Abstract
Background
Spiny mice of the genus Acomys are distributed mainly in dry open habitats in Africa and the Middle East, and they are widely used as model taxa for various biological disciplines (e.g. ecology, physiology and evolutionary biology). Despite their importance, large distribution and abundance in local communities, the phylogeny and the species limits in the genus are poorly resolved, and this is especially true for sub-Saharan taxa. The main aims of this study are (1) to reconstruct phylogenetic relationships of Acomys based on the largest available multilocus dataset (700 genotyped individuals from 282 localities), (2) to identify the main biogeographical divides in the distribution of Acomys diversity in dry open habitats in Afro-Arabia, (3) to reconstruct the historical biogeography of the genus, and finally (4) to estimate the species richness of the genus by application of the phylogenetic species concept.
Results
The multilocus phylogeny based on four genetic markers shows presence of five major groups of Acomys called here subspinosus, spinosissimus, russatus, wilsoni and cahirinus groups. Three of these major groups (spinosissimus, wilsoni and cahirinus) are further sub-structured to phylogenetic lineages with predominantly parapatric distributions. Combination of alternative species delimitation methods suggests the existence of 26 molecular operational taxonomic units (MOTUs), potentially corresponding to separate species. The highest genetic diversity was found in Eastern Africa. The origin of the genus Acomys is dated to late Miocene (ca. 8.7 Ma), when the first split occurred between spiny mice of eastern (Somali-Masai) and south-eastern (Zambezian) savannas. Further diversification, mostly in Plio-Pleistocene, and the current distribution of Acomys were influenced by the interplay of global climatic factors (e.g., Messinian salinity crisis, intensification of Northern Hemisphere glaciation) with local geomorphology (mountain chains, aridity belts, water bodies). Combination of divergence dating, species distribution modelling and historical biogeography analysis suggests repeated “out-of-East-Africa” dispersal events into western Africa, the Mediterranean region and Arabia.
Conclusions
The genus Acomys is very suitable model for historical phylogeographic and biogeographic reconstructions of dry non-forested environments in Afro-Arabia. We provide the most thorough phylogenetic reconstruction of the genus and identify major factors that influenced its evolutionary history since the late Miocene. We also highlight the urgent need of integrative taxonomic revision of east African taxa.
Electronic supplementary material
The online version of this article (10.1186/s12862-019-1380-9) contains supplementary material, which is available to authorized users.
Keywords: Acomys, Savanna, Biogeography, Africa, Arabia, Sahara, Somali-Masai, Zambezian savanna, Plio-Pleistocene
Background
The Old-World savanna biome spans the tropical grasslands, scrublands and wooded savannas of sub-Saharan Africa [1]. These open non-forested habitats represent the most widespread terrestrial environment in Africa [2] and they harbour one of the most abundant and diverse mammalian communities on Earth [3]. In Africa, four major biogeographic regions can be distinguished, which are defined by the geographical distribution of vascular plants and terrestrial vertebrates, where savanna-like ecosystems predominate (Zambezian, Somali, Sudanian and South African [3]).
Numerous geological and climatic events have affected the biological diversity of contemporary savanna-like ecosystems in Africa. In Eastern Africa, the East African Rift System (EARS) started to develop ca. 45–33 Ma [4], which led to a change in the region’s topography and the consequent aridification of East Africa, most intensively since late Miocene [5–8]. These climatic changes are best documented by the shift from C3 (moisture-adapted plants) to C4 (tropical arid-adapted grasses) plants [9–11]. The climatically turbulent Pliocene and especially Pleistocene periods, when arid and humid conditions alternated, resulted in a series of expansions and contractions of climatic zones [12, 13] that influenced the distribution and diversification of biodiversity in this region [14, 15].
Small mammals, especially rodents, are very good model organisms for phylogeographic reconstructions. Most of them are usually habitat specialists, exhibit low dispersal ability and have relatively high substitution rates, at least at mitochondrial DNA (mtDNA). Spiny mice of the genus Acomys I. Geoffroy Saint-Hilaire, 1838 inhabit seasonally dry open habitats in large regions of sub-Saharan Africa, the Eastern Mediterranean and the Arabian Peninsula [16]. Because they usually constitute abundant parts of local small mammal communities and their samples are easy to collect, they potentially represent a suitable group for testing hypotheses pertaining to the biogeography of dry open habitats in Africa and Arabia. Acomys belongs to a handful of rodent taxa that have been extensively studied for decades, and they have been used in several fields of study (e.g. ecology [17–22], physiology [23–26] and evolutionary biology [27–29]). Nevertheless, the vast majority of these studies was performed on taxa from Israel and neighbouring areas of the Middle East, representing only small fragment of the phylogenetic diversity of the genus [30–33].
The genus Acomys was described as a separate taxon at the beginning of the nineteenth century, but there is still no synthesis of diversity across the genus, even though species names and descriptions abound [16, 34–38] (see Additional file 1). There were repeated attempts for systematic classification of Acomys using morphological characters [34, 39] and chromosomes [38, 40] (see Additional file 1). However, many currently recognized species cannot be easily distinguished using morphological characters due to significant intraspecific variability and generalized morphology. Available molecular studies [30–33, 41–46], are all based on limited taxon sampling and/or geographic coverage. Furthermore, earlier studies largely relied only on sequences of mitochondrial genes, which can be misleading in species delimitations (e.g., [47, 48]). To conclude, the estimation of the total number and delimitation of extant Acomys species and their biogeographical history would benefit from a more extensive study based on multiple molecular markers.
Aims
In this study, we focus on phylogeography and biogeography of the genus Acomys by phylogenetic analysis of the largest available dataset and substantially improved geographic and taxon sampling. The aims of this study are (1) to reconstruct the phylogeny of the genus using multilocus dataset; (2) to identify the main biogeographical divides in the distribution of Acomys diversity in seasonally dry open habitats in Afro-Arabia; (3) to test proposed hypotheses of historical biogeography of the genus Acomys (i.e. to disentangle the role of geomorphology and climate changes on their diversification), with particular focus on dispersal events among major dry regions in Africa and between Africa, the Arabian Peninsula and the Eastern Mediterranean; (4) to estimate the species richness of the genus by applying of the phylogenetic species concept to identify the genetic groups and geographical regions that are worth further integrative taxonomic studies.
Results
Phylogeny, species delimitation, and distribution of genetic variability of Acomys
Both Bayesian inference (BI) and Maximum Likelihood (ML) analyses of concatenated multilocus data provided very similar phylogenetic reconstructions and revealed five major groups that we will hereafter call subspinosus, spinosissimus, russatus, wilsoni and cahirinus (Fig. 1a). If the number of nodes that are supported by PP ≥ 0.95 or BP ≥ 70 are considered, BI analyses yielded a slightly more robust topology (79 supported nodes; topology shown at Fig. 1a) compared to the ML tree (64 supported nodes; topology not shown). Five major groups are also supported in separate mitochondrial (not shown) and nuclear (Additional files 2 and 3) gene trees.
Fig. 1.
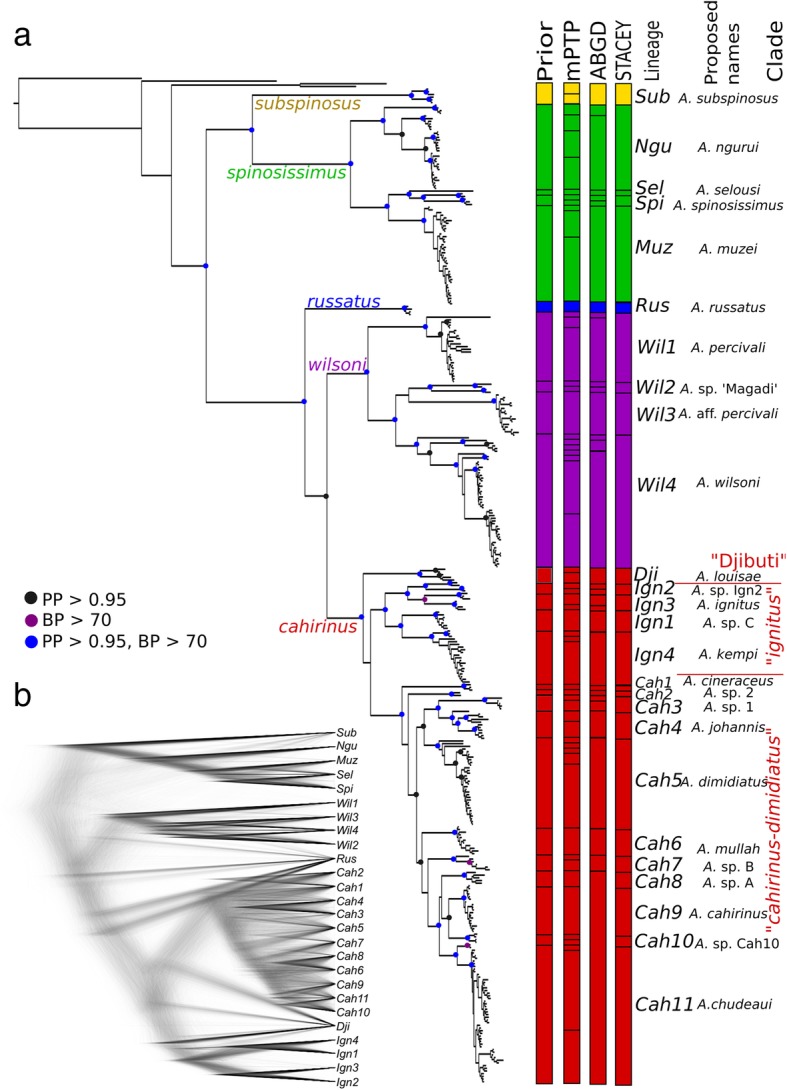
Multilocus phylogeny of the genus Acomys. a Bayesian phylogeny of the concatenated multi-locus matrix calculated in MrBayes. The support from Bayesian analysis in MrBayes (posterior probability, PP) and maximum likelihood analysis in RAxML (bootstrap probability, BP) is indicated by different colours on the nodes (black PP > 0.95, BP < 70; violet PP < 0.95, BP > 70; blue PP > 0.95, BP > 70). Five main Acomys groups (subspinosus, spinosissimus, russatus, wilsoni and cahirinus) are shown by different colours. The results of four different species delimitation approaches (“by-eye” prior; two delimitation approaches based on mtDNA: mPTP, ABGD; and a multilocus species delimitation in STACEY - see more details in the text) are shown in columns on the right, where individual “species” are separated by black lines. Additional information for 26 delimited taxa are provided, abbreviation of the lineages, and previously used taxonomic assignments. b DensiTree cloudogram of coalescent species trees from STACEY (for MCC species tree with PP see Additional file 4)
Three of these major groups (spinosissimus, wilsoni, and cahirinus groups) are further sub-structured to phylogenetic lineages (Fig. 1, Additional files 2 and 3) with predominantly parapatric distributions (Fig. 2). Based on the topology and the shape of the phylogenetic trees and the geographical distribution of genetic variability, we identified 26 distinct genetic lineages as our prior candidate species for next steps of genetic species delimitation (Fig. 1a). Their mutual relationships are relatively well resolved (with three well-defined clades: “Djibuti”, “ignitus” and “cahirinus-dimidiatus”, in the cahirinus group), with the important exception of lineages Cah1-Cah11 representing probably a rapid radiation of the “cahirinus-dimidiatus” clade (Fig. 1a). The multispecies coalescent species tree from Species Tree And Classification Estimation, Yarely (STACEY) also revealed very similar topology to that reconstructed from the concatenated super-matrix (Fig. 1b, Additional file 4). The main differences are in unresolved positions of the russatus group and the “Djibuti” clade and a weakly resolved topology within the three major groups (spinosissimus, wilsoni, cahirinus; Additional file 4).
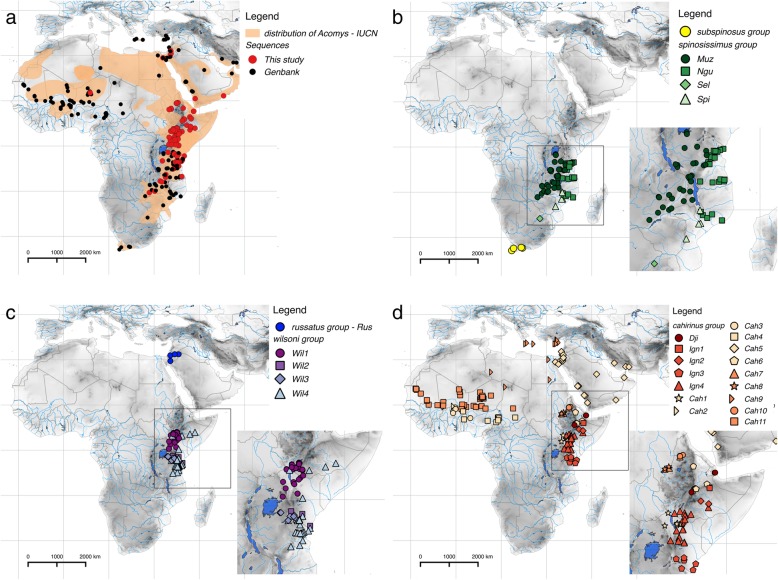
Fig. 2.
Analysed samples and the distribution of genetic variability in the genus Acomys. a Geographical distribution of the genus Acomys according IUCN (orange background); the origin of newly genotyped individuals is shown by red circles, while georeferenced sequences from GenBank are shown by black circles; b distribution of genetic lineages in the subspinosus and spinosissimus groups; c distribution of genetic lineages in the russatus and wilsoni groups; d distribution of genetic lineages in the cahirinus group
Species discovery approaches based on mitochondrial cytochrome b gene (CYTB) variability split the genus Acomys into 57 putative species in multi-rate Poisson Tree Process (mPTP) and 32 putative species in Automatic Barcode Gap Discovery (ABGD; Fig. 1), but ABGD did not find any clear gap between intra- and interspecific distances (“barcoding gap”; not shown). On the other hand, results from multispecies coalescent analysis in STACEY supported all 26 a priori defined species as separate gene pools (Fig. 2a). Taking into account the fact that multispecies coalescent does not statistically distinguish structure associated with population isolation vs. species boundaries [49], we will therefore use the term “species” for genetically distinct lineages or molecular operational taxonomic units (MOTUs).
The subspinosus group with only one lineage (Sub) is limited to South Africa (Fig. 2b). The separate species status was confirmed by all species delimitation analyses, and mPTP even suggested two different mitochondrial sublineages as two separate species (Fig. 1a). Mean intraspecific genetic distance is 1.5%, the distance to the nearest lineage is 21.3% (Table 1).
Table 1.
