Abstract
Roux-en-Y gastric bypass (RYGB) surgery is one of the most effective treatment options for severe obesity and related comorbidities, including hyperlipidemia, a well-established risk factor of cardiovascular diseases. Elucidating the molecular mechanisms underlying the beneficial effects of RYGB may facilitate development of equally effective, but less invasive, treatments. Recent studies have revealed that RYGB increases low-density lipoprotein receptor (LDLR) expression in the intestine of rodents. Therefore, in this study we first examined the effects of RYGB on intestinal cholesterol metabolism in human patients, and we show that they also exhibit profound changes and increased LDLR expression. We then hypothesized that the upregulation of intestinal LDLR may be sufficient to decrease circulating cholesterol levels. To this end, we generated and studied mice that overexpress human LDLR specifically in the intestine. This perturbation significantly affected intestinal metabolism, augmented fecal cholesterol excretion, and induced a reciprocal suppression of the machinery related to luminal cholesterol absorption and bile acid synthesis. Circulating cholesterol levels were significantly decreased and, remarkably, several other metabolic effects were similar to those observed in RYGB-treated rodents and patients, including improved glucose metabolism. These data highlight the importance of intestinal cholesterol metabolism for the beneficial metabolic effects of RYGB and for the treatment of hyperlipidemia.
Improving cardiovascular health through controlling hyperlipidemia (HL) is a major public health challenge, as HL is a significant risk factor for coronary artery disease and stroke. The hallmarks of HL are high levels of low-density lipoprotein (LDL) cholesterol and triglycerides (TGs) and lower levels of high-density lipoprotein (HDL)–cholesterol (1). Treating HL with robust and sustainable results is not easy. Statins, the inhibitors of cholesterol synthesis, are the most commonly used cholesterol-lowering agents, but a significant percentage of patients do not respond to this therapy (2). For obesity-related HL, Roux-en-Y gastric bypass (RYGB) is an effective treatment, and complete 5-year resolution is achieved in one out of two patients undergoing this procedure. Remarkably many patients have normal cholesterol levels soon after the surgery, and a substantial percentage of those who were taking statins or other lipid-lowering medications prior to surgery no longer need them after the surgery (3, 4). Additionally, patients benefit from a sustained increase in HDL cholesterol levels. This healthier metabolic profile leads to a 56% reduction in the chance of death from coronary artery disease and a 57% reduction in the risk of fatal stroke compared with unoperated controls with obesity according to recent studies (5, 6). Furthermore, compared with other treatment options, RYGB leads to the highest improvement in lipid profile for the same degree of weight loss (7). These clinical observations allude to an underlying physiological mechanism that may be specific to RYGB and not consequential to the profound weight loss (8). This hypothesis, however, is controversial, and the exact mechanisms by which RYGB improves metabolic function still remain unknown.
We have recently shown that the anatomically reconfigured intestine following RYGB undergoes remodeling and metabolic reprogramming to meet its increased bioenergetic demands of tissue growth and maintenance (9). Specifically, we have reported that gene along with protein expression of factors and enzymes involved in intestinal cholesterol biosynthesis and uptake are augmented in the reconfigured jejunum (Roux limb) of RYGB-treated rats. This is consistent with the essential role of cholesterol as a staple and a building block for cellular growth and proliferation. Importantly, we have demonstrated that RYGB increases substantially the gene and protein expression levels of LDL receptor (LDLR), which is expressed on the basolateral membrane of the columnar epithelium of the mucosa and mediates the uptake of circulating LDL by the enterocytes. Furthermore, we and others have reported increased fecal fat and cholesterol excretion in human patients and rodents treated with RYGB (10–14).
Based on these observations, our working hypothesis is that the enterocytes after RYGB enhance their cholesterol biosynthesis and the LDLR-mediated circulating cholesterol uptake to serve their increased cholesterol requirements. We further speculate that the increased cholesterol uptake and utilization by the enterocytes could contribute to the improvement of the lipid profile. Thus, the intestine may serve as a “cholesterol depot” to lower circulating cholesterol levels. The elucidation of the molecular mechanisms underlying the RYGB-induced effects on intestinal cholesterol metabolism could offer the opportunity to develop methods to control HL through enhancing intestinal circulating cholesterol uptake and utilization.
To this end, we examined whether the intestine of human patients after RYGB exhibits changes consistent with increased cholesterol uptake and utilization as well as how the intestine-specific enhancement of LDLR expression perturbates intestinal and whole-body cholesterol metabolism.
Materials and Methods
Human patients
In this study, we used human intestinal biopsies. They were collected at the Department of Digestive Surgery of the Pontifical Catholic University of Chile from two groups of patients, who were recruited to participate in this study (i.e., the samples were collected specifically for the purpose of this research study). In the first group, an upper gastrointestinal endoscopy was performed in a group of patients who had undergone RYGB surgery (n = 14). The second group consisted of well-matched patients with obesity (n = 13) who were scheduled for RYGB surgery, whose samples were collected at the time of the surgery. Patients’ characteristics and inclusion criteria are listed in an online repository (15). Importantly, an effort was made to avoid the inclusion of patients with hypercholesterolemia and those taking statins and other lipid-lowering medications, because the effect of these drugs on intestinal metabolism is not well described. Only three patients in the control and one in the RYGB group were treated with statins at the time of the study. The study was approved by the Internal Review Board of the Pontifical Catholic University of Chile.
Reverse transcription–quantitative PCR
Total RNA was extracted using the RNeasy Plus universal tissue mini kit (Qiagen) and concentration was assessed using a Thermo Fisher Scientific NanoDrop spectrophotometer. Genomic DNA was removed using an RNase-free DNase set (Qiagen), and 1 μg of RNA was converted to cDNA with a high-capacity cDNA reverse transcription kit (Applied Biosystems). Primers were designed using NCBI Primer Blast and Primer 3 software (15, 16). Reverse transcription–quantitative PCR (RT-qPCR) was performed using a ViiA™ 7 real-time PCR instrument (Applied Biosystems). cDNA was diluted 20-fold (template) and reactions assembled in 384-well clear optical reaction plates as follows: 1 µL of template, 250 nM each primer, 5 µL of Power SYBR Green master mix (Applied Biosystems), and nuclease-free water to a final volume of 10 µL. Template denaturation at 95°C for 10 minutes was followed by 45 cycles of 15 seconds at 95°C and 1 minute at 60°C. Melting curves were generated and analyzed for the presence of single and specific PCR products. The cycle threshold value of gene targets was normalized with two to four housekeeping genes. Transgene transcriptional levels were assessed with primers detecting both mouse and human LDLR mRNAs, which were then normalized using as a reference gene the endogenous (mouse) LDLR transcript. Relative quantification of gene expression was calculated using the 2−ΔΔCT method.
Western blot
Tissue grinding was performed by mechanical disruption at liquid nitrogen temperature. Using a rotor-stator homogenizer, tissues were homogenized in cold RIPA buffer [10 mM Tris-Cl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl] prepared with a protease inhibitors cocktail (cOmplete™ ULTRA tablets, mini, EDTA-free; Roche Diagnostics). After rotation for 15 minutes, protein extracts were obtained by centrifugation at 11,000g for 20 minutes at 4°C and protein concentration was determined using a Bradford protein assay (Bio-Rad Laboratories). Lysates were denatured at 95°C for 5 minutes in Laemmli buffer and proteins were resolved by SDS-PAGE (10% and 12% gels). After SDS-PAGE, proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P, EMD Millipore) at 400 mA for 75 minutes at 4°C using a wet transfer system (Hoefer). Polyvinylidene difluoride membranes were blocked in 5% nonfat milk dissolved in PBS with 0.1% Tween 20 (PBST) and incubated overnight at 4 °C with the primary antibody of choice. The following day, membranes were washed with PBST and incubated with a secondary antibody diluted 20,000-fold in 5% milk dissolved in PBST at room temperature for 1 hour. Detection of proteins on immunoblots was achieved using a chemiluminescent assay (Pierce ECL Western blotting substrate; Thermo Fisher Scientific). The following primary antibodies were used: 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) and LDLR from BioVision (17, 18), CDK1 from Cell Signaling Technology, and ACTB, β2-microglobulin (B2M), and sterol regulatory (19) element–binding protein 2 (SREBP2) from Abcam (20–22). ACTB and B2M were used as loading controls for intestine and kidney lysates, respectively. Goat anti-rabbit IgG HRP (Abcam) (23) was used as a secondary antibody.
Transgenic animals
All experiments were performed in compliance with, and were approved by, the Institutional Animal Care and Use Committee of Boston Children’s Hospital.
C57BL/6-TG(Vil-LDLR) mice were generated by pronuclear injection at the Mouse Gene Manipulation Core of the Boston Children’s Hospital. Briefly, the open reading frame (ORF) of the human LDLR gene (pCMV6-AC-GFP, catalog no. RG200006; Origene Technologies) was cloned into the 12.4-kb Villin-ΔATG plasmid (Addgene) (24) using MluI and KpnI restriction enzyme sites (New England BioLabs). Recombinant 12.4-kb Villin-ΔATG (human) LDR ORF plasmid was verified by enzymatic digests with MluI and KpnI and Sanger sequencing. To generate C57BL/6-TG(Vil-LDLR) mice, fertilized eggs of C57BL/6 mice were injected with a linearized form of the transgene without prokaryotic sequences. Removal of prokaryotic sequences and linearization of the construct was obtained by digesting the vector with SacII restriction enzyme from New England BioLabs. A founder (F0) mouse of the new transgenic line was identified by PCR and bred with C57BL/6 wild-type mice (The Jackson Laboratory). Experiments were performed with adult male transgenic mice of the F2 generation and wild-type littermates as controls.
