–miR-155 sponge and SAS-pLCE cells. We also performed Western blotting for Pdcd4 for the same set of tumors, and it was found that SAS–miR-155 sponge cells showed higher expression of Pdcd4 protein than SAS-pLCE cells (Fig. 6c). We also performed Western blotting for GFP from lysates of SAS-pLCE and SAS–pLCE–miR-155 sponge xenografts to verify that tumors generated in mice were produced by injected cells (Fig. 6d). The in vivo study clearly demonstrated that miR-155 knockdown suppressed tumor formation of SAS cells in nude mice.
FIG 6.
Effect of the miR-155 sponge on tumor growth in nude mice. (a) Representative images showing tumor formation in nude mice, with a graph showing the changes in tumor volume (n = 3). (b) Changes in the weight of tumor xenografts produced by SAS–pLCE–miR-155 sponge and SAS-pLCE cells (n = 3). (c) Relative expression of miR-155 in tumors as determined by qPCR (n = 3 as experimental triplicates). (d) Western blot for Pdcd4, with vinculin acting as internal control and GFP acting as an indicator of effective inhibition of miR-155 in xenografts. T1, T2, and T3 represent xenografts from mice injected with SAS-pLCE cells, whereas T4, T5 and T6 represent xenografts from mice injected with SAS–pLCE–miR-155 sponge cells. Values are expressed as the means ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
The deregulation of gene expression is often associated with various cancerous phenotypes such as proliferation, invasion, metastasis, and migration. The identification of molecular players that play crucial roles in gene expression, such as miRNAs, is relevant for effective therapeutic strategies. Experimental evidence indicates that some miRNAs, such as miR-21 (37) and miR-139 (38), form feedback regulatory loops with their targets for their constant expression to downregulate their target tumor suppressor mRNA. In this study, we have investigated upregulation of miR-155 in tongue cancer and verified Pdcd4 as its potential target that generates a positive feedback loop with AP-1 to maintain tumorigenic growth in tongue cancer cells. Even though Pdcd4 has recently been shown to be a direct target of miR-155 in non-small-cell lung cancer (16), the data represented in the previous study are not convincing since the miR-155 binding site mutant seemed to be responding to miR-155, and even the title of the report was contrary to the results shown. In contrast, our data provide strong evidence that overexpression of miR-155 in tongue cancer cells is due to the positive feedback loop formed by miR-155/Pdcd4/AP-1. Our study is the first to show that miR-155 modulates its expression by downregulation of Pdcd4 and activation of AP-1-dependent transcription of the BIC promoter in the context of tongue cancer cells (Fig. 3). Through a series of in silico, in vitro, and in vivo approaches, we found that miR-155 plays a crucial role in regulating Pdcd4 by directly targeting its 3′ UTR in tongue cancer cells and downregulating its expression. miR-155 expression is upregulated in solid tumors of diverse origins (23, 39), and consistent with this, we demonstrated that miR-155 expression is significantly higher in tongue cancer tissues and SAS and AWL cells than in adjacent normal tissues and normal FBM cells, respectively (Fig. 1), suggesting that high miR-155 expression is tightly associated with tongue cancer development. A significant negative correlation between miR-155 expression and that of Pdcd4 protein in a maximum number of tongue tumor tissues and SAS and AWL cells observed by us supports the notion that miR-155-mediated control of Pdcd4 is operational in tongue cancer. We have provided several lines of evidence to show for the first time that miR-155 directly targets Pdcd4 in FBM, SCC745, SAS, and AWL cells, and the effect of miR-155 downregulation on cancerous properties of tongue cancer cells was reflected in vitro with a decrease in clonogenic capability and anchorage-independent growth on soft agar (Fig. 4d and e).
Furthermore, coexpression of Pdcd4 and miR-155 and rescue experiments in SAS cells (Fig. 4) confirm that miR-155 acts via the miR-155 binding site present in the Pdcd4 3′ UTR. Further restoration of Pdcd4 by knockdown of miR-155 was accompanied by decreased cell viability, enhanced apoptosis, and reduction of tumor growth in nude mice (Fig. 5 and 6). miRNAs usually regulate a large set of targets, and our data do not rule out the idea that there may be more targets for miR-155 in addition to Pdcd4. Indeed, Pdcd4 has been shown to be targeted by miR-21 in various cancers (hepatocellular carcinoma [40], oral cancer [4], breast cancer [41], and colon carcinoma [42]), by miR-96 and miR-183 in glioma (43, 44), and by miR-499 in oropharyngeal cancer (45).