Genetic distances calculated from BI phylogenetic tree by the Species Delimitation algorithm in Geneious. Intraspecific distances and interspecific distances from the nearest lineage in percents (%)
| Group | Lineage | Nearest lineage | Intra Dist | Inter Dist - Closest |
|---|---|---|---|---|
| subspinosus | Sub | Ngu | 1.5 | 21.3 |
| spinosissimus | Ngu | Muz | 2.9 | 10.7 |
| Sel | Spi | 0.0 | 7.1 | |
| Spi | Sel | 3.2 | 7.1 | |
| Muz | Spi | 1.1 | 8.0 | |
| russatus | Rus | Ign1 | 0.5 | 14.4 |
| wilsoni | Wil1 | Wil2 | 1.5 | 12.3 |
| Wil2 | Wil3 | 4.7 | 10.5 | |
| Wil3 | Wil2 | 1.0 | 10.5 | |
| Wil4 | Wil2 | 3.1 | 9.6 | |
| cahirinus | Dji | Ign1 | 2.2 | 9.6 |
| Ign2 | Ign3 | 2.9 | 6.4 | |
| Ign3 | Ign2 | 2.9 | 6.4 | |
| Ign1 | Ign4 | 0.7 | 5.9 | |
| Ign4 | Ign1 | 1.6 | 5.9 | |
| Cah1 | Cah2 | 0.6 | 7.5 | |
| Cah2 | Cah6 | 0.8 | 6.5 | |
| Cah3 | Cah4 | 0.8 | 6.1 | |
| Cah4 | Cah5 | 2.1 | 6.0 | |
| Cah5 | Cah4 | 1.4 | 6.0 | |
| Cah6 | Cah9 | 0.8 | 5.5 | |
| Cah7 | Cah9 | 1.3 | 5.5 | |
| Cah8 | Cah9 | 1.2 | 4.0 | |
| Cah10 | Cah9 | 0.6 | 3.0 | |
| Cah9 | Cah10 | 0.6 | 3.0 | |
| Cah11 | Cah9 | 1.2 | 4.5 |
The spinosissimus group inhabits the eastern part of the Zambezian bioregion (Fig. 2b). The STACEY approach confirmed four distinct species (Muz, Ngu, Spi, Sel), while mPTP and ABGD suggested 10 and six species, respectively (Fig. 1a). The intraspecific CYTB distances in four species range from 1.1 to 3.2% (excluding Sel, where only a single sequence was available). Interspecific distances to the nearest neighbour are 7.1-27.0% (Table 1).
The distribution of sequenced samples from the russatus group is restricted to arid regions of the Levant (Jordan, Israel; Fig. 2c). A single species (Rus) was supported by all species delimitation analyses. The mean intraspecific distance is low (0.5%), while interspecific distance to the nearest neighbour is 14.4% (Table 1).
The wilsoni group is divided into four well supported lineages, suggested as separate species by STACEY, and they are predominately distributed in the Somali-Masai savanna (Fig. 2c): Wil1 lives on both sides of the Great Rift Valley (GRV) in Kenya and Ethiopia, Wil2 and Wil3 are two lineages with parapatric distribution in southern Kenya and northern Tanzania, and, finally, Wil4 was found east of GRV from north-eastern Tanzania to southern Ethiopia, where it overlaps with Wil1. The mPTP split the wilsoni group into 13 putative species, and ABGD into eight species. The intraspecific distances of four STACEY species ranged from 1.0 to 4.7%, interspecific distances among them are from 10.5 to 12.3% (Table 1).
The highest genetic diversity was found within the cahirinus group (16 lineages delimited as species by STACEY). The group is composed of three significantly supported clades (with unresolved relationships among them), distributed parapatrically, with only small overlap (Fig. 2d): (i) the clade “Djibuti” with a single species (Dji) recorded from Djibuti and Dera National Park in Ethiopia, geographically neighbouring the Ethiopian Afar province; (ii) the clade “ignitus” distributed south-east of GRV with four lineages (Ign1-Ign4); (iii) the clade “cahirinus-dimidiatus” widespread north-west of GRV, including Sahel and Sudanian savanna, eastern Mediterranean, Middle East and Arabian peninsula, with 11 species (Cah1-Cah11) delimited by STACEY (Fig. 1). Intraspecific distances within the cahirinus group range from 0.6 to 2.9%, interspecific from 3.0 to 9.6% (Table 1).
Historical biogeography and divergence dating
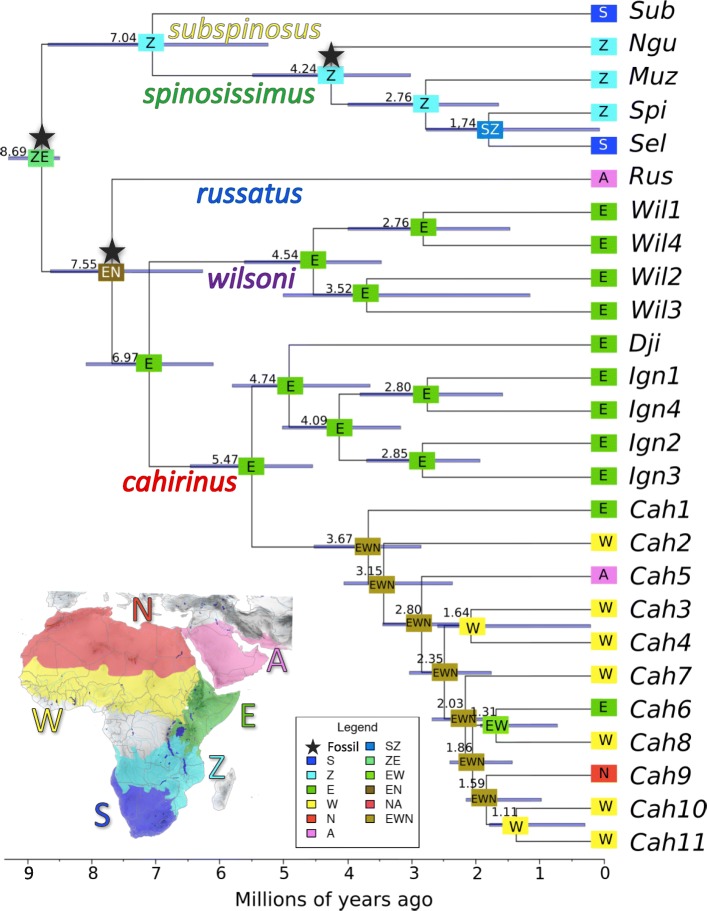
Based on the Dispersal-Extinction Cladogenesis model and the time-calibrated tree, the TMRCA of the genus Acomys is dated to 8.69 Ma (95% HPD = 8.51–9.29 Ma). The ancestral area is predicted to be in Eastern Africa, but it is not resolved whether in the Zambezian or the Somali region (Fig. 3). The first split in the Late Miocene separated southern groups (subspinosus + spinosissimus) from northern groups (russatus + wilsoni + cahirinus). Around 7.04 Ma (95% HPD = 5.25–8.67 Ma) the ancestor of the subspinosus group in South Africa diverged from the ancestor of the spinosissimus group in the Zambezian region, where the latter group started to diverge around 4.24 Ma (95% HPD = 3.02–5.48 Ma). The ancestor of the russatus group in Arabian region separated from Acomys in Somali region around 7.55 Ma (95% HPD = 6.27–8.64 Ma). The split between cahirinus and wilsoni groups occurred in Somali region around 6.97 Ma (HPD = 6.10–8.08 Ma). The wilsoni group started to diversify around 4.54 Ma (95% HPD = 3.48–5.61 Ma), while the first split in the cahirinus group is dated to 5.47 Ma (95% HPD = 4.55–6.46 Ma), in both cases the beginning of radiation is predicted in the Somali-Masai savanna. Subsequent splits in the cahirinus group occurred around 5 Ma, when the ancestors of clades “Djibuti”, “ignitus” and “cahirinus-dimidiatus” diverged most likely in Eastern Africa. The biogeographic history of the “cahirinus-dimidiatus” clade was more complex. While Cah1 stayed in Somali region, the ancestor of lineages Cah2-Cah11 likely dispersed to the north-west (especially Sudanian savanna), where most genetic lineages currently occur. The results suggest one Pleistocene migration back to the Somali region (Cah6 in Afar region, ~ 1.31 Ma) and two independent dispersals to the north, either to the Arabian region (Cah5 dated at 2.80 Ma) and to the Sahara region (Cah9 dated at 1.59 Ma).
Fig. 3.
Divergence dating and the reconstruction of historical biogeography. Numbers on the nodes represent medians of estimated divergence date, and the horizonal bars show 95% highest posterior density of these estimates. Stars indicate the positions of fossil constrains used for the calibration of molecular clock (see Table 3 for more details). Different colours on the nodes represent reconstructed ancestral regions for each clade, according to the map in the frame: South Africa region (S), Zambezian region (Z), Somali region (E), Sudanian region (W), Sahara region (N), Arabian region (A)
Species distribution modelling
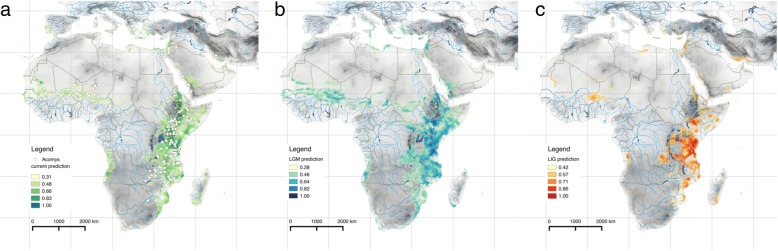
The bioclimatic MaxEnt model for the present shows that the sampling in this study covers almost the complete range of suitable climatic conditions for genus Acomys (Fig. 4a). The most important variables predicting the modelled geographic distribution of spiny mice were the annual range of temperature and annual precipitation. The predicted distribution is relatively continuous in eastern Africa, mainly in the Somali-Masai and eastern Zambezian savanna. On the contrary, climatically unsuitable are moist mountains in Ethiopia, Kenya and Albertine Rift, as well as very arid regions of Horn of Africa and Masai xeric scrublands in north-eastern Kenya. The belt of suitable climatic conditions occurs in West Africa, on the boundary between Sudanian savanna and Sahel, as well as along Mediterranean sea and Arabian Peninsula and southern Iran. Isolated suitable areas are predicted in western Angola and northern Namibia, despite the absence of present-day occurrence of Acomys species. The model for LGM predicts very similar distribution of spiny mice, with more continuous belt of suitable conditions in Sudanian region (Fig. 4b). The predicted distribution during LIG was more fragmented, with highly suitable conditions in southern Ethiopia, Kenya and Tanzania, and isolated patches in the Horn of Africa, southern part of the Sahara desert, and Mozambique (Fig. 4c). Altogether, it seems very likely that the climatic conditions in the core of the present day distribution, in open seasonally dry habitats in East Africa, were apparently favourable at least during the last glacial cycle.
Fig. 4.
The probability of Acomys occurrence based on the MaxEnt modelling of bioclimatic niches. More intensive colour indicates higher probability of suitable conditions. a The model for the present; white dots indicate the sampled localities; b prediction for the last glacial maximum (21 ka); c prediction for the last interglacial period (120–140 ka)
Discussion
Spiny mice of the genus Acomys represent a speciose group of rodents, widely distributed in seasonally dry savanna of Africa, Arabia and Middle East. They often are a dominant component of small mammal assemblages and in some habitats, e.g. in rocky outcrops, they can be even the only rodents captured (our unpubl. data). They were able to colonize wide spectrum of non-forested habitats, from miombo woodlands to rocks in the middle of Sahara, from the sea coast up to 2500 m above sea level (a.s.l.) [e.g., 36]. Despite Acomys abundance and practical importance (e.g., as model taxa in behavioural or biomedical research; [50]), the knowledge of their evolutionary history, taxonomy and biogeography has been very limited and biased to particular regions or intrageneric clades [28, 30, 31, 33, 41, 43, 45]. Here we compiled the largest multilocus genetic dataset to date for the genus Acomys (genotypes of 699 individuals from more than 280 localities covering a majority of the distribution of this genus), reconstructed phylogenetic relationships, described biogeographical patterns and evolutionary history, and estimated the spiny mice species diversity.
Phylogeny and biogeographical patterns in the genus Acomys
Phylogenetic analysis revealed clear evidence for the existence of five major groups, subspinosus, spinosissimus, russatus, wilsoni and cahirinus (Figs. 1 and 2, Additional files 2 and 3), which diverged in the late Miocene (Fig. 3). For the first time we provide nuclear genetic data for the subspinosus group and we refer its sister position with the spinosissimus group, which has never been confirmed [31, 41, 45]. This sister relationship is supported also by the fact that subspinosus and spinosissimus groups share the same triplicate-type X chromosome that is very rare among mammals [51]. For the first time we also sequenced the nuclear markers of the russatus group, but its phylogenetic relationships with the wilsoni and cahirinus groups remained unresolved (Fig. 1), which suggests fast divergence in the late Miocene (see similar results in [45]). Using multilocus genetic data, we unequivocally identified that the first split within the genus occurred between south-eastern and eastern Africa, i.e. (subspinosus + spinosissimus) vs. (russatus + wilsoni + cahirinus) (Fig. 3). Further diversification of the five major groups is dated to Plio-Pleistocene and, interestingly, the diversification rate within them is very unequal. While subspinosus and russatus remained monotypic at the margins of Acomys distribution (in the Cape Region and Middle East, respectively), spinosissimus and wilsoni clades diversified by a comparable rate in Eastern Africa, either in Zambezian or Somali-Masai savannas. The most intensive spatial and cladogenetic expansion occurred in the cahirinus group that colonized large areas of Africa, Arabia and eastern Mediterranean region, even if the most genetic variation is still observed in eastern Africa.
Our phylogenetic analysis revealed at least three independent “out-of-Africa” events, differing significantly by the date of divergence from their African counterparts. The oldest disperser was the ancestor of the russatus group, currently distributed only in the Sinai Peninsula and the Middle East, which split from the ancestors of the wilsoni and cahirinus groups already in late Miocene (Fig. 3, see also [30]). Much more recently, at the beginning of Pleistocene, the ancestors of Cah5 colonized the Arabian Peninsula (and South Iran). It is unclear whether it was through the Bab al-Mandab connection between Africa and Yemen [33, 52] or through Sinai and around the Red Sea coastline [53]. Lastly, the western Mediterranean islands and south coast of Turkey were colonized during Antiquity by possibly commensal populations from the lineage Cah9, likely from Egypt [31, 33, 54].
Spiny mice of the cahirinus group (specifically “cahirinus-dimidiatus” clade) are widely distributed also in the Sudanian savanna. Results of our biogeographical analysis suggest multiple colonization and diversification waves, directed usually from eastern to western Africa. The first split between Cah1 (which stayed restricted in Somali region along the Lake Turkana) and remaining lineages occurred in the late Pliocene/early Pleistocene. The ancestors of lineages Cah2, Cah 3, and Cah4 diverged in southern (more humid) part of Sudanian savanna probably simultaneously with the colonization of Arabian Peninsula by Cah5. Much later (ca. 1–1.5 Mya), the split between northern (Cah9) and southern (Cah10 and Cah11) resulted in current diversity in large arid areas in northern part of the Sudanian region, Sahel and Sahara desert. At the same time the Ethiopian highlands caused the diversification of Cah8 (living in easternmost Sudanian region) and Cah6 (currently limited to Afar triangle). Faunal exchange between Somali-Masai and Sudanian savanna in the Plio-Pleistocene have been described also in other rodents and very often the oldest lineages of Sudanian taxa are found in Eastern Africa [55–57], which well corresponds with the pattern found in spiny mice.
The current distribution of genetic diversity of Acomys is affected by major biogeographic divides, represented by (1) extremely dry open habitats, (2) mountain blocks and (3) large water bodies, e.g. Rift Valley lakes or rivers. Even if the spiny mice are considered as typical inhabitants of (semi-)arid environments [58], they avoid extremely dry habitats. Distribution modelling based on bioclimatic data (Fig. 4) suggests low suitability of habitats in most of Saharan and Arabian deserts, as well as in Masai xeric bushland and shrublands in northernmost Kenya and Horn of Africa. However, even in such habitats the spiny mice can occur, but they have very patchy distribution with isolated populations in rocky areas, considered as remains of more continuous distribution in past humid periods, when savanna-like habitats prevailed [43, 59–63]. Sahara desert seems to work as a barrier between clades Cah9 and Cah11 (albeit the data are missing e.g. from southern Algeria).
