Abstract
C-type lectins are a diverse group of proteins involved in many human physiological and pathological processes. Most C-type lectins are glycan-binding proteins, some of which are pivotal for innate immune responses against pathogens. Other C-type lectins, such as the macrophage galactose-type lectin (MGL), have been shown to induce immunosuppressive responses upon the recognition of aberrant glycosylation on cancer cells. MGL is known to recognize terminal N-acetylgalactosamine (GalNAc), such as the Tn antigen, which is commonly found on malignant cells. Even though this glycan specificity of MGL is well described, there is a lack of understanding of the actual glycoproteins that bind MGL. We present a glycoproteomic workflow for the identification of MGL-binding proteins, which we applied to study MGL ligands on the human Jurkat leukemia cell line. In addition to the known MGL ligands and Tn antigen-carrying proteins CD43 and CD45 on these cells, we have identified a set of novel cell-surface ligands for MGL. Importantly, for several of these, O-glycosylation has hitherto not been described. Altogether, our data provide new insight into the identification and structure of novel MGL ligands that presumably act as modulatory molecules in cancer immune responses.
Keywords: glycoproteomics, lectin, Tn antigen, leukemia, O-glycosylation
Introduction
C-type lectins belong to a family of glycan-binding proteins (GBPs) that interact with their ligands in a Ca2+-dependent manner through a carbohydrate recognition domain (CRD).1,2 They exist as monomers or can oligomerize to enhance the avidity by multivalent glycan interactions. Their activation on immune cells, such as dendritic cells (DCs), triggers different immune responses depending on the crosstalk with other receptors, the ligand/carbohydrate-specific signaling, and the cell subset they are expressed in.3 Within the CLR family, the macrophage galactose-type lectin (MGL) is expressed on tolerogenic DCs and macrophages4 and has the capability to alter DC and macrophage phenotypes.5
MGL is the only lectin that exclusively binds terminal GalNAc residues, including three well-known tumor-associated glycan epitopes:6 the sialylated and nonsialylated Thomsen-nouvelle (Tn) antigens (GalNAcα1-Ser/Thr) and LacdiNAc (GalNAcβ1-4GlcNAc).4 In normal cells, the Tn antigen is usually elongated with other carbohydrate residues, for example, a galactose, to form the core 1 T antigen. This step is mediated by the enzyme T-synthase with the assistance of its chaperone Cosmc. Mutations in Cosmc are responsible for the abortive elongation of O-glycans and higher expression of the Tn antigen,7 as observed in Tn syndrome patients.8 High levels of Tn antigen have also been found in multiple human cancer types such as colon, cervix, stomach, ovary, and breast.9 Moreover, higher levels of this surface-truncated glycan were found in cancer cells with high metastatic behavior and consequently poor prognosis for cancer patients.8 Interestingly, the interaction of MGL with aberrant glycosylation on the cell surface of cancer cells is associated with the activation of immunosuppressive responses, immune tolerance,10 and poor survival of stage-III colorectal cancer patients,11 suggesting a role of MGL in mediating cancer progression.
Notwithstanding our current knowledge on the role of MGL and the glycans it binds, the proteins carrying the MGL ligands are hitherto largely unknown. Therefore, in this article, we sought to identify MGL-binding proteins in a T-cell leukemia model cell line, Jurkat, which is known to have high levels of Tn antigen due to a single nucleotide deletion in Cosmc and has been used as a model system to study immune modulation mediated by MGL.1,12
Experimental Section
Cell Lines Culture and Lysis
Jurkat (provided by S. J. van Vliet, VUMC, Amsterdam, The Netherlands) cells were cultivated in RPMI-1640 medium containing 10% fetal bovine serum (FBS) (Invitrogen) and streptomycin/penicillin (Sigma-Aldrich) at 5% CO2 and 37 °C. Cells were harvested upon ∼70% confluency. A total of 2 × 107 cells were obtained, harvested, washed, and centrifuged at 1500 rpm to obtain cell pellets. Cell pellets were stored at −20 °C until use. Then, cells were lysed as described before12 for 20 min on ice in lysis buffer (10 mM triethanolamine pH 8.2, 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, and 1% (volume/volume) Triton X-100 containing EDTA-free protease inhibitor (Roche Diagnostics)). Protein concentration was quantified by BCA assay (BCA Protein Assay Kit, Pierce).
Antibodies and Reagent
Chimeric MGL-Fc was provided by S. J. van Vliet (VUMC, Amsterdam, The Netherlands). The following antibodies were used for immunoblotting: mouse IgG1 anti-human CD43 (eBio84-3C1, eBioscience) and goat anti-mouse immunoglobulins/HRP (Dako).
Pull-Down Assay and Immunoblot Analysis
MGL ligands were pulled down from 1 mg of protein extracts with 2 μg of chimeric MGL-Fc coupled to 50 μL of Dynabeads protein G (Invitrogen). The specific ligands were then eluted by the addition of 100 mM EDTA and concentrated under vacuum. The addition of 100 mM EDTA, prior to the addition of MGL-Fc, was used as a negative control. Captured products were separated by SDS-PAGE (NuPAGE 4–12% Bis-Tris protein gels, Thermo Fisher Scientific) and transferred to a PVDF membrane (Amersham Hyband P 0.45). Five % low-fat milk (Campina) in 0.1% phosphate-buffered saline with Tween 20 (PBS-T) was used to block the blots for 1 h. Immunoblotting was performed with specific antibodies, followed by secondary antibody peroxidase-conjugated goat anti-mouse (Dako). Immunodetection was done by enhanced chemiluminescence (ECL) using Clarity Western ECL substrate (Bio-Rad) and an Amersham Imager 600.
Mass Spectrometry
The samples obtained with the MGL pull-down assay were cleaned up by a short SDS-PAGE run (NuPAGE 4–12% Bis-Tris protein gels, Thermo Fisher Scientific). For MS analysis, in-gel trypsin digestion was performed using a Proteineer DP digestion robot (Bruker). Prior to digestion, proteins were first reduced and alkylated using dithiothreitol (10 mM) and iodoacetamide (50 mM), respectively.