Concerning the mechanism, our data show that miR-155 acts as a transcriptional regulator of the AP-1 complex by directly targeting Pdcd4, an inhibitor of AP-1 transcription. We found that AP-1 is critical for BIC promoter activity in tongue cancer cells whereas the contribution of NF-κB is relatively minor. AP-1 activation is often paradoxical as it promotes or inhibits apoptosis in different cellular systems, but our data suggest its role in ensuring tongue cancer cell proliferation and emphasize the need to understand the cellular and extracellular contexts within which it functions (46). The inhibition of Pdcd4 by miR-155, in turn, contributes to the increase in AP-1 activity of the BIC promoter that explains the enhanced expression of miR-155 in tongue cancer cells, thus revealing a novel miR-155/Pdcd4/AP-1 feedback regulatory mechanism. Inhibition of AP-1 transcription factor by Pdcd4 involves Jun-Jun homodimers or Jun-Fos heterodimers (32). Our study validates tumor suppressor Pdcd4 as a target of oncogenic miR-155, and the positive feedback loop of miR-155/Pdcd4/AP-1 maintains the miR-155-mediated biological effects of the tongue cancer phenotypic effects (Fig. 7). Our data suggest that molecular strategies manipulate miR-155 expression and/or that the miR-155/Pdcd4/AP-1 feedback loop provides novel avenues to explore in tongue cancer therapeutics.
FIG 7.
Overview of auto-regulatory feedback loop formed by miR-155/Pdcd4/AP-1.
MATERIALS AND METHODS
Cell lines, reagents, and plasmids.
The cell lines Hep3B, SiHa, MCF7, MDA-MB231, and H1299 were obtained from National Centre for Cell Science (NCCS), Pune, India. HCT116 and HCT116P53–/– cells were received as a gift from Bert Vogelstein, Johns Hopkins University School of Medicine, Baltimore, MD. SCC131, SCC745, SCC969, AWL, and SCC172 cell lines were obtained as a gift from Suresh Kumar Rayala, Indian Institute of Technology (IIT) Chennai, India. SAS (tongue carcinoma cell line) was procured from the Japanese Collection of Research Bioresources Cell Bank, Japan. Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) (Invitrogen) containing penicillin and streptomycin as antibiotics. The FBM cell line was a gift from the Milind Vaidya (ACTREC) Tata Memorial Centre, India, and was maintained in Iscove's modified Dulbecco's medium (IMDM) plus 10% FBS supplemented with insulin, hydrocortisone, and epidermal growth factor (EGF). The pcDNA-BIC plasmid was a gift from Iqbal Rather, IISc, Bangalore, India. pLCE and pLCE–miR-155 sponge were gifts of Bryan R. Cullen, Department of Molecular Genetics and Microbiology, Duke University, Durham, NC. pPax2 and pMDL-2 were obtained from Addgene, USA. All the pGL-3 BIC promoter plasmids with their respective AP-1 and NF-κB mutant plasmids were kind gifts from Erik K. Flemington, Tulane University, New Orleans, LA.
Construction of the 3′ UTR reporter for Pdcd4.
The 688 bp of the 3′ UTR of Pdcd4 was amplified using gene-specific primers (forward primer, 5′-ATTACCTAACGTGACATGGCACATAAAATTGGTTAAAAAATTTTG-3′; reverse primer, 5′-TAATGGATATCTTGTGTGACCAGATCCCACCAGTAATG-3′) containing restriction sites for XhoI and NotI to facilitate the directional cloning of the 3′ UTR of Pdcd4 in a psiCheck-2 plasmid. The 3′ UTR for Pdcd4 was mutated at the miR-155 seed recognition region by overlap extension PCR. The inserted sequences for both the wild-type and mutated 3′ UTR of Pdcd4 were verified by DNA sequencing performed by Eurofins Genomics, India.
3′ UTR luciferase assay.
