Abstract
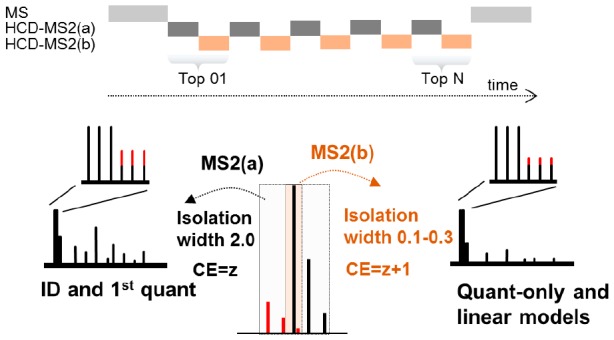
Isobaric labeling is a highly precise approach for protein quantification. However, due to the isolation interference problem, isobaric tagging suffers from ratio underestimation at the MS2 level. The use of narrow isolation widths is a rational approach to alleviate the interference problem; however, this approach compromises proteome coverage. We reasoned that although a very narrow isolation window will result in loss of peptide fragment ions, the reporter ion signals will be retained for a significant portion of the spectra. On the basis of this assumption, we have designed a dual isolation width acquisition (DIWA) method, in which each precursor is first fragmented with HCD using a standard isolation width for peptide identification and preliminary quantification, followed by a second MS2 HCD scan using a much narrower isolation width for the acquisition of quantitative spectra with reduced interference. We leverage the quantification obtained by the “narrow” scans to build linear regression models and apply these to decompress the fold-changes measured at the “standard” scans. We evaluate the DIWA approach using a nested two species/gene knockout TMT-6plex experimental design and discuss the perspectives of this approach.
Keywords: proteomics, isobaric labeling, TMT, iTRAQ, interference, ratio compression, isolation width, Orbitrap
Introduction
Stable isotope labeling of peptides using isobaric reagents such as iTRAQ and TMT enables the multiplexed analysis of proteomes with deep quantitative coverage.1,2 This barcoding strategy has provided comprehensive proteomic portraits of large collections of human cancer tissue samples3,4 and cell lines,5,6 and has enabled the in-depth characterization of protein post-translational modifications in a quantitative fashion.7−10 Isobaric tagging demonstrates high precision11 but imperfect accuracy due to ratio underestimation caused by cofragmentation of ions with mass-to-charge ratios within the isolation window of the targeted precursors.12 Although this problem rarely affects the direction of protein abundance change, many applications can significantly benefit from increased accuracy; examples include the determination of protein localization,13 identification of specific protein–protein interactions in pull down assays,14,15 and verification of protein deletions in gene knockout or knock-down experiments or in samples with natural genomic variation.16 Several groups have proposed solutions to alleviate the interference problem and thereby improve isobaric labeling quantification accuracy. Ow et al. demonstrated that the ratio compression can be decreased using high-resolution HILIC fractionation, which achieves maximum orthogonality and reduces MS sampling complexity.17 An alternative approach by Savitski et al. improves accuracy by fragmentation of the peptides close to their maximum chromatographic peak height.18 Elimination of ratio distortion in a two-proteome model utilizing MS3 for further fragmentation of b- or y- ions specific to the targeted precursor was reported by Ting et al.19 McAlister et al. have further enhanced the sensitivity of the MS3 method using isolation waveforms with multiple frequency notches.20 The latter is currently the method of choice for counteracting interference in isobaric labeling experiments using tribrid mass spectrometry. The QuantMode method developed by Wenger et al. is based on gas-phase purification, by manipulation of either mass or charge through expedient proton-transfer ion–ion reactions and has also been shown to improve quantitative accuracy.21 Using data analysis methods, Wuhr et al. showed that precursor-specific quantitative information can be retrieved at the MS2 level from the complement reporter ion cluster.22 Notably, Shliaha et al. evaluated the utility of ion mobility for additional precursor purification in data-dependent acquisition mode and presented evidence for improved accuracy, especially in combination with narrowed quadrupole isolation window.23 More recently, Niu et al. showed that more accurate quantification can be obtained at the MS2 level with the combination of extensive peptide fractionation, narrow precursor isolation and y1 ion-based interference detection.24
Prompted by empirical observations of isobaric-labeled peptide MS2 spectra, we argue that although a very narrow isolation window will result in severe loss of backbone fragment ions, rendering the spectra unsuitable for peptide identification, the reporter ion signals will remain intense enough to generate quantitative information for a significant portion of the spectra. On the basis of this assumption we have designed a dual isolation width acquisition (DIWA) method, in which each precursor is first fragmented with HCD using a standard isolation width for peptide identification and preliminary quantification, followed by a concomitant MS2 HCD fragmentation using a much narrower isolation width for the acquisition of quantification-only spectra with reduced interference. We leverage the quantitative values obtained by the “narrow” scans to build linear regression models and apply these to decompress the fold-changes measured at the “standard” scans. Here, we evaluate the DIWA method using a nested two species/gene knockout TMT-6plex model and discuss the potential of this approach.
