Abstract
Blockade of the immunoinhibitory PD-1/PD-L1 pathway using monoclonal antibodies has shown impressive results with durable clinical antitumor responses. Anti-PD-1 and anti-PD-L1 antibodies have now been approved for the treatment of a number of tumor types whereas the development of small molecules targeting immune checkpoints lags far behind. Here we characterize two classes of macrocyclic-peptide inhibitors directed at the PD-1/PD-L1 pathway. We show that these macrocyclics act by directly binding to PD-L1 and that they are capable of antagonizing PD-L1 signaling and, similarly to antibodies, can restore the function of T-cells. We also provide the crystal structures of two of these small-molecule inhibitors bound to PD-L1. The structures provide rationales for the checkpoint inhibition by these small molecules and description of their small molecule/PD-L1 interfaces provides a blueprint for design of small-molecule inhibitors of the PD-1/PD-L1 pathway.
Keywords: cancer, immunotherapy, immune checkpoint, PD-1, PD-L1
Graphical Abstract
Peptidic macrocyclic inhibitors can block the PD-1/PD-L1 pathway by directly binding to PD-L1 and, similarly to the antibodies, can restore the function of T-cells. Structures of the macrocycle/PD-L1 interfaces provide foundations for the design of small-molecule inhibitors with antitumor properties.
Anticancer therapies based on the immune checkpoint blockage (ICB) have witnessed spectacular success in the last years. ICB-based immunotherapy using monoclonal antibodies (mAbs) delivers durable antitumor responses and long-term remissions in a subset of patients with a broad spectrum of cancers.[1–7] However, monoclonal antibody therapy is expensive and inherently carries a number of disadvantages such as the immunogenicity of human mAbs (following repeated administration), no oral bioavailability, poor solid tumor tissue penetration and poor control of pharmacokinetics, and thus mAb related toxicities (i.e. immune-related adverse effects, irAEs).[8,9] In contrast, small-molecule therapeutics can have affinity and specificity features rivaling that of antibodies. Importantly, small molecules have been shown to lack immunogenicity and are orally bioavailable.
Development of chemical inhibitors for the PD-1/PD-L1 pathway lags the antibody development. A few series of small-molecules, macrocyclic peptides, peptides and peptidomimetics targeting the PD-1/PD-L1 interaction have been reported, primarily in patent applications, but publicly disclosed validation is almost non-existent.[10–12] We have recently described the binding modes and biological properties of the small-molecule chemical inhibitors of PD-L1 disclosed by Bristol-Myers Squibb.[13] Herein we report the activity and structural characterization of macrocyclic peptides, another class of small molecules, that have recently been reported to inhibit the PD-1/PD-L1 interaction.[10–12,14]
Three classes of macrocyclic peptides were reported by Bristol-Myers Squibb and nanomolar activities in dissociating the PD-1/PD-L1 interaction were determined by the HTRF assay.[14] We selected one representative macrocyclic peptide for each of groups, namely those containing 15, 14 and 13 residues. Respectively, peptides-57 (reported IC50 of 9 nM), peptide-71 (7 nM) and peptide-99 (153 nM) (Supporting Information, Table S1, Figure S1) were synthesized and their affinity towards PD-1 and PD-L1 was evaluated using several methods. First, in the NMR method, titration of the 15N labeled PD-1 with either tested macrocyclic peptide did not result in any significant shifts in 1H-15N signals in 2D HMQC spectra indicating no binding. For all the tested peptides, titration of the 15N labeled PD-L1 resulted in shifts in resonance signals indicating interaction. The shift profile (peak splitting) indicated tight binding (K i< 1uM; Supporting Information, Figures S2 and S3).
Using the differential scanning fluorimetry (DSF),[15] we additionally verified the affinity of peptide-57 and peptide-71 towards the PD-L1 protein. PD-L1 showed low melting temperature (Tm) of 37.6°C (Supporting Information, Figure S4). Peptide-57 stabilized the thermal induced unfolding by 14°C (Tm=51.6°C), whereas peptide-71 by 19°C (Tm=56.6°C). These results confirm the interaction of both peptides with PD-L1 and indicate that peptide-71 shows higher affinity compared to peptide-57.