The expansion of forests during interglacials [64, 65], known as Pleistocene breathing model [66], repeatedly fragmented savanna biome, especially in eastern Africa. The forested mountain blocks thus formed an important biogeographical divide for savanna-dwelling organisms and supported allopatric diversification in this nowadays relatively continuous ecosystem [67]. In agreement with this hypothesis, we can see the coincidence of genetic structure in the spinosissimus group with Eastern Arc Mountains (EAM) and Southern Rift Mountains (SRM). These mountain ranges, even if permeable today for taxa living in open habitats, clearly separate Ngu and Muz lineages (Fig. 2b; see more details in [44]). Similar structure was recently observed in numerous other taxa of sympatric murid and bathyergid rodents living in open habitats of Zambezian region [67–70]. Similarly, the mountains in north-eastern Tanzania (Kilimanjaro, Pare, Usambara) currently delimit distribution for some Acomys lineages (Wil2 vs. Wil3 or cahirinus vs. spinosissimus groups), and, again, this pattern was found in other savanna rodents (e.g. Saccostomus [71], Gerbilliscus [72]). Further to the north, Kenyan highlands could have worked in similar way as they seem to limit distribution of several Acomys lineages (Wil1 vs. Wil2 + Wil3, Ign3 vs. Ign4). Lastly, Ethiopian Highlands, delimit distributions of lineages especially in the cahirinus group. For example lineages Dji and Cah6 are restricted to the Afar province in north-eastern Ethiopia (see also very similar pattern in gerbils of the genus Gerbilliscus, often sympatric with spiny mice; [72]).
Rivers and other more or less linear water bodies (e.g., contemporary rift lakes, or palaeolakes; [73]) are known to play an important role in shaping genetic structure and diversification patterns in organisms living in open habitats. For example the Zambezi-Kafue river system delimits southern border of distribution of Ngu and Muz lineages (Fig. 2b; [44]) and have been implicated in forming genetic diversity in savanna-dwelling organisms as diverse as killifishes [74], gerbils [75], African pouched mouse [71], baboons [76] and several species of antelopes [77]. The largest river in Sudanian savanna is the Niger River, which makes a border between Cah11 and other West African Acomys lineages, is also dominant biogeographical divide in many other rodents (e.g., [43, 55, 67, 78]). On the other hand, the Nile Valley seems to serve rather as a suitable migration corridor for northward spreading of savanna taxa from eastern Africa (see [56] for grass rats, or [79] for shrews). In Acomys, it very likely allowed the lineages Cah5, Cah9 and Rus to colonize the northern Africa and Arabian Peninsula. Trauth et al. [73] suggested that during the humid periods of Pleistocene, the bottom of GRV was filled by water (much more than today), and the so-called palaeolakes formed an important biogeographic barrier causing allopatric diversification or even speciation. The phylogeographic pattern concordant with this hypothesis was recently described in Eastern African gerbils [72] and the role of GRV on genetic structure is visible also in Acomys. For example, Cah1 is separated from “ignitus” clade by GRV and, similarly, more records of Wil1 occur west of GRV, while other lineages of the wilsoni group were found predominantly east of GRV.
Evolutionary scenario of the genus Acomys − interplay of geomorphology and climatic changes
The origin of Acomys is dated in late Miocene, which is in concordance with the first occurrence of savannas that appeared as a result of rifting activity in eastern Africa and climatic changes [4]. Spiny mice belong to the subfamily Deomyinae, whose other members occur mostly in forest or forest margins (i.e. genera Deomys and Lophuromys) or moist savanna (Uranomys) [16]. The phylogenetic relationships among genera of Deomyinae are not sufficiently resolved [46, 80, 81], but it seems likely that the ancestor of Acomys colonized dry open habitats early after their late Miocene appearance profiting from empty niches in this ecosystem. Barome et al. [31] proposed the origin of Acomys ca. 13.7 Ma in East Africa or in South Africa, while Alhajeri et al. [46] placed it more generally to sub-Saharan Africa at 10 Ma. Other authors [40, 82] suggested the origin for the genus in Eastern Africa, mainly in the Ethiopian region [82]. Our historical biogeography reconstruction is in partial agreement with previous studies as the origin of the genus is placed either in Somali and/or Zambezian regions (Fig. 3), i.e. the regions with the highest contemporary genetic diversity of spiny mice. Even if the oldest fossil records of Acomys ancestors are found in Zambezian savanna (†Preacomys griffini and †Preacomys karsticus ca. 9 Ma from Namibia; [83]), they occurred in the late Miocene also in East Africa (†Preacomys kikiae 8.5 Ma from Chorora, Ethiopia; [84]).
Periods of warm humid climate in Late Miocene caused the last occurrence of coast-to-coast belt of tropical forest, which is evidenced by phylogenetic analyses of plants and animals living in nowadays fragmented forests of Congo basin and eastern African montane and coastal forests [85, 86]. This continuous forest was likely one of the most important factors in early evolution of savanna inhabitants, because it separated northern (= Somali-Masai) and southern (= Zambezian) savannas. In Acomys, this resulted in allopatric divergence between the ancestor of subspinosus + spinosissimus group in the Zambezian savanna and the ancestor of wilsoni + russatus + cahirinus in the Somali-Masai savanna (estimated in this study at 8.7 Ma). The same geographical and temporal pattern, i.e. late Miocene split of northern and southern taxa, was observed also in other savanna mammals, e.g. gerbils [57], pouched mice [71], several genera of antellopes [87], warthogs [88] and giraffes [89, 90] .
Later on, but still in Late Miocene, the evolution of savanna’s fauna was significantly influenced mainly by the Messinian salinity crisis (MSC, [91]), dated to 6.0–5.3 Ma. Very little is known about the effect of the MSC on eastern African climate [6], but it is generally held that overall aridification at the Miocene/Pliocene boundary promoted the expansion of very dry habitats [92]. Inhospitable very arid (desert) areas in north-eastern Africa expected at MSC period thus likely interrupted the connection of Acomys populations between Somali-Masai and eastern Mediterranean area. This period corresponds to the split of the russatus group, which remained effectively isolated in the north.
Plio-Pleistocene period (starting 5.3 Ma) is characterized by intensive climatic oscillations. There are several well-known climatic transitions, like the intensification of Northern Hemisphere glaciation (iNHG; 3.2–2.5 Ma, [93, 94]), the development of the Walker circulation (2.0–1.7 Ma; [95]) and the early-middle Pleistocene transition (1.2–0.8 Ma; [96]). These periods of pronounced climate variability significantly affected the distribution of forests, palaeolakes and savannas [97]. For example during more humid periods the currently fragmented montane forests in Eastern Arc Mountains and Kenyan highlands probably expanded into lower altitudes, became more continuous and formed significant barriers to gene flow for taxa living in open dry habitats [67, 71, 72, 98]. The bottom of GRV was filled by the palaeolakes, which prevented gene flow of savanna-dwelling species, leading to diversification, or even speciation [73]. Because of wide confidence intervals of our estimates of divergence times, it is not possible to link particular splits to specific climatic events. However, it is highly probable that a majority of current genetic diversity in spinosissimus, wilsoni and cahirinus groups is a result of repeated fragmentation of savannas in Plio-Pleistocene, caused by climatic changes. This is further supported by parapatric distribution of lineages within major clades, where the distribution borders often correspond to predicted barriers of the gene flow (i.e. too arid or forested areas, and palaeolakes).
Species richness of Acomys − the need of further integrative taxonomic studies
The number of species crucially depends on adopted species concept. Rapidly increasing amount of genetic data now allows to apply the so-called integrative taxonomic approach, which usually complements the widely used typological or biological species concepts by genetic [99] and/or phylogenetic [100] species delimitations. The taxonomy of Acomys has been unresolved since the second half of the twentieth century. In Additional file 1 we listed several taxonomic alternatives, based mostly on morphological traits, used recently for the genus Acomys. The most comprehensive list was provided by Ellerman [34] with 25 species and 17 subspecies. On the other extreme, Setzer [38] recognized only five species that partly correspond to major genetic clades recovered in our study: A. cahirinus (7 subspecies), A. dimidiatus (12 subspecies), A. russatus (one subspecies), A. spinosissimus (one subspecies) and A. subspinosus (9 subspecies, including A. wilsoni). Widely accepted list of Musser and Carleton [37] contained 18 species. Monadjem et al. [36] lists 15 species in sub-Saharan Africa, including three newly delimited species in the spinosissimus group [45]. The most recent and comprehensive Handbook of the Mammals of the World [16] listed 21 species and 12 subspecies.
We used several genetic species delimitation methods and their estimates of spiny mice species richness differ significantly from 57 species (mPTP) to 26 species (STACEY as well as our prior delimitation based on geographical distribution of genetic diversity). The species delimitation based only on mitochondrial markers (ABGD and mPTP) have the tendency to overestimate the number of revealed species and they often identify as separate species also genetic lineages that are traditionally considered as intraspecific phlyogeographic structure. For example ABGD revealed four species within Ngu lineage, while Petružela et al. [44] recently showed on multi-locus dataset that they represent only phylogeographic structure of A. ngurui. Below we use the most conservative estimate (26 MOTUs, here considered as “genetic lineages” and named according Fig. 1a) and compare these species delimitations with previous taxonomic work. Multi-species coalescent approaches to species delimitation (like STACEY) in fact diagnose the genetic structure, with no distinction between structure due to populations or due to species [49]. Therefore, the aim of the following part is not to perform a formal taxonomic revision, but to show the genetic clades and geographic regions where further integrative taxonomic analyses (employing combination of genetic, morphological, ecological and other data) could lead either to new descriptions or synonymization of Acomys taxa.
The subspinosus group
This group is monotypic and contains a single lineage Sub.
(1) Sub
Distribution: South Africa (Cape Province).
Available name: Acomys subspinosus (Waterhouse, 1838).
Type locality: Western Cape Province, Cape of Good Hope, South Africa.
Karyotype: 2n = 64, NF = 70 [101, 102].
Additional information: Based on its unique dental and skull morphology, A. subspinosus has been placed in its own subgenus Subacomys with an “ancestral” karyotype (2n = 64, NF = 70; [51, 101–104]). Its separation from other Acomys was also indicated by phylogenetic analyses of CYTB [31, 32, 45]. Using for the first time the combination of mitochondrial and nuclear markers, we unequivocally showed its sister relationship with spinosissimus group, i.e. it does not represent the first cladogenetic split of Acomys. As a consequence, the validity of the subgenus Subacomys (mentioned erroneously as Preacomys in Denys et al. [16]) is questionable.
The spinosissimus group
This strongly supported monophyletic group has been revised repeatedly [41, 44, 45] and four genetic lineages were distinguished and named. However, the genetic data from the southern part of its distribution are still very limited and especially the taxonomy and distribution of A. selousi and A. spinosissimus should be further explored.
(2) Ngu
Distribution: Lineage Ngu is distributed in three well supported parapatric sublineages from Tanzania (East of EAM) to central Mozambique (north of the Zambezi River; [44]).
Available name: Acomys ngurui Verheyen et al., 2011.
Type locality: Nguru Ya Ndege, Tanzania.
Karyotype: 2n = 60, NFa = 68 [45].
Additional information: This species is very similar to A. muzei from which it differs by relatively shorter tail and non-overlapping distribution [44, 45]. Barome et al. [41] reported it as A. cf. selousi from Berega. Three genetically distinct sublineages probably represent intraspecific variation [44].
(3) Sel
Distribution: Northern part of South Africa, only one genetically confirmed locality from the Kruger National Park [45]. Northern limits of its distribution are not fully resolved.
Available name: Acomys selousi De Winton, 1896.
Type locality: Essex Farm, Zimbabwe.
Karyotype: 2n = 58–62, NFa = 68 [101, 102] .
Additional information: According some authors [35, 37, 38]; A. transvaalensis and A. selousi are synonyms of A. spinosissimus. Here we follow the view of Verheyen et al. [45] and Monadjem et al. [36] and consider this lineage as a separate species, but further taxonomic investigation of the spinosissimus group in South African region is required.
(4) Spi
Distribution: Mozambique, Zimbabwe, southern Malawi.
Available name: Acomys spinosissimus Peters, 1852.
Type locality: Tette and Buio, Mozambique.
Karyotype: 2n = 60, NFa = 68 [101].
Additional information: We follow the view of Verheyen et al. [45] and Monadjem et al. [36] and include only populations from central Mozambique and southern Malawi into this species. Petružela et al. [44] recently showed that its distribution north of the Zambezi River is much more restricted compared to maps in Monadjem et al. [36] and the two well distinct genetic sublineages of this species seem to be separated by the Zambezi river.
(5) Muz
Distribution: Central and western Tanzania, Malawi (west of the Lake Malawi), Zambia.
Available name: Acomys muzei Verheyen et al., 2011.
Type locality: Muze, Tanzania.
Karyotype: 2n = 58–62, NFa = 68 [45].
Additional information: Recent analyses showed that the populations in Malawi and Zambia (reported as A. spinosissimus in Monadjem et al. [36]) belong to this species [44]. Further, the highest genetic diversity of the species was recorded west of the Lake Malawi, while Tanzanian populations represent only relatively recent colonization event [44].
The russatus group
The russatus group is monotypic with only one lineage Rus. The phylogenetic relationships with its sister groups cahirinus and wilsoni are not fully resolved (Fig. 1).
(6) Rus
Distribution: Egypt (separate subspecies aegyptiacus was described in Eastern Desert), Sinai, Jordan, Israel, Saudi Arabia, Yemen and Oman. Genotyped material in this study originates only from Jordan and Israel.
Available name: Acomys russatus (Wagner, 1840).
Type locality: Sinai, Egypt.
Karyotype: 2n = 66, NF ≥ 66 [40].
Additional information: Denys et al. [51] referred that A. russatus is very distinctive in its molar morphology and chromosomal traits and previous CYTB [31, 41, 45] as well as our multilocus genetic analyses showed that it is not closely associated with any other Acomys taxon. Acomys russatus and A. dimidiatus (= Cah5 in this study) can live in sympatry, and their differences in ecology, physiology, and activity patterns (especially in Israel) have been extensively documented (see references in [105]).
The wilsoni group
This group is well supported in all phylogenetic analyses (Fig. 1 in this study, [31, 41]), but its relationships with russatus and cahirinus groups are not completely resolved. Distribution of the wilsoni group is limited to the Somali region [sensu 3]. Based on multilocus species delimitation we recognize four genetic lineages, but the species limits must be further investigated.
(7) Wil1
Distribution: South Ethiopia, Kenya (along GRV).
Available name: Acomys percivali Dollman, 1911.
Type locality: Chanler Fall, Nyiro, Kenya.
Karyotype: 2n = 36 and NF = 68 [40].
Additional information: Phylogenetically the most distinct MOTU within the wilsoni group (Fig. 1). It was not included in previous phylogenetic studies. Distribution of A. percivali reported by Monadjem et al. [36] and Denys et al. [16] is very similar to that of Wil1. Janecek et al. [103] regarded A. percivali as the species genetically most closely related to A. wilsoni (= Wil4). Both clades were found sympatric at several localities in southern Ethiopia, where they can be distinguished by external morphology (our unpublished data) and karyotypes [16].