FBM and SCC745 cells were cotransfected with pcDNA3.1 or pcDNA3.1-BIC and the wild-type or mutant 3′ UTR-luciferase, whereas AWL and SAS cells were transfected only with psiCheck-2 and WT 3′ UTR and MUT 3′ UTR constructs. At 48 h posttransfection, cells were lysed using passive lysis buffer, and Renilla luciferase activity was measured using a Dual-Luciferase assay kit (A2492; Promega) and a luminescence plate reader (Molecular Devices, Inc., Sunnyvale, CA), wherein firefly luciferase acted as the internal control.
BIC and AP-1 promoter luciferase assay.
The regulation of BIC and the 4× AP-1 promoter by Pdcd4 or miR-155 was performed using a different combination of plasmid constructs in HEK293T and SAS cells (see Tables S1 to S4 in the supplemental material). Further downstream processing was performed after 48 h posttransfection by lysing the cells in passive lysis buffer, and firefly luciferase activity was measured using a Dual-Luciferase assay kit (A2492; Promega) in a luminescence plate reader (Molecular Devices, Inc., Sunnyvale, CA), wherein renilla luciferase activity acted as the internal control.
Western blotting and IHC.
All the cell lysates were prepared with radioimmunoprecipitation assay (RIPA) cell lysis buffer, and 30 μg of protein lysates was loaded per well of a 12% SDS-PAGE gel and later transferred to a polyvinylidene difluoride (PVDF) membrane at a constant current of 220 mA for 1.5 h. The membrane was probed by the primary antibody, washed three times with Tris-buffered saline with Tween 20 (TBS-T) and TBS separately and again probed with secondary antibody conjugated with horseradish peroxidase (HRP). All Western blots were developed and analyzed using Image Lab, version 5.1, in a ChemiDoc Western blot apparatus from Bio-Rad. For immunohistochemistry (IHC), the tissue slide was incubated with a 1:500 dilution of primary antibody. The slides were scored by the pathologist as the percentage of positive nuclear or cytoplasmic staining and were graded as follows: negative, ≤5%; weak, 5% to 19%; moderate, 20% to 49%; strong, 50% to 100% (26).
Transfection, RNA extraction, cDNA synthesis, and real-time PCR.
We used linear chain polyethyleneimine (PEI) for plasmid DNA transfections unless mentioned otherwise (27). We optimized transfection efficiency at 2:1 PEI/DNA for FBM, HEK293T, SAS, AWL, and SCC745 cells. For luciferase assays, in each 24-well plate, the number of cells seeded was 8 × 104, and in each six-well plate, the number of cells seeded was 3 × 105. The cells were collected after 48 h of transfection and further processed as described above (the paragraph “3′ UTR luciferase assay”). The tissue samples were stored in RNase Later and at −80°C until further use. Total RNA from cells was collected by a TRIzol reagent (Invitrogen) as per the manufacturer's protocol. For miRNA cDNA synthesis, a stem-loop primer-based PCR protocol was used (28). For cDNA synthesis, oligo(dT)-based conversion of mRNA to cDNA was performed by Moloney murine leukemia virus (MMLV) reverse transcriptase from life technologies. miRNA-155 expression was quantified by performing stem-loop reverse transcription followed by quantitative PCR; reverse transcription by MMLV reverse transcriptase (Life Technologies) was performed using miR-155-specific and U6-specific stem-loop primers. Quantitative reverse transcription-PCR (qRT-PCR) analysis of miR-155 and Pdcd4 was performed by using Sybr green chemistry and quantified by the 2–ΔCT and 2–ΔΔCT (where CT is threshold cycle) methods (29). The expression of miR-155 was normalized to that of U6 snRNA, and expression of Pdcd4 was normalized to that of β-actin. All qRT-PCRs were performed in triplicate.
Production of lentiviruses for stable expression of the miR-155 sponge.
Lentiviruses were produced by cotransfecting HEK293T cells with pPax2, pMDL-2, and pLCE–miR-155 sponge/pLCE. SAS cells were transduced at the optimized virus titer and incubated in the presence of 8 μg/ml Polybrene. The positively transduced cells were enriched by cell sorting in a FACSAria III by using enhanced GFP (eGFP) to sort the positive clones.
Soft-agar assay.