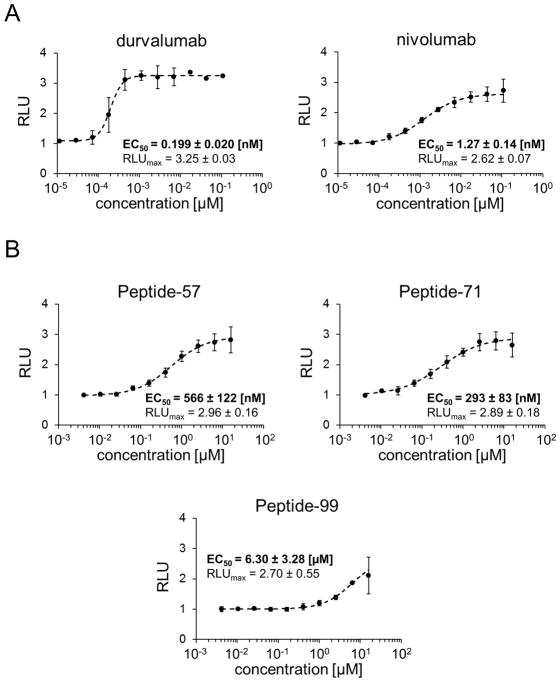
To test if peptides-57, -71 and -99 are capable of inhibiting the PD-1/PD-L1 interaction in the cellular context, we have employed the Jurkat T-like cells carrying a reporter luciferase gene under the control of the NFAT promoter and overexpressing PD-1. These cells were contacted with the surrogate of the antigen presenting cells, a CHO cell-line, which overexpresses a T-cell receptor ligand and PD-L1.[16] In this setup the expression of the reporter is dependent on TCR activation, whereas simultaneous ligation of the PD-1 receptor results in promoter silencing, mimicking the processes within T cells. The promoter is activated only in the presence of the blockers of the PD-1/PD-L1 interaction. To verify this model, the FDA-approved antibodies targeting the PD-1/PD-L1 interaction were used: anti-PD-L1 antibody, durvalumab (AstraZeneca), and anti-PD-1 antibody nivolumab (Bristol-Myers Squibb). Both antibodies dose-dependently restored the activity of the TCR responsive promoter (Figure 1A) suggesting effective inhibition of the PD-1/PD-L1 interaction. The immunomodulatory effects of durvalumab and nivolumab were characterized by EC50 values of 0.199 nM and 1.27 nM, respectively. Peptide-57 and -71 dose dependently restored the activity of the TCR responsive promoter and their activities were characterized by EC50 of 566 nM and 293 nM. Peptide-99 was the least active, being characterized by EC50 of 6.30 μM (Figure 1B). At the maximal activity, all tested antibodies and peptides restored comparable levels of the activity of the tested cells (RLUmax values between 2.62 and 3.25, Figure 1).
Figure 1.
Activities of the macrocyclic peptides in the cell-based PD-1/PD-L1 immune checkpoint assay. Antigen presenting cells (APC) were seeded on culture plates and overlaid with PD-1 Effector Cells in the presence of different concentrations of therapeutic antibodies (A) or macrocyclic peptides (B). The activation of PD-1 Effector Cells, reflected by luciferase activity, was monitored by luminescence measurement. The data represent mean ± SD values from three independent experiments, normalized to the control vehicle-treated cells. For the regression analysis Hill equation was fitted to the experimental data and the half maximal effective concentrations (EC50) and maximal relative luminescence values (RLUmax) were determined.
X-ray crystallography was used to obtain structural insight into the peptide-57 and -71 interactions with PD-L1. The structures of the complexes PD-L1/peptide-71 and PD-L1/peptide-57 were solved at 2.5 and below 1 Å resolutions, respectively (Figure 2, Supporting Information, Table S2, Figures S5 and S6). The structures show the pharmacophore of these macrocycles is not related to the small-molecule chemical inhibitors described recently by us.[13] Thus our structural data provide an important template for the design of new small-molecule inhibitors of the PD-1/PD-L1 pathway.