Tryptic peptides were extracted from the gel slices, lyophilized, dissolved in solvent A (95/3/0.1 water/acetonitrile/formic acid (FA) v/v/v), and subsequently analyzed by online C18 nano-HPLC MS/MS with a system consisting of an Easy nLC 1000 gradient HPLC system (Thermo, Bremen, Germany) and a LUMOS mass spectrometer (Thermo). Fractions were injected onto a homemade precolumn (100 μm × 15 mm; Reprosil-Pur C18-AQ 3 μm, Dr. Maisch, Ammerbuch, Germany) and eluted via a homemade analytical nano-HPLC column (15 cm × 50 μm; Reprosil-Pur C18-AQ 3 μm). The gradient was run from 10 to 40% solvent B (20/80/0.1 water/acetonitrile/FA v/v/v) in 20 min. The nano-HPLC column was drawn to a tip of ∼5 μm and acted as the electrospray needle of the MS source. The LUMOS mass spectrometer was operated in data-dependent MS/MS (top-10 mode) with collision energy at 32 V and recording of the MS2 spectrum in the Orbitrap. In the master scan (MS1) the resolution was 120 000, and the scan range was m/z 400–1500 at an AGC target of 400 000 with maximum fill time of 50 ms. Dynamic exclusion after n = 1 with exclusion duration of 10 s was applied. Charge states 2–5 were included for MS2. For this, precursors were isolated with the quadrupole with an isolation width of 1.2 Da. The MS2 scan resolution was 30 000 with an AGC target of 50 000 with maximum fill time of 60 ms. During acquisition, a product ion trigger was set on the HexNAc oxonium ion at m/z 204.087. Upon the detection of the oxonium ion, three additional data-dependent MS2 scans of the same precursor were executed with higher-energy collisional dissociation (HCD) collision energies of 32, 37, and 41 V, respectively, at an AGC target of 500 000 with a maximum fill time of 200 ms. In addition, a collision-induced dissociation (CID) spectrum of the same precursor was recorded.
In a post-analysis process, raw data were analyzed with MaxQuant 1.5.1.2 using the Andromeda search engine and the Homo sapiens (2015) database. Modifications taken into account in the search were oxidation (M), acetylation (N-term), and HexNAc (ST) as variable and carbamidomethyl (C) as fixed. Trypsin was selected as the enzyme, allowing up to two miss cleavages. A maximum of 12 modifications per peptide was allowed. The mass tolerance used for precursor ions was 10 ppm, whereas it was 0.05 Da for fragment ions. Subsequently, only proteins that were either never found in the negative control experiment but were found in at least two of the MGL pull-downs or proteins that were identified in all three MGL pull-downs and identified a maximum of one time in the negative control samples were subsequently considered MGL-binding proteins. Subsequently, these MGL-binding proteins were further filtered based on data on the subcellular location from the UniProtKB database, thereby selecting only cell-surface membrane proteins. The surface location was subsequently manually curated by a literature search.
Alternatively, MS2 spectra containing the specific HexNAc oxonium ions at m/z 204.087 (HexNAc, [C8H14NO5]+) and 186.076 (HexNAc-H2O, [C8H12NO4]+) were selected and written to an MGF file (in-house software), which was subsequently submitted to Byonic version 2.13.2 (proteinmetrics.com) using default settings. The fixed modification was carbamidomethyl (Cys). Variable modifications set were oxidation (Met) and “N-glycan 309 mammalian no sodium” or “O-glycan 78 mammalian” databases as glycosylation parameters. Only glycopeptides with a Byonic score >200 were further selected.
Manual interpretation of spectra was done using Xcalibur (Thermo) for the spectrum visualization. The corresponding peptide fragment masses were calculated using Protein Prospector (prospector.ucsf.edu).
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE13 partner repository with the data set identifier PXD011307.
Results
Workflow of the MGL Pull-Down Assay
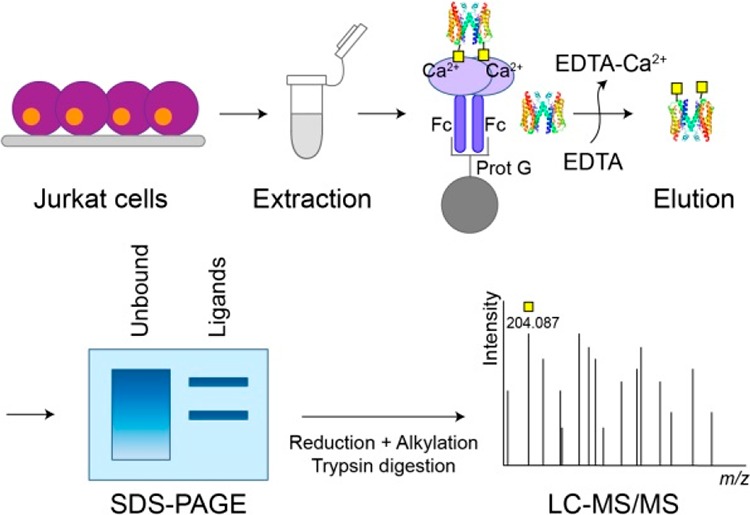
To identify novel binding partners of MGL on Jurkat T-cells, we developed a protocol where we used Fc-coupled MGL as a bait in pull-down assays (Figure 1A). MGL-Fc is a chimeric molecule formed by the extracellular domains of MGL fused to the human immunoglobulin G1 Fc tail,14 allowing binding to magnetic Protein G beads. Because the binding to the CRD of MGL is calcium-dependent, captured proteins were eluted using EDTA and subsequently analyzed by SDS-PAGE and processed for mass-spectrometry-based protein identification. CD43 and CD45 are hitherto the only described MGL-binding proteins on Jurkat cells.12 Therefore, to test the effectiveness of our workflow, we first determined the capturing of CD43 by Western blot. This clearly showed that CD43 was captured by MGL-Fc and could be eluted with EDTA (Figure 1B). Likewise, the addition of EDTA during the pull-down assay prevented the binding of CD43 to MGL (Figure 1B).
Figure 1.