Methods
Experimental Design Overview
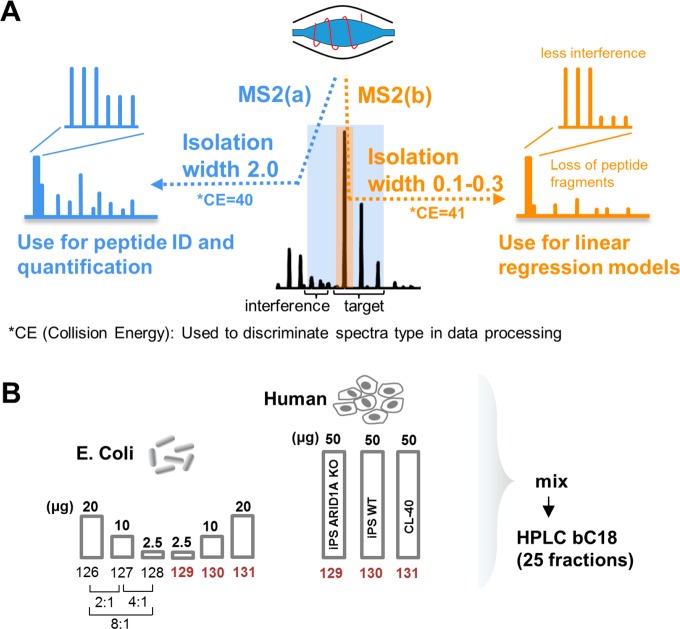
To alleviate the interference problem at the MS2 level we have designed an acquisition method on an LTQ-Orbitrap Velos in which each precursor is fragmented twice back-to-back with HCD using a different isolation width at each scan event (1st MS2 scan 2.0 Th/2nd MS2 scan 0.1 or 0.2 or 0.3 Th) (Figure 1A). Slightly different collision energies are used in each of the two HCD spectra to mark their isolation width of origin. The aim of the method is to collect MS2 spectra devoid of interference in the isobaric tags, which can be used to model the compression effect and to correct all quantitative values obtained at the standard isolation width. To evaluate this approach we have performed a TMT-6plex experiment using the mixed-proteome model19 in combination with the use of a CRISPR-cas9 gene knockout (KO). We analyzed a mixture of Escherichia coli tryptic peptides at ratios 2:1, 4:1, 8:1 (×2 each) and tryptic peptides from three human cell lines (hiPSC ARID1A KO, hiPSC WT and CL-40) at ratios 1:1:1 (Figure 1B). Three TMT channels (129, 130, 131) were overlapping between E. coli and Human peptides, while three channels (126, 127, 128) were used for interference-free E. coli peptides.
Figure 1.
(A) The DIWA method overview. (B) Experimental design for the nested two species/gene knockout TMT-6plex model.
To simulate lower-abundant changing proteins, we spiked the E. coli peptides at 2.5 to 20-fold lower total amount per channel compared to the human peptides. This represents a scenario of high interference; in a real experiment it is more likely that the differentially regulated proteins will cover the entire protein abundance dynamic range rather than the mid-to-low abundant portion only. The labeled peptides were fractionated with high-pH reversed-phase HPLC and the 25 fractions were analyzed on an LTQ Orbitrap Velos. The analysis of the fractions was performed three times at three different “narrow” isolation widths (0.1, 0.2, and 0.3 Th). Finally, linear regression models were generated by plotting the “standard” against the “narrow” isolation peptide logarithmic ratios for each sample comparison. All primary peptide log2ratios were calibrated through these models. We chose to model the compression effect by Deming regression, which is more appropriate when both the dependent and independent variables are measured with error, thus facilitating the comparison of two assays designed to measure the same analyte.
Sample Preparation and LC–MS Analysis
The hiPSC CRISPR-cas9 and the CL-40 cell pellets were obtained as described previously.5 The cell pellets were homogenized in 150 μL 0.1 M triethylammonium bicarbonate (TEAB), 1% sodium deoxycholate (SDC), 10% isopropanol with probe sonication for 3 × 5 s with pulses of 1 s at 40% amplitude (EpiShear) followed by boiling at 90 °C for 5 min. Samples were resonicated and centrifuged at 10 000 rpm for 10 min. Lyophilized Escherichia coli whole protein extract (Bio-Rad) was dissolved in 200 μL 0.1 M TEAB, 0.1% SDS divided into aliquots of 10 μL each and diluted up to 100 μL. The protein content of each aliquot was precipitated by the addition of 30 μL TCA 8 M for 30 min at 4 °C. The protein pellets were washed twice with ice-cold acetone and finally resuspended in 40 μL 0.1 M TEAB, 0.1% SDS with probe sonication, before they were combined in a single E. coli pool. Protein concentration was measured with the Coomassie Plus Bradford Protein Assay (Pierce) according to manufacturer’s instructions. Duplicate aliquots of 20 μg (TMT: 126 and 131), 10 μg (TMT: 127 and 130), and 2.5 μg (TMT: 128 and 129) of E. coli protein and aliquots of 50 μg of hiPSC ARID1A KO (TMT: 129), hiPSC WT (TMT: 130) and CL-40 (TMT: 131) were prepared for trypsin digestion. Cysteines were reduced with 5 mM tris-2-carboxyethyl phosphine (TCEP) for 1 h at 60 °C and blocked by 10 mM iodoacetamide (IAA) for 30 min at room temperature in dark. Trypsin (Pierce, MS grade) solution was added at a final concentration of 70 ng/μL to each sample for overnight digestion. The peptide samples were finally labeled with the TMT-6plex reagents (Thermo Scientific) according to manufacturer’s instructions. The TMT peptide mixture was acidified with 1% formic acid and the precipitated SDC was removed by centrifugation. Offline peptide fractionation was based on high pH Reverse Phase (RP) chromatography using the Waters XBridge C18 column (2.1 × 150 mm, 3.5 μm) on a Dionex Ultimate 3000 HPLC system at a 0.85% gradient with flow rate 0.2 mL/min. Mobile phase A was 0.1% ammonium hydroxide, and mobile phase B was 100% acetonitrile, 0.1% ammonium hydroxide.