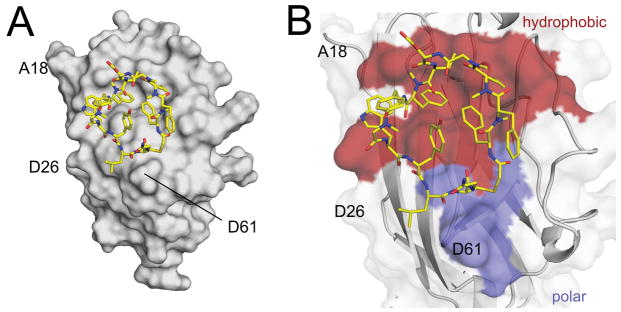
Figure 2.
Crystal structure of the PD-L1/peptide-71 complex. A) Overall view into the PD-L1/peptide-71 interactions. The peptide assumes a ring like shape with its centre filled with the hydroxyphenyl group. B) Close-up view of the PD-L1/peptide-71 interface. Peptide-71 binds on the surface of PD-L1 at the relatively hydrophobic palm. Hydrophobic interactions in the complex are shown in red while hydrophilic in blue.
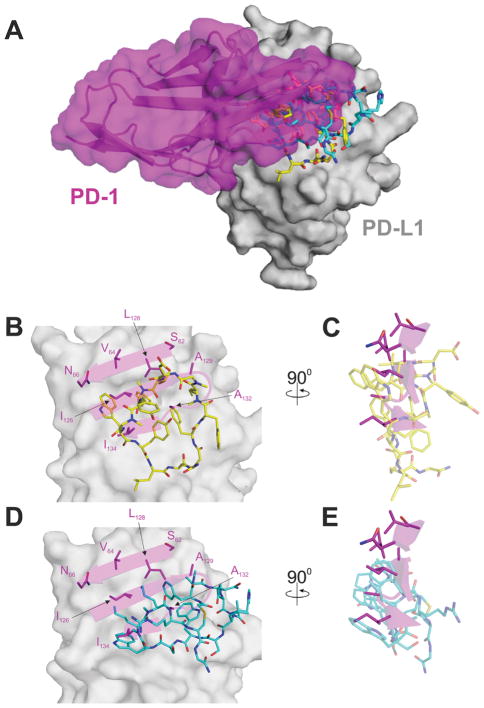
In the structures, the cores of both peptides bind at the interface site of PD-L1 that approximately coincides with the PD-1 binding site of PD-L1 (Figure 3A).[17] However, the detailed realization of the binding of these two macrocyclic peptides to PD-L1 is significantly different from that of PD-1. It differs also in between the peptides - to the extent that not a single residue of one peptide directly mimics the binding of any single residue of the other peptide, as well as PD-1 (Figure 3; detailed features of the interfaces for the complexes peptide-71,-57/PD-L1 are described in the Supporting Information, Results and Discussion, and Figures S7–S13). Peptide-57 extends more towards strand G of PD-L1, which is not observed in peptide-71 (Figure 4 and Supporting Information, Figure S11; the canonical Ig-strand designations are used – Supporting Information, Figure S14). Peptide-71, in turn, extends towards Asp61 and anchors at this residue with the oxygen-sulfur interaction that is not observed in peptide-57. However, the most significant difference in the binding of peptides-57 and -71 to PD-L1 relates to their relative direction of the polypeptide chain. While looking from the top of the G, F, C, C′ β-sheet of PD-L1, peptide-57 is directed clockwise while peptide-71 counterclockwise. Despite this major difference, the physical properties of the interaction surfaces are comparable; this is imposed by the binding landscape at the surface of PD-L1. In terms of the standard view presented in Figure 2B and Supporting Information, Figure S6B the upper part of the binding surface consists of only hydrophobic interactions, while the lower part of the binding surface is dominated by polar interactions.
