Diabetic nephropathy is the leading cause of end‐stage renal failure in Japan. Clarification of its detailed pathogenesis and development of therapeutic methods are urgent issues. In this article in the Journal of the American Society of Nephrology, Pan et al.1 showed the usefulness of weighted gene co‐expression network analysis (WGCNA) and identified potentially important candidate molecules that regulate diabetic nephropathy.
Pan et al. 1 isolated glomeruli from renal biopsy specimens of 41 diabetic nephropathy patients and 20 healthy individuals by microdissection and analyzed gene expression profiles. Furthermore, they carried out WGCNA using clinical information including albuminuria and the estimated glomerular filtration ratio together with the expression profile, and identified 18 co‐expression gene modules. Analysis of the gene module that is most correlated with the estimated glomerular filtration ratio and albuminuria identified a cytoskeleton regulator expressed in podocytes, Slit‐Robo Rho‐guanosine triphosphatase (ρGTPase)‐activating protein 2a (SRGAP2a), as a candidate gene that correlates most with clinical data and works as a hub of various gene expression changes. They continued to carry out the functional analysis for SRGAP2a by using cultured podocytes, mice and zebrafish models, and showed that SRGAP2a works in a protective manner in podocytes under diabetic conditions (Figure 1).
Figure 1.
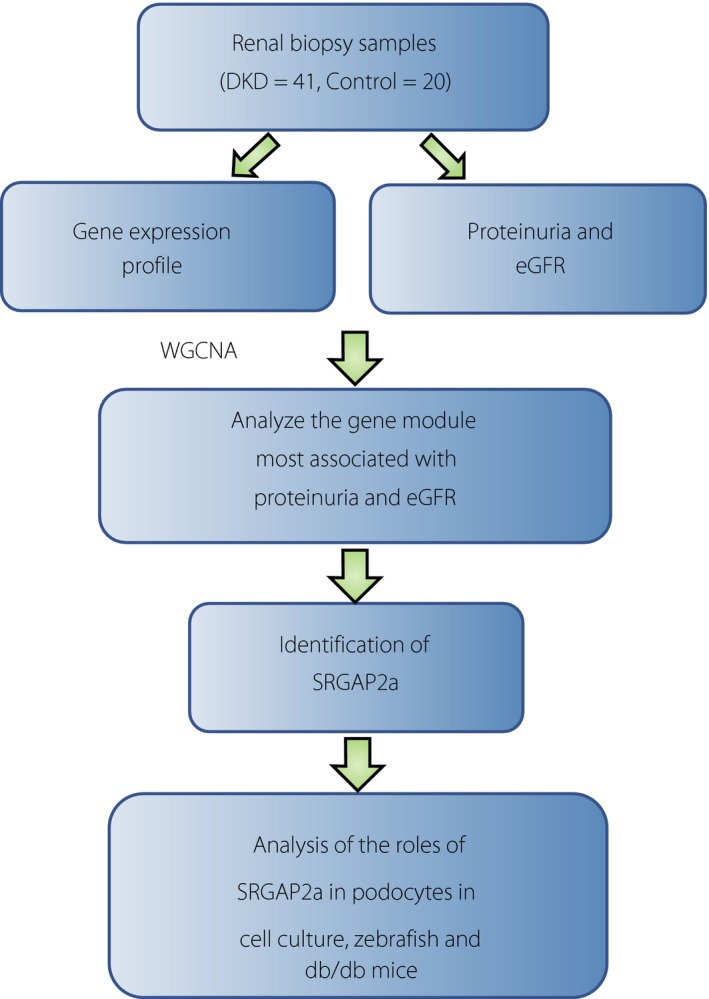
Scheme of the study by Pan et al.1 They carried out a series of analyses including gene expression profiling of renal biopsy samples, identification of Slit‐Robo Rho‐guanosine triphosphatase activating protein 2a (SRGAP2a) that is relevant for diabetic nephropathy and characterization of the molecule using various models. DKD, diabetic kidney disease; eGFR, estimated glomerular filtration ratio; WGCNA, weighted gene co‐expression network analysis.