(8) Wil2
Distribution: Southern Kenya.
Available name: None. Based on Barome et al. [31], we use in Fig. 1 the name A. sp. ‘Magadi’.
Type locality: Not relevant.
Karyotype: Not known.
Additional information: Known only from three localities from southern Kenya, all of them reported by Barome et al. [31]. They mentioned this MOTU as two different species, A. sp. ‘Magadi’ and A. wilsoni, and this structure was reflected also by mPTP and ABGD analyses in our study. The conspecificity with Wil3 and/or Wil4 are plausible hypotheses and should be tested.
(9) Wil3
Distribution: NW Tanzania, southern Kenya, most localities in the bottom of GRV.
Available name: None. Based on Mgode [42], we use in Fig. 1 the name Acomys aff. percivali.
Type locality: Not relevant.
Karyotype: 2n = 58 [42].
Additional information: Mgode [42] named 13 spiny mice from northern Tanzania (localities Tingatinga, Longido, Mt. Gelai-Olikisima and Kilimamoja-Karatu) belonging to this lineage as Acomys cf. percivali. They differ from Wil4 (= A. wilsoni) in skull morphology and karyotype. Because the type locality of A. percivali (Mt. Nyiro, Kenya) is very far from the distribution of Wil3, the name A. percivali more probably belongs to Wil1, while Wil3 likely deserves a formal description as a new species.
(10) Wil4
Distribution: Southern Ethiopia (Somali region), Kenya (east of GRV), north-eastern Tanzania.
Available name: A. wilsoni Thomas, 1892.
Type locality: Mombasa, Kenya.
Karyotype: 2n = 62 and NF = 76 [42, 106].
Additional information: Verheyen et al. [45] suggested that A. wilsoni (meaning Wil2 and Wil4 included in their study of mitochondrial variation) is probably a species complex, which is confirmed by our data. The conspecificity of Wil4 with Wil2 and/or Wil3 should be further investigated as they might represent intraspecific phylogeographic structure (see similar patterns in savanna-dwelling rodent species in southern part of Somali region, e.g. in Gerbilliscus vicinus; [72] or Saccostomus umbriventer; [71]). If Wil2 and Wil4 are different species, the analysis of the type material of A. wilsoni will be required to decide, which of them is true A. wilsoni (both are distributed around the type locality of wilsoni).
The cahirinus group
This is the most diversified Acomys group comprising of three main clades, “Djibuti” (one MOTU Dji), “ignitus” (four MOTUs Ign1 − Ign4) and “cahirinus-dimidiatus” (11 MOTUs Cah1 − Cah11), with unresolved mutual relationships.
(11) Dji
Distribution: Djibuti, Afar province in Ethiopia, probably also Somalia (from where no genetic data are available).
Available name: Acomys louisae Thomas, 1896.
Type locality: 65 km S of Berbera, Somalia.
Karyotype: 2n = 68 and NF = 68 ([107]; our unpubl. data).
Additional information: Acomys louisae was placed in a separate subgenus Peracomys based on dental characters [51]. We have not checked the skull morphology of our material from Dji, but the recently collected animals from eastern Ethiopia (Dire Dawa region) likely assigned to A. lousiae by morphological characters (by C. Denys, unpubl. data) clustered with Dji at CYTB (not included in this study). According to Petter [108], A. louisae cannot be distinguished from the “cahirinus-dimidiatus complex” (sensu [109, 110]) on the basis of skull or external characteristics. This species may co-occur with A. mullah (= Cah6) in the Afar triangle (larger, HB > 100 mm with grey or greyish-brown dorsal pelage). A. louisae should have a bright rufous or brown dorsal pelage and relatively very long tail (> 100% of HB; [36]). In our pilot analysis we were not able to find significant external size differences between individuals from lineages Dji and Cah6 (considered as A. mullah, see below), but more detailed morphological investigation is needed.
(12) Ign1
Distribution: Eastern part of Ethiopia (Babile).
Available name: None. Based on Lavrenchenko et al. [111], we use the name Acomys sp. C in Fig. 1.
Type locality: Not relevant.
Karyotype: 2n = 44, NF = 68 [111, 112].
Additional information: This presumably new species was mentioned for the first time as genetically and cytogenetically very divergent lineage (Acomys sp. C) by Lavrenchenko et al. [111] from the Babile Elephant Sanctuary in Eastern Ethiopia. This lineage is only known from the Babile Elephant Sanctuary, where it is very common and abundant species. It can have wider distribution in poorly sampled regions of southeastern Ethiopia and Somalia (see similar pattern in gerbils, [72]). The conspecificity with other lineages of the ignitus clade should be further tested. The comparison with the type material of A. mullah, described from nearby town Harar, is necessary (see also below).
(13) Ign2
Distribution: South-eastern Ethiopia.
Available name: None.
Type locality: Not relevant.
Karyotype: Not known.
Additional information: This lineage is reported here for the first time. It is known only from two localities in the south-eastern slope of Ethiopian Highlands (Sof Omar caves and Imi; each locality has very distinct mitochondrial haplotypes). It might be more widespread in poorly sampled Somali region of Ethiopia, and in Somalia. Its sister lineage Ign3 (= A. ignitus) is geographically distant, but the conspecificity with other lineages of the ignitus clade are worth of further taxonomic work.
(14) Ign3
Distribution: Southern Kenya, northernmost Tanzania.
Available name: Acomys ignitus Dollman, 1919. In Alhajeri et al. [46] was this species incorrectly mentioned as A. percivali.
Type locality: Voi, Kenya.
Karyotype: 2n = 50, NF = 66–68 [102].
Additional information: A. ignitus has been recorded in and around Tsavo National Park [36], which corresponds to the distribution of this MOTU. Whether or not other lineages of the ignitus clade (especially Ign2) are conspecific with A. ignitus must be investigated by integrative taxonomy approach.
(15) Ign4
Distribution: Kenya and southernmost Ethiopia (east of GRV).
Available name: Acomys kempi Dollman, 1911.
Type locality: Chanler Falls, N Guaso Nyiro, Kenya.
Karyotype: Not known.
Additional information: This species was previously listed as subspecies of A. ignitus (Hollister, 1919) [113] or A. cahirinus (Setzer, 1975) [38], but rehabilitated as clearly distinct species by Janecek [103]. Acomys kempi was found sympatric with A. percivali (= Wil1) at several localities in Kenya and southern Ethiopia and these two taxa can be easily distinguished, e.g. by coat coloration (the latter being usually greyish and darker). There is no evidence of distributional overlap with Ign3 (A. ignitus), but further sampling in southern Kenya would be desirable.
(16) Cah1
Distribution: North-west Kenya (the only region from where the genetic data is available), Sudan, South Sudan. Very probably also in Uganda.
Available name: Acomys cineraceus Heuglin, 1877.
Type locality: Doka, Sudan.
Karyotype: 2n = 48–50 [114].
Additional information: Formerly included in A. cahirinus [38, 108], but Dieterlen (in litt.) noted that A. cineraceus is a distinct species [37]. Separation of A. cineraceus from A. cahirinus is supported by chromosomal data (2n = 48 or 50 for A. cineraceus, 2n = 36 for A. cahirinus; [37, 114]). Limits of the geographic range of A. cineraceus are unresolved, especially its western part [37] . Compared to the distribution maps in Denys et al. [16] and Happold [115], we were not able to confirm its occurrence in western Ethiopia.
(17) Cah2
Distribution: Burkina Faso, Mali.
Available name: None. Based on Barome et al. [30], we call this MOTU as Acomys sp. 2 in Fig. 1.
Type locality: Not relevant.
Karyotype: 2n = 66–68, NF = 66–72 [116].
Additional information: Barome et al. [30] called this MOTU as Acomys sp. 2, Granjon and Duplantier [116] included it in A. johannis species complex. The distribution of Cah2 in West Africa is overlapping with Cah3 and Cah4, and its specific status should be investigated by integrative taxonomy approach using larger material and multi-locus genetic analysis.
(18) Cah3
Distribution: Burkina Faso.
Available name: None. Based on Barome et al. [30], we call this MOTU as Acomys sp. 1 in Fig. 1.
Type locality: Not relevant.
Karyotype: 2n = 66–68, NF = 66–72 [116].
Additional information: Barome et al. [30] called this MOTU as Acomys sp. 1, Granjon and Duplantier [116] included it in A. johannis species complex. The distribution of Cah3 in West Africa is overlapping with Cah2, Cah4 and Cah11. The West African A. johannis species complex (paraphyletic in our study, grouping Cah2, Cah3 and Cah4; see also [116]) requires taxonomic revision.
(19) Cah4
Distribution: Chad, Cameroon, Niger, Nigeria, Benin.
Available name: A. johannis Thomas, 1912.
Type locality: Bauchi Plateau Kabwir, North Nigeria.
Karyotype: 2n = 66–68, NF = 66–72 [116].
Additional information: This MOTU was included in previous phylogenetic studies as A. johannis [30, 31, 37, 43, 46]. Formerly it was included in A. cahirinus [38] or A. cineraceus [104]. Sicard and Tranier [63] provided a detailed report on the geographic distribution of three pelage colour phenotypes of Acomys occurring in Burkina Faso [117], assigned them to A. johannis, and contrasted their external, cranial, and dental morphology with A. chudeaui (= Cah11). Using CYTB sequences, Barome et al. [31] reported the specimens from Burkina Faso and Mali as Acomys sp. 1 (= Cah3) and Acomys sp. 2 (= Cah2) and specimens from Niger, Benin, Cameroon, and Niger as A. johannis (= Cah4), but their conspecificity has never been tested by the combination of multi-locus genetic and phenotypic data.
(20) Cah5
Distribution: Egypt (Sinai only), Arabian Peninsula, South Iran.
Available name: Acomys dimidiatus Cretzschmar, 1826.
Type locality: Sinai, Egypt.
Karyotype: 2n = 36–38, NF = 68–70 [110].
Additional information: Acomys dimidiatus is nearly indistinguishable from A. cahirinus with regard to external morphology, which resulted in great confusions relative to its classification [16]. With few exceptions (e.g., [34, 37, 38, 46, 118, 119]) A. dimidiatus usually has been listed in the synonymy of A. cahirinus [36, 37, 82, 120]. A morphological and cytogenetical review of Acomys species made by Denys et al. [51], who provided the external, skull and dental characteristics of all the type specimens available, validated A. cahirinus as distinct from A. dimidiatus. Frynta et al. [33] referred two major lineages in northern Africa and Middle East, which should represent A. cahirinus and A. dimidiatus, respectively. The type localities of these two species are very close each other (Cairo and Sinai) but their distributions seem to be separated by the Isthmus of Suez.
(21) Cah6
Distribution: Horn of Africa (S Eritrea, Djibouti, E Ethiopia and N Somalia).
Available name: Acomys mullah Thomas, 1904.
Type locality: Harar, Ethiopia.
Karyotype: Not known.
Additional information: Awaiting more detailed taxonomic revision, we assigned the name A. mullah to the lineage Cah6 distributed in the margins of the Afar triangle in Ethiopia. This species is considered a member of the “cahirinus-dimidiatus” complex [16, 121], which is confirmed by our phylogenetic study. This species may co-occur with A. louisae (= Dji). It should be also taken in consideration that the type locality of A. mullah (Harar) is very close to the only known locality of Ign1 and is separated from the Afar lowland by the Chercher Mts. Comparison of Ign1 and Cah6 with the type material of A. mullah (and A. brockmani considered as its synonym) is necessary.
(22) Cah7
Distribution: North-west Ethiopia (Mai Temen and Alatish NP).
Available name: None. Based on Lavrenchenko et al. [112], we use the name Acomys sp. B in Fig. 1.
Type locality: Not relevant.
Karyotype: 2n = 40, NF = 68 [112].
Additional information: Ivlev et al. [25] and Lavrenchenko et al. [112] called this lineage Acomys sp. B. It is sympatric with Cah8, but the two lineages significantly differ by karyotypes, physiological and behavioural traits [112] and thus might represent different biological species.
(23) Cah8
Distribution: Alatish NP, Ethiopia.
Available name: None. Based on Lavrenchenko et al. [112], we use the name Acomys sp. A in Fig. 1.
Type locality: Not relevant.
Karyotype: 2n = 52, NF = 68 [112].
Additional information: Ivlev et al. [25] and Lavrenchenko et al. [112] called this lineage Acomys sp. A. It is also very abundant in the neighbouring Dinder NP in Sudan (J. Bryja et al., unpublished data). The taxonomic revision of Cah7, Cah8, Cah9 and Cah10 is necessary and more intensive sampling in Sudan and northern Ethiopia would be very helpful.
(24) Cah9
Distribution: Egypt, Greece (Crete), Cyprus, Turkey, Libya, northern Chad.
Available name: Acomys cahirinus (É. Geoffroy Saint-Hilaire, 1803).
Type locality: Cairo, Egypt.
Karyotype: 2n = 36–42, NF = 68 [54, 110].
Additional information: The species was described from Cairo (Egypt). It seems very likely that it colonized eastern Mediterranean area during Antiquity. Weak genetic differences revealed also by our multilocus analysis support the view that A. cahirinus should be synonymized with A. minous Bate, 1906 from Crete, A. cilicicus Spitzenberg, 1978 from Turkey and A. nesiotes Bate, 1903 from Cyprus (see [16, 54] and references there). The relationships with A. seurati (a distinct taxon from rocky areas in southern Algeria, differing by karyotype and dental morphology; [51]) should be investigated by using genetic data from the Algerian material.
(25) Cah10
Distribution: Sheraro, Ethiopia.
Available name: None.
Type locality: Not relevant.
Karyotype: Not known.
Additional information: Cah10 is known only from one locality in North Ethiopia. It is a sister MOTU either to Cah9 or Cah11 and its conspecificity with A. cahirinus and/or A. chudeaui should be tested.
(26) Cah11
Distribution: Niger, Mauritania, Mali, Chad.
Available name: Acomys chudeaui Kollman, 1911.
Type locality: Atar, SW of Biskra, Mauritania.
Karyotype: 2n = 40–46, NFa = 66 [116].
Additional information: This taxon has been previously listed as a synonym of A. cahirinus, but most recent works consider it as a distinct species [36, 46, 116]. Nicolas et al. [43] synonymized A. airensis and A. chudeaui and provided a detailed phylogeographic analysis of this taxon.
Conclusions
Using multilocus genetic data, comprehensive geographic sampling and multiple phylogenetic approaches, we revealed that the spiny mice (Acomys) are composed of five main species groups: subspinosus, spinosissimus, russatus, wilsoni and cahirinus. Three of them (spinosissimus, wilsoni and cahirinus) clearly represent species complexes. We delimited 26 genetic lineages as potential Acomys species, and their taxonomic status should now be assessed by multidisciplinary investigations. The origin of the genus is dated to the late Miocene in savannas of eastern Africa, when the first vicariance between “southern” and “northern” groups was probably caused by the development of the coast-to-coast forest belt. The evolutionary history of the genus in Plio-Pleistocene was influenced by global climatic transitions as well as by local geomorphological features (e.g. deserts, mountain blocks and/or large water bodies) and is characterized by repeated cycles of diversifications, especially in eastern Africa, and repeated dispersal events mainly to the North and West. The spiny mice can be thus used as very suitable model for testing specific hypotheses of the role of historical factors on the formation of current biodiversity of seasonally dry environments of Afro-Arabia.