Figure 3.
Rationale for the inhibition of the PD-1/PD-L1 interaction by the macrocyclic peptides. Macrocyclic peptides bind to PD-L1 at the site of PD-1, however, the detailed interactions are different. A) The peptides -57 (blue) and -71 (yellow) bind to PD-L1 partially at the site of the PD-1 interaction (magenta). B)-C) Detailed interactions of peptide-71 at the binding surface of PD-L1 in comparison with the PD-1/PD-L1 interactions. D)-E) Detailed interactions of peptide-57 at the binding surface of PD-L1 in comparison with the PD-1/PD-L1 interactions.
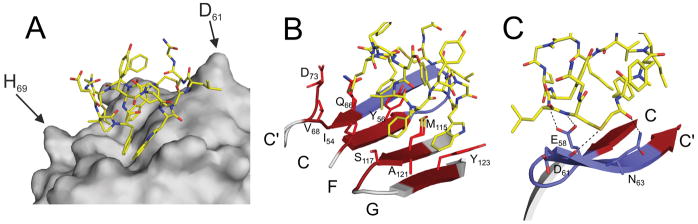
Figure 4.
Detailed view into the PD-L1/peptide-71 interaction. A) Hydrophobic sidechains interact with the cleft characteristic for the “face-on” binding mode. B) Peptide-71 binds PD-L1 at the palm of the β-sheet composed of strands G, F, C and C′ mostly by hydrophobic interactions (red). C) The polar zone of the interaction surface includes two hydrogen bonds contributed by the backbone amines of peptide-71.
Overlay of the structures of PD-L1 determined in complex with peptides-57 and -71 demonstrate that no significant structural changes are induced within the PD-L1 receptor upon the ligand binding. The surfaces that provide hydrophobic interactions are almost identical in both structures (Figure 2B and Supporting Information, Figure S6B) save only for disposition of the Met115 sidechain, which is bent in the PD-L1/peptide-71 complex and thus makes space for the 71NMePhe7 side chain (subscript 71 denotes peptide-71 and the last number indicates the position of the amino acid in the peptide, Figure S1).
Detailed nature of the macrocycle/PD-L1 interactions correspond well with the structure–activity relationship within the groups represented by each of the macrocyclic peptides. In the group of macrocycles containing 14 residues (represented by peptide-71), exchange of the central 71Tyr11 into a small alanine residue (peptide-83) causes fivefold increase of the inhibitory constant value (reported IC50 35 nM) (Supporting Information, Table S1). A much larger decrease in the inhibitory activity is caused by replacement of the residues involved in the hydrophobic interactions by Ala or NMeAla. For example, lack of 71Phe1 and 71NMePhe7 causes the increase of IC50 to 4229 nM for peptide-72 and above IC50 10000 nM for peptide-81. Interestingly, methylations of the side chains in peptide-71 are also necessary to ensure high activity of the macrocycle. Lack of the methylation of 71NMePhe2 or 71NMeNle3 causes again huge increase of the IC50 value above 10000 nM (peptides-74 and -76, respectively).
Closely similar trends are seen for the macrocycles containing 13 residues represented by peptide-57. Replacement each of residues responsible for hydrophobic interactions with smaller side-chain amino acids causes large drop of the activity. This dependence can be seen in the case of the peptides that lack 57Phe1, 57Trp8 or 57Trp10 residues (IC50: 6495 nM, above 30000 nM and 3656 nM for peptides −5, −15 and −63, respectively).