Cancerous cells tend to grow in an anchorage-independent manner and, hence, tend to form colonies in a semisolid medium such as soft agar. The anchorage-independent growth of the SAS-pLCE and SAS–miR-155 sponge cells was analyzed by soft agar-assay in six-well tissue culture plates. DMEM plus 10% FBS with 0.7% soft agar was poured first in each well of the six-well plates with utmost precision not to include air bubbles; later, 3,000 cells were mixed with 1 ml of DMEM plus 10% FBS with 0.35% soft agar and poured over the base agar. The cells were allowed to grow in a 5% CO2 incubator at 37°C for 25 days with the addition of 500 μl of liquid DMEM containing 10% FBS every 3 days.
Clonogenic assay.
For the clonogenic assay, 300 SAS-pLCE and SAS–miR-155 sponge cells were plated per well in six-well plates. Cells were allowed to form colonies for 25 days and later fixed in methanol and stained with crystal violet. A colony of cells was considered prominent if it contained 50 cells or more.
Cell cycle analysis.
The cell cycle analysis of SAS-pLCE and SAS–miR-155 sponge cells was performed by the FACSCalibur flow cytometer after the cells were stained with propidium iodide. We followed an end-to-end protocol found at http://www.abcam.com/protocols/flow-cytometric-analysis-of-cell-cycle-with-propidium-iodide-dna-staining.
Cell viability assay.
SAS cells were first transfected with pLCE and pLCE–miR-155 sponge plasmids in six-well plates. After 24 h of transfection, 2,000 cells were plated in each well of 96-well plates. The MTT reagent, a tetrazole, was added at 24-h time intervals (0 to 72 h) and was reduced to formazan in the mitochondria of living cells. The absorbance of this colored solution was quantified by measuring the absorbance at 570 nm in a spectrophotometer.
Annexin V-APC apoptosis assay.
After cells were transduced with lentivirus expressing miR-155 sponge and a control, apoptosis was measured by annexin V-allophycocyanin (APC) staining. Cells were cultured for another 72 h and then detached by 0.25% trypsin, washed twice with phosphate-buffered saline (PBS), and stained by annexin V-APC for 10 min at room temperature according to the manufacturer’s instructions (BD Biosciences). Subsequently, flow cytometric analysis was performed with annexin V-APC staining.
In vivo assay for tumor formation.
To see the effect of restoration of Pdcd4 expression by inhibition of miR-155 on tumor growth, 5 × 106 SAS-pLCE or SAS–pLCE–miR-155 sponge cells resuspended in PBS with 50% Matrigel matrix were injected subcutaneously into the posterior flank of each female BALB/c athymic 6-week-old nude mouse. Tumor growth was monitored, and the volume was measured every 5 days until the 30th day. Tumor volume (V) was calculated as LW2/2, where L and W represent large and small diameters of the tumor formed, respectively. Tumor weight was measured at the end of the study. All experiments with nude mice were approved by the institutional animal ethics committee.
Statistical analysis.
Experiments were carried out in triplicate. Data are presented as means plus or minus the standard deviations (SD) of the means. The differences between test and control groups were analyzed using the GraphPad Prism program. The statistical analysis was performed by column comparisons between samples of different types. An unpaired t test was performed to compare two samples involved in the experiment. One-way analysis of variance (ANOVA) with multiple comparisons was performed when three or more samples involved in a single experiment were compared. After statistical analysis was performed, P values were calculated and are given in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Department of Biotechnology, Government of India, for the financial support (grant number BT/PR/2672/AGR/36/702/2011) and Indian Institute of Technology Madras (IITM) for all other facilities. We thank the IIT Madras Alumni association for providing funds for travel to perform nude mouse experiments.
We thank Anuj Kaushik for his help with writing the manuscript.
S.Z. performed in vitro and in vivo experiments; V.T. assisted in experiments with nude mice under the supervision of K.S.; V.S. carried out the IHC experiments and scoring under the supervision of R.V.; S.Z. and D.K. analyzed the data; S.Z. and D.K. designed the experiments and analyzed the results; S.Z. and D.K. wrote the manuscript under the overall supervision D.K. All authors approved the submission for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00410-18.