Genotyping of transgenic mice and transgene copy number variation assay
Genotyping of transgenic mice was done by PCR using genomic DNA extracted from tail biopsies. Primers spanning the human LDLR ORF and the villin gene promoter were used (15). PCR mixture reaction was assembled in a final volume of 20 µL containing 10 µL of GoTaq® Green master mix (Promega Corporation), 1 µL of each primer (10 µM concentrated), and 8 µL of 10 to 100 ng of genomic DNA as a template. The PCR reaction thermal profile consisted of an initial step of 1 minute at 95°C followed by 32 cycles of 15 seconds at 95°C, 15 seconds at 64°C, and 30 seconds at 72°C. Final extension was for 7 minutes at 72°C. The amplification fragment generated by PCR was 195 bp. A second PCR was performed using human LDLR-specific primers.
Transgene copy number in the mouse genome of C57BL/6-TG(Vil-LDLR) mice was assessed by droplet digital PCR (ddPCR; Molecular Genetics Core Facility of the Boston Children’s Hospital). ddPCR was performed using an intron-spanning assay for the human LDLR gene (PrimePCR™ ddPCR™ expression probe LDLR; assay ID dHsaCPE5052808; Bio-Rad Laboratories), and an assay for detection of the mouse ribonuclease P/MRP subunit P30 (Rpp30) gene was used as a genomic reference (Bio-Rad Laboratories). Droplet generation, PCR, and data analysis were performed according to the manufacturer’s instructions. Briefly, the ddPCR reaction was set up as follows: 10 µL of ddPCR™ Supermix for probes (Bio-Rad Laboratories), 250 nM both target (human LDLR gene) and reference (mouse Rpp30 gene) probes (labeled with FAM and HEX reporter fluorophores, respectively), and 50 ng of genomic DNA and water to 20 µL. Restriction digestion of genomic DNA was achieved by direct addition of 5 U of MseI enzyme diluted in CutSmart buffer (New England BioLabs) to the ddPCR reaction. PCR mixtures were vortexed thoroughly, loaded into the sample wells of a Bio-Rad automated droplet generator, and dispensed in a 96-well plate. PCR was performed in a Bio-Rad C1000 Touch thermal cycler. The thermal cycling protocol was 95°C for 10 minutes for enzyme activation, 40 cycles of denaturation and annealing/extension achieved at 94°C for 30 seconds and at 60°C for 1 minute, respectively, and a final step of enzyme deactivation at 98°C for 10 minutes. Plates were transferred to a QX200 droplet reader (droplet reader software: QuantaSoft version 1.7.4.0917; Bio-Rad Laboratories). To measure the number of positive and negative droplets for each fluorophore, the fluorescence signal was plotted as a function of event (or droplet) number in QuantaSoft Analysis Pro, and a threshold above which droplets are considered positive was placed at the upper boundary of the negative droplet cluster. Transgene copy number per genomic sample was calculated in QuantaSoft Analysis Pro with the formula T × R/N, where T indicates target molecule concentration, R indicates reference molecule concentration, and N indicates the number of copies of reference loci in the genome (= 2).
Blood lipid profile by fast protein liquid chromatography
Blood lipid analysis was performed at our Center and at the Lipoprotein and Atherosclerosis Analysis Core Laboratory at Wake Forest University School of Medicine (Winston-Salem, NC). Blood was collected from tail vein (survival procedure) or after euthanasia by cardiac puncture and serum was obtained by centrifugation at 2000g for 10 minutes at 4°C. Total cholesterol was measured with a Pointe Scientific total cholesterol reagent. Very-low-density lipoprotein (VLDL)–, LDL-, and HDL-cholesterol fractions were separated from 15 µg of total cholesterol serum samples loaded on a Superose 6 10/300 GL column powered by a LaChrom Elite fast protein liquid chromatography (FPLC) Hitachi system (Chiyoda Corporation). Chromatography was run in the presence of enzymatic reagents releasing free cholesterol and generating from hydrogen peroxide a quinoneimine dye absorbing at 500 nm (cholesterol liquid reagent set, Pointe Scientific). Eluate absorbance was measured at 500 nm and converted to a millivolt response. Curves for analysis were built plotting response values as a function of the time: VLDL-, LDL-, and HDL-cholesterol fractions identified in three major peaks and the area under the curve (AUC) were calculated. Lipoprotein fraction AUC was expressed as a percentage of the total AUC of all the peaks (total cholesterol) and cholesterol concentration of each lipoprotein was calculated. Total and LDL-cholesterol levels in individual samples were also measured by an enzymatic method using a commercially available kit (cholesterol E-test Wako and L-Type LDL-C; Wako Diagnostics total cholesterol reagent, Wako Life Sciences).
Histology and immunohistochemistry
Animals were euthanized and then the intestine was dissected, rinsed in cold PBS, and kept overnight in 10% neutral buffered formalin solution (Sigma-Aldrich). The following day, tissues were washed in cold PBS and stored in ethanol (70%) at 4°C. Tissue was embedded in paraffin wax, and the sectioning and hematoxylin and eosin staining were performed at the Animal Histology Core (Tufts Medical Center, Boston, MA). Blank and serial sections based on the hematoxylin and eosin results were used for experiments of immunohistochemistry. Antigen retrieval, deparaffinization, and rehydration were performed using Trilogy™ solution (Cell Marque, Sigma-Aldrich) in a domestic pressure cooker for 15 minutes. Tissue slides were cooled down to room temperature and incubated for 30 minutes with an endogenous peroxidase blocking reagent (Laboratory Vision™ hydrogen peroxide block; Thermo Fisher Scientific). To prevent nonspecific binding of antibodies to tissues, slides were incubated 1 hour in Tris-buffered saline (Tris-EDTA, pH 7.6)/0.1% Tween 20 with 5% goat serum at room temperature. Blocking solution was removed and tissues slides were incubated with a primary antibody against the cleaved form of caspase-3 (Cell Signaling Technology) (25) overnight at 4°C. Detection and staining of the cleaved form of caspase-3 were performed using Pierce ABC staining (Thermo Fisher Scientific) and Dako North America reagents. Images were obtained with an Olympus BX43 microscope.
Gene expression profiling of the intestine
Gene expression profiling of the intestine was performed at the Molecular Genetics Core Facility of the Boston Children’s Hospital using a GeneChip® mouse gene ST array (Affymetrix, Thermo Fisher Scientific). Briefly, animals were euthanized, the small intestine was dissected in five different segments of equal length, and RNA was extracted from its most proximal portion (duodenum) using the RNeasy Plus universal tissue mini kit (Qiagen). Extracted RNA was checked for quality and integrity via a Bioanalyzer system (Agilent Technologies) and only samples with a RNA integrity number >8 were then labeled and hybridized to arrays. The experiment was conducted using samples from six mice and three arrays per genotype (RNA from the duodenum of two different animals was pooled as one sample). After quality control of hybridizations, signal intensity normalization, and summarization (robust multi-array average), further processing of data was performed and probes with signal intensity <log2(50) were excluded. In case of multiple probes per gene, the probe yielding the smallest SD was used. The log-transformed normalized intensities from duodenum arrays of wild-type and C57BL/6-TG(Vil-LDLR) mice were compared using a two-tailed t test, and homogeneity of variance was tested with the Levene test. The obtained P values were adjusted by Benjamini-Hochberg multiple testing, and the significant level was established at P < 0.05. Analysis of gene ontology (GO) was performed on mRNAs that were associated with a nontrivial GO term (e.g., “biological process”). Heat maps were created in MATLAB (MathWorks) after searching for enriched GO terms with the gene ontology enrichment analysis and visualization (GOrilla) tool and in Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Further functional analysis of gene expression (i.e., pathways maps and pathways distribution) was performed using the GeneGo Metacore™ platform (Thomson Reuters).
Lipid and cholesterol fecal content
Fecal lipid excretion was performed as described by Kraus et al. (26). Briefly, mice were single-housed in wire floor cages prepared with a cotton fiber–based bedding (Nestlets). Body weight and food intake were monitored. Feces were then collected, dried, and weighed until there was no change in weight (for 3 to 4 days). Feces were grinded and 1 g was resuspended in 5 mL of saline; 5 mL of a chloroform-methanol solution was added (2:1 by volume). Then the mixture was thoroughly vortexed and centrifuged at 1000g for 10 minutes at room temperature. The lipid phase was transferred to a glass tube, evaporated to dryness, and weighed on analytical balance. Fecal lipid excretion per day was calculated as follows: (lipid mass × total weight of feces)/days. Fecal cholesterol content was measured from lipid extracts (prepared as above) using an enzymatic colorimetric method (Wako Diagnostics total cholesterol reagent, Wako Life Sciences). Data were normalized for fecal output and expressed as milligrams per gram per day.
