The title compound, the potassium salt of a benzothiazol(methylsulfanyl)pyriminidine, was obtained in a reaction designed to deliver a neutral 2-pyrimidylbenzothiazole. It crystallized with two independent molecular units in the asymmetric unit.
Keywords: crystal structure, benzothiazole, sulfonamide, pyrimidine, hydrogen bonding
Abstract
The title compound, K+·C18H14N5O2S3 −·C3H7NO·0.5H2O, was obtained in a reaction designed to deliver a neutral 2-pyrimidylbenzothiazole. The anion is deprotonated at the sulfonamide nitrogen. The asymmetric unit of the title compound contains two potassium cations, two anions, two molecules of DMF and one of water. The anions display some conformational differences but each contains an intramolecular N—H⋯Nbenzothiazole hydrogen bond. The potassium ions both display a highly irregular six-coordination, different for each potassium ion. The anions, together with the DMF and water molecules, are linked by four classical hydrogen bonds to form chains parallel to the b-axis direction.
Chemical context
Benzothiazoles are versatile heterocyclic biologically active compounds that are common in a variety of pharmaceutical preparations (Azzam et al., 2017a
▸,b
▸). These compounds are of great importance in the field of medicinal chemistry because of their remarkable pharmacological potential (Keri et al., 2015 ▸). Benzothiazole derivatives show a high degree of structural diversity that has proved beneficial in the search for new therapeutic agents (Gill et al., 2015 ▸). Research in benzothiazole-based medicinal chemistry has rapidly become an active topic, since numerous benzothiazole-based compounds have been widely used as clinical drugs to treat various types of diseases with high therapeutic potency (Sharma et al., 2013 ▸). The medicinal properties associated with benzothiazole-related drugs have encouraged medicinal chemists to synthesize a large number of new therapeutics (Elgemeie & Elghandour, 1990 ▸; Elgemeie et al., 2000a
▸,b
▸). In recent years, 2-pyrimidylbenzothiazoles have appeared as an important pharmacological class in the development of anti-tumor agents (Das et al., 2003 ▸); their promising biological profile and synthetic accessibility have been attractive in their design and development as potential chemotherapeutics. In order to access this class of compounds in high yield and to introduce diversity, a variety of new synthetic methods has been invented (Seenaiah et al., 2014 ▸). Recently, we have described the syntheses of various antimetabolites starting from heterocyclic and acyclic cyanoketene dithioacetals (Elgemeie et al., 2015 ▸, 2016 ▸, 2017 ▸). As part of this program the reaction of 2-(benzo[d]thiazol-2-yl)-3,3-bis(methylthio)acrylonitrile (2) with N-(diaminomethylene)benzenesulfonamide (3) was studied (Fig. 1 ▸). The reaction between 2 and 3 in KOH/dioxane gave a product that was crystallized from DMF and identified by X-ray crystallography as the title compound, 5, rather than the expected neutral 2-pyrimidylbenzothiazole derivative, 4. Compound 4 appears to be formed, at least in part, on dissolving 5 in deuterated DMSO; 1H NMR measurements showed the free NH proton at δ 11.50 ppm. However, we have still been unable to isolate and crystallize derivative 4.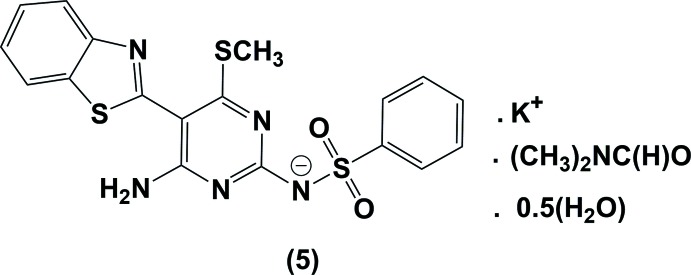
Figure 1.
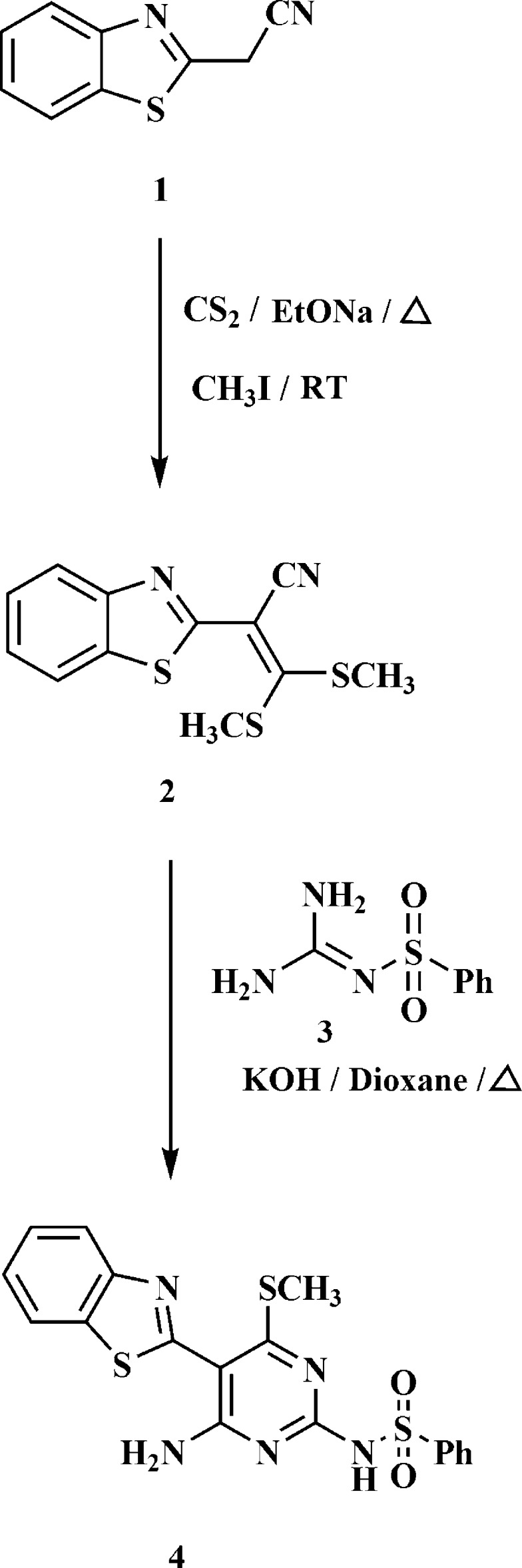
The attempted synthesis of compound 4.
The formation of 5 from the reaction of 2 and 3 is assumed to proceed via initial addition of the amino group of 3 to the double bond of 2, followed by elimination of CH3SH and cyclization via addition of the amino group to the cyano group of benzothiazole to give the product 4, which separated as its potassium salt 5 in the presence of KOH in the reaction medium. The 1H NMR spectra of the product 4, formed in part in solution in deuterated DMSO, revealed the presence of a pyrimidine methylthio group at δ = 2.19 ppm and an amino group at δ = 8.49 ppm in solution. Compound 5 and its derivatives showed interesting preclinical antiviral biological results compared to current antiviral drugs and are currently being patented (Elgemeie et al., 2018 ▸).
Structural commentary
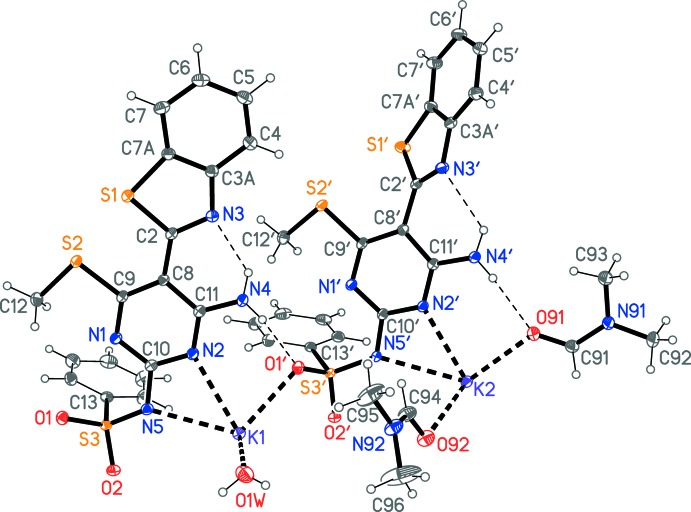
The X-ray crystal structure indicated the exclusive presence of structure 5 in the solid state. The molecular structure of compound 5 is illustrated in Fig. 2 ▸. The asymmetric unit contains two potassium cations, two anions of 4 deprotonated at the sulfonamide nitrogen, two molecules of DMF and one of water; it was chosen arbitrarily in an attempt to maximize the number of weak interactions (bonds to potassium, hydrogen bonds) within this unit.
Figure 2.
The molecular structure of compound 5, with the atom labelling (anion 1 has unprimed atom labels, anion 2 has primed atom labels). Displacement ellipsoids are drawn at the 50% probability level. Dashed lines indicate contacts to the potassium ions (thick) or classical hydrogen bonds (thin). For clarity, the sulfonamide phenyl group is not labelled.
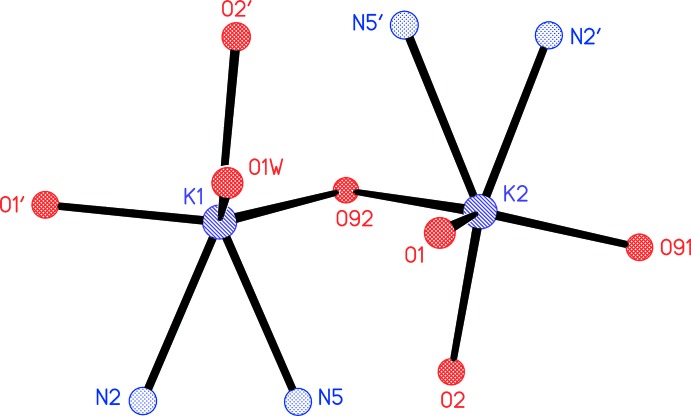
The potassium ions both display a highly irregular six-coordination; all K—N and K—O contacts (Table 1 ▸) are < 2.92 Å, and the next longest are > 3.33 Å. The atom K1 is coordinated by the pyrimidine nitrogen atom N2 and the deprotonated sulfonamide nitrogen N5, the sulfonamide oxygen atom O1′ and the water oxygen O1W within the asymmetric unit, and by the sulfonamide oxygen atom O2′ and the DMF oxygen O92 at (−x + 1, −y + 1, −z + 1). The atom K2 is coordinated by N2′, N5′ and both DMF oxygen atoms within the asymmetric unit, plus O2 at (−x + 1, −y + 1, −z + 1) and O1 at (x, y − 1, z). The angles subtended by the chelating anions via N2/N5 are particularly narrow. The bridging nature of O92 is shown in Fig. 3 ▸.