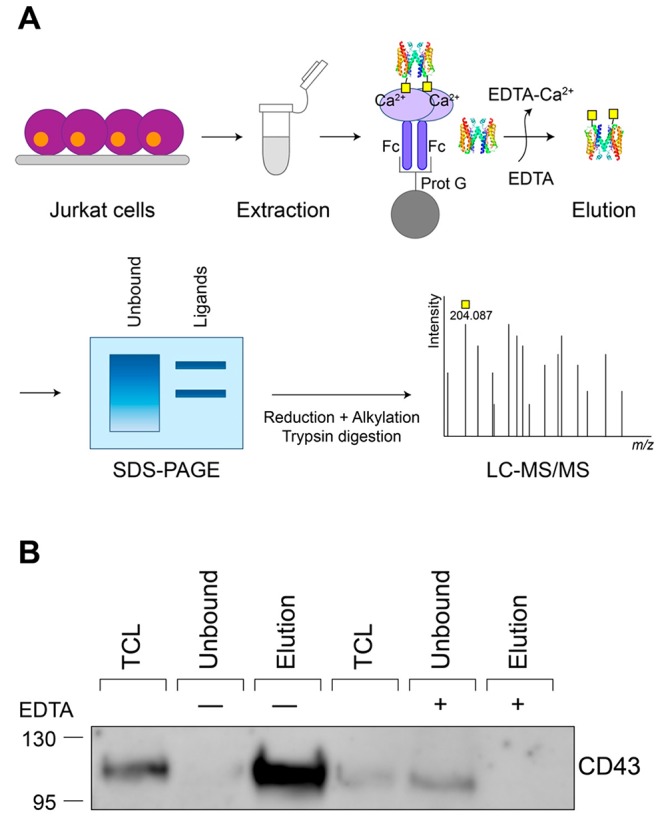
Schematic representation and validation of the experimental workflow for the identification of MGL ligands in Jurkat cells. (A) Pull-down workflow to capture MGL-binding proteins. As negative control, EDTA was added to the sample prior to incubation with MGL-Fc to prevent the binding to MGL. (B) Western blot analysis using monoclonal anti-CD43 staining of the unbound fraction and captured proteins (elution) from the MGL-Fc pull-down assay in the absence (−) and presence (+, negative control) of EDTA (100 mM). Fc: Fragment crystallizable region; Prot G: protein G; TCL: total cell lysate.
MS-Based Identification of MGL-Binding Proteins from Jurkat Cells
Next, we performed three biologically independent pull-down experiments with MGL-Fc and analyzed the bound proteins by LC–MS/MS following trypsin digestion. As a negative control, pull-down assays in the presence of EDTA were performed. Altogether, these experiments resulted in the identification of 775 proteins (data not shown), of which 540 were identified in at least two experiments. To filter for proteins specifically binding to MGL, we selected proteins that were not observed in the negative controls and at least two times in the pull-downs with MGL or proteins that were found in all three pull-downs with MGL and at most once in a negative control. This resulted in a list of 85 proteins (Table S1). The candidate MGL-binding proteins included intracellular, plasma membrane, and predicted secreted proteins. Because we used total cell lysates for the pull-down assays, some of these proteins may be physiologically less relevant. To filter for cell-surface proteins, which may be expected to be in direct contact with MGL, we next selected only those proteins that were annotated as cell-surface proteins in the UniProtKB database, resulting in a final list of 17 MGL-binding cell-surface proteins (Table 1). Importantly, these proteins were not found in the negative control samples (see also Table S1), making them strong candidate MGL-binding proteins. As expected, CD45 (PTPRC) and CD43 (SPN) were the two main MGL-binding proteins identified based on the total number of peptide-spectrum matches (summed for the three biological replicates) for these two proteins, respectively.
Table 1. MGL-Binding Cell-Surface Proteins from Jurkat Cellsa.
| protein name | gene name | UniProt entry | mol. weight (kDa) | exp 1: #pep | exp 2: #pep | exp 3: #pep | sum MS/MS count | glycopeptide |
|---|---|---|---|---|---|---|---|---|
| receptor-type tyrosine-protein phosphatase C (CD45) | PTPRC | P08575 | 147 | 73 | 60 | 62 | 1125 | yes |
| leukosialin (CD43) | SPN | P16150 | 40 | 11 | 9 | 6 | 145 | yes |
| leucine-rich repeat-containing protein 8D | LRRC8D | Q7L1W4 | 98 | 21 | 22 | 10 | 73 | yes |
| podocalyxin | PODXL | O00592 | 59 | 7 | 8 | 6 | 66 | yes |
| leucine-rich repeat-containing protein 8A | LRRC8A | Q8IWT6 | 94 | 15 | 16 | 6 | 53 | no |
| protein EVI2B | EVI2B | P34910 | 49 | 7 | 10 | 4 | 35 | yes |
| dyslexia-associated protein KIAA0319-like protein | KIAA0319L | E7EN73 | 113 | 12 | 9 | 7 | 30 | no |
| leucine-rich repeat-containing protein 8C | LRRC8C | Q8TDW0 | 92 | 8 | 11 | 4 | 28 | no |
| Protein tyrosine phosphatase receptor type C-associated protein | PTPRCAP | Q14761 | 21 | 5 | 3 | 0 | 11 | no |
| semaphorin-4D | SEMA4D | Q92854 | 96 | 7 | 2 | 2 | 10 | yes |
| TREM-like transcript 2 protein | TREML2 | Q5T2D2 | 35 | 3 | 2 | 2 | 6 | yes |
| protein HEG homologue 1 | HEG1 | Q9ULI3 | 147 | 1 | 2 | 1 | 6 | no |
| lysosome-associated membrane glycoprotein 3 | LAMP3 | E7ETP9 | 42 | 3 | 1 | 1 | 5 | yes |
| chloride intracellular channel protein 1 | CLIC1 | O00299 | 27 | 2 | 3 | 0 | 5 | no |
| low-density lipoprotein receptor | LDLR | H0YM92 | 34 | 2 | 1 | 1 | 4 | no |
| leucine-rich repeat-containing protein 8B | LRRC8B | Q6P9F7 | 92 | 1 | 3 | 0 | 4 | no |
| tumor necrosis factor receptor superfamily member 8 | TNFRSF8 | P28908 | 64 | 2 | 3 | 0 | 3 | yes |
Using total cell lysates from Jurkat cells, three independent pull-down assays with MGL-Fc were performed, and captured proteins were identified by LC–MS/MS. Shown are the MGL-binding cell-surface proteins; for a full list, see Table S1. Each protein is represented with the number of unique peptides identified in the three pull-down assays and the summed spectral count from all three assays. mol. weight: molecular weight; exp: experiment number; #pep: number of unique peptides per protein; sum MS/MS count: summed spectral count from the three assays; and glycopeptide: O-glycopeptide(s) of the respective protein found (yes) or not (no).