To the best of our knowledge, the first report on transcriptome analysis of diabetic nephropathy was published in 2004 by Susztak et al.2 Using microarray analysis of whole kidneys, they identified 303 and 272 genes that were altered in db/db mice and streptozotocin‐induced diabetic mice, respectively. After that study, several studies reported transcriptome analyses including microarray or ribonucleic acid sequencing using rodent diabetes models. Recently, studies on mice took advantage of cell isolation techniques and reported transcriptomes of specific cell types or even single cell analysis. These include gene expression profiling of isolated glomerular endothelium from diabetic mice, parietal epithelium or single cell analysis of mesangial cells in normal mice. These advances have provided enormous knowledge on the involvement of specific pathways in each cell type, and also the variation of gene expressions even within one cell type.
In contrast, it is known that mouse and human diabetes are different. Hodgin et al.3 compared glomerular expression profiles with human diabetes, streptozotocin‐induced diabetic mice, db/db and db/db + endothelial nitric oxide synthase knockout mice, and among 277 altered genes there were just 26 common variable genes. Therefore, in addition to sophisticated rodent studies, direct evaluation of human samples is critically important. In this regard, Woroniecka et al.4 collected glomeruli and tubules from diabetic kidney samples in 2011. In microarray and pathway analysis, rat sarcoma homolog gene family member A (RhoA), cell division cycle 42, integrin and vascular endothelial growth factor signaling were highlighted in glomeruli. In the tubulointerstitial compartment, inflammation‐related pathways were enriched, indicating that the mechanisms of disease progression are different between glomeruli and tubulointerstitium. Later, integrative analysis that included genome‐wide association study candidate genes, renal biopsy‐derived transcriptional profiles and clinical information was carried out on various chronic kidney diseases. In that trial, all the pathways that were associated with chronic kidney disease aggregated into metabolism‐related pathways and oxidative stress responses through nuclear factor erythroid 2‐related factor 2. Recently, Nair et al.5 carried out a sophisticated study of 49 Pima Indian type 2 diabetes renal biopsy specimens. They combined compartment‐specific gene expression profiling and morphometric analysis, and showed that cortical interstitium volume, a marker for tubulointerstitial damage, is most associated with transcriptional regulation. Then, they carried out WGCNA, and identified gene modules that are associated with glomerular filtration ratio, urinary albumin and cortical interstitium volume. The transcripts showed enrichment in mitochondrial dysfunction, inflammation, migratory mechanisms and tubular metabolism.
Even though several transcriptome analyses have provided great advances in the mechanisms of human diabetic nephropathy, as described above, they are still rare because of the restrictions of human sample availability, which include ethical approval procedures, limited access to the human samples and the small sample amount of renal biopsy. Furthermore, functional analysis of the genes provided by transcriptome analysis requires additional experimental settings. In this regard, the article from Pan et al 1 provided valuable information on the human glomerular transcriptome, and showed a model of a series of experiments from WGCNA, candidate gene identification and their functional analysis. In addition, the data of this study are registered in the public database and will be useful data for many researchers working on diabetic nephropathy.
Podocytes have been considered to play a central role in diabetic nephropathy6. Since the 1990s, studies using Pima Indian specimens have shown that podocytes detach from the glomerular basement membrane into urine by apoptosis in diabetic nephropathy7. Podocytes constitute the final barrier of glomerular filtration by interdigitated foot processes and the slit membrane present there between. In chronic kidney disease with urine protein, humans and mice show foot processes effacement. For this reason, how podocytes maintain the architecture of these foot processes has been of great interest.