Table 1. Selected geometric parameters (Å, °).
| K1—N2 | 2.8371 (10) | K2—N2′ | 2.8320 (10) |
| K1—N5 | 2.9038 (11) | K2—N5′ | 2.9133 (10) |
| K1—O1′ | 2.6918 (9) | K2—O1ii | 2.6846 (10) |
| K1—O2′i | 2.7811 (9) | K2—O2i | 2.6782 (9) |
| K1—O1W | 2.8991 (13) | K2—O91 | 2.6594 (10) |
| K1—O92i | 2.6779 (11) | K2—O92 | 2.8193 (10) |
| O92i—K1—O1′ | 122.10 (3) | O91—K2—O1ii | 122.24 (3) |
| O92i—K1—O2′i | 89.19 (3) | O2i—K2—O1ii | 104.71 (3) |
| O1′—K1—O2′i | 94.53 (3) | O91—K2—O92 | 116.05 (3) |
| O92i—K1—N2 | 117.13 (3) | O2i—K2—O92 | 74.59 (3) |
| O1′—K1—N2 | 72.98 (3) | O1ii—K2—O92 | 120.48 (3) |
| O2′i—K1—N2 | 153.65 (3) | O91—K2—N2′ | 78.44 (3) |
| O92i—K1—O1W | 125.24 (3) | O2i—K2—N2′ | 145.92 (3) |
| O1′—K1—O1W | 105.49 (3) | O1ii—K2—N2′ | 108.90 (3) |
| O2′i—K1—O1W | 59.75 (3) | O92—K2—N2′ | 92.77 (3) |
| N2—K1—O1W | 100.51 (3) | O91—K2—N5′ | 124.93 (3) |
| O92i—K1—N5 | 80.48 (3) | O2i—K2—N5′ | 145.82 (3) |
| O1′—K1—N5 | 116.98 (3) | O1ii—K2—N5′ | 83.45 (3) |
| O2′i—K1—N5 | 147.63 (3) | O92—K2—N5′ | 72.82 (3) |
| N2—K1—N5 | 46.67 (3) | N2′—K2—N5′ | 46.50 (3) |
| O1W—K1—N5 | 101.98 (3) | K1i—O92—K2 | 87.35 (3) |
| O91—K2—O2i | 79.15 (3) | ||
| S1—C2—C8—C9 | 22.06 (18) | S1′—C2′—C8′—C9′ | −42.51 (16) |
| N1—C10—N5—S3 | 15.18 (17) | N1′—C10′—N5′—S3′ | −15.33 (16) |
| C13—S3—N5—C10 | 62.49 (11) | C13′—S3′—N5′—C10′ | 65.28 (11) |
Symmetry codes: (i) 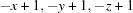 ; (ii)
; (ii)  .
.
Figure 3.
Coordination of the two potassium ions, showing the bridging nature of the DMF oxygen atom O92. Atoms O2′, O91, O92, K2, N2′, N5′ have been transformed to (−x + 1, −y + 1, −z + 1) and O1 to (−x + 1, −y + 2, −z + 1). The K1⋯K2 distance is 3.7975 (4) Å.
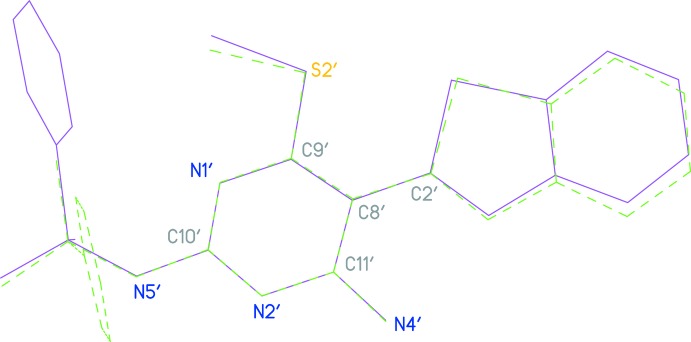
Each anion displays an intramolecular hydrogen bond (N4—H01⋯N3 and N4′—H01′⋯N3′; Table 2 ▸), forming an S(6) ring motif. The anions display some differences in conformation; the angle between the benzothiazole ring (seven atoms) and the pyrimidine ring plus immediate substituents (ten atoms) is 20.56 (5)° for anion 1 (unprimed atoms) but 42.20 (2)° for anion 2 (primed atoms). Comparing the torsion angles in Table 1 ▸, it may be seen that the signs of the torsion angles C9—C8—C2—S1 are different for the two anions [22.06 (18) and −42.51 (16)°]. A molecular fit of the ten atoms of the pyrimidine ring, inverting one anion, gives an r.m.s. deviation of 0.03 Å (Fig. 4 ▸); the benzothiazole rings are then a better fit, but the phenyl rings of the sulfonamide groups then point to opposite sides in the two anions, cf. torsion angle C10—N5—S3—C13 is 62.49 (11)°, and 65.28 (11)° in the non-inverted system.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4—H01⋯N3 | 0.85 (2) | 2.01 (2) | 2.6859 (15) | 136 (2) |
| N4′—H01′⋯N3′ | 0.87 (2) | 2.26 (2) | 2.8781 (15) | 129 (2) |
| N4—H02⋯O1′ | 0.87 (2) | 2.06 (2) | 2.9205 (14) | 168 (2) |
| N4′—H02′⋯O91 | 0.847 (19) | 2.180 (19) | 3.0207 (14) | 172 (2) |
| O1W—H03⋯O2′i | 0.81 (2) | 2.07 (2) | 2.8314 (15) | 157 (2) |
| O1W—H04⋯O1iii | 0.82 (3) | 2.00 (3) | 2.8211 (15) | 176 (3) |
Symmetry codes: (i) 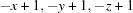 ; (iii)
; (iii) 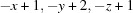 .
.
Figure 4.
A view of the molecular fit of the two independent anions of compound 5. Fitted atoms are labelled; anion 1 (inverted from the refined coordinates) is green, anion 2 (primed atoms) is purple.
Supramolecular features
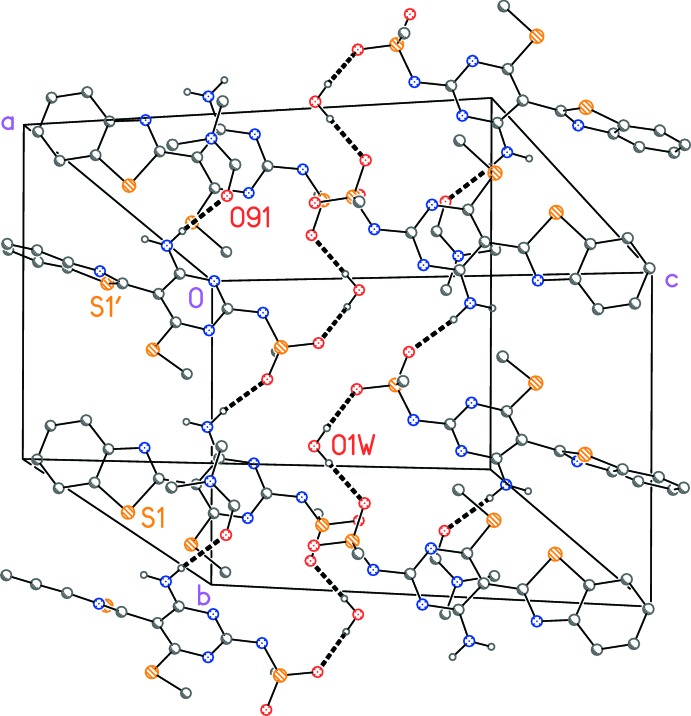
Classical hydrogen bonds are shown in Table 2 ▸. These four hydrogen bonds combined with the contacts at the potassium ions give a highly complex packing pattern. If the potassium ions are omitted, a much more simple pattern emerges; the residues are linked via the water molecules to form chains parallel to the b-axis direction, two of which are shown in Fig. 5 ▸.
Figure 5.
A view normal to plane (101) of the crystal packing of compound 5. Hydrogen bonds (Table 2 ▸) are shown as dashed lines; hydrogen atoms not involved in hydrogen bonding have been omitted for clarity. Also omitted is the DMF molecule based on atom O92 (which does not form any hydrogen bonds). For clarity, the phenyl ring at C13 is reduced to the ipso carbon.
Database survey
A search of the Cambridge Structural Database (CSD, V5.40, November 2018; Groom et al., 2016 ▸) gave 47 hits (including ten duplicated structures) for the fragment consisting of a pyrimidine ring system bearing a two-coordinate, and thus negatively charged, nitrogen substituent at the ring carbon between the two nitrogen atoms, with the nitrogen substituent forming part of a sulfonamide system. The hits included a silver salt (ASULDZ; Cook & Turner, 1975 ▸) and a sodium salt (JUBGUI/01; Hannan & Talukdar, 1992 ▸; Patel, 1995 ▸).