Structural characteristics of therapeutic antibodies can guide the design of non-antibody drugs that would mimic key antibodies residues.[18] The binding surfaces of peptides -57 and -71 within PD-L1 overlap partially with the epitopes of anti-PD-L1 antibodies (atezolizumab, avelumab, durvalumab, and BMS-936559 (Supporting Information, Figure S15)).[19,20] Analysis of the interactions of the residues of the antibodies avelumab and BMS-936559 and the peptides shows that several residues of the peptides and the antibodies interact similarly. A number of the residues of peptide-71 mimic the amino acids of the VH domain of avelumab responsible for the interactions with PD-L1 (Supporting Information, Figures S16 and S17): for example, the hydrophobic side chains of peptide-71: 71Phe1, 71NMeNle3 and 71NMePhe7, interact similarly to AIle33, APro53 and AIle57, respectively (subscript A indicates the avelumab residues). In the case of the PD-L1/peptide-57 structure, the overlapping is smaller; however, the main chain of 57Phe1 and the sidechain of 57NMeNle2 mimic the avelumab APro53 and AIle57. Comparison of the structures of both PD-L1/peptide complexes with that of the PD-L1/BMS-936559 complex shows that both peptides mimic the main hydrophobic interactions of the antibody residues: BIle54 and BPhe55 (subscript B indicates the BMS-936559 residues) by locating in the same clefts residues: 71Trp10, 71NMePhe7 and 57NMeNle11, 57NMeNle12 (Supporting Information, Figures S16 and S18). Peptide-71 again better mimics the BMS-936559 antibody and additionally interacts by using 71NMeNle3 similarly to BHis59. Overall, however, the peptides mimic only of about the 37% of the PD-L1/antibodies interactions and the binding interface of the anti-PD-L1 avelumab and BMS-936559 may provide additional information onto the direction of the further peptide modifications to enhance their potency.
Supplementary Material
Acknowledgments
This research has been supported by Grants UMO-2012/06/A/ST5/00224 and UMO-2014/12/W/NZ1/00457 from the National Science Centre, Poland and co-financed by the EU European Regional Development Fund and the Marie Curie FP7-Reintegration-Grant within the seventh European Community Framework Programme (to T.A. Holak). L. Skalniak was supported Grant UMO-2016/21/D/NZ7/00596 and G. Dubin by Grant UMO-2012/07/E/NZ1/01907 both from the National Science Centre, Poland. L. Berlicki would like to acknowledge the financial support by a statutory activity subsidy from the Polish Ministry of Science and Higher Education for the Faculty of Chemistry of Wrocław University of Technology. Moreover, funding was received (to A. Domling) from NIH 2R01GM097082-05, COFUND (grant agreement No 665250) and KWF grant (grant agreement No 10504). The research was carried out with the equipment purchased thanks to the financial support of the European Union structural funds (contract nos. POIG.02.01.00-12-064/08 and POIG.02.01.00-12-167/08) and European Regional Development Fund in the framework of the Polish Innovation Economy Operational Program (contract no. POIG.02.01.00-582 12-023/08). P. Grudnik was supported by grant UMO-2015/19/D/NZ1/02009 and K. M. Zak by grant UMO-2016/20/T/NZ1/00519 both from the National Science Centre, Poland.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Conflict of Interests
The authors declare no conflicts of interests.
Contributor Information
Dr. Katarzyna Magiera-Mularz, Department of Organic Chemistry, Faculty of Chemistry, Jagiellonian University, Ingardena 3, 30-060 Krakow, Poland
Dr. Lukasz Skalniak, Department of Organic Chemistry, Faculty of Chemistry, Jagiellonian University, Ingardena 3, 30-060 Krakow, Poland
Krzysztof M. Zak, Malopolska Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, Poland., Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland
Dr. Bogdan Musielak, Department of Organic Chemistry, Faculty of Chemistry, Jagiellonian University, Ingardena 3, 30-060 Krakow, Poland
Dr. Ewa Rudzinska-Szostak, Department of Bioorganic Chemistry, Faculty of Chemistry, Wrocław University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wrocław, Poland
Dr. Łukasz Berlicki, Department of Bioorganic Chemistry, Faculty of Chemistry, Wrocław University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wrocław, Poland
Justyna Kocik, Department of Organic Chemistry, Faculty of Chemistry, Jagiellonian University, Ingardena 3, 30-060 Krakow, Poland.
Dr. Przemyslaw Grudnik, Malopolska Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, Poland., Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland
Dominik Sala, Department of Organic Chemistry, Faculty of Chemistry, Jagiellonian University, Ingardena 3, 30-060 Krakow, Poland.