LC–MS analysis was performed on the Dionex Ultimate 3000 UHPLC system coupled with the LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). Samples were analyzed with the Acclaim PepMap RSLC C18 capillary column (75 μm × 50 cm, 2 μm, 100 Å). Mobile phase A was 0.1% formic acid and mobile phase B was 80% acetonitrile, 0.1% formic acid. The gradient separation method was as follows: for 85 min gradient up to 38% B, for 10 min up to 95% B, for 10 min isocratic at 95% B, re-equilibration to 5% B in 5 min, for 10 min isocratic at 5% B. For the DIWA method, the five most abundant multiply charged precursors within 380–1500 m/z were selected with FT mass resolution of 30 000 and isolated for HCD fragmentation twice with isolation width 2.0 and 0.1 or 0.2 or 0.3 Th in an Nth order double play method (henceforth “standard” and “narrow” scans, respectively). Normalized collision energy was set at 40 for the standard scans and at 41 for the narrow scans. Tandem mass spectra were acquired at 7500 FT resolution with 40 s dynamic exclusion and 10 ppm mass tolerance. FT maximum ion time for full MS experiments was set at 200 ms and FT MSn maximum ion time was set at 50 ms. The AGC target vales were 3 × 106 for full FTMS and 5 × 105 for MSn FTMS. For optimum ion transmission efficiency, we performed an S-lens cleaning prior to the analysis. Additionally, we evaluated the effect of the narrow isolation widths on the total ion current at the HCD-MS2 level (CE = 40, max IT = 100) of the m/z 1422 precursor peak of the ESI positive calibration mix. We noted an 1.7, 2.3, 2.7, and 6-fold signal decrease as the isolation width was decreased from 2.0 to 1.0, 0.5, 0.3, and 0.1 Th, respectively, in a linear fashion (R2 = 0.98).
Data Processing
The MS2 spectra collected with collision energy 40 (“standard” scans) were searched against Uniprot human (reviewed only) and Escherichia coli entries using SequestHT in Proteome Discoverer 2.2. The precursor mass tolerance was set at 30 ppm and the fragment ion mass tolerance was set at 0.02 Da. Static modifications were as follows: TMT6plex at N-termini/K, and carbamidomethyl at C. Dynamic modifications included oxidation of M and deamidation of N/Q. The data were processed twice, first with maximum collision energy 40 for quantification using the “standard” scans and second with minimum collision energy 41 for quantification using the “narrow” scans (Reporter Ions Quantifier node). Quantification was based on un-normalized signal-to-noise (S/N) values. Peptide confidence was estimated with the Percolator node. Peptide FDR was set at 0.01 and validation was based on q-value and decoy database search. Deming regression was performed in R Studio with the “deming” package. A matrix containing the log2 ratios: 131/129, 131/130 and 130/129 for the standard (IW2) and narrow (IW01) scans was used as input. The R code and the regression models are provided in Supporting Information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE25 partner repository with the data set identifier PXD010571.