Characterization of Glycopeptides
MGL is known to bind terminal residues of N-acetylgalactosamine. To validate the specificity of the interaction of the proteins listed in Table 1 with MGL, we next evaluated our data for the presence of glycopeptides carrying this motif. During our LC–MS/MS analyses of the tryptic digests, we used a method that triggered additional MS/MS acquisitions once the MS/MS spectrum showed the presence of the characteristic HexNAc oxonium ion at m/z 204.087 ([C8H14NO5]+). First of all, for these additional MS/MS events, we used a higher number of ions for fragmentation, resulting in a higher signal-to-noise for the fragment ions. Moreover, to improve the confidence of the glycopeptide identifications, we collected Orbitrap spectra using different HCD collision energies in addition to an ion trap CID spectrum.
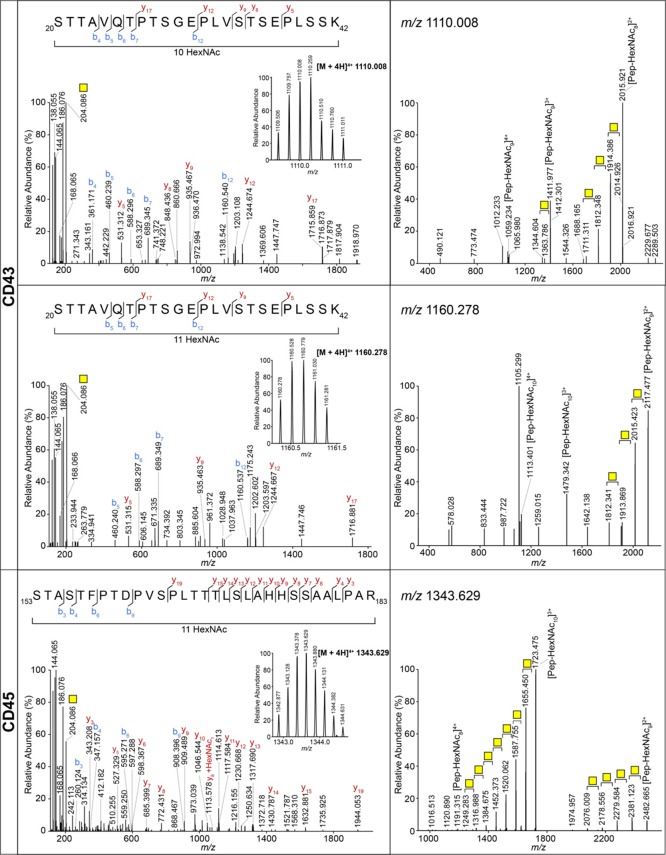
Next, we searched for glycopeptides carrying the Tn antigen following database searches using MaxQuant and Byonic. With these approaches, we could confirm the presence of the Tn antigen on CD45 and CD43 (Table 1) with peptides containing a maximum of four HexNAcs (Table 2). However, it is known that certain regions in these two proteins contain a high density of O-glycans, which may have been missed by the automatic data analysis due to limitations of the particular software used, for example, due to the high number of occupied glycosylation sites in one individual peptide. Indeed, upon manual inspection of our data, we observed peptides containing up to 11 HexNAcs (Figure 2). Of note, one peptide with an extended O-glycan on the CD45 tryptic peptide LNPTPGSNAISDVPGER (HexNAc2Hex1NeuAc1) was found using Byonic, which, considering the Cosmc mutation in Jurkat cells, could correspond to a core 6 O-glycan, a specific MGL-binder (GlcNAcβ1-6GalNAcαSer/Thr).14
Table 2. Identified O-Glycopeptides from MGL-Binding Cell-Surface Proteins in Jurkat Cellsa.
| protein name | gene name | UniProt entry | glycopeptide sequence | MaxQuant # HexNAc | byonic # HexNAc | manual # HexNAc |
|---|---|---|---|---|---|---|
| lysosome-associated membrane glycoprotein 3 | LAMP3 | E7ETP9 | QAPHQTLAAR | 1 | 1 | 1 |
| DYSQPTAAATVQDIK | 3 | 3 | ||||
| leukosialin (CD43) | SPN | P16150 | MYTTSITSDPK | 3, 4 | 3 | 3, 4, 5 |
| MOxYTTSITSDPK | 3, 4 | |||||
| STTAVQTPTSGEPLVSTSEPLSSK | 9, 10, 11 | |||||
| podocalyxin | PODXL | O00592 | TPSPTVAHESNWAK | 2 | 2, 3 | 2, 3 |
| ANEILASVK | 1 | 1 | ||||
| CEDLETQTQSEK | 1 | 1 | ||||
| protein EVI2B | EVI2B | P34910 | QLPSAR | 1 | 1 | |
| STPGFILDTTSNK | 4 | 4, 5 | ||||
| STPGFILDTTSNKQTPQK | 4, 5 | |||||
| QITVHNPSTQPTSTVKNSPR | 7 | |||||
| P-selectin glycoprotein ligand 1 | SELPLG | Q14242 | GLFIPFSVSSVTHK | 3 | 3 | 3 |
| receptor-type tyrosine-protein phosphatase C (CD45) | PTPRC | P08575 | LNPTPGSNAISDVPGER | 2 | 1, 2, 3 | 2, 3 |
| LNPTPGSNAISDVPGER | HexNAc(2) | |||||
| Hex(1)Neu | ||||||
| Ac(1) | ||||||
| STASTFPTDPVSPLTTTLSLAHHSSAALPAR | 10, 11 | |||||
| NGSAAMOxCHFTTK | 1 | 1 | ||||
| semaphorin-4D | SEMA4D | Q92854 | VVPKPVVAPTLSVVQTEGSR | 2 | 1, 2 | 1, 2 |
| IVINTVPQLHSEK | 1 | 1, 2 | 1,2 | |||
| TREM-like transcript 2 protein | TREML2 | Q5T2D2 | LLASTRPASK | 3 | 2, 3 | 3 |
| NIPFTHLDNILK | 2 | 2 | ||||
| tumor necrosis factor receptor superfamily member 8 | TNFRSF8 | P28908 | LAQEAASKLTR | 2 | 2 | 2 |
| transferrin receptor protein 1 | TFRC | P02786 | LAGTESPVREEPGEDFPAAR | 1 | 1 | |
| voltage-dependent calcium channel subunit alpha-2/delta-4 | CACNA2D4 | Q7Z3S7 | YKDVESSLK | 2 | 2 | |
| glycosyltransferase 8 domain-containing protein 1 | GLT8D1 | Q68CQ7 | YTEISNIK | 2 | 2 | |
| volume-regulated anion channel subunit LRRC8D | LRRC8D | Q7L1W4 | TDFALPNQEAK | 1 | 1 |
O-glycosylated peptides were annotated using three different approaches (Maxquant, Byonic, manual annotation). # HexNAc: number of N-acetylhexosamine carried by each glycopeptide; Hex: hexose; NeuAc: N-acetylneuraminic acid; and Ox: oxidation of methionine.