Synthesis and crystallization
Synthesis of potassium [4-amino-5-(benzo[ d ]thiazol-2-yl)-6-(methylsulfanyl)pyrimidin-2-yl](phenylsulfonyl)azanide dimethylformamide monosolvate hemihydrate (5):
The reaction pathway is illustrated in Fig. 1 ▸. 2-(Benzo[d]thiazole-2-yl)-3,3-bis(methylthio)acrylonitrile (2) (0.01 mol) was added to a stirred solution of the N-(diaminomethylene)benzenesulfonamide (3) (0.01 mol) in dry dioxane (20 ml) containing potassium hydroxide (0.01 mol); the reaction mixture was refluxed for 2 h. After completion of the reaction (TLC), the solid precipitate was filtered off, and then recrystallized from DMF/H2O to give colourless block-like crystals of compound 5, the potassium salt of compound 4, in 75% yield (m.p. = 517 K). IR (KBr, cm−1): ν 3431 and 3874 (NH, NH2). 1H NMR (400 MHz, DMSO-d6): δ 2.19 (s, 3H, SCH3), 7.32–7.39 (m, 4H, 3CH-phenyl, CH benzothiazole), 7.46 (t, 1H, J = 8.0 Hz, CH benzothiazole), 7.83–7.85 (m, 2H, 2CH-phenyl), 7.92 (d, 1H, J = 8.0 Hz, CH benzothiazole), 8.01 (d, 1H, J = 8.0 Hz, CH benzothiazole), 8.49 (s, br, 2H, NH2), 11.50 (s, br, 1H, NH).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The NH and OH hydrogen atoms were identified in difference-Fourier maps and refined freely. Methyl groups were identified from difference-Fourier maps, idealized and refined as rigid groups [C—H = 0.98 Å, H—C—H = 109.5° with U iso(H) = 1.5U eq(C-methyl)], and allowed to rotate but not to tip (AFIX 137). Other hydrogen atoms were included using a riding model starting from calculated positions: C—Haromatic = 0.95 Å with U iso(H) = 1.2U eq(C).
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | K+·C18H14N5O2S3 −·C3H7NO·0.5H2O |
| M r | 549.72 |
| Crystal system, space group | Triclinic, P
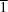
|
| Temperature (K) | 100 |
| a, b, c (Å) | 11.8407 (2), 12.6001 (4), 18.8671 (5) |
| α, β, γ (°) | 90.160 (2), 102.361 (2), 117.933 (3) |
| V (Å3) | 2411.88 (13) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.52 |
| Crystal size (mm) | 0.35 × 0.25 × 0.25 |
| Data collection | |
| Diffractometer | Oxford Diffraction Xcalibur Eos |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) |
| T min, T max | 0.994, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 256622, 13912, 11954 |
| R int | 0.047 |
| (sin θ/λ)max (Å−1) | 0.704 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.030, 0.077, 1.05 |
| No. of reflections | 13912 |
| No. of parameters | 652 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.49, −0.40 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989019002275/su5477sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019002275/su5477Isup2.hkl
CCDC reference: 1896740
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| K+·C18H14N5O2S3−·C3H7NO·0.5H2O | Z = 4 |
| Mr = 549.72 | F(000) = 1140 |
| Triclinic, P1 | Dx = 1.514 Mg m−3 |
| a = 11.8407 (2) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 12.6001 (4) Å | Cell parameters from 60104 reflections |
| c = 18.8671 (5) Å | θ = 2.4–30.6° |
| α = 90.160 (2)° | µ = 0.52 mm−1 |
| β = 102.361 (2)° | T = 100 K |
| γ = 117.933 (3)° | Block, colourless |
| V = 2411.88 (13) Å3 | 0.35 × 0.25 × 0.25 mm |
Data collection
| Oxford Diffraction Xcalibur Eos diffractometer | 13912 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 11954 reflections with I > 2σ(I) |
| Detector resolution: 16.1419 pixels mm-1 | Rint = 0.047 |
| ω–scan | θmax = 30.0°, θmin = 2.2° |
| Absorption correction: multi-scan (CrysAlis PRO; Rigaku OD, 2015) | h = −16→16 |
| Tmin = 0.994, Tmax = 1.000 | k = −17→17 |
| 256622 measured reflections | l = −26→26 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.030 | Hydrogen site location: mixed |
| wR(F2) = 0.077 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0331P)2 + 1.360P] where P = (Fo2 + 2Fc2)/3 |
| 13912 reflections | (Δ/σ)max = 0.002 |
| 652 parameters | Δρmax = 0.49 e Å−3 |
| 0 restraints | Δρmin = −0.40 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. Least-squares planes (x,y,z in crystal coordinates) and deviations from them (* indicates atom used to define plane) 7.5026 (0.0024) x + 0.9321 (0.0047) y + 8.4090 (0.0051) z = 6.8996 (0.0032) * -0.0075 (0.0009) C2 * 0.0127 (0.0009) N3 * 0.0119 (0.0012) C3A * -0.0103 (0.0011) C4 * -0.0129 (0.0011) C5 * 0.0036 (0.0011) C6 * 0.0057 (0.0010) C7 * 0.0136 (0.0012) C7A * -0.0168 (0.0007) S1 Rms deviation of fitted atoms = 0.0113 5.7089 (0.0026) x + 5.0746 (0.0027) y + 5.8404 (0.0040) z = 8.9926 (0.0015) Angle to previous plane (with approximate esd) = 20.556 ( 0.046 ) * 0.0683 (0.0011) C8 * 0.0132 (0.0010) C9 * 0.0105 (0.0010) C10 * -0.0228 (0.0011) C11 * -0.0190 (0.0009) N1 * -0.0234 (0.0010) N2 * 0.1204 (0.0009) C2 * -0.1062 (0.0006) S2 * 0.0646 (0.0008) N5 * -0.1055 (0.0009) N4 -0.1967 (0.0016) C12 -0.2593 (0.0011) S3 Rms deviation of fitted atoms = 0.0689=============================================================================== - 2.2187 (0.0034) x + 11.9323 (0.0012) y - 3.9110 (0.0057) z = 2.1027 (0.0024) * 0.0421 (0.0009) C2' * -0.0030 (0.0009) N3' * -0.0322 (0.0011) C3A' * -0.0032 (0.0011) C4' * 0.0107 (0.0011) C5' * 0.0163 (0.0011) C6' * 0.0101 (0.0010) C7' * -0.0308 (0.0011) C7A' * -0.0099 (0.0007) S1' Rms deviation of fitted atoms = 0.0220 4.0185 (0.0027) x + 7.7469 (0.0023) y + 2.9794 (0.0039) z = 6.0513 (0.0008) Angle to previous plane (with approximate esd) = 42.200 ( 0.021 ) * -0.0154 (0.0011) C8' * 0.0164 (0.0010) C9' * 0.0066 (0.0010) C10' * 0.0299 (0.0010) C11' * 0.0483 (0.0009) N1' * 0.0253 (0.0009) N2' * -0.1309 (0.0009) C2' * 0.0515 (0.0006) S2' * -0.1005 (0.0008) N5' * 0.0688 (0.0008) N4' -0.2532 (0.0017) C12' 0.1802 (0.0010) S3' Rms deviation of fitted atoms = 0.0625=============================================================================== |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.67977 (4) | 1.02828 (3) | 0.09802 (2) | 0.01751 (7) | |
| S2 | 0.47162 (3) | 1.04245 (3) | 0.15477 (2) | 0.01496 (6) | |
| S3 | 0.31676 (3) | 0.90940 (3) | 0.39554 (2) | 0.01143 (6) | |