Regarding the cytoskeleton of podocytes, the intracellular domain of nephrin, a major constituent of the slit membrane, is bound to the actin cytoskeleton of the foot processes through adaptor proteins, including CD2‐associated protein and podocin. It is considered that tyrosine phosphorylation by Fyn proto‐oncogene Src family tyrosine kinase on these molecules leads to rearrangement of the actin cytoskeleton and preserves the viability of podocytes. In addition, it is clear that cytoskeletal abnormality is important for podocyte function, because hereditary focal segmental glomerulosclerosis also occurs as a result of mutations of α‐actinin 4, which is a reinforcing protein of podocyte's actin, inverted formin‐2 and Myo1E, which controls stretching of actin. Furthermore, mutation of RhoA regulator Rho‐guanosine triphosphatase (GTPase) activating protein 24 and Rho‐guanosine diphosphate dissociation inhibitor 1, which is a small GTPase for the actin cytoskeleton, is recognized in focal segmental glomerulosclerosis families. Among the Rho family GTPases important for cytoskeleton control, mutant mice show nephrotic syndrome from birth when cell division cycle 42 is knocked out specifically in podocytes, but for the same family RhoA, Rac1, steady‐state podocyte damage does not occur. In contrast, when RhoA is constitutively activated, podocyte disorder and proteinuria occur, showing that it is important for podocyte function to control the Rho family GTPase to a normal range. Regarding the cytoskeleton of podocytes in diabetic nephropathy, the above‐mentioned slit membrane‐related proteins decrease. Also, Coward et al 8 reported a model where insulin directly regulates the cytoskeleton of podocytes through the RhoA signal. According to this hypothesis, in response to insulin secretion after meals, it is assumed that podocytes periodically and dynamically change their morphology.
In this article by Pan et al.1, they identified SRGAP2a, a GTPase associated with the Slit‐ROBO signal, as a novel protein that regulates the podocyte cytoskeleton. SRGAP2a co‐localizes with synaptopodin of podocytes, and decreases in diabetes patient glomeruli and db/db mice. They showed that SRGAP2a binds to both RhoA/cell division cycle 42 and inhibits its activity, keeping the podocytes quiescent, and protecting the podocytes using a mouse and zebrafish model. In clinical practice, inhibitors of Rho‐associated kinase, a downstream target of Rho, are used to control glaucoma and cerebral vasospasm, but several studies have reported that these are also useful in diabetic nephropathy in rodents9. In contrast, although mammalian loss of function experiments were not carried out, upregulation of SRGAP2a using adenovirus significantly alleviated podocyte injury and proteinuria in db/db mice, suggesting that SRGAP2a might be a potential target for future therapy.
In conclusion, the article by Pan et al. provides good additional insights to the glomerular transcriptome analysis in diabetes patients and thus revealed a potential candidate pathway, the Rho family signaling, as a future therapeutic target. Although the effectiveness of Rho‐associated kinase inhibitors has only been reported in the experimental diabetic models as yet, future drug repositioning and development of novel therapeutic methods are expected.
Disclosure
The authors declare no conflict of interest.
References
- 1. Pan Y, Jiang S, Hou Q, et al Dissection of glomerular transcriptional profile in patients with diabetic nephropathy: SRGAP2a protects podocyte structure and function. Diabetes 2018; 67: 717–730. [DOI] [PubMed] [Google Scholar]
- 2. Susztak K, Bottinger E, Novetsky A, et al Molecular profiling of diabetic mouse kidney reveals novel genes linked to glomerular disease. Diabetes 2004; 53: 784–794. [DOI] [PubMed] [Google Scholar]
- 3. Hodgin JB, Nair V, Zhang H, et al Identification of cross‐species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes 2013; 62: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woroniecka KI, Park AS, Mohtat D, et al Transcriptome analysis of human diabetic kidney disease. Diabetes 2011; 60: 2354–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nair V, Komorowsky CV, Weil EJ, et al A molecular morphometric approach to diabetic kidney disease can link structure to function and outcome. Kidney Int 2018; 93: 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakamura T, Ushiyama C, Suzuki S, et al Urinary excretion of podocytes in patients with diabetic nephropathy. Nephrol Dial Transplant 2000; 15: 1379–1383. [DOI] [PubMed] [Google Scholar]
- 7. Verzola D, Gandolfo MT, Ferrario F, et al Apoptosis in the kidneys of patients with type II diabetic nephropathy. Kidney Int 2007; 72: 1262–1272. [DOI] [PubMed] [Google Scholar]
- 8. Coward R, Fornoni A. Insulin signaling: implications for podocyte biology in diabetic kidney disease. Curr Opin Nephrol Hypertens 2015; 24: 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matoba K, Kawanami D, Okada R, et al Rho‐kinase inhibition prevents the progression of diabetic nephropathy by downregulating hypoxia‐inducible factor 1alpha. Kidney Int 2013; 84: 545–554. [DOI] [PubMed] [Google Scholar]