Figure 2.
Highly O-glycosylated peptides of CD43 and CD45. MS/MS spectra of glycopeptides from CD43 carrying 10 and 11 GalNAcs and CD45 carrying 11 GalNAcs. Left panels: HCD MS/MS spectra. b and y ions represent fragments without the GalNAc, unless indicated otherwise. The inset shows the MS spectrum of the corresponding precursor with charge state and m/z value. Right panels: CID MS/MS spectra. Yellow square: GalNAc.
In addition to CD43 and CD45, our database searches confirmed the presence of the Tn antigen on another 11 proteins (Table 2). We confirmed the Tn antigen on all of these peptides by manual interpretation of the data. As an example, two glycopeptides from EVI2B and TREML2, respectively, are shown in Figure 3. Between the peptides presented in Table 2, one peptide (GLFIPFSVSSVTHK) with three HexNAcs was identified from P-selectin glycoprotein ligand 1 (SELPLG), which was not identified as a specific binder in the proteomics data (Table 1). Inspection of the data showed that, unexpectedly, it was filtered out due to the fact that normal tryptic peptides of SELPLG were observed in only one of the three MGL pull-downs.
Figure 3.
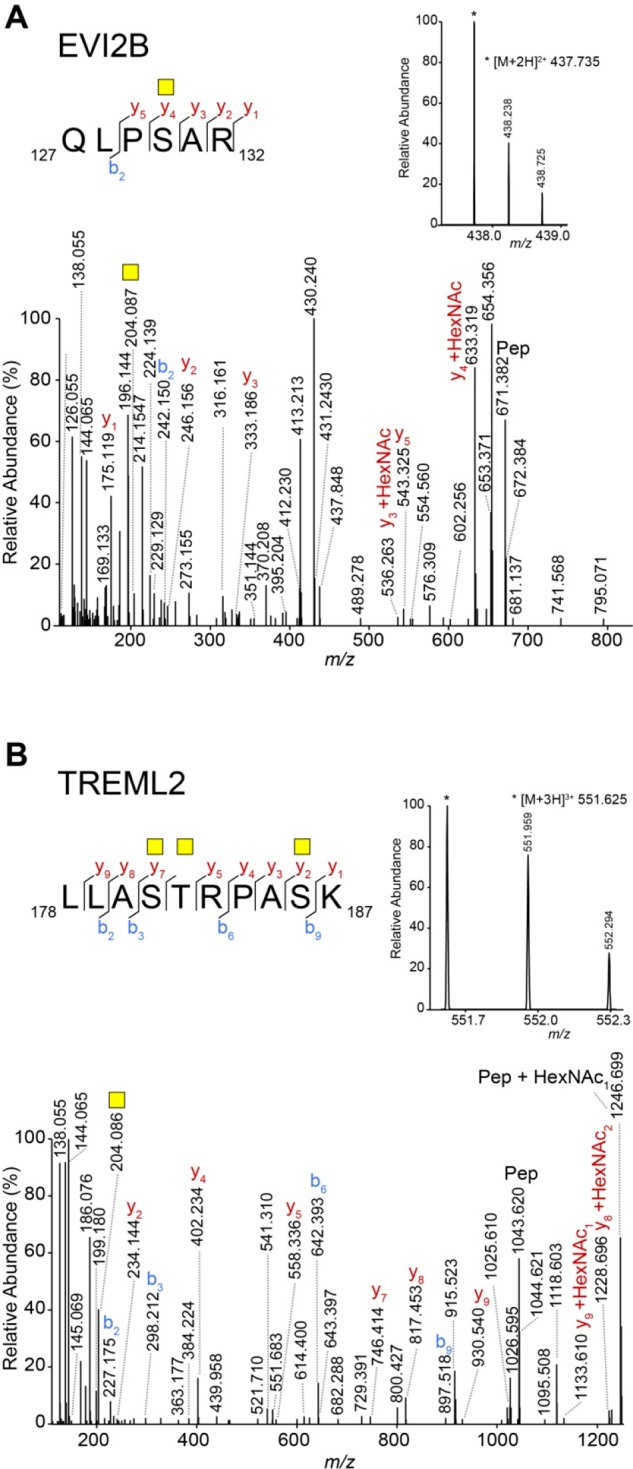
Tn-bearing O-glycosylated peptides of EVI2B and TREML2. (A) Manually assigned HCD MS/MS spectrum of the tryptic peptide QLPSAR from EVI2B carrying one GalNAc. The inset shows the MS spectrum of the precursor ion at m/z 437.735 [M + 2H]2+. (B) Manually assigned HCD MS/MS spectrum of the tryptic peptide LAQEAASKLTR from TREML2 carrying three GalNAcs. The inset shows the MS spectrum of the precursor ion at m/z 531.950 [M + 3H]3+.Yellow square: GalNAc. b and y ions represent fragments without the GalNAc, unless otherwise indicated.