Indirect calorimetry
Indirect calorimetry was performed in transgenic mice and controls using the Oxymax comprehensive laboratory animal monitoring system (CLAMS; Columbus Instruments International), as previously described by Nestoridi et al. (10). Briefly, the temperature of the CLAMS room was set to thermoneutrality (27 to 30°C). Animals were single-housed and oxygen consumption and carbon dioxide production were measured during a 3-day period and after 1 day of acclimation. Mice had ad libitum access to diet and water except for the last day of the experiment, in which they were fasted by removing the food from the cage the previous night (at ∼5:00 pm).
Phenotypic assessments and diet studies
Food intake, body weight, body composition, and body length measurements
Mice were single-housed and food intake was measured for a period of 3 days. A wire floor was placed in each cage and the bedding was removed. We observed that food grinding was increased in transgenic mice compared with wild-type littermates. Thus, the amount of ground food spilled on the cage floors was taken into account in both C57BL/6-TG(Vil-LDLR) and control mice. Body weight was measured weekly between 11:00 pm and 4:00 pm for a period of 16 and 10 weeks for animals on chow and high-fat, high-cholesterol (HFHC) diets, respectively. Body mass composition was assessed by dual-energy x-ray absorptiometry (DEXA) as previously reported (10). Body length measurements (nose to anus) were performed in animals after euthanasia at 3 to 5 months of age. With regard to the HFHC diet, animals were given ad libitum for 10 weeks a purified atherogenic diet containing 1% cholesterol and 0.5% sodium cholate and providing 49.1% of total energy as carbohydrate, 34.2% as fat, 16.6% as protein (Teklad Custom Diet TD.09237; Envigo, Life Sciences Research).
Diet preference testing
Mice were single-housed and had access, for 3 days, to chow and HFCH diets. Each day, each cage had 15 g per diet and diet position was switched daily to avoid any bias related to side preference. For each diet, the intake was measured separately and expressed as grams per day.
Blood glucose measurements
For fasting blood glucose measurements, mice were fasted overnight (by removing the food at ∼5:00 pm) and blood was obtained from the tail of the mice the following day (∼9:00 am). Glucose levels were measured using a blood glucometer (LifeScan). Glucose was also measured in animals at weaning and the measurements were performed without fasting, but always at the same time of the day (∼1 pm). A blood glucose tolerance test was performed as previously reported (9).
Statistical analysis
The tests used to analyze the data are also stated in the legend of each figure. Data are presented as mean ± SD. A Student t test for independent samples was used to assess statistically significant differences between C57BL/6-TG(Vil-LDLR) and control mice. Two-way repeated measures ANOVA was used to assess genotype-dependent differences over time (e.g., body weight curves, energy expenditure). The differences between means were considered statistically significant with a P value ≤0.05. Analyses were performed with Prism 7 (GraphPad Software) and SPSS 24. Data analysis of RT-qPCR was conducted using a one-sample t test and the mean of gene expression fold change values compared with 1 (control).
Supplemental Data
Supplemental tables and figures can be found in an online repository (15).
Results
The reconfigured jejunum in human patients after RYGB augments the machinery of cholesterol utilization
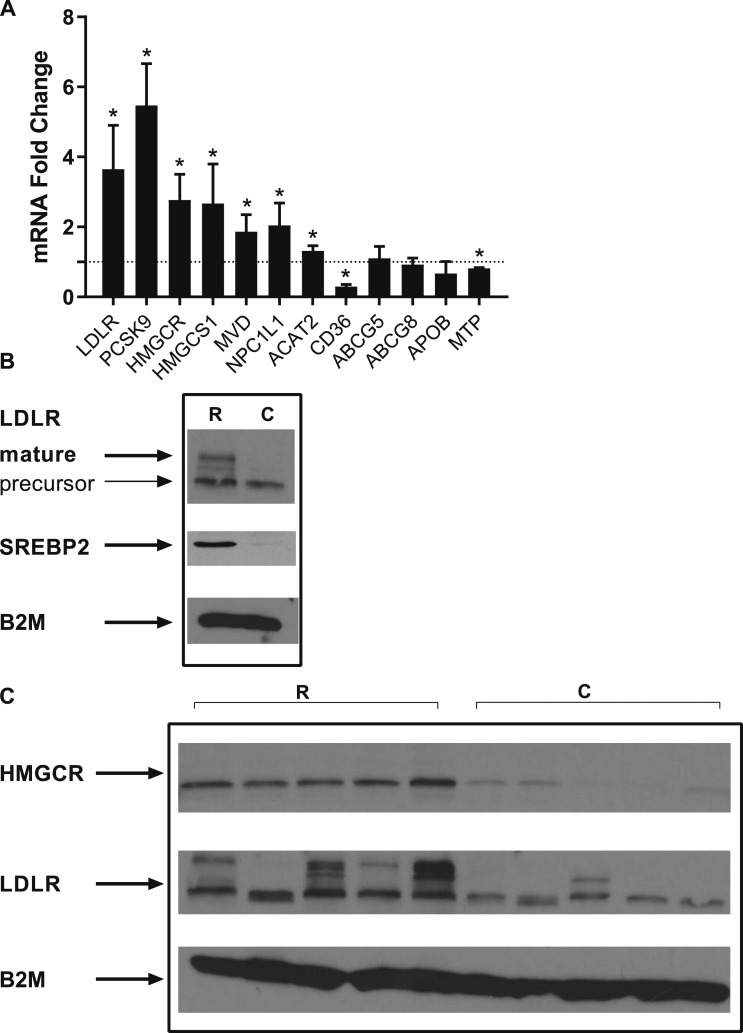
To determine whether RYGB in human patients induces changes consistent with increased circulating cholesterol uptake and de novo cholesterol synthesis, we analyzed jejunal biopsies that were collected from patients who had undergone RYGB and from controls with obesity. Patient characteristics are summarized in an online repository (15). RYGB upregulated the gene expression levels of LDLR (Fig. 1). Consistently, we observed an increase in proprotein convertase subtilisin/kexin type 9 (PCSK9) levels. Several enzymes related to cholesterol biosynthesis were increased, including HMGCR, the enzyme of the rate-limiting step; and 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1), which provides the substrate for the reductase and the mevalonate diphosphate decarboxylase (MVD), catalyzing the conversion of mevalonate pyrophosphate into isopentenyl pyrophosphate. Whereas sterol efflux transporters ATP-binding cassette subfamily G members 5 and 8 (ABCG5 and ABCG8) were unchanged, the Niemann-Pick C1-like protein 1 (NPC1L1), which plays a major role in luminal cholesterol absorption by the enterocytes, was significantly upregulated. In agreement with a potential increase in intestinal cholesterol influx and thus augmented cellular stores of cholesterol esters, acetyl-coenzyme A acetyltransferase 2 (ACAT2), the enzyme responsible for cholesterol esterification was increased. Microsomal TG transfer protein (MTP), playing a central role in the assembly and secretion of lipoproteins important for the synthesis of chylomicrons and VLDL, was downregulated. There was also a trend toward decreased expression of apolipoprotein B (APOB). These findings are consistent with the significant decrease in TG levels that is observed in patients after RYGB, including those participating in this study (15). Additionally, CD36, which is expressed in the luminal surface of enterocytes and is involved in the transport of fatty acids and cholesterol from the intestinal lumen to the lymphatic system, was downregulated. We then confirmed that LDLR and HMGCR were upregulated at the protein level (Fig. 1B and 1C). We also found that the protein levels of SREBP2, the master regulator of cholesterol metabolism in cells, were increased. Collectively, these results suggest that RYGB may trigger an intestinal metabolic response, which enhances the intestinal machinery involved in circulating cholesterol absorption, uptake, and biosynthesis while downregulating the machinery of intestinal cholesterol and TG efflux.
Figure 1.
RYGB induces changes in the expression of genes related to cholesterol metabolism in the transposed intestine (Roux limb) of human patients. (A) RNA levels of several metabolic targets (n = 3 to 13 per group). Values represent mean ± SD and are shown in arbitrary units after normalization to housekeeping genes (see “Materials and Methods”). (B) Protein levels in pooled samples from RYGB-treated patients (n = 14; time after surgery, 18.9 ± 5 mo) and control patients (n = 13). (C) Protein levels in individual tissue lysate samples (n = 5 per group; 13.2 ± 1.30 mo after the surgery). For SREBP2, in Western blot the protein was detected in its active form. *P < 0.05, by one-sample t test. Loading control: ABCG5, ATP-binding cassette subfamily G member 5; ABCG8, ATP-binding cassette subfamily G member 8; ACAT2, acetyl-coenzyme A acetyltransferase 2; APOB, apolipoprotein B; B2M, β2-microglobulin; C, control; HMGCS1, 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1; MTP, microsomal triglyceride transfer protein; MVD, mevalonate diphosphate decarboxylase; R, RYGB.