Results
To evaluate the depth of proteome coverage and the accuracy level that can be achieved by the dual isolation width acquisition method, we performed a proof-of-concept TMT6plex-based analysis of varying amounts of spike-in E. coli protein extract into human protein lysates of equal total amounts representing different cell types. For straightforward implementation of the method, we used an LTQ-Orbitrap Velos platform in which the method editor already allows the setup of dual acquisitions (e.g., CID-HCD). The application of the DIWA method in the two-proteome model resulted in the quantification of 6724 total human and E. coli unique protein groups at isolation width (IW) 2.0. This level of proteome coverage is in line with previous studies using similar instrumentation.9 A subset of 6132 (91%) proteins were also fully quantified at the narrowest isolation width 0.1, confirming that the isobaric tags remain at quantifiable levels for the majority of the peptides despite the significant loss of the peptide fragment ion signals (Table S1). As expected, the percentages of quantified PSMs, peptides and protein groups increased as the narrow scan windows were opened to 0.2 and 0.3 (Table 1, Tables S2 and S3). The overall lower quantification coverage of E. coli peptides and proteins is due to their lower abundances compared to human proteins and due to the 8-fold lower sample load in two of the TMT channels. These low-intensity TMT channels were more frequently below the quantification limit compared to the human samples, resulting in smaller number of fully quantified E. coli PSMs. Notably, quantification at narrow isolation was more efficient for doubly charged peptides (Figure S1A) with medium to high precursor intensity (Figure S1B). The charge state dependency is possibly due to the fact that the monoisotopic peak of doubly charged peptides is the most intense within their isotopic cluster yielding more efficient isolation. However, multiply charged peptides appeared to have overall lower precursor intensities (Figure S1C) suggesting that isolation efficiency depends on both the isotopic cluster pattern and precursor intensity. Overall, at isolation width 0.1, we observed a median 10-fold reduction in the mean S/N for the human peptides with 87% of these retaining a signal-to-noise ratio greater than 5.
Table 1. Number and Comparative Percentages of PSMs, Unique Peptides, and Protein Groups Quantified at Each Isolation Width for Human and E. coli Samples.
| isol. width 2.0 | isol. width 0.1 | overlap (%) | isol. width 2.0 | isol. width 0.2 | overlap (%) | isol. width 2.0 | isol. width 0.3 | overlap (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Human | PSMs | 62 450 | 43 932 | 70 | 62 335 | 48 566 | 78 | 59 249 | 50 524 | 85 |
| unique peptides | 38 739 | 31 264 | 81 | 38 353 | 33 167 | 86 | 35 784 | 32 732 | 91 | |
| protein groups | 5 479 | 5 080 | 93 | 5 460 | 5 158 | 94 | 5 338 | 5 106 | 96 | |
| E. coli | PSMs | 12 141 | 5 007 | 41 | 11 998 | 6 510 | 54 | 11 710 | 7 473 | 64 |
| unique peptides | 6 576 | 3 640 | 55 | 6 323 | 4 389 | 69 | 6 084 | 4 749 | 78 | |
| protein groups | 1 245 | 1 052 | 84 | 1 234 | 1 129 | 91 | 1 198 | 1 119 | 93 |
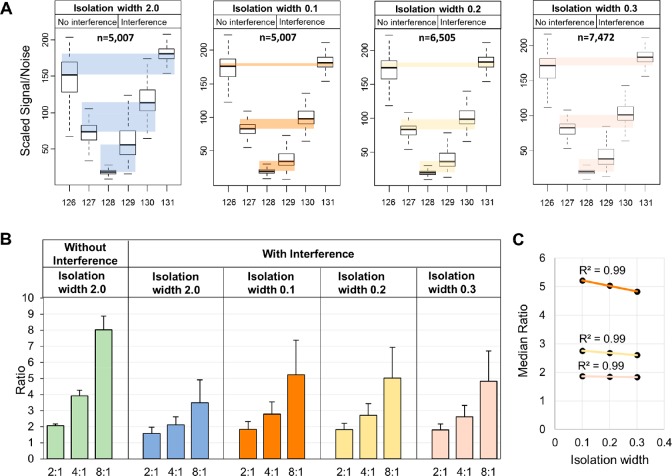
Next, we evaluated the correction efficiency of the DIWA method using the E. coli PSMs fully quantified in the different isolation widths. Significantly less TMT signal distortion was found using the narrow isolation widths as shown by the difference between the medians of the expected (without interference) and measured (with interference) scaled quantitative values between the replicate channels (Figure 2A). Although the compression effect was not eliminated, the narrow isolations yielded significantly improved ratios (Figure 2B).
Figure 2.
Reduction of interference by the DIWA method. (A) Box plots of the scaled TMT signal-to-noise values of E. coli PSMs across the six TMT channels at the different isolation widths. The embedded colored boxes highlight the difference of the medians between the expected (No interference) and measured (Interference) values for each replicate pair. (B) Bar plots of the E. coli PSM ratios for each theoretical ratio at the different isolation widths without and with interference. Error bars represent standard deviation. (C) Scatter plot of the isolation width setting (x-axis) against the median ratio (y-axis) for each theoretical ratio.