REFERENCES
- 1.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. 2006. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. 2008. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci U S A 105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reis PP, Tomenson M, Cervigne NK, Machado J, Jurisica I, Pintilie M, Sukhai MA, Perez-Ordonez B, Grénman R, Gilbert RW, Gullane PJ, Irish JC, Kamel-Reid S. 2010. Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol Cancer 9:238. doi: 10.1186/1476-4598-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Day E, Lal A. 2010. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res 12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Knösel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, Ozaki I, Petersen I. 2003. Loss of PDCD4 expression in human lung cancer correlates with tumor progression and prognosis. J Pathol 200:640–646. doi: 10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J, Qin Y, Sun Z, Zheng X. 2010. miR-183 inhibits TGF-β1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer 10:354. doi: 10.1186/1471-2407-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao F, Zhang P, Zhou C, Li J, Wang Q, Zhu F, Ma C, Sun W, Zhang L. 2007. Frequent loss of PDCD4 expression in human glioma: possible role in the tumorigenesis of glioma. Oncol Rep 17:123–128. [PubMed] [Google Scholar]
- 9.Allgayer H. 2010. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol 73:185–191. doi: 10.1016/j.critrevonc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Young MR, Yang H-S, Colburn NH. 2003. Promising molecular targets for cancer prevention: AP-1, NF-κB and Pdcd4. Trends Mol Med 9:36–41. [DOI] [PubMed] [Google Scholar]
- 11.Vikhreva P, Shepelev M, Korobko E, Korobko I. 2010. Pdcd4 tumor suppressor: properties, functions, and their application to oncology. Mol Gen Mikrobiol Virusol 2:3–11. (In Russian.) [PubMed] [Google Scholar]
- 12.Lankat‐Buttgereit B, Göke R. 2003. Programmed cell death protein 4 (pdcd4): a novel target for antineoplastic therapy? Biol Cell 95:515–519. [DOI] [PubMed] [Google Scholar]
- 13.Yang H-S, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. 2001. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-κB or ODC transactivation. Oncogene 20:669–676. [DOI] [PubMed] [Google Scholar]
- 14.Yang H-S, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomsen KG, Terp MG, Lund RR, Søkilde R, Elias D, Bak M, Litman T, Beck HC, Lyng MB, Ditzel HJ. 2015. miR-155, identified as anti-metastatic by global miRNA profiling of a metastasis model, inhibits cancer cell extravasation and colonization in vivo and causes significant signaling alterations. Oncotarget 6:29224. doi: 10.18632/oncotarget.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Song D, Wu Y, Liu X, Zhu J, Tang Y. 2017. MiR‐155 inhibits proliferation and invasion by directly targeting PDCD 4 in non‐small cell lung cancer. Thorac Cancer 8:613–619. doi: 10.1111/1759-7714.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. 2008. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer 123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang S, Zhang H-W, Lu M-H, He X-H, Li Y, Gu H, Liu M-F, Wang E-D. 2010. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 19.Carinci F, Lo Muzio L, Piattelli A, Rubini C, Chiesa F, Ionna F, Palmieri A, Maiorano E, Pastore A, Laino G, Favia G, Dolci M, Pezzetti F. 2005. Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunopathol Pharmacol 18:513–524. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]
- 20.Gironella M, Seux M, Xie M-J, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang K-T, Chaix A, Fazli L, Motoo Y, Wang Q, Rocchi P, Russo A, Gleave M, Dagorn J-C, Iovanna JL, Carrier A, Pebusque M-J, Dusetti NJ. 2007. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A 104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. 2007. MicroRNA expression profiling in prostate cancer. Cancer Res 67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 22.Calin GA, Croce CM. 2006. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 23.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. 2009. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Li M, Zuo K, Li D, Ye M, Ding L, Cai H, Fu D, Fan Y, Lv Z. 2013. Upregulated miR-155 in papillary thyroid carcinoma promotes tumor growth by targeting APC and activating Wnt/β-catenin signaling. J Clin Endocrinol Metab 98:E1305–E1313. doi: 10.1210/jc.2012-3602. [DOI] [PubMed] [Google Scholar]