Methods
Sampling
The genetic dataset is based on 700 individuals of spiny mice. We produced original genetic data from 421 individuals collected at more than one hundred localities, and complemented them by 279 georeferenced mitochondrial sequences from GenBank. This material covers large part of the distribution of the genus as predicted by the IUCN [122] (see Fig. 2a). All individuals were DNA-barcoded at mitochondrial markers to get as precise distributional maps of genetic clades as possible, but part of sequences was removed as redundant from subsequent phylogenetic analyses (see Additional file 5). All fieldwork performed in the frame of this study complied with legal regulations in particular countries and sampling was in accordance with local legislation (see more details in Ethics approval section). Rodents were trapped in Sherman live traps (H.B. Sherman Traps Inc., Tallahassee, USA) and snap traps baited with a mixture of peanut butter, maize flour and dried fish. Mice caught in live traps were euthanized by cervical dislocation or an overdose of Isoflurane prior to dissection (Directive 2010/63/EU). When present, the spiny mice are generally the most abundant component of the small mammal communities and are not listed as endangered. Each individual was identified to the genus by the external features and the tissue sample (tail, toe, spleen, etc.) was stored in 96% ethanol until DNA extraction. GPS coordinates of each locality were recorded. For more details on particular specimens, localities and collectors, see Additional file 5.
DNA extraction, amplification and sequencing
DNA from 96% ethanol-preserved tissue samples was extracted using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. For phylogenetic analysis we selected four genetic markers; two mitochondrial fragments, cytochrome b (CYTB) and control region (D-loop) and two nuclear exons, Interphotoreceptor Binding Protein gen (IRBP) and Recombination activating gene 1 (RAG1). Individual markers were amplified by the polymerase chain reaction (PCR) using following combination of primers: L14723 and H15915 [123] for CYTB; ‘primers 1–4’ [124] for D-loop; IRBP217 and IRBP1531 [125] for IRBP and RAG1F1705 and RAG1R2951 [126] for RAG1. Each locus was amplified using a final concentration of 3 mM of MgCl2 (for IRBP only 2 mM), 0.2 mM of each dNTP, 0.4 μM of each primer, 1 x Taq buffer (Thermo Fisher Scientific, Waltham, USA), 0.2 μl of Taq polymerase (5 U/μl, Thermo Fisher Scientific), 30 ng/μl of genomic DNA, and ddH2O to a total volume of 15 μl. PCR products were purified with Calf Intestine Alkaline Phosphatase and Exonuclease I (New England Biolabs, Ipswich, USA), and Sanger-sequenced in both directions using the BigDye® Terminator chemistry (Thermo Fisher Scientific) either at the Institute of Vertebrate Biology CAS on an ‘Applied Biosystems® 3130xl Genetic Analyzer’ or commercially through the GATC Biotech company (Konstanz, Germany). All corresponding sequences were deposited in GenBank under accession numbers MH044731-MH045045 (see Additional file 5).
Phylogenetic analysis
The final dataset for phylogenetic analyses consisted of 373 unique sequences of CYTB, 71 sequences of IRBP, 59 sequences of RAG1 and 96 sequences of D-loop. The remaining mitochondrial sequences (usually identical and/or shorter sequences and/or from the same or neighbouring localities) were unambiguously assigned to particular mtDNA lineages by preliminary phylogenetic analysis and they were removed as redundant (see Additional file 5). These data were used only to increase the precision by which we mapped the geographical distribution of phylogenetic clades. Nuclear exons were sequenced only in the representative subset of 102 individuals, covering the geographic distribution and mitochondrial diversity as much as possible (see Additional files 5 and 6). As outgroups we used four taxa from subfamily Deomyinae, to which Acomys belong (Deomys ferrugineus, Lophuromys flavopunctatus, Lophuromys sikapusi and Uranomys ruddi; see Additional file 5). Sequences were aligned in MUSCLE [127] and the concatenated dataset with total length 4005 bp was created in Mesquite. For all three protein-coding genes (CYTB, IRBP, RAG1), we used Mesquite 3.03 [128] to check the coding frame for possible errors or stop codons.
Phylogenetic reconstructions were conducted using maximum likelihood (ML) and Bayesian inference (BI). For both phylogenetic approaches were carried out partitioned analyses to improve phylogenetic accuracy [129]. The molecular dataset was divided into ten partitions: we used three partitions for each of the protein-coding genes, and one partition for the control region (D-loop). The best partitioning scheme and substitution models were determined with PartitionFinder v1 [130] using a greedy heuristic algorithm with ‘linked branch lengths’ option. The Bayesian information criterion (BIC) was used to compare partitioning schemes and substitution models following the recommendation of Ripplinger and Sullivan ([131]; Table 2).
Table 2.
The substitution models used in particular phylogenetic analyses. They were selected by PartitionFinder using BIC model selection, greedy search, linked branch length
| Partitions | RAxML | MrBayes | BEAST | |
|---|---|---|---|---|
| 1 | CYTB_pos1 | GTR + I + G | GTR + I + G | GTR + I + G |
| 2 | CYTB_pos2 | GTR + I + G | HKY + I + G | HKY + I + G |
| 3 | CYTB_pos3 | GTR + I + G | GTR + I + G | GTR + I + G |
| 4 | IRBP_pos1, RAG1_pos2 | GTR + G | HKY + I | TrN + I |
| 5 | IRBP_pos2, RAG1_pos3 | GTR + G | HKY + G | TrN + G |
| 6 | IRBP_pos3, RAG1_pos1 | GTR + I + G | HKY + I + G | HKY + I + G |
| 7 | D-loop | GTR + I + G | GTR + I + G | GTR + I + G |
Maximum likelihood analyses were performed using RAxML v8.2.8 [132] for separate gene trees (CYTB and D-loop not shown same topology as Fig. 1, IRBP Additional file 2, RAG1 Additional file 3) as well as for concatenate matrix. Based on the BIC results in PartitionFinder we used seven partitions for concatenate matrix, five partitions with GTR + I + G model and two with GTR + G substitution model (Table 2). The ML tree was obtained using heuristic searches with 100 random addition replicates and the clade support was then assessed using a non-parametric bootstrap procedure with 1000 replicates. Following Hillis and Bull [133], nodes supported by bootstrap values (BP) ≥ 70 were considered strongly supported.
Bayesian inference analyses were carried out using MrBayes v3.2.6 [134] with seven partitions (Table 2). Two independent runs with four MCMC (one cold and three incrementally heated) were conducted: they ran for 50 million generations, with trees sampled every 1000 generations. A conservative 25% burn-in was applied after checking for stability on the log-likelihood curves and the split-frequencies of the runs. Support of nodes for MrBayes analyses was provided by clade posterior probabilities (PP) as directly estimated from the majority-rule consensus topology. Following Erixon et al. [135], nodes supported by PP ≥ 0.95 were considered strongly supported.
Estimates of species richness
For estimation of Acomys species richness we applied multiple species delimitation methods (as suggested by Carstens et al. [136]): (1) “by-eye” analysis of genetic structure (based primarily on CYTB barcodes) and geographical distribution of genetic lineages; (2) species discovery approach to assign individuals to putative groups based on the variability of CYTB sequences [137, 138]; (3) species delimitation based on multi-locus data and multispecies coalescent methods [139].
In the first simplest approach, we produced ML tree based only on CYTB sequences. We then compared the revealed clades with the species names used in previous studies (e.g. [31, 32, 41, 43, 45]), and the distribution of particular clades with positions of type localities of nominal species. By using this approach we newly identified several highly supported phylogenetic clades with parapatric distribution to previously analysed and named species, which might represent new species and are worth of further taxonomic studies. Genetic distances (within species and to the genetically nearest lineage) from BI tree were additionally also computed using the species delimitation package [140] implemented in Geneious v9.1.8 [141].
Second, we performed two analyses of species discovery based on diversity of CYTB marker. The Automatic Barcode Gap Discovery approach (ABGD; [137]) was used to identify barcode gap between intraspecific and interspecific genetic distances. An alternative Poisson Tree process (PTP) approach models intra- and interspecies processes by directly using the number of substitutions [138]. We used a recently improved algorithm based on PTP, the so-called multi-rate PTP (mPTP; [142]), which works better for phylogenies that have different rates of speciation-coalescence and allows to account for the different rates of branching events within each delimited species [142]. Both analyses (ABGD and mPTP) were performed using the ultrametric CYTB phylogeny produced by Bayesian method with strict clock in BEAST v2.4.7 [143].
The last used species delimitation approach, Species Tree And Classification Estimation, Yarely (STACEY; [139]), is a Bayesian method based on the multispecies coalescent model and estimates the probability of distinct species delimitation hypotheses given multilocus data. By utilizing multi-species coalescent theory and phylogenetic inference under a full probability Bayesian network, STACEY simultaneously estimates gene trees, the species tree and species delimitations under the assumption that all individuals that are affected by the same coalescent process, also belong to the same species/clade. We assumed conspecificity of individuals bearing mtDNA of the same lineage, identified by the first arbitrary approach described above. All but one lineages were represented at least by two individuals genotyped minimally at 3–4 markers (see Additional file 5). To relax the prior assumptions about species delimitation, we estimated a species tree using the birth-death-collapse model [144] as implemented in STACEY for BEAST 2 [145]. STACEY does not require guide tree, therefore errors resulting from a priori phylogenetic assumptions are avoided. Parameters and priors for the analysis were set according to the recommendations of STACEY manual [139]. Sequence alignments were imported into BEAUTI where they were assigned separate and unlinked substitution, clock and tree models. For mitochondrial markers (CYTB + D-loop) the ploidy was set of 0.5, for nuclear genes (IRBP, RAG1) 2.0. For the species tree prior the collapse height was set to 10− 3. Three independent MCMC chains were run for 10− 7 generations and log every 5000 generations. The burn-in was 25%, and the outputs from three runs were combined in LogCombiner 2.4.7 [146]. The similarity matrix was created using SpeciesDelimationAnalyser version 1.8.9 [147] with 25% burn-in and collapse height of 10− 3. Species tree was visualised as a cloudogram using DensiTree [148].
Divergence dating
To calibrate a molecular clock, we compiled the set of usable fossils for the genus Acomys and its ancestors: (1) an extinct genus †Preacomys with three described species: †P. griffini Mein et al., 2004 and †P. karsticus Mein et al., 2004 from Harasib, Namibia (9 Ma; [83, 149]) and †P. kikiae Geraads, 2001 from Chorora, Ethiopia (8.5 Ma; [150, 151]). Because the position of these three fossils on phylogenetic tree is not unequivocally clear, we used a minimum age 8.5 Ma as a root for the genus Acomys. (2) Acomys from Lemudong’o locality in Kenya (6.08–6.12 Ma; [152, 153]) is the oldest Acomys and we considered it as the most recent common ancestor (MRCA) for taxa living currently in the northern part of eastern Africa (with the centre of their distribution in Somali-Masai savanna) and in Arabia, i.e. the clade encompassing cahirinus + wilsoni + russatus groups. (3) The oldest fossil of the spinosissimus group (sensu Verheyen et al. [45]) was discovered in Transvaal, South Africa (3 Ma; [154]), and we used it as MRCA for this group. Bayesian analyses of divergence dating were conducted on a species tree in *BEAST v2.4.7 [143]. The species were defined based on STACEY results. The mitochondrial (CYTB + D-loop) and nuclear genes (IRBP, RAG1) were imported in BEAUTI where they were assigned separate and unlinked substitution, clock and tree models. Bayesian analysis run with uncorrelated lognormal relaxed clocks [155], birth-death tree prior [156] and selected fossil constraints were defined by using lognormal statistical distributions (see Table 3 for more details). Two independent runs were carried out for 107 generations with sampling every 1000 generations in BEAST. We discarded first 25% as burn-in and the resulting parameter and tree files were examined for convergence and effective sample sizes (> 200) in Tracer 1.6 [157]. The two runs were combined in LogCombiner and the species tree was visualized in TreeAnotator.
Table 3.
List of fossils associated with the genus Acomys used in the divergence dating. The offset and mean represent the specification of lognormal priors used for the calibration of molecular clock. All fossil constrains were used as a crown
| Fossil | MRCA | Locality | Author | Age (Ma) | Offset | Mean |
|---|---|---|---|---|---|---|
| †Preacomys kikiae | Acomys | Chorora, Ethiopia | Geraads (2001, 2002); Suwa et al., (2015) | 8.5 | 8.5 | 1.0 |
| †Acomys I. Geoffroy | cahirinus + wilsoni + russatus | Lemudong’o, Kenya | Manthi (2007); Manthi and Ambrose (2007) | 6.08–6.12 | 6.08 | 1.0 |
| †Acomys spinosissimus Peters | spinosissimus | Transvaal, South Africa | Denys (1999) | 3 | 3 | 1.0 |
Biogeographical reconstructions
The BioGeoBEARS approach [158] was used to reconstruct the ancestral distributions and diversification patterns. Six major biogeographic regions with Acomys occurrence were defined on the basis of Holt et al. [159] and Linder et al. [3]: South Africa region (S – South Africa), Zambezian region (Z – Zambezian region), Somali region (E – East Africa), Sudanian region (W – West Africa), Sahara region (N – North Africa) and the Arabian region (A – Arabian region). Dispersal rate between adjacent areas (S-Z, Z-E, E-W, E-N, N-A) was fixed to 1, whereas the dispersal of 0.5 (S-E, Z-W, Z-N, E-A) was defined for long distance dispersal (i.e. biogeographical areas separated by another region) or whenever a geographical barrier had to be crossed (e.g. multiple water bodies). Dispersal was disallowed between geographical areas separated by two or more areas (S-W, S-A, S-N, Z-N, Z-A, W-A). Biogeographic reconstruction relied on the Dispersal-Extinction Cladogenesis model (DEC) of range evolution [160]. DEC model estimates geographical range evolution using a phylogenetic tree with branch lengths scaled to time, geographical (habitat) areas for all tips, and an adjacent matrix of plausibly connected areas [85]. Because of concerns with its statistical validity [161] we did not use the +J model of Matzke [158] in our analyses.
Species distribution modelling
Assuming phylogenetic niche conservatism [162] and generally similar ecological requirements for all taxa of spiny mice, we modelled the present and past distribution of suitable climatic conditions for the genus Acomys by the maximum entropy approach [163]. We used 282 presence records (unique localities, Additional file 5, Fig. 2) as the input data to train the model. We modelled the suitable conditions in the recent, but we also produced paleoclimatic projections for the last glacial maximum (21 ka; MIROC resolution 2.5 min [164]) and for the last interglacial (120–140 ka; resolution 30 s [165]) using 19 bioclimatic variables from the WorldClim database [166]. The background was restricted to whole Africa and Arabia (Figs. 2 and 3). The species distribution modelling (SDM) analysis was performed using MaxEnt v3.3.3 k [167]. We used 10 replicates and the importance of environmental variables was tested using jackknife option, and for the regularization multiplier we used the default value of 1. The SDM results were converted in a map using QGIS with a maximized sum threshold [168, 169].