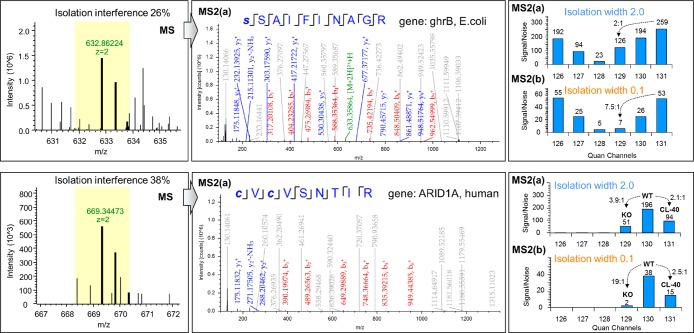
Specifically, in isolation width 0.1, the percent error was decreased from 59% to 35% for the higher ratio 8:1 and from 21% to 11% for the lower ratio 2:1. For example, while 67% of the ratios measured at isolation width 2.0 were below 4 for the expected ratio 8:1, 73% of these were above the 4-fold threshold using the narrow acquisition. Additionally, the ratio compression effect showed a linear correlation (R2 = 0.99) with the isolation width, as found by the median ratios at each one of the narrow acquisitions (Figure 2C). This suggests that the compression effect could be modeled by the isolation width gradient to predict the ratios at isolation widths close to zero. Because the use of the narrowest isolation width yielded the smallest interference, for all downstream analysis we use only the data obtained at isolation width 0.1. Two example identification and quantification spectra matched to ghrB (E. coli) and ARID1A (human) peptides are shown in Figure 3. Both peptides suffered significant ratio compression at the “standard” isolation width due to high precursor interference, however the second HCD scan at isolation width 0.1 provided more accurate quantification (Figure 3, right panels). Specifically, at the “standard” scan, the ARID1A peptide displayed a 3.9-fold down-regulation in the ARID1A knockout cells suggesting an in-complete silencing of the gene. However, the narrow MS2 scan of the same precursor ion revealed a 19-fold reduction of the protein product. The latter is more likely to reflect a complete knockout, particularly when the previously described spatial constraints on protein quantification by TMT26 that do not permit proper discrimination of truly missing proteins are taken into consideration. Interestingly, both MS2 spectra of these Arginine-ending peptides contain a peak at m/z 376.27 which is a characteristic y1 ion from Lysine-ending interfering peptide as suggested by Niu et al.24 This suggests that additional correction could be achieved using the y1 ion-based interference detection method.24
Figure 3.
Example spectra for E. coli and human peptides. Precursor ion cluster, annotated HCD MS2 fragment ion spectrum and TMT signal-to-noise values at “standard” and “narrow” isolation widths for a peptide matched to E. coli ghrB gene (top panel) and for a peptide matched to human ARID1A (bottom panel). Unassigned peaks are shown in gray font on the MS2 spectra.
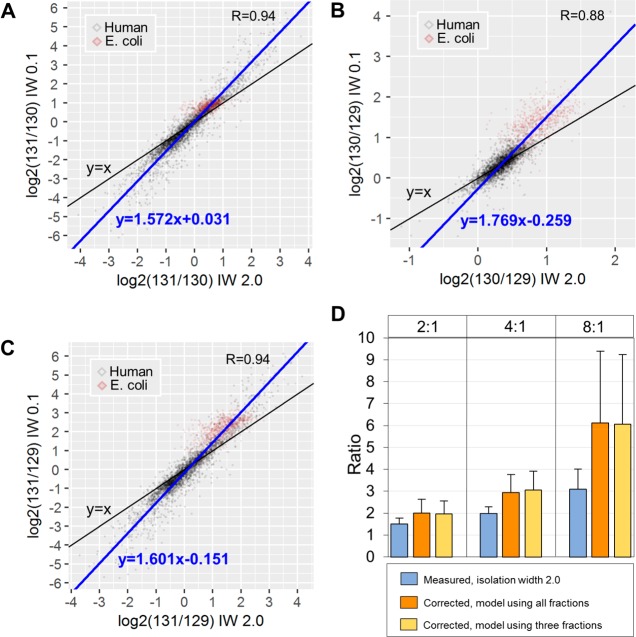
As the narrow isolation widths did not provide quantification for about 25–35% of the respective “standard” scans, we next aimed to model the compression effect using linear regression and to calibrate all primary quantifications obtained at the “standard” scans. Upon manual examination of the quantification spectra, we found that the narrow isolation widths were not always effective in reducing isolation interference. To identify the E. coli spectra with significant reduction of the interference upon the application of narrower precursor selection and therefore to model the compression effect more accurately, we computed the ratio 129(IW 0.1)/129(IW 2.0) of the scaled abundances as a metric for the magnitude of correction. In this instance, a low ratio (large difference in signal intensity) would suggest effective correction by the narrow scan, whereas a high ratio would suggest insufficient correction or spectra with originally low interference. Examples of quantification spectra with low and high 129(IW 0.1)/129(IW 2.0) ratio are shown in Figure S2A. To identify which peptide features are associated with effective correction by the narrow scans, we next correlated this ratio metric with peptide m/z, charge, precursor intensity, and isolation interference (the percentage of ion signal not attributed to the targeted precursor within a specified isolation window as reported by ProteomeDiscoverer software) (Figure S2B). We found that peptides with lower m/z, charge and precursor intensity (positively correlated) and high isolation interference (negatively correlated) are more effectively corrected by the narrow scans. Therefore, we can enrich for peptides that are effectively corrected by the narrow scans in samples with unknown protein abundances, by applying cutoffs to these features. Consequently, we selected doubly charged PSMs, with m/z and precursor intensity smaller than the median of all PSMs (<698.4 and <2.4 × 106 respectively) as well as isolation interference greater than the median of all PSMs (>18.2%) as input for the Deming regression analysis (n = 3616). To model the compression effects we generated the scatterplots of the selected PSMs using the logarithmic ratios from IW 2.0 scans against their counterparts at IW 0.1. We observed a linear response with Pearson’s R > 0.88 and Deming regression slope >1.57 for three comparisons (131/130, 130/129, and 131/129) indicative of the compression effect (Figure 4A,B,C). Using the slope and the intercept of these linear models we calibrated the log2ratios acquired at IW 2.0 for all human and E. coli PSMs. To evaluate the decompression efficiency, we retrieved the calibrated E. coli PSM log2ratios, converted to 2log2 ratio and computed the mean ratio per protein. This