Terminal N-acetylgalactosamines can also be part of N-glycans, for example, as part of the LacdiNAc epitope. Hence we also searched our data for the presence of N-glycopeptides using Byonic. Only for CD45 presented in Table 1 we could identify N-glycopeptides (on N234, 278, 337, 380, 421, and 470, respectively), but none of these appear to contain a terminal N-acetylgalactosamine, as judged on the basis of the glycan compositions as well as the absence of the LacdiNAc (GalNAcβ1-4GlcNAcβ1) marker ion at m/z 407.166 (Table S2).
Discussion
We developed a workflow for the identification of MGL ligands, which resulted in the identification of 17 cell-surface proteins of Jurkat cells that bind to MGL. For most of these proteins, we confirmed the specificity of the interaction with MGL through the identification of O-glycopeptides carrying the Tn antigen.
The glycopeptide, mediating the interaction with MGL, could not be identified for all MGL ligands. These peptides may have been missed due to insufficient sensitivity or low MS/MS spectrum quality, which would hamper identification by both search algorithms as well as manual inspection. Although in the past years, we and others have made considerable progress in the mass-spectrometry-based identification of glycopeptides,15−20 several issues related to their fragmentation behavior still often impede straightforward (data) analysis, even though the Tn antigen represents a relatively simple post-translational modification. We combined multiple strategies for data acquisition and analysis to maximize our glycopeptide identification. Other methods, such as electron-transfer dissociation (ETD), have also been valuable tools for the identification of glycopeptides,16 especially in cases where the localization of the GalNAc(s) was ambiguous. However, for this study, we were primarily interested in identifying MGL ligands, and not necessarily the site-specific glycan assignment. Moreover, for several peptides, we had full occupancy of all serine and threonine residues present in the identified glycopeptide.
From the 17 potential MGL-binding proteins (Table 1), some could have been obtained through an interaction with a genuine MGL ligand. For example, protein tyrosine phosphatase receptor type C-associated protein (PTPRCAP), for which we could not find the corresponding glycopeptide, was captured during the MGL pull-down probably because it is a binding partner of CD45.21 Similarly, for only one of the four LRRC8 subunits (LRRC8D) that we identified could a specific O-glycopeptide be found. Because it is known that the LRRC8 subunits form heterodimers,22,23 it is conceivable that the whole LRRC8 complex was captured.
As expected, CD43 and CD45 were among the top proteins carrying the Tn antigen in Jurkat cells. A recent study presented a workflow where a combination of enzymatic and chemical methods was used to selectively tag terminal GalNAc, and not GlcNAc residues,24 allowing the enrichment of the corresponding glycopeptides. To study proteins carrying the Tn antigen, this method was applied to total cell lysates of Jurkat cells. As a result, a total of 97 proteins harboring the Tn antigen were identified, of which 27 were consistently found in all three independent biological replicates. From the total of 97 proteins, 11 were transmembrane signaling molecules, including CD45, Semaphorin-4D, and TNFRSF8, which were also identified in our study. Some other Tn-antigen-bearing glycoproteins were not found in our experiments. The observed differences may relate to the different experimental approaches because several Tn-bearing glycoproteins were clearly identified in our experiments, such as, CD43, EVI2B, and TREML2, but not in the above-mentioned study.24 On the contrary, it could be due to the fact that for MGL binding the presence of Tn is necessary but not sufficient.12,25 For example, all CD45 isoforms (CD45ABC/AB/BC/B) except for one (CD45RO) bind MGL. Unlike the others, which are highly glycosylated, CD45RO has only two O-linked glycan epitopes.
Although it is known that MGL can have immune-modulatory activities, the specific proteins involved in the cellular response elicited by MGL are largely unknown. Tolerogenic antigen presenting cells (APCs) express high levels of MGL on the cell surface, and the binding of MGL to CD45 suppresses TCR-mediated T-cell activation. This results in an anti-inflammatory response characterized by the reduced production of pro-inflammatory cytokines and lower proliferation of T-cells and induction of cell death in Jurkat.12
For the new MGL ligands identified in this study, the outcome of the interaction with MGL remains to be determined. Semaphorin 4D, also known as CD100, is a homodimeric transmembrane protein of 150 kDa that is highly expressed in secondary lymphoid organs and constitutively on naïve T-cells. It binds its ligand CD72, a C-type lectin expressed on the surface of APCs, such as B cells and DCs,26 but also macrophages and some subpopulation of T-cells.27 Our data show that, at least in tumor cells with aberrant glycosylation, Semaphorin 4D also binds to MGL.
For several of the newly identified MGL ligands, literature provides limited information about their function. For example, although KIAA0319L has been demonstrated to be an essential receptor for adeno-associated virus infection, the cellular function, especially in immune responses, is unknown. However, our data support previous findings that suggested high levels of mucin-type O-glycosylation on KIAA0319L.28 Our data also provide evidence of O-glycosylation on proteins that hitherto were unknown to be O-glycosylated, such as EVI2B and TREML2. Although the cellular function of these proteins is largely unknown, it has been demonstrated that EVI2B is the target for the transcription factor CCAAT/enhancer-binding protein alpha (C/EBPα),29 and TREML2 binding by CD276 leads to enhanced T-cell responses.30 Altogether, our data warrant further exploration of the functional implications of the interaction of MGL with the newly identified ligands. Notwithstanding the importance of this, some responses elicited by MGL might also be the result of the interaction of MGL with glycolipids,14 which was not the topic of our study.
The high level of Tn antigen is not restricted to leukemias but is frequently observed in other tumors as well. The best characterized ligand carrying Tn or sialyl-Tn (sTn, Neu5Acα2,6-GalNAc-O-Ser/Thr) in epithelial cells is the glycoprotein MUC1. MUC1-derived glycopeptides bind to MGL on DCs and induce the activation of the extracellular signal-regulated kinases 1 and 2 (ERK1,2) and the nuclear factor-κB (NF-κB) pathways.31 In another study, it was demonstrated that these pathways are crucial for IL-10 production, which regulates the DC maturation phenotype.32
The higher level of Tn in other cancers is not necessarily due to Cosmc mutations, as in the Jurkat T-cell used in our study, but may also be the result of other genetic alterations. For example, in colorectal cancer, higher levels of several GALNTs have recently been linked to the BRAFV600E mutation.33 A positive correlation between this oncogenic mutation and MGL ligands on tumor cells was previously identified.11 Hence, it will be interesting to study the proteins carrying MGL ligands in these tumor cells as well.