Intestine-specific LDLR overexpression leads to decreased blood cholesterol levels
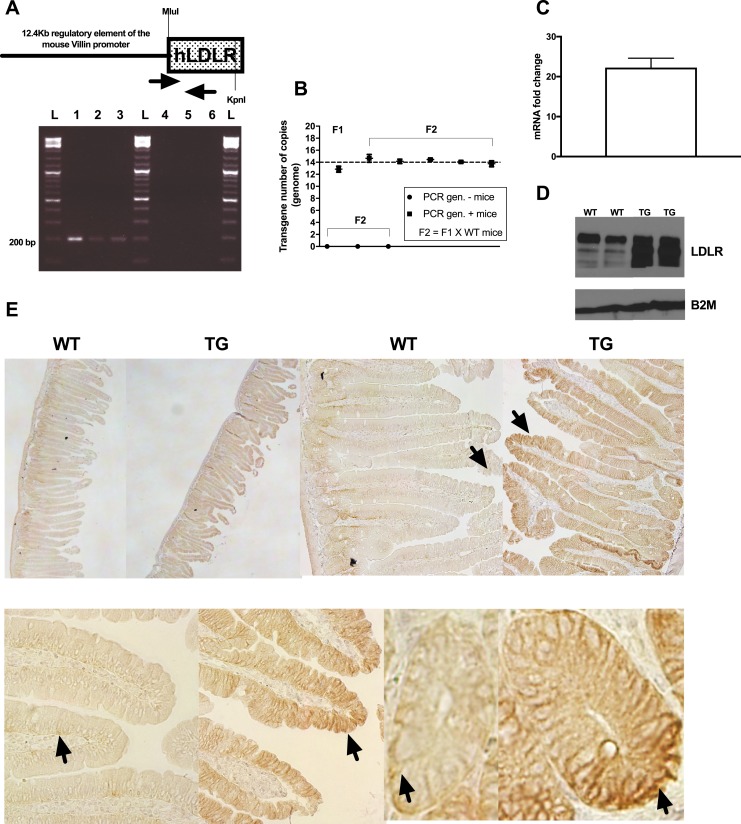
We then wanted to determine whether increasing LDLR expression specifically in the intestine could be sufficient to replicate the effects of RYGB on whole-body cholesterol metabolism and induce a decrease in circulating cholesterol levels. To test this hypothesis, we generated mice that overexpress the human LDLR specifically in the intestine, under the control of a 12.4-kb mouse villin regulatory sequence (15, 24) (Fig. 2A). C57BL/6-TG(Vil-LDLR) mice were viable and healthy and the transgene was stably transmitted over the generations (Fig. 2B and 2D). By analyzing intestinal sections with immunohistochemistry against LDLR, we found that the transgene expressed specifically in the columnar epithelium and crypt cells, and it closely paralleled the endogenous gene expression pattern (Figure 2E). Because villin is also expressed in renal epithelium, we analyzed kidney lysates and we ruled out any ectopic expression of the transgene (15). With Western blotting, we observed a continuous pattern of overexpression of the transgene along the different segments of the small intestine (duodenum through ileum) (15). The intestine did not present any apparent structural abnormality (15), but interestingly its total length was significantly and consistently increased (15). Additionally, overexpression of LDLR in the intestine did not interfere with normal mouse growth (15).
Figure 2.
Molecular characterization of the transgenic mouse line [C57BL/6-TG(Vil-LDLR)] of intestine-specific LDLR overexpression. (A) Schematic representation of the construct we used to generate the mice. MluI and KpnI restriction enzyme sites, which were used to clone the open reading frame of the human (h)LDLR gene, are shown. Primer location and representative genotyping results [(A), lower panel; anticipated PCR product, 195 bp] are shown. (B) Droplet PCR results obtained using F1 and F2 animals of known genotype. Transgene was stably transmitted over the generations in multiple (13, 14) copies. (C) Representative transgene and (D) LDLR protein expression levels in the duodenum of F2 animals (transgenic mice, n = 4; wild-type mice, n = 3). Data shown in (C) represent mean ± SD. (E) Immunohistochemical staining detected intestinal LDLR overexpression and located the transgene in the columnar epithelium and crypt cells. Representative sections of proximal intestinal sections stained against LDLR are shown. Upper panel: left to right, ×4 and ×10 original magnification of sections. Lower panel: details of villi and crypt cell staining (original magnification, ×40). Arrows indicate examples of areas with positive staining. 1, positive control (12.4-kb Villin-ΔATG-hLDLR plasmid DNA); 2, colony founder (F0); 3, F1 mouse (obtained by crossing F0 with C57BL/6 wild-type mice); 4, negative control (C57BL/6 wild-type mouse); 5, F1 mouse; 6, H2O; L, ladder; TG, C57BL/6-TG(Vil-LDLR) transgenic mice; WT, wild-type littermate controls.
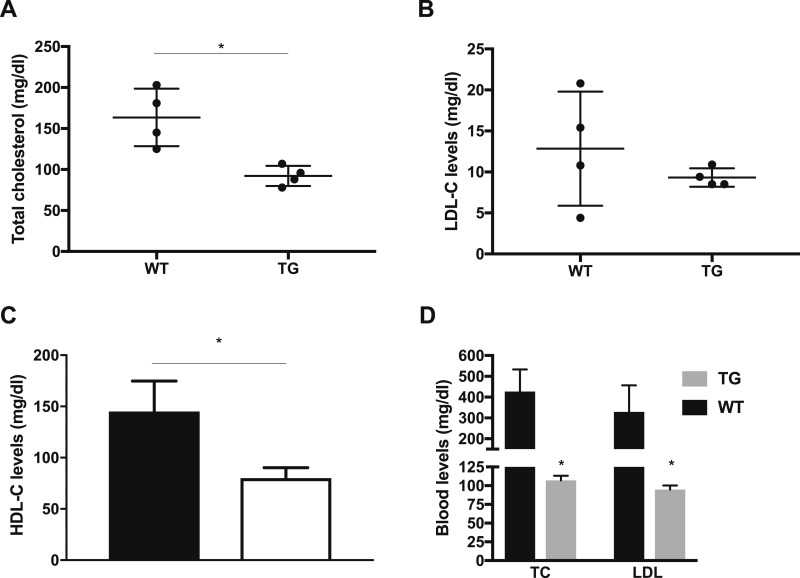
Compared with wild-type littermate controls, the blood levels of total cholesterol were significantly reduced by almost 50% in C57BL/6-TG(Vil-LDLR) mice (Fig. 3A). LDL, HDL, and VLDL serum lipoproteins were then separated by FPLC. All cholesterol fractions associated with the different lipoproteins tended to be decreased, and we also observed a significant decrease in HDL-cholesterol (Fig. 3B and 3C). Mice carry most plasma cholesterol in HDL, and this result is consistent with previous observations of reduced circulating HDL levels in murine models of global LDLR overexpression (27). Collectively, the lipid profile of C57BL/6-TG(Vil-LDLR) mice suggested that the expression of a functional LDLR transgene likely mediates a constitutively increased flux of cholesterol across the intestine.
Figure 3.
Intestine-specific LDLR overexpression improves cholesterol metabolism. (A) Decreased levels of total circulating cholesterol in transgenic mice overexpressing the human LDLR in the intestine. Each black point represents a pooled serum sample of multiple animals, which consumed a normal chow diet. (B) We performed FPLC separation of plasma lipoproteins from pooled mouse serum samples. C57BL/6-TG(Vil-LDLR) mice on normal chow had decreased blood LDL-cholesterol levels compared with wild-type mice. Each black point represents a pooled serum sample of multiple animals, which consumed a normal chow diet. (C) We observed a significant decrease in HDL-cholesterol, which is consistent with previous observations in murine models of global LDLR overexpression. (D) Total cholesterol and LDL-cholesterol levels in animals that consumed an HFHC diet for 8 wk. Statistics: n = 8 per group per data point for (A)–(C), n = 9 individual mice for (D). *P < 0.05, by t test. TG, C57BL/6-TG(Vil-LDLR) transgenic mice; WT, wild-type littermate controls.
We hypothesized that this may also confer a metabolic advantage in conditions of increased exposure to factors that increase the risk of hypercholesterolemia. Therefore, we challenged a group of C57BL/6-TG(Vil-LDLR) mice, from weaning and for many weeks, with an atherogenic diet containing 1% cholesterol and 0.5% cholate. As shown in Fig. 3D, the intestine-specific LDLR overexpression led to complete normalization of serum total cholesterol and LDL-cholesterol levels.