analysis showed that the regression-based calibration could decompress the original ratios up to 1.9-fold on average (Figure 4D). For example, in the expected ratio 8:1 only 28 proteins (2%) had a ratio greater than 5 at the standard isolation width; however, in the calibrated values, 820 proteins (66%) were above this threshold. This improvement can have important implications in statistical analysis and identification of differentially expressed proteins when specific cut-offs are applied. As the dual acquisition method is associated with longer cycle times, we tested whether efficient predictive models could be built using a smaller subset of PSMs from only three randomly selected fractions. Indeed, a very similar degree of decompression could be achieved from only a subset of the fractions (Figure 4D and Supporting Information). This suggests that all peptide fractions could be analyzed with a usual TMT method for maximum proteome coverage, followed by the DIWA rerun of only a few fractions for regression analysis and retrospective decompression of all the original ratios. Additionally, correlation analysis showed that the percent error at IW 2.0 is positively correlated with charge state and isolation interference and negatively correlated with precursor intensity, m/z and Sequest cross-correlation score (Xcorr) (Figure S2C). These characteristic features could be utilized to build more accurate predictive models using machine learning approaches (e.g., Support Vector Regression). Overall, our feasibility experiment shows that the acquisition of an additional HCD scan event at a narrow isolation width immediately after the acquisition of a standard MS2 scan for the same precursor, can be used to enhance accuracy of quantification for the majority of the identified peptides. Moreover, the dual isolation data can be used to model the compression effect by linear regression extending the coverage of the ratio decompression.
Figure 4.
Deming regression models and ratio decompression. Scatter plots of the (A) 131/130, (B) 130/129, and (C) 131/129 logarithmic ratios at isolation width 2.0 (x-axis) versus isolation width 0.1 (y-axis). (D) Bar plots of the E. coli protein ratios (average of PSM ratios) before and after decompression at the different theoretical ratios. Error bars represent standard deviation.
Conclusions
The selection of precursor ions using narrow isolation windows is a rational approach to reduce peptide cofragmentation and therefore to improve isobaric labeling-based quantification at the MS2 level. However, this approach yields very low proteome coverage as the narrow precursor selection results in poor peptide fragment ion spectra. To overcome this limitation, we have designed a novel method based on sequential HCD-HCD activation in a dual isolation width mode followed by modeling of ratio compression and correction. We have tested the method on an LTQ Orbitrap Velos system with a common Nth-order double play method. Importantly, apart from some additional data analysis steps, the DIWA approach does not require major changes in sample preparation protocols or specialized instrument configuration adjustments of the LTQ-Orbitrap systems (Velos and Elite). Using a two-proteome model and a CRISPR-cas9 gene knockout, we show that the method achieves comprehensive proteome coverage and preliminary quantification of all peptides, while the additional narrow isolation width can improve the quantitative accuracy for a significant portion of these. Furthermore, the low-interference spectra can be used as “pseudo-internal standards” to model the compression effect by linear regression in a sample-specific manner. With appropriate factory-level tuning of the minimum isolation width setting, the enhancement offered by the DIWA method has the potential to be universal to other platforms, such as benchtop Q-Exactives or Q-TOFs, that can only perform isobaric labeling quantification at the MS2 level. We expect a variation in the DIWA performance depending on the geometries of different spectrometry platforms and their isolation efficiencies, and therefore further tests and optimizations are warranted. Moreover, the combination of DIWA with previously described approaches such as gas-phase purification or the recently described high-field asymmetric waveform ion mobility spectrometry (FAIMS)27 could further improve the accuracy of isobaric labeling at the MS2 level. Future developments in mass spectrometry technology, which improve isolation efficiency and analytical speed in combination with intelligent precursor selection decision trees, could further boost the sensitivity and accuracy of the method. We conclude that the DIWA approach can provide significant ratio decompression in isobaric labeling at the MS2 level, and that the current implementation offers the foundations for further developments and offers universal applicability.
Acknowledgments
We thank Ultan McDermott and Stacey Price for donating the CL-40 cell pellet, David J. Adams for donating the human iPS WT and ARID1A KO cell pellets, Lu Yu for her help with mass spectrometry quality control, and Daniel Bode for discussions about data analysis. This work was funded by a core grant from the Wellcome Trust (098051).