Additional files
Taxonomic classifications of the genus Acomys. Bold italic names represent species reported in particular lists, standard italics represent subspecies (in Ellerman [34]; Setzer [38]; Denys et al. [16]). (XLSX 41 kb)
ML phylogenetic tree based on IRBP sequences (nexus file). (NEXUS 6 kb)
ML phylogenetic tree based on RAG1 sequences (nexus file). (NEXUS 5 kb)
Maximum clade credibility tree from STACEY with PP support (nexus file). (NEXUS 24 kb)
Complete list of individuals used in this study, with details on localities and genetic data. (XLSX 97 kb)
Alignment of 369 ingroup and 4 outgroup concatenated sequences of CYTB, IRBP, RAG1 and D-loop. (NEXUS 1470 kb)
Acknowledgements
For help during the field work we acknowledge J. Komárková-Vrbová, S. Gryssels, J. Šklíba, T. Vlasatá, E. Hrouzková, S. Šafarčíková, H. Konvičková, M. Lövy, V. Mazoch, J. Zima, P. Kaňuch, F. Sedláček, O. Mikula, A. Konečný, A. Hánová, L. Cuypers, A. Ribas, J. Krásová, M. Kamler, J. Kopečná, P. Široký, T. Mazuch, A. Kassahun, C. Sabuni, A. Katakweba, A. Massawe, K. Welegerima, A.A. Warshavsky, Yu. F. Ivlev, D. Yu. Alexandrov, D. S. Kostin, A. A. Martynov, A. R. Gromov, Z. Tomass and all local colaborators. We would like to thank also M. M. McDonough (Field Museum of Natural History, Chicago), S. C. Peurach (Smithsonian Institution, National Museum of Natural History, Washington), V. Nicolas (Muséum National d’Histoire Naturelle, Paris) and P. Benda (National Museum, Prague) for providing additional samples. For help with genotyping we acknowledge A. Bryjová, H. Konvičková, D. Mizerovská and A. Hánová. Most phylogenetic analyses were run on the Czech-grid infrastructure METACENTRUM, minor part then on CIPRES Gateway or CBGP cluster.
Funding
This study was supported by the Czech Science Foundation, projects no. 15-20229S and 18-17398S, the Ministry of Culture of the Czech Republic (DKRVO 2018/14, National Museum, 00023272) and the Russian Foundation for Basic Research (project no. 18–04-00563-a). This funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All corresponding sequences were deposited in GenBank under accession numbers MH044731-MH045045 (see Additional file 5). The final alignment of concatenated sequences used in phylogenetic analyses is in Additional file 6.
Abbreviations
- 2n
Diploid number of chromosomes
- a.s.l.
Above sea level
- ABGD
Automatic Barcode Gap Discovery
- BI
Bayesian Inference
- BIC
Bayesian information criterion
- bp
Base pairs
- BP
Bootstrap value
- CYTB
Cytochrome b gene
- DEC
Dispersal-Extinction Cladogenesis model
- D-loop
Control region
- EAM
Eastern Arc Mountains
- EARS
The East African Rift System
- GRV
The Great Rift Valley
- HPD
Highest posterior density
- iNHG
The intensification of Northern Hemisphere glaciation
- IRBP
Interphotoreceptor Binding Protein gen
- ka
Thousand years
- LGM
Last Glacial Maximum
- LIG
Last Interglacial
- Ma
Million of years ago
- MaxEnt
Maximum entropy
- MCMC
Markov chain Monte Carlo
- ML
Maximum likelihood
- MOTU(s)
Molecular operational taxonomic unit(s)
- mPTP
Multi-rate Poisson Tree process
- MRCA
The most recent common ancestor
- MSC
Messinian salinity crisis
- Mt. (s)
Mountain(s)
- mtDNA
Mitochondrial DNA
- NF
Fundamental number of chromosome arms
- NFa
Fundamental number of autosomal chromosome arms
- NP
National park
- PCR
The polymerase chain reaction
- PP
Posterior probability
- PTP
Poisson Tree process
- RAG1
Recombination activating gene 1
- SDM
Species distribution modelling
- SRM
Southern Rift Mountains
- STACEY
Species Tree And Classification Estimation, Yarely
Authors’ contributions
DF, RS and JB conceived and designed the study; TA, DF, RS, LL, YM, JS, JV, JM, DM, JB collected important part of samples; TA and KP performed laboratory analysis; TA analysed data; TA, JB, KP drafted the first version of the manuscript. All authors made substantial contribution on acquisition of data, revised the draft, gave final approval of the version to be published and agreed to be accountable for all aspects of the work.
Ethics approval and consent to participate
We are indebted to many local authorities for providing permits to carry out the research (including the ethics approval, when applicable), especially the Zambia Wildlife Authority (ZAWA), the National Directorate for Protected Areas (DINAC – Mozambique), the National Research Council and Forestry Department in Malawi, Sokoine University of Agriculture in Morogoro (Tanzania), the Kenyan Forest Service and the Kenyan Wildlife Service (Kenya), the Ethiopian Wildlife Conservation Authority (EWCA), Government of Ethiopia and the Oromia Forest and Wildlife Enterprise (OFWE) in Ethiopia. Furthermore, Josef Bryja is holder of the certificate of competency on Protection of Animals against Cruelty (reg. no. V/1/2005/05) and all manipulations with animals, i.e. when captured in life traps, followed the specific rules for working with alive animals as specified by the Central Commission for Animal Welfare of Ministry of Agriculture of the Czech Republic, and the general protocols were approved by the Committee for Animal Welfare of the Czech Academy of Sciences. Newly analysed material from other countries has been collected in last decades within other research projects and we are indebted to museum curators for allowing us to study the tissue samples in their care.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
T. Aghová, Email: tatiana.aghova@gmail.com
K. Palupčíková, Email: klpr@post.cz
R. Šumbera, Email: sumbera@prf.jcu.cz
D. Frynta, Email: frynta@centrum.cz
L. A. Lavrenchenko, Email: llavrenchenko@gmail.com
Y. Meheretu, Email: meheretu.yonas@mu.edu.et
J. Sádlová, Email: jovanas@seznam.cz
J. Votýpka, Email: jan.votypka@natur.cuni.cz
J. S. Mbau, Email: jsyombua04@yahoo.com
D. Modrý, Email: modryd@vfu.cz
J. Bryja, Email: bryja@brno.cas.cz
References
- 1.Mayaux P, Bartholomé E, Fritz S, Belward A. A new land-cover map of Africa for the year 2000. J Biogeogr. 2004;31:861–877. [Google Scholar]
- 2.Sayre R, Comer P, Hak J, Josse C, Bow J, Warner H, Larwanou M, Kelbessa E, Bekele T, Kehl H, Amena R, Andriamasimanana R, Ba T, Benson L, Boucher T, Brown M, Cress J, Dassering O, Friesen B, Gachathi F, Houcine S, Keita M, Khamala E, Marangu D, Mokua F, Morou B, Mucina L, Mugisha S, Mwavu E, Rutherford M, Sanou P, Syampungani S, Tomor B, Vall A, Weghe JV, Wangui E, Waruingi L. A new map of standardized terrestrial ecosystems of Africa. Washington DC: Association of American Geographers; 2013. [Google Scholar]
- 3.Linder HP, de Klerk HM, Born J, Burgess ND, Fjeldså J, Rahbek C. The partitioning of Africa: statistically defined biogeographical regions in sub-Saharan Africa. J Biogeogr. 2012;39:1189–1205. [Google Scholar]
- 4.Maslin MA, Trauth MH. Plio-Pleistocene east African pulsed climate variability and its influence on early human evolution. In: Grine FE, Leakey RE, Fleagle JG, editors. The first humans - origins of the genus homo. New York: Springer Verlag; 2009. pp. 151–158. [Google Scholar]
- 5.Kingston JD, Deino AL, Edgar RK, Hill A. Astronomically forced climate change in the Kenyan Rift Valley 2.7–2.55 Ma: implications for the evolution of early hominin ecosystems. J Hum Evol. 2007;53:487–503. doi: 10.1016/j.jhevol.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Maslin MA, Christensen B. Tectonics, orbital forcing, global climate change, and human evolution in Africa: introduction to the African paleoclimate special volume. J Hum Evol. 2007;53:443–464. doi: 10.1016/j.jhevol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Pickford M. Uplift of the roof of Africa and its bearing on the evolution of mankind. Hum Evol. 1990;5:1–20. [Google Scholar]
- 8.Prömmel K, Cubasch U, Kaspar F. A regional climate model study of the impact of tectonic and orbital forcing on African precipitation and vegetation. Palaeogeogr Palaeoclimatol Palaeoecol. 2013;369:154–162. [Google Scholar]
- 9.Cerling TE, Harris JM, Macfadden BJ, Leakey MG, Quadek J, Eisenmann V, Ehleringer JR. Global vegetation change through the Miocene/Pliocene boundary. Nature. 1997;389:153–158. [Google Scholar]
- 10.Cerling TE, Ehleringer JR, Harris JM. Carbon dioxide starvation, the development of C4 ecosystems, and mammalian evolution. Philos Trans R Soc B Biol Sci. 1998;353:159–171. doi: 10.1098/rstb.1998.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W, Takebayashi N, Qi Y, Hickerson MJ. MTML-msBayes: approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity. BMC Bioinf. 2011;12:1. doi: 10.1186/1471-2105-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Houérou HN. Outline of the biological history of the Sahara. J Arid Environ. 1992;22:3–30. [Google Scholar]
- 13.Swezey CS. Cenozoic stratigraphy of the Sahara. Northern Africa J African Earth Sci. 2009;53:89121. [Google Scholar]
- 14.Brito JC, Godinho R, Martínez-Freiría F, Pleguezuelos JM, Rebelo H, Santos X, Vale CG, Velo-Antón G, Boratyński Z, Carvalho SB, Ferreira S, Gonçalves DV, Silva TL, Tarroso P, Campos JC, Leite JV, Nogueira J, Alvares F, Sillero N, Sow AS, Fahd S, Crochet P-A, Carranza S. Unravelling biodiversity, evolution and threats to conservation in the Sahara-Sahel. Biol Rev Camb Philos Soc. 2014;89:215–231. doi: 10.1111/brv.12049. [DOI] [PubMed] [Google Scholar]
- 15.Gonçalves DV, Martínez-Freiría F, Crochet PA, Geniez P, Carranza S, Brito JC. The role of climatic cycles and trans-Saharan migration corridors in species diversification: biogeography of Psammophis schokari group in North Africa. Mol Phylogenet Evol. 2018;118:64–74. doi: 10.1016/j.ympev.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Denys C, Taylor PJ, Aplin KP. Family Muridae. In: Wilson DE, Lacher TE Jr, Mittermeier RA, editors. Hadbook of the mammals of the world, volume 7: rodents II. Barcelona: Lynx Edicions; 2017. [Google Scholar]
- 17.Carere C, Casetti R, De Acetis L, Perretta G, Cirulli F, Alleva E. Behavioural and nociceptive response in male and female spiny mice (Acomys cahirinus) upon exposure to snake odour. Behav Process. 1999;47:1–10. doi: 10.1016/s0376-6357(99)00048-0. [DOI] [PubMed] [Google Scholar]
- 18.Frynta D, Fraňková M, Čížková B, Skarlandtová H, Galeštoková K, Průšová K, Šmilauer P, Šumbera R. Social and life history correlates of litter size in captive colonies of precocial spiny mice (Acomys) Acta Theriol (Warsz) 2011;56:289–295. [Google Scholar]
- 19.Gutman R, Dayan T. Temporal partitioning: an experiment with two species of spiny mice. Ecol. 2005;86:164–173. [Google Scholar]
- 20.Haim A, Yedidia I, Haim D, Zisapel N. Photoperiodicity in daily rhythms of body temperature, food and energy intake of the golden spiny mouse (Acomys russatus) Isr J Zool. 1994;40:145–150. [Google Scholar]
- 21.Kronfeld-Schor N, Shargal E, Haim A, Dayan T, Zisapel N, Heldmaier G. Temporal partitioning among diurnally and nocturnally active desert spiny mice: energy and water turnover costs. J Therm Biol. 2001;26:139–142. doi: 10.1016/s0306-4565(00)00034-6. [DOI] [PubMed] [Google Scholar]
- 22.Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, Maden M. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489:561–565. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrhardt N, Heldmaier G, Exner C. Adaptive mechanisms during food restriction in Acomys russatus: the use of torpor for desert survival. J Comp Physiol B. 2005;175:193–200. doi: 10.1007/s00360-005-0475-3. [DOI] [PubMed] [Google Scholar]
- 24.Horowitz M, Borut A. The spiny mous (Acomys cahirinus) - a rodent prototype for studying plasma volume regulation during thermal dehydration. Isr J Zool. 1994;40:117–125. [Google Scholar]
- 25.Ivlev YF, Lavrenchenko LA, OF Chernova, Bekele A. Response to overheating in spiny mice (the genus Acomys) from arid regions in northwestern Ethiopia. Dokl Biol Sci. 2011;440:335–9. [DOI] [PubMed]
- 26.Weissenberg S, Shkolnik A. Metabolic rate and water economy in the desert and Mediterranean populations of the common spiny mouse (Acomys cahirinus) in Israel. Isr J Zool. 1994;40:135–143. [Google Scholar]
- 27.Dieterlen F. Vergleichende Untersuchungen zur Ontogenese von Stachelmaus (Acomys) und Wanderratte (Rattus norvegicus). Beiträge zum Nesthocker-Nestflüchter-Prolem bei Negetieren. Z Säugetierkd. 1963;28:193–227. [Google Scholar]
- 28.Hadid Y, Pavlicek T, Beiles A, Ianovici R, Raz S, Nevo E. Sympatric incipient speciation of spiny mice Acomys at “Evolution Canyon,” Israel. Proc Natl Acad Sci. 2014;111:1043–1048. doi: 10.1073/pnas.1322301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tučková V, Šumbera R, Čížková B. Alloparental behaviour in Sinai spiny mice Acomys dimidiatus: a case of misdirected parental care? Behav Ecol Sociobiol. 2016;70:437–447. [Google Scholar]
- 30.Barome PO, Monnerot M, Gautun JC. Intrageneric phylogeny of Acomys (Rodentia, Muridae) using mitochondrial gene cytochrome b. Mol Phylogenet Evol. 1998;9:560–566. doi: 10.1006/mpev.1998.0507. [DOI] [PubMed] [Google Scholar]
- 31.Barome PO, Monnerot M, Gautun JC. Phylogeny of the genus Acomys (Rodentia, Muridae) based on the cytochrome b mitochondrial gene: implications on taxonomy and phylogeography. Mammalia. 2000;64:423–438. [Google Scholar]
- 32.Barome PO, Lymberakis P, Monnerot M, Gautun JC. Cytochrome b sequences reveal Acomys minous (Rodentia, Muridae) paraphyly and answer the question about the ancestral karyotype of Acomys dimidiatus. Mol Phylogenet Evol. 2001;18:37–46. doi: 10.1006/mpev.2000.0859. [DOI] [PubMed] [Google Scholar]
- 33.Frynta D, Palupčiková K, Bellinvia E, Benda P, Skarlantová H, Schwarzová L, Modrý D. Phylogenetic relationships within the cahirinus-dimidiatus group of the genus Acomys (Rodentia : Muridae): new mitochondrial lineages from Sahara, Iran and the Arabian Peninsula. Zootaxa. 2010;2660:46–56. [Google Scholar]
- 34.Ellerman JR. Family muridae, volume II. London: British Museum (Natural History); 1941. The Families and Genera of Living Rodents. [Google Scholar]
- 35.Honacki JH, Kinman KE, Koeppl JW. Mammal species of the world: a taxonomic and geographic reference. Kansas: Allen Press Inc. and The Association of Systematics Collections; 1982. [Google Scholar]
- 36.Monadjem A, Taylor P, Denys C, Cotterill FPD. Rodents of sub-Saharan Africa. A biogeographic and taxonomic synthesis. Berlin, Boston: De Gruyter; 2015. [Google Scholar]
- 37.Musser GG, Carleton MD. Superfamily Muroidea. In: Wilson DE, DAM R, editors. Mammal Species of the World. A Taxonomic and Geographic Reference. Baltimore: The Johns Hopkins University Press; 2005. pp. 894–1531. [Google Scholar]
- 38.Setzer HW. Genus Acomys. In: Meester J, Setzer HW, editors. The mammals of Africa: an identification manual. Washington: Smithsonian Institution Press; 1975. pp. 1–2. [Google Scholar]
- 39.Chevret P, Denys C, Jaeger JJ, Michaux J, Catzeflis FM. Molecular evidence that the spiny mouse (Acomys) is more closely related to gerbils (Gerbillinae) than to true mice (Murinae) Proc Natl Acad Sci U S A. 1993;90:3433–3436. doi: 10.1073/pnas.90.8.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthey R. Cytogenetique et taxonomie du genre Acomys A. percivali Dollman et A. wilsoni Thomas, especes D’Abyssinie. Mammalia. 1968;32:621–627. [Google Scholar]
- 41.Barome PO, Volobouev V, Monnerot M, Mfune JK, Chitaukali W, Gautun JC, Denys C. Phylogeny of Acomys spinosissimus (Rodentia, Muridae) from North Malawi and Tanzania: evidence from morphological and molecular analysis. Biol J Linn Soc. 2001;73:321–340. [Google Scholar]
- 42.Mgode GF. Application of a multidisciplinary approach to the systematics of Acomys (Rodentia: Muridae) from northern Tanzania. PhD thesis: University of Pretoria, Zoology and Entomology; 2006. http://www.suaire.sua.ac.tz:8080/xmlui/bitstream/handle/123456789/1433/Mgode18.pdf?sequence=1&isAllowed=y. Accessed 8 Feb 2019.