The intestine of C57BL/6(Vil-LDLR) mice exhibits a molecular signature of cholesterol overload, an adaptive protective response against cytotoxicity, and increased glucose transport and consumption
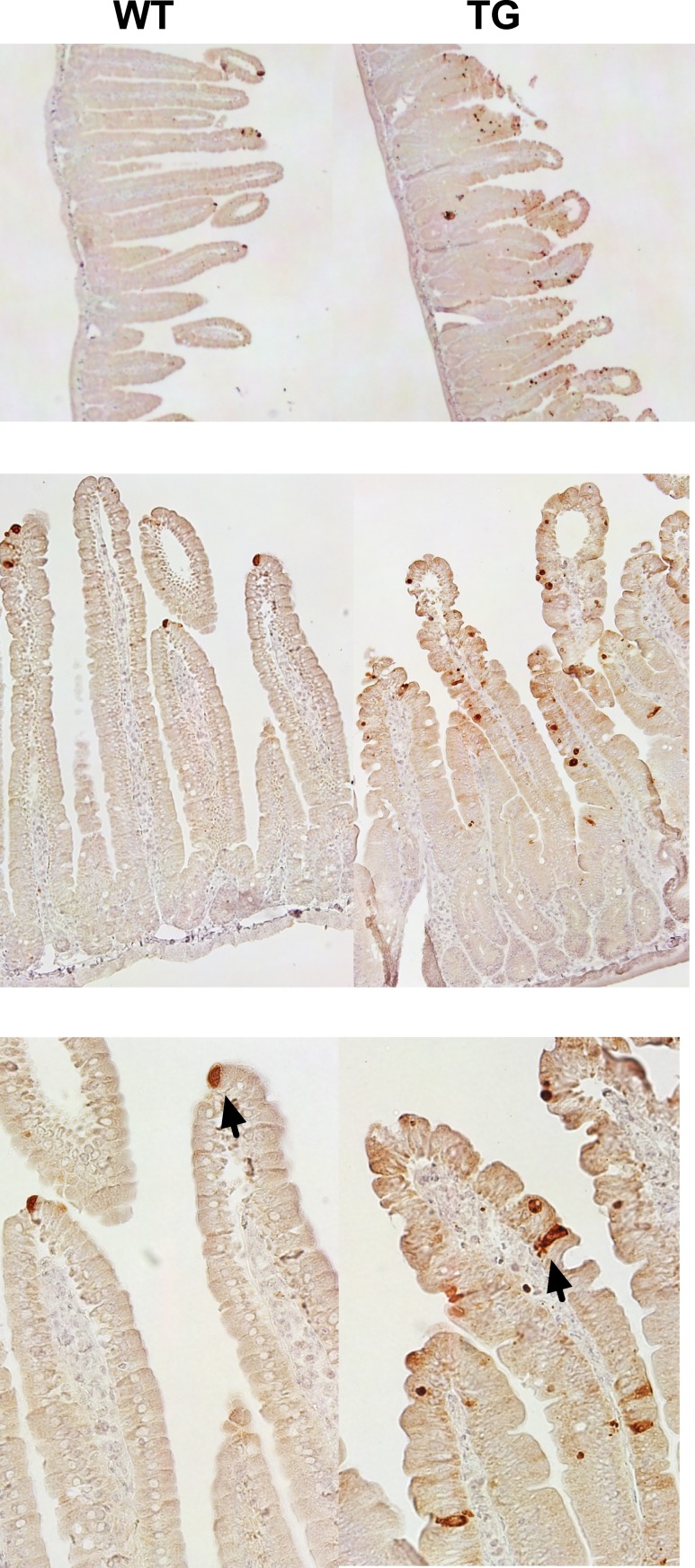
To characterize the changes in intestinal metabolism of C57BL/6-TG(Vil-LDLR) mice, we performed a transcriptome analysis of the duodenum and we compared the gene expression signatures of the transgenic animals to those of wild-type controls. C57BL/6-TG(Vil-LDLR) mice exhibited a marked enrichment of pathways ascribable to immune response mediated by interferon-α and interferon-β (15). Specifically, we observed a significant increase of several key components of the interferon response, such as signal transducers and activator of transcription (Stat) 1 and 2. Consistently, the most significantly upregulated gene was the interferon-induced 2′,5′-oligoadenylate synthetase (Oas) 2 (fold change, 34.45; false discovery rate, 0.012). We speculated that the enrichment of this process may be secondary to changes in intestinal permeability. Specifically, we hypothesized that the excess of intracellular cholesterol could enhance mechanisms of cell apoptosis (intestinal shedding), alter the barrier function of the intestine, and eventually promote a protective response (28). Using immunohistochemistry, we looked at the cleaved form of caspase-3 (activated caspase-3), a marker of intestinal epithelial shedding, and we found a strong increase in the proximal intestine of C57BL/6-TG(Vil-LDLR) mice compared with controls (Fig. 4). Importantly, there was no histological evidence of mucosal damage on histology and the intestinal architecture was normal (15). In agreement with our hypothesis of compromised intestinal barrier function, C57BL/6-TG(Vil-LDLR) mice presented with a substantial downregulation of components of innate defense mechanisms (29, 30), such as defensins, mucins, and various Paneth cell–associated genes (15).
Figure 4.
Epithelial cell shedding is increased in the proximal intestine of C57BL/6-TG(Vil-LDLR) mice. Representative staining of intestinal sections from control and transgenic mice obtained using a cleaved caspase-3 antibody. From top to bottom, original magnification of sections was ×4, ×20, and ×40. Arrows indicate examples of apoptotic cells.
The transcriptome analysis also revealed a concurrent enrichment of cell cycle–related pathways and an upregulation of several cyclins, suggesting an increase in the rate of epithelial cell turnover (15).
We also noticed that genes belonging to the superfamily of solute-carrier (SLC), membrane-bound transporters were generally downregulated (15). Among these, there were many transporters mediating luminal absorption and resorption of several factors and nutrients, including monocarboxylates (e.g., Slc5a12), amino acids and oligopeptides (e.g., Slc5a4b, Slc36a1, Slc7a15, Slc15a1), ascorbate and folate (e.g., Slc23a1, Slc46a1), and glucose and other sugars (e.g., Slc5a4a, Slc5a11, Slc5a1). Collectively, these results point to a phenotype of decreased macronutrient and micronutrient absorption.
Additionally, genes of the proteasome family were substantially upregulated (15). Proteasome is involved in several functions, including maintenance of protein homeostasis, cell cycle regulation (e.g., apoptosis), and immune response. Thus, it was not surprising that its activation emerged as one of the main metabolic features of the gene signature of the intestine of these transgenic mice.
We also looked for any potential consequences of LDLR overexpression on the regulation of endogenous intestinal cholesterol metabolism. Endogenous Ldlr expression levels were unchanged in the intestine of transgenic mice (Fig. 5A). There was also upregulation of several genes related to cholesterol synthesis, whereas Cd36 was downregulated (15). These data seemed to rather reflect a complex balance of cell proliferation and death and, consequently the balance between cholesterol requirement and utilization (31), which occur to a different extent in distinct intestinal compartments (i.e., along the crypt/villus axis).
Figure 5.
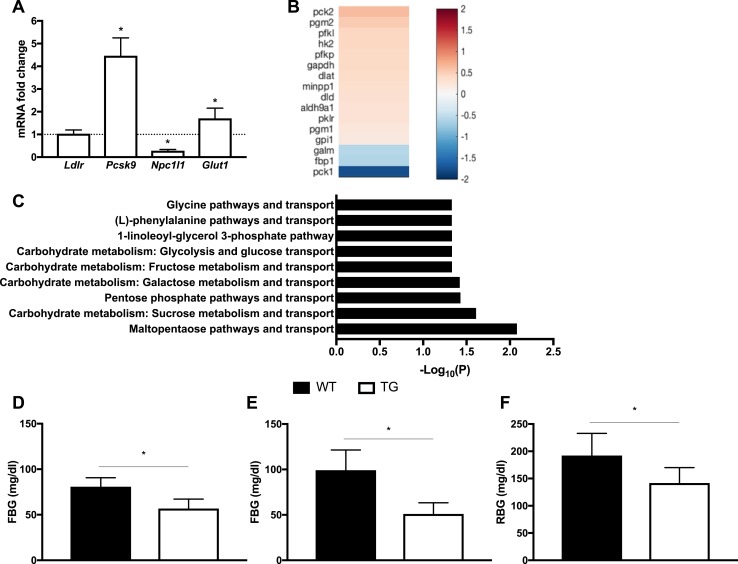
Intestine-specific overexpression of LDLR improves glucose metabolism. (A) Real-time PCR was used to validate several targets that we selected based on a microarray study of the duodenum. The targets included Glut1 and several key factors and enzymes of cholesterol metabolism: Ldlr, Npc1l1, and Pcsk9. (B) Heat map and color key for significantly regulated genes related to glucose metabolism in the duodenum. A comparison of transgenic vs wild-type mice is shown. Each cell represents the average log2 fold change. The list was based on the microarray gene expression analysis of the duodenum. (C) List of the statistically significant metabolic networks in the duodenum. A comparison of transgenic vs wild-type mice is shown. These pathways were determined using Metacore™ (Thompson Reuters). Most of these networks are related to the metabolism of glucose and other carbohydrates, suggesting that LDLR overexpression affects glucose metabolism of the intestine. (D) Chow-fed C57BL/6-TG(Vil-LDLR) mice had lower fasting blood glucose (FBG) levels compared with controls. (E) Fasting blood glucose levels normalized in transgenic mice fed with a HFHC diet. (F) Blood glucose levels were lower at weaning. These measurements were performed without fasting [random blood glucose (RBG) levels]. Statistics: n = 3 to 15 per group; results are expressed as mean ± SD; one-sample t test for real-time PCR, t test for other comparisons between groups. *P < 0.05. TG, C57BL/6-TG(Vil-LDLR) transgenic mice; WT, wild-type littermate controls.