| C2 | 0.63692 (12) | 0.90552 (11) | 0.15096 (6) | 0.0128 (2) | |
| C3A | 0.73099 (13) | 0.85659 (11) | 0.07477 (7) | 0.0142 (2) | |
| C4 | 0.77389 (14) | 0.78717 (12) | 0.04155 (7) | 0.0191 (3) | |
| H4 | 0.762989 | 0.712529 | 0.057998 | 0.023* | |
| C5 | 0.83241 (14) | 0.82984 (13) | −0.01570 (7) | 0.0202 (3) | |
| H5 | 0.862215 | 0.784025 | −0.038767 | 0.024* | |
| C6 | 0.84838 (14) | 0.93991 (13) | −0.04020 (7) | 0.0198 (3) | |
| H6 | 0.889986 | 0.967721 | −0.079226 | 0.024* | |
| C7 | 0.80507 (14) | 1.00885 (12) | −0.00895 (7) | 0.0189 (3) | |
| H7 | 0.814762 | 1.082683 | −0.026224 | 0.023* | |
| C7A | 0.74648 (13) | 0.96563 (11) | 0.04907 (7) | 0.0149 (2) | |
| C8 | 0.57484 (12) | 0.89569 (10) | 0.21128 (6) | 0.0121 (2) | |
| C9 | 0.50259 (12) | 0.95499 (10) | 0.22093 (6) | 0.0121 (2) | |
| C10 | 0.46477 (12) | 0.87455 (11) | 0.32734 (6) | 0.0123 (2) | |
| C11 | 0.57609 (12) | 0.81497 (11) | 0.26460 (6) | 0.0126 (2) | |
| C12 | 0.37416 (14) | 1.08905 (12) | 0.19405 (7) | 0.0172 (2) | |
| H12A | 0.417727 | 1.120507 | 0.245597 | 0.026* | |
| H12B | 0.286843 | 1.019598 | 0.190301 | 0.026* | |
| H12C | 0.364720 | 1.152476 | 0.167707 | 0.026* | |
| C13 | 0.17010 (12) | 0.82029 (11) | 0.32687 (6) | 0.0134 (2) | |
| C14 | 0.11572 (15) | 0.69479 (12) | 0.32221 (8) | 0.0217 (3) | |
| H14 | 0.159533 | 0.658842 | 0.352897 | 0.026* | |
| C15 | −0.00368 (15) | 0.62278 (13) | 0.27195 (8) | 0.0260 (3) | |
| H15 | −0.041467 | 0.537174 | 0.268056 | 0.031* | |
| C16 | −0.06766 (14) | 0.67590 (14) | 0.22751 (8) | 0.0232 (3) | |
| H16 | −0.149850 | 0.626424 | 0.193940 | 0.028* | |
| C17 | −0.01210 (14) | 0.80064 (13) | 0.23196 (8) | 0.0219 (3) | |
| H17 | −0.055603 | 0.836490 | 0.200925 | 0.026* | |
| C18 | 0.10750 (13) | 0.87374 (12) | 0.28186 (7) | 0.0172 (2) | |
| H18 | 0.145806 | 0.959352 | 0.285031 | 0.021* | |
| N1 | 0.44791 (11) | 0.94544 (9) | 0.27717 (6) | 0.0132 (2) | |
| N2 | 0.52377 (11) | 0.80718 (9) | 0.32239 (6) | 0.0135 (2) | |
| N3 | 0.67040 (11) | 0.82571 (10) | 0.13235 (6) | 0.0148 (2) | |
| N4 | 0.63003 (12) | 0.74264 (10) | 0.26054 (6) | 0.0176 (2) | |
| H02 | 0.6135 (19) | 0.6870 (18) | 0.2900 (11) | 0.033 (5)* | |
| H01 | 0.651 (2) | 0.7365 (18) | 0.2208 (11) | 0.034 (5)* | |
| N5 | 0.41916 (11) | 0.86521 (10) | 0.38931 (6) | 0.0137 (2) | |
| O1 | 0.35584 (9) | 1.03538 (8) | 0.38428 (5) | 0.01534 (17) | |
| O2 | 0.28269 (9) | 0.87920 (8) | 0.46485 (5) | 0.01566 (18) | |
| S1' | 0.61437 (3) | 0.29511 (3) | 0.01676 (2) | 0.01725 (7) | |
| S2' | 0.54376 (3) | 0.46946 (3) | 0.09428 (2) | 0.01721 (7) | |
| S3' | 0.46192 (3) | 0.42692 (3) | 0.35846 (2) | 0.01176 (6) | |
| C2' | 0.72463 (12) | 0.34847 (11) | 0.10370 (6) | 0.0122 (2) | |
| C3A' | 0.85618 (13) | 0.34467 (11) | 0.03647 (7) | 0.0145 (2) | |
| C4' | 0.97234 (14) | 0.36291 (13) | 0.01881 (7) | 0.0193 (3) | |
| H4' | 1.051620 | 0.390641 | 0.055865 | 0.023* | |
| C5' | 0.97023 (15) | 0.34001 (13) | −0.05340 (8) | 0.0209 (3) | |
| H5' | 1.048490 | 0.351042 | −0.065639 | 0.025* | |
| C6' | 0.85444 (15) | 0.30083 (12) | −0.10871 (7) | 0.0209 (3) | |
| H6' | 0.855166 | 0.285520 | −0.157894 | 0.025* | |
| C7' | 0.73941 (15) | 0.28421 (12) | −0.09258 (7) | 0.0198 (3) | |
| H7' | 0.661289 | 0.259304 | −0.130140 | 0.024* | |
| C7A' | 0.74080 (13) | 0.30496 (11) | −0.01957 (7) | 0.0150 (2) | |
| C8' | 0.68045 (12) | 0.36129 (10) | 0.16870 (6) | 0.0115 (2) | |
| C9' | 0.59510 (12) | 0.40880 (10) | 0.17095 (6) | 0.0120 (2) | |
| C10' | 0.58805 (12) | 0.36622 (10) | 0.28789 (6) | 0.0114 (2) | |
| C11' | 0.72036 (12) | 0.32086 (10) | 0.23520 (6) | 0.0117 (2) | |
| C12' | 0.41439 (15) | 0.48795 (13) | 0.11841 (7) | 0.0206 (3) | |
| H12D | 0.348305 | 0.410299 | 0.129017 | 0.031* | |
| H12E | 0.372818 | 0.515917 | 0.077628 | 0.031* | |
| H12F | 0.451606 | 0.547696 | 0.161728 | 0.031* | |
| C13' | 0.31457 (13) | 0.37337 (12) | 0.28815 (6) | 0.0144 (2) | |
| C14' | 0.24643 (13) | 0.25133 (12) | 0.26169 (7) | 0.0163 (2) | |
| H14' | 0.280816 | 0.199034 | 0.279043 | 0.020* | |
| C15' | 0.12741 (14) | 0.20642 (13) | 0.20955 (7) | 0.0205 (3) | |
| H15' | 0.081078 | 0.123450 | 0.190146 | 0.025* | |
| C16' | 0.07620 (15) | 0.28272 (15) | 0.18583 (7) | 0.0234 (3) | |
| H16' | −0.006040 | 0.251404 | 0.150880 | 0.028* | |
| C17' | 0.14437 (15) | 0.40440 (15) | 0.21284 (8) | 0.0242 (3) | |
| H17' | 0.108590 | 0.456059 | 0.196406 | 0.029* | |
| C18' | 0.26492 (15) | 0.45107 (13) | 0.26390 (7) | 0.0199 (3) | |
| H18' | 0.312684 | 0.534651 | 0.281964 | 0.024* | |
| N1' | 0.55003 (11) | 0.41385 (9) | 0.22932 (5) | 0.01266 (19) | |
| N2' | 0.67301 (10) | 0.32232 (9) | 0.29374 (5) | 0.01215 (19) | |
| N3' | 0.84368 (11) | 0.36751 (10) | 0.10580 (6) | 0.0146 (2) | |
| N4' | 0.80664 (11) | 0.27784 (10) | 0.24374 (6) | 0.0154 (2) | |
| H02' | 0.8068 (18) | 0.2361 (17) | 0.2788 (10) | 0.026 (5)* | |
| H01' | 0.8381 (19) | 0.2724 (17) | 0.2073 (11) | 0.032 (5)* | |
| N5' | 0.53703 (11) | 0.35582 (9) | 0.34781 (5) | 0.01244 (19) | |
| O1' | 0.53653 (10) | 0.55695 (8) | 0.35774 (5) | 0.01774 (19) | |
| O2' | 0.41688 (10) | 0.39421 (8) | 0.42515 (5) | 0.01618 (18) | |
| O1W | 0.74945 (12) | 0.84359 (11) | 0.54810 (6) | 0.0277 (2) | |
| H03 | 0.721 (2) | 0.779 (2) | 0.5641 (12) | 0.043 (6)* | |
| H04 | 0.722 (3) | 0.881 (2) | 0.5691 (14) | 0.059 (7)* | |
| K1 | 0.49406 (3) | 0.68951 (3) | 0.45040 (2) | 0.01683 (6) | |
| K2 | 0.61675 (3) | 0.18387 (2) | 0.41184 (2) | 0.01349 (6) | |
| C91 | 0.80325 (13) | 0.04442 (12) | 0.39544 (7) | 0.0162 (2) | |
| H91 | 0.767210 | 0.011982 | 0.435682 | 0.019* | |
| C92 | 0.90128 (15) | −0.08901 (13) | 0.40894 (8) | 0.0220 (3) | |
| H92A | 0.859007 | −0.110674 | 0.449887 | 0.033* | |
| H92B | 0.996418 | −0.057876 | 0.427120 | 0.033* | |
| H92C | 0.864741 | −0.160831 | 0.373445 | 0.033* | |
| C93 | 0.93416 (16) | 0.04719 (14) | 0.31230 (8) | 0.0245 (3) | |
| H93A | 0.914558 | 0.111216 | 0.294568 | 0.037* | |
| H93B | 0.896673 | −0.019652 | 0.273044 | 0.037* | |
| H93C | 1.029863 | 0.079505 | 0.327664 | 0.037* | |