- 43.Nicolas V, Granjon L, Duplantier JM, Cruaud C, Dobigny G. Phylogeography of spiny mice (genus Acomys, Rodentia: Muridae) from the south-western margin of the sahara with taxonomic implications. Biol J Linn Soc. 2009;98:29–46. [Google Scholar]
- 44.Petružela J, Šumbera R, Aghová T, Bryjová A, Katakweba AS, Sabuni CA, Chitaukali WN, Bryja J. Spiny mice of the Zambezian bioregion–phylogeny, biogeography and ecological differentiation within the Acomys spinosissimus complex. Mamm Biol. 2018;91:79–90. [Google Scholar]
- 45.Verheyen W, Hulselmans J, Wendelen W, Leirs H, Corti M, Backeljau T, Verheyen E. Contribution to the systematics and zoogeography of the east-African Acomys spinosissimus Peters 1852 species complex and the description of two new species (Rodentia: Muridae) Zootaxa. 2011;3059:1–35. [Google Scholar]
- 46.Alhajeri BH, Hunt OJ, Steppan SJ. Molecular systematics of gerbils and deomyines (Rodentia: Gerbillinae, Deomyinae) and a test of desert adaptation in the tympanic bulla. J Zool Syst Evol Res. 2015;53:312–30.
- 47.Bryja J, Kostin D, Meheretu Y, Šumbera R, Bryjová A, Kasso M, Mikula O, Lavrenchenko LA. Reticulate Pleistocene evolution of Ethiopian rodent genus along remarkable altitudinal gradient. Mol Phylogenet Evol. 2018;118:75–87. doi: 10.1016/j.ympev.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 48.Correa C, Vásquez D, Castro-Carrasco C, Zúñiga-Reinoso Á, Ortiz JC, Palma RE. Species delimitation in frogs from south American temperate forests: the case of Eupsophus, a taxonomically complex genus with high phenotypic variation. PLoS One. 2017;12:e0181026. doi: 10.1371/journal.pone.0181026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sukumaran J, Knowles LL. Multispecies coalescent delimits structure, not species. Proc Natl Acad Sci U S A. 2017;114:1607–1612. doi: 10.1073/pnas.1607921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haughton CL, Gawriluk TR, Seifert AW. The biology and husbandry of the African spiny mouse (Acomys cahirinus) and the research uses of a laboratory Colony. J Am Assoc Lab Anim Sci. 2016;55:9–17. [PMC free article] [PubMed] [Google Scholar]
- 51.Denys C, Gautun J-C, Tranier M, Volobouev V. Evolution of the genus Acomys (Rodentia, Muridae) from dental and chromosomal patterns. Isr J Zool. 1994;40:215–246. [Google Scholar]
- 52.Bray TC, Bennett NC, Mohammed OB, Alagaili AN. On the genetic diversity of spiny mice (genus Acomys) and gerbils (genus Gerbillus) in the Arabian peninsula. Zool Middle East. 2013;59:283–288. [Google Scholar]
- 53.Fernandes CA, Rohling EJ, Siddall M. Absence of post-Miocene Red Sea land bridges: biogeographic implications. J Biogeogr. 2006;33:961–966. [Google Scholar]
- 54.Giagia-Athanasopoulou EB, Rovatsos MTH, Mitsainas GP, Martimianakis S, Lymberakis P, Angelou LXD, Marchal JA, Sánchez A. New data on the evolution of the Cretan spiny mouse, Acomys minous (Rodentia: Murinae), shed light on the phylogenetic relationships in the cahirinus group. Biol J Linn Soc. 2011;102:498–509. [Google Scholar]
- 55.Brouat C, Tatard C, Bã K, Cosson J-F, Dobigny G, Fichet-Calvet E, Granjon L, Lecompte E, Loiseau A, Mouline K, Piry S, Duplantier J-M. Phylogeography of the Guinea multimammate mouse (Mastomys erythroleucus): a case study for Sahelian species in West Africa. J Biogeogr. 2009;36:2237–2250. [Google Scholar]
- 56.Dobigny G, Tatard C, Gauthier P, Ba K, Duplantier J, Granjon L, Kergoat GJ. Mitochondrial and Nuclear Genes-Based Phylogeography of Arvicanthis niloticus (Murinae) and Sub-Saharan Open Habitats Pleistocene History. PLoS One. 2013; 10.1371/journal.pone.0077815. [DOI] [PMC free article] [PubMed]
- 57.Granjon L, Colangelo P, Tatard C, Colyn M, Dobigny G, Nicolas V. Intrageneric relationships within Gerbilliscus (Rodentia, Muridae, Gerbillinae), with characterization of an additional west African species. Zootaxa. 2012;25:1–25. [Google Scholar]
- 58.Kingdon J. The Kingdon field guide to African mammals. 2. London: Bloomsbury Publishing Plc; 2015. [Google Scholar]
- 59.Douady CJ, Catzeflis F, Raman J, Springer MS, Stanhope MJ. The Sahara as a vicariant agent, and the role of Miocene climatic events, in the diversification of the mammalian order Macroscelidea (elephant shrews) Proc Natl Acad Sci U S A. 2003;100:8325–8330. doi: 10.1073/pnas.0832467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franck P, Garnery L, Loiseau A, Oldroyd BP, Hepburn HR, Solignac M, Cornuet JM. Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity. 2001;86:420–430. doi: 10.1046/j.1365-2540.2001.00842.x. [DOI] [PubMed] [Google Scholar]
- 61.Guillaumet A, Crochet PA, Pons JM. Climate-driven diversification in two widespread Galerida larks. BMC Evol Biol. 2008;8:32. doi: 10.1186/1471-2148-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosevear D. The rodents of West Africa. London: Trustees of the British Museum Natural History; 1969. [Google Scholar]
- 63.Sicard B, Tranier M. Caractères et répartition de trois phénotypes d’Acomys (Rodentia, Muridae) au Burkina Faso. Mammalia. 1996;60:53–68. [Google Scholar]
- 64.Diamond JM, Hamilton AC. The distribution of forest passerine birds and quaternary climatic change in tropical Africa. J Zool. 1980;191:379–402. [Google Scholar]
- 65.Kingdon J. Where have the colonists come from? A zoogeographical examination of some mammalian isolates in eastern Africa. Afr J Ecol. 1981;19:115–124. [Google Scholar]
- 66.Jesus FF, Wilkins JF, Solferini VN, Wakeley J. Expected coalescence times and segregating sites in a model of glacial cycles. Genet Mol Res. 2006;5:466–474. [PubMed] [Google Scholar]
- 67.Colangelo P, Verheyen E, Leirs H, Tatard C, Denys C, Dobigny G, Duplantier JM, Brouat C, Granjon L, Lecompte E. A mitochondrial phylogeographic scenario for the most widespread African rodent, Mastomys natalensis. Biol J Linnean Soc. 2013;108:901–916. [Google Scholar]
- 68.Bryja J, Mikula O, Šumbera R, Meheretu Y, Aghová T, Lavrenchenko LA, Mazoch V, Oguge N, Mbau JS, Welegerima K, Amundala N, Colyn M, Leirs H, Verheyen E. Pan-African phylogeny of Mus (subgenus Nannomys) reveals one of the most successful mammal radiations in Africa. BMC Evol Biol. 2014;14:256–276. doi: 10.1186/s12862-014-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bryja J, Konvičková H, Bryjová A, Mikula O, Makundi R, Chitaukali WN, Šumbera R. Differentiation underground: range-wide multilocus genetic structure of the silvery mole-rat does not support current taxonomy based on mitochondrial sequences. Mamm Biol. 2018;93:82–92. [Google Scholar]
- 70.Mazoch V, Mikula O, Bryja J, Konvičková H, Russo IR, Verheyen E, Šumbera R. Phylogeography of a widespread sub-Saharan murid rodent Aethomys chrysophilus: the role of geographic barriers and paleoclimate in the Zambezian bioregion. Mammalia. 2018;82:373–387. [Google Scholar]
- 71.Mikula O, Šumbera R, Aghova T, Mbau JS, Katakweba AS, Sabuni CA, Bryja J. Evolutionary history and species diversity of African pouched mice (Rodentia: Nesomyidae: Saccostomus) Zool Scr. 2016;45:595–617. [Google Scholar]
- 72.Aghová T, Šumbera R, Piálek L, Mikula O, Mcdonough MM, Lavrenchenko LA, Meheretu Y, Mbau JS, Bryja J. Multilocus phylogeny of east African gerbils (Rodentia, Gerbilliscus) illuminates the history of the Somali-Masai savanna. J Biogeogr. 2017;44:2295–2307. [Google Scholar]
- 73.Trauth MH, Maslin MA, Deino AL, Junginger A, Lesoloyia M, Odada EO, Olago DO, Olaka LA, Strecker MR, Tiedemann R. Human evolution in a variable environment: the amplifier lakes of eastern Africa. Quat Sci Rev. 2010;29:2981–2988. [Google Scholar]
- 74.Bartáková V, Reichard M, Blažek R, Polačik M, Bryja J. Terrestrial fishes: rivers are barriers to gene flow in annual fishes from the African savanna. J Biogeogr. 2015;42:1832–1844. [Google Scholar]
- 75.McDonough MM, Šumbera R, Mazoch V, Ferguson AW, Phillips CD, Bryja J. Multilocus phylogeography of a widespread savanna-woodland-adapted rodent reveals the influence of Pleistocene geomorphology and climate change in Africa’s Zambezi region. Mol Ecol. 2015;24:5248–5266. doi: 10.1111/mec.13374. [DOI] [PubMed] [Google Scholar]
- 76.Keller C, Roos C, Groeneveld LF, Fischer J, Zinner D. Introgressive hybridization in southern African baboons shapes patterns of mtDNA variation. Am J Phys Anthropol. 2010;142:125–136. doi: 10.1002/ajpa.21209. [DOI] [PubMed] [Google Scholar]
- 77.Cotterill FPD. A biogeographic review of tsessebe antelopes Damaliscus lunatus (Bovidae: Alcelaphini) in south-Central Africa. Durban Mus Novit. 2003;28:45–55. [Google Scholar]
- 78.Bryja J, Granjon L, Dobigny G, Patzenhauerová H, Konečný A, Duplantier J-M, Gauthier P, Colyn M, Durnez L, Lalis A, Nicolas V. Plio-Pleistocene history of west African Sudanian savanna and the phylogeography of the Praomys daltoni complex (Rodentia): the environment/geography/genetic interplay. Mol Ecol. 2010;19:4783–4799. doi: 10.1111/j.1365-294X.2010.04847.x. [DOI] [PubMed] [Google Scholar]
- 79.Jacquet F, Denys C, Verheyen E, Bryja J, Hutterer R, Peterhans JCK, Stanley WT, Goodman SM, Couloux A, Colyn M, Nicolas V. Phylogeography and evolutionary history of the Crocidura olivieri complex (Mammalia, Soricomorpha): from a forest origin to broad ecological expansion across Africa. BMC Evol Biol. 2015;15:1–15. doi: 10.1186/s12862-015-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aghová T, Kimura Y, Bryja J, Dobigny G, Granjon L, Kergoat GJ. Fossils know it best: using a new set of fossil calibrations to improve the temporal phylogenetic framework of murid rodents (Rodentia: Muridae) Mol Phylogenet Evol. 2018;128:98–111. doi: 10.1016/j.ympev.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 81.Steppan SJ, Schenk JJ. Muroid rodent phylogenetics: 900-species tree reveals increasing diversification rates. PLoS One. 2017;12:e0183070. doi: 10.1371/journal.pone.0183070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ellerman JR, TCS M-S. A Checklist of Palaeartic and Indian Mammals, 1758-1946. London: Trustees of the British museum (natural history); 1951. [Google Scholar]
- 83.Pickford M. Age of supergene ore bodies at berg Aukas and Harasib 3a, Namibia. Commun Geol Surv Namibia. 1992;8:147–150. [Google Scholar]
- 84.Geraads D. Rongeurs du Miocene superieur de Chorora (Ethiopie): Dendromuridae, Muridae et conclusions. Palaeovertebrata. 2001;30:89–109. [Google Scholar]
- 85.Bryja J, Šumbera R, Peterhans K, Julian C, Aghová T, Bryjová A, Mikula O, Nicolas V, Denys C, Verheyen E. Evolutionary history of the thicket rats (genus Grammomys) mirrors the evolution of African forests since late Miocene. J Biogeogr. 2017;44:182–94.