Surprisingly, intestinal glucose metabolism also appeared to be profoundly affected in C57BL/6-TG(Vil-LDLR) mice. A pathway analysis using the Metacore™ analytical software suite (Thompson Reuters) showed that most of the metabolic pathways that were statistically significant (false discover rate, <0.05) were related to glucose and carbohydrate metabolism. Additionally, one of the most significantly enriched pathways appeared to be related to the regulation and function of sirtuin 6 (15), which includes key glucose metabolic targets, such as phosphoenolpyruvate carboxykinase 1 (Pck1) and the SLC family 2 member 1 (Slc2a1) known also as glucose transporter 1 (Glut1). As shown in Fig. 5A–5C, Pck1, which is the cytosolic variant of the enzyme catalyzing the conversion of oxaloacetate to phosphoenolpyruvate and the main control of gluconeogenesis, was significantly downregulated. Glut1, whose expression in the intestine gradually declines during fetal life was significantly upregulated. Importantly, this increase was confirmed by RT-qPCR. Overall, the microarray signatures pointed to an enhancement of glycolysis, with genes coding for enzymes involved in its key steps being significantly upregulated. These genes included hexokinase 2 (Hk2), glyceraldehyde-3-phosphate (Gapdh), pyruvate kinase (Pklr), and phosphofructokinase, liver type (Pfkl), which catalyzes the rate-limiting step of glycolysis. SLC family 2 member 2 [Slc2a2 or glucose transporter 2 (Glut2)] expression levels decreased, suggesting a reduction in luminal glucose absorption. Collectively, these data suggest that intestinal glucose homeostasis is shifted toward increased circulating glucose uptake and utilization, in addition to a reciprocal decrease in luminal absorption and gluconeogenesis. Consistently, we observed that glucose metabolism was also substantially affected in C57BL/6-TG(Vil-LDLR) mice. Chow-fed transgenic animals exhibited significantly lower fasting blood glucose levels (Fig. 5D). Additionally, the fasting blood glucose levels of a group of C57BL/6-TG(Vil-LDLR) animals that were placed on an HFHC diet were significantly lower than those of the HFCH wild-type controls, and no different than the levels of the wild-type, chow-fed animals (Fig. 5E). These transgenic mice displayed also improved glucose tolerance (15). The beneficial changes in glucose metabolism occurred early in the life of the transgenic mice, and these animals had better random glucose levels compared with controls, even at weaning (Fig. 5F).
C57BL/6(Vil-LDLR) mice exhibit increased fecal cholesterol content
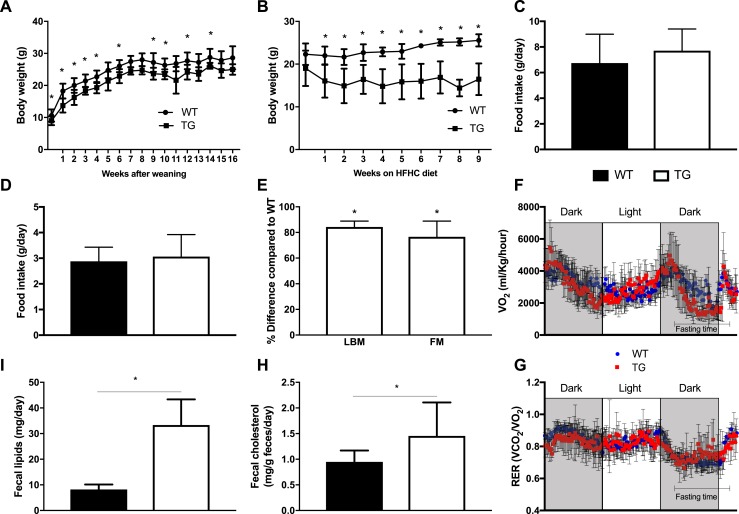
We then hypothesized that the gene signatures of augmented cellular turnover and intestinal shedding may be associated with an increase in fecal lipid content. To test this hypothesis, we first characterized some key metabolic traits. We observed that C57BL/6-TG(Vil-LDLR) mice had significantly lower body weight compared with wild-type controls on both normal chow and HFHC diets (Fig. 6A and 6B). This occurred despite the lack of difference in food intake between the two groups (Fig. 6C and 6D). Additionally, there was no difference in diet preferences (HFHC or normal chow) (15). To characterize the nature of body weight loss in transgenic mice, we assessed body fat and lean tissue contents by DEXA scanning. Interestingly, we found a decrease in both lean and fat masses in C57BL/6-TG(Vil-LDLR) mice as compared with controls (Fig. 6E). To further examine the physiological mechanism underlying the decreased body weight of C57BL/6-TG(Vil-LDLR) mice, we assessed their energy expenditure with indirect calorimetry. We did not observe any difference in resting and total oxygen consumption or heat production and respiratory exchange ratio compared with controls (Fig. 6F and 6G) (15). Finally, we measured cholesterol and lipid content in the fecal output of chow-fed C57BL/6-TG(Vil-LDLR) and normal control mice, and we found that both were increased in the transgenic mice (Fig. 6H and 6I). Based on these results, we concluded that the increased cholesterol and lipid excretion may be the primary underlying cause for the decreased body weight.
Figure 6.
Intestine-specific LDLR overexpression improves metabolic function. (A) Transgenic mice on normal chow diet have lower body weight compared with wild-type controls. (B) When challenged with an HFHC diet, transgenic animals exhibit a dramatic loss of body weight, but the animals do not become sick or cachectic. (C and D) There was no difference in food intake either on a (C) normal chow or (D) HFHC diet. (E) Body composition was performed with DEXA and showed a decrease in both fat and lean body mass in transgenic animals compared with controls. Both groups were on an HFHC diet from weaning and for 10 wk. (F and G) There was no difference in (F) oxygen consumption and (G) respiratory exchange ratio (RER) between the two groups on an HFHC diet. (H) Fecal cholesterol and (I) total lipid contents were increased in transgenic mice. Statistics: n = 4 to 15 per group; t test, two-way repeated measures ANOVA for assessment of differences of body weight curves and energy expenditure data. *P < 0.05. TG, C57BL/6-TG(Vil-LDLR) transgenic mice; WT, wild-type littermate controls.
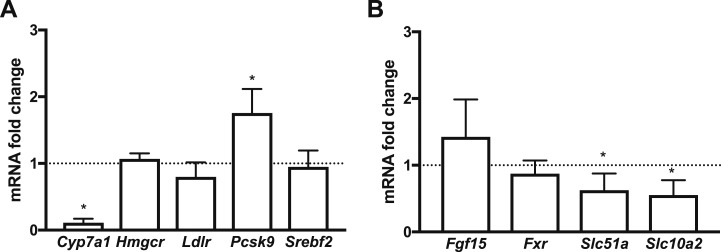
Because under normal physiological conditions, biliary excretion is the main route for cholesterol elimination, we sought to examine the effects of intestine-specific LDLR overexpression on bile acid metabolism. We first looked at the hepatic gene expression levels of the cytochrome P450 family 7 subfamily A member 1 (Cyp7a1), the rate-limiting enzyme in bile acid synthesis. Compared with controls, they were dramatically decreased (Fig. 7A). We then continued to further examine bile metabolism by analyzing some of the key components of enterohepatic circulation and bile acid transport in the ileum of the animals (Fig. 7B). Expression levels of the nuclear bile acid receptor farnesoid X receptor (Fxr), which senses bile acid levels, and fibroblast growth factor 15 (Fgf15), which represses Cyp7a1 in liver (32, 33), were unchanged. However, SLC family 10 member 2 (Slc10a2 or Asbt), which actively transports bile acids from the lumen of the small intestine across the apical brush border membrane, and SLC family 51 α subunit (Slc51a or Osta), which mediates the efflux of bile into the portal circulation, were both significantly downregulated in the ileum of transgenic mice. Collectively, these results suggested that hepatic biliary synthesis and bile acid transport in the ileum may be reduced in C57BL/6-TG(Vil-LDLR) mice.
Figure 7.
Gene expression changes (assessed by real-time PCR) point to a reduction of enterohepatic circulation in C57BL/6-TG(Vil-LDLR) mice. (A) RNA expression levels of genes related to bile acid and cholesterol metabolism in the liver of C57BL/6-TG(Vil-LDLR) and control mice. (B) RNA expression levels of key regulators of bile acid metabolism and transport in the ileum. Statistics: n = 3 to 4 per group; one-sample t test. *P < 0.05. Fgf15, fibroblast growth factor 15; Fxr, farnesoid X receptor; Slc51a, solute carrier family 51 α subunit; Slc10a2, solute carrier family 10 member 2; Srebf2, sterol regulatory element–binding protein 2.
We also examined whether there may be any changes in the machinery of luminal cholesterol absorption that could explain the increased fecal cholesterol content. Transcriptional levels of the cell surface cholesterol-sensing receptor Npc1l1 were significantly reduced in the intestine of transgenic mice (Fig. 5A). This finding suggests a reciprocal relationship between LDLR and NPC1L1, which likely contributes to reduced luminal efficiency of C57BL/6-TG(Vil-LDLR mice in absorbing cholesterol, thereby possibly increasing further the fecal cholesterol content.