Tryfonas Zizigas-Zarganis, Department for Drug Design, University of Groningen, A. Deusinglaan 9, AV 9713 Groningen, the Netherlands.
Shabnam Shaabani, Department for Drug Design, University of Groningen, A. Deusinglaan 9, AV 9713 Groningen, the Netherlands.
Prof. Dr. Alexander Dömling, Department for Drug Design, University of Groningen, A. Deusinglaan 9, AV 9713 Groningen, the Netherlands
Dr. Grzegorz Dubin, Malopolska Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, Poland., Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland
Prof. Dr. Tad A. Holak, Department of Organic Chemistry, Faculty of Chemistry, Jagiellonian University, Ingardena 3, 30-060 Krakow, Poland., Max Planck Institute for Biochemistry, Am Klopferspitz 18a, 82152 Martinsried, Germany
References
- 1.Dömling A, Holak TA. Angew Chemie Int Ed. 2014;53:2286–2288. doi: 10.1002/anie.201307906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoos A. Nat Rev Drug Discov. 2016;15:235–247. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 3.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. Nat Rev Clin Oncol. 2016;13:273–290. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahoney KM, Rennert PD, Freeman GJ. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 5.Sharma P, Allison JP. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 6.Shin DS, Ribas A. Curr Opin Immunol. 2015;33:23–35. doi: 10.1016/j.coi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Topalian SL, Drake CG, Pardoll DM. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harding FA, Stickler MM, Razo J, DuBridge RB. MAbs. 2010;2:256–65. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson AL, Dhimolea E, Reichert JM. Nat Rev Drug Discov. 2010;9:767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 10.Weinmann H. ChemMedChem. 2016;11:450–466. doi: 10.1002/cmdc.201500566. [DOI] [PubMed] [Google Scholar]
- 11.Zarganes-Tzitzikas T, Konstantinidou M, Gao Y, Krzemien D, Zak K, Dubin G, Holak TA, Dömling A. Expert Opin Ther Pat. 2016;26:973–977. doi: 10.1080/13543776.2016.1206527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan M-M, Hu X-Q, Liu X-X, Ruan B-F, Xu J, Liao C. Drug Discov Today. 2016;21:1027–1036. doi: 10.1016/j.drudis.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Zak KM, Grudnik P, Guzik K, Zieba BJ, Musielak B, Dömling A, Dubin G, Holak TA. Oncotarget. 2016;7:30323–35. doi: 10.18632/oncotarget.8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MM, Mapelli C, Allen MP, Bowshe MS, Boy KM, Gillis EP, Langley DR, Mull E, Poirier MA, Sanghvi N, Sun L-Q, Tenney DJ, Yeung K-S, Zhu J, Reid PC, Scola PM, Cornelius LA. US 20140294898 A1. Bristol-Myers Squibb Company. 2014
- 15.Niesen FH, Berglund H, Vedadi M. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 16.Cheng Z-JJ, Karassina N, Grailer J, Hartnett J, Fan F, Cong M. Cancer Res; Proceedings of the 106th Annual Meeting of the AACR; 2015. [abstract] Abstract nr 5440. [Google Scholar]
- 17.Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B, Dömling A, Dubin G, Holak TA. Structure. 2015;23:2341–2348. doi: 10.1016/j.str.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawson ADG. Nat Rev Drug Discov. 2012;11:519–525. doi: 10.1038/nrd3756. [DOI] [PubMed] [Google Scholar]
- 19.Lee JY, Lee HT, Shin W, Chae J, Choi J, Kim SH, Lim H, Won Heo T, Park KY, Lee YJ, et al. Nat Commun. 2016;7:13354. doi: 10.1038/ncomms13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, Tan S, Chai Y, Chen D, Song H, Zhang CWH, Shi Y, Liu J, Tan W, Lyu J, et al. Cell Res. 2017;27:151–153. doi: 10.1038/cr.2016.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.