| N91 | 0.87702 (12) | 0.00353 (10) | 0.37388 (6) | 0.0171 (2) | |
| O91 | 0.77808 (10) | 0.12169 (9) | 0.36692 (5) | 0.01859 (19) | |
| C94 | 0.80107 (14) | 0.47812 (13) | 0.48372 (8) | 0.0227 (3) | |
| H94 | 0.776435 | 0.467817 | 0.431813 | 0.027* | |
| C95 | 0.96954 (19) | 0.68087 (16) | 0.47527 (10) | 0.0424 (5) | |
| H95A | 0.931121 | 0.654844 | 0.422790 | 0.064* | |
| H95B | 1.062670 | 0.701335 | 0.486721 | 0.064* | |
| H95C | 0.962128 | 0.752100 | 0.488784 | 0.064* | |
| C96 | 0.9427 (3) | 0.60669 (19) | 0.59444 (10) | 0.0733 (9) | |
| H96A | 1.030708 | 0.614032 | 0.610117 | 0.110* | |
| H96B | 0.880906 | 0.539661 | 0.615825 | 0.110* | |
| H96C | 0.945776 | 0.682111 | 0.610803 | 0.110* | |
| N92 | 0.89949 (12) | 0.58360 (10) | 0.51602 (6) | 0.0225 (2) | |
| O92 | 0.73809 (10) | 0.39159 (9) | 0.51491 (5) | 0.0222 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.02517 (18) | 0.01570 (14) | 0.01810 (14) | 0.01219 (13) | 0.01195 (12) | 0.00747 (11) |
| S2 | 0.01847 (16) | 0.01588 (14) | 0.01479 (13) | 0.01093 (12) | 0.00581 (11) | 0.00621 (11) |
| S3 | 0.01119 (14) | 0.01133 (12) | 0.01105 (12) | 0.00464 (11) | 0.00318 (10) | 0.00208 (9) |
| C2 | 0.0127 (6) | 0.0121 (5) | 0.0126 (5) | 0.0052 (5) | 0.0028 (4) | 0.0032 (4) |
| C3A | 0.0134 (6) | 0.0159 (5) | 0.0133 (5) | 0.0071 (5) | 0.0031 (4) | 0.0019 (4) |
| C4 | 0.0214 (7) | 0.0201 (6) | 0.0193 (6) | 0.0118 (5) | 0.0074 (5) | 0.0033 (5) |
| C5 | 0.0201 (7) | 0.0250 (7) | 0.0181 (6) | 0.0121 (6) | 0.0065 (5) | 0.0004 (5) |
| C6 | 0.0167 (7) | 0.0280 (7) | 0.0141 (6) | 0.0096 (6) | 0.0050 (5) | 0.0032 (5) |
| C7 | 0.0198 (7) | 0.0219 (6) | 0.0156 (6) | 0.0095 (5) | 0.0065 (5) | 0.0057 (5) |
| C7A | 0.0155 (6) | 0.0171 (6) | 0.0133 (5) | 0.0083 (5) | 0.0046 (4) | 0.0026 (4) |
| C8 | 0.0133 (6) | 0.0112 (5) | 0.0120 (5) | 0.0055 (5) | 0.0041 (4) | 0.0030 (4) |
| C9 | 0.0115 (6) | 0.0110 (5) | 0.0126 (5) | 0.0047 (4) | 0.0023 (4) | 0.0029 (4) |
| C10 | 0.0101 (6) | 0.0125 (5) | 0.0123 (5) | 0.0040 (5) | 0.0023 (4) | 0.0024 (4) |
| C11 | 0.0114 (6) | 0.0118 (5) | 0.0129 (5) | 0.0047 (5) | 0.0019 (4) | 0.0024 (4) |
| C12 | 0.0183 (7) | 0.0189 (6) | 0.0180 (6) | 0.0120 (5) | 0.0038 (5) | 0.0028 (5) |
| C13 | 0.0117 (6) | 0.0146 (5) | 0.0124 (5) | 0.0048 (5) | 0.0040 (4) | 0.0013 (4) |
| C14 | 0.0213 (7) | 0.0154 (6) | 0.0223 (6) | 0.0057 (5) | 0.0015 (5) | 0.0037 (5) |
| C15 | 0.0230 (8) | 0.0161 (6) | 0.0260 (7) | 0.0008 (6) | 0.0017 (6) | 0.0003 (5) |
| C16 | 0.0143 (7) | 0.0268 (7) | 0.0191 (6) | 0.0036 (6) | 0.0009 (5) | −0.0039 (5) |
| C17 | 0.0184 (7) | 0.0274 (7) | 0.0200 (6) | 0.0130 (6) | −0.0002 (5) | −0.0008 (5) |
| C18 | 0.0165 (7) | 0.0176 (6) | 0.0184 (6) | 0.0095 (5) | 0.0030 (5) | 0.0005 (5) |
| N1 | 0.0135 (5) | 0.0141 (5) | 0.0130 (5) | 0.0072 (4) | 0.0040 (4) | 0.0038 (4) |
| N2 | 0.0143 (5) | 0.0143 (5) | 0.0137 (5) | 0.0076 (4) | 0.0047 (4) | 0.0039 (4) |
| N3 | 0.0169 (6) | 0.0157 (5) | 0.0143 (5) | 0.0088 (4) | 0.0061 (4) | 0.0036 (4) |
| N4 | 0.0261 (7) | 0.0183 (5) | 0.0174 (5) | 0.0154 (5) | 0.0109 (5) | 0.0087 (4) |
| N5 | 0.0133 (5) | 0.0166 (5) | 0.0131 (5) | 0.0082 (4) | 0.0046 (4) | 0.0041 (4) |
| O1 | 0.0151 (5) | 0.0109 (4) | 0.0176 (4) | 0.0043 (3) | 0.0039 (3) | 0.0015 (3) |
| O2 | 0.0170 (5) | 0.0183 (4) | 0.0124 (4) | 0.0081 (4) | 0.0059 (3) | 0.0033 (3) |
| S1' | 0.01307 (16) | 0.02409 (16) | 0.01137 (13) | 0.00726 (13) | 0.00081 (11) | −0.00157 (11) |
| S2' | 0.02224 (17) | 0.02347 (15) | 0.01330 (14) | 0.01611 (14) | 0.00615 (12) | 0.00867 (11) |
| S3' | 0.01608 (15) | 0.01188 (13) | 0.01004 (12) | 0.00854 (11) | 0.00399 (10) | 0.00284 (9) |
| C2' | 0.0135 (6) | 0.0116 (5) | 0.0099 (5) | 0.0053 (5) | 0.0016 (4) | 0.0022 (4) |
| C3A' | 0.0174 (6) | 0.0149 (5) | 0.0126 (5) | 0.0084 (5) | 0.0047 (5) | 0.0037 (4) |
| C4' | 0.0193 (7) | 0.0250 (6) | 0.0171 (6) | 0.0127 (6) | 0.0060 (5) | 0.0054 (5) |
| C5' | 0.0249 (8) | 0.0252 (7) | 0.0207 (6) | 0.0156 (6) | 0.0122 (5) | 0.0075 (5) |
| C6' | 0.0290 (8) | 0.0218 (6) | 0.0150 (6) | 0.0131 (6) | 0.0093 (5) | 0.0030 (5) |
| C7' | 0.0224 (7) | 0.0214 (6) | 0.0128 (6) | 0.0088 (6) | 0.0031 (5) | −0.0011 (5) |
| C7A' | 0.0165 (6) | 0.0141 (5) | 0.0139 (5) | 0.0065 (5) | 0.0045 (5) | 0.0009 (4) |
| C8' | 0.0105 (6) | 0.0116 (5) | 0.0108 (5) | 0.0042 (4) | 0.0020 (4) | 0.0021 (4) |
| C9' | 0.0119 (6) | 0.0099 (5) | 0.0121 (5) | 0.0042 (4) | 0.0016 (4) | 0.0030 (4) |
| C10' | 0.0113 (6) | 0.0096 (5) | 0.0118 (5) | 0.0043 (4) | 0.0014 (4) | 0.0013 (4) |
| C11' | 0.0102 (6) | 0.0112 (5) | 0.0118 (5) | 0.0041 (4) | 0.0011 (4) | 0.0008 (4) |
| C12' | 0.0236 (7) | 0.0284 (7) | 0.0179 (6) | 0.0194 (6) | 0.0041 (5) | 0.0058 (5) |
| C13' | 0.0161 (6) | 0.0213 (6) | 0.0110 (5) | 0.0123 (5) | 0.0053 (4) | 0.0039 (4) |
| C14' | 0.0164 (7) | 0.0211 (6) | 0.0141 (5) | 0.0102 (5) | 0.0058 (5) | 0.0030 (5) |
| C15' | 0.0165 (7) | 0.0272 (7) | 0.0152 (6) | 0.0075 (6) | 0.0064 (5) | 0.0014 (5) |
| C16' | 0.0161 (7) | 0.0421 (8) | 0.0150 (6) | 0.0159 (6) | 0.0048 (5) | 0.0061 (5) |
| C17' | 0.0267 (8) | 0.0400 (8) | 0.0186 (6) | 0.0256 (7) | 0.0069 (5) | 0.0092 (6) |
| C18' | 0.0254 (8) | 0.0259 (7) | 0.0161 (6) | 0.0185 (6) | 0.0055 (5) | 0.0043 (5) |
| N1' | 0.0136 (5) | 0.0137 (5) | 0.0117 (4) | 0.0074 (4) | 0.0031 (4) | 0.0035 (4) |
| N2' | 0.0122 (5) | 0.0135 (5) | 0.0112 (4) | 0.0069 (4) | 0.0019 (4) | 0.0022 (4) |
| N3' | 0.0145 (5) | 0.0179 (5) | 0.0118 (5) | 0.0080 (4) | 0.0039 (4) | 0.0034 (4) |
| N4' | 0.0169 (6) | 0.0217 (5) | 0.0126 (5) | 0.0131 (5) | 0.0042 (4) | 0.0048 (4) |
| N5' | 0.0154 (5) | 0.0140 (5) | 0.0114 (4) | 0.0097 (4) | 0.0035 (4) | 0.0035 (4) |
| O1' | 0.0255 (5) | 0.0119 (4) | 0.0171 (4) | 0.0094 (4) | 0.0065 (4) | 0.0032 (3) |
| O2' | 0.0212 (5) | 0.0196 (4) | 0.0117 (4) | 0.0118 (4) | 0.0067 (3) | 0.0040 (3) |
| O1W | 0.0339 (7) | 0.0209 (5) | 0.0306 (6) | 0.0110 (5) | 0.0177 (5) | 0.0049 (4) |
| K1 | 0.02118 (15) | 0.01917 (13) | 0.01416 (12) | 0.01203 (11) | 0.00636 (10) | 0.00500 (9) |
| K2 | 0.01341 (13) | 0.01430 (12) | 0.01346 (11) | 0.00694 (10) | 0.00394 (9) | 0.00372 (9) |