Glossary
Abbreviations
- CID
collision-induced dissociation
- DIWA
dual isolation width acquisition
- HCD
higher-energy collisional dissociation
- HILIC
hydrophilic interaction chromatography
- iTRAQ
isobaric tags for relative and absolute quantification
- IW
isolation width
- KO
knockout
- PSM
peptide spectrum matches
- RP
reversed phase
- TMT
tandem mass tags.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00870.
Author Contributions
Conceptualization: T.I.R., J.S.C.; Experiments: T.I.R.; Data analysis: T.I.R., H.W.; Writing original draft: T.I.R., J.S.C.; Final draft/editing: all authors.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE25 partner repository with the data set identifier PXD010571.
Supplementary Material
References
- Rauniyar N.; Yates J. R. 3rd Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 2014, 13 (12), 5293–309. 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J. D.; Paulo J. A.; O’Brien J. J.; Gygi S. P. Proteome-Wide Evaluation of Two Common Protein Quantification Methods. J. Proteome Res. 2018, 17 (5), 1934–1942. 10.1021/acs.jproteome.8b00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Liu T.; Zhang Z.; Payne S. H.; Zhang B.; McDermott J. E.; Zhou J. Y.; Petyuk V. A.; Chen L.; Ray D.; Sun S.; Yang F.; Chen L.; Wang J.; Shah P.; Cha S. W.; Aiyetan P.; Woo S.; Tian Y.; Gritsenko M. A.; Clauss T. R.; Choi C.; Monroe M. E.; Thomas S.; Nie S.; Wu C.; Moore R. J.; Yu K. H.; Tabb D. L.; Fenyo D.; Bafna V.; Wang Y.; Rodriguez H.; Boja E. S.; Hiltke T.; Rivers R. C.; Sokoll L.; Zhu H.; Shih I. M.; Cope L.; Pandey A.; Zhang B.; Snyder M. P.; Levine D. A.; Smith R. D.; Chan D. W.; Rodland K. D.; Investigators C. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166 (3), 755–765. 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P.; Mani D. R.; Ruggles K. V.; Gillette M. A.; Clauser K. R.; Wang P.; Wang X.; Qiao J. W.; Cao S.; Petralia F.; Kawaler E.; Mundt F.; Krug K.; Tu Z.; Lei J. T.; Gatza M. L.; Wilkerson M.; Perou C. M.; Yellapantula V.; Huang K. L.; Lin C.; McLellan M. D.; Yan P.; Davies S. R.; Townsend R. R.; Skates S. J.; Wang J.; Zhang B.; Kinsinger C. R.; Mesri M.; Rodriguez H.; Ding L.; Paulovich A. G.; Fenyo D.; Ellis M. J.; Carr S. A.; Nci C. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534 (7605), 55–62. 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumeliotis T. I.; Williams S. P.; Goncalves E.; Alsinet C.; Del Castillo Velasco-Herrera M.; Aben N.; Ghavidel F. Z.; Michaut M.; Schubert M.; Price S.; Wright J. C.; Yu L.; Yang M.; Dienstmann R.; Guinney J.; Beltrao P.; Brazma A.; Pardo M.; Stegle O.; Adams D. J.; Wessels L.; Saez-Rodriguez J.; McDermott U.; Choudhary J. S. Genomic Determinants of Protein Abundance Variation in Colorectal Cancer Cells. Cell Rep. 2017, 20 (9), 2201–2214. 10.1016/j.celrep.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapek J. D. Jr.; Greninger P.; Morris R.; Amzallag A.; Pruteanu-Malinici I.; Benes C. H.; Haas W. Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat. Biotechnol. 2017, 35 (10), 983–989. 10.1038/nbt.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svinkina T.; Gu H.; Silva J. C.; Mertins P.; Qiao J.; Fereshetian S.; Jaffe J. D.; Kuhn E.; Udeshi N. D.; Carr S. A. Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyl-lysine Antibodies and an Optimized Proteomic Workflow. Mol. Cell. Proteomics 2015, 14 (9), 2429–40. 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C. M.; Isasa M.; Ordureau A.; Prado M. A.; Beausoleil S. A.; Jedrychowski M. P.; Finley D. J.; Harper J. W.; Gygi S. P. Highly Multiplexed Quantitative Mass Spectrometry Analysis of Ubiquitylomes. Cell Syst 2016, 3 (4), 395–403. 10.1016/j.cels.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P.; Udeshi N. D.; Clauser K. R.; Mani D. R.; Patel J.; Ong S. E.; Jaffe J. D.; Carr S. A. iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol. Cell. Proteomics 2012, 11 (6), M111.014423. 10.1074/mcp.M111.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrebe A.; von Stechow L.; Bekker-Jensen D. B.; Weinert B. T.; Kelstrup C. D.; Olsen J. V. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat. Commun. 2018, 9 (1), 1045. 10.1038/s41467-018-03309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong P. K.; Gan C. S.; Pham T. K.; Wright P. C. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: Implication of multiple injections. J. Proteome Res. 2006, 5 (5), 1232–40. 10.1021/pr060018u. [DOI] [PubMed] [Google Scholar]