We then wanted to know how the intestine-specific overexpression of LDLR affects key enzymes and factors of cholesterol metabolism in the liver. We found that expression levels of Srebf2 were unchanged in transgenic mice (Fig. 7A). Consistently, we did not observe any significant change in Hmgcr and Ldlr transcriptional regulation. However, Pcsk9 expression levels were significantly increased in the liver of transgenic mice, suggesting that mechanisms counteracting intestinal LDLR overexpression at the posttranslational level may have been enacted (34). Collectively, our results suggest that in C57BL/6-TG(Vil-LDLR) mice, the suppression of the biliary route may not be paralleled by other major changes in cholesterol hepatic metabolism and may primarily occur to compensate for the loss of cholesterol mediated by the intestine.
Overall, these data show that owing to the overexpression of LDLR in the intestinal epithelium, whole-body cholesterol homeostasis is shifted toward an enhancement of luminal cholesterol excretion, which, in turn, appears to improve whole-body lipid and glucose metabolism and decrease body weight, affecting, however, both lean and fat body mass.
Discussion
The foundational finding of this study is that rodents and patients treated with RYGB exhibit an increase in the intestinal machinery of cholesterol metabolism. Factors that orchestrate cellular cholesterol metabolism (SREBP2) or play an important role in the regulation of cholesterol synthesis (HMGCR) and uptake (LDLR) appear to be upregulated in the reconfigured intestine (Roux limb). These results corroborate our recent rodent studies (9) suggesting that the RYGB-induced changes in intestinal cholesterol metabolism are conserved across species and thus highly likely play a fundamental role in the beneficial metabolic effects of this procedure. Because cholesterol is an essential building block for cellular growth and function, the observed changes are consistent with a metabolic remodeling process aiming to meet the increased structural and bioenergetic demands of tissue growth and maintenance of the reconfigured intestine.
Based on these findings, our working hypothesis is that the overexpression of LDLR in the intestinal mucosa leads to disposal of circulating cholesterol into intestinal cells, and the increased cholesterol uptake and utilization could contribute to the improvement of the lipid profile of RYGB-treated rodents and human patients. The study of our new mouse transgenic line, which overexpresses the receptor specifically in the intestinal mucosa [C57BL/6-TG(Vil-LDLR)], confirms that this mechanism may be sufficient to reduce blood cholesterol levels. Intestinal overexpression of human LDLR induced a systemic rearrangement of cholesterol metabolism. C57BL/6-TG(Vil-LDLR) mice exhibited a favorable blood lipid profile on both low-cholesterol and high-cholesterol diets, likely driven by an increase in fecal cholesterol output. Remarkably, these mice had significantly lower body weight than did littermate controls. However, their energy expenditure and food intake were not different, which suggests that the increased lipid fecal output and the consequent loss of energy may be responsible for the lower body weight.
Several sources contribute to total fecal cholesterol content (35), and the overexpression of intestinal LDLR in C57BL/6-TG(Vil-LDLR) mice appears to lead to enrichment of the fractions that come from intestinal mucosa shedding and nonabsorbed cholesterol. Unexpectedly, there was a decrease in Npc1l1 transcriptional levels and an increase of the cleaved caspase-3 protein in the distal intestinal epithelium of C57BL/6-TG(Vil-LDLR) mice. Thus, intestine-specific LDLR overexpression may substantially enhance the otherwise scarce LDL-cholesterol influx from the circulation into the distal intestine (36), which in turn may reduce the absorption of cholesterol from the lumen. This is further supported by other studies suggesting a reciprocal relationship between intestinal luminal cholesterol absorption and basolateral uptake via LDLR (37). Additionally, several studies have shown that the accumulation of cholesterol inside the cells may trigger cellular programs of apoptosis (28).
To compensate for the luminal loss of cholesterol, C57BL/6-TG(Vil-LDLR) mice appear to suppress CYP7A1 and hepatic bile synthesis. In mouse models with either reduced or abrogated biliary cholesterol secretion, including mice deficient for Cyp7a1, neutral fecal sterol excretion is preserved (38–40). This observation has provided evidence for the hypothesis that there is a mechanism mediating the active transport of cholesterol from the bloodstream into the lumen known as reverse, transintestinal cholesterol excretion (TICE). According to some studies, LDLR may participate in TICE (41), and this mechanism may account for up to 70% of fecal neutral sterol excretion in mice (42). Our finding that transgenic mice exhibit increased cholesterol fecal content despite the reduced transcriptional levels of hepatic Cyp7a1 and the suppression of the cholesterol biliary route may suggest that LDLR overexpression may enhance TICE. Although it is difficult to determine whether the increased cholesterol luminal content is due to enhanced TICE or enhanced cellular apoptosis, it is clear that our study supports the hypothesis that the biliary route may not be the only pathway that substantially contributes to reverse cholesterol excretion into the intestinal lumen. In agreement, Blanchard et al. (43) have recently reported that RYGB decreased plasma cholesterol levels in mice by enhancing TICE-related mechanisms, with a small, however, upregulation of LDLR expression, which did not reach statistical significance.
Surprisingly, as in RYGB-treated rodent models, the C57BL/6-TG(Vil-LDLR) mice revealed an increase in intestinal cell proliferation. This likely emerged as a response to counteract apoptosis and epithelial cell loss. Interestingly, a recent study has shown that intestinal polyps in a mouse model of colorectal and intestinal cancer (Apc-deficient Min mouse model) exhibit overexpression of LDLR, and it has been speculated that lipid accumulation may play an important role in polyp formation (44). Consistently, another recent study showed that the excess cellular cholesterol increases intestinal stem cell proliferation (45).
Several other noteworthy findings that are similar to what we have observed in the intestine of RYGB-treated rodents include the upregulation of the machinery responsible for glucose utilization, the increased expression of the basolateral glucose transporter Glut1, and the reduction in transcriptional levels of enzymes of gluconeogenesis. Remarkably, C57BL/6-TG(Vil-LDLR) mice had decreased blood glucose levels, which is also another similarity with RYGB. These data support the hypothesis that independently of the causes triggering cell proliferation, intestinal growth requires glucose, and the transporter GLUT1 may play a role. Additionally, they further support the concept that glucose utilization by the intestine may play an important role in whole-body glucose metabolism.
In conclusion, the commonalities between rodents and human patients treated with RYGB and the systemic rearrangement of cholesterol metabolism that occurs in mice with intestine-specific overexpression of LDLR suggest that the perturbation of intestinal cholesterol metabolism may be a key mechanism underlying the beneficial metabolic effects of RYGB. Additionally, they highlight that the manipulation of intestinal cholesterol metabolism may be an alternative therapeutic target for the treatment of HL. It appears, however, that loading the intestinal mucosa with intestinal cholesterol may lead to increased intestinal cellular turnover, emphasizing the need to better understand the control and release of intestinal cholesterol load.
Acknowledgments
We thank N. Li of Tufts Comparative Pathology Services Core for assistance with histomorphological studies.
Financial Support: This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R01-DK108642 (to N.S.), as well as funding from the Edward Mallinckrodt Jr. Foundation (to N.S.), the Diabetes Action Research and Education Foundation (to N.S.), a Rothschild fellowship (to D.B.-Z.), a Human Frontiers in Science postdoctoral fellowship (to D.B.-Z.), and National Fund for Scientific and Technological Development of the Government of Chile (FONDECYT) Grant 11160688 (to R.M.).