| C91 | 0.0155 (6) | 0.0176 (6) | 0.0155 (5) | 0.0076 (5) | 0.0047 (5) | 0.0019 (4) |
| C92 | 0.0273 (8) | 0.0219 (6) | 0.0239 (7) | 0.0167 (6) | 0.0081 (6) | 0.0079 (5) |
| C93 | 0.0295 (8) | 0.0312 (7) | 0.0257 (7) | 0.0208 (7) | 0.0165 (6) | 0.0128 (6) |
| N91 | 0.0193 (6) | 0.0188 (5) | 0.0182 (5) | 0.0118 (5) | 0.0077 (4) | 0.0064 (4) |
| O91 | 0.0206 (5) | 0.0208 (5) | 0.0190 (4) | 0.0130 (4) | 0.0062 (4) | 0.0039 (4) |
| C94 | 0.0196 (7) | 0.0230 (7) | 0.0179 (6) | 0.0054 (6) | 0.0012 (5) | 0.0036 (5) |
| C95 | 0.0359 (10) | 0.0305 (9) | 0.0324 (9) | −0.0041 (8) | 0.0014 (7) | 0.0163 (7) |
| C96 | 0.097 (2) | 0.0295 (9) | 0.0224 (9) | −0.0215 (11) | 0.0030 (10) | −0.0015 (7) |
| N92 | 0.0224 (6) | 0.0163 (5) | 0.0193 (5) | 0.0029 (5) | 0.0023 (5) | 0.0050 (4) |
| O92 | 0.0209 (5) | 0.0163 (4) | 0.0219 (5) | 0.0036 (4) | 0.0037 (4) | 0.0033 (4) |
Geometric parameters (Å, º)
| S1—C7A | 1.7299 (13) | S3'—N5' | 1.5672 (10) |
| S1—C2 | 1.7716 (12) | S3'—C13' | 1.7778 (13) |
| S2—C9 | 1.7600 (12) | C2'—N3' | 1.3058 (17) |
| S2—C12 | 1.7957 (13) | C2'—C8' | 1.4685 (16) |
| S3—O2 | 1.4500 (9) | C3A'—N3' | 1.3915 (15) |
| S3—O1 | 1.4611 (9) | C3A'—C4' | 1.3986 (19) |
| S3—N5 | 1.5770 (11) | C3A'—C7A' | 1.4046 (18) |
| S3—C13 | 1.7767 (13) | C4'—C5' | 1.3843 (18) |
| C2—N3 | 1.3126 (16) | C4'—H4' | 0.9500 |
| C2—C8 | 1.4575 (16) | C5'—C6' | 1.401 (2) |
| C3A—N3 | 1.3859 (16) | C5'—H5' | 0.9500 |
| C3A—C7A | 1.4008 (17) | C6'—C7' | 1.379 (2) |
| C3A—C4 | 1.4020 (18) | C6'—H6' | 0.9500 |
| C4—C5 | 1.3822 (19) | C7'—C7A' | 1.3964 (17) |
| C4—H4 | 0.9500 | C7'—H7' | 0.9500 |
| C5—C6 | 1.403 (2) | C8'—C9' | 1.4024 (17) |
| C5—H5 | 0.9500 | C8'—C11' | 1.4256 (16) |
| C6—C7 | 1.3818 (19) | C9'—N1' | 1.3360 (15) |
| C6—H6 | 0.9500 | C10'—N2' | 1.3432 (15) |
| C7—C7A | 1.3966 (17) | C10'—N1' | 1.3519 (15) |
| C7—H7 | 0.9500 | C10'—N5' | 1.3708 (15) |
| C8—C9 | 1.4100 (17) | C11'—N2' | 1.3451 (15) |
| C8—C11 | 1.4354 (16) | C11'—N4' | 1.3462 (16) |
| C9—N1 | 1.3346 (15) | C12'—H12D | 0.9800 |
| C10—N2 | 1.3419 (16) | C12'—H12E | 0.9800 |
| C10—N1 | 1.3507 (15) | C12'—H12F | 0.9800 |
| C10—N5 | 1.3726 (15) | C13'—C14' | 1.3877 (18) |
| C11—N4 | 1.3454 (16) | C13'—C18' | 1.3929 (17) |
| C11—N2 | 1.3474 (15) | C14'—C15' | 1.3893 (19) |
| C12—H12A | 0.9800 | C14'—H14' | 0.9500 |
| C12—H12B | 0.9800 | C15'—C16' | 1.387 (2) |
| C12—H12C | 0.9800 | C15'—H15' | 0.9500 |
| C13—C18 | 1.3877 (17) | C16'—C17' | 1.386 (2) |
| C13—C14 | 1.3943 (18) | C16'—H16' | 0.9500 |
| C14—C15 | 1.393 (2) | C17'—C18' | 1.390 (2) |
| C14—H14 | 0.9500 | C17'—H17' | 0.9500 |
| C15—C16 | 1.390 (2) | C18'—H18' | 0.9500 |
| C15—H15 | 0.9500 | N4'—H02' | 0.847 (19) |
| C16—C17 | 1.385 (2) | N4'—H01' | 0.87 (2) |
| C16—H16 | 0.9500 | O1W—H03 | 0.81 (2) |
| C17—C18 | 1.3938 (19) | O1W—H04 | 0.82 (3) |
| C17—H17 | 0.9500 | C91—O91 | 1.2380 (15) |
| C18—H18 | 0.9500 | C91—N91 | 1.3291 (17) |
| K1—N2 | 2.8371 (10) | C91—H91 | 0.9500 |
| K1—N5 | 2.9038 (11) | C92—N91 | 1.4548 (16) |
| K1—O1' | 2.6918 (9) | C92—H92A | 0.9800 |
| K1—O2'i | 2.7811 (9) | C92—H92B | 0.9800 |
| K1—O1W | 2.8991 (13) | C92—H92C | 0.9800 |
| K1—O92i | 2.6779 (11) | C93—N91 | 1.4497 (17) |
| K2—N2' | 2.8320 (10) | C93—H93A | 0.9800 |
| K2—N5' | 2.9133 (10) | C93—H93B | 0.9800 |
| K2—O1ii | 2.6846 (10) | C93—H93C | 0.9800 |
| K2—O2i | 2.6782 (9) | C94—O92 | 1.2333 (17) |
| K2—O91 | 2.6594 (10) | C94—N92 | 1.3177 (18) |
| K2—O92 | 2.8193 (10) | C94—H94 | 0.9500 |
| N4—H02 | 0.87 (2) | C95—N92 | 1.4516 (19) |
| N4—H01 | 0.85 (2) | C95—H95A | 0.9800 |
| S1'—C7A' | 1.7299 (13) | C95—H95B | 0.9800 |
| S1'—C2' | 1.7612 (12) | C95—H95C | 0.9800 |
| S2'—C9' | 1.7639 (12) | C96—N92 | 1.439 (2) |
| S2'—C12' | 1.8002 (14) | C96—H96A | 0.9800 |
| S3'—O1' | 1.4541 (9) | C96—H96B | 0.9800 |
| S3'—O2' | 1.4566 (9) | C96—H96C | 0.9800 |
| C7A—S1—C2 | 89.78 (6) | N2'—C11'—N4' | 115.39 (10) |
| C9—S2—C12 | 100.78 (6) | N2'—C11'—C8' | 121.93 (11) |
| O2—S3—O1 | 113.35 (5) | N4'—C11'—C8' | 122.68 (11) |
| O2—S3—N5 | 106.98 (5) | S2'—C12'—H12D | 109.5 |
| O1—S3—N5 | 116.05 (6) | S2'—C12'—H12E | 109.5 |
| O2—S3—C13 | 106.02 (6) | H12D—C12'—H12E | 109.5 |
| O1—S3—C13 | 106.14 (6) | S2'—C12'—H12F | 109.5 |
| N5—S3—C13 | 107.73 (6) | H12D—C12'—H12F | 109.5 |
| N3—C2—C8 | 122.89 (11) | H12E—C12'—H12F | 109.5 |
| N3—C2—S1 | 113.58 (9) | C14'—C13'—C18' | 121.11 (12) |
| C8—C2—S1 | 123.50 (9) | C14'—C13'—S3' | 118.40 (9) |
| N3—C3A—C7A | 114.97 (11) | C18'—C13'—S3' | 120.36 (10) |
| N3—C3A—C4 | 125.16 (11) | C13'—C14'—C15' | 119.30 (12) |
| C7A—C3A—C4 | 119.86 (12) | C13'—C14'—H14' | 120.3 |
| C5—C4—C3A | 118.46 (12) | C15'—C14'—H14' | 120.3 |
| C5—C4—H4 | 120.8 | C16'—C15'—C14' | 120.01 (13) |
| C3A—C4—H4 | 120.8 | C16'—C15'—H15' | 120.0 |
| C4—C5—C6 | 120.89 (13) | C14'—C15'—H15' | 120.0 |
| C4—C5—H5 | 119.6 | C17'—C16'—C15' | 120.38 (13) |
| C6—C5—H5 | 119.6 | C17'—C16'—H16' | 119.8 |
| C7—C6—C5 | 121.59 (12) | C15'—C16'—H16' | 119.8 |
| C7—C6—H6 | 119.2 | C16'—C17'—C18' | 120.21 (13) |
| C5—C6—H6 | 119.2 | C16'—C17'—H17' | 119.9 |
| C6—C7—C7A | 117.32 (12) | C18'—C17'—H17' | 119.9 |
| C6—C7—H7 | 121.3 | C17'—C18'—C13' | 118.96 (13) |
| C7A—C7—H7 | 121.3 | C17'—C18'—H18' | 120.5 |
| C7—C7A—C3A | 121.87 (12) | C13'—C18'—H18' | 120.5 |
| C7—C7A—S1 | 128.54 (10) | C9'—N1'—C10' | 116.17 (10) |
| C3A—C7A—S1 | 109.57 (9) | C10'—N2'—C11' | 117.31 (10) |
| C9—C8—C11 | 113.91 (11) | C10'—N2'—K2 | 101.07 (7) |
| C9—C8—C2 | 125.26 (10) | C11'—N2'—K2 | 136.87 (8) |
| C11—C8—C2 | 120.71 (11) | C2'—N3'—C3A' | 111.15 (10) |
| N1—C9—C8 | 124.03 (11) | C11'—N4'—H02' | 116.1 (13) |
| N1—C9—S2 | 115.19 (9) | C11'—N4'—H01' | 119.3 (13) |
| C8—C9—S2 | 120.71 (9) | H02'—N4'—H01' | 121.2 (18) |
| N2—C10—N1 | 125.23 (11) | C10'—N5'—S3' | 120.32 (8) |
| N2—C10—N5 | 113.88 (10) | C10'—N5'—K2 | 96.61 (7) |
| N1—C10—N5 | 120.89 (11) | S3'—N5'—K2 | 143.06 (5) |
| N4—C11—N2 | 115.53 (11) | S3'—O1'—K1 | 114.51 (5) |
| N4—C11—C8 | 122.42 (11) | K1—O1W—H03 | 74.2 (16) |
| N2—C11—C8 | 122.05 (11) | K1—O1W—H04 | 93.9 (18) |