- 25.Rather MI, Nagashri MN, Swamy SS, Gopinath KS, Kumar A. 2013. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics. J Biol Chem 288:608–618. doi: 10.1074/jbc.M112.425736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsolier J, Pineau S, Medjkane S, Perichon M, Yin Q, Flemington E, Weitzman MD, Weitzman JB. 2013. OncomiR addiction is generated by a miR-155 feedback loop in Theileria-transformed leukocytes. PLoS Pathog 9:e1003222. doi: 10.1371/journal.ppat.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. 2010. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. 2010. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-β. J Biol Chem 285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin Q, Wang X, McBride J, Fewell C, Flemington E. 2008. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem 283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohmann D, Bos TJ, Admon A, Nishimura T, Vogt PK, Tjian R. 1987. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238:1386–1393. [DOI] [PubMed] [Google Scholar]
- 31.Karin M, Liu Z-g, Zandi E. 1997. AP-1 function and regulation. Curr Opin Cell Biol 9:240–246. [DOI] [PubMed] [Google Scholar]
- 32.Shaulian E, Karin M. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390–2400. [DOI] [PubMed] [Google Scholar]
- 33.Shukla GC, Singh J, Barik S. 2011. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol 3:83. [PMC free article] [PubMed] [Google Scholar]
- 34.Franken NA, Rodermond HM, Stap J, Haveman J, Van Bree C. 2006. Clonogenic assay of cells in vitro. Nat Protoc 1:2315. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 35.Borowicz S, Van Scoyk M, Avasarala S, Mk KR, Tauler J, Bikkavilli RK, Winn RA. 2014. The soft agar colony formation assay. J Vis Exp 92:e51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Göke R, Barth P, Schmidt A, Samans B, Lankat-Buttgereit B. 2004. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21 Waf1/Cip1. Am J Physiol Cell Physiol 287:C1541–C1546. doi: 10.1152/ajpcell.00025.2004. [DOI] [PubMed] [Google Scholar]
- 37.Sun Q, Miao J, Luo J, Yuan Q, Cao H, Su W, Zhou Y, Jiang L, Fang L, Dai C, Zen K, Yang J. 2018. The feedback loop between miR-21, PDCD4 and AP-1 functions as a driving force for renal fibrogenesis. J Cell Sci 131:jcs202317. doi: 10.1242/jcs.202317. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Shen W-L, Shi M-L, Zhang L-Z, Zhang Z, Li P, Xing L-Y, Luo F-Y, Sun Q, Zheng X-F, Yang X, Zhao Z-H. 2015. Involvement of aberrant miR-139/Jun feedback loop in human gastric cancer. Biochim Biophys Acta 1853:481–488. doi: 10.1016/j.bbamcr.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Moles R. 2017. MicroRNAs-based therapy: a novel and promising strategy for cancer treatment. MicroRNA 6:102–109. doi: 10.2174/2211536606666170710183039. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Q, Wang Z, Hu Y, Li J, Li X, Zhou L, Huang Y. 2012. miR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncol Rep 27:1660–1668. doi: 10.3892/or.2012.1682. [DOI] [PubMed] [Google Scholar]
- 41.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. 2008. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 42.Asangani I, Rasheed S, Nikolova D, Leupold J, Colburn N, Post S, Allgayer H. 2008. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 43.Gu W, Gao T, Shen J, Sun Y, Zheng X, Wang J, Ma J, Hu X-Y, Li J, Hu M-J. 2014. MicroRNA-183 inhibits apoptosis and promotes proliferation and invasion of gastric cancer cells by targeting PDCD4. Int J Clin Exp Med 7:2519. [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Q-q, Huang J-t, Xiong Y-g, Yang X-y, Han R, Zhu W-w. 2017. MicroRNA-96 regulates apoptosis by targeting PDCD4 in human glioma cells. Technol Cancer Res Treat 16:92–98. doi: 10.1177/1533034616629260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Gee H, Rose B, Lee CS, Clark J, Elliott M, Gamble JR, Cairns MJ, Harris A, Khoury S, Tran N. 2016. Regulation of the tumor suppressor PDCD4 by miR-499 and miR-21 in oropharyngeal cancers. BMC Cancer 16:86. doi: 10.1186/s12885-016-2109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ameyar M, Wisniewska M, Weitzman J. 2003. A role for AP-1 in apoptosis: the case for and against. Biochimie 85:747–752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.