In conclusion, here we provide an optimized method to capture MGL-binding proteins followed by glycoproteomic analysis. The application of this procedure on the Jurkat cell line provides important novel insights into previously unknown MGL ligands. However, further investigations should evaluate the functional immune responses triggered by MGL-specific recognition of those proteins, as has been previously reported for the already known interaction partner CD45 by van Vliet et al.12 This will provide a deeper understanding of the MGL involvement in cancer progression and the glycan-specific immune responses mediated by this lectin.
Acknowledgments
This work was supported by the European Commissions Horizon 2020 programme “GlyCoCan” project, grant number 676421, and by the research programme Investment Grant NWO Medium with project number 91116004, which is (partially) financed by ZonMw.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00796.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE13 partner repository with the data set identifier PXD011307.
Supplementary Material
References
- Brown G. D.; Willment J. A.; Whitehead L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18 (6), 374–389. 10.1038/s41577-018-0004-8. [DOI] [PubMed] [Google Scholar]
- Cummings R. D.; McEver R. P.. C-Type Lectins. In Essentials of Glycobiology; Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., Seeberger P. H., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2015; pp 435–452. [PubMed] [Google Scholar]
- Geijtenbeek T. B.; Gringhuis S. I. Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 2009, 9 (7), 465–79. 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kooyk Y.; Ilarregui J. M.; van Vliet S. J. Novel insights into the immunomodulatory role of the dendritic cell and macrophage-expressed C-type lectin MGL. Immunobiology 2015, 220 (2), 185–92. 10.1016/j.imbio.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Zizzari I. G.; Napoletano C.; Battisti F.; Rahimi H.; Caponnetto S.; Pierelli L.; Nuti M.; Rughetti A. MGL Receptor and Immunity: When the Ligand Can Make the Difference. J. Immunol. Res. 2015, 2015, 1. 10.1155/2015/450695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis C. A.; Osorio H.; Silva L.; Gomes C.; David L. Alterations in glycosylation as biomarkers for cancer detection. J. Clin. Pathol. 2010, 63 (4), 322–9. 10.1136/jcp.2009.071035. [DOI] [PubMed] [Google Scholar]
- Ju T.; Lanneau G. S.; Gautam T.; Wang Y.; Xia B.; Stowell S. R.; Willard M. T.; Wang W.; Xia J. Y.; Zuna R. E.; Laszik Z.; Benbrook D. M.; Hanigan M. H.; Cummings R. D. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008, 68 (6), 1636–46. 10.1158/0008-5472.CAN-07-2345. [DOI] [PubMed] [Google Scholar]
- Ju T.; Otto V. I.; Cummings R. D. The Tn antigen-structural simplicity and biological complexity. Angew. Chem., Int. Ed. 2011, 50 (8), 1770–91. 10.1002/anie.201002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C.; Zhao H.; Wang Y.; Cai H.; Xiao Y.; Zeng Y.; Chen H. Tumor-associated antigens: Tn antigen, sTn antigen, and T antigen. HLA 2016, 88 (6), 275–286. 10.1111/tan.12900. [DOI] [PubMed] [Google Scholar]
- Rabinovich G. A.; Croci D. O. Regulatory circuits mediated by lectin-glycan interactions in autoimmunity and cancer. Immunity 2012, 36 (3), 322–35. 10.1016/j.immuni.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Lenos K.; Goos J. A.; Vuist I. M.; den Uil S. H.; Delis-van Diemen P. M.; Belt E. J.; Stockmann H. B.; Bril H.; de Wit M.; Carvalho B.; Giblett S.; Pritchard C. A.; Meijer G. A.; van Kooyk Y.; Fijneman R. J.; van Vliet S. J. MGL ligand expression is correlated to BRAF mutation and associated with poor survival of stage III colon cancer patients. Oncotarget 2015, 6 (28), 26278–90. 10.18632/oncotarget.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet S. J.; Gringhuis S. I.; Geijtenbeek T. B.; van Kooyk Y. Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 2006, 7 (11), 1200–8. 10.1038/ni1390. [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A.; Csordas A.; Del-Toro N.; Dianes J. A.; Griss J.; Lavidas I.; Mayer G.; Perez-Riverol Y.; Reisinger F.; Ternent T.; Xu Q. W.; Wang R.; Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44 (22), 11033. 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet S. J.; van Liempt E.; Saeland E.; Aarnoudse C. A.; Appelmelk B.; Irimura T.; Geijtenbeek T. B.; Blixt O.; Alvarez R.; van Die I.; van Kooyk Y. Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int. Immunol. 2005, 17 (5), 661–9. 10.1093/intimm/dxh246. [DOI] [PubMed] [Google Scholar]
- Hinneburg H.; Stavenhagen K.; Schweiger-Hufnagel U.; Pengelley S.; Jabs W.; Seeberger P. H.; Silva D. V.; Wuhrer M.; Kolarich D. The Art of Destruction: Optimizing Collision Energies in Quadrupole-Time of Flight (Q-TOF) Instruments for Glycopeptide-Based Glycoproteomics. J. Am. Soc. Mass Spectrom. 2016, 27 (3), 507–19. 10.1007/s13361-015-1308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen K.; Hinneburg H.; Kolarich D.; Wuhrer M. Site-Specific N- and O-Glycopeptide Analysis Using an Integrated C18-PGC-LC-ESI-QTOF-MS/MS Approach. Methods Mol. Biol. 2017, 1503, 109–119. 10.1007/978-1-4939-6493-2_9. [DOI] [PubMed] [Google Scholar]