| S2—C12—H12A | 109.5 | H03—O1W—H04 | 103 (2) |
| S2—C12—H12B | 109.5 | O92i—K1—O1' | 122.10 (3) |
| H12A—C12—H12B | 109.5 | O92i—K1—O2'i | 89.19 (3) |
| S2—C12—H12C | 109.5 | O1'—K1—O2'i | 94.53 (3) |
| H12A—C12—H12C | 109.5 | O92i—K1—N2 | 117.13 (3) |
| H12B—C12—H12C | 109.5 | O1'—K1—N2 | 72.98 (3) |
| C18—C13—C14 | 120.90 (12) | O2'i—K1—N2 | 153.65 (3) |
| C18—C13—S3 | 120.94 (10) | O92i—K1—O1W | 125.24 (3) |
| C14—C13—S3 | 118.07 (10) | O1'—K1—O1W | 105.49 (3) |
| C15—C14—C13 | 119.14 (13) | O2'i—K1—O1W | 59.75 (3) |
| C15—C14—H14 | 120.4 | N2—K1—O1W | 100.51 (3) |
| C13—C14—H14 | 120.4 | O92i—K1—N5 | 80.48 (3) |
| C16—C15—C14 | 120.15 (13) | O1'—K1—N5 | 116.98 (3) |
| C16—C15—H15 | 119.9 | O2'i—K1—N5 | 147.63 (3) |
| C14—C15—H15 | 119.9 | N2—K1—N5 | 46.67 (3) |
| C17—C16—C15 | 120.28 (13) | O1W—K1—N5 | 101.98 (3) |
| C17—C16—H16 | 119.9 | O91—K2—O2i | 79.15 (3) |
| C15—C16—H16 | 119.9 | O91—K2—O1ii | 122.24 (3) |
| C16—C17—C18 | 120.15 (13) | O2i—K2—O1ii | 104.71 (3) |
| C16—C17—H17 | 119.9 | O91—K2—O92 | 116.05 (3) |
| C18—C17—H17 | 119.9 | O2i—K2—O92 | 74.59 (3) |
| C13—C18—C17 | 119.37 (12) | O1ii—K2—O92 | 120.48 (3) |
| C13—C18—H18 | 120.3 | O91—K2—N2' | 78.44 (3) |
| C17—C18—H18 | 120.3 | O2i—K2—N2' | 145.92 (3) |
| C9—N1—C10 | 116.76 (10) | O1ii—K2—N2' | 108.90 (3) |
| C10—N2—C11 | 117.63 (10) | O92—K2—N2' | 92.77 (3) |
| C10—N2—K1 | 101.67 (7) | O91—K2—N5' | 124.93 (3) |
| C11—N2—K1 | 140.70 (8) | O2i—K2—N5' | 145.82 (3) |
| C2—N3—C3A | 112.08 (10) | O1ii—K2—N5' | 83.45 (3) |
| C11—N4—H02 | 116.3 (13) | O92—K2—N5' | 72.82 (3) |
| C11—N4—H01 | 116.8 (13) | N2'—K2—N5' | 46.50 (3) |
| H02—N4—H01 | 122.6 (18) | O91—C91—N91 | 124.77 (12) |
| C10—N5—S3 | 121.03 (9) | O91—C91—H91 | 117.6 |
| C10—N5—K1 | 97.74 (7) | N91—C91—H91 | 117.6 |
| S3—N5—K1 | 137.67 (6) | N91—C92—H92A | 109.5 |
| S3—O1—K2iii | 115.26 (5) | N91—C92—H92B | 109.5 |
| S3—O2—K2i | 136.13 (5) | H92A—C92—H92B | 109.5 |
| C7A'—S1'—C2' | 89.38 (6) | N91—C92—H92C | 109.5 |
| C9'—S2'—C12' | 101.82 (6) | H92A—C92—H92C | 109.5 |
| O1'—S3'—O2' | 113.04 (5) | H92B—C92—H92C | 109.5 |
| O1'—S3'—N5' | 114.63 (6) | N91—C93—H93A | 109.5 |
| O2'—S3'—N5' | 107.06 (5) | N91—C93—H93B | 109.5 |
| O1'—S3'—C13' | 107.07 (6) | H93A—C93—H93B | 109.5 |
| O2'—S3'—C13' | 104.46 (6) | N91—C93—H93C | 109.5 |
| N5'—S3'—C13' | 110.11 (6) | H93A—C93—H93C | 109.5 |
| N3'—C2'—C8' | 123.71 (11) | H93B—C93—H93C | 109.5 |
| N3'—C2'—S1' | 114.91 (9) | C91—N91—C93 | 121.04 (11) |
| C8'—C2'—S1' | 121.23 (9) | C91—N91—C92 | 122.18 (11) |
| N3'—C3A'—C4' | 125.82 (12) | C93—N91—C92 | 116.75 (11) |
| N3'—C3A'—C7A' | 114.92 (11) | C91—O91—K2 | 120.87 (8) |
| C4'—C3A'—C7A' | 119.19 (11) | O92—C94—N92 | 125.77 (13) |
| C5'—C4'—C3A' | 119.11 (13) | O92—C94—K2 | 52.85 (7) |
| C5'—C4'—H4' | 120.4 | N92—C94—K2 | 163.54 (11) |
| C3A'—C4'—H4' | 120.4 | O92—C94—H94 | 117.1 |
| C4'—C5'—C6' | 121.01 (13) | N92—C94—H94 | 117.1 |
| C4'—C5'—H5' | 119.5 | K2—C94—H94 | 67.1 |
| C6'—C5'—H5' | 119.5 | N92—C95—H95A | 109.5 |
| C7'—C6'—C5' | 120.77 (12) | N92—C95—H95B | 109.5 |
| C7'—C6'—H6' | 119.6 | H95A—C95—H95B | 109.5 |
| C5'—C6'—H6' | 119.6 | N92—C95—H95C | 109.5 |
| C6'—C7'—C7A' | 118.25 (13) | H95A—C95—H95C | 109.5 |
| C6'—C7'—H7' | 120.9 | H95B—C95—H95C | 109.5 |
| C7A'—C7'—H7' | 120.9 | N92—C96—H96A | 109.5 |
| C7'—C7A'—C3A' | 121.65 (12) | N92—C96—H96B | 109.5 |
| C7'—C7A'—S1' | 128.70 (11) | H96A—C96—H96B | 109.5 |
| C3A'—C7A'—S1' | 109.58 (9) | N92—C96—H96C | 109.5 |
| C9'—C8'—C11' | 114.77 (11) | H96A—C96—H96C | 109.5 |
| C9'—C8'—C2' | 124.58 (10) | H96B—C96—H96C | 109.5 |
| C11'—C8'—C2' | 120.62 (11) | C94—N92—C96 | 120.61 (13) |
| N1'—C9'—C8' | 123.92 (11) | C94—N92—C95 | 122.44 (13) |
| N1'—C9'—S2' | 116.05 (9) | C96—N92—C95 | 116.95 (14) |
| C8'—C9'—S2' | 120.02 (9) | C94—O92—K1i | 132.77 (10) |
| N2'—C10'—N1' | 125.70 (11) | C94—O92—K2 | 106.74 (9) |
| N2'—C10'—N5' | 113.44 (10) | K1i—O92—K2 | 87.35 (3) |
| N1'—C10'—N5' | 120.83 (11) | ||
| C7A—S1—C2—N3 | −0.17 (11) | C3A'—C4'—C5'—C6' | 1.0 (2) |
| C7A—S1—C2—C8 | 177.68 (11) | C4'—C5'—C6'—C7' | 0.1 (2) |
| N3—C3A—C4—C5 | 179.77 (13) | C5'—C6'—C7'—C7A' | −1.4 (2) |
| C7A—C3A—C4—C5 | 0.8 (2) | C6'—C7'—C7A'—C3A' | 1.59 (19) |
| C3A—C4—C5—C6 | −0.1 (2) | C6'—C7'—C7A'—S1' | 178.38 (11) |
| C4—C5—C6—C7 | −0.8 (2) | N3'—C3A'—C7A'—C7' | 176.71 (12) |
| C5—C6—C7—C7A | 1.1 (2) | C4'—C3A'—C7A'—C7' | −0.55 (19) |
| C6—C7—C7A—C3A | −0.3 (2) | N3'—C3A'—C7A'—S1' | −0.63 (14) |
| C6—C7—C7A—S1 | −178.52 (11) | C4'—C3A'—C7A'—S1' | −177.89 (10) |
| N3—C3A—C7A—C7 | −179.64 (12) | C2'—S1'—C7A'—C7' | −175.59 (13) |
| C4—C3A—C7A—C7 | −0.6 (2) | C2'—S1'—C7A'—C3A' | 1.51 (9) |
| N3—C3A—C7A—S1 | −1.16 (14) | N3'—C2'—C8'—C9' | 142.01 (13) |
| C4—C3A—C7A—S1 | 177.90 (11) | S1'—C2'—C8'—C9' | −42.51 (16) |
| C2—S1—C7A—C7 | 179.08 (13) | N3'—C2'—C8'—C11' | −40.08 (17) |
| C2—S1—C7A—C3A | 0.73 (10) | S1'—C2'—C8'—C11' | 135.40 (10) |
| N3—C2—C8—C9 | −160.28 (13) | C11'—C8'—C9'—N1' | −1.83 (17) |
| S1—C2—C8—C9 | 22.06 (18) | C2'—C8'—C9'—N1' | 176.19 (11) |
| N3—C2—C8—C11 | 15.40 (19) | C11'—C8'—C9'—S2' | 176.82 (9) |
| S1—C2—C8—C11 | −162.25 (10) | C2'—C8'—C9'—S2' | −5.15 (17) |
| C11—C8—C9—N1 | 4.82 (18) | C12'—S2'—C9'—N1' | −12.29 (11) |
| C2—C8—C9—N1 | −179.23 (12) | C12'—S2'—C9'—C8' | 168.96 (10) |
| C11—C8—C9—S2 | −171.83 (9) | C9'—C8'—C11'—N2' | 3.63 (17) |
| C2—C8—C9—S2 | 4.12 (18) | C2'—C8'—C11'—N2' | −174.47 (11) |
| C12—S2—C9—N1 | 1.16 (11) | C9'—C8'—C11'—N4' | −176.93 (11) |
| C12—S2—C9—C8 | 178.09 (10) | C2'—C8'—C11'—N4' | 4.96 (18) |
| C9—C8—C11—N4 | 174.05 (12) | O1'—S3'—C13'—C14' | 159.94 (10) |
| C2—C8—C11—N4 | −2.09 (19) | O2'—S3'—C13'—C14' | −79.93 (11) |
| C9—C8—C11—N2 | −6.32 (17) | N5'—S3'—C13'—C14' | 34.72 (12) |
| C2—C8—C11—N2 | 177.53 (11) | O1'—S3'—C13'—C18' | −24.15 (12) |
| O2—S3—C13—C18 | 113.30 (11) | O2'—S3'—C13'—C18' | 95.97 (11) |
| O1—S3—C13—C18 | −7.52 (12) | N5'—S3'—C13'—C18' | −149.37 (10) |
| N5—S3—C13—C18 | −132.44 (11) | C18'—C13'—C14'—C15' | 0.69 (19) |
| O2—S3—C13—C14 | −63.18 (12) | S3'—C13'—C14'—C15' | 176.57 (10) |