- 86.Couvreur TL, Chatrou LW, Sosef MS, Richardson JE. Molecular phylogenetics reveal multiple tertiary vicariance origins of the African rain forest trees. BMC Biol. 2008;6:54. doi: 10.1186/1741-7007-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lorenzen ED, Heller R, Siegismund HR. Comparative phylogeography of African savannah ungulates. Mol Ecol. 2012;21:3656–3670. doi: 10.1111/j.1365-294X.2012.05650.x. [DOI] [PubMed] [Google Scholar]
- 88.Randi E, D’Huart JP, Lucchini V, Aman R. Evidence of two genetically deeply divergent species of warhthog, Phacochoerus africanus and P. aethiopicus (Artiodactyla: Suiformes) in eastern Africa. Mamm Biol. 2002;67:91–96. [Google Scholar]
- 89.Brown DM, Brenneman RA, Koepfli K-P, Pollinger JP, Milá B, Georgiadis NJ, Louis EE, Grether GF, Jacobs DK, Wayne RK. Extensive population genetic structure in the giraffe. BMC Biol. 2007;5:57. doi: 10.1186/1741-7007-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fennessy J, Bidon T, Reuss F, Kumar V, Elkan P, Nilsson MA, Vamberger M, Fritz U, Janke A. Multi-locus analyses reveal four giraffe species instead of one. Curr Biol. 2016;26:1–7. doi: 10.1016/j.cub.2016.07.036. [DOI] [PubMed] [Google Scholar]
- 91.Hsü KJ, Giovanoli F. Messinian event in the black sea. Palaeogeogr Palaeoclimatol Palaeoecol. 1979;29:75–93. [Google Scholar]
- 92.Hodell DA, Curtis JH, Sierro FJ, Raymo ME. Correlation of Late Miocene to early Pliocene sequences between the Mediterranean and North Atlantic. Paleoceanography. 2001;16:164–178. [Google Scholar]
- 93.Maslin MA, Haug GH, Sarnthein M, Tiedemann R, Erlenkeuser H, Stax R. 21. Northwest pacific site 882: The initiation of Northern Hemisphere Glaciation. In: Rea DK, Basov IA, Scholl DW, Allan JF, editors. Proceedings of the Ocean Drilling Program Vol 145. 1995. pp. 179–194. [Google Scholar]
- 94.Maslin MA, Li XS, Loutre MF, Berger A. The contribution of orbital forcing to the progressive intensification of northern hemisphere glaciation. Quat Sci Rev. 1998;17:411–426. [Google Scholar]
- 95.Ravelo AC, Andreasen DH, Lyle M, Lyle AO, Wara MW. Regional climate shifts caused by gradual global cooling in the Pliocene epoch. Nature. 2004;429:263–270. doi: 10.1038/nature02567. [DOI] [PubMed] [Google Scholar]
- 96.Berger WH, Jansen E. Mid-Pleistocene climate shift: the Nansen connection. Geophys Monogr Ser. 1994;85:295–311. [Google Scholar]
- 97.Potts R. Hominin evolution in setings of strong environmental variability. Quat Sci Rev. 2013;73:1–13. [Google Scholar]
- 98.Kebede F, Rosenbom S, Khalatbari L, Moehlman PD, Beja-Pereira A, Bekele A. Genetic diversity of the Ethiopian Grevy’s zebra (Equus grevyi) populations that includes a unique population of the Alledeghi plain. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27:397–400. doi: 10.3109/19401736.2014.898276. [DOI] [PubMed] [Google Scholar]
- 99.Baker RJ, Bradley RD. Speciation in mammals and the genetic species concept. J Mammal. 2006;87:643–662. doi: 10.1644/06-MAMM-F-038R2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Groves CP. The nature of species: a rejoinder to Zachos et al. Mamm Biol. 2013;78:7–9.
- 101.Dippenaar NJ, Rautenbach IL. Morphometrics and karyology of the southern African species of the genus Acomys I. Geoffroy saint-Hilaire, 1838 (Rodentia: Muridae) Ann Transvaal Museum. 1986;34:129–183. [Google Scholar]
- 102.Matthey R. Etudes de cytogenetique Sur des Murinae Africains appartenant aux genres Arvicanthis, Acomys et Mastomys (Rodentia) Mammalia. 1965;29:228–249. [Google Scholar]
- 103.Janecek LL, Schlitter DA, Rautenbach IL. A genetic comparison of spiny mice, genus Acomys. Am Soc Mammal. 1991;72:542–552. [Google Scholar]
- 104.Musser G, Carleton M. Family Muridae. In: Wilson DE, Reeder DM, editors. Mammal species of the world. Washington DC: Smithsonian Institution Press; 1993. [Google Scholar]
- 105.Nevo E. Natural selection of body size differentiation in spiny mice, Acomys. Z Säugetierkd. 1989;54:81–99. [Google Scholar]
- 106.Corti M, Castiglia R, Colangelo P, Capanna E, Beolchini F, Bekele A, Oguge N, Makundi RH, Sichilima AM, Leirs H, Verheyen W, Vergagen R. Cytotaxonomy of rodent species from Ethiopia, Kenya, Tanzania and Zambia. Belgian J Zool. 2005;135:197–216. [Google Scholar]
- 107.Sokolov VE, Orlov VN, Baskevich MI, Bekele A, Mebrate A. A karyological study of the spiny mouse Acomys Geoffroy 1838 (Rodentia, Muridae) along the Ethiopian Rift Valley. Trop Zool. 1993;6:227–235. [Google Scholar]
- 108.Petter F. Eléments d’une révision des Acomys africains. Un sous-genre nouveau, Peracomys Petter and Roche, 1981 (Rongeurs, Muridés) Ann Mus R Afr Cent Sci Zool. 1983;237:109–119. [Google Scholar]
- 109.Volobouev VT, Gautun JC, Tranier M. Chromosome evolution in the genus Acomys (Rodentia, Muridae): chromosome banding analysis of Acomys cahirinus. Mammalia. 1996;60:217–222. [Google Scholar]
- 110.Volobouev V, Auffray JC, Debat V, Denys C, Gautun JC. Species delimitation in the Acomys cahirinus – dimidiatus complex (Rodentia, Muridae) inferred from chromosomal. Biol J Linn Soc. 2007;91:203–214. [Google Scholar]
- 111.Lavrenchenko LA, Kruskop SV, Bekele A, Belay G, Morozov PN, Ivlev YF, Warshavsky AA. Mammals of the Babille elephant sanctuary (eastern Ethiopia) Russ J Theriol. 2010;9:47–60. [Google Scholar]
- 112.Lavrenchenko LA, Nadjafova RS, Bulatova NS. Three new karyotypes extend a Robertsonian fan in Ethiopian spiny mice of the genus Acomys I. Geoffroy, 1838 (Mammalia, Rodentia) Comp Cytogenet. 2011;5:423–431. doi: 10.3897/CompCytogen.v5i5.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hollister N. East African mammals in the United States National Museum. II. Rodentia, Lagomorpha and Tubulidentata. Bull U S Natn Mus. 1919;99:1–184. [Google Scholar]
- 114.Kunze B, Dieterlen F, Traut W, Winking H. Karyotype relationship among four species of spiny mice (Acomys, Rodentia) Z Säugetierkd. 1999;64:220–229. [Google Scholar]
- 115.Happold DCD, editor. Volume 3: Rodents, Hares and Rabbits. Mammals of Africa. London: Bloomsbury Publishing; 2013. p. 789.
- 116.Granjon L, Duplantier JM. Les Rongeurs de l’Afrique sahélo-soudanienne. Marseille: Editions de l’IRD (Collection Faune et Flore tropicales); 2009. [Google Scholar]
- 117.Gautun JC, Sankhon I, Tranier M. Nouvelle contribution à la connaissance des rongeurs du massif guinéen des monts Nimba (Afrique occidentale). Systématique et aperçu quantitatif. Mammalia. 1986;50:205–217. [Google Scholar]
- 118.Morrison-Scott TCS. The identity of the Acomys megalotis (Lichtenstein), described from Arabia. Ann Mag Nat Hist. 1939;11:238–240. [Google Scholar]
- 119.Setzer HW. The spiny mice (Acomys) of Egypt. J Egypt Public Health Assoc. 1959;34:93–101. [Google Scholar]
- 120.Ellerman JR, Morrison-Scott TCS, Hayman RW. Southern African mammals. London: British Museum; 1953. [Google Scholar]
- 121.Yalden DW, Largen MJ, Kock D. Catalogue of the mammals of Ethiopia. Ital J Zool. 1976;1:1–118. [Google Scholar]
- 122.IUCN The IUCN red list of threatened species. Version 2017–3. 2017. http://www.iucnredlist.org. Accessed 17 Dec 2017.
- 123.Ducroz JF, Volobouev V, Granjon L. A molecular perspective on the systematics and evolution of the genus Arvicanthis (Rodentia, Muridae): inferences from complete cytochrome b gene sequences. Mol Phylogenet Evol. 1998;10:104–117. doi: 10.1006/mpev.1997.0477. [DOI] [PubMed] [Google Scholar]
- 124.Bellinvia E. A phylogenetic study of the genus Apodemus by sequencing the mitochondrial DNA control region. J Zool Syst Evol Res. 2004;42:289–297. [Google Scholar]
- 125.Stanhope MJ, Czelusniak J, Si JS, Nickerson J, Goodman M. A molecular perspective on mammalian evolution from the gene encoding interphotoreceptor retinoid binding protein, with convincing evidence for bat monophyly. Mol Phylogenet Evol. 1992;1:148–160. doi: 10.1016/1055-7903(92)90026-d. [DOI] [PubMed] [Google Scholar]
- 126.Teeling EC, Scally M, Kao DJ, Romagnoli ML, Springer MS, Stanhope MJ. Molecular evidence regarding the origin of echolocation and flight in bats. Nature. 2000;403:188–192. doi: 10.1038/35003188. [DOI] [PubMed] [Google Scholar]
- 127.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 3.31. 2017. [Google Scholar]
- 129.Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- 130.Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 131.Ripplinger J, Sullivan J. Does choice in model selection affect maximum likelihood analysis? Syst Biol. 2008;57:76–85. doi: 10.1080/10635150801898920. [DOI] [PubMed] [Google Scholar]
- 132.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 134.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol. 2003;52:665–673. doi: 10.1080/10635150390235485. [DOI] [PubMed] [Google Scholar]
- 136.Carstens BC, Pelletier TA, Reid NM, Satler JD. How to fail at species delimitation. Mol Ecol. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- 137.Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol. 2012;21:1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- 138.Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jones G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J Mat Biol. 2017;74:447–467. doi: 10.1007/s00285-016-1034-0. [DOI] [PubMed] [Google Scholar]
- 140.Masters BC, Fan V, Ross HA. Species delimitation – a geneious plugin for the exploration of species boundaries. Mol Ecol Resoursces. 2011;11:154–7. [DOI] [PubMed]
- 141.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kapli P, Lutteropp S, Zhang J, Kobert K, Pavlidis P, Stamatakis A, Flouri T. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics. 2017;33:1630–1638. doi: 10.1093/bioinformatics/btx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Drummond AJ, Bouckaert RR. Bayesian evolutionary analysis with BEAST. Cambridge: Cambridge University Press; 2015. [Google Scholar]
- 144.Jones G, Aydin Z, Oxelman B. DISSECT: an assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics. 2015;31:991–998. doi: 10.1093/bioinformatics/btu770. [DOI] [PubMed] [Google Scholar]
- 145.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rambaut A, Drummond AJ. LogCombiner v2.4.7. 2017. [Google Scholar]
- 147.Jones G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J Math Biol. 2017;74:447–467. doi: 10.1007/s00285-016-1034-0. [DOI] [PubMed] [Google Scholar]
- 148.Bouckaert RR. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. doi: 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- 149.Senut B, Pickford M, Mein P, Conroy G, Van Couvering J. Discovery of 12 new Late Cainozoic fossiliferous sites in palaeokarsts of the Otavi Mountains, Namibia. Comptes rendus de l’Académie des sciences. Série 2, Mécanique, Physique, Chimie, Sciences de l'univers. Sci Terre. 1992;314:727–733. [Google Scholar]
- 150.Geraads D. Plio-pleistocene mammalian biostratigraphy of Atlantic Morocco. Quaternaire. 2002;13:43–53. [Google Scholar]
- 151.Suwa G, Beyene Y, Nakaya H, Bernor RL, Boisserie JR, Bibi F, Ambrose SH, Sano K, Katoh S, Asfaw B. Newly discovered cercopithecid, equid and other mammalian fossils from the Chorora formation, Ethiopia. Anthropol Sci. 2015;123:19–39. [Google Scholar]
- 152.Manthi FK. A preliminary review of the rodent fauna from Lemudong’o, southwestern Kenya, and its implication to the late Miocene paleoenvironments. Kirtlandia. 2007;56:92–105. [Google Scholar]
- 153.Deino AL, Ambrose SH. 40Ar/39Ar dating of the Lemudong’o late Miocene fossil assemblages, southern Kenya rift. Kirtlandia. 2007;56:65–71. [Google Scholar]
- 154.Denys C. Of mice and men. Evolution in East and South Africa during Plio-Pleistocene times. In: Bromage GT, Schrenk F, editors. African Biogeography, Climate Change and Human Evolution. New York: Oxford University Press; 1999. pp. 226–252. [Google Scholar]
- 155.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gernhard T. The conditioned reconstructed process. J Theor Biol. 2008;253:769–778. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 157.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014. [Google Scholar]
- 158.Matzke NJ. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst Biol. 2014;63:951–970. doi: 10.1093/sysbio/syu056. [DOI] [PubMed] [Google Scholar]
- 159.Holt BG, Lessard JP, Borregaard MK, Fritz SA, Araújo MB, Dimitrov D, Fabre PH, Graham CH, Graves GR, Jønsson KA, Nogués-Bravo D, Wang Z, Whittaker RJ, Fjeldså J, Rahbek C. An update of Wallace’s zoogeographic regions of the world. Science. 2013;339:74–78. doi: 10.1126/science.1228282. [DOI] [PubMed] [Google Scholar]
- 160.Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 161.Ree RH, Sanmartín I. Conceptual and statistical problems with the DEC+J model of founder-event speciation and its comparison with DEC via model selection. J Biogeogr. 2018;45:741–749. [Google Scholar]
- 162.Wiens JJ, Donoghue MJ. Historical biogeography, ecology, and species richness. Trends Ecol Evol. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 163.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006;190:231–259. [Google Scholar]
- 164.Braconnot P, Otto-Bliesner B, Harrison S, Joussaume S, Peterchmitt JY, Abe-Ouchi A, Crucifix M, Driesschaert E, Fichefet T, Hewitt CD, Kageyama M, Kitoh A, Laîné A, Loutre MF, Marti O, Merkel U, Ramstein G, Valdes P, Weber SL, Yu Y, Zhao Y. Results of PMIP2 coupled simulations of the mid-Holocene and last glacial maximum - part 1: experiments and large-scale features. Clim Past. 2007;3:261–277. [Google Scholar]
- 165.Otto-Bliesner BL. Simulating arctic climate warmth and icefield retreat in the last interglaciation. Science. 2006;311:1751–1753. doi: 10.1126/science.1120808. [DOI] [PubMed] [Google Scholar]
- 166.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 167.Phillips SJ, Dudík M, Schapire RE. A maximum entropy approach to species distribution modeling. Canada: Banff; 2004. pp. 655–662. [Google Scholar]
- 168.Jiménez-Valverde A, Lobo JM. Threshold criteria for conversion of probability of species presence to either-or presence-absence. Acta Oecol. 2007;31:361–369. [Google Scholar]
- 169.Manel S, Williams HC, Ormerod SJ. Evaluating presence absence models in ecology; the need to count for prevalence. J Appied Ecol. 2001;38:921–931. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxonomic classifications of the genus Acomys. Bold italic names represent species reported in particular lists, standard italics represent subspecies (in Ellerman [34]; Setzer [38]; Denys et al. [16]). (XLSX 41 kb)
ML phylogenetic tree based on IRBP sequences (nexus file). (NEXUS 6 kb)
ML phylogenetic tree based on RAG1 sequences (nexus file). (NEXUS 5 kb)
Maximum clade credibility tree from STACEY with PP support (nexus file). (NEXUS 24 kb)
Complete list of individuals used in this study, with details on localities and genetic data. (XLSX 97 kb)
Alignment of 369 ingroup and 4 outgroup concatenated sequences of CYTB, IRBP, RAG1 and D-loop. (NEXUS 1470 kb)
Data Availability Statement
All corresponding sequences were deposited in GenBank under accession numbers MH044731-MH045045 (see Additional file 5). The final alignment of concatenated sequences used in phylogenetic analyses is in Additional file 6.