- Ow S. Y.; Salim M.; Noirel J.; Evans C.; Rehman I.; Wright P. C. iTRAQ underestimation in simple and complex mixtures: ″the good, the bad and the ugly″. J. Proteome Res. 2009, 8 (11), 5347–55. 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- Christoforou A.; Mulvey C. M.; Breckels L. M.; Geladaki A.; Hurrell T.; Hayward P. C.; Naake T.; Gatto L.; Viner R.; Martinez Arias A.; Lilley K. S. A draft map of the mouse pluripotent stem cell spatial proteome. Nat. Commun. 2016, 7, 8992. 10.1038/ncomms9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y.; Markham K.; Chen F.; Weerasekera R.; Watts J.; Horne P.; Wakutani Y.; Bagshaw R.; Mathews P. M.; Fraser P. E.; Westaway D.; St George-Hyslop P.; Schmitt-Ulms G. The in vivo brain interactome of the amyloid precursor protein. Mol. Cell. Proteomics 2008, 7 (1), 15–34. 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- Papachristou E. K.; Kishore K.; Holding A. N.; Harvey K.; Roumeliotis T. I.; Chilamakuri C. S. R.; Omarjee S.; Chia K. M.; Swarbrick A.; Lim E.; Markowetz F.; Eldridge M.; Siersbaek R.; D’Santos C. S.; Carroll J. S. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat. Commun. 2018, 9 (1), 2311. 10.1038/s41467-018-04619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.; Candille S. I.; Choi Y.; Xie D.; Jiang L.; Li-Pook-Than J.; Tang H.; Snyder M. Variation and genetic control of protein abundance in humans. Nature 2013, 499 (7456), 79–82. 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow S. Y.; Salim M.; Noirel J.; Evans C.; Wright P. C. Minimising iTRAQ ratio compression through understanding LC–MS elution dependence and high-resolution HILIC fractionation. Proteomics 2011, 11 (11), 2341–6. 10.1002/pmic.201000752. [DOI] [PubMed] [Google Scholar]
- Savitski M. M.; Sweetman G.; Askenazi M.; Marto J. A.; Lang M.; Zinn N.; Bantscheff M. Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on Orbitrap-type mass spectrometers. Anal. Chem. 2011, 83 (23), 8959–67. 10.1021/ac201760x. [DOI] [PubMed] [Google Scholar]
- Ting L.; Rad R.; Gygi S. P.; Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 2011, 8 (11), 937–40. 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister G. C.; Nusinow D. P.; Jedrychowski M. P.; Wuhr M.; Huttlin E. L.; Erickson B. K.; Rad R.; Haas W.; Gygi S. P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014, 86 (14), 7150–8. 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger C. D.; Lee M. V.; Hebert A. S.; McAlister G. C.; Phanstiel D. H.; Westphall M. S.; Coon J. J. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Methods 2011, 8 (11), 933–5. 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhr M.; Haas W.; McAlister G. C.; Peshkin L.; Rad R.; Kirschner M. W.; Gygi S. P. Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Anal. Chem. 2012, 84 (21), 9214–21. 10.1021/ac301962s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shliaha P. V.; Jukes-Jones R.; Christoforou A.; Fox J.; Hughes C.; Langridge J.; Cain K.; Lilley K. S. Additional precursor purification in isobaric mass tagging experiments by traveling wave ion mobility separation (TWIMS). J. Proteome Res. 2014, 13 (7), 3360–9. 10.1021/pr500220g. [DOI] [PubMed] [Google Scholar]
- Niu M.; Cho J. H.; Kodali K.; Pagala V.; High A. A.; Wang H.; Wu Z.; Li Y.; Bi W.; Zhang H.; Wang X.; Zou W.; Peng J. Extensive Peptide Fractionation and y1 Ion-Based Interference Detection Method for Enabling Accurate Quantification by Isobaric Labeling and Mass Spectrometry. Anal. Chem. 2017, 89 (5), 2956–2963. 10.1021/acs.analchem.6b04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J. A.; Csordas A.; Del-Toro N.; Dianes J. A.; Griss J.; Lavidas I.; Mayer G.; Perez-Riverol Y.; Reisinger F.; Ternent T.; Xu Q. W.; Wang R.; Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44 (22), 11033. 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J. J.; O’Connell J. D.; Paulo J. A.; Thakurta S.; Rose C. M.; Weekes M. P.; Huttlin E. L.; Gygi S. P. Compositional Proteomics: Effects of Spatial Constraints on Protein Quantification Utilizing Isobaric Tags. J. Proteome Res. 2018, 17 (1), 590–599. 10.1021/acs.jproteome.7b00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert A. S.; Prasad S.; Belford M. W.; Bailey D. J.; McAlister G. C.; Abbatiello S. E.; Huguet R.; Wouters E. R.; Dunyach J. J.; Brademan D. R.; Westphall M. S.; Coon J. J. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Anal. Chem. 2018, 90 (15), 9529–9537. 10.1021/acs.analchem.8b02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.