| O1—S3—C13—C14 | 175.99 (10) | C13'—C14'—C15'—C16' | −1.62 (19) |
| N5—S3—C13—C14 | 51.07 (12) | C14'—C15'—C16'—C17' | 1.2 (2) |
| C18—C13—C14—C15 | −0.6 (2) | C15'—C16'—C17'—C18' | 0.1 (2) |
| S3—C13—C14—C15 | 175.93 (11) | C16'—C17'—C18'—C13' | −1.0 (2) |
| C13—C14—C15—C16 | −0.4 (2) | C14'—C13'—C18'—C17' | 0.6 (2) |
| C14—C15—C16—C17 | 1.1 (2) | S3'—C13'—C18'—C17' | −175.16 (10) |
| C15—C16—C17—C18 | −1.0 (2) | C8'—C9'—N1'—C10' | −2.00 (18) |
| C14—C13—C18—C17 | 0.7 (2) | S2'—C9'—N1'—C10' | 179.30 (9) |
| S3—C13—C18—C17 | −175.65 (10) | N2'—C10'—N1'—C9' | 4.65 (18) |
| C16—C17—C18—C13 | 0.0 (2) | N5'—C10'—N1'—C9' | −173.44 (11) |
| C8—C9—N1—C10 | 0.33 (18) | N1'—C10'—N2'—C11' | −2.95 (18) |
| S2—C9—N1—C10 | 177.15 (9) | N5'—C10'—N2'—C11' | 175.27 (10) |
| N2—C10—N1—C9 | −4.84 (18) | N1'—C10'—N2'—K2 | −162.67 (10) |
| N5—C10—N1—C9 | 175.93 (11) | N5'—C10'—N2'—K2 | 15.55 (11) |
| N1—C10—N2—C11 | 3.35 (19) | N4'—C11'—N2'—C10' | 179.06 (11) |
| N5—C10—N2—C11 | −177.38 (11) | C8'—C11'—N2'—C10' | −1.46 (17) |
| N1—C10—N2—K1 | −177.35 (11) | N4'—C11'—N2'—K2 | −30.78 (17) |
| N5—C10—N2—K1 | 1.93 (12) | C8'—C11'—N2'—K2 | 148.69 (9) |
| N4—C11—N2—C10 | −177.73 (11) | C8'—C2'—N3'—C3A' | 178.01 (11) |
| C8—C11—N2—C10 | 2.63 (18) | S1'—C2'—N3'—C3A' | 2.26 (13) |
| N4—C11—N2—K1 | 3.35 (19) | C4'—C3A'—N3'—C2' | 176.01 (12) |
| C8—C11—N2—K1 | −176.30 (9) | C7A'—C3A'—N3'—C2' | −1.04 (15) |
| C8—C2—N3—C3A | −178.31 (11) | N2'—C10'—N5'—S3' | 166.36 (9) |
| S1—C2—N3—C3A | −0.45 (14) | N1'—C10'—N5'—S3' | −15.33 (16) |
| C7A—C3A—N3—C2 | 1.06 (16) | N2'—C10'—N5'—K2 | −14.92 (10) |
| C4—C3A—N3—C2 | −177.94 (13) | N1'—C10'—N5'—K2 | 163.39 (10) |
| N2—C10—N5—S3 | −164.13 (9) | O1'—S3'—N5'—C10' | −55.50 (11) |
| N1—C10—N5—S3 | 15.18 (17) | O2'—S3'—N5'—C10' | 178.27 (9) |
| N2—C10—N5—K1 | −1.86 (11) | C13'—S3'—N5'—C10' | 65.28 (11) |
| N1—C10—N5—K1 | 177.45 (10) | O1'—S3'—N5'—K2 | 126.62 (9) |
| O2—S3—N5—C10 | 176.11 (10) | O2'—S3'—N5'—K2 | 0.38 (11) |
| O1—S3—N5—C10 | −56.26 (12) | C13'—S3'—N5'—K2 | −112.60 (9) |
| C13—S3—N5—C10 | 62.49 (11) | O2'—S3'—O1'—K1 | −20.89 (7) |
| O2—S3—N5—K1 | 22.74 (10) | N5'—S3'—O1'—K1 | −143.97 (5) |
| O1—S3—N5—K1 | 150.37 (7) | C13'—S3'—O1'—K1 | 93.58 (6) |
| C13—S3—N5—K1 | −90.88 (9) | O91—C91—N91—C93 | −0.8 (2) |
| O2—S3—O1—K2iii | 99.65 (6) | O91—C91—N91—C92 | −178.96 (13) |
| N5—S3—O1—K2iii | −24.76 (7) | N91—C91—O91—K2 | 176.30 (10) |
| C13—S3—O1—K2iii | −144.38 (5) | O92—C94—N92—C96 | −0.2 (3) |
| O1—S3—O2—K2i | −113.35 (8) | K2—C94—N92—C96 | −79.3 (4) |
| N5—S3—O2—K2i | 15.85 (9) | O92—C94—N92—C95 | 179.47 (17) |
| C13—S3—O2—K2i | 130.61 (7) | K2—C94—N92—C95 | 100.3 (4) |
| C7A'—S1'—C2'—N3' | −2.26 (10) | N92—C94—O92—K1i | 98.16 (17) |
| C7A'—S1'—C2'—C8' | −178.12 (10) | K2—C94—O92—K1i | −102.27 (11) |
| N3'—C3A'—C4'—C5' | −177.66 (12) | N92—C94—O92—K2 | −159.57 (14) |
| C7A'—C3A'—C4'—C5' | −0.73 (19) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y−1, z; (iii) x, y+1, z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H01···N3 | 0.85 (2) | 2.01 (2) | 2.6859 (15) | 136.0 (18) |
| N4′—H01′···N3′ | 0.87 (2) | 2.26 (2) | 2.8781 (15) | 128.5 (16) |
| N4—H02···O1′ | 0.87 (2) | 2.06 (2) | 2.9205 (14) | 167.5 (18) |
| N4′—H02′···O91 | 0.847 (19) | 2.180 (19) | 3.0207 (14) | 172.0 (17) |
| O1W—H03···O2′i | 0.81 (2) | 2.07 (2) | 2.8314 (15) | 157 (2) |
| O1W—H04···O1iv | 0.82 (3) | 2.00 (3) | 2.8211 (15) | 176 (3) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (iv) −x+1, −y+2, −z+1.
References
- Azzam, R. A., Elgemeie, G. H., Elsayed, R. E. & Jones, P. G. (2017a). Acta Cryst. E73, 1041–1043. [DOI] [PMC free article] [PubMed]
- Azzam, R. A., Elgemeie, G. H., Elsayed, R. E. & Jones, P. G. (2017b). Acta Cryst. E73, 1820–1822. [DOI] [PMC free article] [PubMed]
- Cook, D. S. & Turner, M. F. (1975). J. Chem. Soc. Perkin Trans. 2, pp. 1021–1025.
- Das, J., Moquin, R. V., Lin, J., Liu, C., Doweyko, A. M., DeFex, H. F., Fang, Q., Pang, S., Pitt, S., Shen, D. R., Schieven, G. L., Barrish, J. C. & Wityak, J. (2003). Bioorg. Med. Chem. Lett. 13, 2587–2590. [DOI] [PubMed]
- Elgemeie, G. H., Abou-Zeid, M., Alsaid, S., Hebishy, A. & Essa, H. (2015). Nucleosides Nucleotides Nucleic Acids, 34, 659–673. [DOI] [PubMed]
- Elgemeie, G. H., Abu-Zaied, M. & Azzam, R. A. (2016). Nucleosides Nucleotides Nucleic Acids, 35, 211–222. [DOI] [PubMed]
- Elgemeie, G. H., Altalbawy, F., Alfaidi, M., Azab, R. & Hassan, A. (2017). Drug Design, Development & Therapy, 11, 3389–3399. [DOI] [PMC free article] [PubMed]
- Elgemeie, G. H., Azzam, R. A. & Ramadan, R. (2018). Egyptian Academy of Scientific Research. Patent No. 292/2018.
- Elgemeie, G. H. & Elghandour, A. H. (1990). Phosphorus Sulfur Silicon, 48, 281–284.
- Elgemeie, G. H., Shams, Z., Elkholy, M. & Abbas, N. S. (2000a). Heterocycl. Commun. 6, 363–268.
- Elgemeie, G. H., Shams, H. Z., Elkholy, Y. M. & Abbas, N. S. (2000b). Phosphorus Sulfur Silicon, 165, 265–272.
- Gill, R. K., Rawal, R. K. & Bariwal, J. (2015). Arch. Pharm. Chem. Life Sci. 348, 155–178. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hannan, S. S. & Talukdar, A. N. (1992). Acta Cryst. C48, 2021–2023.
- Keri, R. S., Patil, M. R., Patil, S. A. & Budagumpi, S. (2015). Eur. J. Med. Chem. 89, 207–251. [DOI] [PubMed]
- Patel, U. (1995). Cryst. Res. Technol. 30, 381–387.
- Rigaku OD (2015). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, UK.
- Seenaiah, D., Reddy, P. R., Reddy, G. M., Padmaja, A., Padmavathi, V. & Krishna, N. S. (2014). Eur. J. Med. Chem. 77, 1–7. [DOI] [PubMed]
- Sharma, P. C., Sinhmar, A., Sharma, A., Rajak, H. & Pathak, D. P. J. (2013). J. Enzyme Inhib. Med. Chem. 28, 240–266. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Siemens (1994). XP. Siemens Analytical X–Ray Instruments, Madison, Wisconsin, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989019002275/su5477sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019002275/su5477Isup2.hkl
CCDC reference: 1896740
Additional supporting information: crystallographic information; 3D view; checkCIF report