Author Contributions: All authors designed experiments and analyzed and interpreted data. L.M. and N.S. conceptualized, designed, performed, and/or analyzed the animal studies. L.M., P.P., R.M., and N.S. conceptualized, designed, performed, or analyzed human patient studies. S.K. participated in the design of the transgenic mouse line and, along with C.P., performed and analyzed animal studies. L.M. and N.S. wrote the manuscript with contributions from all authors.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- AUC
area under the curve
- CYP7A1
cytochrome P450 family 7 subfamily A member 1
- ddPCR
droplet digital PCR
- DEXA
dual-energy x-ray absorptiometry
- FPLC
fast protein liquid chromatography
- GLUT1
glucose transporter 1
- GO
gene ontology
- HDL
high-density lipoprotein
- HFHC
high-fat, high-cholesterol
- HL
hyperlipidemia
- HMGCR
3-hydroxy-3-methylglutaryl-coenzyme A reductase
- LDL
low-density lipoprotein
- LDLR
low-density lipoprotein receptor
- NPC1L1
Niemann-Pick C1-like protein 1
- ORF
open reading frame
- PBST
PBS with 0.1% Tween 20
- PCSK9
proprotein convertase subtilisin/kexin type 9
- RT-qPCR
reverse transcription–quantitative PCR
- RYGB
Roux-en-Y gastric bypass
- SLC
solute carrier
- SREBP2
sterol regulatory element–binding protein 2
- TG
triglyceride
- TICE
transintestinal cholesterol excretion
- VLDL
very-low-density lipoprotein
References
- 1. National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143–3421. [PubMed] [Google Scholar]
- 2. Thompson GR, O’Neill F, Seed M. Why some patients respond poorly to statins and how this might be remedied. Eur Heart J. 2002;23(3):200–206. [DOI] [PubMed] [Google Scholar]
- 3. Nguyen NT, Varela E, Sabio A, Tran CL, Stamos M, Wilson SE. Resolution of hyperlipidemia after laparoscopic Roux-en-Y gastric bypass. J Am Coll Surg. 2006;203(1):24–29. [DOI] [PubMed] [Google Scholar]
- 4. Vix M, Diana M, Liu KH, D’Urso A, Mutter D, Wu HS, Marescaux J. Evolution of glycolipid profile after sleeve gastrectomy vs. Roux-en-Y gastric bypass: results of a prospective randomized clinical trial. Obes Surg. 2013;23(5):613–621. [DOI] [PubMed] [Google Scholar]
- 5. Adams TD, Gress RE, Smith SC, Halverson RC, Simper SC, Rosamond WD, Lamonte MJ, Stroup AM, Hunt SC. Long-term mortality after gastric bypass surgery. N Engl J Med. 2007;357(8):753–761. [DOI] [PubMed] [Google Scholar]
- 6. Cohen RV, Pinheiro JC, Schiavon CA, Salles JE, Wajchenberg BL, Cummings DE. Effects of gastric bypass surgery in patients with type 2 diabetes and only mild obesity. Diabetes Care. 2012;35(7):1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cunha FM, Oliveira J, Preto J, Saavedra A, Costa MM, Magalhães D, Lau E, Bettencourt-Silva R, Freitas P, Varela A, Carvalho D. The effect of bariatric surgery type on lipid profile: an age, sex, body mass index and excess weight loss matched study. Obes Surg. 2016;26(5):1041–1047. [DOI] [PubMed] [Google Scholar]
- 8. Stefater MA, Wilson-Pérez HE, Chambers AP, Sandoval DA, Seeley RJ. All bariatric surgeries are not created equal: insights from mechanistic comparisons. Endocr Rev. 2012;33(4):595–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saeidi N, Meoli L, Nestoridi E, Gupta NK, Kvas S, Kucharczyk J, Bonab AA, Fischman AJ, Yarmush ML, Stylopoulos N. Reprogramming of intestinal glucose metabolism and glycemic control in rats after gastric bypass. Science. 2013;341(6144):406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nestoridi E, Kvas S, Kucharczyk J, Stylopoulos N. Resting energy expenditure and energetic cost of feeding are augmented after Roux-en-Y gastric bypass in obese mice. Endocrinology. 2012;153(5):2234–2244. [DOI] [PubMed] [Google Scholar]
- 11. Pihlajamäki J, Grönlund S, Simonen M, Käkelä P, Moilanen L, Pääkkönen M, Pirinen E, Kolehmainen M, Kärjä V, Kainulainen S, Uusitupa M, Alhava E, Miettinen TA, Gylling H. Cholesterol absorption decreases after Roux-en-Y gastric bypass but not after gastric banding. Metabolism. 2010;59(6):866–872. [DOI] [PubMed] [Google Scholar]
- 12. Odstrcil EA, Martinez JG, Santa Ana CA, Xue B, Schneider RE, Steffer KJ, Porter JL, Asplin J, Kuhn JA, Fordtran JS. The contribution of malabsorption to the reduction in net energy absorption after long-limb Roux-en-Y gastric bypass. Am J Clin Nutr. 2010;92(4):704–713. [DOI] [PubMed] [Google Scholar]
- 13. Kumar R, Lieske JC, Collazo-Clavell ML, Sarr MG, Olson ER, Vrtiska TJ, Bergstralh EJ, Li X. Fat malabsorption and increased intestinal oxalate absorption are common after Roux-en-Y gastric bypass surgery. Surgery. 2011;149(5):654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carswell KA, Vincent RP, Belgaumkar AP, Sherwood RA, Amiel SA, Patel AG, le Roux CW. The effect of bariatric surgery on intestinal absorption and transit time. Obes Surg. 2014;24(5):796–805. [DOI] [PubMed] [Google Scholar]
- 15. Meoli L, Ben-Zvi D, Panciotti C, Kvas S, Pizarro P, Munoz R, Stylopoulos N. Data from: Intestine-specific overexpression of LDLR improves lipid and glucose metabolism by enhancing cholesterol excretion in male mice. Mendeley 2019. Deposited 14 January 2019. https://data.mendeley.com/datasets/7jkxprm9mx/1.
- 16. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.RRID:AB_10990337.
- 18.RRID:AB_2281168.
- 19.RRID:AB_2074654.
- 20.RRID:AB_2305186.
- 21.RRID:AB_1523204.
- 22.RRID:AB_779079.
- 23.RRID:AB_10679369.
- 24. Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277(36):33275–33283. [DOI] [PubMed] [Google Scholar]
- 25.RRID:AB_2341188.
- 26. Kraus D, Yang Q, Kahn BB. Lipid extraction from mouse feces. Bio Protoc. 2015;5(1):e1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci USA. 2005;102(15):5374–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J Clin Invest. 2002;110(7):905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wehkamp J, Koslowski M, Wang G, Stange EF. Barrier dysfunction due to distinct defensin deficiencies in small intestinal and colonic Crohn’s disease. Mucosal Immunol. 2008;1(Suppl 1):S67–S74. [DOI] [PubMed] [Google Scholar]
- 30. Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12(5):319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abumrad NA, Davidson NO. Role of the gut in lipid homeostasis. Physiol Rev. 2012;92(3):1061–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–225. [DOI] [PubMed] [Google Scholar]
- 33. Flynn CR, Albaugh VL, Cai S, Cheung-Flynn J, Williams PE, Brucker RM, Bordenstein SR, Guo Y, Wasserman DH, Abumrad NN. Bile diversion to the distal small intestine has comparable metabolic benefits to bariatric surgery. Nat Commun. 2015;6(1):7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, Horton JD, Cohen JC, Hobbs HH. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282(25):18602–18612. [DOI] [PubMed] [Google Scholar]
- 35. van der Veen JN, van Dijk TH, Vrins CL, van Meer H, Havinga R, Bijsterveld K, Tietge UJ, Groen AK, Kuipers F. Activation of the liver X receptor stimulates trans-intestinal excretion of plasma cholesterol. J Biol Chem. 2009;284(29):19211–19219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fong LG, Fujishima SE, Komaromy MC, Pak YK, Ellsworth JL, Cooper AD. Location and regulation of low-density lipoprotein receptors in intestinal epithelium. Am J Physiol. 1995;269(1 Pt 1):G60–G72. [DOI] [PubMed] [Google Scholar]
- 37. Engelking LJ, McFarlane MR, Li CK, Liang G. Blockade of cholesterol absorption by ezetimibe reveals a complex homeostatic network in enterocytes. J Lipid Res. 2012;53(7):1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7α-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J Lipid Res. 1998;39(9):1833–1843. [PubMed] [Google Scholar]
- 39. Kruit JK, Plösch T, Havinga R, Boverhof R, Groot PH, Groen AK, Kuipers F. Increased fecal neutral sterol loss upon liver X receptor activation is independent of biliary sterol secretion in mice. Gastroenterology. 2005;128(1):147–156. [DOI] [PubMed] [Google Scholar]
- 40. Temel RE, Tang W, Ma Y, Rudel LL, Willingham MC, Ioannou YA, Davies JP, Nilsson LM, Yu L. Hepatic Niemann-Pick C1-like 1 regulates biliary cholesterol concentration and is a target of ezetimibe. J Clin Invest. 2007;117(7):1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le May C, Berger JM, Lespine A, Pillot B, Prieur X, Letessier E, Hussain MM, Collet X, Cariou B, Costet P. Transintestinal cholesterol excretion is an active metabolic process modulated by PCSK9 and statin involving ABCB1. Arterioscler Thromb Vasc Biol. 2013;33(7):1484–1493. [DOI] [PubMed] [Google Scholar]
- 42. van der Velde AE, Brufau G, Groen AK. Transintestinal cholesterol efflux. Curr Opin Lipidol. 2010;21(3):167–171. [DOI] [PubMed] [Google Scholar]
- 43. Blanchard C, Moreau F, Ayer A, Toque L, Garçon D, Arnaud L, Borel F, Aguesse A, Croyal M, Krempf M, Prieur X, Neunlist M, Cariou B, Le May C. Roux-en-Y gastric bypass reduces plasma cholesterol in diet-induced obese mice by affecting trans-intestinal cholesterol excretion and intestinal cholesterol absorption. Int J Obes. 2018;42(3):552–560. [DOI] [PubMed] [Google Scholar]
- 44. Mutoh M, Komiya M, Teraoka N, Ueno T, Takahashi M, Kitahashi T, Sugimura T, Wakabayashi K. Overexpression of low-density lipoprotein receptor and lipid accumulation in intestinal polyps in Min mice. Int J Cancer. 2009;125(11):2505–2510. [DOI] [PubMed] [Google Scholar]
- 45. Wang B, Rong X, Palladino END, Wang J, Fogelman AM, Martín MG, Alrefai WA, Ford DA, Tontonoz P. Phospholipid remodeling and cholesterol availability regulate intestinal stemness and tumorigenesis. Cell Stem Cell. 2018;22(2):206–220.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]