- Stavenhagen K.; Kayili H. M.; Holst S.; Koeleman C. A. M.; Engel R.; Wouters D.; Zeerleder S.; Salih B.; Wuhrer M. N- and O-glycosylation Analysis of Human C1-inhibitor Reveals Extensive Mucin-type O-Glycosylation. Mol. Cell. Proteomics 2018, 17 (6), 1225–1238. 10.1074/mcp.RA117.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.; Cheng K.; Lo C. Y.; Li J.; Qu J.; Neelamegham S. A Comprehensive, Open-source Platform for Mass Spectrometry-based Glycoproteomics Data Analysis. Mol. Cell. Proteomics 2017, 16 (11), 2032–2047. 10.1074/mcp.M117.068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlmann J.; Taubenschmid J.; Wenzel D.; Gattinger A.; Durnberger G.; Dusberger F.; Elling U.; Mach L.; Mechtler K.; Penninger J. M. Comparative glycoproteomics of stem cells identifies new players in ricin toxicity. Nature 2017, 549 (7673), 538–542. 10.1038/nature24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi H. J.; Jorgensen A.; Schjoldager K. T.; Halim A.; Dworkin L. A.; Steentoft C.; Wandall H. H.; Clausen H.; Vakhrushev S. Y. GlycoDomainViewer: a bioinformatics tool for contextual exploration of glycoproteomes. Glycobiology 2018, 28 (3), 131–136. 10.1093/glycob/cwx104. [DOI] [PubMed] [Google Scholar]
- Leitenberg D.; Falahati R.; Lu D. D.; Takeda A. CD45-associated protein promotes the response of primary CD4 T cells to low-potency T-cell receptor (TCR) stimulation and facilitates CD45 association with CD3/TCR and lck. Immunology 2007, 121 (4), 545–54. 10.1111/j.1365-2567.2007.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradogna A.; Gavazzo P.; Boccaccio A.; Pusch M. Subunit-dependent oxidative stress sensitivity of LRRC8 volume-regulated anion channels. J. Physiol. 2017, 595 (21), 6719–6733. 10.1113/JP274795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss F. K.; Ullrich F.; Munch J.; Lazarow K.; Lutter D.; Mah N.; Andrade-Navarro M. A.; von Kries J. P.; Stauber T.; Jentsch T. J. Identification of LRRC8 heteromers as an essential component of the volume-regulated anion channel VRAC. Science 2014, 344 (6184), 634–8. 10.1126/science.1252826. [DOI] [PubMed] [Google Scholar]
- Zheng J.; Xiao H.; Wu R. Specific Identification of Glycoproteins Bearing the Tn Antigen in Human Cells. Angew. Chem., Int. Ed. 2017, 56 (25), 7107–7111. 10.1002/anie.201702191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelo F.; Garcia-Martin F.; Matsushita T.; Sardinha J.; Coelho H.; Oude-Vrielink A.; Koller C.; Andre S.; Cabrita E. J.; Gabius H. J.; Nishimura S.; Jimenez-Barbero J.; Canada F. J. Delineating binding modes of Gal/GalNAc and structural elements of the molecular recognition of tumor-associated mucin glycopeptides by the human macrophage galactose-type lectin. Chem. - Eur. J. 2014, 20 (49), 16147–55. 10.1002/chem.201404566. [DOI] [PubMed] [Google Scholar]
- Duran-Struuck R.; Tawara I.; Lowler K.; Clouthier S. G.; Weisiger E.; Rogers C.; Luker G.; Kumanogoh A.; Liu C.; Ferrara J. L.; Reddy P. A novel role for the semaphorin Sema4D in the induction of allo-responses. Biol. Blood Marrow Transplant. 2007, 13 (11), 1294. 10.1016/j.bbmt.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumanogoh A.; Kikutani H. The CD100-CD72 interaction: a novel mechanism of immune regulation. Trends Immunol. 2001, 22 (12), 670–6. 10.1016/S1471-4906(01)02087-7. [DOI] [PubMed] [Google Scholar]
- Velayos-Baeza A.; Toma C.; Paracchini S.; Monaco A. P. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum. Mol. Genet. 2008, 17 (6), 859–71. 10.1093/hmg/ddm358. [DOI] [PubMed] [Google Scholar]
- Zjablovskaja P.; Kardosova M.; Danek P.; Angelisova P.; Benoukraf T.; Wurm A. A.; Kalina T.; Sian S.; Balastik M.; Delwel R.; Brdicka T.; Tenen D. G.; Behre G.; Fiore F.; Malissen B.; Horejsi V.; Alberich-Jorda M. EVI2B is a C/EBPalpha target gene required for granulocytic differentiation and functionality of hematopoietic progenitors. Cell Death Differ. 2017, 24 (4), 705–716. 10.1038/cdd.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi M.; Kobori H.; Ritprajak P.; Kamimura Y.; Kozono H.; Azuma M. Triggering receptor expressed on myeloid cell-like transcript 2 (TLT-2) is a counter-receptor for B7-H3 and enhances T cell responses. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (30), 10495–500. 10.1073/pnas.0802423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoletano C.; Zizzari I. G.; Rughetti A.; Rahimi H.; Irimura T.; Clausen H.; Wandall H. H.; Belleudi F.; Bellati F.; Pierelli L.; Frati L.; Nuti M. Targeting of macrophage galactose-type C-type lectin (MGL) induces DC signaling and activation. Eur. J. Immunol. 2012, 42 (4), 936–45. 10.1002/eji.201142086. [DOI] [PubMed] [Google Scholar]
- van Vliet S. J.; Bay S.; Vuist I. M.; Kalay H.; Garcia-Vallejo J. J.; Leclerc C.; van Kooyk Y. MGL signaling augments TLR2-mediated responses for enhanced IL-10 and TNF-alpha secretion. J. Leukocyte Biol. 2013, 94 (2), 315–23. 10.1189/jlb.1012520. [DOI] [PubMed] [Google Scholar]
- Sahasrabudhe N. M.; Lenos K.; van der Horst J. C.; Rodriguez E.; van Vliet S. J. Oncogenic BRAFV600E drives expression of MGL ligands in the colorectal cancer cell line HT29 through N-acetylgalactosamine-transferase 3. Biol. Chem. 2018, 399 (7), 649–659. 10.1515/hsz-2018-0120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE13 partner repository with the data set identifier PXD011307.